Abstract
In order for human microbiome studies to translate into actionable outcomes for health, meta-analysis of reproducible data from population-scale cohorts is needed. Achieving sufficient reproducibility in microbiome research has proven challenging. We report a baseline investigation of variability in taxonomic profiling for the Microbiome Quality Control (MBQC) project baseline study (MBQC-base). Blinded specimen sets from human stool, chemostats and artificial microbial communities were sequenced by 15 laboratories and analyzed using nine bioinformatics protocols. Variability depended most on biospecimen type and origin, followed by DNA extraction, sample handling environment, and bioinformatics. Analysis of artificial community specimens particularly revealed differences in extraction efficiency and bioinformatic classification. These results may guide researchers in experimental design choices for gut microbiome studies.
Introduction
Translation of basic microbiome research to population studies and the clinic requires reproducibile experimental and computational methods for analysing human-associated microbial communities. The Human Microbiome Project (HMP)1, MetaHIT2, and other large consortia3–5 have produced protocols that can be used to characterize human microbiomes, but relatively few epidemiological studies have evaluated the role of the human microbiota in health with the degree of standardization necessary for translational applications. Inter-study technical variability in microbiome measurements can sometimes, for example, outweigh biological effects6–9. Basic scientists, clinicians, epidemiologists, microbiologists, statisticians, and bioinformaticians should thus collaborate to develop best practices and to identify potential measurement variability in each step of microbial profiling.
Inspired by studies such as the Microarray Quality Control (MAQC) and Sequencing Quality Control (SEQC) projects10, we initiated the Microbiome Quality Control (MBQC) project to address this need. The MBQC aims to evaluate methods for measuring the human microbiome, including protocols for handling human microbiome samples and computational pipelines for microbial data processing. The goal of the MBQC is not to define the ‘best’ protocols for human microbiome studies. Instead, the aim is to inform experimental design by identifying the relative effect sizes of individual variables and to provide a "menu" of context-dependent protocol choices. This will enable the community to quantify protocol variables' inter-study variation, provide a set of context-dependent protocol choices, help develop guidelines for minimum reporting standards, develop methods for normalization and systematic meta-analysis, facilitate consensus on best practice in epidemiological and translational human microbiome studies, and promote open sharing of standard operating procedures. We expand on many previous studies that have evaluated aspects of microbiome data generation protocols, although typically neither in the context of population epidemiology nor across multiple labs11–14 (Table 1; full list in Supplementary Dataset 1; Supplementary Note 1).
Table 1. Selected microbiome protocol evaluation studies.
A summary of findings from a subset of previous studies of protocols for human and/or artificial microbial community 16S amplicon data analysis (Supplementary Dataset 1 contains all protocol evaluation studies published since 2010). Publications in this table were required to compare multiple protocols, target whole communities (not single microbial genomes), and to be neither phylogenetically-targeted (i.e. as bacterially universal as possible) nor specific to 454 pyrosequencing (although comparisons to such data were included). In addition to computational analyses, we subdivide the sample handling / data generation process here into approximate stages: positive controls in the form of one or more mock communities; sample collection and storage or fixation; nucleotide extraction; PCR primer or variable region selection; other aspects of PCR amplification (e.g. enzyme, cycles, index design); sequencing platform; and negative controls or contaminant analysis. Variables broadly tested in each study are marked with “+” if a significant finding was reported or “−” if the variable’s effects were non-significant or described as low effect size (non-tested variables are unmarked). Mock community codes indicate “+” if a positive control was included but sequencing results diverged substantially from the expected reference, “−” if sequencing results for all included positive controls agreed with the expected gold standard.
| Study | Year | PMID | Mock(s) | Storage | Extraction | Primer / Region |
PCR | Platform | Contam. | Bioinf. |
|---|---|---|---|---|---|---|---|---|---|---|
| Degnan | 2012 | 21677692 | + | + | − | + | + | |||
| Schloss | 2011 | 22194782 | + | + | + | + | ||||
| Biesbroek | 2012 | 22412957 | + | + | ||||||
| Yuan | 2012 | 22457796 | + | + | ||||||
| Gaspar | 2013 | 23536909 | + | |||||||
| Kozich | 2013 | 23793624 | + | + | − | + | ||||
| Kennedy | 2014 | 24586470 | + | |||||||
| Kennedy | 2014 | 25002428 | + | |||||||
| Schmidt | 2015 | 25156547 | + | + | ||||||
| Hang | 2014 | 25228989 | + | + | + | + | + | + | ||
| Salter | 2014 | 25387460 | + | + | ||||||
| Koskinen | 2015 | 25525895 | + | + | ||||||
| Jeon | 2015 | 25557481 | + | + | + | |||||
| Brooks | 2015 | 25880246 | + | + | + | − | − | |||
| Walker | 2015 | 26120470 | + | + | ||||||
| Tremblay | 2015 | 26300854 | − | + | − | + | − | |||
| D'Amore | 2016 | 26763898 | + | + | + | + | ||||
| Clooney | 2016 | 26849217 | + | + | ||||||
| Hiergeist | 2016 | 27052158 | + | + | + | − | − | |||
| Schloss | 2016 | 27069806 | + | + | − | + | ||||
| Jovel | 2016 | 27148170 | − | + | + | + | ||||
| Lauder | 2016 | 27338728 | + | + | ||||||
| Fouhy | 2016 | 27342980 | + | − | + | + | ||||
| Gohl | 2016 | 27454739 | + | + | + | |||||
| Song | 2016 | 27822526 | + | − | ||||||
| Schloss | 2016 | 27832214 | + | + | ||||||
| MBQC-base | + | + | + | − | − | + | − |
We report the baseline (MBQC-base) results, in which we evaluate the variability in taxonomic profiling of the gut microbiome introduced during amplicon data generation. Specifically, the steps evaluated included experimental sample handling (DNA extraction15, 16S rRNA gene amplification16 and sequencing) and bioinformatic data processing. This baseline MBQC was carried out by volunteer laboratories; all samples were provided by a central repository, blinded, and handled by 15 laboratories. Re-blinded data were analyzed by nine bioinformatics labs, to produce more than 16,000 taxonomic profiles (approximately three times the size of the HMP17). The MBQC analysed the detailed protocols carried out by handling and bioinformatic laboratories to identify factors that contribute to variability.
Results
The experiments we report find that differences in DNA extraction, amplification protocol, and bioinformatics sometimes (but not always) result in effects comparable to those of biological differences; differences induced by bioinformatics pipelines were typically smaller than those arising from data generation; both types of effects can differ per sample (and sometimes interact); contamination is a frequent cause of variation, from exogenous, endogenous, and computational sources; and that relative (rather than absolute) computational measures were typically more reliable than absolute measures in resolving these differences among protocols. Additional body sites, sample sources, and specimen collection methods will be incorporated in future phases of the project to aid in designing studies that have the reproducibility needed to develop and test microbiome therapeutics and diagnostics.
MBQC-base study design
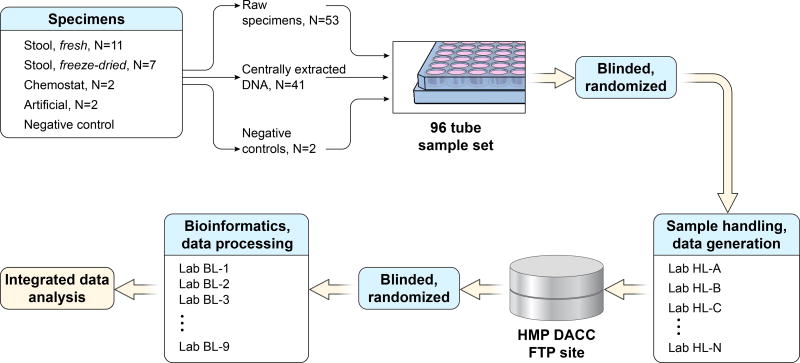
The MBQC baseline study comprised two modules: sample handling (extraction, amplification, and sequencing) and bioinformatic processing (Fig. 1) (see also Life Sciences Reporting Summary). Five types of physical specimens were included: fresh (n=11) and freeze-dried human stool (n=7); "robogut" or chemostat stool-derived communities (n=2); positive gut- and oral-derived artificial communities (Supplementary Table 1); and negative reagent (Tris buffer) controls (23 total specimens). A subset of specimens were centrally extracted and replicated or triplicated into 96-sample aliquot sets that were shipped to handling laboratories (each 96-sample aliquot set contained 53 raw samples and 43 centrally extracted samples Supplementary Table 2). Fifteen laboratories carried out extraction and/or 16S amplicon sequencing, and the raw sequence data were re-blinded and distributed for processing to nine bioinformatics labs (Supplementary Table 3). All data were deposited at the HMP Data Analysis and Coordination Center (DACC), were pooled for integrated analysis, and are available at http://mbqc.org and SRA PRJNA260846).
Figure 1. Microbiome Quality Control Project study design.
MBQC laboratories were provided with at least one blinded set of 96 aliquots including extracted DNA, raw fecal (frozen or freeze-dried) aliquots, and positive and negative control aliquots (22 specimens with replication). Each lab extracted DNA from the raw fecal aliquots, which was then amplified and sequenced DNA samples in tandem with pre-extracted aliquots using Illumina platforms targeting the 16S rRNA gene. Sequencing datasets were re-blinded and distributed for bioinformatic analysis, resulting in an integrated table of operational taxonomic units (OTUs) that were called against the Greengenes 13.5 database18 and made publicly available through the Human Microbiome Project Data Analysis and Coordinating Center1 at http://mbqc.org/ and PRJNA260846).
Data generation protocols were not prescribed; instead, handling and bioinformatics laboratories recorded their protocols using standardized forms (Table 2, Supplementary Datasets 2–4, Online Methods). Handling laboratories each received one or more blinded copies of the 96-aliquot set and processed them within a small number of fixed constraints. Sequences of 16S rRNA gene amplicons were generated using an Illumina platform (MiSeq or HiSeq), although the variable region and primers to be used were not fixed. Data were submitted as demultiplexed FASTQ files and provided to bioinformatics labs, who then generated operational taxonomic unit (OTU) counts for each sample using Greengenes 13.518 identifiers, optional additional open-reference OTUs, and a corresponding phylogenetic reconstruction of representative sequences as the final output.
Table 2. Major sample handling and bioinformatic protocol variables.
In both the handling and bioinformatics modules, each laboratory recorded its own preferred procedures using standardized metadata capture forms. Counts or mean +/− standard deviations of major variables are shown here; see Supplementary Datasets 2–3 for complete data.
| N or Mean±Std. | |
|---|---|
| Handling | |
| DNA extraction kit manufacturerA | |
| Chemagen | 1 |
| GeneRite | 1 |
| MO-BIO | 7 |
| Omega BioTek | 1 |
| Promega | 1 |
| Qiagen | 2 |
| Zymo Research | 1 |
| Not reported/Custom | 2 |
| Homogenizer used? | |
| Yes | 12 |
| No | 3 |
| PCR primer design | |
| 318F/806R | 1 |
| EMP V4 515F/806R | 12 |
| Schloss 2013 | 2 |
| Sequencing machine model | |
| HiSeq 2500 | 1 |
| MiSeq | 14 |
| Paired end sequencing? | |
| Yes | 13 |
| No | 2 |
| Sequencing read length | |
| 150 | 5 |
| 175 | 1 |
| 210 | 1 |
| 250 | 6 |
| 300 | 2 |
| Fraction of PhiX (range 0–0.3) | 0.11 ± 0.08 |
| Fraction quality bases (range 0.57–0.96) | 0.86 ± 0.12 |
| Bioinformatics | |
| Performed QC using…B | |
| QIIME | 4 |
| Trimmomatic | 2 |
| UPARSE | 3 |
| None/other | 2 |
| Called OTUs using… | |
| QIIME | 5 |
| UPARSE | 4 |
| Other/custom | 1 |
| Taxonomic assignment strategy | |
| Classification | 4 |
| Clustering | 4 |
| Mapping | 2 |
| Post-assignment QC by…B | |
| Taxonomy | 2 |
| Sample | 2 |
| OTU | 5 |
| None | 4 |
One lab used two different methods
Will not add up to 10 because some groups used multiple methods
Specimen selection, set design, and blinding
To facilitate assessment of variation among handling laboratories introduced during DNA extraction, aliquots of the 22 specimens were extracted at a central facility using the MO-BIO PowerSoil kit. The resulting combination of specimens and extracted DNA comprised 96 aliquots with 60% duplicates and 40% triplicates. The final aliquot set included 41 aliquots of centrally extracted DNA, 53 aliquots of raw specimens (frozen and freeze-dried feces, chemostat, and artificial communities), and two negative control aliquots of storage buffer (Supplementary Table 2). Each handling lab received one or more of these 96-aliquot sets. The number of unique specimens, raw versus extracted aliquots, and replicates/triplicates was chosen to maximize power in quantifying variance introduced by protocol choice in DNA extraction, sequencing, and bioinformatics, as well as that from intrinsic sample characteristics such as subject and specimen type (Online Methods).
Briefly, data from balanced, triplicate, or replicate-enriched designs were simulated using different proportions of raw versus extracted aliquots. Fixed (to estimate effect size) and random effects (to estimate variance) models were fit and their accuracy assessed by sum of squared deviation over all effects and datasets. Near-balanced replicates/triplicates (54 vs. 42 aliquots, respectively) and raw aliquots (53 raw vs. 43 pre-extracted) were the best-powered in our simulations and hence yielded the final aliquot set design. All aliquots in the set were assigned blinded identifiers prior to distribution, such that handling labs received no information linking aliquots either to replicates or to specimens of origin.
Sample handling and bioinformatics protocols
From these aliquots, handling laboratories generated a total of ~215M sequence reads spanning 2,250 samples (aliquot-laboratory combinations, Supplementary Dataset 5), available at http://mbqc.org and SRA PRJNA260846. Handling protocols included nine different extraction kits (predominantly MO-BIO, Carlsbad, CA), and most incorporated a bead-beating homogenization step (Table 2, please see Supplementary Dataset 2–3 and http://mbqc.org for detailed protocols). All handling laboratories except one used V4 primers from one of two protocols19, 20; one lab used a V3-4 primer pair21. One laboratory used Illumina HiSeq, all the others used MiSeq, and almost all protocols (13) [how many of the 15?] used paired end overlapping reads. Finally, one laboratory deposited data after datasets were re-distributed, yielding a total of 14 handling labs’ data which were distributed for bioinformatic analyses (all raw data are available at the DACC
Each sample handling laboratory deposited demultiplexed FASTQ files containing the resulting data at the DACC, which were then re-blinded for distribution to bioinformatics participants (Online Methods). Bioinformatics protocols (Table 2, please see Supplementary Dataset 2–3 and http://mbqc.org for detailed protocols), but broadly included steps for read filtering and quality control (QC), overlapped read stitching, OTU calling, and OTU table QC. Read QC, when performed, generally comprised removal of low-quality bases and, subsequently, reads below a length threshold. Read stitching (or assembly) merged mates from paired end samples, discarding reads that failed to meet mismatch or base quality thresholds. OTU QC most often included feature or row QC, i.e. removal of OTUs below an abundance or prevalence threshold; some labs also performed sample or column QC to omit samples with too few reads (Supplementary Note 2).
Taxonomic profiles
The combination of read-level QC and OTU assignment yielded an average of ~40M reads per sample handling laboratory and ~35,000 ±43,700 reads/sample, for a total of 15,749 filtered samples (each sample being a unique combination of aliquot, handling laboratory and bioinformatics laboratory; Supplementary Table 4). These comprised 11,991 human-derived samples, 2,033 control artificial communities, and 1,725 chemostat-derived samples used for subsequent analysis; all combined OTU analyses excluded two labs that provided only open-reference OTU calls without mergeable Greengenes identifiers (BL-5 and BL-7). After quality control, most bioinformatics labs generated sequence datasets with highly negatively skewed distributions, that is, low-count outlier samples were discarded, and high-count samples retained. The two labs using fully open-reference OTU calling found high variance in their per-sample QC criteria, because in some, but not all, samples many reads were captured only by de novo OTUs.
Reads and QC together produced 27,211 OTUs with Greengenes v13.5 identifiers that are detailed in the summary MBQC-base table (Supplementary Table 5). This table excludes de novo OTUs, although they are available on the DACC for further analysis. Human-derived samples averaged 383 ±326 OTUs, with 325 ±244 in chemostat-derived samples, 187 ±200 in positive control artificial communities, and 302 ±283 in negative controls Supplementary Dataset 6). These last two values are somewhat surprising and are discussed in greater depth below. These statistics include only samples processed during the MBQC-base using blinded samples; an additional mothur-based protocol was added post-hoc (Online Methods). This produced comparable taxonomic profiles to MBQC-base bioinformatics, shifted significantly (but with modest effect size) to deplete some clades that were dominant in protocols BL-1 to 9 (genus Fusobacterium, unclassified Enterobacteriaceae, most species-level assignments, etc.) and enriching the otherwise less-abundant Firmicutes (Paenibacillus, Blautia, Phascolarctobacterium, Oscillospira, and other genera; Supplementary Fig. 1).
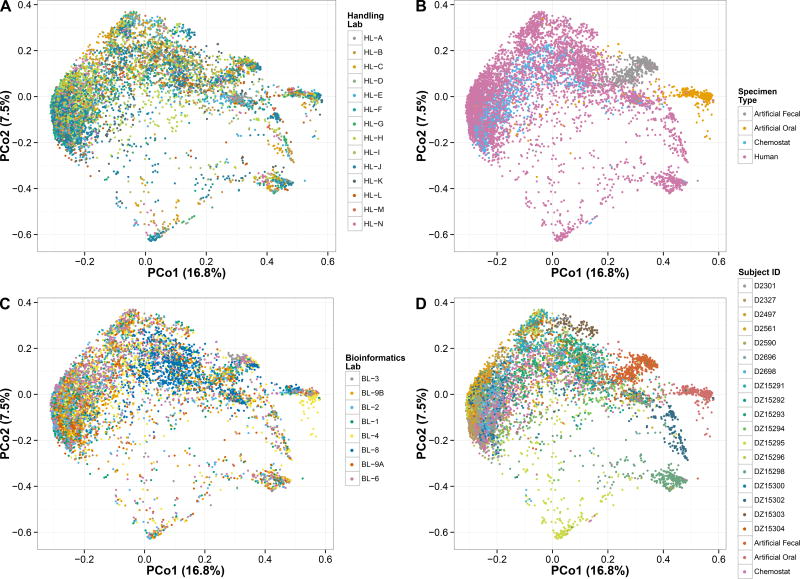
A qualitative overview of sample beta-diversity values (Fig. 2) revealed that biological, experimental, and bioinformatics variables all contribute to facets of between-sample variation (Supplementary Fig. 1). Most human- and chemostat-derived samples cluster together, with the biologically distinct artificial communities and a subset of human-derived samples forming outgroups. Differences induced by a subset of computational protocols are readily visible (e.g. BL-8), with those arising from sample handling protocol choices more difficult to see in this visualization but nevertheless present (e.g. HL-D or HL-E, see Fig. 3). Interestingly, human- and chemostat-derived samples do not segregate substantially in this high-level overview, highlighting the latter’s value as biologically-realistic positive controls.
Figure 2. Beta-diversity of MBQC-base microbial community analyses.
Ordination of 16,554 samples corresponding to 2,237 replicated sequencing results on 22 originating physical specimens (human-derived, chemostat, and oral and gut artificial communities) using multidimensional scaling of Bray-Curtis dissimilarities. Labels indicate stratification by A) sample handling laboratory, B) specimen type, C) bioinformatics laboratory, or D) subject. Major contributors to between-sample diversity thus include biological origin, handling protocol differences, and bioinformatics protocol variables (Supplementary Fig. 1).
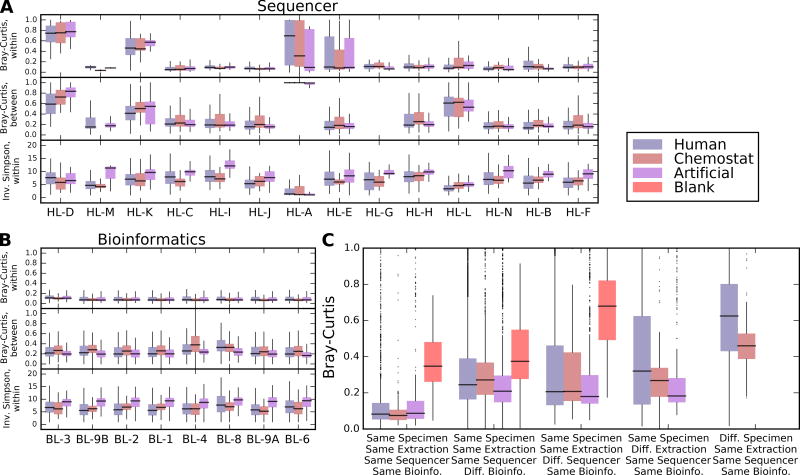
Figure 3. Individual and aggregate effects of sample handling and bioinformatics labs on microbial profiles.
A) Distributions of within- and between-sample alpha and beta diversities, respectively, stratified by sample type (n=2,033 for artificial communities, n=11,991 for human-derived samples, and n=1,725 for chemostat samples) and by handling or B) bioinformatics lab. Raw data, including sample sizes, are included in Supplementary Dataset 7. Bray-Curtis dissimilarities within labs are computed only between technical replicates handled and extracted identically; between lab distributions compare only replicates from the same originating specimen as processed by one lab to all others. Outlier values outside 1.5 times the interquartile range are omitted for clarity. Within-lab comparisons thus assess the consistency of each lab between replicates; between-lab comparisons assess how (dis)similar each lab’s results are to all others. C) Effect size distributions of technical variation (between identically handled replicate samples), differences only due to bioinformatics lab, sequencing lab, extraction (local vs. central), and between different biological specimens. In general, biological differences were largest, followed by extraction (particularly for heterogeneous human-derived samples), sequencing protocol, and computational protocol effects were smallest. Omnibus tests for differences among specimen type, handling laboratory, and bioinformatics laboratory are all significant at Kruskal-Wallis p<0.05; pairwise Wilcoxon tests for the effects of most individual handling laboratories are significant, while most bioinformatics laboratories are not (Supplementary Table 6).
Variation in taxonomic profiles
To quantify the effects of protocol differences on microbial community composition (Supplementary Fig. 1; Supplementary Note 3), we evaluated within- and between-sample diversity for bioinformatics, sample handling, and DNA extraction protocol variation (separating each basic specimen type, Fig. 3, Supplementary Table 6). Quantitative alpha diversity was relatively robust to protocol differences (Fig. 3A), although both bioinformatics and sample handling protocols tended to affect the median observed diversity more than biological specimen type. Absolute levels of diversity varied among laboratories (Supplementary Fig. 2–3), although relative correlations between community profiles in the 22 primary specimens across laboratories remained high (min. Spearman r=0.9; Supplementary Note 4).
For each specimen, beta diversity distributions summarize the difference between replicate samples within one variable e.g. dry lab, wet lab, or extraction kit, and all other values in the variable set (Fig. 3B). For example, when comparing bioinformatics labs, each boxplot summarizes the set of beta diversities between each specimen’s replicate samples as analyzed by one lab and the same specimen’s replicates as analyzed by all other labs. Comparisons were also stratified by sample type (human-derived, chemostat, or artificial community). While each bioinformatics laboratory induced a modest overall shift in a samples’ microbial profile, only one laboratory (BL-8) resulted in a consistent, large shift in the sample composition. Differences owing to sample handling in laboratories were generally larger than the effect size of sample type, especially in those labs that handled samples extracted centrally and in other locations (e.g. HL-E processed DNA samples extracted by the HL-F, HL-J, and HL-A labs). Grossly different beta-diversity profiles were generally caused by a subset of samples that were strikingly dissimilar, rather than consistent differences in all of the samples, indicating that a handful of samples may have been contaminated, mislabeled, or otherwise altered in these laboratories.
We observed that any protocol variable can produce large technical differences, as evidenced by a small number of outlying, large effect size contrasts in all of the extraction, sequencing, and bioinformatics comparisons (Fig. 3C). However, in almost all cases, the range of differences between samples was greatest for the 18 human-derived specimens. This means that the overall effect size of biological variation outweighed that of computational or experimental protocol choices (Supplementary Fig. 4, Supplementary Table 7).
Variation among biospecimens
We found that different types of specimen were differentially affected by laboratory protocol choices (Supplementary Fig. 5). Some samples, e.g. artificial communities, were minimally affected by different sample handling protocols, and there was no statistically significant variation in measures of diversity (e.g. Simpson’s diversity) associated with a handling laboratory. For all other samples, however, there was up to 5-fold variation between communities profiled in different laboratories This agrees with our observation that relative diversity levels remain consistent (Supplementary Fig. 2–4), because each protocol will have a set of biases that affect all samples equally. To summarize, estimates of absolute diversity are not comparable among laboratories even when processed using the same bioinformatics pipeline, whereas relative diversity levels are on average consistent among similar sample types.
Sample-specific effects on diversity occurred due to bioinformatics protocol choices as well. There was no single bioinformatics pipeline that was consistently either the least or most conservative in estimating the effects of handling laboratory on measures of community structure such as alpha diversity (Supplementary Fig. 6). This resulted in complex interactions between handling and bioinformatics protocol choices: some sample diversity estimates agreed across handling labs and bioinformatics protocols, while others differed significantly across handling, bioinformatics, or both (Supplementary Fig. 7). While some of these interactions might arise from technical sources such as sample mislabeling, our results suggest that microbial physiology, nucleotide composition, protocol biochemistry, and computational assumptions can all interact to induce variation in microbiome assay results9.
More broadly, our findings suggest that standardization efforts themselves need to be scaled up, because our data show that experiments on only one or a few samples can produce results that are not fully generalizable among distinct experiments or populations.
Variation due to extraction protocol
DNA extraction protocols are a known source of variation between microbial community characterization experiments7–9. We separated variation caused by DNA extraction from variation owing to PCR and sequencing by comparing centrally- versus locally-extracted samples (Supplementary Fig. 8). 22 unique specimens had replicate aliquots sequenced from DNA that was extracted either by individual handling laboratories (locally) or centrally using a single MO-BIO-based protocol. Beta diversities between locally- versus centrally-extracted samples were specimen-specific, with some samples showing little effect and others differing significantly primarily based on this single handling protocol variable (Supplementary Note 5; Supplementary Fig. 9).
Positive controls
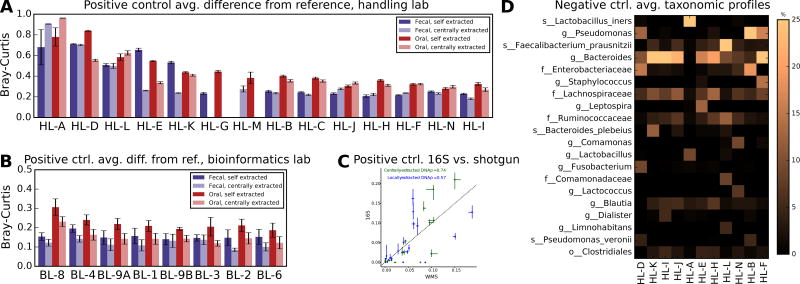
We included positive (artificial community) and negative (buffer blank) control samples so that we could assess the overall accuracy of different protocols (Fig. 4). Specifically, each sample set included two artificial communities, one comprising 20 gut microbiota species and one comprising 22 oral species (Supplementary Table 1). These were blinded and processed alongside fecal and chemostat specimens during data generation and processing. Sample handling and bioinformatics protocols had an effect on community profile accuracy (Fig. 4A–B), but the largest contributor to variability was contamination or omissions owing to microbes whose DNA was not extracted (Supplementary Fig. 10). Bioinformatics protocol differences had less impact (Fig. 4A–B).
Figure 4. Detection of abundant taxa in positive and negative control samples is affected by sample handling.
Average distance (genus-level Bray-Curtis beta-diversity) from two reference positive control communities (20 fecal and 22 oral isolates, respectively), stratified between centrally and locally extracted samples and by A) sample handling laboratory (averaging each over all bioinformatics) and by B) bioinformatics lab (averaging over each over all sample handlers). Error bars show standard error; no data were provided by combinations that are missing bars. Raw data, including sample sizes, are included in Supplementary Dataset 8. Sample handling had a greater overall effect on distance from truth, and showed greater variation, than did bioinformatics; some effects were specific only to locally or centrally extracted samples and appeared to be driven by contamination of only these respective sample subsets (see Supplementary Fig. 9–10). C) Spearman correlation between whole metagenome shotgun (WMS) and 16S amplicon sequence data on centrally and locally extracted gut-derived artificial communities. Points indicate each of 17 species that were jointly identifiable in both data types, due to uniquely identifiable species-level agreement between the Greengenes and MetaPhlAn taxonomies (see Methods, Supplementary Fig. 11). Error bars represent interquartile ranges (IQRs) across 43 and 36 artificial community 16S amplicon samples for gut centrally and locally extracted DNA samples, respectively, intersecting at medians; three WMS samples were used in each comparison (six total). Dashed line indicates the diagonal. D) Mean taxa observed in negative control samples containing only Tris-HCl buffer (see Methods). Most apparent contamination was handling lab-specific (see Supplementary Fig. 9–10), thus averages are per handling lab over all bioinformatics.
Nearly every combination of extraction, sequencing and bioinformatics protocols affected artificial community taxonomic profiles in at least one sample (Supplementary Fig. 10). Fecal and oral microbes were depleted in all bioinformatics protocols in the handling lab using V3-4 primers (HL-A), in favor of genus Lactobacillus. However, lab-specific effects also interacted with the bioinformatics protocols. Fusobacterium was only substantively detected in HL-A samples by the BL-2 and BL-6 pipelines, for example, or by BL-1, BL-4, and BL-9A in the sole HiSeq laboratory, HL-L. Some handling laboratories introduced different biases in locally versus centrally extracted samples (e.g. HL-E), which then produced different profiles in bioinformatics labs owing to the systematic differences in input sequences. Overall taxonomic profiles were also sensitive to handling protocol, bioinformatics, and to the interaction of these variables, although all estimates of OTU richness were high (means ~30–35 for centrally and locally extracted samples, respectively) and neared 100 OTUs in one case (HL-E handling, BL-6 bioinformatics), despite an expectation of 20 and 22 (fecal and oral, respectively) and low rarefaction depth of 1,000 sequences (Supplementary Fig. 12).
To check artificial community composition, six gut and six oral artificial community samples were centrally subjected to metagenomic sequencing (half pre- and half re-extracted), profiled with MetaPhlAn222, and compared with 16S amplicon-derived abundances (Fig. 4C, Supplementary Fig. 11). Although factors such as PCR, re-extraction, and database representation (16S versus whole genome) all prevent perfect correlations between amplicon and metagenomic data, this experiment supported the expected ground truth and further confirmed the variability of laboratory and bioinformatics protocols when reading out even these low-complexity communities.
Finally, raw reads deposited by each handling lab, prior to any bioinformatics, were analyzed to assess the potential causes of artifacts (Supplementary Fig. 13). Approximately 50–80% of 16S amplicon reads matched one of the 20 or 22 fecal/oral reference sequences exactly, with most labs varying only slightly (typically <10%) between samples. Single base error accounted for many of the remainder, at a rate of 0.15–0.2% per nucleotide, while two labs (HL-C and HL-N, two of only three labs using single-indexed 8nt barcodes) also generated 1–3% mismatched sequences due to single base gaps at the barcode/linker. Chimeric reads represented another ~1–5% of mismatched reads across all labs, although this rate did not vary monotonically with number of PCR thermal cycles (Supplementary Dataset 2). Interestingly, read-level error rates did not correlate with handling labs’ diversity averages. Instead, inflated diversity arose from other handling (e.g. contamination) or bioinformatics sources, although such errors do decrease absolute accuracy in taxonomic profile assessment (Fig. 4).
A substantial source of errant sequences in the positive control samples (and, below, in negative controls as well) proved to be reads originating from other samples or, possibly, from external contaminants (Supplementary Fig. 14). Artificial community samples from all labs included substantial reads identical to those most abundant in other MBQC specimens. Three labs that used single 8nt barcodes (HL-C, HL-N, and HL-I) had the highest proportion of these reads, although this factor was not directly associated with diversity rates (see Supplementary Fig. 12) or absolute accuracy (see Fig. 4). A per-barcode synthesis error rate comparable to the per-read errors above (~0.2%) is sufficient to explain much of this effect, leading to a non-negligible fraction of reads being assigned to the wrong sample through barcode hopping (“bar-hopping”). While the MBQC-base sample size is too small to quantify the relative sizes of these effects, sequencing error, extraction efficiency, computational stringency, bar-hopping, and contamination may all have detectable and combinatorial roles in taxonomic profiling errors.
Negative controls
We included Tris-HCl buffer aliquots as negative control samples (two per 96-sample set) (Online Methods, Fig. 4D). In total, 345 negative controls were fully processed. At least one OTU was reported in 342 of these samples, with the observed number ranging from 1 to ~130 at low (1,000 sequences) rarefaction and almost 200 in all sequences (Supplementary Fig. 12). Sources of contamination varied, including one lab with samples comprising mainly Lactobacillus (HL-A), three labs with Pseudomonas (possibly reagent contamination23 , HL-B, HL-F, and HL-D), and sequence, bioinformatic, and/or physical contamination from gut-derived samples in all laboratories. The most abundant contaminant OTU was an unidentified Bacteroides at 1.6 s.d. 7.0%, which was detected in 22.6% of negative control samples and was also abundant in stool samples (average 1.2 s.d. 3.7%). Of the top 100 most abundant false positive Greengenes identifiers (average relative abundances ranging from 0.2% to 1.5%), 34% were from genera Bacteroides; Faecalibacterium prausnitzii also comprised 9% of the top 100 IDs, genus Pseudomonas 7%, and family Enterobacteriaceae 6% (Supplementary Note 6).
Multivariate quantification of variation
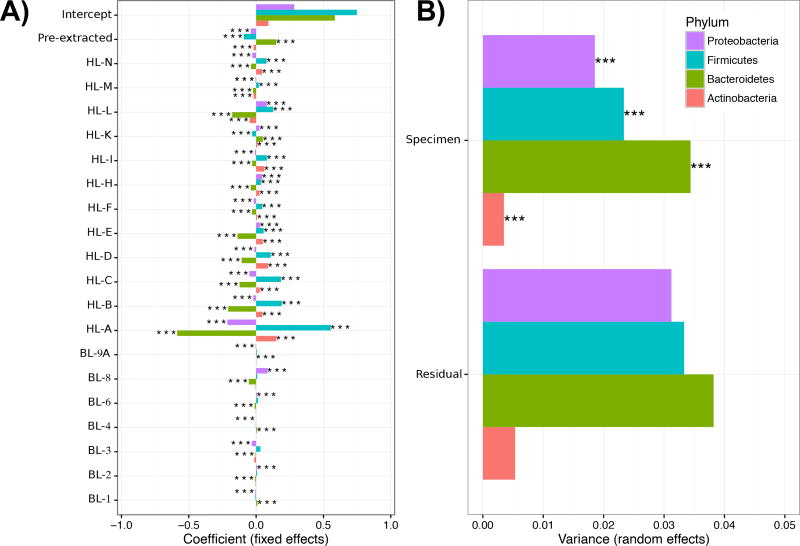
As a final, joint quantification of the multiple aspects of variance among both sample handling and bioinformatics protocols, we fit two mixed effects models to the transformed abundances of the four major gut phyla (Actinobacteria, Bacteroides, Firmicutes, and Proteobacteria; see Methods; Supplementary Table 8, Supplementary Fig. 15, Fig. 5). Using arcsine-square root transformed phylum-level taxonomic abundances as a readout, the greatest variability in microbiome profiling was assigned to biological differences between specimen sources and handling laboratories, as in the univariate whole-community models above. Additionally, DNA extraction method was the individual protocol variable of greatest effect, also as observed in univariate analyses. Centrally extracted (pre-extracted) samples were associated with higher abundances of Bacteroidetes; since all pre-extracted samples were extracted using a MO-BIO kit, these results are consistent with reports showing that the MO-BIO based HMP protocol generated data enriched for genera within the Bacteroidetes24. Finally, our full model was unable to assign significance to any specific fixed effects (i.e. individual protocol variables), since in the small MBQC-base these were in large part confounded with individual handling and bioinformatics laboratories (Supplementary Fig. 15). Because this leaves a large proportion of taxonomic variation unaccounted for, systematic assessment of individual protocol variables (e.g. individual extraction kits, amplification primers, or sequence filtering parameters) will be crucial for the next phase of the MBQC.
Figure 5. Variation in community profiling analyzed using a multivariate model of experimental and bioinformatic protocol variables.
Significant A) fixed and B) random effects on phylum-level variation in taxonomic abundance, derived from a simplified model of handling and bioinformatics laboratory variables for which there were sufficient data available for evaluation (see Supplementary Table 8, Supplementary Fig. 15 for full model). Variability in taxonomic profiling is dominated by systematic differences between handling laboratory protocols in addition to choice of DNA extraction kit, while bioinformatics protocol choices were much smaller at the phylum level (see Results). Bar length indicates the magnitude of A) average differences in abundance contributed by each lab or B) variation contributed by different specimens or by noise, while stars indicate significance at p<0.001. All parameters were tested using a likelihood ratio test with Benjamini-Hochberg-Yekutieli FDR correction across all outcomes.
Discussion
We report the baseline study from the MBQC project consortium. This project has been set up to improve reproducibility and rigor of microbiome analyses, so that human microbiome studies can be used for population epidemiology and translation into therapeutics9, 25. We found that each microbiome protocol step, including sample handling environment, DNA extraction, and bioinformatic processing has the potential to introduce variation of comparable effect size to that of biological differences. Within these broad categories, almost any data generation or analysis protocol choice has the potential to yield divergent results. This is visible, for example, in the long tails of outliers in Fig. 3, and in the finding that almost half of the participating wet-laboratories produced variable results for positive control artificial communities. Conversely, however, many potential sources of variation (sequencing platform, chemistry, sequence-level bioinformatics, and others) were, when detectable, typically of smaller effect size than phenotypes of clinical interest.
The goal of the MBQC project is to provide information that will allow microbiome researchers and regulators to make informed design choices, rather than to prescribe specific protocols (Supplementary Note 7). Our results indicate that carrying out meta-analyses of microbiome studies is challenging at present because individual experiments frequently include incompatible protocol variables. Of note, there are no batch normalization approaches for microbiome data, unlike in transcriptional or genetic meta-analyses26. Previous detailed investigations of specific variables in microbiome experiments such as DNA extraction method have, in common with the results we report, enriched for broad classes of Gram-negative rather than Gram-positive bacteria due to technical rather than biological variables9, 24, 27, 28. Likewise, choices of 16S rRNA gene variable region primers may enable improved detection or differentiation of microbes16; for example, Propionibacterium spp. abundant on the skin are often undetected by V4 primers29, and Lactobacillus spp. in the vagina are better differentiated by including V321. Furthermore, different human host cohorts may harbor remarkably different Gram-negative versus Gram-positive diversity3, 30, and a protocol appropriate for one type of community may work well in one study but not in another. Some bioinformatics protocol choices can mitigate these differences; for example, relative (rather than absolute) diversity measures, phylogenetic (rather than taxonomic) analyses, and quantitative (rather than presence-absence based) measures all tended to be more robust to inter-protocol variation.
In our view, the wide variation in biological questions, coupled with technical variability, precludes the recommendation of a single “best” protocol for all studies, although standards should be agreed upon that capture a consensus of compatible protocol choices and document their applicability in large-scale studies. Sequencing data for positive controls diverge in almost every published study28, including ours (Supplementary Dataset 1), which confirms that relative, not absolute, measures are comparable between protocols. Likewise, many studies, including ours, agree that DNA extraction (particularly bead beating), 16S rRNA gene primer selection, and negative control contamination have large effects on variation. Previous results have been mixed in their characterization of sequencing platform effects; we included Illumina MiSeq and HiSeq 16S data, but earlier evaluations capture these, Roche 454, Ion Torrent, Pacific Biosciences, and other technologies (including comparisons of amplicon and shotgun metagenomic sequencing). Almost all cases in which platforms differed31–33 specifically investigated Ion Torrent 16S amplicon sequencing (Supplementary Note 8).
In the next phase of the MBQC project, we expect to carry out systematic surveys of microbiome assay protocols (Supplementary Note 9). Among other variables of interest, the community will need a shared library of positive control standards for different microbial habitats, such as “typical” gut, skin, oral, or other microbial mixtures of defined composition, as well as guidelines for including these in addition to negative controls for simultaneous within- and between-study standardization. The results we present here set the scene for the next phase of addressing variability in microbiome studies.
Online Methods
Biological specimens
Fresh specimens
Fresh stool samples were collected in a plastic commode (Fisherbrand Commode Specimen Collection System, Fisher 02-544-208), ziplock bag, diaper, or clinical urine collection container from volunteers under the IRB protocol 0409.13 at the University of Colorado, Boulder. These were frozen at −20°C if immediate aliquoting was not possible, and any initially frozen samples were thawed at room temperature for aliquoting. Bulk fecal samples for DNA extraction were scooped into a sampling tube (Fecal Collection Containers, Globe Scientific – Polystyrene, VWR 60820-100) using a plastic spatula (Fisherbrand Disposable Sterile Spatula, Fisher 14-375-253), frozen, and sent to the Allen-Vercoe lab at the University of Guelph, Canada, on dry ice. To make fresh aliquots, 10 g of thawed fecal matter was added to a 250 ml disposable sterile bottle (Corning™, Fisher 09-761-4) and combined with 85 ml of EB buffer (Qiagen 19086). Specimens were vortexed until the mixture was as homogenous as possible. One hundred µl of fresh stool samples were aliquoted into 1.2 ml cryovials (Fisher 12567500) at room temperature. The aliquots were stored overnight at −20°C, and then transferred to −80°C until they were shipped to the NCI repository, in Frederick MD, on dry ice for specimen set construction.
Freeze-dried specimens
Freeze-dried specimen collection procedures and donors have been outlined previously1. Briefly, newly diagnosed cases with adenocarcinoma of the colon or rectum were recruited prior to surgery and treatment during 1985–19891, 2. Controls were patients awaiting elective surgery for non-oncologic, non-gastrointestinal conditions at these hospitals during the same period. A median of 6 days (IQR 3–13 days) prior to hospitalization and surgery, participants completed dietary and demographic questionnaires and provided two-day fecal samples that were frozen at home on dry ice and subsequently lyophilized. Some of the participants provided an additional two-day fecal sample four to six months later. The two-day lyophilates were pooled, mixed and stored at −40°C. This study was reviewed and approved by an Institutional Review Board at the National Cancer Institute1, 2 and approval from the the NCI Office of Human Subjects Research No. 11147. Fecal samples from three controls and four cases in the original study were included in the MBQC study.
Chemostat (robogut) specimens
A single-stage chemostat model of the human distal gut ecosystem was used to develop controlled fecal communities with realistic ecological diversity, as outlined previously3. One healthy donor (male, 25 years-old) provided fresh fecal samples on two separate occasions, 3 months apart that were used to inoculate two separate chemostat runs. Briefly, fresh fecal samples were placed into a Concept 300 anaerobic chamber (Ruskinn, Sanford, ME) supporting an atmosphere of H2:CO2:N2, 10:10:80, within 5–10 min of voiding. A 10% (w/v) fecal slurry was prepared by homogenizing 5 g of feces in 50 mL of pre-reduced growth medium for 1 min using a stomacher (Tekmar Stomacher Lab Blender, Seward; Worthing, West Sussex, UK). The resulting slurry was centrifuged for 10 min at 175 ×g to remove large food particles4, and 100 mL of the supernatant slurry was added to 300 mL of sterile growth medium in each vessel. Cultures were gently agitated and vessel pH was adjusted to 6.9–7.0. Medium feed was started 24 h post-inoculation, and the culture was grown for 22 days before harvesting and aliquoting. The Research Ethics Board of the University of Guelph approved this study (REB#10JL002).
Artificial (mock) communities
Artificial communities were developed from in-house isolates representative of the range of bacterial genera found within the oral or gut habitats, including both Gram positive and Gram negative bacteria with a range of genomic G+C ratios (Supplementary Table 1). All strains were originally isolated from human subjects, and HMP genome reference strains were included where available5. Each strain was separately cultured on Fastidious Anaerobe Agar (FAA) (Acumedia®, Lansing, MI) supplemented with 5% defibrinated sheep’s blood (Hemostat, Dixon, CA) for 72 hrs at 37°C under anaerobic conditions in a Concept 300 anaerobe chamber as above, H2:CO2:N2, 10:10:80, with the exception of 1_1_55 (K. pneumoniae), 30_1 (Ent. saccharolyticus), 1_1_43 (Esch. coli), and 5_7_47FAA (R. pickettii) that were grown on FAA under aerobic conditions (the latter with no blood supplementation and at 30°C), and GT4ACT1 (N. mucosae) and CC94D (G. adiacens) that were grown on FAA under an atmosphere of 5% CO2. Biomass for each community was scraped from relevant plates using 10 µL inoculating loops (Globe Scientific Inc., Paramus, NJ) and transferred to 500 mL filter-sterilized saline (0.9% w/v NaCl) to a final OD600nm of 0.191 for oral and 0.131 for gut community suspensions. Similar numbers of loops were inoculated for most species and inoculated cell densities were estimated as multiples of 7.5×1010 (approximate CFU/mL of a 10 µL inoculating loop). Some species grew poorly and harvesting biomass was difficult; hence for these isolates (e.g. CD1 D5 FAA 3, M. timidum, and CD1 D5 FAA 6,D. pneumosintes), as much biomass as possible was included in the final suspension. Samples were aliquotted after gentle mixing to ensure an even suspension, and immediately frozen at −20°C prior to shipping to the NCI repository, or prepared for DNA extraction.
Negative control blanks
Blank samples consisted of 10 mM Tris-HCl pH 8.5 (Qiagen 19086).
Sample sets
Upon registering, each of the 15 sample handling laboratories was sent a minimum of one set containing 96 samples in Fisher 1.2 mL Cryovial (Product Code 12567500) tubes on dry ice. Each set contained, first, non-extracted replicates (at least duplicate, some triplicate) totaling 25 fresh stool, 16 freeze-dried stool, 6 chemostat-derived, and 6 artificial community samples (Supplementary Table 2). Each set further contained centrally extracted DNA: 17 from fresh stool, 14 from freeze-dried stool, 4 from chemostat, and 6 from the artificial communities. Finally, each sample set also included 2 negative control blank samples. Frozen fresh stool were shipped frozen as described above, freeze-dried stool were shipped in 1 mL sterile Elution Buffer (Qiagen 19086), and extracted DNA was shipped in 10 µl aliquots of 10 mM Tris. The samples were placed in the same order for each laboratory. Participating labs were blinded to sample collection methods or sample types. All sample types, metadata, and phenotypes remained blinded to participants until consolidated data release at the end of the baseline experiment.
Sample sets were designed by simulation under the constraints that, of 96 total samples, 40–60% should be pre-extracted DNA (the remainder raw specimens), 40–60% should represent duplicates, and 40–60% triplicates. Specimens 3, 11, 13, and 14 were required to be represented at least once (as these represented potential outlier phenotypes from samples captured in the ICU), and simulations aimed to select as many different subjects as possible in addition to these. Of non-control specimens, 40–60% of samples were required to be fresh (not freeze-dried) stool, and 40–60% of samples were required to derive from healthy subject specimens. Finally, all control samples (chemostat, artificial, and negative) were required to be included, the positive controls (two chemostat and two artificial) as both pre-extracted and raw specimens. This parameter space was searched exhaustively in R and one of the several sample configurations that near-equally maximized power to partition variance (as described below by multivariate modeling) was selected for construction and distribution.
DNA extraction
Central DNA extraction
The sample subset with centrally extracted DNA was generated at the University of Guelph using the PowerMax® Soil DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA) according to manufacturer’s instructions. A range of sample DNA concentrations and Abs 260/280 values were obtained, as measured using a NanoDrop® ND-8000 instrument (Wilmington, DE) and are detailed in Supplementary Dataset 2. DNA samples were aliquoted to cryotubes and frozen at −20°C prior to shipping to the NCI repository.
Individual handling laboratories
Upon receipt of the samples, all laboratories froze the sample sets at −20 °C or −80 °C prior to DNA extraction (Supplementary Dataset 2). The samples remained frozen anywhere from 4 days to 92 days. Most laboratories thawed samples at room temperature, but a few laboratories thawed the samples on ice and one lab reported thawing on dry ice. About half of the laboratories spun the samples down prior to processing. The majority of laboratories (N=6) used the MO-BIO PowerSoil kit for DNA extraction and one used the MO-BIO PowerMag kit. One laboratory used two methods (MO-BIO PowerSoil and Qiagen QIAamp) and one laboratory used an in-house protocol. One laboratory each used Qiagen QIAsymphony, Zymo fecal DNA miniprep, Promega Maxwell, Omega BioTek EZ-96, Chemagen Chemagic, and GeneRite DNA-EZ RW02 for DNA extraction. The majority of the laboratories used a homogenizer including MO-BIO, MP Bio, Qiagen, Scientific Industries, BioSpec, and Chemagen. See Supplementary Dataset 2 and 4 for per-lab protocol details.
Most participating handling laboratories received samples in sets of 96 as above, extracted the extracted subset, and sequenced these in combination with pre-extracted DNA, with some exceptions. The HL-G laboratory conducted the extraction portion of the handling module and shipped the extracted DNA to the HL-F laboratory for 16S rRNA gene amplification and sequencing. HL-I spanned two physical laboratories, one that extracted the DNA while the other conducted 16S rRNA gene amplification and sequencing. Finally, while HL-F, HL-J and HL-A laboratories each independently conducted all portions of the handling module, they also sent residual extracted DNA for 16S rRNA gene amplification and sequencing to the HL-E laboratory.
16S rRNA gene amplification primers
Fourteen of the fifteen laboratories used V4 forward primer 515F (GTGCCAGCMGCCGCGGTAA) and all the laboratories used reverse primer number 806R (GGACTACHVGGGTWTCTAAT); the remaining laboratory used V3-4 forward primer 318F (ACTCCTACGGGAGGCAGCAG)6–8. After DNA was extracted, most laboratories froze the samples at 4 °C, −20 °C, or −80 °C prior to amplification, except for one which began amplification the day of DNA extraction. In the laboratories that froze the extracted DNA, it remained frozen anywhere from 1 day to 45 days. Some laboratories reported their PCR reagent manufacturer and these included Invitrogen, 5 Prime, Promega GoTaq, Promega, NEB, and TaKaRa. Ten laboratories reported desalting as a part of the PCR primer purification process (see again Supplementary Dataset 2).
Sequencing
After PCR amplification, samples were frozen in all laboratories except for two at 4 °C or −20 °C for anywhere from 1 day to 39 days. The Illumina MiSeq was used for sequencing in all laboratories except for one, which used the HiSeq; only one laboratory did not use paired end reads. Four laboratories used a sequencing read length of 150 base pairs, 1 used a sequencing read length of 175 base pairs, 1 used a sequencing read of 210 base pairs, 6 used a sequencing read length of 250 base pairs, and 2 used a sequencing read length of 300 base pairs. For the MiSeq runs, Illumina chemistry V2 was used most often, with 2 laboratories using V3 chemistry and one laboratory using V4 chemistry. During sequencing, the fraction of PhiX ranged from 0 to 0.25 and the fraction of quality bases (using the Illumina default Q=15) ranged from 0.573 to 0.961 (Supplementary Dataset 2).
Whole metagenome shotgun sequencing of artificial communities
12 artificial community replicate samples were also sequenced using shotgun metagenomics by Baylor College of Medicine, six pre-extracted DNA and six raw, and half of each from the oral and fecal positive controls. The raw samples were stored in ~30ul volumes and were extracted using the MoBio PowerSoil kit (manual extraction). DNA was quantified using the Quant-iT™ PicoGreen® dsDNA reagent and fluorescence measured with a Synergy 2 spectrophotometer. DNA ranged from <5 ng to >300 ng. Samples were stratified by quantity and those >100 ng were processed via automated liquid handlers; samples with insufficient DNA (from <5 ng to 17 ng) were processed manually (Supplementary Table 9). Shearing, adapter ligation, and the remainder of library construction was then the same for both sample sets.
Illumina paired-end libraries were constructed by shearing each sample into fragments of approximately 300–400 base pairs using a Covaris E210 (Covaris, Inc. Woburn, MA) followed by end-repair, A-tailing and ligation of Illumina multiplexing PE adaptors. Products were then amplified through Ligation Mediated-PCR (LM-PCR), which was performed using the KAPA HiFi DNA Polymerase (Kapa Biosystems, Inc., Cat. no. KM2602). Samples with at least 100 ng input were processed robotically and received 6 cycles of amplification, while samples with lower input required a total of 12 cycles to achieve sufficient yield for sequencing. Purification was performed with Agencourt AMPure XP beads after enzymatic reactions. Following the final XP bead purification, quantification and size distribution of the LM-PCR product was determined using an Agilent Bioanalyzer 7500.
The libraries had an average final size of 402 bp (including adapter and barcode) and were pooled in equimolar amounts to achieve a final concentration of 10 nM. The library templates were prepared for sequencing using Illumina’s cBot cluster generation system with TruSeq PE Cluster Generation Kits. Briefly, this library was denatured with sodium hydroxide and diluted to 7 pM in hybridization buffer in order to achieve a load density of 766K clusters/mm2. The library pool was loaded in one lane of a HiSeq 2000 flow cell which was spiked with 1% PhiX for run quality control. The sample then underwent bridge amplification to form clonal clusters followed by hybridization with the sequencing primer. Sequencing runs were performed in paired-end mode using TruSeq SBS kits for 101 cycles from each end, with an additional 7 cycles for the index read. After sequencing, .bcl files were processed through Illumina’s analysis software (CASAVA), which demultiplexes pooled samples and generates sequence reads and base-call confidence values (qualities).
Bioinformatics
Data coordination
Data files for this project were stored at the DCC and made available via encrypted FTP using the open-source pure-ftpd FTP server software package. Separate accounts were created for each bioinformatics laboratory with separate document roots so that each laboratory would not have visibility into the data uploaded by its peers. After the data uploads were completed and an initial round of quality control performed, the document root names belonging to each laboratory were renamed to randomly generated strings so that each dataset was anonymized. A shared FTP account with read-only privileges was then created so that all participants could view and download the complete and anonymized dataset.
Individual bioinformatics laboratories
Each bioinformatics laboratory downloaded re-blinded, de-multiplexed samples from all handling labs from the DCC, and they re-deposited at the least an OTU table (using Greengenes 13.5 identifiers when possible) and, for open-reference OTUs, a Newick-formatted phylogeny. Optionally, transcripts and/or analysis code was also deposited by some bioinformatics labs. Detailed protocol information was also deposited separately (Supplementary Dataset 3). Bioinformatic protocol details were substantially more diverse than were handling protocols, and we roughly divided sequence processing into four stages: raw sequence quality control, mate pair stitching, OTU quantification, and OTU quality control.
All bioinformatic protocols removed a subset of raw reads based on low-quality base trimming, although exact criteria varied (Q threshold, from end vs. within window, etc.) Most (N=7) also included a minimum (or percent) length threshold to retain reads, and many (N=5) performed explicit chimera checking as well. Overlapping reads were stitched (and often filtered) using QIIME9, UPARSE10, FLASH11, PANDAseq12, or custom software. Only methods included in QIIME (N=6) or UPARSE (N=4) were used for OTU creation, although taxonomic assignments in some cases also used the RDP classification 13 or custom software. Three labs reported filtering OTUs from the resulting table for quality control after creation, and one reported filtering samples. See Supplementary Dataset 3 for per-lab protocol details.
As a follow-up to the main set of MBQC bioinformatic experiments and protocols, the complete dataset was re-processed centrally using mothur14 (v1.39.3 x64 Linux), which was recorded in Supplementary Dataset 3 as BL-10. Since this protocol was run centrally post-hoc, it was not recorded formally nor included in most systematic, blinded analyses. Briefly, it consisted of the mothur MiSeq SOP (https://www.mothur.org/wiki/MiSeq_SOP) with several modifications. First, after SILVA15 alignment, filtering, and RDP13 classification the split.abund command was used with cutoff=10 to remove all unique representative sequences associated with fewer than 10 reads. This was necessary to make sequence classification within mothur computationally feasible. Second, instead of performing de novo OTU calling, the classify.seqs command was used to assign abundant unique sequences to Greengenes16 13.5.99 OTUs. Finally, handling labs producing unpaired reads were analyzed using a "dummy" paired end, reverse-complemented from the single (forward) read. Detailed methods are available online at https://pastebin.com/CmXvidSu.
Artificial community metagenomic validation sequence quality control and taxonomic profiling
For the positive control artificial communities sequenced metagenomically for validation, quality control and removal of human “contaminant” sequences was performed with the KneadDATA v0.3 pipeline (http://huttenhower.sph.harvard.edu/kneaddata), which incorporates Trimmomatic17 and BMTagger18 for filtering and decontamination, respectively. Reads were scanned with a four-base wide sliding window and trimmed when the average base Phred score dropped below 20. Trimmed reads that were shorter than 70 nt were discarded. Human genome assembly version hg38 was downloaded from https://genome.ucsc.edu/ and used as reference for removal of human ‘contaminant’ sequences from sequence data. Taxonomic profiles of filtered shotgun sequence datasets were determined with MetaPhlAn219 (http://huttenhower.sph.harvard.edu/metaphlan2).
Data Integration
OTU tables were re-deposited at the DCC by all bioinformatics labs and made available for integrative analysis by the consortium. Raw sequences are available at BioProject SRP047083 and processed data products at http://mbqc.org. Integrative analyses that relied on OTU matching were performed either using only the subset of OTUs assigned to Greengenes identifiers (although open reference OTUs are available on the DCC) or, for taxonomic analyses, using the lowest non-OTU level taxonomic assignment (species, genus, or otherwise). The taxonomic features and diversity measures used for each analysis are noted on the relevant figures and tables.
Ecological diversity measures and univariate effect modeling
In most cases, Bray-Curtis dissimilarity was used for beta-diversity analyses and inverse Simpson for alpha-diversity. This includes the main univariate assessment of biological and technical effect sizes (Fig. 3), in which four tests were performed: 1) beta-diversity comparisons of replicate samples (i.e. samples from identical specimens) within each lab, without varying any parameters, to identify which labs were most internally consistent (Fig. 3A–B); 2) beta-diversity comparisons of replicate samples between labs, varying only the bioinformatics (Fig. 3A) or handling lab, to identify which labs agreed with each specimen type’s consensus readout (Fig. 3B); 3) alpha-diversity of all stratified samples within lab, to identify which performed more complete extractions (inducing higher diversity) or inflated positive control diversity (inducing lower diversity); and 4) beta-diversity across all samples, varying only one parameter at a time (bioinformatics lab, extracter, sequencer, or specimen, Fig. 3C). This identified, on average, the degree of change induced in microbial community readout when considering each variable in isolation.
Multivariate effect modeling
We fit two linear mixed models to identify protocol variables significantly associated with magnitude (fixed effects) or variability (random effects) in microbiome measures. We first fit a full model with fixed effects for pre-extraction, collapsed specimen type, collapsed health status, PCR primer, read length, collapsed sequencing chemistry version, PhiX fraction, fraction quality bases, log read count, collapsed OTU software, OTU clustering, taxonomic assignment method, and OTU filtering; and random effects for specimen number, handling laboratory, and bioinformatics laboratory. We also fit a simplified model with fixed effects for handling laboratory, bioinformatics laboratory, and pre-extraction; and a random effect for specimen number. The simple model used handling laboratory HL-J and bioinformatics laboratory BL-9B for the reference categories. Arcsine-square root transformed abundances (for variance stabilization) were used as outcomes, from the four phyla present in at least half of all subjects (Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria). We fit each model using restricted maximum likelihood (REML) estimation in the lme4 R package (version 1.1.11). P-values for fixed and random effects were calculated using a likelihood-ratio test and adjusted using the Benjamini-Hochberg-Yekutieli method across all four phyla per model.
Data Availability
Raw sequences are available at BioProject SRP047083 and processed data products, copies of supplementary materials, and computational workflows are available at http://mbqc.org.
Supplementary Material
Acknowledgments
The authors are grateful to the many additional lab members and scientists who contributed to the Microbiome Quality Control Project baseline study, particularly during the sample handling and data generation processes. We would also like to extend our thanks to the participants in the original studies who generously provided specimens to support this and other research. This work was funded in part by the National Institutes of Health NIDDK U54DE023798 (CH), NHGRI R01HG005969 (CH), NHGRI R01 HG005220 (CH, to Rafael Irizarry), NHGRI U01HG004866 (OW), NHGRI U01HG006537 (RK), R01HG004872 (RK), U01HG004866 (RK), the NCI Intramural Research Program (RS), NSF DBI-1053486 (CH), ARO W911NF-11-1-0473 (CH), the W. M. Keck Foundation (RK), John Templeton Foundation (RK), and Alfred P. Sloan Foundation (RK). RK was a Howard Hughes Medical Institute Early Career Scientist.
The Microbiome Quality Control Project Consortium
Gail Ackermann, BioFrontiers Institute, University of Colorado - Boulder (glackermann@ucsd.edu)
Nadim J Ajami, Alkek Center of Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine (nadim.ajami@bcm.edu)
Tulin Ayvaz, Alkek Center for Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine (tayvaz@bcm.edu)
Jordan E Bisanz, Microbiology and Immunology/ Lawson Health Research Institute, Western University (Jordan.Bisanz@gmail.com)
Ian Brown, Molecular and Cellular Biology, University of Guelph (ihlbrown1@Gmail.com)
Zigui Chen, Department of Pediatrics, Albert Einstein College of Medicine (zigui.chen@gmail.com)
Michelle C Daigneault, Molecular and Cellular Biology, University of Guelph (mdaignea@uoguelph.ca)
Mike S Humphrys, School of Medicine, Institute for Genome Sciences, University of Maryland (mhumphrys@som.umaryland.edu)
Catherine A Kelty, ORD, NRMRL, WSWRD, MCCB, USEPA (kelty.catherine@epa.gov)
Randy S Longman, Pathology, Skirball Institute of Biomolecular Medicine (ral2006@med.cornell.edu)
Bing Ma, Institute for Genome Sciences, Department of Microbiology and Immunology, University of Maryland (bma@som.umaryland.edu)
Corinne F Maurice, FAS Center for Systems Biology, Harvard University (corinne.maurice@mcgill.ca)
Julie AK McDonald, Molecular and Cellular Biology, University of Guelph (juliea.k.mcdonald1@gmail.com)
Michael Minson, Chemistry & Biochemistry, University of Colorado at Boulder (mikeminson@gmail.com)
Tiffany W Poon, MPG, Broad Institute (tpoon@broadinstitute.org)
Joshua N Sampson, Biostatistics Branch, DCEG, National Cancer Institute (joshua.sampson@nih.gov)
Daniel A Victorio, Jill Roberts Center for Inflammatory Bowel Disease, Weill Cornell Medical College (dvictorio13@gmail.com)
Matthew C Wong, Alkek Center for Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine (mcwong@bcm.edu)
Xiaolin Wu, Cancer Research Technology Program, Ledois Biomedical Research Inc., Frederick National Laboratory for Cancer Research (forestwu@mail.nih.gov)
Guoqin Yu, Division of Cancer Epidemiology and Genetics, National Cancer Institute (yug3@mail.nih.gov)
Emma Allen-Vercoe, Molecular and Cellular Biology, University of Guelph (eav@uoguelph.ca)
Robert D Burk, Pediatrics; Microbiology & Immunology; Epidemiology & Population Health, Albert Einstein College of Medicine (robert.burk@einstein.yu.edu)
J Gregory Caporaso, Department of Biological Sciences, Northern Arizona University (gregcaporaso@gmail.com)
Nicholas Chia, Surgery, Biomedical Engineering and Physiology, Mayo College (Chia.Nicholas@mayo.edu)
Roberto Flores, Nutritional Science Research Group / Division of Cancer Prevention, National Cancer Institute (floresr2@mail.nih.gov)
Dirk Gevers, Broad Institute of MIT and Harvard (dgevers3@its.jnj.com)
Gregory B Gloor, Biochemistry, University of Western Ontario (ggloor@uwo.ca)
Andrew L Goodman, Department of Microbial Pathogenesis and Microbial Sciences Institute, Yale University School of Medicine (andrew.goodman@yale.edu)
Dan R Littman, Molecular Pathogenesis Program, Kimmel Center for Biology and Medicine of the Skirball Institute, New York University School of Medicine (Dan.Littman@med.nyu.edu)
David A Mills, Food Science and Technology, Viticulture and Enology, and Foods for Health Institute, University of California, Davis (damills@ucdavis.edu)
Joseph F Petrosino, Alkek Center for Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine (petrosi@bcm.edu)
Jacques Ravel, Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD, USA, 21201 (jravel@som.umaryland.edu)
Orin C Shanks, Office of Research and Development, United States Environmental Protection Agency (shanks.orin@epa.gov)
Peter J Turnbaugh, FAS Center for Systems Biology, Harvard University (peter.turnbaugh@ucsf.edu)
Footnotes
Author Contributions
AA, GA, NJA, RDB, JGC, NC, ZC, AAF, GBG, CH, MSH, RK, BM, JFP, BR, JR, MCW, XW, and GY contributed bioinformatics; AA, GA, JGC, AAF, CH, RK, BR, and ES contributed cross-lab data handling or analysis; EA, NJA, TA, RDB, IB, ZC, MCD, RF, DG, ALG, MSH, CAK, RK, DRL, RSL, DAM, CFM, JFP, JR, OCS, PJT, DAV, and XW contributed dna extraction; AA, CCA, GA, AAF, CH, BR, ES, RS, EV, and OW contributed manuscript preparation/writing; EA, GA, NJA, TA, RDB, JEB, IB, ZC, RF, GBG, DG, ALG, MSH, CAK, RK, DRL, RSL, DAM, CFM, JAKM, MM, JFP, JR, OCS, PJT, DAV, and XW contributed other sample handling (sequencing library preparation, sequencing, etc.); EA, RDB, JGC, NC, AAF, RF, GBG, DG, ALG, CH, RK, DRL, DAM, JFP, JR, OCS, RS, and PJT contributed as PIs; GA, TWP, and EV contributed project management; CCA, CH, RK, RS, and OW contributed to the steering committee; CCA, CH, RK, ES, JNS, RS, and EV contributed study design.
Competing Financial Interests
NJA, JFP, and MCW own shares in Diversigen, Inc.
References
- 1.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Integrative H.M.P.R.N.C. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe. 2014;16:276–289. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vatanen T, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozupone CA, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evaluation of 16S rDNA-Based Community Profiling for Human Microbiome Research. PLoS One. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCafferty J, et al. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–2125. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks JP, et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium, S.M.-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014;32:903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson CK, Brotman RM, Ravel J. Intricacies of assessing the human microbiome in epidemiologic studies. Ann Epidemiol. 2016;26:311–321. doi: 10.1016/j.annepidem.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu BC, et al. Characterization of the gut microbiome in epidemiologic studies: the multiethnic cohort experience. Ann Epidemiol. 2016;26:373–379. doi: 10.1016/j.annepidem.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas V, Clark J, Dore J. Fecal microbiota analysis: an overview of sample collection methods and sequencing strategies. Future Microbiol. 2015;10:1485–1504. doi: 10.2217/fmb.15.87. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy NA, et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 2014;9:e88982. doi: 10.1371/journal.pone.0088982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner Mackenzie B, Waite DW, Taylor MW. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Frontiers in microbiology. 2015;6:130. doi: 10.3389/fmicb.2015.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soergel DA, Dey N, Knight R, Brenner SE. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 2012;6:1440–1444. doi: 10.1038/ismej.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fadrosh DW, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wesolowska-Andersen A, et al. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome. 2014;2:19. doi: 10.1186/2049-2618-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huttenhower C, et al. Advancing the microbiome research community. Cell. 2014;159:227–230. doi: 10.1016/j.cell.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan JL, Darling AE, Eisen JA. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS One. 2010;5:e10209. doi: 10.1371/journal.pone.0010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS One. 2014;9:e94249. doi: 10.1371/journal.pone.0094249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Amore R, et al. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genomics. 2016;17:55. doi: 10.1186/s12864-015-2194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clooney AG, et al. Comparing Apples and Oranges?: Next Generation Sequencing and Its Impact on Microbiome Analysis. PLoS One. 2016;11:e0148028. doi: 10.1371/journal.pone.0148028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD. 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016;16:123. doi: 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Schiffman MH, et al. Case-control study of colorectal cancer and fecapentaene excretion. Cancer Res. 1989;49:1322–1326. [PubMed] [Google Scholar]
- 2.Schiffman MH, et al. Case-control study of colorectal cancer and fecal mutagenicity. Cancer Res. 1989;49:3420–3424. [PubMed] [Google Scholar]
- 3.McDonald JA, et al. Evaluation of microbial community reproducibility, stability and composition in a human distal gut chemostat model. J Microbiol Methods. 2013;95:167–174. doi: 10.1016/j.mimet.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 4.De Boever P, Deplancke B, Verstraete W. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder. J Nutr. 2000;130:2599–2606. doi: 10.1093/jn/130.10.2599. [DOI] [PubMed] [Google Scholar]
- 5.Nelson KE, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fadrosh DW, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 11.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilmaz P, et al. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences are available at BioProject SRP047083 and processed data products, copies of supplementary materials, and computational workflows are available at http://mbqc.org.