Mutations in the ALS5/SPG11/ KIAA1840 gene cause autosomal recessive hereditary spastic paraplegia or autosomal recessive juvenile amyotrophic lateral sclerosis. Montecchiani et al . show that KIAA1840 mutations can manifest also as recessive Charcot-Marie-Tooth disease. They describe 12 kindreds with 15 different mutations, two of which have not been reported previously.
Keywords: ALS5/SPG11/ KIAA1840 mutations , axonal degeneration, Charcot–Marie–Tooth disease, spatacsin
Mutations in the ALS5/SPG11/ KIAA1840 gene cause autosomal recessive hereditary spastic paraplegia or autosomal recessive juvenile amyotrophic lateral sclerosis. Montecchiani et al . show that KIAA1840 mutations can manifest also as recessive Charcot-Marie-Tooth disease. They describe 12 kindreds with 15 different mutations, two of which have not been reported previously.
Abstract
Charcot–Marie–Tooth disease is a group of hereditary peripheral neuropathies that share clinical characteristics of progressive distal muscle weakness and atrophy, foot deformities, distal sensory loss, as well as diminished tendon reflexes. Hundreds of causative DNA changes have been found, but much of the genetic basis of the disease is still unexplained. Mutations in the ALS5/SPG11/ KIAA1840 gene are a frequent cause of autosomal recessive hereditary spastic paraplegia with thin corpus callosum and peripheral axonal neuropathy, and account for ∼40% of autosomal recessive juvenile amyotrophic lateral sclerosis. The overlap of axonal Charcot–Marie–Tooth disease with both diseases, as well as the common autosomal recessive inheritance pattern of thin corpus callosum and axonal Charcot–Marie–Tooth disease in three related patients, prompted us to analyse the ALS5/SPG11/ KIAA1840 gene in affected individuals with autosomal recessive axonal Charcot–Marie–Tooth disease. We investigated 28 unrelated families with autosomal recessive axonal Charcot–Marie–Tooth disease defined by clinical, electrophysiological, as well as pathological evaluation. Besides, we screened for all the known genes related to axonal autosomal recessive Charcot–Marie-Tooth disease (CMT2A2/HMSN2A2/ MFN2 , CMT2B1/ LMNA , CMT2B2/ MED25 , CMT2B5/ NEFL , ARCMT2F/dHMN2B/ HSPB1 , CMT2K/ GDAP1 , CMT2P/ LRSAM1 , CMT2R/ TRIM2 , CMT2S/ IGHMBP2 , CMT2T/ HSJ1 , CMTRID/ COX6A1 , ARAN-NM/ HINT and GAN/ GAN ), for the genes related to autosomal recessive hereditary spastic paraplegia with thin corpus callosum and axonal peripheral neuropathy (SPG7/ PGN , SPG15/ ZFYVE26, SPG21/ ACP33 , SPG35/ FA2H , SPG46/ GBA2 , SPG55/ C12orf65 and SPG56/ CYP2U1 ), as well as for the causative gene of peripheral neuropathy with or without agenesis of the corpus callosum ( SLC12A6 ) . Mitochondrial disorders related to Charcot–Marie–Tooth disease type 2 were also excluded by sequencing POLG and TYMP genes. An additional locus for autosomal recessive Charcot–Marie–Tooth disease type 2H on chromosome 8q13-21.1 was excluded by linkage analysis. Pedigrees originated in Italy, Brazil, Canada, England, Iran, and Japan. Interestingly, we identified 15 ALS5/SPG11/ KIAA1840 mutations in 12 families (two sequence variants were never reported before, p.Gln198* and p.Pro2212fs*5). No large deletions/duplications were detected in these patients. The novel mutations seemed to be pathogenic since they co-segregated with the disease in all pedigrees and were absent in 300 unrelated controls. Furthermore, in silico analysis predicted their pathogenic effect. Our results indicate that ALS5/SPG11/ KIAA1840 is the causative gene of a wide spectrum of clinical features, including autosomal recessive axonal Charcot–Marie–Tooth disease.
Introduction
Mutations in the ALS5/SPG11/ KIAA1840 gene, located on chromosome 15q21.1, cause autosomal recessive hereditary spastic paraplegia (ARHSP; OMIM #604360), frequently associated with thin corpus callosum and axonal peripheral neuropathy ( Stevanin et al. , 2007 a , b ; Lo Giudice et al. , 2014 ), as well as autosomal recessive juvenile amyotrophic lateral sclerosis (ARJALS; OMIM %602099), characterized by slowly progressive spasticity of limb and facial muscles with distal amyotrophy of hands and feet ( Orlacchio et al. , 2010 ). Charcot–Marie–Tooth (CMT) disease is a hereditary peripheral neuropathy characterized by slowly progressive distal muscle weakness, atrophy, and mild sensory loss, primarily in lower limbs. It is the most common degenerative disorder of the peripheral nervous system, with a global prevalence of 1 in 2500 ( Tazir et al. , 2013 ). Phenotypes are grouped within the hereditary motor and sensory neuropathies (HMSNs; Dyck and Thomas, 2005 ) and can be divided into demyelinating (CMT1), axonal (CMT2), and intermediate forms, on the basis of electrophysiological criteria as well as pathological features ( Dyck et al. , 1968 ). In CMT2, peripheral nerves are neither enlarged nor hypertrophic, with normal or near-normal conduction velocities; the disease course is highly variable, ranging from mild to severe forms ( Feely et al. , 2011 ; Tazir et al. , 2014 ). The subtypes belonging to ARCMT2 share common and distinctive clinical features, e.g. CMT2B2 is characterized by sensory deficits, whereas CMT2K is characterized by hoarse voice and vocal cord paresis ( Claramunt et al. , 2005 ; Leal et al. , 2009 ). To date, ARCMT2 has been associated with mutations in only a few genes, and many forms are still unrecognized ( Bird, 1993-2015 ). Recent advances in the molecular genetics of Charcot–Marie–Tooth disease have contributed to the classification and diagnosis of this heterogeneous disease ( Jerath and Shy, 2015 ).
The overlap between CMT2 and hereditary spastic paraplegia ( Timmerman et al. , 2013 ; Fridman and Murphy, 2014 ), as well as the fact that the ALS5/SPG11/ KIAA1840 gene product (spatacsin) is implicated in axonal maintenance and cargo trafficking ( Pérez-Brangulí et al. , 2014 ), and the common autosomal recessive inherited findings of thin corpus callosum and CMT2 in three related patients, motivated us to investigate the ALS5/SPG11/ KIAA1840 gene in families with ARCMT2 with no causative gene identified.
Materials and methods
Patients
This study was performed according to a protocol reviewed and approved by the Ethics Committee of the Istituto di Ricovero e Cura a Carattere Scientifico Santa Lucia , Rome, Italy. After obtaining informed consent, patients were recruited by a network of neurologists in Italy, Brazil, Canada, England, and Japan, with a significant background in peripheral neuropathy.
The study focused on 28 unrelated pedigrees with ARCMT2 and without genetic assessment. The diagnosis of ARCMT2 was based on neurological findings, familial history, along with neurophysiological characteristics ( Bird, 1993-2015 ). Flexor and extensor muscle strength was assessed manually using the standard Medical Research Council Scale ( Medical Research Council, 1981 ). The Functional Disability Scale and the Charcot–Marie–Tooth Neuropathy Score assessed the staging of the disease ( Birouk et al. , 1997 ; Murphy et al. , 2011 ).
All affected subjects under the age of 18 were investigated for IQ through the Wechsler Intelligence Scales for Children IV, including the Wechsler Memory Scale ( http://alpha.fdu.edu/psychology/WISCIV_Index.htm ). According to the criteria of the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition ( American Psychiatric Association, 2013 ), mental retardation was considered when the IQ was ≤70 before the age of 18. Adults were evaluated by Mental Deterioration Battery ( Carlesimo et al. , 1996 ) to assess cognitive impairment. Brain MRI was obtained in all affected individuals, with the exception of eight subjects. Haematological and biochemical profiles, as well as lysosomal enzyme assay of β-hexosaminidase A and B in peripheral blood leucocytes, were performed in at least one patient within each family. CSF analysis was available in 21 patients. Vitamins B 1 , B 2 , B 6 , B 9 , B 12 , and E were measured in all patients.
Nerve conduction study was carried out with a surface skin temperature between 32°C and 34°C. Nerves were analysed bilaterally in upper and lower limbs using percutaneous stimulation and surface recording electrodes. Concentric needle EMG was performed in muscles of upper and lower limbs: spontaneous activity, duration, and amplitude of motor unit action potentials, as well as motor unit action potentials, interference pattern during maximal voluntary contraction, were recorded.
Sural nerve biopsy was available in 25 patients. Light microscopy preparations, and the electron microscopic analysis in one patient, were performed according to standard methods ( Palumbo et al. , 2002 ).
Genetic analyses
Genomic DNA was extracted from peripheral blood using the Promega Wizard® Genomic DNA isolation kit. Linkage study of the 28 families with ARCMT2 was performed using microsatellite markers flanking the ALS5/SPG11 locus, as previously described ( Orlacchio et al. , 2010 ). In the probands, all the coding exons of ALS5/SPG11/ KIAA1840 and at least 50 bp of flanking intronic sequence were PCR-amplified by primer pairs, as previously described ( Stevanin et al. , 2007 a ), and by Roche FastStart™ PCR Master Mix polymerase. All the PCR-amplified products were purified using a Qiagen PCR purification kit. Purified products were sequenced with respective forward and reverse primers by an Applied Biosystems 3130 Genetic Analyzer. Sequence analysis was performed using SeqScape software (v2.6; Applied Biosystems). Large genomic rearrangements of ALS5/SPG11/ KIAA1840 gene were investigated by multiplex ligation-dependent probe amplification method with the P306-B1 SPG11 probe mix, containing probes for each of the 40 KIAA1840 exons. Data were collected and analysed with GeneScan software (v.3.1.2; Applied Biosystems). For each sample, peak areas were calculated and compared with three wild-type controls, using Coffalyser software (v.7.0; MRC Holland).
The segregation analysis within the family and the surveillance of mutations in the control population were performed using a PCR-restriction fragment length polymorphism method. Three hundred control chromosomes were obtained from healthy volunteers of mixed ethnic origins, including Caucasian, Brazilian and Japanese.
An in silico pathogenicity prediction tool (Mutation Taster, http:www.mutationtaster.org ) was used to predict the effect of the novel mutations identified, as previously described ( Carosi et al. , 2015 ).
In all probands, we performed direct sequencing of all exons and flanking introns of CMT2A2/HMSN2A2/ MFN2 (OMIM: #609260; Nicholson et al. , 2008 ) , CMT2B1/ LMNA (OMIM: #605588; De Sandre-Giovannoli et al. , 2002 ), CMT2B2/ MED25 (OMIM: #605589; Leal et al. , 2009 ), CMT2B5/ NEFL (OMIM: #607734; Yum et al. , 2009 ), ARCMT2F/dHMN2B/ HSPB1 (OMIM: #608634; Houlden et al. , 2008 ), CMT2K/ GDAP1 (OMIM: #607831; Claramunt et al. , 2005 ), CMT2P/ LRSAM1 (OMIM: #614436; Guernsey et al. , 2010 ), CMT2R/ TRIM2 (OMIM: #615490; Pehlivan et al. , 2015 ), CMT2S/ IGHMBP2 (OMIM: #616155; Cottenie et al. , 2014 ), CMT2T/ HSJ1 (OMIM: #616233; Gess et al. , 2014 ), CMTRID/ COX6A1 (OMIM: #616039; Tamiya et al. , 2014 ), ARAN-NM/ HINT (OMIM: #137200; Zimoń et al. , 2012 ), GAN/ GAN (OMIM: #256850; Bomont et al. , 2000 ), SPG7/ PGN (OMIM: #607259; Casari et al. , 1998 ), SPG15/ ZFYVE26 (OMIM: #270700; Hanein et al. , 2008 ), SPG21/ ACP33 (OMIM: #248900; Simpson et al. , 2003 ), SPG35/ FA2H (OMIM: #612319; Pierson et al. , 2012 ), SPG46/ GBA2 (OMIM: #614409; Hammer et al. , 2013 ; Martin et al. , 2013 ), SPG55/ C12orf65 (OMIM: #615035; Spiegel et al. , 2014 ), SPG56/ CYP2U1 (OMIM: #610670; Tesson et al. , 2012 ), and SLC12A6 (OMIM: #604878; Howard et al. , 2002 ). Direct sequencing of POLG (OMIM: #174763; Van Goethem et al. , 2001 ) and TYMP (OMIM: #131222; Nishino et al. , 2000 ) genes was also carried out to exclude mitochondrial disorders related to CMT2 ( Cassereau et al. , 2014 ). Furthermore, linkage to chromosome 8q13-21.1 was performed to investigate autosomal recessive CMT2H, as previously reported ( Barhoumi et al. , 2001 ).
Results
Genetic findings
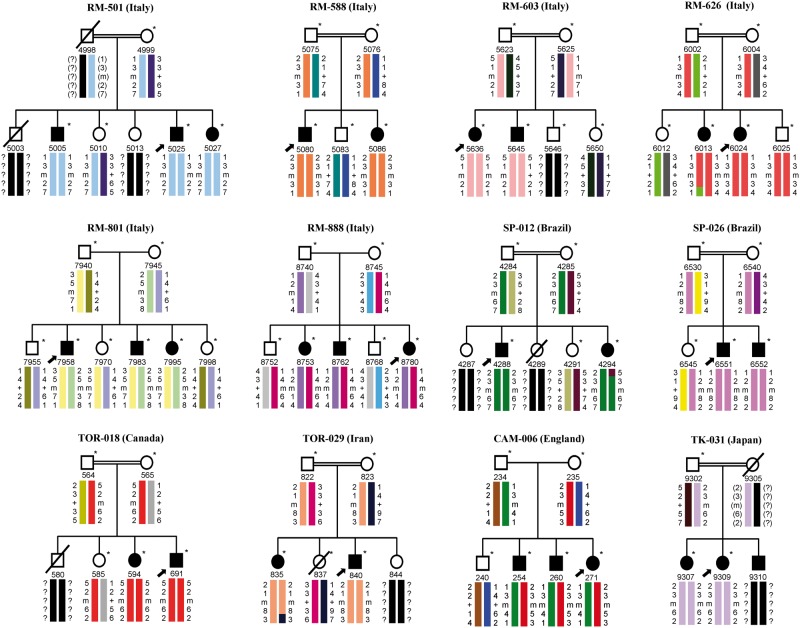
Linkage analysis in 28 unrelated pedigrees with ARCMT2 highlighted homozygous haplotypes with positive logarithm of odds score in all affected subjects from consanguineous parents of nine families (Families RM-501, RM-588, RM-603, RM-626, SP-012, SP-026, TOR-018, TOR-029 and TK-031), showing a cumulative two-point logarithm of odds score of 11.76 at the recombination fraction θ = 0.0 for marker D15S537 ( Fig. 1 ). The other ARCMT2 families showed heterozygous haplotypes and three of these kindred (Families RM-801, RM-888, and CAM-006) had positive logarithm of odds score in all affected individuals, showing a cumulative two-point logarithm of odds score of 6.29 at the recombination fraction θ = 0.0 for marker D15S537 ( Fig. 1 ). All other ARCMT2 pedigrees with heterozygous haplotypes had a negative or borderline significant cumulative logarithm of odds score by the genetic program HOMOG ( Ott, 1983 ).
Figure 1.
Pedigrees and segregation chart of 12 ARCMT2 families linked to the ALS5/SPG11 locus . Black solid symbols indicate affected individuals carrying ALS5/SPG11/ KIAA1840 mutations; white symbols represent unaffected subjects; square symbols are males, circles are females, and slashed symbols are deceased individuals. Individuals are reported as code numbers below the symbols. Probands are marked by arrows. Colour barcodes define the haplotypes created with markers D15S146, D15S537, D15S659, and D15S123, from top to bottom . Reconstructed genotypes are indicated by parentheses and question marks symbolize alleles that could not be reconstructed. The ALS5/SPG11/ KIAA1840 gene is placed between markers D15S537 and D15S659; key recombination events are observed between markers D15S659 and D15S123 in Patients 6013 (Family RM-626) and 835 (Family TOR-029), as well as between markers D15S146 and D15S537 in Patient 4294 (Family SP-012). * = sample subjects; m = mutation; + = wild-type.
We detected 15 different ALS5/SPG11/ KIAA1840 mutations in 29 affected individuals from 12 of 28 unrelated families with ARCMT2 ( Table 1 ). Nine of these nucleotide changes were homozygous and six others, detected in the affected subjects of Families RM-801, RM-888 and CAM-006, were heterozygous compounds. All variants but one were truncating, including eight frameshift mutations, and six nonsense mutations. One nucleotide change (p.Arg945Gly) was a missense mutation. Variants segregated with the disease in all pedigrees and were absent in the panel of control chromosomes. Thirteen mutations had already been reported, while two (p.Gln198* and p.Pro2212Serfs*5) were novel and segregated with the disease ( Table 1 and Fig. 2 ). Their pathogenic effect was predicted by bioinformatics analysis. In these families, the ALS5/SPG11/ KIAA1840 gene had no large deletions/duplications.
Table 1.
Mutations identified in the ALS5/SPG11/ KIAA1840 gene
| Family | Location | Mutation (cDNA) | Effect on protein | RFLP | Neuropathy a | Reference study |
|---|---|---|---|---|---|---|
| RM-501 | Exon 2 | c.398delG | p.Cys133Leufs*22 | SfcI (loss) | SM | Paisan-Ruiz et al ., 2008 |
| RM-588 | Exon 15 | c.2678G>A | p.Trp893* | – | M | Bettencourt et al ., 2014 |
| RM-603 | Exon 4 | c.704_705delAT | p.His235Argfs*12 | BsiI (gain) | SM | Stevanin et al ., 2007 b |
| RM-626 | Exon 1 | c.118C>T | p.Gln40* | – | SM | Stevanin et al ., 2007 a |
| RM-801 | Exon 3 | c.592C>T | p.Gln198* | BsgI (loss) | – | This study |
| Exon 3 | c.529_533delATATT | p.Ile177Serfs*2 | AseI b (loss) | SM | Stevanin et al ., 2007 a | |
| RM-888 | Exon 36 | c.6632dupG | p.Pro2212fs*5 | Cfr10I (gain) | – | This study |
| Exon 32 | c.6100C>T | p.Arg2034* | Cac8I b (loss) | M/SM c | Stevanin et al ., 2007 a | |
| SP-012 | Exon 37 | c.6832_6833delAG | p.Ser2278Leufs*61 | – | M | Stevanin et al ., 2007 b |
| SP-026 | Exon 38 | c.6856C>T | p.Arg2286* | MaeIII (gain) | SM | Denora et al ., 2009 |
| TOR-018 | Exon 15 | c.2697G>A | p.Trp899* | – | SM | Denora et al ., 2009 |
| TOR-029 | Exon 15 | c.2833A>G | p.Arg945Gly | MscI (loss) | SM | Stevanin et al ., 2007 b |
| CAM-006 | Exon 7 | c.1549_1550delCT | p.Leu518Leufs*39 | AhdI (gain) | SM | Stevanin et al ., 2007 b |
| Exon 36 | c.6739_6742delGAGT | p.Glu2247Leufs*14 | TfiI (gain) | SM | Stevanin et al ., 2007 b | |
| TK-031 | Exon 1 | c.165del19 | p.Ser56Alafs*7 | Cac8I (loss) | SM | Pensato et al ., 2014 |
Gene nucleotide numbering was based on the reference sequence NM_025137.3, with the A of the ATG start codon as position 1.
del = deletion; fs = frameshift; ins = insertion; M = motor; SM = sensorimotor; RFLP = restriction fragment length polymorphism.
a Axonal peripheral neuropathy refers to previously reported features; b mismatch primer used; c axonal and demyelinating peripheral neuropathy.
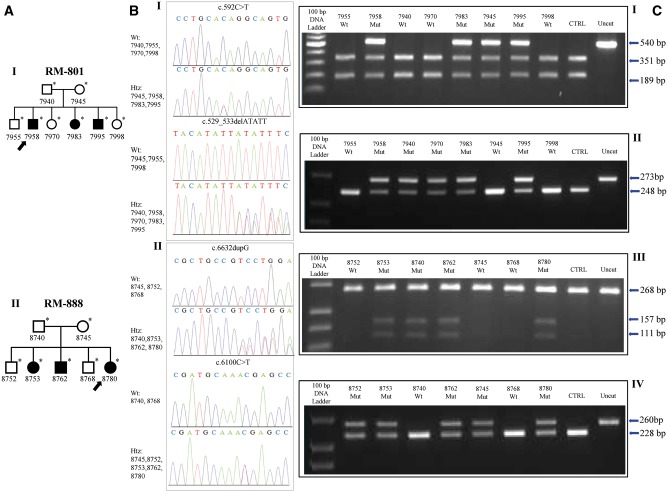
Figure 2.
Pedigree charts, electropherograms and PCR-restriction fragment length polymorphism assays of families with novel ALS5/SPG11/ KIAA1840 mutations. ( A ) Pedigree charts RM-801 ( I ) and RM-888 ( II ). ( B ) Electropherogram of exon 3 of the ALS5/SPG11/ KIAA1840 gene in individuals of Family RM-801 ( I ), showing compound heterozygous status in the affected subjects (ID 7958, ID 7983, and ID 7995), as well as heterozygous status in the relatives (ID 7940, ID 7945, and ID 7970). ( II ) Electropherogram of exon 36 and exon 32 of the ALS5/SPG11/ KIAA1840 gene in individuals of Family RM-888 ( II ), displaying compound heterozygous status in the affected subjects (ID 8753, ID 8762, and ID 8780), and heterozygous status in the relatives (ID 8740, ID 8745, and ID 8752). ( C ) PCR-restriction fragment length polymorphism assays. After BsgI digestion, heterozygous individuals of Family RM-801 show three bands at 540, 351, and 189 base pairs (bp), while wild-type subjects display two bands at 351 and 189 bp ( I ). AseI digestion proves one band in wild-type components at 248 bp, as well as two bands at 273 and 248 bp in mutated individuals ( II ). Cfr 10I digestion (c.6632dupG) leads to three bands at 268, 157, and 111 bp in heterozygous samples and one band at 268 bp (uncleaved) in wild-type components of Family RM-888 ( III ). Assay with enzyme Cac8I displays one band in wild-type individuals at 228 bp, as well as two bands at 260 and 228 bp in mutated subjects ( IV ). Wt = wild-type; Htz = heterozygous; Mut = mutated.
Genetic analysis of the other candidate genes (CMT2A2/HMSN2A2/ MFN2 , CMT2B1/ LMNA , CMT2B2/ MED25 , CMT2B5/ NEFL , ARCMT2F/dHMN2B/ HSPB1 , CMT2K/ GDAP1 , CMT2P/ LRSAM1 , CMT2R/ TRIM2 , CMT2S/ IGHMBP2 , CMT2T/ HSJ1 , CMTRID/ COX6A1 , ARAN-NM/ HINT , GAN/ GAN , SPG7/ PGN , SPG15/ ZFYVE26 , SPG21/ ACP33 , SPG35/ FA2H , SPG46/ GBA2 , SPG56/ CYP2U1 , and SLC12A6 ) did not reveal any coding mutations in the unrelated 28 families with ARCMT2. Direct sequencing of POLG and TYMP genes excluded mitochondrial disorders related to CMT2. Linkage analysis showed no linkage to chromosome 8q13-21.1 in all families (cumulative logarithm of odds score was −2.11 for marker D15S537 at θ = 0.0).
Clinical findings of the ALS5/SPG11/ KIAA1840 -related ARCMT2 families
Among the 12 ALS5/SPG11/ KIAA1840 mutated pedigrees, nine showed consanguinity marriage (first-degree cousins) in their parents. Six families were from Italy, two from Brazil, one from England, and one from Japan. Moreover, two families were from Canada, one of which was an Iranian family who emigrated to Ontario ( Fig. 1 ).
Affected subjects had a mainly distal, slowly progressive sensory and motor axonal neuropathy. The age at onset of the first motor symptoms in the 29 mutated ALS5/SPG11/ KIAA1840 patients oscillated from 4 to 35, with a mean of 11.4 ± 5.9 years. The mean age at examination was 25.1 ± 7.8 years (range 9–52).
A wide variation of phenotypic expression was detected ( Table 2 ). Asymmetrical onset in lower limbs was present in 12 patients. Distal lower limb weakness was found in all affected individuals; 16 patients showed muscular weakness in upper limbs too. Wasting was frequent in lower limb muscles distally (69%) and in intrinsic hand muscles (48%). Lower limb fasciculations were detected in eight patients. No patient showed pontobulbar signs, such as jaw spasticity, poor palatal elevation, increased facial reflexes, masseter, pterygoids, sternocleidomastoid, tongue muscle weakness, or muscle atrophy with fasciculations. Four patients had proximal lower limb weakness. Loss of pinprick and light touch sensation was common and involved lower limbs in most cases (86%).
Table 2.
Clinical features of 29 patients with ALS5/SPG11/ KIAA1840 mutations
| Onset a in the second decade of life | 21 (72) |
| Disease duration >10 years | 18 (62) |
| Males | 15 (52) |
| Females | 14 (48) |
| Muscle weakness | |
| Distal lower limbs | 29 (100) |
| Proximal lower limbs | 4 (14) |
| Distal upper limbs | 16 (55) |
| Proximal upper limbs | 0 (0) |
| Muscle wasting | |
| Distal lower limbs | 20 (69) |
| Proximal lower limbs | 1 (3) |
| Hands | 14 (48) |
| Loss of pinprick and light touch in lower limbs | 25 (86) |
| Loss of proprioception in lower limbs | 15 (52) |
| Loss of pinprick and light touch in upper limbs | 8 (28) |
| Loss of proprioception in upper limbs | 0 (0) |
| Total absence of deep tendon reflexes | |
| Lower limbs | 8 (28) |
| Upper limbs | 2 (7) |
| Asymmetrical onset | 12 (41) |
| Positive Romberg test | 7 (24) |
| Gait impairment | 18 (62) |
| Babinski sign | 2 (7) |
| Postural tremor | 10 (34) |
| Wrist contracture | 3 (10) |
| Elbow contractures | 1 (3) |
| Ankle contracture | 14 (48) |
| Calf hypertrophy | 4 (14) |
| Foot deformities | 23 (79) |
| Fasciculations | |
| Lower limbs | 8 (28) |
| Upper limbs | 1 (3) |
| Foot drop | 11 (38) |
| Hand deformities | 7 (24) |
| Kyphoscoliosis | 17 (59) |
| Mental retardation | 3 (10) |
| Thin corpus callosum | 3 (10) |
| Cerebellar signs | 0 (0) |
| Retinal disease | 0 (0) |
| Grey matter atrophy on brain MRI | 1 (3) |
| White matter abnormalities on brain MRI | 0 (0) |
| Bladder disturbances | 9 (31) |
| Sexual dysfunctions | 7 (24) |
| Spasticity | 0 (0) |
| FDS 0-2 | 14 (48) |
| FDS 3-5 | 13 (45) |
| FDS 6-8 | 2 (7)* |
| CMTNS mild | 9 (31) |
| CMTNS moderate | 16 (55) |
| CMTNS severe | 4 (14)# |
Values are n (%).
a Age at onset was calculated as the time when motor symptoms appeared. FDS (Functional Disability Scale) from 0 to 8 as follows: 0 = normal; 1 = normal, but with cramps and fatigability; 2 = inability to run; 3 = walking difficult, but still possible unaided; 4 = able to walk with a cane; 5 = able to walk with crutches; 6 = able to walk with a walker; 7 = wheelchair bound; 8 = bedridden. CMTNS = Charcot-Marie-Tooth Neuropathy Score: mild (≤10), moderate (11–20), and severe (>20). *Ages at onset were 4 and 9 years; # ages at onset were 4, 8, 9, and 12 years.
The most frequent additional signs were foot deformities (79%), mainly pes cavus and kyphoscoliosis (59%). Hand deformities were present in seven patients with longer disease duration. Ankle contracture was present in 14 patients (48%) and tremor in 10 (34%). Bladder disturbances and sexual dysfunctions were detected in about one-third of the affected subjects. Babinski’s sign was present bilaterally in two unrelated patients. No affected subject showed cerebellar signs or retinal disease.
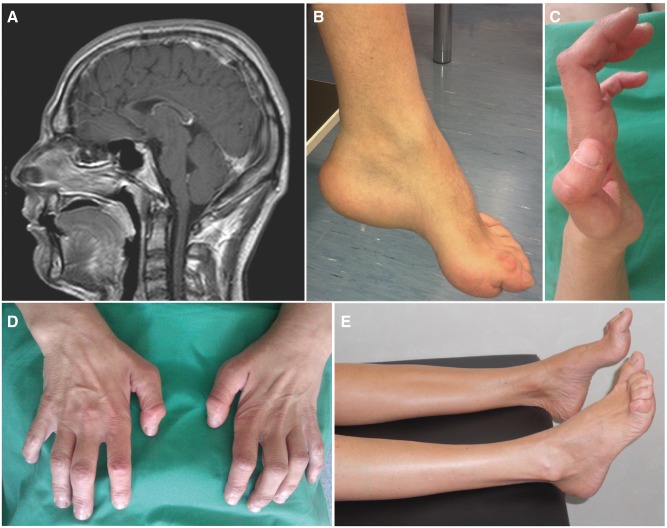
Brain MRI images were available for all mutated KIAA1840 patients. Thin corpus callosum was evident in the three affected siblings of Family RM-801, with severe thinning of the anterior portion and mild thinning of the splenium ( Fig. 3 A). These patients had low scores in the Wechsler Intelligence Scales for Children IV Intelligence Quotient (ID 7958: 51, ID 7983: 49, and ID 7995: 62; at 13, 11, and 9 years, respectively) with poor school performance and deficit in memory and calculation tests. Normal corpus callosum and no mental retardation or cognitive deficits were observed in the other ARCMT2 KIAA1840 -mutated subjects.
Figure 3.
Clinical features of Patients 7958 (Family RM-801) and 8780 (Family RM-888) . ( A ) T 1 -weighted MRI showing thin corpus callosum in the proband of Family RM-801. ( B ) Pes cavovarus of the same patient. ( C ) Deformities and ( D ) global atrophy of hands in the proband of Family RM-888. ( E ) Atrophied legs with pes cavus of the same patient.
Haematological and biochemical profiles, including serum creatine kinase, were unremarkable in all subjects. Lysosomal enzyme assay of peripheral blood leucocytes revealed that both β-hexosaminidase A and B isoenzymes were present at normal levels. Vitamins were within normal range. The CSF analysis was within normal range in the five patients analysed.
The disease was slowly progressive and most patients (86%) expressed mild-to-moderate phenotypes (Charcot–Marie–Tooth neuropathy score < 20); only two patients, one of which was wheelchair bound, reached Functional Disability Scale scores > 5 ( Table 2 ).
Motor and sensory conduction studies showed motor and sensory axonal neuropathy, more prominent in lower limbs, with low amplitudes of compound motor and sensory nerve action potentials. Motor and sensory nerve conduction velocities were normal or slightly reduced, according to the secondary demyelinating processes. All motor nerve conduction velocities of median nerves were >38 m/s, differentiating CMT2 from CMT1. In 29 affected individuals, EMG of the tibialis anterior muscle detected predominant findings of chronic denervation/reinnervation (motor unit action potentials with long duration and large amplitude, or polyphasic, along with single or mixed interference patterns), and/or active denervation signs (fibrillation potentials, positive sharp waves, as well as fasciculation potentials). In 11 affected subjects, EMG of the abductor pollicis brevis muscle showed similar neurophysiological characteristics ( Table 3 ).
Table 3.
Electroneurography and EMG characteristics in the 29 ALS5/SPG11/ KIAA1840 –mutated individuals
| Mean ± SD | Range | Reference lower value a | UR (subjects) | Pathological (subjects) b | Chronic denervation | Active denervation | |
|---|---|---|---|---|---|---|---|
| Median nerve | |||||||
| CMAP (mV) | 3.6 ± 2.4 | 0.1–9.2 | 5 | 5 | 15 | – | – |
| SNAP (µV) c | 6.8 ± 5.3 | 0.4–21.7 | 9 | 6 | 16 | – | – |
| MNCV (m/s) | 48.1 ± 7.5 | 39–67 | 50 | – | 12 | – | – |
| SNCV (m/s) | 51.1 ± 8.2 | 35–64 | 54 | – | 13 | – | – |
| Peroneal nerve | |||||||
| CMAP (mV) | 2.1 ± 3.8 | 0.1–11.8 | 3 | 13 | 26 | – | – |
| MNCV (m/s) | 38.3 ± 5.1 | 31–44 | 42 | – | 20 | – | – |
| Tibial nerve | |||||||
| CMAP (mV) | 2.4 ± 3.9 | 0.1–16.5 | 4 | 14 | 27 | – | – |
| MNCV (m/s) | 38.1 ± 4.8 | 32–46 | 42 | – | 21 | – | – |
| Sural nerve | |||||||
| SNAP (µV) | 2.3 ± 1.5 | 0.3–3.8 | * | 16 | 29 | – | – |
| SNCV (m/s) | 37.3 ± 5.2 | 31–47 | 42 | – | 22 | – | – |
| TA muscle | – | – | – | – | 29 | 23 | 8 |
| APB muscle | – | – | – | – | 11 | 9 | 3 |
APB = abductor pollicis brevis; CMAP = compound nervous motor action potential (amplitude); MNCV = motor nerve conduction velocity; SD = standard deviation; SNAP = sensory nerve action potential (amplitude); SNCV = sensory nerve conduction velocity; TA = tibialis anterior; UR = unrecordable (action potentials). a Reference lower value is the lower limit assessed by our laboratories; b The ‘Pathological’ column includes non–recordable items; c Orthodromic evaluation. *SNAP reference lower value of sural nerve is calculated as: 11.34 − (0.092 × age in years) − 5.04 µV ( Bienfait, 2007 ).
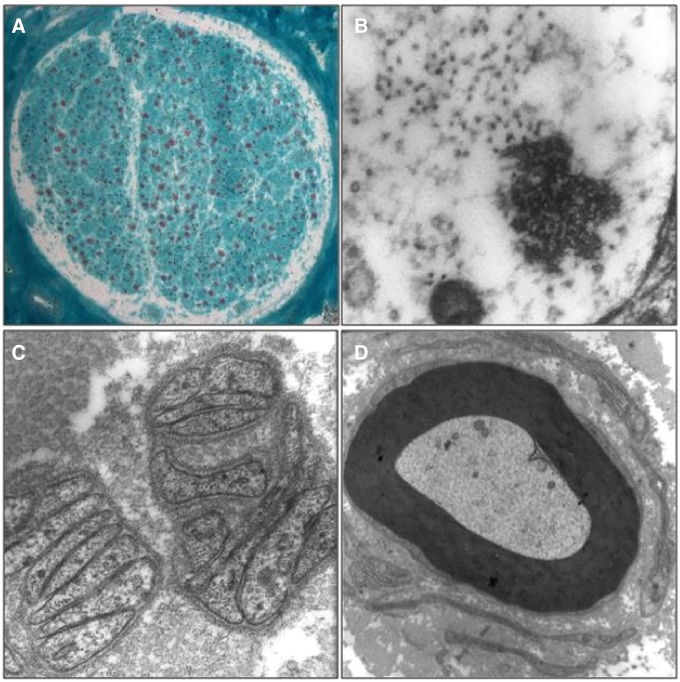
Neuropathological analyses of the sural nerve biopsies were performed in 17 mutated patients and showed loss of myelinated fibres ( Fig. 4 A), mainly in fibres of large calibre, in line with the diagnosis of CMT2.
Figure 4.
Sural nerve biopsy of Patient 7958 (Family RM-801). ( A ) Transverse cryostat section showing a nerve fascicle with medium grade myelinated fibre loss. Modified Gomori Trichrome Stain, ×80. ( B–D ) Transmission electron micrographs showing: ( B ) an unmyelinated fibre displaying intra-axonal aggregates, seemingly composed by neurotubules and neurofilaments, ×85 000; ( C ) flattened Schwann cell processes indicating unmyelinated axon loss, ×30 000; ( D ) a rudimentary onion bulb surrounding an apparently intact myelinated fibre, possibly indicating secondary Schwann cell pathology, ×7000.
See the online Supplementary material for indicative case reports of patients with novel ALS5/SPG11/ KIAA1840 mutations.
Discussion
The ALS5/SPG11/ KIAA1840 gene was analysed in ARCMT2 patients from 28 unrelated pedigrees and a high frequency of pathogenic mutations, two of which were novel, was found in 12 families (43%), appearing to be a significant cause of ARCMT2. Our study showed novel genotype–phenotype correlation, which further broadens the clinical spectrum associated with ALS5/SPG11/ KIAA1840 mutations ( Fig. 5 ). Therefore, genetic screening of the ALS5/SPG11/ KIAA1840 gene should be considered not only in patients with ARHSP with thin corpus callosum and in affected individuals with ARJALS, but also in affected subjects with ARCMT2. Due to the complexity and heterogeneity of these neurodegenerative diseases, next-generation sequencing techniques are the most effective diagnostic tool to identify the genetic cause in patients with myelopathy or neuropathy, either by targeted sequencing panel approach or by whole-genome sequencing.
Figure 5.
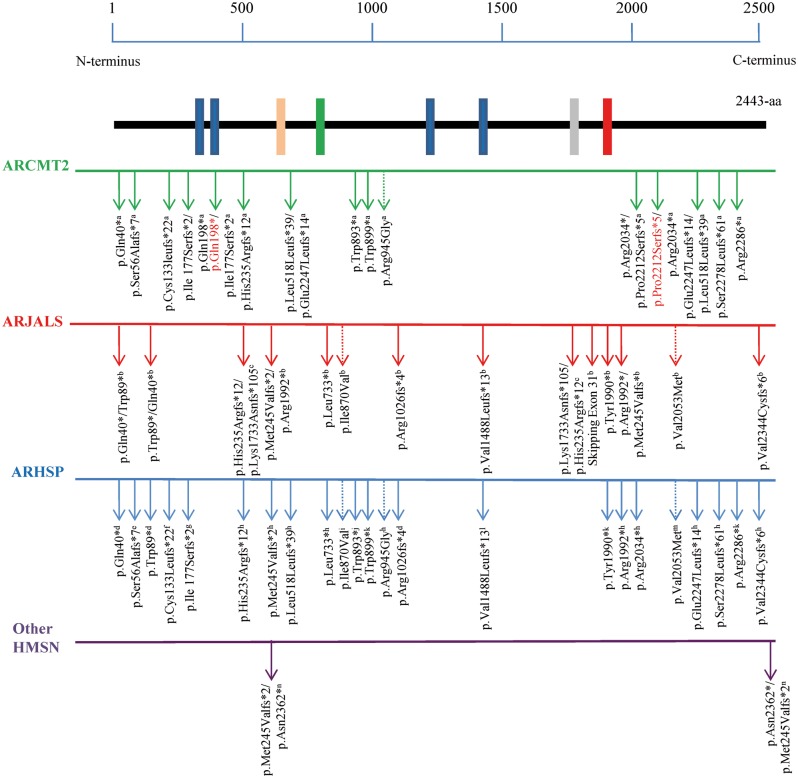
Schematic representation of the ALS5/SPG11 protein and position of previously reported (black) and novel (red) mutations, stratified by distinctive phenotypes . Specific mutations for clinical heterogeneity are represented. Putative functional domains are depicted as rectangles, and their positions within the amino acid sequence are indicated: the transmembrane domain (blue box; positions, 163–194, 200–240, 1239–1267, and 1471–1493), glycosyl hydroxylase F1 signature (pink box; position, 482–490), leucine zipper (green box; position, 611–632), coil-coil domain (grey box; position, 1556–1590), and Myb domain (red box; position 1766–1774). Arrows indicate truncating mutations; dotted arrows indicate missense mutations. Aa = amino acids. a This study; b Orlacchio et al. , 2010 ; c Daoud et al. , 2012 ; d Hehr et al. , 2007 ; e Pensato et al. , 2014 ; f Paisan-Ruiz et al. , 2008 ; g Stevanin et al. , 2007 a ; h Stevanin et al. , 2007 b ; i Pippucci et al. , 2009 ; j Bettencourt et al. , 2014 ; k Denora et al. , 2009 ; l Lee et al. , 2008 ; m Del Bo et al. , 2007 ; n Querin et al. , 2014 .
This clinical–genetic entity shows a worldwide distribution, since the pedigrees carrying mutations in ALS5/SPG11/ KIAA1840 were from Italy, Brazil, Canada, England, Iran, and Japan. The variants are scattered throughout the entire amino acid sequence, without evidence of ‘hot spots’, and 93% were truncating mutations. This finding is consistent with previous studies describing mostly mutations leading to the truncation of the ALS5/SPG11 protein and consequent loss of function mechanism ( Pensato et al. , 2014 ). Furthermore, a probable mechanism of nonsense-mediated mRNA decay might also be hypothesized ( Paisan-Ruiz et al. , 2008 ).
The two unrelated ARCMT2 patients with bilateral Babinski’s sign further enlarge the clinical spectrum of the disorder, showing that KIAA1840 -related diseases might be clinically suspected if neuropathy is present in addition to upper motor neuron signs or symptoms, e.g. deep tendon reflexes are symmetrically diminished in the lower extremities with bilateral positive Babinski’s sign. Moreover, the impairment of higher mental functions impairment and brain alterations at MRI in three affected siblings of one ARCMT2 family should be noted. Indeed, the presence of clinical signs of leukoencephalopathy, as well as white matter alterations at brain MRI, has been previously reported in demyelinating, axonal, and intermediate forms of the disorder ( Genari et al. , 2011 ; Reyes-Marin et al. , 2011 ; Sagnelli et al. , 2014 ). Therefore, we believe that brain MRI and neuropsychological testing are useful tools in the clinical work-up of patients with Charcot–Marie–Tooth disease. To our knowledge, this is the first report of ALS5/SPG11/ KIAA1840 mutations in subjects with ARCMT2 and all of our families can be classified as CMT2X, according to the classification of the Charcot–Marie–Tooth disease currently used in OMIM ( http://www.ncbi.nlm.nih.gov/omim ).
The disease has a slow progression and affected subjects with earlier onset show a more severe phenotype compared to those with late onset. Overall, the age at onset is the second decade of life, in line with both ARCMT2 and ARHSP ( Tazir et al. , 2013 ; Lo Giudice et al. , 2014 ). Clinical heterogeneity is well documented in ALS5/SPG11/ KIAA1840 mutations causing ARHSP, ranging from the most frequent association of thin corpus callosum, white matter alterations, mental retardation, cerebellar signs, and axonal peripheral neuropathy, to less common clinical findings, such as seizures, abnormal eye signs, maculopathy, amyotrophy, and parkinsonism ( Tessa et al. , 2014 ). In addition, ALS5/SPG11/ KIAA1840 mutations have been often found in patients with ARJALS ( Orlacchio et al. , 2010 ). Interestingly, no specific differences in the type of variants, or position within the primary amino acid sequence, were observed. Even identical mutations causing different phenotypes were found ( Fig. 5 ), confirming that variants at the same locus can lead to distinct phenotypes ( Hegele, 2005 ). This aspect is not uncommon in monogenic diseases and demonstrates how genotype–phenotype associations can be often unpredictable.
The complexity of a correct clinical classification of motor neuron diseases linked to spatacsin is further increased by the presence of ALS5/SPG11/ KIAA1840 mutations in patients with an overlapping phenotype, exhibiting features of amyotrophic lateral sclerosis as well as spastic paraplegia, as described by Querin et al. (2014) .
As previously hypothesized, the ALS5/SPG11/ KIAA1840 phenotype results from the combined degeneration of central and peripheral axons ( Hehr et al. , 2007 ). In particular, peripheral neuropathy might affect both sensory and motor fibres and might lead to pure distal amyotrophy, as well as to CMT2. The roles of spatacsin in the cellular pathway of axonal maintenance, including cargo trafficking, is becoming clearer: it is located in axons and dendrites, co-localized with cytoskeletal and synaptic vesicles, and it is present in synaptosomes ( Chiurchiù et al. , 2014 ; Pérez-Brangulí et al. , 2014 ). Other forms of hereditary spastic paraplegia linked to proteins involved in axon maintenance, such as kinesin SPG10/ KIF5A and SPG17/ BSCL2 , are known to be allelic to CMT2 ( Goizet et al. , 2009 ; Ito and Suzuki, 2009 ; Crimella et al. , 2012 ; Timmerman et al. , 2013 ). Furthermore, mutations in SPG4/ SPAST , SPG3A /ATL1 , SPG17/ BSCL2 , as well as SPG43/ C19orf12 might cause spastic paraplegia and axonal peripheral neuropathy with atrophy of small hand muscles (Silver syndrome) ( Windpassinger et al. , 2004 ; Orlacchio et al. , 2008 ; Fusco et al. , 2010 ; Landouré et al. , 2013 ). Besides, mutations in ATL1 , whose protein is implicated in neurite outgrowth, intracellular membrane trafficking, and axon elongation during neuronal development ( Byrnes and Sondermann, 2011 ), might cause hereditary sensory neuropathy type I (HSN-I) with or without pyramidal tract features ( Guelly et al. , 2011 ; Leonardis et al. , 2012 ).
Remarkably, the previously reported mutations in ALS5/SPG11/ KIAA1840 were often associated with ARHSP complicated by axonal peripheral neuropathy, mainly motor, but also sensory ( Table 1 ). In KIAA1840 mutations, a preferential involvement of the CNS or the peripheral nervous system may be due to different causes. First of all, the blood–nerve barrier might influence protein expression in peripheral axons, as well as the blood–brain barrier in central axons. Moreover, ALS5/SPG11/ KIAA1840 knockdown in Zebrafish ( Danio rerio ) reveals a compromised outgrowth of spinal motor axons, thus indicating that spatacsin is involved in the formation of neuromuscular junctions during development ( Martin et al. , 2012 ). The association in synaptosomes of spatacsin with spastizin (SPG15/ ZFYVE26 ) and with the adaptor protein complex 5 (SPG48/ KIAA0415 ) ( Hirst et al. , 2013 ) suggests that the loss of function of spatacsin could not be the only mechanism for axonal dysfunction in ALS5/SPG11/ KIAA1840 -mutated patients. Indeed, it may be hypothesized that a variable phenotypic expression of KIAA1840 mutations results from the interaction of gene modifier factors. Further studies are necessary to clarify whether allelic heterogeneity depends on environmental or other genetic/epigenetic factors. In conclusion, the presence of common molecular, pathological, and genetic features in KIAA1840 -related diseases, suggests that the different phenotypes of axonal degeneration induced by ALS5/SPG11/ KIAA1840 mutations may be targeted by common therapeutic strategies.
Supplementary Material
Acknowledgements
We thank the patients and their family members for taking part in this study. We thank Michela Renna (MA) for her language advice and assistance, Martina Di Lullo (BSc) and Valerio Battisti (BSc) for the technical support, and the members of our laboratories for the stimulating discussions and helpful comments on this manuscript. We are extremely grateful to the Genetic Bank of the Laboratorio di Neurogenetica, CERC - IRCCS Santa Lucia , Rome, Italy ( http://www.hsantalucia.it/neurogen/index_en.htm ) for the service provided.
Glossary
Abbreviations
- ARCMT2
autosomal recessive Charcot–Marie–Tooth disease type 2
- ARHSP
autosomal recessive hereditary spastic paraplegia
- CMT
Charcot–Marie–Tooth
Funding
This work was supported by the Italian Ministero della Salute (Grant no. GR09.109 to A.O.), the Comitato Telethon Fondazione Onlus , Italy (Grant no. GGP10121 to A.O.), the Università di Roma “Tor Vergata” , Rome, Italy (Grant no. E82I15000190005 to A.O.), the Rotary Club Sanluri Medio Campidano , Sanluri (VS), Italy (Grant Noi per Voi to A.O.), the Japan Society for the Promotion of Science (JSPS KAKENHI Grant no. 26461294 to T.K.), the Ministry of Health, Labour, and Welfare of Japan (Grant for research on rare and intractable diseases and establishment of novel treatments for amyotrophic lateral sclerosis to T.K.), and the Brain Science Foundation, Japan (Grant to T.K.).
Supplementary material
Supplementary material is available at Brain online.
References
- American Psychiatric Association . Diagnotic and statistical manual of mental disorders . 5th edn. Arlington, VA: : American Psychiatric Association; ; 2013. 10.1176/appi.books.9780890425596 [DOI] [Google Scholar]
- Barhoumi C, Amouri R, Ben Hamida C, Ben Hamida M, Machghoul S, Gueddiche M, et al. . Linkage of a new locus for autosomal recessive axonal form of Charcot-Marie-Tooth disease to chromosome 8q21.3 . Neuromuscul Disord 2001. ; 11 : 27 – 34 . [DOI] [PubMed] [Google Scholar]
- Bettencourt C, López-Sendón JL, García-Caldentey J, Rizzu P, Bakker IM, Shomroni O, et al. . Exome sequencing is a useful diagnostic tool for complicated forms of hereditary spastic paraplegia . Clin Genet 2014. ; 85 : 154 – 8 . [DOI] [PubMed] [Google Scholar]
- Bienfait HME . Axonal phenotypes in Charcot-Marie-Tooth disease . Amsterdam, The Netherlands: : Faculty of Medicine, University of Amsterdam (AMC-UvA) ; 2007. . http://dare.uva.nl/document/2/47043 [Google Scholar]
- Bird DT . GeneReviews [Internet] In: Pagon RA, Adam MP, Ardinger HH, Bird DT, Dolan CR, Fong CT, et al. ., editors. Charcot-Marie-Tooth neuropathy type 2 . Seattle, WA: : University of Washington, Seattle; ; 1993-2015. . http://www.ncbi.nlm.nih.gov/books/NBK1285/ [Google Scholar]
- Birouk N, Gouider R, Le Guern E, Gugenheim M, Tardieu S, Maisonobe T, et al. . Charcot-Marie-Tooth disease type 1A with 17p11.2 duplication. Clinical and electrophysiological phenotype study and factors influencing disease severity in 119 cases . Brain 1997. ; 120 : 813 – 23 . [DOI] [PubMed] [Google Scholar]
- Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, et al. . The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy . Nat Genet 2000. ; 26 : 370 – 4 . [DOI] [PubMed] [Google Scholar]
- Byrnes LJ, Sondermann H . Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A . Proc Natl Acad Sci USA 2011. ; 108 : 2216 – 21 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlesimo GA, Caltagirone C, Gainotti G . The mental deterioration battery: normative data, diagnostic reliability and qualitative analyses of cognitive impairment. The group for the standardization of the mental deterioration battery . Eur Neurol 1996. ; 36 : 378 – 84 . [DOI] [PubMed] [Google Scholar]
- Carosi L, Lo Giudice T, Di Lullo M, Lombardi F, Babalini C, Gaudiello F, et al. . Hereditary spastic paraplegia: a novel mutation and expansion of the phenotype variability in SPG10 . J Neurol Neurosurg Psychiatry 2015. ; 86 : 702 – 4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, et al. . Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease . Cell 1998. ; 93 : 973 – 83 . [DOI] [PubMed] [Google Scholar]
- Cassereau J, Codron P, Funalot B . Inherited peripheral neuropathies due to mitochondrial disorders . Rev Neurol (Paris) 2014. ; 170 : 366 – 74 . [DOI] [PubMed] [Google Scholar]
- Chiurchiù V, Maccarrone M, Orlacchio A . The role of reticulons in neurodegenerative diseases . Neuromolecular Med [Review] 2014. ; 16 : 3 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claramunt R, Pedrola L, Sevilla T, López de Munain A, Berciano J, Cuesta A, et al. . Genetics of Charcot-Marie-Tooth disease type 4A: mutations, inheritance, phenotypic variability, and founder effect . J Med Genet 2005. ; 42 : 358 – 65 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottenie E, Kochanski A, Jordanova A, Bansagi B, Zimon M, Horga A, et al. . Truncating and missense mutations in IGHMBP2 cause Charcot-Marie Tooth disease type 2 . Am J Hum Genet 2014. ; 95 : 590 – 601 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimella C, Baschirotto C, Arnoldi A, Tonelli A, Tenderini E, Airoldi G, et al. . Mutations in the motor and stalk domains of KIF5A in spastic paraplegia type 10 and in axonal Charcot-Marie-Tooth type 2 . Clin Genet 2012. ; 82 : 157 – 64 . [DOI] [PubMed] [Google Scholar]
- Daoud H , Zhou S , Noreau A , Sabbagh M , Belzil V , Dionne-Laporte A , et al. . Exome sequencing reveals SPG11 mutations causing juvenile ALS . Neurobiol Aging 2012. ; 33 : 839. e5–9 . [DOI] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Chaouch M, Kozlov S, Vallat JM, Tazir M, Kassouri N, et al. . Homozygous defects in LMNA , encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse . Am J Hum Genet 2002. ; 70 : 726 – 36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bo R, Di Fonzo A, Ghezzi S, Locatelli F, Stevanin G, Costa A, et al. . SPG11: a consistent clinical phenotype in a family with homozygous spatacsin truncating mutation . Neurogenetics 2007. ; 8 : 301 – 5 . [DOI] [PubMed] [Google Scholar]
- Denora PS, Schlesinger D, Casali C, Kok F, Tessa A, Boukhris A, et al. . Screening of ARHSP-TCC patients expands the spectrum of SPG11 mutations and includes a large scale gene deletion . Hum Mutat 2009. ; 30 : 500 – 19 . [DOI] [PubMed] [Google Scholar]
- Dyck PJ, Thomas PK , editors. Peripheal neuropathy . 4th edn. Philadelphia, PA: : Elsevier Saunders; ; 2005. . http://www.scribd.com/doc/243563627/Peripheral-Neuropathy-2005-Dyck-pdf#scribd [Google Scholar]
- Dyck PJ, Gutrecht JA, Bastron JA, Karnes WE, Dale AJ . Histologic and teased-fiber measurements of sural nerve in disorders of lower motor and primary sensory neurons . Mayo Clin Proc 1968. ; 43 : 81 – 123 . [PubMed] [Google Scholar]
- Feely S, Laura M, Siskind CE, Sottile S, Davis M, Gibbons VS, et al. . MFN2 mutations cause severe phenotypes in most patients with CMT2A . Neurology 2011. ; 76 : 1690 – 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman V, Murphy SM . The spectrum of axonopathies: from CMT2 to HSP . Neurology 2014. ; 83 : 580 – 1 . [DOI] [PubMed] [Google Scholar]
- Fusco C, Frattini D, Farnetti E, Nicoli D, Casali B, Fiorentino F, et al. . Hereditary spastic paraplegia and axonal motor neuropathy caused by a novel SPG3A de novo mutation . Brain Dev 2010. ; 32 : 592 – 4 . [DOI] [PubMed] [Google Scholar]
- Genari AB, Borghetti VH, Gouvêa SP, Bueno KC, dos Santos PL, dos Santos AC, et al. . Characterizing the phenotypic manifestations of MFN2 R104W mutation in Charcot-Marie-Tooth type 2 . Neuromuscul Disord 2011. ; 21 : 428 – 32 . [DOI] [PubMed] [Google Scholar]
- Gess B, Auer-Grumbach M, Schirmacher A, Strom T, Zitzelsberger M, Rudnik-Schöneborn S, et al. . HSJ1 -related hereditary neuropathies: novel mutations and extended clinical spectrum . Neurology 2014. ; 83 : 1726 – 32 . [DOI] [PubMed] [Google Scholar]
- Goizet C, Boukhris A, Mundwiller E, Tallaksen C, Forlani S, Toutain A, et al. . Complicated forms of autosomal dominant hereditary spastic paraplegia are frequent in SPG10 . Hum Mutat 2009. ; 30 : 376 – 85 . [DOI] [PubMed] [Google Scholar]
- Guelly C, Zhu PP, Leonardis L, Papić L, Zidar J, Schabhüttl M, et al. . Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I . Am J Hum Genet 2011. ; 88 : 99 – 105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Bedard K, Evans SC, Ferguson M, Matsuoka M, et al. . Mutation in the gene encoding ubiquitin ligase LRSAM1 in patients with Charcot-Marie-Tooth disease . PLoS Genet 2010. ; 6 : e1001081 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MB, Eleuch-Fayache G, Schottlaender LV, Nehdi H, Gibbs JR, Arepalli SK, et al. . Mutations in GBA2 cause autosomal-recessive cerebellar ataxia with spasticity . Am J Hum Genet 2013. ; 92 : 245 – 51 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein S, Martin E, Boukhris A, Byrne P, Goizet C, Hamri A, et al. . Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome . Am J Hum Genet 2008. ; 82 : 992 – 1002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele R . LMNA mutation position predicts organ system involvement in laminopathies . Clin Genet 2005. ; 68 : 31 – 4 . [DOI] [PubMed] [Google Scholar]
- Hehr U, Bauer P, Winner B, Schüle R, Olmez A, Koehler W, et al. . Long-term course and mutational spectrum of spatacsin-linked spastic paraplegia . Ann Neurol 2007. ; 62 : 656 – 65 . [DOI] [PubMed] [Google Scholar]
- Hirst J, Borner GH, Edgar J, Hein MY, Mann M, Buchholz F, et al. . Interaction between AP-5 and the hereditary spastic paraplegia proteins SPG11and SPG15 . Mol Biol Cell 2013. ; 24 : 2558 – 69 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H, Laura M, Wavrant-De Vrièze F, Blake J, Wood N, Reilly MM . Mutations in the HSP27 ( HSPB1 ) gene cause dominant, recessive, and sporadic distal HMN/CMT type 2 . Neurology 2008. ; 71 : 1660 – 8 . [DOI] [PubMed] [Google Scholar]
- Howard HC, Mount DB, Rochefort D, Byun N, Dupré N, Lu J, et al. . The K-Cl cotransporter KCC3 is mutant in a severe peripheral neuropathy associated with agenesis of the corpus callosum . Nat Genet 2002. ; 32 : 384 – 92 . Erratum in Nat Genet 2002; 32: 681 . [DOI] [PubMed] [Google Scholar]
- Ito D, Suzuki N . Seipinopathy: a novel endoplasmic reticulum stress-associated disease [Review] . Brain 2009. ; 132 : 8 – 15 . [DOI] [PubMed] [Google Scholar]
- Jerath NU, Shy ME . Hereditary motor and sensory neuropathies: Understanding molecular pathogenesis could lead to future treatment strategies . Biochim Biophys Acta 2015. ; 1852 : 667 – 78 . [DOI] [PubMed] [Google Scholar]
- Landouré G, Zhu PP, Lourenço CM, Johnson JO, Toro C, Bricceno KV, et al. . Hereditary spastic paraplegia type 43 (SPG43) is caused by mutation in C19orf12 . Hum Mutat 2013. ; 34 : 1357 – 60 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal A, Huehne K, Bauer F, Sticht H, Berger P, Suter U, et al. . Identification of the variant Ala335Val of MED25 as responsible for CMT2B2: molecular data, functional studies of the SH3 recognition motif and correlation between wild-type MED25 and PMP22 RNA levels in CMT1A animal models . Neurogenetics 2009. ; 10 : 275 – 87 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Cheng TW, Hua MS, Pan MK, Wang J, Stephenson DA, et al. . Mutations of the SPG11 gene in patients with autosomal recessive spastic paraparesis and thin corpus callosum . J Neurol Neurosurg Psychiatry 2008. ; 79 : 607 – 9 . [DOI] [PubMed] [Google Scholar]
- Leonardis L, Auer-Grumbach M, Papić L, Zidar J . The N355K atlastin 1 mutation is associated with hereditary sensory neuropathy and pyramidal tract features . Eur J Neurol 2012. ; 19 : 992 – 8 . [DOI] [PubMed] [Google Scholar]
- Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A . Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms [Review] . Exp Neurol 2014. ; 261 : 518 – 39 . [DOI] [PubMed] [Google Scholar]
- Martin E, Schüle R, Smets K, Rastetter A, Boukhris A, Loureiro JL, et al. . Loss of function of glucocerebrosidase GBA2 is responsible for motor neuron defects in hereditary spastic paraplegia . Am J Hum Genet 2013. ; 92 : 238 – 44 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E, Yanicostas C, Rastetter A, Naini SM, Maouedj A, Kabashi E, et al. . Spatacsin and spastizin act in the same pathway required for proper spinal motor neuron axon outgrowth in Zebrafish . Neurobiol Dis 2012. ; 48 : 299 – 308 . [DOI] [PubMed] [Google Scholar]
- Medical Research Council . Aids to the examination of the peripheral nervous system . Her Majesty's Stationery Office; . London, UK: ; 1981. , Memorandum no. 45 . [Google Scholar]
- Murphy SM, Herrmann DN, McDermott MP, Scherer SS, Shy ME, Reilly MM, et al. . Reliability of the CMT neuropathy score (second version) in Charcot-Marie-Tooth disease . J Peripher Nerv Syst 2011. ; 16 : 191 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson GA, Magdelaine C, Zhu D, Grew S, Ryan MM, Sturtz F, et al. . Severe early-onset axonal neuropathy with homozygous and compound heterozygous MFN2 mutations . Neurology 2008. ; 70 : 1678 – 81 . [DOI] [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Papadimitriou A, Hammans S, Steiner I, Hahn CD, et al. . Mitochondrial neurogastrointestinal encephalomyopathy: an autosomal recessive disorder due to thymidine phosphorylase mutations . Ann Neurol 2000. ; 47 : 792 – 800 . [PubMed] [Google Scholar]
- Orlacchio A, Patrono C, Gaudiello F, Rocchi C, Moschella V, Floris R, et al. . Silver syndrome variant of hereditary spastic paraplegia: a locus to 4p and allelism with SPG4 . Neurology 2008. ; 70 : 1959 – 66 . [DOI] [PubMed] [Google Scholar]
- Orlacchio A, Babalini C, Borreca A, Patrono C, Massa R, Basaran S, et al. . Spatacsin mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis . Brain 2010. ; 133 : 591 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J . Linkage analysis and family classification under heterogeneity . Ann Hum Genet 1983. ; 47 : 311 – 20 . [DOI] [PubMed] [Google Scholar]
- Palumbo C, Massa R, Panico MB, Di Muzio A, Sinibaldi P, Bernardi G, et al. . Peripheral nerve extracellular matrix remodelling in Charcot-Marie-Tooth type I disease . Acta Neuropathol 2002. ; 104 : 287 – 96 . [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Nath P, Wood NW, Singleton A, Houlden H . Clinical heterogeneity and genotype-phenotype correlations in hereditary spastic paraplegia because of spatacsin mutations (SPG11) . Eur J Neurol 2008. ; 15 : 1065 – 70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehlivan D, Coban Akdemir Z, Karaca E, Bayram Y, Jhangiani S, Yildiz EP, et al. . Exome sequencing reveals homozygous TRIM2 mutation in a patient with early onset CMT and bilateral vocal cord paralysis . Hum Genet 2015. ; 134 : 671 – 3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensato V, Castellotti B, Gellera C, Pareyson D, Ciano C, Nanetti L, et al. . Overlapping phenotypes in complex spastic paraplegias SPG11, SPG15, SPG35 and SPG48 . Brain 2014. ; 137 : 1907 – 20 . [DOI] [PubMed] [Google Scholar]
- Pérez-Brangulí F, Mishra HK, Prots I, Havlicek S, Kohl Z, Saul D, et al. . Dysfunction of spatacsin leads to axonal pathology in SPG11-linked hereditary spastic paraplegia . Hum Mol Genet 2014. ; 23 : 4859 – 74 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TM, Simeonov DR, Sincan M, Adams DA, Markello T, Golas G, et al. . Exome sequencing and SNP analysis detect novel compound heterozygosity in fatty acid hydroxylase-associated neurodegeneration . Eur J Hum Genet 2012. ; 20 : 476 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippucci T, Panza E, Pompilii E, Donadio V, Borreca A, Babalini C, et al. . Autosomal recessive hereditary spastic paraplegia with thin corpus callosum: a novel mutation in the SPG11 gene and further evidence for genetic heterogeneity . Eur J Neurol 2009. ; 16 : 121 – 6 . [DOI] [PubMed] [Google Scholar]
- Querin G, Bertolin C, Martinuzzi A, Bassi MT, Arnoldi A, Polo A, et al. . The blurred scenario of motor neuron disorders linked to spatacsin mutations: a case report . Eur J Neurol 2014. ; 21 : 85 – 6 . [DOI] [PubMed] [Google Scholar]
- Reyes-Marin K, Jimenez-Pancho J, Pozo L, Garcia-Villanueva M, de Blas G, Vazquez JM, Jimenez-Escrig A . A novel myelin protein zero (V136G) homozygous mutation causing late onset demyelinating polyneuropathy with brain white matter lesions . Clin Neurol Neurosurg 2011. ; 113 : 243 – 4 . [DOI] [PubMed] [Google Scholar]
- Sagnelli A, Piscosquito G, Chiapparini L, Ciano C, Salsano E, Saveri P, et al. . X-linked Charcot-Marie-Tooth type 1: stroke-like presentation of a novel GJB1 mutation . J Peripher Nerv Syst 2014. ; 19 : 183 – 6 . [DOI] [PubMed] [Google Scholar]
- Simpson MA, Cross H, Proukakis C, Pryde A, Hershberger R, Chatonnet A, et al. . Maspardin is mutated in mast syndrome, a complicated form of Hereditary Spastic Paraplegia associated with dementia . Am J Hum Genet 2003. ; 73 : 1147 – 56 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel R, Mandel H, Saada A, Lerer I, Burger A, Shaag A, et al. . Delineation of C12orf65 -related phenotypes: a genotype-phenotype relationship . Eur J Hum Genet 2014. ; 22 : 1019 – 25 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS, et al. . Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum . Nat Genet 2007a. ; 39 : 366 – 72 . [DOI] [PubMed] [Google Scholar]
- Stevanin G, Azzedine H, Denora P, Boukhris A, Tazir M, Lossos A, et al. . Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration . Brain 2007b. ; 131 : 772 – 84 . [DOI] [PubMed] [Google Scholar]
- Tamiya G, Makino S, Hayashi M, Abe A, Numakura C, Ueki M, et al. . A Mutation of COX6A1 causes a recessive axonal or mixed form of Charcot-Marie-Tooth Disease . Am J Hum Genet 2014. ; 95 : 294 – 300 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazir M, Bellatache M, Nouioua S, Vallat JM . Autosomal recessive Charcot-Marie-Tooth disease: from genes to phenotypes . J Peripher Nerv Syst 2013. ; 18 : 113 – 29 . [DOI] [PubMed] [Google Scholar]
- Tazir M, Hamadouche T, Nouioua S, Mathis S, Vallat JM . Hereditary motor and sensory neuropathies or Charcot-Marie-Tooth diseases: an update . J Neurol Sci 2014. ; 347 : 14 – 22 . [DOI] [PubMed] [Google Scholar]
- Tessa A, Denora PS, Racis L, Storti E, Orlacchio A, Santorelli FM . Bridging over the troubled heterogeneity of SPG-related pathologies: mechanisms unite what genetics divide [Review] . Curr Mol Med 2014. ; 14 : 1034 – 42 . [DOI] [PubMed] [Google Scholar]
- Tesson C, Nawara M, Salih MAM, Rossignol R, Zaki MS, Al Balwi M, et al. . Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia . Am J Hum Genet 2012. ; 91 : 1051 – 64 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman V, Clowes VE, Reid E . Overlapping molecular pathological themes link Charcot-Marie-Tooth neuropathies and hereditary spastic paraplegias [Review] . Exp Neurol 2013. ; 246 : 14 – 25 . [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Löfgren A, Martin JJ, Van Broeckhoven C . Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions . Nat Genet 2001. ; 28 : 211 – 2 . [DOI] [PubMed] [Google Scholar]
- Yum SW, Zhang J, Mo K, Li J, Scherer SS . A novel recessive NEFL mutation causes a severe, early-onset axonal neuropathy . Ann Neurol 2009. ; 66 : 759 – 70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windpassinger C, Auer-Grumbach M, Irobi J, Patel H, Petek E, Hörl G, et al. . Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome . Nat Genet 2004. ; 36 : 271 – 6 . [DOI] [PubMed] [Google Scholar]
- Zimoń M1, Baets J, Almeida-Souza L, De Vriendt E, Nikodinovic J, Parman Y, et al. ., Loss-of-function mutations in HINT1 cause axonal neuropathy with neuromyotonia . Nat Genet 2012. : 44 : 1080 – 3 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.