Supplemental digital content is available in the text.
Key Words: Ovarian cancer, Surveillance, Biomarkers, Second look, Next-generation sequencing, Genetics
Abstract
Objectives
The objectives of this study were to assess if targeted investigation for tumor-specific mutations by ultradeep DNA sequencing of peritoneal washes of ovarian cancer patients after primary surgical debulking and chemotherapy, and clinically diagnosed as disease free, provides a more sensitive and specific method to assess actual treatment response and tailor future therapy and to compare this “molecular second look” with conventional cytology and histopathology-based findings.
Methods/Materials
We identified 10 patients with advanced-stage, high-grade serous ovarian cancer who had undergone second-look laparoscopy and for whom DNA could be isolated from biobanked paired blood, primary and recurrent tumor, and second-look peritoneal washes. A targeted 56 gene cancer-relevant panel was used for next-generation sequencing (average coverage, >6500×). Mutations were validated using either digital droplet polymerase chain reaction (ddPCR) or Sanger sequencing.
Results
A total of 25 tumor-specific mutations were identified (median, 2/patient; range, 1–8). TP53 mutations were identified in at least 1 sample from all patients. All 5 pathology-based second-look positive patients were confirmed positive by molecular second look. Genetic analysis revealed that 3 of the 5 pathology-based negative second looks were actually positive. In the 2 patients, the second-look mutations were present in either the original primary or recurrent tumors. In the third, 2 high-frequency, novel frameshift mutations in MSH6 and HNF1A were identified.
Conclusions
The molecular second look detects tumor-specific evidence of residual disease and provides genetic insight into tumor evolution and future recurrences beyond standard pathology. In the precision medicine era, detecting and genetically characterizing residual disease after standard treatment will be invaluable for improving patient outcomes.
Ovarian cancer (OvCa) is the leading cause of gynecologic cancer death in the United States with an estimated 22,280 new cases and 14,240 deaths in 2017.1 Most women with epithelial OvCa present with advanced-stage disease and undergo primary cytoreductive surgery followed by combination platinum-based adjuvant chemotherapy. Despite the initial clinical and diagnostic impression of response to primary treatment, the majority will recur and ultimately die of their disease within 5 years.2 Without methods to accurately and rapidly detect residual disease, an early critical window to modify treatment is therefore lost.
Currently, a combination of serial physical examinations, serum cancer antigen 125 (CA-125) levels, and imaging modalities including computed tomography (CT) and positron emission tomography are used for posttreatment surveillance and each has sensitivity and specificity limitations.3 For example, studies have shown that up to 60% of patients with normal CA-125 levels after primary treatment nonetheless have pathologic evidence of disease.4 Similarly, CT and positron emission tomography imaging lack sensitivity for detection of subcentimeter disease and CT may be inconclusive for diagnosis of disease on many sites of interest.3,5
Seventy years ago, Wangensteen et al6,7 first suggested and then demonstrated the concept of second-look surgery (SLS) for detecting and treating early recurrent colorectal cancer in asymptomatic, high-risk patients. The procedure was soon adopted for OvCa patients and evolved through its original goal of therapeutic effect to determination of optimal treatment duration to diagnostic utility and therapeutic value (reviewed in Ref.8). Second-look surgery in the setting of OvCa consists of a systematic surgical exploration of the abdomen and pelvis as well as obtaining peritoneal washes and directed biopsies for cytologic and histopathologic examination in asymptomatic patients who have completed a planned course of chemotherapy. The limited sensitivity and specificity of testing strategies for determining disease status after surgery and chemotherapy were evident in that despite an apparent complete clinical response, about half of patients had persistent, usually microscopic disease at the time of SLS.8–10 Against the background of the potential advantages of SLS, practically, and calling into question the sensitivity and false-negative rate of the cytology and histopathology-based SLS procedure, up to 50% of patients with a negative second look ultimately had disease recurrence.11–13 Although some evidence suggested prognostic value for specific subsets of patients with positive and negative second looks,10,13–15 a clear survival advantage has never been established.9,11,12,15–17 Thus, although the philosophy underpinning SLS retains merit, the current procedure remains controversial and without conclusive evidence-based data to support its routine use in OvCa.
Today, the exquisite sensitivity and specificity afforded by a number of DNA-based technologies coupled with the promise of precision medicine to move beyond a “one-size-fits-all” treatment approach suggest that the methodologic underpinnings of SLS should be reevaluated. Moreover, recent advances in our understanding of tumor heterogeneity and its direct relationship to treatment failure18–20 and the mutational landscape of OvCa21 coupled with increasing availability of sequencing technologies, leading to precision-based biomarkers in gynecologic cancers for disease diagnostics and surveillance, including the use of circulating tumor DNA22,23 and the molecular analysis of peritoneal washings,24,25 not only provide the impetus for these studies but also suggest a framework for their implementation.
We hypothesized that molecular analysis of second-look washings using ultradeep targeted sequencing of cancer-relevant genes even in asymptomatic patients could detect tumor-specific mutations and afford a more comprehensive, sensitive, and specific picture of persistent disease than currently available. Given the above, we sought to provide the proof of concept for a more personalized approach to disease surveillance, a “molecular second look.”
METHODS
Patients and Samples
We selected paired blood, second-look peritoneal washings, and surgical tumor samples from 10 patients with high-grade serous ovarian, fallopian tube, or primary peritoneal cancer (HGSOC) previously enrolled in our biobanking study. Patients received initial treatment from January 2010 to December 2014. Written informed consent had been obtained for each patient in accordance with the institutional review board at the Icahn School of Medicine at Mount Sinai. Importantly, all laparoscopic second-look patients were asymptomatic and clinically defined as having had a complete clinical response after primary surgical cytoreduction and platinum-based combination chemotherapy. Complete clinical response was defined as a normal physical examination, no evidence of disease on CT imaging, and a normal serum CA-125 level (<35 U/mL). For inclusion in this study, genomic DNA from blood, the cellular fraction of second-look peritoneal washings, and primary and/or recurrent tumor had to be available.
DNA Isolation and Extraction
Genomic tumor DNA was isolated and extracted from tissue using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Germline DNA was isolated from blood samples (ArchivePure DNA Kit, 5 Prime, Gaithersburg, Md). All frozen and stored peritoneal wash samples were thawed and centrifuged at 2000g for 30 minutes. The acellular supernatant was separated, and the remaining cell pellet was recentrifuged at 9000g for 10 minutes at 4°C. DNA was extracted from the centrifuged cell pellets (ArchivePure DNA Kit, 5 Prime, Gaithersburg, Md) with a modified protocol as previously described.26 DNA concentrations from all samples were determined by QuBit fluorometry (Thermo-Fischer Scientific, Waltham, Mass).
Identification of Somatic Mutations Using Next-Generation Sequencing
For each patient, all DNA samples were sequenced using a targeted amplicon panel. DNA sample quantity and integrity were first assessed using an ALU repeat quantitative PCR assay (Swift Biosciences, Ann Arbor, Mich), and 10 ng quantitative PCR quantified DNA was used as input into the Accel-Amplicon Panel.26 The sequencing panel used consisted of 56 cancer-related genes with hotspot coverage using a total of 263 amplicons (average size, 138 bp) to cover hotspots across this gene set along with coverage of the complete TP53 coding region (Swift Biosciences, Ann Arbor, Mich). Next-generation sequencing (NGS) was performed using an Illumina MiSeq with v2 chemistry. Average coverage depth was greater than 6500 times. Somatic variant calling was performed using GATK Best Practices and LoFreq to identify variants with a predetermined allele frequency threshold of 1.0%. The average performance metrics for each sample was 96% on target and 99% coverage uniformity as defined by 20% of the mean. Variants were manually curated and removed if there was evidence of strand bias, clustering, or sequencing error. Raw read alignments were inspected using Integrative Genomics Viewer.27 Using tumor-specific mutation profiles, which we defined for each patient, and when adequate sample was available, we interrogated the paired second-look peritoneal fluid for evidence of these mutations using digital droplet PCR (ddPCR).
Validation of NGS-Identified Mutations
Next-generation sequencing findings were validated using 1 of 2 orthogonal technologies. Digital droplet PCR was used for validation of NGS-identified somatic mutations with allele frequencies less than 10%, and Sanger sequencing, with allele frequencies greater than or equal to 10%. Custom TaqMan probe assays were designed using Life Technologies' web-based tool (www.lifetechnologies.com/order/custom-genomic-products) for ddPCR. Assays were created with probes labeled with VIC or FAM for wild-type and mutant variants, respectively. Quantitative PCR was used to first validate specificity. Sensitivity and lower limits of detection were established using ddPCR (RainDance Technologies, Billerica, Mass), as previously described.22 As per our protocol, ddPCR probes were also generated based on a patient's cumulative mutation fingerprint and used to screen for the presence of mutations in a sample if not originally identified by panel-based sequencing.
RESULTS
Patient Demographics and Conventional Pathology-Based Second-Look Findings
Ten HGSOC patients with complete clinical responses, as defined by clinical examination, serum CA-125 and radiology, after debulking surgery and planned chemotherapy and who had undergone SLS were selected for study inclusion (Table S1, http://links.lww.com/IGC/A598). The cohort median age was 63 years (range, 50–68 years). All patients had advanced stage disease. At the time of primary surgery, all patients were either optimally cytoreduced or had no gross residual disease. Based on their original cytology or histopathology at the time of SLS, 5 (50%) of the 10 patients had been diagnosed as second-look positive and 5 patients (50%) were second-look negative.
Tumor-Specific Mutations Are Detectable in Second-Look Washings and Establish a Genetic Link Between Primary and Recurrent Tumors
Because this was a retrospective study and dependent on sample availability (Table S2, http://links.lww.com/IGC/A599), we were able to simultaneously sequence genomic DNA isolated from blood, primary tumor, second-look peritoneal washings, and recurrent tumor. Genomic DNA from blood was used as germline control. In total, and using a 56-gene panel, we identified 25 mutations across the entire sample set (Table 1). All patients had at least 1 mutation detected from their samples (median, 2; range, 1–8). Nine of 10 patients had tumor samples that were informative for mutations. In these 9 patients, a total of 19 mutations were identified. Allele frequencies ranged from 1% (our predetermined lower level threshold cut-off) to 89%. In the single patient without tumor-identified mutations, PT169, only the primary tumor sample was available; no material was available from the recurrence.
TABLE 1.
Somatic mutation results from targeted NGS
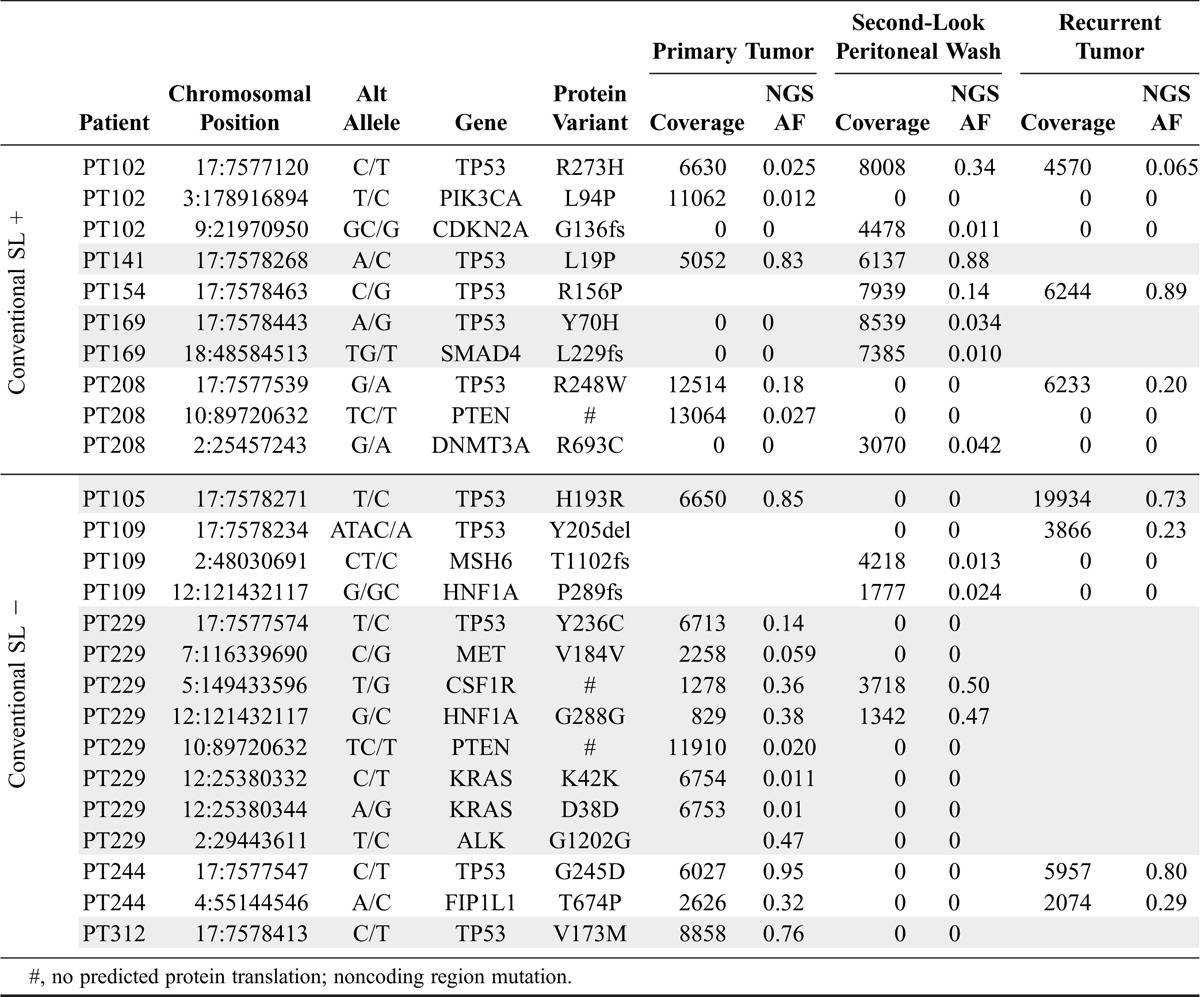
In accord with TCGA findings that all HGSOCs possess TP53 mutations,24 TP53 was the most frequently mutated gene in our cohort. All patients had a unique somatic TP53 mutation detected in at least 1 of their samples. The second most frequently mutated genes in the cohort were PTEN and HNF1A, each mutated in 2 patients. The following 10 genes had mutations in 1 patient: MSH6, PIK3CA, KRAS, CSF1R, MET, FIP1L1, CDKN2A, SMAD4, ALK, and DNMT3A (Table 1). No combination of mutated genes was noted to be overlapping between patients.
All Conventional Second-Look Positive Patients Are Also Molecular Second-Look Positive and Harbor Tumor-Specific Mutations Across Time
All 5 patients originally diagnosed as second-look positive by conventional pathology were also found to be molecular second-look positive (top panel, Fig. 1). Moreover, in 4 patients, the second-look mutations matched those identified in at least 1 of their paired primary or recurrent tumors (Fig. 1). In the 2 patients where both primary and recurrent tumor were available, PT102 and PT208, the TP53 mutations matched in all samples, thus providing a direct genetic link across these 3 distinct points of clinical interaction across time. In PT169, the 2 second-look mutations, TP53 Y70H (allele fraction [AF], 3.4%) and SMAD4 L229fs (AF, 1.0%), were not detected in the patient's primary tumor sample (Table 1). Unfortunately, we did not have recurrent tumor available for molecular analysis and thus could not rule out the possibility that these mutations are also present in the recurrence.
FIGURE 1.
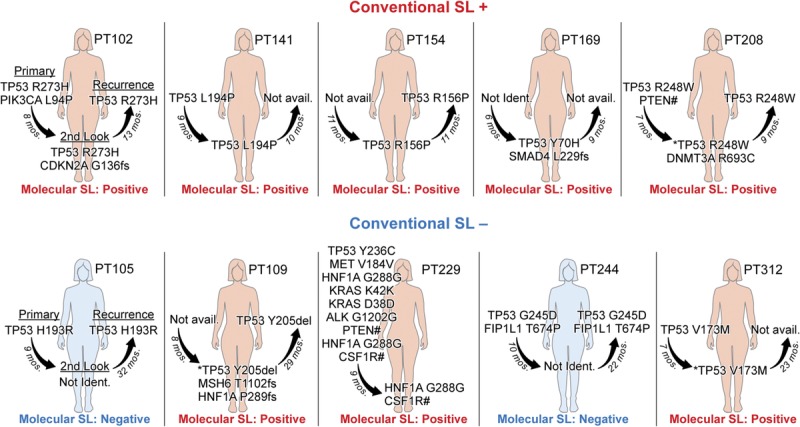
Overview of somatic DNA sequencing results organized by patient and sample. *, detected by ddPCR, but not NGS. #, no predicted protein translation; noncoding region mutation.
For PT208, the tumor-specific TP53 R248W mutation was identified by NGS in both primary and recurrent tumor samples at the same robust allele frequency of ~20%. Interestingly, the mutation was only detected in the second-look peritoneal wash by ddPCR with a much lower allele fraction (AF, 0.08%). A second mutation was identified by NGS in the second-look sample, DNMT3A R693C, which was not present in either tumor sample. Similarly, PT102 primary tumor had a PIK3CA mutation not detected in either second look or the recurrent tumor and a CKDN2A mutation in second look not detected in either paired tumor sample. Taken together, these results are consistent with the existence of tumor heterogeneity, clonal expansion, and selection.
Molecular Second Look Identified Tumor-Specific Mutations in Patients Diagnosed as Free of Residual Disease by Conventional Pathology
Three of the 5 patients who had originally been diagnosed as second-look negative by conventional methods were found to have mutations detectable in their second-look washes (Fig. 1, bottom panel; Table 1). In this sample set, second-look mutations were tumor-specific for 2 of the 3 patients, PT229 and PT312, matching those identified in at least 1 of their paired associated tumors. In PT229, NGS detected a noncoding, high allele frequency mutation in CSF1R (AF, 49%) and a synonymous HNF1A mutation (AF, 47%) in the second-look wash. These mutations were also present in the primary tumor, albeit at lower allele frequencies. Mutations in TP53 (Y236C; AF, 14%), noncoding mutations in MET (AF, 6%) and PTEN (AF, 2%), and synonymous mutations in KRAS (AF, 2% and 1%) and ALK (AF, 47%) that were detected in the primary tumor by NGS were not detected in second-look wash. The increased allele frequency of the CSF1R and HNF1A mutations with concomitant loss of TP53, MET, PTEN, KRAS, and ALK mutations in the second look could be suggestive of clonal selection.
For PT312, NGS identified a high allele frequency TP53 V173M mutation in primary tumor (AF, 76%). Recurrent tumor was not available. Originally, no mutations were detected by NGS in this patient's second-look peritoneal wash. Given the near universal presence of TP53 mutations in HGSOC, we suspected that the TP53 mutation identified in primary tumor may in fact have been present in her second-look peritoneal wash, but for some technical reason not detected by NGS. We generated and validated the sensitivity and specificity of a ddPCR probe to detect this TP53 mutation (Figure S1, http://links.lww.com/IGC/A597). Using ddPCR, the TP53 mutation was identified in the second-look wash but at an extremely low AF threshold (0.009%).
Panel-based sequencing of PT109's recurrent tumor and second-look wash identified mutations in 3 genes not shared between the samples. No primary tumor sample was available for sequencing. A TP53 mutation was present in the recurrent tumor (AF, 23%), but not in second-look wash. Two mutations in the second-look wash, MSH6 (AF, 1%) and HNF1A (AF, 2%), were not detected in recurrent tumor. Similar to PT312, we developed a ddPCR assay to probe for the TP53 mutation. As suspected, the tumor-specific TP53 mutation was confirmed to be present in the peritoneal wash (AF, 6%; Table S3, http://links.lww.com/IGC/A600).
Validation of NGS-Identified Mutations
To confirm the NGS results and exclude the possibility of sequencing artifacts, we initially selected 14 mutations for validation using 1 of 2 orthogonal technologies, either ddPCR or Sanger sequencing, as previously described.23 Each patient had at least 1 mutation represented in the validation set (Table S3, http://links.lww.com/IGC/A600). Ultimately, 12 assays passed all quality metrics and all 12 mutations tested were validated and confirmed. In addition, allele frequencies for each ddPCR-determined mutation were almost all within 10% of allele frequencies determined by NGS.
DISCUSSION
With the ultimate complementary goals of earlier detection of residual disease, defining tumor heterogeneity, optimizing OvCa treatment, and improving survival, the theoretical advantages of SLS would seem self-evident. Despite this optimism, and most possibly hobbled by sensitivity and specificity limitations of cytology and histopathology-based detection technologies, the benefit of SLS has never been conclusively established. We sought to use ultradeep targeted gene sequencing to provide a more sensitive means of detection and genetic characterization of persistent disease. Using this molecular approach, we demonstrated that this NGS-based approach was not only as sensitive as traditional techniques but also suggestive of even greater sensitivity and depth of genetic characterization and we identified tumor-specific mutations in patients wherein classic pathology techniques had failed.
Somatic mutations in peritoneal and pleural effusion fluid from OvCa patients have been previously identified using NGS.24,28,29 Our current study, using only 10 ng of DNA, provides further evidence that cytology specimens from OvCa patients are suitable for molecular analysis. Furthermore, our proposed molecular approach to second look could allow for an easier, more minimally invasive method in which percutaneous peritoneal lavage is obtained in an outpatient setting as now proposed for other cancers.30 Such an alternative NGS-based method would also bypass morbidities associated with general anesthesia and diagnostic laparoscopy and provide an even less invasive procedure.
The clinicopathologic findings in our sample set with regards to second-look results and survival are consistent with previous studies: women with positive second looks have shorter progression-free survivals (PFSs) than those with negative second looks.10,14,15 The 5 patients who had a positive conventional second look had a median PFS of 10 months (range, 9–13 months). All ultimately recurred and died from their disease with a median OS of 28 months (range, 15–44 months). By comparison, the 5 patients with negative conventional second looks had a median PFS of 26 months (range, 22–32 months). Only PT109 died of disease, after 69 months. PT229 has no evidence of disease recurrence after 44 months. PT312 is alive with disease. The remaining 2 patients, both with negative conventional and molecular second looks, are both currently alive with disease (Table 2). Given the variable treatment strategies between the patients, no conclusions regarding OS differences should be drawn at this time.
TABLE 2.
Clinical correlations with molecular analysis
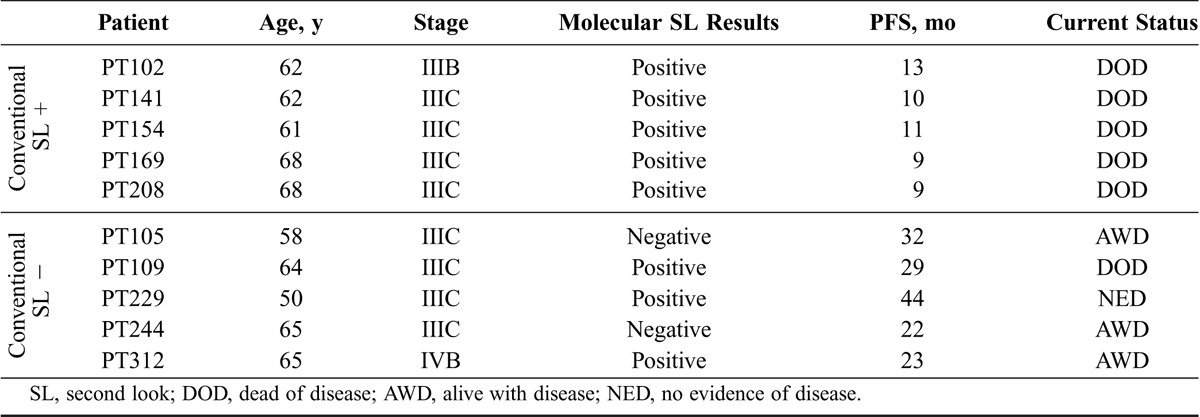
Nonetheless, 2 molecular positive/cytopathology negative patients invite particular attention. First, it initially seems counterintuitive that PT229 would be no evidence of disease. Notably, despite 2 tumor-specific second-look mutations, the known oncogenic hotspot TP53 Y236C mutation31,32 present in her primary tumor was undetectable. Intriguingly, the 2 detected mutations were noncoding (CSF1R) and synonymous (HNF1A, G288G; Table 1; Table S3, http://links.lww.com/IGC/A600). Thus, 1 hypothesis for her exceptional survival response is that her primary treatment successfully eradicated TP53-containing tumor clones while leaving residual, less aggressive, or indolent tumor clones.
In PT109, molecular second look identified a relatively low frequency (AF, 1.3%) truncating mutation in the MSH6 gene (T1102fs), which normally encodes for a protein involved in DNA damage repair. Given that heterogeneous loss of MSH6 has been associated with microsatellite instability (MSI),33 the molecular second look may have had therapeutic implications. Pembrolizumab is the first tissue-agnostic drug recently approved for treating solid tumors that are MSI-high or mismatch repair-deficient.34 Although the clinical value of this drug for this patient, who eventually died of her disease, is unknown, it could be argued that knowledge of this mutation would today almost certainly result in testing for MSI status and possible use of a targeted therapeutic.
Tumor heterogeneity likely plays an important role in both OvCa tumor spread and treatment failure.19,20 As demonstrated by PT229, molecular second look offers a chance to identify clones being selected for or against during treatment. PT169 offers additional potential insight into tumor heterogeneity. Although 2 somatic mutations were identified by NGS in the patient's second-look peritoneal wash, TP53 Y70H and SMAD4 L229fs, neither was identified in the primary tumor specimen. Given the nearly universal presence of TP53 mutations in HGSOC,21 it is of particular interest that no TP53 mutations were identified in the primary tumor but only identified at the time of the second look. To account for the lack of a TP53 mutation in the primary tumor, 3 likely scenarios are possible: (1) the TP53 mutation was originally present in the bulk tumor but below our NGS threshold level of detection (1.0% cut-off); (2) mutation-containing clones were not represented in our tissue sampling; or (3) the mutation represented a de novo mutation.
Future studies are planned and will address some of the limitations of our proof-of-principle study. For example, this would include having primary and recurrent tumor samples from all patients with recurrent disease as part of a prospective, multi-institution study, interrogation of a broader gene panel for identification of additional mutations, the addition of a more sensitive initial sequencing technology, and more standardized treatment regimens for correlating molecular findings to patient outcomes.
Currently, postoperative residual tumor burden is the strongest predictor of survival in HGSOC.35 The molecular second look, which theoretically provides for resolution on the single cell level, may provide new diagnostic and predictive insights and therapeutic guidance in a disease, which clinical experience has repeatedly demonstrated is most responsive when disease volume is lowest. The future question to be addressed is the degree of clinical relevance provided by the added genetic information derived from the molecular second look. Theoretically, understanding the molecular genetic makeup of residual OvCa after standard primary treatment will be invaluable to our understanding of chemoresistance, tumor evolution, and prediction of response to chemotherapy. Such information should aid in the development of more effective posttreatment surveillance strategies, which are critical to early detection of residual cancer or recurrence, and could improve the treatment of OvCa. As we move further into an era of personalized medicine and the increasing availability of targeted therapies, the additional information provided by a molecular second look should become another powerful tool in treating patients and improving their outcomes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the patients and their families for participation in the study. J.A.M. and P.D. thank the Gordon family, the Ruttenberg families, and the Varadi Ovarian Initiative in Cancer Education for their financial support, which funded, in part, these studies.
Footnotes
The authors declare no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citation appears in the printed text and is provided in the HTML and PDF versions of this article on the journal’s Web site (www.ijgc.net).
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.SEER Cancer Statistics Factsheets. Ovary Cancer. National Cancer Institute. Bethesda, MD. Available at: http://seer.cancer.gov/statfacts/html/ovary.html.
- 3.Gu P, Pan LL, Wu SQ, et al. CA 125, PET alone, PET-CT, CT and MRI in diagnosing recurrent ovarian carcinoma: a systematic review and meta-analysis. Eur J Radiol. 2009;71:164–174. [DOI] [PubMed] [Google Scholar]
- 4.Niloff JM, Bast RC, Jr, Schaetzl EM, et al. Predictive value of CA 125 antigen levels in second-look procedures for ovarian cancer. Am J Obstet Gynecol. 1985;15:981–986. [DOI] [PubMed] [Google Scholar]
- 5.Bell DJ, Pannu HK. Radiological assessment of gynecologic malignancies. Obstet Gynecol Clin North Am. 2011;38:45–68 vii. [DOI] [PubMed] [Google Scholar]
- 6.State D, Moore G, Wangensteen OH. Carcinoma of the stomach; a ten year survey (1936 to 1945 inclusive) of early and late results of surgical treatment at the University of Minnesota hospitals. J Am Med Assoc. 1947;135:262–267. [DOI] [PubMed] [Google Scholar]
- 7.Wangensteen OH, Lewis FJ, Tongen LA. The “second-look” in cancer surgery; a patient with colic cancer and involved lymph nodes negative on the “sixth-look”. J Lancet. 1951;71:303–307. [PubMed] [Google Scholar]
- 8.Cass I, Karlan BY. Second-Look Surgery. Surgery for Ovarian Cancer: Principles and Practice. CRC Press; 2005:289–304. [Google Scholar]
- 9.Luesley D, Lawton F, Blackledge G, et al. Failure of second-look laparotomy to influence survival in epithelial ovarian cancer. Lancet. 1988;2:599–603. [DOI] [PubMed] [Google Scholar]
- 10.Chambers SK, Chambers JT, Kohorn EI, et al. Evaluation of the role of second-look surgery in ovarian cancer. Obstet Gynecol. 1988;72:404–408. [PubMed] [Google Scholar]
- 11.Podratz KC, Cliby WA. Second-look surgery in the management of epithelial ovarian carcinoma. Gynecol Oncol. 1994;55:S128–S133. [DOI] [PubMed] [Google Scholar]
- 12.Rubin SC, Randall TC, Armstrong KA, et al. Ten-year follow-up of ovarian cancer patients after second-look laparotomy with negative findings. Obstet Gynecol. 1999;93:21–24. [DOI] [PubMed] [Google Scholar]
- 13.Rahaman J, Dottino P, Jennings TS, et al. The second-look operation improves survival in suboptimally debulked stage III ovarian cancer patients. Int J Gynecol Cancer. 2005;15:19–25. [DOI] [PubMed] [Google Scholar]
- 14.Lippman SM, Alberts DS, Slymen DJ, et al. Second-look laparotomy in epithelial ovarian carcinoma: prognostic factors associated with survival duration. Cancer. 1988;61:2571–2577. [DOI] [PubMed] [Google Scholar]
- 15.Greer BE, Bundy BN, Ozols RF, et al. Implications of second-look laparotomy in the context of optimally resected stage III ovarian cancer: a non-randomized comparison using an explanatory analysis: a Gynecologic Oncology Group study. Gynecol Oncol. 2005;99:71–79. [DOI] [PubMed] [Google Scholar]
- 16.Tuxen MK, Strauss G, Lund B, et al. The role of second-look laparotomy in the long-term survival in ovarian cancer. Ann Oncol 1997;8:643–648. [DOI] [PubMed] [Google Scholar]
- 17.Dowdy SC, Constantinou CL, Hartmann LC, et al. Long-term follow-up of women with ovarian cancer after positive second-look laparotomy. Gynecol Oncol. 2003;91:563–568. [DOI] [PubMed] [Google Scholar]
- 18.Patch AM, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. [DOI] [PubMed] [Google Scholar]
- 19.McPherson A, Roth A, Laks E, et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat Genet. 2016;48:758–767. [DOI] [PubMed] [Google Scholar]
- 20.Paracchini L, Mannarino L, Craparotta I, et al. Regional and temporal heterogeneity of epithelial ovarian cancer tumor biopsies: implications for therapeutic strategies. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira E, Camacho-Vanegas O, Anand S, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. 2015;10:e0145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkinson CA, Gale D, Piskorz AM, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016;13:e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krimmel JD, Schmitt MW, Harrell MI, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci U S A. 2016;113:6005–6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi YJ, Rhee JK, Hur SY, et al. Intraindividual genomic heterogeneity of high-grade serous carcinoma of the ovary and clinical utility of ascitic cancer cells for mutation profiling. J Pathol. 2017;241:57–66. [DOI] [PubMed] [Google Scholar]
- 26.Nair N, Camacho-Vanegas O, Rykunov D, et al. Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: a prospective cross-sectional study. PLoS Med. 2016;13:e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swisher EM, Wollan M, Mahtani SM, et al. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol. 2005;193:662–667. [DOI] [PubMed] [Google Scholar]
- 29.Shah RH, Scott SN, Brannon AR, et al. Comprehensive mutation profiling by next-generation sequencing of effusion fluids from patients with high-grade serous ovarian carcinoma. Cancer Cytopathol. 2015;123:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pak LM, Coit DG, Eaton AA, et al. Percutaneous peritoneal lavage for the rapid staging of gastric and pancreatic cancer. Ann Surg Oncol. 2017;24:1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang MT, Asthana S, Gao SP, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J, Chang MT, Johnsen HC, et al. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med. 2017;9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graham RP, Kerr SE, Butz ML, et al. Heterogenous MSH6 loss is a result of microsatellite instability within MSH6 and occurs in sporadic and hereditary colorectal and endometrial carcinomas. Am J Surg Pathol. 2015;39:1370–1376. [DOI] [PubMed] [Google Scholar]
- 34.First tissue-agnostic drug approval issued. Cancer Discov. 2017;7:656. [DOI] [PubMed] [Google Scholar]
- 35.Nick AM, Coleman RL, Ramirez PT, et al. A framework for a personalized surgical approach to ovarian cancer. Nat Rev Clin Oncol. 2015;12:239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


