Abstract
Growth plate chondrocytes undergo sequential differentiation to form the resting zone, the proliferative zone (PZ), and the hypertrophic zone (HZ). The important role of microRNAs (miRNAs) in the growth plate was previously revealed by cartilage-specific ablation of Dicer, an enzyme essential for biogenesis of many miRNAs. To identify specific miRNAs that regulate differentiation of PZ chondrocytes to HZ chondrocytes, we microdissected individual growth plate zones from juvenile rats and performed miRNA profiling using a solution hybridization method and miRNA sequencing. Thirty-four miRNAs were differentially expressed between the PZ and the HZ, and we hypothesized that some of the miRNAs that are preferentially expressed in the PZ may promote proliferation and inhibit hypertrophic differentiation. Consistent with this hypothesis, transfection of inhibitors for four of these miRNAs (mir-369-3p, mir-374-5p, mir-379-5p, and mir-503-5p) decreased proliferation in primary epiphyseal chondrocytes. The inhibitors for three of these miRNAs (mir-374-5p, mir-379-5p, and mir-503-5p) also increased expression of multiple genes that are associated with chondrocyte hypertrophic differentiation. We next hypothesized that preferential expression of these miRNAs in the PZ is driven by the parathyroid hormone–related protein (PTHrP) concentration gradient across the growth plate. Consistent with this hypothesis, treatment of primary chondrocytes with a parathyroid hormone (PTH)/PTHrP receptor agonist, PTH1-34, increased expression of mir-374-5p, mir-379-5p, and mir-503-5p. Taken together, our findings suggest that the PTHrP concentration gradient across the growth plate induces differential expression of mir-374-5p, mir-379-5p, and mir-503-5p between the PZ and the HZ. In the PZ, the higher expression levels of these miRNAs promote proliferation and inhibit hypertrophic differentiation. In the HZ, downregulation of these miRNAs inhibits proliferation and promotes hypertrophic differentiation.
Specific miRNAs that are differentially expressed in proliferative vs hypertrophic zones of the growth plate regulate chondrocyte proliferation and hypertrophic differentiation in vitro.
The growth plate is a thin layer of cartilage that is responsible for longitudinal bone growth and shows a high degree of spatial regulation (1–4). The growth plate is divided into three histologically distinct zones: the resting zone (RZ), the proliferative zone (PZ), and the hypertrophic zone (HZ). The RZ, which lies closest to the end of the bone, contains progenitor chondrocytes, which can differentiate into PZ chondrocytes (3, 4). The proliferative chondrocytes undergo rapid proliferation to form columnar cell clones. The PZ cells farthest from the RZ then cease proliferation and undergo hypertrophic differentiation, which includes physical enlargement of the cell and expression of collagen X, to form the HZ (1, 2).
The molecular mechanisms regulating hypertrophic differentiation are complex. One of the principal molecular switches involves parathyroid hormone–related protein (PTHrP) (4). In the postnatal growth plate, PTHrP is produced in the resting zone (4, 5). PTHrP is thought to diffuse into the proliferative zone, where it inhibits hypertrophic differentiation (4, 5). The PZ chondrocytes farthest from the RZ are exposed to lower concentrations of PTHrP, initiating hypertrophic differentiation (5). As the PZ chondrocytes differentiate into prehypertrophic chondrocytes, they begin to secrete Indian hedgehog (Ihh), which positively regulates PTHrP, thus forming a negative feedback loop (4). In addition to PTHrP, other extracellular signaling proteins, including Bmp2 and Bmp6, appear to contribute to hypertrophic differentiation, involving complex interactions with the PTHrP/Ihh system (2, 6).
Growth plate chondrocyte differentiation is also regulated by microRNAs (miRNAs) (7–9). miRNAs are short noncoding RNAs that bind to target sequences in the 3′ untranslated region of multiple messenger RNAs to inhibit translation or promote messenger RNA degradation (7–9). miRNAs function as a general and potent mechanism to regulate gene expression in many tissues (7–9). The important role of miRNAs in the growth plate was revealed by cartilage-specific ablation of Dicer, an enzyme essential for biogenesis of many miRNAs (9). The mice showed lethal skeletal growth failure due to critical defects in the spatial regulation of growth plate chondrocytes, including decreased chondrocyte proliferation and accelerated differentiation into postmitotic hypertrophic chondrocytes (9). In subsequent studies, mir-140 and let-7 were found to promote resting to proliferative differentiation in the RZ and to promote proliferation in the PZ, respectively (9).
We hypothesized that specific miRNAs are differentially expressed in the PZ vs the HZ and contribute to the spatial regulation of proliferation and hypertrophic differentiation. We also hypothesized that these miRNAs would be regulated by PTHrP and thus help mediate the effect of PTHrP on hypertrophic differentiation. To test our hypothesis, we microdissected the three principal zones from juvenile rat growth plates and assessed miRNA expression profiles. We then focused on specific miRNAs that were preferentially expressed in the PZ compared with the HZ. In cultured chondrocytes, we found that inhibitors of these miRNAs decreased chondrocyte proliferation and stimulated expression of multiple genes involved in hypertrophic differentiation. Finally, we found that treatment of growth plate chondrocytes with parathyroid hormone (PTH)1-34, which acts as a PTH/PTHrP receptor agonist, induced expression of these miRNAs. Thus, the findings suggest that specific miRNAs help mediate the effects of PTHrP on chondrocyte proliferation and differentiation in the growth plate.
Materials and Methods
Animal procedures and tissue processing
Sprague-Dawley rats (Harlan, Indianapolis, IN) were maintained and used in accordance with the Guide for the Care and Use of Laboratory Animals (10). All animals received standard rodent chow (Zeigler Bros, Gardners, PA) and water ad libitum. Tibial epiphyses of 4-day-old male rats were rapidly excised, trimmed of cortical bone, embedded in Tissue-Tek O.C.T. Compound (Electron Microscopy Sciences, Hatfield, PA), and stored at –80°C until processing. The protocol was approved by the Animal Care and Use Committee, National Institute of Child Health and Human Development, National Institutes of Health.
Microdissection and isolation of RNA
Frozen sections (60 μm) were obtained and stained as previously described. Under a dissecting microscope, each zone was identified by histological features, microdissected with a razor, and placed in lysis buffer for RNA extraction (11, 12). The transitional area between the RZ and the PZ and the prehypertrophic zone were not included. RNA was extracted using a mirVana kit (Thermo Fisher Scientific, Waltham, MA) per the manufacturer’s instructions. Total RNA (50 ng) was used for NanoString, and 1 μg of total RNA was used for miRNA sequencing (miRNA-seq).
Multiplex solution hybridization miRNA assay
Three growth plate zones were microdissected from each of four animals, yielding four samples for each zone. RNA concentration was estimated by spectrophotometry (Spectrophotometer 2000; Nanodrop), and the quality was evaluated by bioanalyzer (Agilent 2100 Bioanalyzer, Agilent). Samples with an RNA integrity number >7 were used for assay. Then, miRNAs were quantified in each of the 12 total RNA samples (50 ng) by multiplex solution hybridization (nCounter Analysis, Nanostring Technologies, Seattle, WA) according to the manufacturer’s instructions. The Nanostring method was designed to measure 420 miRNAs. The mean of the negative controls was used as a threshold value for transcript detection and was subtracted from all raw values. The data in each sample were normalized to the geometric mean of the 420 miRNAs in that sample and log-transformed. Partek Genomics Suite 6.6 (Partek Inc., St. Louis, MO) was used to compare the expression levels between zones for each miRNA. The false discovery rate (FDR) was set at 0.05.
miRNA-seq
Three growth plate zones were microdissected from each of six animals. Each sample for miRNA-seq was derived by pooling RNA from two animals, yielding three samples for each zone. Libraries were constructed from the RNA samples using the SOLiD Total miRNA-Seq kit (Thermo Fisher Scientific) following the manufacturer’s instructions for small RNA library preparation. Briefly, adapters were ligated onto the RNA, which was then reverse transcribed. The complementary DNA was size selected and amplified, which incorporated barcodes. The nine libraries were pooled and sequenced on two lanes using a SOLiD 5500xl sequencer (Thermo Fisher Scientific) with Exact Call Chemistry. Sequence data analysis used miRanalyzer, using the rn4 build.
miRNA profiling data analysis
miRNAs were considered to be differentially expressed between the RZ and the PZ or between the PZ and the HZ if they showed a difference greater than twofold and FDR <0.05 by the Nanostring analysis and if the expression differences were confirmed to be greater than twofold by miRNA-seq. To prioritize miRNAs for study, we took advantage of a prior study in which we had microdissected individual zones from rat growth plates and used microarray to identify genes that are differentially expressed in the PZ vs the HZ (13). We then prioritized miRNAs with predicted target sites overrepresented among these differentially expressed genes (using Partek Genomics Suite) and particularly miRNAs that were predicted (using Targetscan, miRDB, and microRNA.org) to target well-established markers of hypertrophic differentiation (Bmp2, Bmp6, Col10a1, Ihh, and Vegfa).
Primary chondrocyte culture and transfection
Epiphyseal cartilage from femurs and tibias was collected from 4-day-old male Sprague Dawley rats using a dissecting microscope as previously described, and the perichondrium was removed (11–13). The epiphyseal cartilage was washed twice in ice-cold phosphate-buffered saline (PBS) containing 1% penicillin/streptomycin and 250 ng/mL of fungizone. Epiphyseal cartilage was then incubated for 15 minutes at 37°C in 15 mL of 0.1% EDTA in PBS followed by incubation for 30 minutes at 37°C in 14 mL of 0.125% trypsin (Thermo Fisher Scientific) in PBS. The epiphyseal cartilage was resuspended and incubated in 20 mL of 0.3% collagenase (Sigma-Aldrich, St. Louis, MO) for 30 minutes at 37°C. The supernatant was collected and centrifuged at 300 g for 5 minutes, and the chondrocyte pellet was resuspended in chondrogenic medium. Collagenase digestion was repeated until all cartilage was digested. Chondrocytes were pooled and filtered through a cell strainer (70 µm). The cells were washed with 20 mL of PBS containing 1% of penicillin/streptomycin and 250 ng/mL of fungizone, and 1 million cells per well were plated in six-well plates in chondrocyte culture medium [Dulbecco’s modified Eagle medium (DMEM)/F12 Glutamax (Gibco, Gaithersburg, MD) + 10% heat-inactivated fetal bovine serum (Gibco), 50 µg/mL l-ascorbic acid] without antibiotics or fungizone. After 24 hours, at ∼50% confluency, the cells were washed twice with PBS, and the medium was exchanged for Opti-MEM (Thermo Fisher Scientific). Specific miRNA inhibitors (200 pmol) and Lipofectamin (Lipofectamine 2000 Reagent; Invitrogen, Carlsbad, CA) complex in 500 μL of Opti-Mem (Thermo Fisher Scientific) was added to each well (n = 5 per inhibitor); rno-mir-369-3p inhibitor (IH-320484-04-0005), rno-mir-374-5p inhibitor (IH-320485-03-0005), rno-mir-379-5p inhibitor (IH-320469-04-0005), and rno-mir-503-5p (IH-320490-04-0005) (Invitrogen). After 8 hours, the medium was changed to the chondrogenic medium including penicillin/streptomycin and fungizone. Primary chondrocytes without transfection and transfected with a scrambled miRNA inhibitor (Invitrogen) were used as controls (n = 5 each).
Real-time reverse transcription polymerase chain reaction for messenger RNAs
The transfected cells and controls were collected 48 hours after transfection, and the RNA was extracted using a mirVana kit following the manufacturer’s instructions. Total RNA was reverse transcribed using random primers (150 ng/mL) and the SuperScript IV Reverse Transcription Kit (Invitrogen) following the manufacturer’s instructions, and quantitative polymerase chain reaction (PCR) was performed using the following Taqman gene assay probes (Applied Biosystems, Foster City, CA): Col10a1 (Rn01408030_m1), Ihh (Rn03810376_m1), Bmp2 (Rn00567818_m1), Bmp6 (Rn00432095_m1), Mmp13 (Rn01448194_m1), Vegfa (Rn01511601_m1), Pth1r (Rn01447066_m1), Nppc (Rn00587070_m1), Npr2 (Rn00587693_m1), and Gdf10 (Rn00666937_m1). Expression was normalized to 18S ribosomal RNA. Each reaction was performed in triplicate.
Proliferation rate
Primary chondrocytes were plated in six-well plates as described previously. Forty-eight hours after transfection with a miRNA inhibitor or a scrambled miRNA inhibitor, primary chondrocytes were incubated with 1 mL of fresh culture medium containing 1 μCi of 3H-thymidine (63 Ci/mmol) (MP Biomedicals, Solon, OH) at 37°C and 5% CO2 for 8 hours and then vigorously washed three times with PBS. Radioactivity was measured by liquid scintillation counting using Bio-Safe II Complete Counting Cocktail (Research Products International, Mt. Prospect, IL).
PTH1-34 treatment and real-time PCR for miRNA
Primary chondrocytes were isolated as described previously, and 0.5 million cells/well were plated in 12-well plates in chondrogenic medium with penicillin/streptomycin and fungizone. After 6 hours, PTH1-34 (10−7 M) (R&D Systems, Minneapolis, MN) or vehicle (PBS) was added to the culture medium. After 6 hours of treatment, cells were collected, and total RNA was extracted using the mirVana kit. miRNAs were reverse transcribed and amplified with the following steps according to the manufacturer’s instructions (TaqManAdvanced miRNA Assays; Thermo Fisher Scientific): (1) Poly(A) tail reaction, (2) adaptor ligation, (3) reverse transcription, and (4) miRNA amplification reaction. Quantitative PCR was performed using a ViiA7 Real-Time PCR system (Thermo Fisher Scientific) with the following Taqmangene assay probes (Applied Biosystems): mir-374-5p (rno481140_mir), mir-379-5p (rno481147_mir), and mir-503-5p (rno481403_mir). Six biological replicates were performed.
Statistics
The effects of each miRNA inhibitor on proliferation (thymidine uptake) and on messenger RNA (mRNA) expression [real-time reverse transcription polymerase chain reaction (RT-PCR)] were assessed by comparing with a scrambled miRNA inhibitor by Student t test with Bonferroni correction for multiple comparisons. The effect of PTH1-34 treatment vs vehicle on miRNA expression (real-time RT-PCR) was assessed by Student t test.
Results
Differential expression of miRNAs between the PZ and the HZ
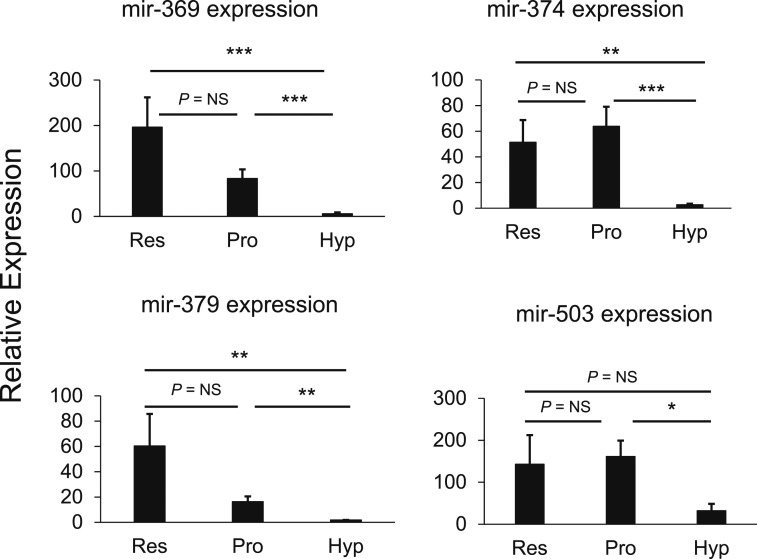
To assess differential miRNA expression, individual growth plate zones (RZ, PZ, and HZ) were microdissected from frozen sections of growth plates of 4-day-old rats. RNA was isolated from each sample, and miRNA levels were assessed by two methods: a multiplex solution hybridization method (Nanostring) and miRNA-seq. miRNA-seq detected 476 known miRNAs (Supplemental Table 1) and 61 candidates for new miRNAs (data not shown). A total of 279 miRNAs were found to be expressed by both Nanostring and miRNA-seq (Supplemental Table 1), and the miRNA levels measured by the two methods were correlated (Supplemental Fig. 1). No miRNAs were found to be differentially expressed between the RZ and the PZ by both Nanostring and miRNA-seq (Table 1). However, 34 miRNAs were differentially expressed between the PZ and the HZ by Nanostring (fold difference, >2; FDR < 0.05) and confirmed by miRNA-seq (fold difference, >2). Of these, 29 miRNAs showed greater expression in the PZ than in the HZ, and five miRNAs showed greater expression in the HZ than in the PZ (Table 1). We focused on the 29 miRNAs that were more highly expressed in the PZ and selected four miRNAs (mir-369-3p, mir-374-5p, mir-379-5p, and mir-503-5p) (Fig. 1) for further study based on the bioinformatic prioritization criteria (see Materials and Methods). All four of these miRNAs had predicted target sites strongly overrepresented among genes differentially expressed in the PZ vs the HZ and were predicted to target at least one of the following hypertrophic markers: Bmp2, Bmp6, Col10a1, Ihh, and Vegfa. Because these specific miRNAs were more highly expressed in the PZ than in the HZ, we hypothesized that in the PZ they promote proliferation and suppress hypertrophic differentiation and that the downregulation of these miRNAs in the HZ contributes to the suppression of proliferation and promotion of hypertrophic differentiation that occurs in that region of the growth plate.
Table 1.
miRNA Profiling
| miRNAa | Differential Expression |
Fold Difference |
P Valued | |
|---|---|---|---|---|
| Nanostringb | RNA-seqc | |||
| RZ vs PZ | ||||
| rno-miR-667-3p | RZ > PZ | 4.7 | 1.2 | 3.98 × 10−5 |
| PZ vs HZ | ||||
| rno-let-7b-5p | PZ > HZ | 2.6 | 2.0 | 0.01 |
| rno-let-7d-5p | PZ > HZ | 2.9 | 1.6 | 0.007 |
| rno-let-7e-5p | PZ > HZ | 3.1 | 2.4 | 0.02 |
| rno-let-7i-5p | PZ > HZ | 3.1 | 1.2 | 0.01 |
| rno-miR-100-5p | PZ > HZ | 7.7 | 2.3 | 0.007 |
| rno-miR-125a-5p | PZ > HZ | 3.0 | 1.7 | 0.008 |
| rno-miR-125b-5p | PZ > HZ | 3.7 | 1.9 | 0.02 |
| rno-miR-127-3p | PZ > HZ | 12.7 | 8.7 | 0.002 |
| rno-miR-130a-3p | PZ > HZ | 3.3 | 1.9 | 0.003 |
| rno-miR-134-5p | PZ > HZ | 20.7 | 7.8 | 0.0001 |
| rno-miR-136-5p | PZ > HZ | 7.0 | 11.6 | 0.0009 |
| rno-miR-140-5p | PZ > HZ | 4.4 | 1.5 | 0.02 |
| rno-miR-154-5p | PZ > HZ | 4.1 | 9.8 | 0.008 |
| rno-miR-15b-5p | PZ > HZ | 6.1 | 1.9 | 0.002 |
| rno-miR-188-5p | PZ > HZ | 4.6 | 1.4 | 0.007 |
| rno-miR-191a-5p | PZ > HZ | 5.0 | 1.2 | 0.01 |
| rno-miR-195-5p | PZ > HZ | 6.4 | 1.0 | 0.008 |
| rno-miR-196a-5p | PZ > HZ | 4.8 | 2.5 | 0.001 |
| rno-miR-196b-5p | PZ > HZ | 4.5 | 2.5 | 0.001 |
| rno-miR-199a-3p | PZ > HZ | 3.6 | 2.0 | 0.006 |
| rno-miR-199a-5p | PZ > HZ | 3.1 | 1.9 | 0.02 |
| rno-miR-214-3p | PZ > HZ | 3.3 | 1.6 | 0.01 |
| rno-miR-224-5p | PZ > HZ | 6.2 | 4.5 | 0.02 |
| rno-miR-23b-3p | PZ > HZ | 8.9 | 1.4 | 0.01 |
| rno-miR-25-3p | PZ > HZ | 5.0 | 1.9 | 0.003 |
| rno-miR-26b-5p | PZ > HZ | 5.0 | 1.5 | 0.02 |
| rno-miR-27b-3p | PZ > HZ | 3.4 | 1.5 | 0.008 |
| rno-miR-28-5p | PZ > HZ | 5.7 | 1.3 | 0.02 |
| rno-miR-300-3p | PZ > HZ | 10.5 | 9.7 | 0.0004 |
| rno-miR-301a-3p | PZ > HZ | 14.6 | 2.1 | 0.0007 |
| rno-miR-30b-5p | PZ > HZ | 9.0 | 1.6 | 0.001 |
| rno-miR-30d-5p | PZ > HZ | 2.5 | 1.3 | 0.01 |
| rno-miR-328a-3p | PZ > HZ | 5.1 | 0.7 | 0.02 |
| rno-miR-329-3p | PZ > HZ | 10.4 | 4.6 | 0.001 |
| rno-miR-335 | PZ > HZ | 4.1 | 2.7 | 0.01 |
| rno-miR-342-3p | PZ > HZ | 17.5 | 0.8 | 0.003 |
| rno-miR-345-5p | PZ > HZ | 4.1 | 1.5 | 0.02 |
| rno-miR-365-3p | PZ > HZ | 4.3 | 1.2 | 0.02 |
| rno-miR-369-3p | PZ > HZ | 18.3 | 4.5 | 0.0005 |
| rno-miR-374-5p | PZ > HZ | 23.9 | 2.1 | 6.60 × 10−5 |
| rno-miR-376b-3p | PZ > HZ | 15.7 | 14.2 | 0.0001 |
| rno-miR-376c-3p | PZ > HZ | 26.1 | 4.4 | 0.0001 |
| rno-miR-379-5p | PZ > HZ | 8.2 | 10.5 | 0.002 |
| rno-miR-382-5p | PZ > HZ | 9.0 | 5.9 | 0.0005 |
| rno-miR-410-3p | PZ > HZ | 14.1 | 7.9 | 0.0009 |
| rno-miR-411-5p | PZ > HZ | 4.4 | 6.4 | 0.004 |
| rno-miR-423-3p | PZ > HZ | 10.9 | 1.0 | 0.003 |
| rno-miR-425-5p | PZ > HZ | 8.0 | 1.1 | 0.02 |
| rno-miR-431 | PZ > HZ | 6.5 | 5.8 | 0.002 |
| rno-miR-433-3p | PZ > HZ | 9.0 | 4.6 | 0.005 |
| rno-miR-434-3p | PZ > HZ | 8.3 | 7.4 | 0.002 |
| rno-miR-487b-3p | PZ > HZ | 18.3 | 4.2 | 0.0003 |
| rno-miR-494-3p | PZ > HZ | 9.4 | 5.8 | 0.003 |
| rno-miR-503-5p | PZ > HZ | 8.9 | 2.1 | 0.03 |
| rno-miR-652-3p | PZ > HZ | 7.5 | 1.5 | 0.003 |
| rno-miR-872-5p | PZ > HZ | 3.9 | 1.1 | 0.02 |
| rno-miR-93-5p | PZ > HZ | 6.2 | 1.2 | 0.002 |
| rno-miR-99b-5p | PZ > HZ | 4.5 | 1.2 | 0.02 |
| rno-miR-144-3p | PZ < HZ | −5.1 | −7.9 | 0.01 |
| rno-miR-741-3p | PZ < HZ | −4.8 | ND | 0.01 |
| rno-miR-206-3p | PZ < HZ | −3.3 | −11.6 | 0.02 |
| rno-miR-223-3p | PZ < HZ | −2.2 | −10.4 | 0.01 |
| rno-miR-742-3p | PZ < HZ | −2.2 | ND | 0.01 |
| rno-miR-568 | PZ < HZ | −2.9 | ND | 0.01 |
| rno-miR-1-3p | PZ < HZ | −3.5 | −11.0 | 0.01 |
| rno-miR-124-3p | PZ < HZ | −2.2 | −3.0 | 0.01 |
| rno-miR-878 | PZ < HZ | −2.2 | ND | 0.02 |
miRNAs that showed differential expression between RZ vs PZ and PZ vs HZ were included.
Abbreviations: ND, not detected by RNA-seq; RNA-seq, RNA sequencing.
miRNAs that were confirmed with RNA-seq are in bold.
Fold changes calculated from Nanostring data.
Fold changes calculated from RNA-seq.
P values calculated from Nanostring data.
Figure 1.
mir-369-3p, mir-374-5p, mir-379-5p, and mir-503-5p show greater expression of growth plate cartilage in the PZ than in the HZ. Growth plate cartilage from 4-day-old rats was separated into specific zones by microdissection. RNA was purified, and miRNA levels were assessed by solution hybridization (Nanostring, mean ± standard error of the mean of four biological replicates each from an individual animal). *P < 0.05; **P < 0.01; ***P < 0.001. NS, not significant.
Inhibition of specific miRNAs (mir-369-3p, mir-374-5p, mir-379-5p, and mir-503-5p) decreased chondrocyte proliferation
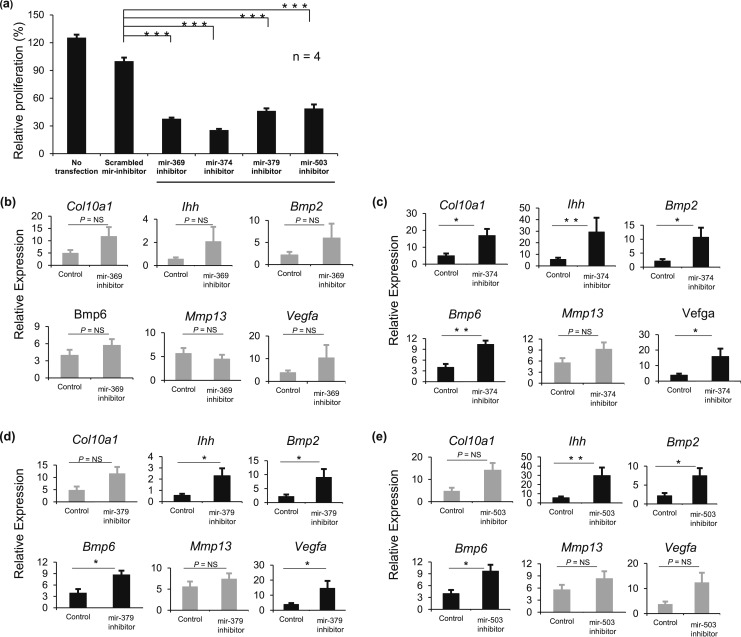
To study the effect of the selected miRNAs on proliferation, cultured rat primary growth plate chondrocytes were transfected with inhibitors for the selected four miRNAs, and proliferation was measured by 3H-thymidine incorporation. All four miRNA inhibitors (mir-369-3p, mir-374-5p, mir-379-5p, and mir-503-5p) decreased the proliferation rate of cultured chondrocytes to 38, 26, 46, and 49%, respectively, compared with a scrambled miRNA inhibitor (P < 0.001; n = 4) [Fig. 2(a)], suggesting that these miRNAs normally promote proliferation of growth plate chondrocytes, consistent with our hypothesis.
Figure 2.
Inhibitors of mir-369-3p, mir-374-5p, mir-379-5p, and mir-503-5p decrease growth plate chondrocyte proliferation and stimulate hypertrophic differentiation. Primary chondrocytes from 4-day-old rat growth plates were transfected with inhibitors for mir-369-3p, mir-374-5p, mir-379-5p, and mir-503-5p or with a scrambled miRNA inhibitor. (a) Proliferation rates (mean ± standard error of the mean) were measured by 3H-thymidine incorporation beginning 48 hours after transfection. (b–e) In separate experiments, RNA was isolated from the primary chondrocytes 48 hours after transfection with inhibitors for (b) mir-369-3p, (c) mir-374-5p, (d) mir-379-5p, or (e) mir-503-5. Real-time RT-PCR was used to measure mRNA levels (mean ± standard error of the mean) for specific hypertrophy-associated genes. The scrambled miRNA inhibitor served as the control. *P < 0.05; **P < 0.01; ***P < 0.001. NS, not significant.
Inhibition of specific miRNAs (mir-374-5p, mir-379-5p, and mir-503-5p) stimulated hypertrophic differentiation
Rat primary growth plate chondrocytes were individually transfected with miRNA inhibitors, and real-time RT-PCR was used to measure the effect on expression of multiple genes involved in hypertrophic differentiation. The mir-369-3p inhibitor did not cause statistically significant changes in gene expression [Fig. 2(b)], but inhibitors for mir-374-5p, mir-379-5p, and mir-503-5p increased the expression of genes that are associated with hypertrophic differentiation. The mir-374-5p inhibitor increased the expression of Col10a1, Ihh, Bmp2, Bmp6, and Vegfa by 5.3-, 7.5-, 7.6-, 3.5-, and 4.6-fold, respectively (P < 0.05, 0.01, 0.05, 0.01, and 0.05, respectively) [Fig. 2(c)]. The mir-379-5p inhibitor increased the expression of Ihh, Bmp2, Bmp6, and Vegfa by 6.9-, 5.4-, 3.0-, and 3.8-fold, respectively (P < 0.05, 0.05, 0.05, and 0.05, respectively) [Fig. 2(d)]. The mir-503-5p inhibitor increased the expression of Ihh, Bmp2, and Bmp6 by 8.0-, 5.6-, and 3.2-fold, respectively (P < 0.01, 0.05, and 0.05, respectively) [Fig. 2(e)]. However, none of these miRNA inhibitors affected the expression of Mmp13, which, like Vegfa, is expressed in the lower HZ (14). These miRNA inhibitors did not significantly change the expression of other genes thought to play important roles in the growth plate but not in the proliferative-to-hypertrophic transition, such as Gdf10 (highly expressed in the PZ), Pth1r (parathyroid hormone 1 receptor), Nppc (encodes C-natriuretic peptide), or Npr2 (receptor for C-natriuretic peptide) (data not shown). These findings suggest that mir-374-5p, mir-379-5p, and mir-503-5p normally suppress chondrocyte hypertrophic differentiation.
PTH1-34 increases the expression of mir-374-5p, mir-379-5p, and mir-503-5p
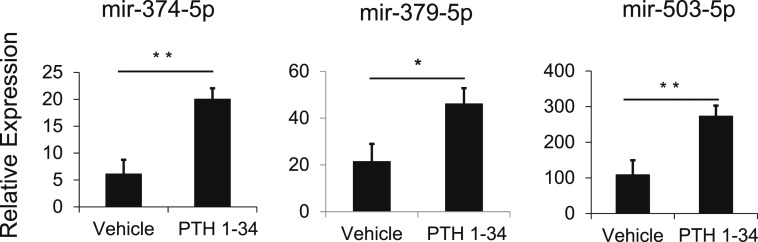
PTHrP is expressed in the RZ of the postnatal growth plate, establishing a concentration gradient of PTHrP across the growth plate that regulates hypertrophic differentiation (4, 5). We therefore hypothesized that the greater concentration of PTHrP in the PZ stimulates expression of mir-374-5p, mir-379-5p, and mir-503-5p and that the lower concentration of PTHrP in the HZ leads to the observed lower expression of these miRNAs in that zone. To test this hypothesis, we treated primary rat growth plate chondrocytes with PTH1-34, which acts as an agonist of the PTH/PTHrP receptor, and measured expression of specific miRNAs by real-time PCR. Treatment with PTHrP for 6 hours increased the expression of mir-374-5p, mir-379-5p, and mir-503-5p (Fig. 3), suggesting that these miRNAs are upregulated by PTHrP.
Figure 3.
Regulation of miRNA expression by PTH1-34. Primary chondrocytes from 4-day-old rat growth plates were treated with PTH1-34, which acts as a PTH/PTHrP receptor agonist. After 6 hours of treatment, the expression (mean ± standard error of the mean) of mir-374-5p, mir-379-5p, and mir-503-5p was measured by real-time RT-PCR. Six biological replicates were performed. *P < 0.05; **P < 0.01.
Combined treatment of transfected primary chondrocytes with miRNA inhibitors and PTH 1-34
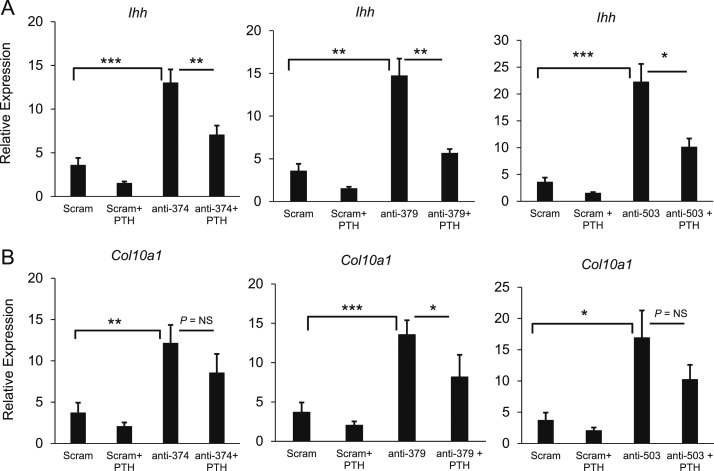
Primary chondrocytes were transfected with a scrambled miRNA inhibitor or one of three miRNA inhibitors (mir-374-5p, mir-379-5p, or mir-503-5p). After 42 hours, the cells were treated with 10−7 M PTH 1-34 or vehicle for 6 hours. In the absence of PTH, treatment with each miRNA inhibitor increased expression of Ihh and Col10, confirming the findings shown in Fig. 2 (Fig. 4). In the absence of miRNA inhibitors, treatment with PTH 1-34 decreased expression of Ihh and Col10, consistent with the known effect of PTH/PTHrP receptor agonists to inhibit hypertrophic differentiation (Fig. 4). In the combined treatment, the presence of any one miRNA inhibitor did not completely abolish the effects of PTH, suggesting that none of these three miRNAs is the sole mediator of PTH action (Fig. 4).
Figure 4.
Combined treatment of transfected primary chondrocytes with miRNA inhibitors and PTH 1-34. Primary chondrocytes were transfected with a scrambled RNA inhibitor or one of three miRNA inhibitors (mir-374-5p, mir-379-5p, or mir-503-5p). After 42 hours, the cells were treated with PTH 1-34 or vehicle for 6 hours, and mRNA levels of (a) Ihh and (b) Col10a1 were measured by real-time PCR. Six biological replicates were performed. *P < 0.05; **P < 0.01; ***P < 0.001. NS, not significant.
Discussion
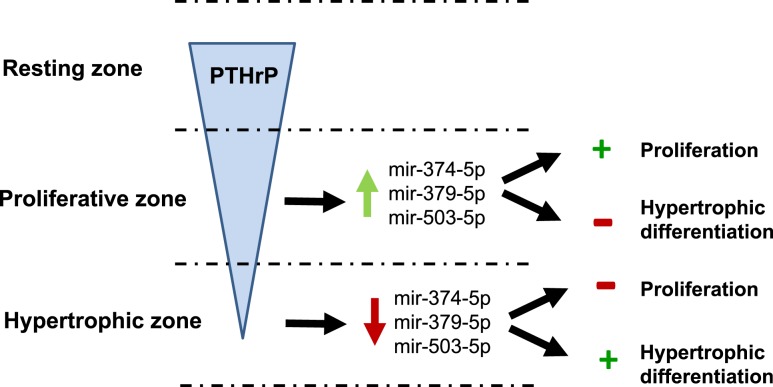
We microdissected growth plate cartilage into its constituent zones to study the spatial regulation of miRNAs in the growth plate. We identified specific miRNAs that were differentially expressed in the PZ vs the HZ and found evidence that three of the miRNAs that showed higher expression in the PZ than in the HZ promote growth plate chondrocyte proliferation and inhibit hypertrophic differentiation. We also found that PTH1-34, an agonist of the PTH/PTHrP receptor, stimulates expression of these miRNAs in growth plate chondrocytes. Taken together, our data suggest the following model (Fig. 5): (1) PTHrP is expressed in the RZ, setting up a protein concentration gradient such that the PTHrP concentration is higher in the PZ than in the HZ (5, 6); (2) the greater concentration of PTHrP in the PZ induces greater expression of specific miRNAs in the PZ compared with the HZ; (3) in the PZ, these miRNAs promote proliferation and inhibit hypertrophic differentiation of growth plate chondrocytes; (4) in the HZ, the lower concentration of PTHrP leads to downregulation of these specific miRNAs; and (5) in the HZ, the lower expression of these miRNAs leads to inhibition of proliferation and stimulation of hypertrophic differentiation. Thus, these specific miRNAs appear to help mediate the known potent inhibitory effect of PTHrP on hypertrophic differentiation of growth plate chondrocytes. However, our findings do not preclude the likely possibility that other independent pathways also mediate the effects of PTHrP on chondrocyte hypertrophy.
Figure 5.
Proposed model for miRNA regulation of growth plate chondrocyte proliferation and hypertrophy. PTHrP is expressed in the RZ, establishing a concentration gradient with higher concentrations of PTHrP in the PZ than in the HZ (4, 5). In the PZ, higher concentrations of PTHrP upregulate mir-374-5p, mir-379-5p, and mir-503-5p, which promote proliferation and inhibit hypertrophic differentiation. In the HZ, the lower concentrations of PTHrP cause downregulation of these miRNAs, which inhibits proliferation and promotes hypertrophic differentiation. Black arrows indicate causation (+, stimulation; −, inhibition). Red and green arrows represent upregulation and downregulation, respectively.
Our findings also suggest possible functional redundancy in the miRNA control of growth plate chondrocyte differentiation. Our in vitro experiments indicate that individual inhibition of mir-374-5p, mir-379-5p, or mir-503-5p was sufficient to inhibit proliferation and to stimulate hypertrophy-associated gene expression, suggesting that all three miRNAs are required to maintain the full proliferative phenotype. Our in vivo expression data suggest that all three of these miRNAs are downregulated in the hypertrophic zone compared with the proliferative zone. Therefore, taken together, the in vitro and in vivo data suggest that the observed in vivo downregulation of all three miRNAs in hypertrophic zone might be redundant because downregulation of any one of these miRNAs might be sufficient to induce the hypertrophic program.
One limitation of this study is that we measured expression of hypertrophy-associated genes at the mRNA level but not at the protein level. miRNAs can increase degradation of target mRNAs and inhibit the translation of target mRNAs. Thus, our approach may miss important direct inhibition of protein expression at the translational level and indirect effects that are secondary to miRNA downregulation of genes that affect protein synthesis or degradation.
Our findings are consistent with a previous study of mice with cartilage-specific ablation of Dicer (9) and further advance the understanding of the underlying mechanisms in several ways. First, the growth plates from Dicer knockout mice showed a striking reduction in the number of proliferating chondrocytes due to decreased proliferation and accelerated differentiation into postmitotic hypertrophic chondrocytes (9), suggesting that miRNAs as a group promote proliferation and inhibit hypertrophic differentiation. Our findings confirm these effects on proliferation and hypertrophic differentiation and help identify the specific miRNAs involved. Second, in the Dicer knockout mice, the defect causing accelerated hypertrophic differentiation appeared to be either downstream or independent of PTHrP (9). Our findings indicate that specific miRNAs lie downstream of PTHrP in the molecular pathways regulating hypertrophic differentiation. Third, the growth plate in Dicer knockout mice showed accelerated chondrocyte hypertrophy but showed no changes in the expression of Mmp13, which is a marker of late hypertrophic differentiation. None of the miRNA inhibitors tested altered the expression of Mmp13, suggesting that these miRNAs are primarily involved in the proliferative to hypertrophic switch that occurs near the upper hypertrophic zone but not in the ossification of hypertrophic cartilage that occurs near the lower hypertrophic zone (15).
Because mir-374-5p, mir-379-5p, and mir-503-5p showed potent effects on proliferation and hypertrophic differentiation in vitro, one might have expected that the combined loss of these three miRNAs and other similarly acting miRNAs in the Dicer knockout mice would have halted proliferation and induced near-universal hypertrophic differentiation. The observed phenotype was not that absolute. However, in the Col2-Cre:Dicer mice, the loss of Dicer expression and miRNA expression appeared not to be complete (9), and the residual expression may have moderated the observed effects.
Previous studies have shown evidence that two miRNAs, miR-140 and let-7, regulate growth plate chondrocytes (16–18). The data suggest that let-7 promotes proliferation and that miR-140 inhibits hypertrophic differentiation. Our findings implicate additional miRNAs that affect proliferation and hypertrophic differentiation and suggest that these miRNAs are differentially expressed in the PZ under the control of PTHrP, thus fitting these miRNAs into the known PTHrP-Ihh pathway regulating growth plate chondrocyte differentiation. Our miRNA profiling showed that let-7b-5p and let-7e-5p were expressed at higher levels in the PZ than in the HZ and thus provide indirect evidence that they promote proliferation of PZ chondrocytes in vivo.
The specific mRNA targets of mir-374-5p, mir-379-5p, and mir-503-5p in growth plate chondrocytes remain unknown. For example, the observed regulation of Col10a1, Ihh, Bmp2, Bmp6, and Vegfa could be due to direct regulation of mRNA stability and/or translation by these miRNAs or indirect regulation. The bioinformatic tools TargetScan, miRDB, and microRNA.org suggest that many of these genes are not predicted targets of mir-374-5p, mir-379-5p, and mir-503-5p, suggesting indirect regulation. Similarly, the mRNA targets responsible for promotion of proliferation remain to be determined. One possibility is that these miRNAs target transcription factors that are important for chondrocyte hypertrophy. Consistent with this possibility, bioinformatic tools to predict miRNA targets suggest that mir-374-5p may target transcription factors c-Maf, Mafb, and Mef2d, which are highly expressed in the HZ and were previously implicated in the regulation of chondrocyte proliferation and differentiation (13). Similarly, mir-379-5p is predicted to target the transcription factors Atf3, which is highly expressed in the HZ, and Jun, which shows no differential expression between zones (13). However, bioinformatic tools used to identify miRNA targets show limited predictive specificity, and thus further investigation is required to test this hypothesis.
In summary, our findings suggest that the PTHrP concentration gradient across the growth plate regulates the expression of mir-374-5p, mir-379-5p, and mir-503-5p, which in turn regulates chondrocyte proliferation and hypertrophic differentiation. The data support a model in which higher concentrations of PTHrP in the PZ upregulate mir-374-5p, mir-379-5p, and mir-503-5p to promote proliferation and inhibit hypertrophic differentiation, whereas in the HZ, lower concentrations of PTHrP cause downregulation of these miRNAs to inhibit cell proliferation and promote hypertrophic differentiation.
Supplementary Material
Acknowledgments
We thank the Eunice Kennedy Shriver National Institute of Child Health and Human Development Molecular Genetics Core Laboratory for performing the miRNA-seq.
Financial Support: This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (to J.B.), the Intramural Research Program of the National Human Genome Research Institute, and the Common Fund of the National Institutes of Health. O.N. was supported by grants from the Swedish Research Council (Project K2015–54X-22 736-01-4 and 2015-02227); the Swedish Governmental Agency for Innovation Systems (Vinnova) (2014-01438); the Marianne and Marcus Wallenberg Foundation; the Stockholm County Council; Byggmästare Olle Engkvist Stiftelse; the Novo Nordisk Foundation; Sällskapet Barnavård; Stiftelsen Frimurare Barnhuset i Stockholm; Karolinska Institutet, Stockholm, Sweden; and Örebro University, Örebro, Sweden.
Disclosure Summary:
The authors have nothing to disclose.
Glossary
Abbreviations:
- FDR
false discovery rate
- HZ
hypertrophic zone
- Ihh
Indian hedgehog
- mRNA
messenger RNA
- miRNA
microRNA
- miRNA-seq
microRNA sequencing
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PTH
parathyroid hormone
- PTHrP
parathyroid hormone–related protein
- PZ
proliferative zone
- RT-PCR
reverse transcription polymerase chain reaction
- RZ
resting zone
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. [DOI] [PubMed] [Google Scholar]
- 2.De Luca F, Barnes KM, Uyeda JA, De-Levi S, Abad V, Palese T, Mericq V, Baron J, Baron J. Regulation of growth plate chondrogenesis by bone morphogenetic protein-2. Endocrinology. 2001;142(1):430–436. [DOI] [PubMed] [Google Scholar]
- 3.Abad V, Meyers JL, Weise M, Gafni RI, Barnes KM, Nilsson O, Bacher JD, Baron J. The role of the resting zone in growth plate chondrogenesis. Endocrinology. 2002;143(5):1851–1857. [DOI] [PubMed] [Google Scholar]
- 4.Kronenberg HM. PTHrP and skeletal development. Ann N Y Acad Sci. 2006;1068(1):1–13. [DOI] [PubMed] [Google Scholar]
- 5.Chau M, Forcinito P, Andrade AC, Hegde A, Ahn S, Lui JC, Baron J, Nilsson O. Organization of the Indian hedgehog--parathyroid hormone-related protein system in the postnatal growth plate. J Mol Endocrinol. 2011;47(1):99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon BS, Lyons KM. Multiple functions of BMPs in chondrogenesis. J Cell Biochem. 2004;93(1):93–103. [DOI] [PubMed] [Google Scholar]
- 7.Mirzamohammadi F, Papaioannou G, Kobayashi T. MicroRNAs in cartilage development, homeostasis, and disease. Curr Osteoporos Rep. 2014;12(4):410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lui JC. Regulation of body growth by microRNAs. Mol Cell Endocrinol. 2017;456:2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008;105(6):1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Research Council. Guide for the Care and Use of Laboratory Animals. 8th ed. Washington, DC: The National Academies Press; 2011.
- 11.Nilsson O, Parker EA, Hegde A, Chau M, Barnes KM, Baron J. Gradients in bone morphogenetic protein-related gene expression across the growth plate. J Endocrinol. 2007;193(1):75–84. [DOI] [PubMed] [Google Scholar]
- 12.Andrade AC, Chrysis D, Audi L, Nilsson O. Methods to study cartilage and bone development. Endocr Dev. 2011;21:52–66. [DOI] [PubMed] [Google Scholar]
- 13.Lui JC, Andrade AC, Forcinito P, Hegde A, Chen W, Baron J, Nilsson O. Spatial and temporal regulation of gene expression in the mammalian growth plate. Bone. 2010;46(5):1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golovchenko S, Hattori T, Hartmann C, Gebhardt M, Gebhard S, Hess A, Pausch F, Schlund B, von der Mark K. Deletion of beta catenin in hypertrophic growth plate chondrocytes impairs trabecular bone formation. Bone. 2013;55(1):102–112. [DOI] [PubMed] [Google Scholar]
- 15.Inada M, Wang Y, Byrne MH, Rahman MU, Miyaura C, López-Otín C, Krane SM. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci U S A. 2004;101(49):17192–17197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papaioannou G, Inloes JB, Nakamura Y, Paltrinieri E, Kobayashi T. let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proc Natl Acad Sci U S A. 2013;110(35):E3291–E3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barter MJ, Tselepi M, Gómez R, Woods S, Hui W, Smith GR, Shanley DP, Clark IM, Young DA. Genome-wide MICRORNA and gene analysis of mesenchymal stem cell chondrogenesis identifies an essential role and multiple targets for miR-140-5p. Stem Cells. 2015;33(11):3266–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura Y, Inloes JB, Katagiri T, Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31(14):3019–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.