SUMMARY
MicroRNA-223 is known as a myeloid-enriched anti-inflammatory microRNA that is dysregulated in numerous inflammatory conditions. Here, we report that neutrophilic inflammation (wound response) is augmented in miR-223-deficient zebrafish, due primarily to elevated activation of the canonical nuclear factor κB (NF-κB) pathway. NF-κB over-activation is restricted to the basal layer of the surface epithelium, although miR-223 is detected throughout the epithelium and in phagocytes. Not only phagocytes but also epithelial cells are involved in miR-223-mediated regulation of neutrophils’ wound response and NF-κB activation. Cul1a/b, Traf6, and Tab1 are identified as direct targets of miR-223, and their levels rise in injured epithelium lacking miR-223. In addition, miR-223 is expressed in cultured human bronchial epithelial cells, where it also downregulates NF-κB signaling. Together, this direct connection between miR-223 and the canonical NF-κB pathway provides a mechanistic understanding of the multifaceted role of miR-223 and highlights the relevance of epithelial cells in dampening neutrophil activation.
In Brief

microRNA-223 is dysregulated in many inflammatory conditions such as cancer and asthma, yet its physiological role is not clear. Zhou et al. demonstrate that microRNA-223 in both phagocytes and epithelial cells cooperate to suppress the canonical NF-κB pathway in epithelial cells to restrict the magnitude of inflammation.
INTRODUCTION
Neutrophilic inflammation is critical for host defense and tissue repair. Neutrophils are recruited by multiple “intermediate” and “end-target” chemoattractants released by tissue-resident sentinel cells upon activation by pathogen or damage-associated molecular pattern molecules (Kim and Luster, 2015; Kolaczkowska and Kubes, 2013). Activated neutrophils release oxidants, proteases, and antimicrobial proteins to eliminate pathogens or damaged cells, which also causes collateral tissue damage (Nathan, 2006). As a result, chronic or rampant neutrophilic inflammation drives the immunopathology involved in numerous human diseases, including those directly involving an immune component such as rheumatic arthritis and those that are not obviously linked such as diabetes, neurodegenerative disease, and cancer (Borregaard, 2010; Nathan, 2006). Therefore, fine-tuning the magnitude and resolution of neutrophilic inflammation is critical for the host to restore homeostasis.
It is estimated that 30%–80% of genes in humans are regulated by microRNAs (miRNAs) (Lu and Clark, 2012). miRNAs are small (20–22 nucleotides) non-coding RNA molecules that typically suppress the translation and the stability of transcripts through partial complementarity (Ha and Kim, 2014; Jonas and Izaurralde, 2015). miRNAs are fine-tuners that suppress target gene expression at modest levels, yet master regulators that suppress multiple genes in the same pathways (Gurol et al., 2016; Orellana and Kasinski, 2015). These features suggest that miRNAs may be suitable modulators for the duration and the magnitude of inflammation. Several miRNAs, including miR-223, miR-155, miR-146, and miR-125b, have a role in the differentiation and function of the innate immune system (Johnnidis et al., 2008; Lindsay, 2008; Lu and Liston, 2009). However, as a result of the complex and dynamic interactions between multi-tissues that coordinate an inflammatory response, the functions of these miRNAs in immune response are not fully understood.
MiR-223 has previously been observed primarily in the myeloid lineage, especially neutrophils (Chen et al., 2004; Johnnidis et al., 2008), and is implicated in many inflammatory disorders, infections, and cancers (Haneklaus et al., 2013). Global miRNA expression profiling revealed dysregulation of miR-223 in numerous conditions, however, no consensus between the level of miR-223 and the type or progress of the diseases can be concluded (reviewed in Haneklaus et al., 2013). Opposing roles of miR-223 have been reported, suggesting that miR-223 plays parts of a complex regulatory network with profound impact from the surrounding tissues.
The first in vivo characterization of miR-223 was performed in a miR-223 loss-of-function mouse model (miR-223−/Y), where an increase in granulopoiesis, as well as hyper-mature and hyper-responsive neutrophils were observed (Johnnidis et al., 2008). This strain displayed increased susceptibility to Mycobacterial infection, which is due to increased neutrophil accumulation in the lung (Dorhoi et al., 2013), where a cell-autonomous role of miR-223 in neutrophil recruitment was suggested. NLR family pyrin domain containing 3 (NLRP3) inflammasome activity was also regulated by miR-223 in primary murine neutrophils (Bauernfeind et al., 2012). In addition to the immune system, Harraz et al. (2012) revealed that miR-223, delivered by adenovirus to the brain, protected mice from an ischemic reperfusion brain injury.
In addition, there is a body of literature related to the function of miR-223 in myeloid cells in vitro, with yet to be verified in vivo relevance. As one of the most abundant miRNAs in macrophages, miR-223 responds to stimuli to control the production of interleukin (IL)-6 and IL-1β (Chen et al., 2012) or promotes alternative macrophage activation to inhibit diet-induced adipose tissue inflammation and insulin resistance by targeting Pknox1, Rasa1, and Nfat5 in murine macrophages (Ying et al., 2015; Zhuang et al., 2012). MiR-223 is also associated with macrophage differentiation through targeting IKKα (Li et al., 2010). Furthermore, macrophages and monocytes secrete miR-223 in microvesicles or associated high-density lipoproteins, which are delivered into non-immune cells such as endothelial cells (Ismail et al., 2013; Tabet et al., 2014). Those transferred miRNAs are functionally active, indicating the potential of miR-223 as a central mediator for intercellular cross-talk.
Aside from the immune cells, miR-223 is required for stem cell differentiation, such as the differentiation of mesenchymal stem cells into adipocytes and osteoblasts (Guan et al., 2015). In cancers, the role of miR-223 is conflicting. For example, miR-223 is upregulated in acute lymphoblastic leukemia and bladder cancers, but downregulated in chronic lymphoid leukemia and hepatocellular carcinoma (Chiaretti et al., 2010; Gottardo et al., 2007; Stamatopoulos et al., 2009; Wong et al., 2008). Elevated miR-223 supports migration and invasion in gastric cancer cells but has opposite effect in esophageal cancer cells and human cervical cancer (Li et al., 2011a, 2011b; Tang et al., 2015).
In this study, we took advantage of the zebrafish system, which offers a unique genetic and imaging platform to dissect the interplay of various tissues during inflammation. Moreover, zebrafish are a suitable model for studying innate immunity as they possess conserved innate immune cells and signaling molecules as human (Renshaw and Trede, 2012). We discovered an unexpected importance of nuclear factor κ (NF-κB) activation in epithelial cells at the center of miR-223 regulated neutrophilic inflammation, which provides insights into the multifaceted role of miR-223 in various inflammatory conditions.
RESULTS
miR-223 Deficiency Delays the Resolution of Neutrophilic Inflammation
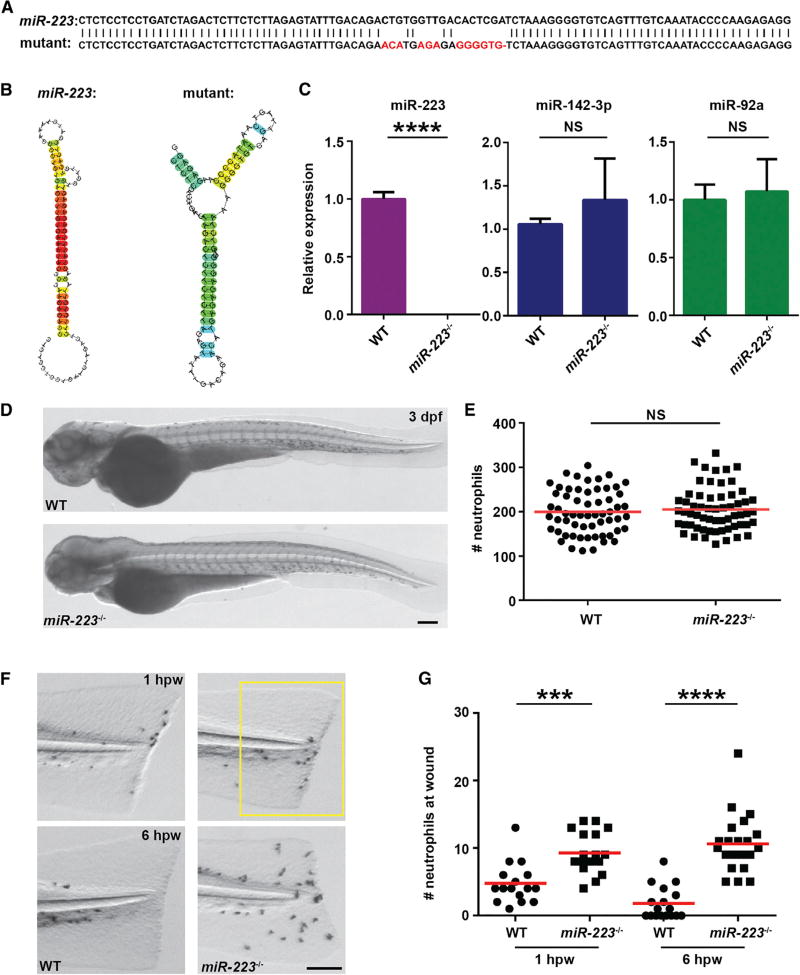
The gene structure and mature sequence of miR-223 are conserved within vertebrates (Roberto et al., 2015). Similar to mice and human (Johnnidis et al., 2008), miR-223 expression was significantly higher in neutrophils in zebrafish (Figures S1A and S1B), supporting zebrafish as a suitable model for miR-223 research. We then generated a miR-223-deficient (miR-223−/−) zebrafish line using the CRISPR/Cas9 system. An allele containing an 18-bp mismatch to the wild-type (WT), which is predicted to disrupt the hairpin structure of pre-mir-223 was selected (Figures 1A and 1B). A complete loss of miR-223, with no alterations of two other miRNAs expressed in neutrophils (miR-142-3p and miR-92a), was detected in the miR-223−/− fish (Figure 1C). No apparent abnormalities in development, viability, or life-span were noted in the miR-223−/− fish, similarly to the miR-223−/Y mice (Johnnidis et al., 2008) and a recently generated miR-223−/− zebrafish line (Kasper et al., 2017). In addition, no differences in total neutrophil numbers, spontaneous inflammation, or cell death in 3-day post fertilization (dpf) embryos were noted (Figures 1D, 1E, and S1C–S1F).
Figure 1. miR-223-Deficient Embryos Display Augmented Neutrophil Response to Tissue Injury.
(A) Sequences of pre-mir-223 in WT and miR-223 mutant embryos. Mutated nucleotides are labeled in red.
(B) The hairpin structures of miR-223 in WT and mutant embryos predicted by Centroidfold.
(C) Expression of miR-223, miR-142-3p, and miR-92a in WT and miR-223−/− embryos determined by qRT-PCR.
(D and E) Representative images (D) and quantification (E) of total neutrophil numbers in WT and miR-223−/− embryos.
(F and G) Representative images (F) and quantification (G) of neutrophil recruitment to tail transection sites in WT and miR-223−/− embryos at 1 hpw and 6 hpw.
The number of neutrophils in the boxed region were quantified. Scale bars, 100 µm.
Data are representative of three independent experiments (E and G) or are pooled from three independent experiments (C). Mean ± SD. ***p < 0.001 and ****p < 0.0001, unpaired Student’s t test. See also Figures S1 and S2.
We next investigated the function of miR-223 in a self-resolving inflammation model. Specifically, a tailfin transection injury in 3 dpf embryos results in a rapid recruitment of neutrophils that peaks at 1 hr post wounding (1 hpw), which is spontaneously resolved by 6 hpw. Significant increases in neutrophils at the wound region in miR-223−/− embryos were detected at both time points (Figures 1F and 1G). The miR-223+/− embryos has a phenotype comparable to miR-223−/−, possibly due to the self-reinforced expression of miR-223 (Fazi et al., 2005). As a result, adult fish (miR-223+/+ and miR-223−/−) from the same parents (miR-223+/−) were used to produce embryos for experiments. To better identify the source of neutrophils at the wound vicinity at 6 hpw, photoconversion-enabled neutrophil fate-mapping was performed (Deng et al., 2011) (Figure S2A). Green neutrophils present at the wound at 1 hpw were photo-converted into red neutrophils. Five hours later, the percentage of red neutrophils remaining at the wound in miR-223−/− was modestly higher than that in WT, indicating a defect in neutrophil reverse migration (migration away from the wound) (Figures S2B and S2C). Furthermore, the number of green neutrophils at the wound in 6 hpw miR-223−/− was significantly higher than that in WT, suggesting a continuous recruitment of neutrophils in the miR-223−/− (Figure S2D). Altogether, the excessive neutrophilic inflammation in miR-223−/− embryos was primarily a result of continuous neutrophil recruitment, with a minor defects in reverse migration.
Neutrophil-Intrinsic miR-223 Is Required to Control Neutrophilic Inflammation
Because miR-223 has been known as a myeloid-specific miRNA, we next determined whether miR-223 regulates neutrophil wound response cell-autonomously. To specifically knock down miR-223 in neutrophils, a miR-223 sponge, containing six bulged binding sites of miR-223, was expressed under the lysozyme C (lyzC) promoter (Hall et al., 2007) (Figure 2A). Indeed, embryos expressing the miR-223 sponge accumulated more neutrophils at the wound at 6 hpw (Figures 2B and 2C; Movie S1). To further confirm the neutrophil-intrinsic role of miR-223, a neutrophil-specific miR-223-overexpression line, Tg(lyzC: miR-223-RFP)pu9 was generated and crossed into the miR-223−/− background (Figure 2D). The neutrophil-specific rescue partially restored the miR-223 expression level and partially rescued the over-inflammation phenotype (Figures 2E and 2F), suggesting that miR-223 regulates inflammation, at least partially, inside neutrophils. A control rescue with RFP alone did not yield any phenotype (Figures 2D, 2E, and 2G).
Figure 2. Neutrophil-Intrinsic miR-223 Regulates Neutrophilic Inflammation.
(A) Schematics of Tol2-lyzC-Gal4-crys-CFP construct, injected into WT embryos to generate the transgenic line Tg(lyzC:Gal4-Vp16, crys:CFP)pu8; Tol2-UAS-miR-223 sponge that contains 6 bulged miR-223 binding sites after the UAS element and the Dendra2 control plasmid.
(B and C) miR-223 sponge or Dendra2 control plasmids in (A) were injected into embryos from Tg(lyzC: Gal4-Vp16, crys:CFP)pu8 and Tg(lyzC: mCherry-H2B) cross. Tailfins were transected at 3 dpf. Representative images at indicating time points are shown in (B), and neutrophil recruitment at 6 hpw is shown in (C).
(D) Schematics of Tol2-lyzC-miR-223/RFP constructs, injected into WT embryos to generate the transgenic line Tg(lyzC: RFP-miR-223)pu9 and the control line Tg(lyzC: RFP)pu10.
(E) The transgenic lines illustrated in (D) were crossed into the miR-223−/− background. The siblings without RFP were used as negative control. miR-223 expression in indicated groups was quantified by qRT-PCR (mean ± SD).
(F and G) Quantification of neutrophil recruitment to tail transection sites in embryos with miR-223 (F) or RFP control (G) expressed in neutrophils in the miR-223−/− background at 1 hpw and 6 hpw.
Scale bars, 100 µm. One representative experiment of three independent repeats is shown. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001, unpaired Student’s t test (C, F, and G) or one-way ANOVA (E). See also Movie S1.
NF-κB Pathway Is Activated in miR-223−/− Embryos
To fully understand how miR-223 regulates neutrophilic inflammation, microarray analysis of both WT and miR-223−/− embryos, unwounded or at 1 and 6 hpw, was performed. Transcriptome analysis revealed that 166, 115, and 114 genes (that have known human orthologs) were upregulated over 1.5-fold in miR-223−/− at indicated time points (Figure S3A). This cutoff was selected according to a pervious report that miR-223 modulates gene expression at modest levels (Baek et al., 2008). The top four pathways altered are inflammatory response, cancer, organismal injury and abnormalities, and reproductive system disease. Among the genes involved in inflammatory response, those in the interferon pathways and NF-κB pathways were enriched.
To identify the biological relevant pathway, a CRISPR screen was performed, aiming to identify genes whose suppression would rescue the over-inflammation phenotype in miR-223−/− embryos. The genes initially screened were: Irf3 and Irf7 (interferon regulatory transcription factors), Ifng1 and Ifng2 (type 2 interferons), Crfb5 and Crfb17 (type 1 or type 2 interferon receptor subunits (Aggad et al., 2010), Stat1a/b and Jak2a (signaling molecules), Myd88 (the adaptor of Toll-like receptor [TLR] and IL1R), Caspa (Caspase1 ortholog), and Il1b (pro-inflammatory cytokine) (Figure S3B). A single guide RNA (sgRNA) targeting GFP that knocked out GFP expression in zebrafish with high efficiency was used as a control (Figures S4A and S4B). Among all the genes screened, only the Myd88 sgRNAs injected embryos exhibited a rescue of the over-inflammation phenotype (Figure S4C). The Crfb17 sgRNAs injected embryos displayed an opposite phenotype, suggesting that interferon-γ may provide a feed-back inhibition of over-inflammation induced with miR-223 loss of function, but this requires further characterization. This result of Myd88 knockout was confirmed in three independent experiments (Figure 3A). When the Myd88 sgRNA target sites were sequenced, 98.6% of the alleles contain indels (Figure S5A). Further validation was done with a specific inhibitor of the canonical NF-κB pathway, BAY (11-7085), which blocks the phosphorylation of the inhibitory I-κB (Figure 3B). Collectively, miR-223 suppresses neutrophilic inflammation, at least partially, through downregulating the NF-κB pathway.
Figure 3. miR-223 Regulates NF-κB Pathway by Suppressing Cul1a, Cul1b, Traf6, and Tab1.
(A) Quantification of neutrophil recruitment at 6 hpw in miR-223−/− embryos injected with sgRNAs targeting Myd88, using gfp sgRNAs as the control.
(B) Quantification of neutrophil recruitment at 6 hpw in miR-223−/− embryos treated with BAY (1 µM) or DMSO (1%).
(C) Schematics of the canonical NF-κB signaling pathway.
(D) The expression of Cul1a, Cul1b, Traf6, and Tab1 in WT and miR-223−/− embryos in unwounded embryos or at 1 hpw as determined by qRT-PCR.
(E) Dual luciferase reporter assay showing specific suppression of Renilla luciferase depending on the 3′UTRs of Cul1a, Cul1b, Traf6, and Tab1 by miR-223.
(F) Quantification of neutrophil recruitment at 6 hpw in miR-223−/− embryos injected with sgRNAs targeting both Cul1a and Cul1b or the gfp sgRNAs control. Data are representative of three independent experiments (A, B, and F) or are pooled from three independent experiments (D and E).
Mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001, unpaired Student’s t test and paired Student’s t test in (E). See also Figures S3–S5.
miR-223 Directly Suppresses Cul1a/b, Traf6, and Tab1
Despite the established importance of the canonical NF-κB pathway in inflammation, a direct link of this pathway with miR-223 has not been reported. We used three algorithms (Target scan, PicTar, and miRanda) (Grimson et al., 2007; John et al., 2004; Krek et al., 2005) and identified four genes in the NF-κB pathway: Cul1a, Cul1b, Traf6, and Tab1 as potential miR-223 targets (Figure 3C). CUL1 is an essential component of the SCF (complex of SKP1, CUL1, and F-box protein) E3 ubiquitin ligase. The SCF E3 ligase complex (SKP1-Cul1-β-TrCP1) ubiquitinates I-κB and activates the canonical NF-κB pathway (Villeneuve et al., 2010). Cul1a and Cul1b are duplicated orthologous in zebrafish. TRAF6 (tumor necrosis factor [TNF] receptor-associated factor protein 6) and TAB1 (transforming growth factor (TGF)-β activated kinase 1 binding protein 1) are signaling components in the canonical NF-κB pathway. The mRNA levels of Cul1a and Traf6 were significantly elevated in miR-223−/− at 1 hpw, suggesting that they are regulated by miR-223 (Figure 3D). However, there is a lack of reduction in Cu1b and Tab1 transcripts, which may reflect the fact that miRNAs do not always degrade the target mRNAs (Cipolla, 2014). To verify the direct suppression of Cul1a, Cul1b, Traf6, and Tab1 by miR-223, dual-luciferase reporter assays were performed. As expected, miR-223 significantly inhibited the expression of luciferase reporters fused with the 3′ untranslated regions (3′UTRs) of Cul1a, Cul1b, Traf6, or Tab1, but not with a control 3′UTR (Figure 3E). Additionally, transient knocking out of Cul1a and Cul1b reduced neutrophil recruitment in miR-223−/− embryos at 6 hpw, further validating Cul1a and Cul1b as physiological relevant miR-223 targets (Figure 3F). The mutation efficiency of Cul1a and Cul1b was 79.6% and 60.8%, respectively (Figures S5B and S5C). Taken together, miR-223 negatively regulates the canonical NF-κB signaling through suppressing the expression of Cul1a, Cul1b, Traf6, and Tab1.
Loss of miR-223 Elevates NF-κB Activation in Basal Epithelial Cells
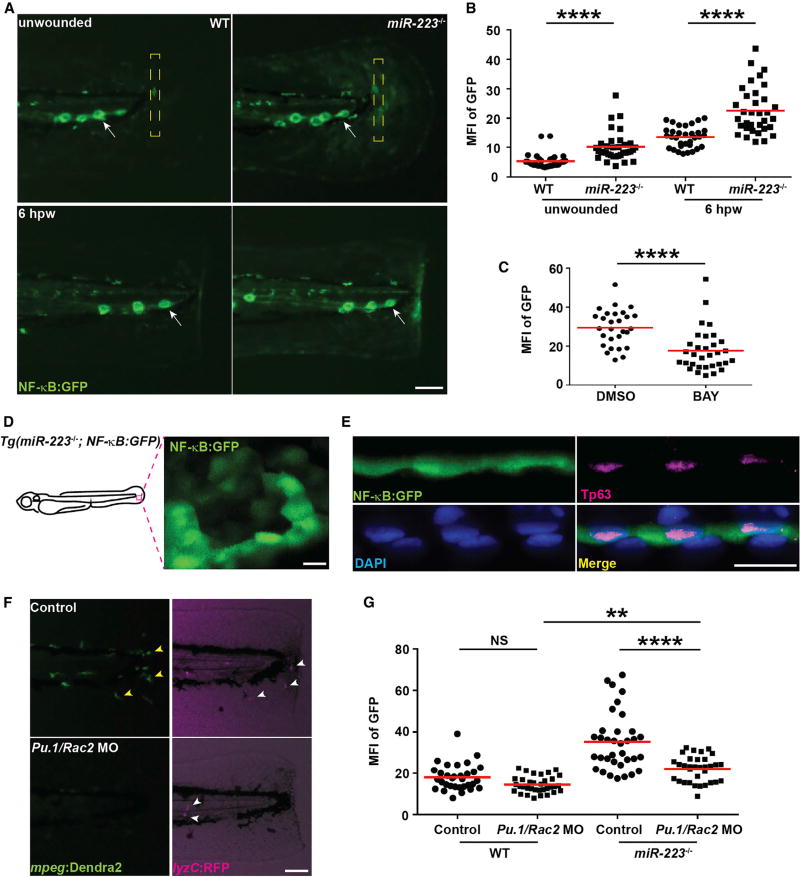
To determine the dynamics of the NF-κB activation, we utilized a NF-κB reporter line Tg(NFκB:GFP), where GFP transcription is controlled by NF-κB recognition sequences (Kanther et al., 2011). miR-223−/− embryos displayed elevated GFP signal in the tail fin at steady state, as well as at the wound margin at 6 hpw (Figures 4A and 4B; Movie S2), which is consistent with our microarray results (Figure S6). As expected, the NF-κB inhibitor BAY, significantly downregulated GFP expression at wound (Figure 4C). Interestingly, the GFP+ cells in the fin were immobile with an epithelium-like morphology (Figure 4D). The tailfin epithelium in zebrafish larva are composed of one apical layer and one basal layer. With immunofluorescence, we discovered that the GFP signal was restricted to the Tp63-positive (Lee and Kimelman, 2002) basal layer (Figure 4E). Collectively, NF-κB pathway was elevated in basal epithelial cells in miR-223−/− embryos.
Figure 4. NF-κB Pathway Is Elevated in Basal Epithelial Cells in miR-223−/− Embryos.
The NF-κB reporter line Tg(NF-κB:GFP) was crossed into the miR-223−/− and matched WT background.
(A and B) Representative images (A) and quantification (B) of GFP signal. Mean fluorescence intensity (MFI) in the yellow square (A) in unwounded embryos or at the wound edge at 6 hpw was quantified. White arrows, neuromast cells constitutively expressing NF-κB signal.
(C) Quantification of GFP signal at the wound edge at 6 hpw in miR-223−/− embryos treated with DMSO or BAY.
(D) Representative confocal image of GFP+ cells in miR-223−/− embryos.
(E) Immunofluorescence of GFP and Tp63 (basal cell marker) in Tg(miR-223−/−, NF-κB:GFP) embryos. Nucleus were stained with DAPI. Representative confocal images of vertical view are shown in (E).
(F and G) Embryos from WT and miR-223−/− were injected with Pu.1 (200 µM) and Rac2 (100 µM) morpholinos. (F). Representative images showing the efficiency of the two morpholinos. Yellow arrowhead, macrophage; white arrowhead, neutrophils.
(G) Quantification of GFP signal at the wound edge at 6 hpw in WT and miR-223−/− embryos.
Scale bars, 100 µm (A and F), 20 µm (D and E). Data are representative of three independent experiments. **p < 0.01 and ****p < 0.0001, unpaired Student’s t test (B and C) or two-way ANOVA (G). See also Figure S6 and Movie S2.
miR-223 Suppresses NF-κB Activation in Basal Epithelial Cells in a Phagocyte-Dependent and -Independent Fashion
Because miR-223 is highly expressed in phagocytes, we then sought to determine whether the elevated wound signal is a result of hyperactive phagocytes. Morpholinos that disrupt macrophages development (Pu.1) (Rhodes et al., 2005) and inhibit neutrophil and macrophage motility (Rac2) (Deng et al., 2011; Rosowski et al., 2016) were used to remove all phagocytes at the wound margin (Figure 4F). The elevated NF-κB signal in wounded epithelial cells was partially inhibited in the phagocyte-deficient miR-223−/− embryos (Figure 4G), suggesting both intra- and extra-phagocyte regulation.
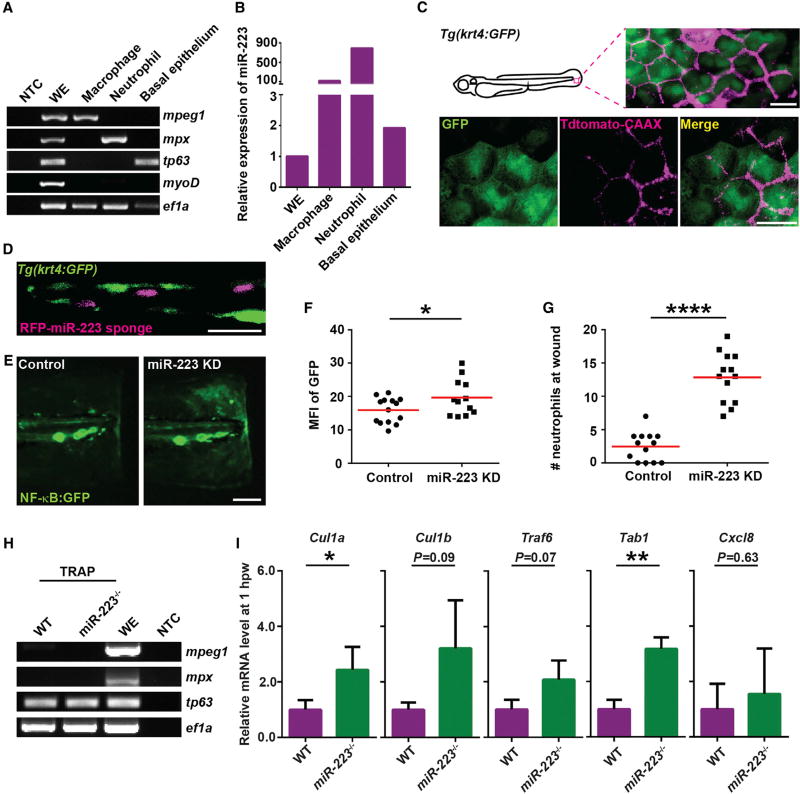
Furthermore, the elevated NF-κB signal in unwounded fin was not affected by phagocytes (Figure S6), suggesting a surprising possibility that miR-223 directly regulates NF-κB pathway in epithelial cells, whereas no, or very low, expression of miR-223 in epithelial cells was reported. Thus, whether miR-223 is expressed in the epithelium was determined. Neutrophils, macrophages, and basal epithelial cells were sorted from the 3 dpf embryos. The quality of cell sorting was validated by RT-PCR using lineage-specific markers (Figure 5A). Mature miR-223 was enriched for 2-fold in basal epithelial cells compared with the whole embryo (Figure 5B).
Figure 5. miR-223 Suppresses NF-κB Activation in Basal Epithelial Cells in a Cell-Intrinsic Manner.
(A and B) Macrophages, neutrophils, and basal epithelial cells were sorted from 3 dpf embryos. (A) RT-PCR of lineage-specific markers. mpeg, macrophage marker; mpx, neutrophil marker; tp63, basal epithelial cell marker; myoD, muscle cell marker; ef1a, loading control; WE, whole embryo; NTC, non-template control. (B) qRT-PCR of miR-223.
(C) Tdtomato-CAAX mRNA was injected into Tg(krt4: GFP)pu11 embryos at 4-cell stage. Representative confocal images from the lateral view of 3 dpf larvae are shown.
(D) RFP-miR-223 sponge mRNA was injected into Tg(krt4: GFP)pu11 embryos at 4-cell stage. A representative confocal image from the vertical view of 2 dpf larvae is shown.
(E and F) Tg(NF-κB:GFP) were injected with miR-223 sponge or the RFP control mRNA at 4-cell stage. Representative images and quantification of GFP signal at the wound edge at 6 hpw are shown.
(G) Quantification of neutrophil recruitment to the wound in embryos injected with miR-223 sponge or RFP mRNA at 4-cell stage.
(H and I) Tol2-tp63-GFP-L10a was injected into WT or miR-223−/− embryos. At 3 dpf, cell-specific mRNA-ribosome complexes were isolated by anti-GFP antibodies at 1 hpw.
(H) RT-PCR of lineage-specific markers as described in (A).
(I) Real-time qPCR of indicated genes. Data are pooled from three independent experiments (mean + SD).
Scale bars, 20 um (C and D), 100 µm (E). Data are representative of three independent experiments (F and G) or of two independent experiments (A and B). *p < 0.05, **p < 0.01, and ****p < 0.0001, unpaired Student’s t test.
To demonstrate that basal epithelium-intrinsic miR-223 regulates the NF-κB pathway, an RFP-miR-223 sponge mRNA was injected into embryos at the 4-cell stage. It was previously reported that injecting mRNAs at 4- to 8-cell stage led to mosaic expression predominantly in basal epithelial cells (Gault et al., 2014), and the same observation was confirmed using Tdtomato-CAAX mRNA (Figure 5C). When the RFP-miR-223 sponge mRNA was delivered this way, its expression was also restricted to the close proximity to the apical layer (Figure 5D). Embryos expressing RFP-miR-223 sponge had elevated NF-κB signals at the wound edge and enhanced neutrophil wound response at 6 hpw, compared to embryos receiving the RFP control (Figures 5E–5G). To further validate that miR-223 downregulates the target gene expression in basal epithelial cells, transcripts associated with the ribosomes in the basal epithelium were isolated. We have optimized the published protocol (Heiman et al., 2014) that no contamination from the phagocytes were detected (Figure 5H). The translation of Cul1a/b, Traf6, and Tab1, but not Cxcl8, was indeed enhanced in the miR-223-deficient larvae after wounding (Figure 5I). Therefore, miR-223 in basal epithelial cells regulates neutrophilic inflammation and NF-κB pathway in a cell autonomous manner.
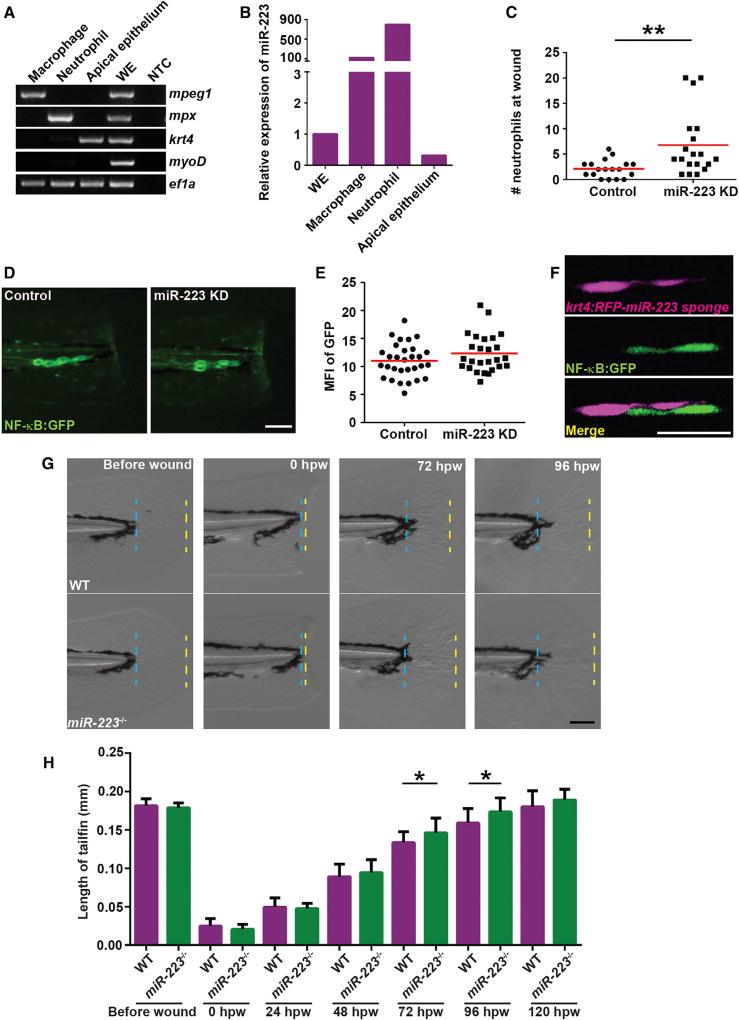
To be noted, apical epithelial cells also contain modest, yet detectable levels of miR-223 (Figures 6A and 6B). Therefore, a transgenic line that expresses the miR-223 sponge specifically in apical epithelial cells, Tg(krt4: RFP-miR-223 sponge)pu12 was generated, in which a significantly enhanced recruitment of neutrophils was also observed (Figure 6C). Interestingly, the NF-κB signal was not present in the apical epithelial cells, but again was restricted to the basal cells, although not significantly elevated (Figures 6D–6F), suggesting that miR-223 in the apical layer regulates neutrophil recruitment in a way separable from the NF-κB activation in the basal layer.
Figure 6. Fin Regeneration Is Accelerated in miR-223-Deficient Embryos.
(A) Apical epithelial cell were sorted from the Tg(krt4:GFP)pu11 and RT-PCR of lineage-specific markers were performed as described in Figure 5A. krt4, apical epithelial cell marker.
(B) Real-time qPCR of miR-223.
(C) Quantification of neutrophil recruitment at 6 hpw in embryos from Tg(krt4: RFP-miR-223 sponge)pu12 and Tg(krt4: RFP)pu13.
(D and E) Tg(krt4: RFP-miR-223 sponge)pu12 and Tg(krt4: RFP)pu13 were crossed with Tg(NF-κB:GFP). Representative images and quantification of GFP signal at the wound edge at 6 hpw.
(F) Representative confocal images of embryos from Tg(NF-κB:GFP/krt4: RFP-miR-223 sponge)pu12 at 6 hpw.
(G and H) Tailfin regeneration in WT and miR-223−/− embryos. Tailfin transection was performed at 3 dpf. The length of regenerated tailfin was measured as the distance between the blue and yellow dash lines.
(G) Representative images of tailfins at indicated time points.
(H) Quantification of the length of tailfin at different time points (mean ± SD; n > 20 in each group).
Scale bars, 100 µm (D and G), 20 um (F). Data are representative of three independent experiments (C, E, and H) or of two independent experiments (A and B). *p < 0.05, **p < 0.01, and ****p < 0.0001, unpaired Student’s t test.
Furthermore, an accelerated tailfin regeneration was observed in miR-223-deficient larvae, especially at 72 hpw and 96 hpw (Figures 6G and 6H), possibly due to the enhanced NF-κB activation that is known to promote cell proliferation (Brantley et al., 2001). This result further supports that miR-223 regulates physiological processes in epithelial cells.
miR-223 Regulates NF-κB Pathway in Human Bronchial Epithelial Cells
Zebrafish skin is a suitable model for human mucosal epithelium (Enyedi et al., 2016). To extend our findings to humans, the expression of miR-223 was evaluated in human cells. Indeed, we detected miR-223 in the immortalized human bronchial epithelial cells (HBECs), but not in the human embryonic kidney 293 cells (HEK293T). MiR-223 expression was further elevated in the H441 human lung cancer cell line (Figure 7A). Moreover, human CUL1 and TAB2 were bioinformatically identified as potential targets of miR-223, which was further confirmed in luciferase reporter assays (Figure 7B). Additionally, overexpression of miR-223 in HEK293T cells suppressed NF-κB activation with or without stimulation using heat-killed P. aeruginosa. In HBECs, miR-223 overexpression suppressed, whereas its inhibition enhanced the activation of NF-κB after P. aeruginosa stimulation (Figure 7C). Together, our results indicated that miR-223 regulates NF-κB pathway in human cells.
Figure 7. miR-223 Regulates NF-κB Activation in Human Bronchial Epithelial Cells.
(A) Real-time qPCR of miR-223 expression in human neutrophils, human immortalized bronchial epithelial cells (HBEC), transformed lung cancer epithelial cells (H441), and HEK293T.
(B) Dual luciferase reporter assay showing specific suppression of Renilla luciferase dependent on 3′UTRs of human CUL1 and TAB2 by miR-223. (C) NF-κB activity in HEK293T cells expressing miR-223 or control with or without P. aeruginosa stimulation. NF-κB activity in HBEC cells expressing miR-223/control or miR-223-sponge/control with or without P. aeruginosa stimulation. Data are pooled from three independent experiments (mean ± SD).
*p < 0.05, unpaired Student’s t test (A, C, and D) and paired Student’s t test (B).
DISCUSSION
Here, we have reported that miR-233 regulates distinct signaling pathways in multiple tissues that coordinate the resolution of neutrophilic inflammation. In basal epithelial cells, miR-223 suppresses the canonical NF-κB signaling by directly targeting multiple components in the signaling cascades. Phagocytes also contribute to suppress basal cell NF-κB activation during inflammation. In the apical epithelium, miR-223 promotes the resolution of neutrophilic inflammation in a separate mechanism. Our observation is in line with the seminal work performed by McDonald et al. (2010) that complex overlapping chemokines and lipid mediators, produced by the phagocytes and injured tissue coordinate to recruit neutrophils to a sterile injury.
The most surprising observation is that miR-223 modulates the inflammatory signaling, visualized by the NF-κB reporter, primarily in epithelial cells in a cell-intrinsic manner. This phenomenon is in line with the report that the reactive oxygen species, which are essential for neutrophil wound response, are primarily restricted to the injured epithelium (Niethammer et al., 2009), whereas the phagocytes were not the primary source as previously speculated. Neutrophils are recruited to tissue injury sites by a hierarchical system of chemoattractive signals. Other signaling molecules possibly produced by the injured epithelium are the metalloproteases, such as mmp9 (LeBert et al., 2015) and mmp13 (Lisse et al., 2016), as well as ATP and proinflammatory lipid mediators, released as a consequence of osmotic swelling of the epithelial cells and their nucleus (Enyedi et al., 2016; Gault et al., 2014). In addition, the formyl-peptide signals produced by injured hepatocytes recruits neutrophils to the necrotic core in mice (McDonald et al., 2010). It remains to be determined how these signals are modulated by miR-223 and the related NF-κB pathway. It remains to be determined whether neutrophil maturation is also regulated by the function of miR-223 outside the phagocytes. Neutrophils in miR-223 knockout mice are hypermature and hyperactive (Johnnidis et al., 2008), however, the mechanism is not clear.
Indeed, the importance of the epithelial cells in inflammation is increasingly recognized. A good example is that epidermal cells utilize fatty acid β-oxidation, which again was previously investigated in the immune cells, to fuel the production of matrix metalloproteinase and coordinated the immune response during cutaneous inflammation (Hall et al., 2014). Because the expression level of miR-223 changes under many disease conditions, further animal or human work is still needed to elucidate the physiological significance of miR-223-NF-κB in various disease conditions. It is speculated that zebrafish skin is a good model for human mucosal epithelium (Enyedi et al., 2016). Here, we detected the presence of miR-223 in human bronchial epithelial cells, cells in the lower respiratory track that are critical for asthma pathogenesis (Erle and Sheppard, 2014). In line with our observation, miR-223 is upregulated in both bronchial and alveolar epithelial cells upon lipopolysaccharide (LPS) challenge in mice (Sugatani and Hruska, 2007). In addition, a recent study (Maes et al., 2016) reported a significant upregulation of miR-223 in sputum of patients with severe neutrophilic asthma, supporting a physiological role of miR-223 in mucosal epithelial cells.
Our result also indicates a neutrophil-intrinsic role of miR-223 in regulating neutrophil recruitment, which is consistent with the previous observation that miR-223 regulates neutrophil recruitment in a blood cell-intrinsic manner (Dorhoi et al., 2013). Neutrophils isolated from miR-223-deficient mice produce more Cxcl2 and Ccl3 upon Mycobacterial infection. Cxcl2 is a neutrophil chemoattractant. Ccl3 stimulates neutrophils to produce proinflammatory mediates such as platelet-activation factor and lipid leukotriene B4 (LTB4) to recruit other neutrophils (Reichel et al., 2009). In an elegant study, Lämmermann et al. (2013) have shown that neutrophils secret LTB4 in exosomes to amplify the range of neutrophil recruitment during neutrophil swarming (Majumdar et al., 2016).
Here, we visualized that miR-223-deficient phagocytes also have a direct impact on the injured epithelium. Many molecules produced by the phagocytes, including reactive oxygen species, granule enzymes, inflammatory mediates, and neutrophil extracellular traps can lead to further tissue damage and amplifies inflammation. Many of these molecules are regulated by the NF-κB transcription factor or are direct miR-223 targets in human neutrophils such as the cysteine proteases cathepsin L and Z (Baek et al., 2008). It is also possible that activated phagocytes deliver additional miR-223 into the epithelium via exosomes or microvesicles, as demonstrated recently in mice (Neudecker et al., 2017). Inhibiting the Cxcl8 receptor Cxcr2 or the high-affinity LTB4 receptor Ltb4r1 did not reduce neutrophil recruitment in the miR-223 knockout (data not shown). It is likely that miR-223 regulates redundant or other inflammatory signals to coordinate neutrophil recruitment. Further work is required to fully dissect the mechanism.
The adaptor protein Myd88 was detected as a suppressor to the over-inflammation induced by miR-223 deficiency. Both the TLRs and the IL-1/18 receptor can activate the canonical NF-kB signaling through Myd88. Due to the poor characterization of the zebrafish IL-1/18 pathway and the high functional redundancy of the TLRs (Li et al., 2017; Sepulcre et al., 2009), it is difficult at present to pinpoint the receptor(s) that activates the NF-κB pathway during tissue injury. The nuclear factor NF-κB plays a complex role in inflammation and cancer (Hoesel and Schmid, 2013; Lawrence, 2009). On one hand, it promotes inflammation by producing pro-inflammatory cytokines and chemokines. On the other hand, it resolves inflammation by promoting leukocytes apoptosis, the production of anti-inflammatory cytokines and survival of somatic tissue under stress. Despite the vast number of predicted and validated miR-223 targets, here, we report that miR-223 directly targets four different components in the canonical NF-κB pathway in zebrafish and human, providing a significant advance in miR-223 biology. In human cells, besides CUL1 and TAB2 (Figure 6), TRAF6 and TAB1 also harbor miR-223 binding sites in their longest splice variants. Furthermore, miR-223 can suppress IKKα, downregulating the alternative NF-κB pathway (Li et al., 2010). Together, miR-223 possibly regulates the NF-κB activation differentially in various tissues, as disease progress, as a result of alternative splicing, or balancing the canonical and the alternative pathways. In light of the complexed biological function of the NF-κB pathway, our work provides a mechanistic understanding of the multifaceted and multilayered role of miR-223 in inflammatory diseases and cancer.
EXPERIMENTAL PROCEDURES
Animals
The zebrafish experiment was conducted in accordance to the internationally accepted standards. The Animal Care and Use Protocol was approved by The Purdue Animal Care and Use Committee (PACUC), adhering to the Guidelines for Use of Zebrafish in the NIH Intramural Research Program (protocol number: 1401001018). Embryos at 3 days post fertilization, when the sex is not determined, were used for experiments if not otherwise noted.
The miR-223-deficient line and other transgenic lines were generated as described in the Supplemental Information.
Cell Culture
HEK293T cells, HBEC, and H441 were obtained from American Type Culture Collection (ATCC) and maintained as described in the Supplemental Information.
Zebrafish Handling
Microinjection, tailfin wounding, Sudan black staining, fluorescence-activated cell sorting, and immunofluorescent staining were carried out as described in the Supplemental Information.
qRT-PCR
RNA purification and qRT-PCR were performed as described in the Supplemental Information.
Live Imaging
Time-lapse fluorescence images, NF-κB:GFP reporter imaging, confocal imaging, and photoconversion assay were performed as described in the Supplemental Information.
Microarray
Microarray analysis was performed following MIAME guidelines as described in the Supplemental Information.
CRISPR Screening
Transient gene knockout were performed as descried in the Supplemental Information.
NF-κB Reporter Assay
A NF-κB reporter plasmid were generated and dual luciferase reporter assay was performed as described in the Supplemental Information.
Translating Ribosome Affinity Purification
WT or miR-223−/− embryos were injected with Tol2-tp63-GFP-L10a plasmid and translating ribosome affinity purification (TRAP) was performed as described in the Supplemental Information.
Statistical Analysis
Statistical analysis was performed with Prism 6 (GraphPad). The statistical significance of differences between groups was compared with an unpaired two-tailed Student’s t test, paired Student’s t test, or ANOVA. For dual luciferase reporter assays, each psiCHECK2 reporter was normalized to the control pCDNA vector and evaluated with paired Student’s t test. A p value of <0.05 was accepted as statistically significant.
Supplementary Material
Highlights.
miR-223-deficient zebrafish display augmented neutrophilic inflammation
Elevated NF-κB activation increases inflammation in miR-223−/− embryos
miR-223 suppresses canonical NF-κB pathway in basal epithelial cells
Both epithelial and phagocytic miR-223 regulate inflammation
Acknowledgments
The authors thank Dr. Anna Huttenlocher (UW-Madison) for the human neutrophil RNA, Dr. Sandra Rieger (MDI Biological Laboratory) for zebrafish Tp63 promoter, Dr. John Rawl (Duke) for Tg(NF-κB:GFP), Dr. Zhaoqing Luo (Purdue) for assistance with luciferase assays, and Dr. Donna Fekete (Purdue) for critical reading of the manuscript. The work is supported by NIH R35GM119787 and the Jim and Diann Robbers Cancer Research Grant for New Investigators to Q.D., NIH R00CA178091 and R01CA205420 to A.L.K., and NIH P30CA023168 to the Purdue Center for Cancer Research.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the microarray results reported in this paper is GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94996): GSE94996.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and two movies and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.058.
AUTHOR CONTRIBUTIONS
Q.D. and W.Z. designed research and wrote the manuscript. W.Z., A.Y.H.-H., T.G., and X.Z. performed experiments and analyzed data. S.E.W.-H. and J.L.F. performed and analyzed microarray. A.S.P. and A.L.K. performed experiments with human cells.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
References
- Aggad D, Stein C, Sieger D, Mazel M, Boudinot P, Herbomel P, Levraud JP, Lutfalla G, Leptin M. In vivo analysis of Ifn-γ1 and Ifn-γ2 signaling in zebrafish. J. Immunol. 2010;185:6774–6782. doi: 10.4049/jimmunol.1000549. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. J. Immunol. 2012;189:4175–4181. doi: 10.4049/jimmunol.1201516. [DOI] [PubMed] [Google Scholar]
- Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Brantley DM, Chen CL, Muraoka RS, Bushdid PB, Bradberry JL, Kittrell F, Medina D, Matrisian LM, Kerr LD, Yull FE. Nuclear factor-kappaB (NF-kappaB) regulates proliferation and branching in mouse mammary epithelium. Mol. Biol. Cell. 2001;12:1445–1455. doi: 10.1091/mbc.12.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wang H, Liu Y, Song Y, Lai L, Han Q, Cao X, Wang Q. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3. PLoS ONE. 2012;7:e42971. doi: 10.1371/journal.pone.0042971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaretti S, Messina M, Tavolaro S, Zardo G, Elia L, Vitale A, Fatica A, Gorello P, Piciocchi A, Scappucci G, et al. Gene expression profiling identifies a subset of adult T-cell acute lymphoblastic leukemia with myeloid-like gene features and over-expression of miR-223. Haematologica. 2010;95:1114–1121. doi: 10.3324/haematol.2009.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolla GA. A non-canonical landscape of the microRNA system. Front. Genet. 2014;5:337. doi: 10.3389/fgene.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Yoo SK, Cavnar PJ, Green JM, Huttenlocher A. Dual roles for Rac2 in neutrophil motility and active retention in zebrafish hematopoietic tissue. Dev. Cell. 2011;21:735–745. doi: 10.1016/j.devcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A, Iannaccone M, Farinacci M, Faé KC, Schreiber J, Moura-Alves P, Nouailles G, Mollenkopf HJ, Oberbeck-Müller D, Jörg S, et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Invest. 2013;123:4836–4848. doi: 10.1172/JCI67604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi B, Jelcic M, Niethammer P. The cell nucleus serves as a mechanotransducer of tissue damage-induced inflammation. Cell. 2016;165:1160–1170. doi: 10.1016/j.cell.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erle DJ, Sheppard D. The cell biology of asthma. J. Cell Biol. 2014;205:621–631. doi: 10.1083/jcb.201401050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Gault WJ, Enyedi B, Niethammer P. Osmotic surveillance mediates rapid wound closure through nucleotide release. J. Cell Biol. 2014;207:767–782. doi: 10.1083/jcb.201408049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Gao Y, Zhou J, Wang J, Zheng F, Guo F, Chang A, Li X, Wang B. miR-223 regulates adipogenic and osteogenic differentiation of mesenchymal stem cells through a C/EBPs/miR-223/FGFR2 regulatory feedback loop. Stem Cells. 2015;33:1589–1600. doi: 10.1002/stem.1947. [DOI] [PubMed] [Google Scholar]
- Gurol T, Zhou W, Deng Q. MicroRNAs in neutrophils: potential next generation therapeutics for inflammatory ailments. Immunol. Rev. 2016;273:29–47. doi: 10.1111/imr.12450. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CJ, Boyle RH, Sun X, Wicker SM, Misa JP, Krissansen GW, Print CG, Crosier KE, Crosier PS. Epidermal cells help coordinate leukocyte migration during inflammation through fatty acid-fuelled matrix metalloproteinase production. Nat. Commun. 2014;5:3880. doi: 10.1038/ncomms4880. [DOI] [PubMed] [Google Scholar]
- Haneklaus M, Gerlic M, O’Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J. Intern. Med. 2013;274:215–226. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harraz MM, Eacker SM, Wang X, Dawson TM, Dawson VL. MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc. Natl. Acad. Sci. USA. 2012;109:18962–18967. doi: 10.1073/pnas.1121288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat. Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, Shah P, Wisler J, Eubank TD, Tridandapani S, et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121:984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- Kanther M, Sun X, Mühlbauer M, Mackey LC, Flynn EJ, 3rd, Bagnat M, Jobin C, Rawls JF. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. 2011;141:197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper DM, Moro A, Ristori E, Narayanan A, Hill-Teran G, Fleming E, Moreno-Mateos M, Vejnar CE, Zhang J, Lee D, et al. MicroRNAs establish uniform traits during the architecture of vertebrate embryos. Dev. Cell. 2017;40:552–565. doi: 10.1016/j.devcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ND, Luster AD. The role of tissue resident cells in neutrophil recruitment. Trends Immunol. 2015;36:547–555. doi: 10.1016/j.it.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lämmermann T, Afonso PV, Angermann BR, Wang JM, Kastenmüller W, Parent CA, Germain RN. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 2013;498:371–375. doi: 10.1038/nature12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBert DC, Squirrell JM, Rindy J, Broadbridge E, Lui Y, Zakrzewska A, Eliceiri KW, Meijer AH, Huttenlocher A. Matrix metalloproteinase 9 modulates collagen matrices and wound repair. Development. 2015;142:2136–2146. doi: 10.1242/dev.121160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat. Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li Z, Guo F, Qin X, Liu B, Lei Z, Song Z, Sun L, Zhang HT, You J, Zhou Q. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J. Biomed. Sci. 2011a;18:24. doi: 10.1186/1423-0127-18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol. Cancer Res. 2011b;9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- Li Y, Li Y, Cao X, Jin X, Jin T. Pattern recognition receptors in zebrafish provide functional and evolutionary insight into innate immune signaling pathways. Cell. Mol. Immunol. 2017;14:80–89. doi: 10.1038/cmi.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Lisse TS, Middleton LJ, Pellegrini AD, Martin PB, Spaulding EL, Lopes O, Brochu EA, Carter EV, Waldron A, Rieger S. Paclitaxel-induced epithelial damage and ectopic MMP-13 expression promotes neurotoxicity in zebrafish. Proc. Natl. Acad. Sci. USA. 2016;113:E2189–E2198. doi: 10.1073/pnas.1525096113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012;22:1243–1254. doi: 10.1101/gr.132514.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology. 2009;127:291–298. doi: 10.1111/j.1365-2567.2009.03092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes T, Cobos FA, Schleich F, Sorbello V, Henket M, De Preter K, Bracke KR, Conickx G, Mesnil C, Vandesompele J, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J. Allergy Clin. Immunol. 2016;137:1433–1446. doi: 10.1016/j.jaci.2016.02.018. [DOI] [PubMed] [Google Scholar]
- Majumdar R, Tavakoli Tameh A, Parent CA. Exosomes mediate LTB4 release during neutrophil chemotaxis. PLoS Biol. 2016;14:e1002336. doi: 10.1371/journal.pbio.1002336. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CCM, Beck PL, Muruve DA, Kubes P. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- Nathan C. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- Neudecker V, Brodsky KS, Clambey ET, Schmidt EP, Packard TA, Davenport B, Standiford TJ, Weng T, Fletcher AA, Barthel L, et al. Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci. Transl. Med. 2017;9:eaah5360. doi: 10.1126/scitranslmed.aah5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana EA, Kasinski AL. MicroRNAs in cancer: a historical perspective on the path from discovery to therapy. Cancers (Basel) 2015;7:1388–1405. doi: 10.3390/cancers7030842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel CA, Rehberg M, Lerchenberger M, Berberich N, Bihari P, Khandoga AG, Zahler S, Krombach F. Ccl2 and Ccl3 mediate neutrophil recruitment via induction of protein synthesis and generation of lipid mediators. Arterioscler. Thromb. Vasc. Biol. 2009;29:1787–1793. doi: 10.1161/ATVBAHA.109.193268. [DOI] [PubMed] [Google Scholar]
- Renshaw SA, Trede NS. A model 450 million years in the making: zebrafish and vertebrate immunity. Dis. Model. Mech. 2012;5:38–47. doi: 10.1242/dmm.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev. Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Roberto VP, Tiago DM, Gautvik K, Cancela ML. Evidence for the conservation of miR-223 in zebrafish (Danio rerio): Implications for function. Gene. 2015;566:54–62. doi: 10.1016/j.gene.2015.04.022. [DOI] [PubMed] [Google Scholar]
- Rosowski EE, Deng Q, Keller NP, Huttenlocher A. Rac2 Functions in Both Neutrophils and Macrophages To Mediate Motility and Host Defense in Larval Zebrafish. J. Immunol. 2016;197:4780–4790. doi: 10.4049/jimmunol.1600928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre MP, Alcaraz-Pérez F, López-Muñoz A, Roca FJ, Meseguer J, Cayuela ML, Mulero V. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J. Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113:5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J. Cell. Biochem. 2007;101:996–999. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G, Catherinet C, Prado-Lourenco L, Levin MG, Thacker S, et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wang Y, Chen Q, Qiu N, Zhao Y, You X. MiR-223 inhibited cell metastasis of human cervical cancer by modulating epithelial-mesenchymal transition. Int. J. Clin. Exp. Pathol. 2015;8:11224–11229. [PMC free article] [PubMed] [Google Scholar]
- Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into Cullin-Ring ubiquitin ligases. Antioxid. Redox. Signal. 2010;13:1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Ying W, Tseng A, Chang RC, Morin A, Brehm T, Triff K, Nair V, Zhuang G, Song H, Kanameni S, et al. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J. Clin. Invest. 2015;125:4149–4159. doi: 10.1172/JCI81656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125:2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.