Abstract
Sex-specific temporal patterns of pituitary growth hormone (GH) secretion determine the sex-biased transcription of hundreds of genes in the liver and impart important sex differences in liver physiology, metabolism, and disease. Sex differences in hepatic gene expression vary widely, ranging from less than twofold to >1000-fold in the mouse. Here, we use small RNA sequencing to discover 24 sex-biased mouse liver microRNAs (miRNAs), and then investigate the roles of two of these miRNAs in GH-regulated liver sex differences. Studies in prepubertal and young adult mice, and in mice in which pituitary hormones are ablated or where sex-specific hepatic GH signaling is dysregulated, demonstrated that the male-biased miR-1948 and the female-biased miR-802 are both regulated by sex-specific pituitary GH secretory patterns, acquire sex specificity at puberty, and are dependent on the GH-activated transcription factor STAT5 for their sex-specific expression. Both miRNAs are within genomic regions characterized by sex-biased chromatin accessibility. miR-1948, an uncharacterized miRNA, has essential features for correct Drosha/Dicer processing, generates a bona fide mature miRNA with strong strand bias for the 5p arm, and is bound by Argonaute in liver tissue, as is miR-802. In vivo studies using inhibitory locked nucleic acid sequences revealed that miR-1948-5p preferentially represses female-biased messenger RNAs (mRNAs) and induces male-biased mRNAs in male liver; conversely, miR-802-5p preferentially represses male-biased mRNAs and increases levels of female-biased mRNAs in female liver. Cytochrome P450 mRNAs were strongly enriched as targets of both miRNAs. Thus, miR-1948-5p and miR-802-5p are functional components of the GH regulatory network that shapes sex-differential gene expression in mouse liver.
Small RNA-seq identified two sex-biased, plasma GH pattern–regulated liver miRNAs. Locked nucleic acid anti-miR experiments established their functional roles in regulating >100 downstream sex-biased mouse liver RNAs.
Growth hormone (GH) has a wide range of physiological effects, including stimulation of longitudinal bone growth, regulation of hepatic drug and steroid metabolism, and induction of insulinlike growth factor-1 (1, 2). In humans and other species, GH is secreted in a rhythmic manner by anterior pituitary somatotrophs under the control of two hypothalamic peptides, GH-releasing hormone and somatostatin, an inhibitory factor (3, 4). The temporal pattern of pituitary GH release is sexually dimorphic (5) and regulates sex differences in liver function, metabolism, and disease (6). In mice and rats, male pituitary GH secretion is characterized by discrete, high-amplitude pulses every 3 to 4 hours with well-defined, GH-free intervals between pulses, whereas, in females, pituitary GH secretion is more frequent and at a lower amplitude, resulting in the persistent presence of GH in circulation (7–9). Hepatic GH signaling and downstream transcriptional responses are particularly sensitive to these sex differences in GH secretion. GH binds to its cell surface receptor and stimulates tyrosine phosphorylation of the transcription factors STAT5a and STAT5b (collectively, STAT5), which translocate to the nucleus and bind STAT5 motifs at thousands of discrete genomic locations and induce transcription of STAT5 target genes (10). In male liver, STAT5 is activated, translocates to the nucleus, and binds DNA intermittently in direct response to each plasma GH pulse, whereas in female liver, nuclear STAT5 activity is more persistent (11–13). These sex differential patterns of liver STAT5 activation and DNA binding are essential for the sex differential transcription of hundreds of genes, including many cytochromes (Cyps) P450 and other genes active in steroid and drug metabolism (6). Sex-biased liver gene expression is largely abolished when plasma GH profiles are ablated by hypophysectomy (14, 15) or are feminized when males are infused with GH continuously for several days (16).
GH regulates sex-biased liver gene expression through the direct stimulatory effects of GH-activated STAT5 (10, 14) as well as indirectly through the actions of downstream GH/STAT5-regulated liver transcription factors, including the transcriptional repressors BCL6 (male-biased) and CUX2 (female-specific) (10, 16, 17). Many strongly sex-biased genes bind STAT5 and other GH-responsive transcription factors directly, consistent with a direct regulatory mechanism. However, many other sex-biased genes lack nearby binding sites for GH-dependent transcription factors and exhibit comparatively small sex differences in expression (twofold or less) (18, 19), suggesting indirect mechanisms of regulation. Sex-differences of lower than twofold also characterize many sex-biased genes in human liver (20).
MicroRNAs (miRNAs) often impart modest (lower than twofold) effects on target messenger RNAs (mRNAs) (21) and could conceivably regulate a subset of the moderately sex-biased genes through posttranscriptional mechanisms, conferring robustness to the sex-biased liver transcriptional networks (22). miRNAs repress translation, in part by targeting mRNAs for degradation based on partial complementary pairing of the miRNA seed region [nucleotides (nts) 2 to 8] to the seed match site in the 3′ untranslated region (UTR) of the target mRNA (23–25). miRNAs are initially transcribed as extended primary transcripts that harbor the miRNA hairpin structure. The primary transcript is sequentially cleaved by the RNase III enzymes Drosha and Dicer to generate a mature ∼22 nt miRNA duplex whose guide strand is incorporated into the RNA-induced silencing complex, enabling Argonaute-catalyzed degradation of target mRNAs (26). miRNAs have widespread effects on gene expression; 60% of human genes are proposed to be regulated by miRNAs (27). Comparatively few of the miRNAs cataloged in miRBase database (version 21) (28) have been validated experimentally and characterized (29).
Previous studies used microarrays to identify several sex-biased, developmentally regulated rat liver miRNAs (30, 31). However, the mechanisms regulating the sex-biased expression of these miRNAs were not identified and their functional roles, if any, were not determined. Computational studies and luciferase reporter assays were used to predict miRNA regulators of Cyp2b9, a female-specific testosterone 16α-hydroxylase in mouse liver (32); however, the functional roles of the miRNAs identified could only be inferred on the basis of weak inverse correlation data, and their role in regulating other sex-biased mouse liver mRNAs was not investigated. Here, we use small RNA sequencing (RNA-seq) to discover the global repertoire of sex-biased miRNAs in mouse liver. Further, we characterize the role of pituitary GH secretory patterns and STAT5 signaling in regulating two miRNAs that exhibit robust sex-biased expression. Finally, we use inhibitory locked nucleic acids (LNAs) (33) in mouse liver to test the hypothesis that these sex-biased miRNAs regulate a subset of genes that show a sex bias in expression, through posttranscriptional mechanisms.
Materials and Methods
Experimental animals
All animal procedures were conducted in accord with accepted standards of humane animal care under protocols approved by the Boston University Institutional Animal Care and Use Committee. Livers were collected from young adult (7 to 9 weeks) male and female CD1 mice [Crl:CD1(ICR) strain, Charles River Laboratories; IMSR catalog no. CRL:22, Research Resource Identifier (RRID): IMSR_CRL:22] that were untreated or were treated as described later. In some cases, mice were hypophysectomized or were sham-operated on at 8 weeks of age and euthanized ∼3 weeks later (n = 4 biological replicates per group) (15). Other livers were collected from male and female CD1 mice euthanized at 3, 4, and 8 weeks of age (postnatal developmental series, n = 4 livers per group), as described previously (34). Continuous GH infusion in 8- to 10-week-old male CD1 mice (35) was achieved using ALZET osmotic pumps (7-day pump, model 1007D; Durect Corp., Cupertino, CA) filled with recombinant rat GH dissolved in 30 mM NaHCO3 (pH 8.3) buffer containing 0.15 M NaCl and 100 μg/mL rat albumin. Pumps were implanted subcutaneously into n = 5 male mice and GH was infused at a rate of 20 ng/g body weight per hour for 7 days. Livers were snap frozen in liquid nitrogen and stored at −80°C until use.
Livers from 8- to 12-week-old male and female hepatocyte-specific STAT5a/STAT5b-knockout (KO) mice and floxed controls were obtained from Dr. Lothar Hennighausen (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health) (n = 6 mice per sex and genotype). Mice were generated by mating mice having a floxed Stat5a-Stat5b locus with mice harboring a Cre recombinase transgene regulated by the Albumin promoter (36). Livers from somatostatin-KO mice and wild-type controls (n = 5 to 6 males and n = 3 to 5 females per genotype) were a gift of Dr. Rhonda D. Kineman (University of Illinois at Chicago) (37).
Quantitative polymerase chain reaction measurements of miRNA
Total RNA was extracted from individual livers and reverse transcribed using Qiagen miScript II RT kit (Qiagen catalog no. 218160) and 5× miScript HiSpec buffer to selectively reverse transcribe mature miRNA species. Quantitative polymerase chain reaction (qPCR) was performed using ABI SYBR Green reagents. PCR primers were purchased from Qiagen miScript Primer Assay for Hs_RNU6B (MS00029204), mmu-miR-122a_1 (MS00001526), mmu-mir-802-1 (MS00003101), and mmu-mir-1948-1 (MS00017143). qPCR threshold cycle values were normalized based on the content of a nuclear small RNA housekeeping gene RNU6B (Hs_RNU6B_2), which is conserved in human, mouse, and rat. Statistical significance was determined by Student t test. qPCR was performed for mouse miR-1948-3p rather than the more abundant miR-1948-5p (see the following section) because the sequence of the mature miR-1948-5p exhibited low CG content, which precluded optimal primer design and did not give reliable results or PCR products with symmetric, single-product melting curves.
Mapping miRNAs to DHS
Mouse miRNAs were mapped to the set of ∼72,000 mouse liver DNase hypersensitive sites (DHSs) defined previously (38), using bedtools (39) to identify all DHS within 5 kb of an miRNA. Supplemental Table 1 lists all miRNA-proximal DHS whose chromatin accessibility was previously shown to be significantly greater in male than female liver or vice versa (sex-biased DHS).
Small RNA sequencing and data analysis
Total RNA was extracted from 8-week-old CD1 strain mouse livers using TRIzol (n = 6 males, n = 6 females). Small RNA-seq libraries were created using NEBNext small RNA kit (New England Biolabs) per the manufacturer's instructions, and sequenced on an Illumina HiSeq instrument to give single-end 50-bp sequences. Raw sequence reads were trimmed by TrimGalore! (version 0.4.3) to remove adapter sequences; the trimmed reads were then mapped to the mouse mm9 genome using Bowtie2 with default parameters, allowing for mapping of multimapped sequence reads to ensure that mature miRNA sequence reads arising from miRNAs found at two or more genomic locations (e.g., mmu-mir-3084-1, mmu-mir-3084-2) are included in the counting and differential expression analysis. Coordinates of mature mouse miRNAs were downloaded from MiRBase, version 21 (28), and the bedtools intersect command was used to determine the number of reads at each mature miRNA locus (bedtools intersect parameters: -c -f 0.8 -r). Differential expression analysis was implemented using EdgeR (40) to obtain a list of sex differential miRNAs, with differential expression defined by cutoff values of sex difference >1.8-fold, false discovery rate (FDR) <5E-2, and log2 counts per million (CPM) > 1 (Supplemental Table 2). To analyze 5′-processing heterogeneity, we used custom Python scripts to extract from Bam files all sequence reads falling within the primary sequence of each miRNA of interest and then analyze their sequences. RNA secondary structure analysis and visualization was carried out using RNAfold (41).
Argonaute pull-down
One day before the Argonaute pull-down experiment, Protein A beads (200 μL; Thermo Fisher Scientific 10001D) were mixed with 50 μL of rabbit antimouse bridging immunoglobulin G (IgG; Millipore; catalog no. 06-371; RRID: AB_390146) in 200 μL 1× BWB buffer [1× phosphate-buffered saline (PBS), 0.02% volume-to-volume ratio Tween-20], then rotated for 30 minutes at room temperature and washed three times with 1 mL of 1× BWB buffer. The washed beads were then bound to 2 μg of Ago2A8 Millipore MABE56 antibody (Millipore; catalog no. MABE56; RRID:AB_10807962) in 200 μL of 1× BWB buffer overnight at 4°C with gentle agitation, and washed three times with 1× PXL buffer (see later) before use. Whole frozen liver from 8-week-old untreated male CD1 mice was pulverized to a fine powder with a mortar and pestle on dry ice. The resultant liver powder (≤200 mg liver powder spread thinly on petri dish placed on dry ice) was crosslinked by ultraviolet irradiation using a ultraviolet Stratalinker 2400 instrument (Stratagene, Inc.), first with 400 mJ/cm2, and then with 200 mJ/cm2. The crosslinked liver powder (∼200 mg) was homogenized on ice in 1 mL of 1× PXL buffer supplemented with 2 mM 2-mercaptoethanol [where 5× PXL buffer = 5× PBS, 1% (volume-to-volume ratio) Ipegal, 0.5% (weight-to-volume ratio) sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)] in a Dounce homogenizer. Thirty units of DNase (Promega; catalog no. M6101) were added followed by incubation at 37°C for 5 minutes with gentle mixing. The lysate was centrifuged at 18,000 relative centrifugal force for 20 minutes at 4°C, the supernatant was recovered, and a 100 μL aliquot was flash frozen and saved as the input sample. The supernatant (∼800 μL) was then added to Protein A beads preloaded with antibody to Argonaute (Ago2A8) and rotated end over end for 2 hours at 4°C. The beads were then washed three times with 1 mL of 1× PXL buffer, twice with 1 mL high-salt buffer (1× PBS, 1 M NaCl, 1% Ipegal, 0.5% sodium deoxycholate, 0.1% SDS) (Santa Cruz Biotechnology, Santa Cruz, CA), twice with 1 mL of high-stringency buffer (1 mM Tris-HCl pH 7.5, 5 mM EDTA, 2.5 mM EGTA, 1% Ipegal, 1% sodium deoxycholate, 0.1% SDS, 120 mM NaCl, 25 mM KCl), and once with 1 mL of low-salt wash buffer (15 mM Tris pH 7.5, 5 mM EDTA). The washed beads were suspended in 250 μL of proteinase K buffer (300 mM NaCl, 200 mM Tris pH 7.5, 25 mM EDTA, 2% SDS) containing 100 μg of proteinase K (Bioline; catalog no. 37084) and incubated at 65°C for 15 minutes with gentle agitation. Trizol LS was added directly to the proteinase K-digested beads to extract RNA. qPCR was performed on complementary DNA prepared from the input RNA, and from the immunoprecipitation (IP) and IgG control RNA, to determine the enrichment of each miRNA of interest.
Ectopic expression of primary miR-1948 sequence
HEK293 cells were cultured to 80% confluence in 12 well plates and infected with 4 to 7 × 106 infectious units of adenovirus harboring a cytomegalovirus-driven GFP reporter fused with ∼400 nt sequence flanking the miR-1948 hairpin structure, or with GFP as control only. These adenoviral constructs were designed and expressed in adenovirus serotype 5 by Applied Biological Materials (British Columbia, Canada; control adenovirus, catalog no. m009; and custom-designed Adeno-mmu-miR-1948). Adenovirus seed stocks were propagated in HEK293A cells and titered using plaque assays and standard protocols (42). Cells were infected with Adenovirus for 48 to 55 hours to induce a cytopathic effect and strong GFP fluorescence. Total RNA was then extracted and used for small RNA-seq library preparation.
In vivo inhibition of miR-1948 and miR-802
To inhibit miR-1948-5p and miR-802-5p function in mouse liver in vivo, LNA sequences complementary to each miRNA, and a negative control LNA, were purchased from Exiqon, Inc. and injected into 9-week-old male and female CD1 mice (LNAs: anti–mmu-miR-1948-5p, GGCAGAATACTCATA; anti–mmu-miR-802-5p, GAATCTTTGTTACTG; negative control, ACGTCTATACGCCCA). A pilot experiment showed that these LNAs did not induce liver toxicity, as determined by monitoring mouse behavior, body weight, and serum alanine transaminase activity 6 days after treatment of male mice with LNA miR-1948-5p and female mice with LNA miR-802-5p. Alanine transaminase was measured using a colorimetric activity assay kit (Cayman Chemical, catalog no. 700260; normal serum activity range, 8 to 40 U/L). In a follow up, large-scale study, mice were given two subcutaneous injections of LNA dissolved in 1× PBS at 25 mg LNA/kg mouse body weight on day 0, followed by repeat LNA injection on day 1. Injections were carried out between 10 am and noon. Mice were euthanized on day 3 or 6, resulting in a total of 3 or 6 days of exposure to each LNA. Male mice were injected with LNA-miR-1948-5p; female mice were injected with LNA-miR-802-5p. Both male and female mice were injected with LNA-negative control, for both the 3- and the 6-day time points. Mice were euthanized by cervical dislocation between 10 am and noon, and livers were harvested for RNA extraction and RNA-seq analysis. For each sex and at each time point, n = 5 mice received miRNA inhibitory LNA sequence (LNA-antimiR), and n = 3 mice received the negative control LNA (LNA-negative). All 32 liver RNA samples were analyzed by RNA-seq,
RNA-seq of LNA antimiR-treated liver RNA
Paired-end (50 bp) RNA-seq raw FASTQ files were mapped by TopHap (43) to the mouse mm9 genome using default parameters. FeatureCounts (44) was used to quantify the reads within gene bodies of RefSeq genes; EdgeR (40) was used to identify differentially expressed genes. Sex-biased genes were defined by differential expression analysis from the following comparison: [male mouse liver treated with LNA-negative for 3 or for 6 days, n = 6] / [female mouse liver treated with LNA-negative for 3 or for 6 days, n = 6], using a cutoff for significance of |fold-change| >1.5 at FDR <0.05, and a signal intensity >0.25 fragments per read per kilobase (FPKM) in either LNA-negative male or LNA-negative female mouse liver. A total of 990 sex-biased genes (356 male-biased, 634 female-biased) and 11,132 sex-independent genes met these FPKM thresholds (Supplemental Table 3A). LNA-responsive genes were identified from the following comparisons at each time point: [LNA-antimiR, n = 5] / [LNA-negative, n = 3], with a significance cutoff of |fold-change| > 1.5, P value (unadjusted) < 0.05, with an FPKM > 0.25 filter for expression level in either LNA-negative male or LNA-negative female mouse liver, as above. The male-female RNA-seq comparisons for LNA-negative treated mice (above) were used to designate individual LNA-responsive mRNAs with regard to their sex-bias, as follows: 1) male-biased and female-biased, |fold-change| > 1.5 at FDR<0.05; 2) weakly male-biased and weakly female-biased, P value (unadjusted) < 0.1; 3) and stringent sex-independent, p-value (unadjusted) ≥ 0.1. Genes whose mRNAs were responsive to each LNA are listed in Supplemental Table 3B–3E.
Enrichment scores and TargetScan predictions
The enrichment of LNA-responsive genes in sex-biased genes was calculated as follows: enrichment score (ES) = [number of sex-biased LNA-responsive genes / number of sex-independent LNA-responsive genes] / [number of sex-biased liver-expressed genes / number of sex-independent liver-expressed genes], where sex-independent genes include weakly sex-biased genes and liver expressed is defined by FPKM >0.25. Significance was assessed by χ2 test with Yates correction, implemented in GraphPad Prism. miRNA target gene predictions were carried out using TargetScan Mouse Version 7.1 (www.targetscan.org/) (45). The top 300 predicted targets of each miRNA, ranked by TargetScan’s cumulative weighted context score, were used in our analysis.
Gene Expression Omnibus accession numbers
Raw sequencing data and processed files are available under the following accession numbers: small RNA-seq of male and female mouse liver (GSE103879); RNA-seq expression analysis of mouse liver treated with LNAs that inhibit miR-802-5p and miR-1948-5p (GSE103880); and small RNA-seq of human HEK293 cells overexpressing mmu-miR-1948 primary sequence (GSE103881).
Results
Sex-biased liver expression of miR-1948 and miR-802
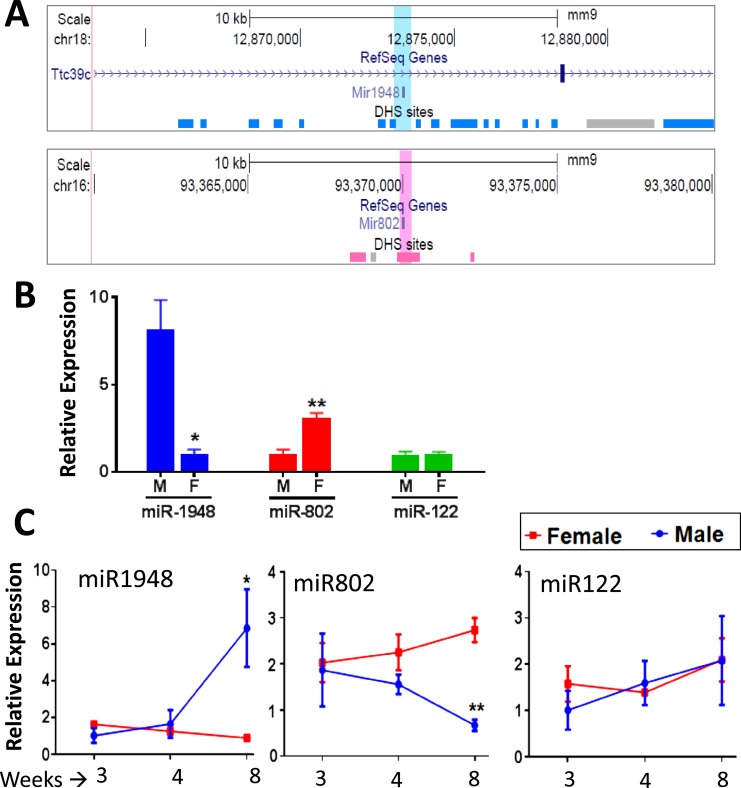
Initially, we searched for sex-specific mouse liver miRNAs by screening MiRBase (version 21) to identify mouse miRNA loci located within 5 kb of any of the 4179 mouse genomic regions that show substantial sex differences in chromatin accessibility in mouse liver, as determined in DHS assays (38). These sex-differential DHS are enriched for key regulatory regions and transcription factor binding sites controlling sex-biased liver gene expression (38, 46). Several miRNAs located in genomic regions with multiple sex-differential DHS regions were identified, most strikingly miR-1948, which is flanked by 13 male-biased DHS and is found in an intron of the male-biased gene Ttc39c, and miR-802, which overlaps a prominent female-biased DHS and is near two other female-biased DHS (Fig. 1A; Table 1).
Figure 1.
Localization of miR-1948 and miR-802 nearby sex-biased DHS and their sex differential expression in mouse liver. (A) University of California Santa Cruz browser screenshot of genomic regions surrounding miR-1948 (top; blue highlight) and miR-802 (bottom; pink highlight). DHS peak regions are shown as horizontal blocks in dark blue (male-biased DHS), pink (female-biased DHS), and gray (sex-independent DHS) based on Ling et al. (38). (B) Sex-biased expression of miR-1948 and miR-802, compared with the sex-independent expression of the abundant liver miR-122, as determined by qPCR analysis of male and female mouse liver RNA (mean ± SE, n = 5 livers per sex). The sex with the lower expression was set to a y-axis value of 1 for each miRNA. Significance between sexes was determined by Student t test: *P < 0.05; **P < 0.005. (C) Developmental profile for the expression of miR-1948, miR-802, and miR-122 in livers of male and female mice at 3, 4, and 8 weeks of age (mean ± SE, n = 4 livers per group). The lowest expression sample group in each time point was set to 1. Sex-differential expression is first seen after puberty (8 weeks of age), when miR-1948 is upregulated and miR-802 is downregulated in male liver. Significance comparing male with female in each age group was determined by Student t test: *P < 0.05; **P < 0.005. F, female; M, male; SE, standard error.
Table 1.
miRNA Nearby Genomic Regions Showing Male- or Female-Biased Chromatin Accessibility in Mouse Liver, as Indicated by the Presence of Sex-Biased DHS Peak Regions
| Mouse miRNA Near Female-Biased DHS | Female-Biased DHS Within 5 kb of miRNA Gene, No. | Mouse miRNA Near Male-Biased DHS | Male-Biased DHS Within 5 kb of miRNA Gene, No. |
|---|---|---|---|
| mmu-miR-33 | 1 | mmu-miR-21a | 1 |
| mmu-miR-802 | 3 | mmu-miR-28b | 1 |
| mmu-miR-1247 | 1 | mmu-miR-455 | 2 |
| mmu-miR-1902 | 1 | mmu-miR-1948 | 13 |
| mmu-miR-6366 | 1 | mmu-miR-5128 | 1 |
| mmu-miR-6392 | 1 | mmu-miR-6397 | 1 |
| mmu-miR-6928 | 1 | mmu-miR-6965 | 1 |
| mmu-miR-6993 | 1 | mmu-miR-7224 | 1 |
| mmu-miR-7078 | 1 | mmu-miR-7684 | 1 |
| mmu-miR-7662 | 1 | ||
| mmu-miR-8105 | 1 |
The number of sex-specific DHS within 5 kb upstream or 5 kb downstream of each miRNA gene is summarized (see Supplemental Table 1 for complete details). miR-802 and miR-1948 were within 5 kb of multiple female- and male-biased DHS, respectively (Fig. 1A). Small RNA-seq analysis (Table 2) confirmed the female-biased expression of miR-802 and the male-biased expression of miR-21a, miR-455, and miR-1948 in mouse liver.
The strong sex-specific chromatin environment surrounding miR-1948 and miR-802 suggests these miRNAs are expressed in a sex-dependent manner. Indeed, qPCR analysis showed that miR-1948 is expressed at a ∼sevenfold higher level in young adult (8 week) male compared with female liver, and that miR-802 showed ∼threefold higher expression female liver (Fig. 1B). No sex difference was seen for miR-122 (47), the most abundant liver miRNA (33% of all 2044 mouse miRNAs examined; Supplemental Table 2). Each miRNA locus produces two mature miRNAs, designated 5p and 3p, which come from the 5′ strand and the 3′ strand of the miRNA precursor, respectively (48). For qPCR, we used primers targeting the mature transcripts of miR-1948-3p and miR-802-5p, which represent the more biologically active guide strand, according to miRBase annotation (28), and generated symmetrical, single-product melting curves in our qPCR analysis.
Regulation of miR-1948 and miR-802 by GH and STAT5
Next, we examined the expression of miR-1948-3p and miR-802-5p during postnatal liver development. These analyses revealed that the sex differences in miR-1948 and miR-802 expression emerge around puberty (∼4 weeks of age), at which time miR-1948 was induced and miR-802 was suppressed in male liver. In contrast, neither miRNA showed a substantial change in expression in female liver over this period (Fig. 1C). This pattern of a change in gene expression at puberty in male but not female liver characterizes many sex-biased protein coding genes (34). miR-122 did not show any sex-dependent changes in expression over this period.
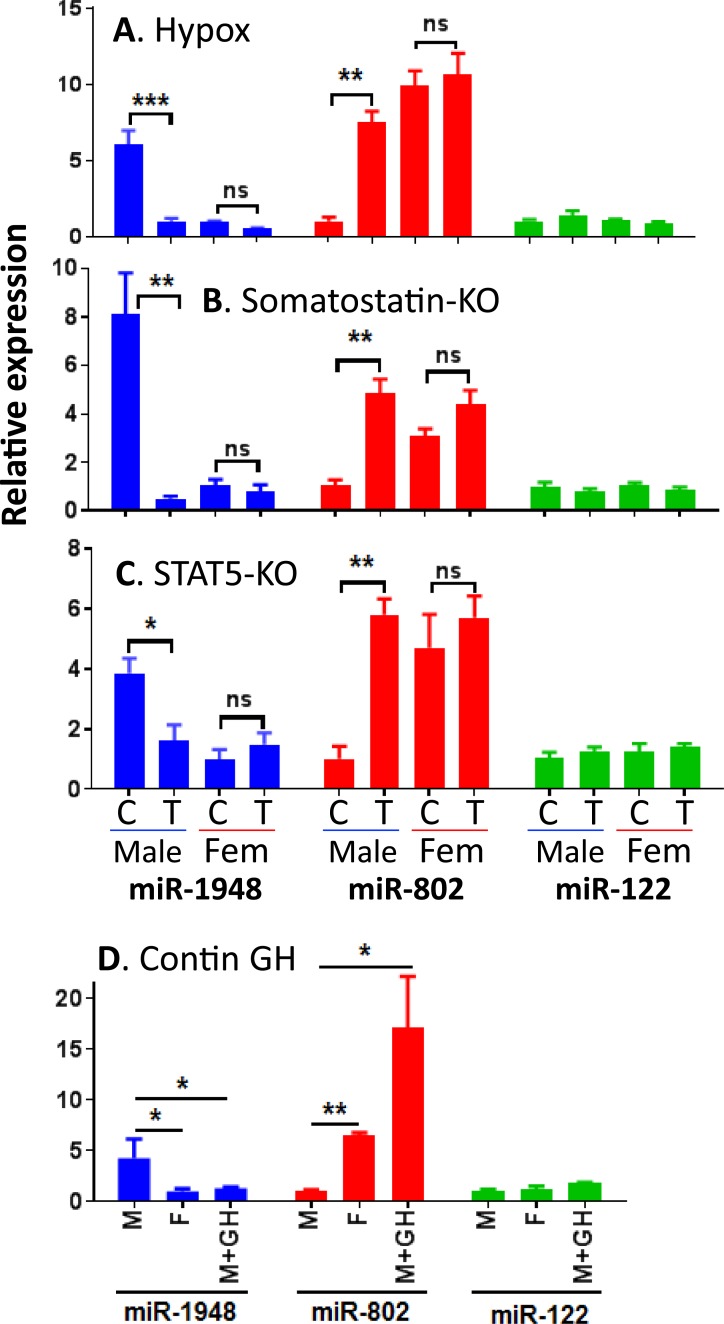
To determine whether pituitary hormones regulate the expression of miR-1948 or miR-802, we measured the levels of each miRNA in livers of hypophysectomized mice. Hypophysectomy (i.e., surgical removal of the pituitary gland) ablates circulating GH and essentially all other pituitary-derived hormones and results in a near-global loss of liver sex differences (15). Following hypophysectomy of male mice, liver miR-1948 levels decreased and miR-802 levels increased, indicating that both miRNAs are regulated by pituitary hormones in male liver (Fig. 2A). In contrast, hypophysectomy of female mice had no impact on the expression of either miRNA. Further, we examined livers from somatostatin-KO mice (37, 49) to characterize the dependence of miR-1948 and miR-802 on the sex-specific pattern of pituitary GH secretion. Somatostatin, a hypothalamic peptide hormone, antagonizes GH-releasing hormone-stimulated pituitary GH release, thereby ensuring that each pulse of pituitary GH release is followed by a GH-free interval. Loss of somatostatin leads to a persistently elevated basal GH level and, consequently, feminization of male mouse liver (49). In the absence of somatostatin, male liver expression of miR-1948 was downregulated to the lower, female liver level, and miR-802 expression was upregulated to female liver levels (Fig. 2B). miR-122 levels were unaffected by hypophysectomy or by somatostatin deficiency.
Figure 2.
Regulation of liver miR-1948 and miR-802 expression by GH and STAT5. Expression levels of each miRNA were measured by qPCR in four mouse models where the GH-STAT5 axis is perturbed: (A) hypophysectomy, (B) somatostatin-KO, (C) hepatocyte-specific STAT5-KO, and (D) continuous GH infusion for 7 days. Groups of male and female livers are marked C to indicate intact or wild-type mice (control group) or T to indicate hypophysectomized or KO livers (treatment group). Data shown are mean ± SE values, with the numbers of livers per group specified in the Materials and Methods section. The lowest expressed group in each analysis was set to 1. Significance between groups: *P < 0.05; **P < 0.005; ***P < 0.001. contin, continuous; Fem, female; M+GH, male mice given 7-day GH infusion; ns, not significant; SE, standard error.
STAT5 is a GH-activated transcription factor and major regulator of a substantial fraction of sex-biased genes in mouse liver. Sex differential gene expression is largely abolished in mice with a hepatocyte-specific deletion of the STAT5a/STAT5b locus (STAT5-KO) (19, 35, 36). To test whether miR-1948 and miR-802 are dependent on STAT5, we measured their expression in STAT5-KO male mouse liver (36). We observed downregulation of miR-1948 and upregulation of miR-802, but no change in miR-122 (Fig. 2C). No changes in expression were seen in STAT5-KO female liver. Thus, pituitary hormones, a pulsatile plasma GH pattern, and the GH pulse-activated STAT5 are required in male but not female mouse liver for the sex-biased expression of miR-1948 and miR-802. These responses are consistent with miR-1948 being a class I male-biased gene (14, 15) (i.e., one that is positively regulated in male liver by male plasma GH pulse-activated STAT5) and that miR-802 is class II female-biased gene (14, 15) (i.e., one that is negatively regulated in male liver by male plasma GH pulse-activated STAT5).
Finally, we examined miR-1948 and miR-802 expression in livers of continuous GH-infused male mice, in which exogenous GH infusion using an ALZET osmotic minipump overrides the normal male plasma GH pulses and leads to downregulation of a large fraction of male-biased genes and upregulation of female-biased genes (16, 35). Consistent with these responses, miR-1948 expression was decreased and miR-802 expression was induced in male liver following GH infusion, confirming that both miRNAs are dependent on the temporal pattern of plasma GH (Fig. 2D). Together, these studies establish that the sex-dependent expression of miR-1948 and miR-802 is primarily regulated in male liver, where it is dependent on the male profile of plasma GH pulse-activated STAT5, which onsets at puberty (11).
Sex-biased liver miRNAs identified by small RNA-seq
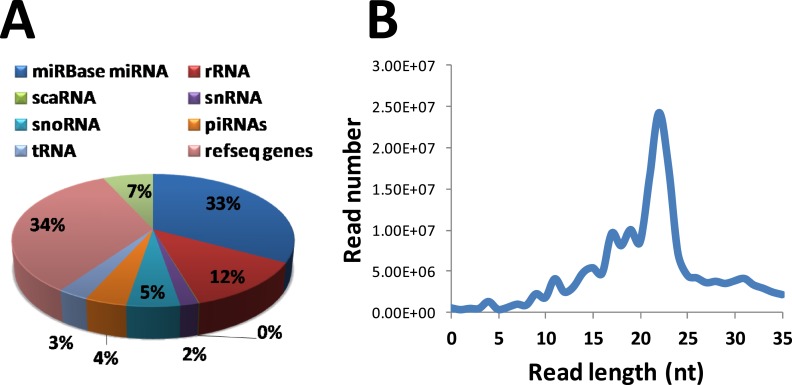
We performed small RNA-seq analysis of 8-week-old male and female mouse liver to identify the global repertoire of sex-biased liver miRNAs. The small RNA-seq dataset was strongly enriched for miRNAs among other noncoding RNA types, with the read length distribution profile peaking at a length of 23 nt (Fig. 3). Approximately 34% of small RNA-seq reads did not map to annotated RNA sequences, but overlapped protein-coding RefSeq genes; these sequence reads likely include unannotated miRNAs and mRNA degradation intermediates. Differential expression analysis identified 24 unique miRNAs derived from 19 miRNA loci that showed significant sex-differential expression (fold difference >1.8, FDR <5E-2, log2 CPM >1), including miR-1948 and miR-802 (Table 2). miR-21a, which is a highly conserved oncogenic miRNA whose expression is elevated in a variety of cancers (50). The elevated expression of miR-21a in male liver could contribute to the greater predisposition of males to hepatocellular carcinoma (51). These analyses also confirmed the female-biased expression of miR-802-5p, whose overexpression has been implicated in obesity-associated glucose deregulation (52). In addition to miR-1948 and miR-802, two other sex-biased miRNAs identified (miR-21a, miR-455) were nearby corresponding sex-biased DHS (Table 1). Thus, all four sex-biased miRNAs are within genomic regions with sex-biased chromatin accessibility.
Figure 3.
Small RNA-seq statistics. (A) Distribution of small RNA-seq reads among different RNA classes. The two largest sources of small RNA reads are annotated miRNAs (33%) and unannotated reads from Ref-Seq loci (34%). (B) Read-length distribution of small-RNA sequencing peaked at 23 nt, which corresponds to the length of mature miRNAs. piRNA, PIWI-interacting RNA; rRNA, ribosomal RNA; scaRNA, small Cajal body-specific RNA; snoRNA, small nucleolar RNA; tRNA, transfer RNA.
Table 2.
Top Sex-Differentially Expressed miRNAs (Fold-change > 2, log2 CPM >1, and FDR < 0.05; Supplemental Table 2) Determined by Small RNA-seq Analysis of Male and Female Mouse Liver
| Mouse miRNA | Chromosome | miRNA Length | Sex Difference (M/F Fold Change) | CPM | FDR |
|---|---|---|---|---|---|
| miR-1948-5p | chr18 | 21 | 6.52 | 209 | 1.56E-38 |
| miR-1948-3p | chr18 | 22 | 6.28 | 6 | 7.19E-12 |
| miR-592-5p | chr6 | 23 | 5.09 | 122 | 1.38E-02 |
| miR-3084-1-3p | chr19 | 21 | 4.73 | 17 | 5.51E-06 |
| miR-101b-5p | chr19 | 23 | 4.67 | 5 | 5.62E-08 |
| miR-3084-2-3p | chr19 | 21 | 4.47 | 18 | 2.64E-06 |
| miR-881-3p | chrX | 22 | 3.08 | 3 | 5.26E-03 |
| miR-455-5p | chr4 | 22 | 2.51 | 616 | 5.55E-07 |
| miR-3084-5p | chr19 | 22 | 2.24 | 4 | 1.38E-02 |
| miR-455-3p | chr4 | 21 | 2.24 | 437 | 2.86E-08 |
| miR-21a-5p | chr11 | 22 | 2.14 | 127,428 | 5.42E-08 |
| miR-21a-3p | chr11 | 22 | 2.07 | 13 | 5.63E-03 |
| miR-182-5p | chr6 | 25 | 2.06 | 203 | 4.04E-02 |
| miR-149-5p | chr1 | 23 | 2.04 | 19 | 9.86E-03 |
| miR-221-5p | chrX | 26 | 1.98 | 28 | 1.94E-04 |
| miR-677-5p | chr10 | 22 | 1.89 | 58 | 1.27E-04 |
| miR-6239 | chr14 | 20 | −1.89 | 49 | 2.65E-02 |
| miR-218-5p | chr5 | 21 | −1.93 | 8 | 3.66E-02 |
| miR-6238 | chr7 | 22 | −2.00 | 7 | 7.83E-03 |
| miR-674-3p | chr2 | 22 | −2.03 | 45 | 2.50E-05 |
| miR-802-3p | chr16 | 22 | −2.06 | 24 | 7.39E-05 |
| miR-3968 | chr11 | 21 | −2.21 | 3 | 2.29E-02 |
| miR-802-5p | chr16 | 22 | −2.23 | 173 | 8.99E-07 |
| miR-2137 | chrX | 21 | −2.35 | 4 | 1.99E-02 |
| miR-296-5p | chr2 | 21 | −3.37 | 5 | 1.45E-06 |
Shown is the male/female fold difference in expression in which positive values indicate male bias and negative values indicate female bias. Also shown is the expression level of each miRNA (CPM) and the significance of the sex bias, indicated by FDR. mir-3084-1 and mir-3084-2 refer to the two locations on mouse chromosome 19 with the identical mature miR-3084 sequence. Results are shown for the reads mapped to each genomic location using the multimapping settings of Bowtie 2 (see Materials and Methods). Boldface type indicates isoforms of the two major sex-biased miRNAs characterized in this study.
Characterization of miR-1948
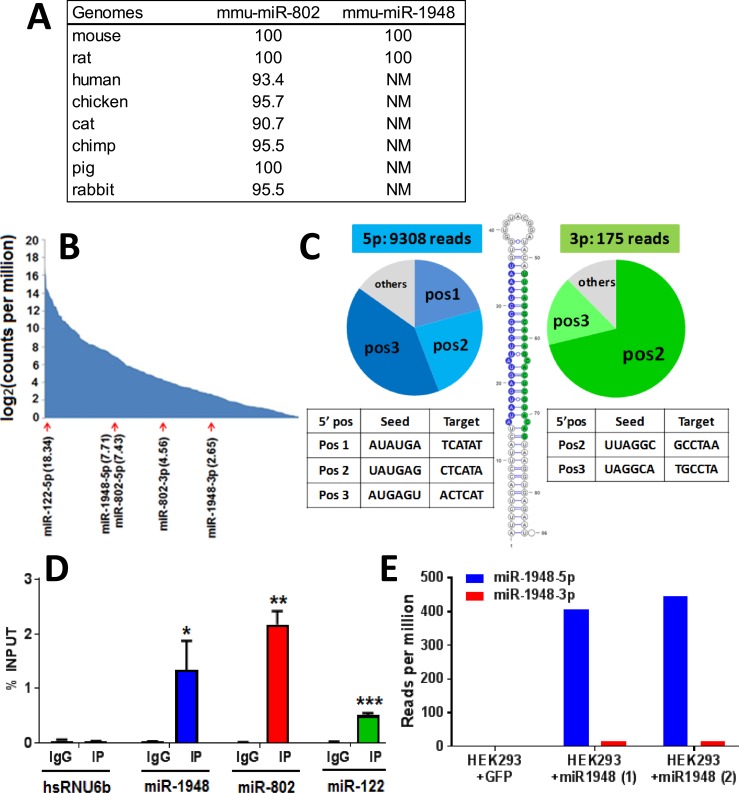
The majority of cataloged miRNAs lack functional validation; therefore, it is unclear whether they represent functional genes or fortuitously acquired hairpin RNA structures. Most functional studies have focused on broadly conserved miRNA families, such as that of miR-802-5p, whose expression and functional role has been confirmed in several studies (52, 53). It is often assumed that poorly conserved miRNAs, such as miR-1948 (Fig. 4A), which TargetScan lists as a random, misannotated miRNA, are likely to be nonfunctional (29). To determine if miR-1948 is a bona fide miRNA, we characterized miR-1948 with regard to the following six characteristics: abundance, pairing characteristics of the predicted hairpin, presence of corresponding miRNA* species, 5′ heterogeneity, association with Argonaute proteins, and correct processing to mature miRNA following ectopic expression.
Figure 4.
Computation and experimental validation of miR-1948. (A) Conservation of mouse miR-802 and miR-1948. Values shown are percent sequence identity in mouse compared with the other seven genomes listed, determined by BLAT analysis of each miRNA’s ∼80 to 100 bp primary DNA sequence. (B) Abundance of miR-1948 and miR-802 in male and female mouse liver, respectively, compared with the full set of 603 liver-expressed miRNAs [defined as miRNAs with log2 (CPM) > 0], which are ranked by their expression levels. Log2 (CPM) values are as shown in parentheses. The star strands, miR-1948-3p and miR-802-3p, showed much lower expression than the primary strands, miR-1948-5p and miR-802-5p. (C) 5′ processing heterogeneity of miR-1948. The 84 nt primary sequence miR-1948 is predicted to form a duplex, as shown. The ∼23 nt mature 5p (blue) and 3p (green) arms of miR-1948 were examined, and the number of sequence reads at each 5′ start position at the 5′ ends of the 5p and the 3p miRNAs were counted, as shown. miR-1948-5p showed three dominant 5′ start positions, and the 3′ arm showed two start sites, as indicated in the pie charts. Because the 5′ start site of a miRNA determines its seed sequence, this 5′ heterogeneity indicates the potential to generate multiple miRNA isoforms. (D) miR-1948 and miR-802 associate with Argonaute protein. Shown is the enrichment of miR-1948, miR-802, miR-122, and nuclear small RNA HsRNU6b (negative control) following RNA immunoprecipitation of mouse liver cytoplasmic extract with anti-Argonaute antibody (IP) or IgG (control), based on qPCR analysis (mean ± SE, n = 3 biological replicates). Student t test was used to assess the significance of difference between IP and control (*P < 0.05, **P < 0.01; ***P < 0.005). (E) Human HEK293 cells, which do not express the mouse-specific miR-1948 sequence, were infected with adenovirus carrying a cytomegalovirus-driven expression cassette for the primary miR-1948 sequence plus ∼400-bp sequence flanking the hairpin sequence. Shown is the normalized number of small RNA-seq reads for each mature miRNA in cells infected with adenovirus expressing GFP (HEK293 + GFP; control group), or adenovirus expressing miR-1948 (HEK293 + miR-1948; two biological replicates). Mature miR-1948 was absent from cells infected with adeno-GFP. Adeno-miR-1948–infected cells contained high levels of correctly processed mature miR-1948 with strong 5p over 3p bias, as indicated. NM, no match found; pos, position; SE, standard error.
Mouse miR-1948 is within an intron of the male-biased protein coding gene Ttc39c. Based on the University of California Santa Cruz browser whole genome alignment (54), the primary sequence of miR-1948 is limited to the mouse lineage, suggesting its relatively recent evolutionary emergence. Supporting the proposal that miR-1948 is functional is its comparatively high expression (Fig. 4B). Secondary structure prediction by RNAfold (55) for the precursor sequence of miR-1948 indicated a robust hairpin structure (Fig. 4C). Further, our small RNA-seq results showed overwhelming discrimination in arm usage, with the expression of 5p arm miRNA being 53-fold higher than the 3p arm miRNA (Fig. 4C). This finding contradicts the annotation in miRBase (28) that miR-1948-3p is the predominant form.
Examination of the 5′ sequence heterogeneity of miR-1948 revealed three tandem cleavage positions for miR-1948-5p and two for miR-1948-3p. Thus, five mature miRNA isoforms are generated from the miR-1948 locus in mouse liver, with miR-1948-5p contributing most to the heterogeneity (Fig. 4C). Because the 5′ start position of a miRNA determines its seed sequence at nt positions 2 through 7, there is selective pressure for the majority of conserved miRNAs to enforce precise cleavage of the 5′ terminus to prevent the formation of miRNAs with off-target seed sequences (23). However, miRNAs with multiple functional forms can be generated by using both arms of the hairpin and/or by 5′ processing heterogeneity (56, 57); miR-1948 may be such a case.
To validate the authenticity of miR-1948, we examined its association with the Argonaute complex (58) by performing Argonaute RNA immunoprecipitation and found that mature miR-1948 and miR-802 are significantly associated with Argonaute (Fig. 4D). This finding suggests that both miRNAs are functional. Next, we used adenovirus to ectopically express in human HEK293 cells the primary sequence of miR-1948 together with ∼400 nt sequence flanking the miRNA hairpin. Small RNA-seq analysis of the adeno-miR-1948-infected cells resulted in high levels of the mature 22 nt miR-1948-5p and very low levels of miR-1948-3p (Fig. 4E), consistent with the strong preference of 5p arm usage seen in mouse liver (Fig. 4C). No miR-1948 sequences were detected in HEK293 cells infected with adenoviral-GFP (negative control), consistent with the mouse (and rat) lineage-specific nature of miR-1948 (Fig. 4A). Thus, miR-1948 and its flanking sequences contain the correct features for Drosha/Dicer processing into the mature miRNA.
Assessment of sex-biased miRNA function using LNA antimiRs
We initially investigated the potential targets of miR-1948-5p and miR-802-5p using TargetScan (45), a computational tool that predicts miRNA-gene interactions by combination of conservation and sequence match between the miRNA seed sequence and an mRNA target’s 3′ UTR sequence (59). Gene targets predicted by TargetScan for each miRNA showed no enrichment for liver sex-biased genes (P > 0.1, Fisher exact test). Although miRNA target prediction tools, such as TargetScan, are useful for identifying mRNAs with specific canonical seed sequences in the 3′ UTR, they exhibit high false-positive rates and do not capture the complexity of miRNA targeting in vivo (60, 61). As such, we investigated the impact of inhibition of miR-1948-5p and miR-802-5p in vivo using LNA sequences. LNAs are modified RNAs with a phosphorothioate backbone that are nuclease resistant, form highly stable heteroduplexes with their target miRNAs, and are well tolerated in vivo with minimal toxicity (62–64). When injected systemically, LNAs preferentially accumulate in clearance organs, specifically liver and kidneys, where they bind their target mature miRNA and inhibit miRNA-Argonaute interactions (65).
We delivered LNAs antisense to miR-1948-5p and miR-802-5p to livers of male and female mice, respectively (i.e., to the sex where each miRNA is more highly expressed). We then used RNA-seq to identify mRNAs whose expression is specifically dysregulated by each LNA after 3 and 6 days of LNA exposure (Fig. 5A). LNAs with random sequences not found in the mouse lineage were used as negative controls. We found that the mRNAs responsive to each LNA antimiR were significantly enriched for sex-biased genes, which comprised 40% (126/317) of LNA-miR-802 responsive genes and 55% (131/239) of LNA-miR-1948 responsive genes compared with 8.2% of all liver-expressed genes (LNA-miR-802, ES = 7.4, P < 0.0001; LNA-miR-1948, ES = 13.6, P < 0.0001). However, the sets of LNA-responsive genes shared little overlap with the targets of each miRNA predicted by TargetScan (Fig. 5B). Further, we observed a striking, statistically significant difference in the distribution of male- vs female-biased mRNAs dysregulated by each LNA (Fig. 5C and 5D). Thus, at the 3-day time point, LNA-miR-1948 upregulated (derepressed) 96 female- but only 4 male-biased mRNAs in male liver. Further, LNA-miR-1948 treatment led to downregulation of 27 male- but only 4 female-biased mRNAs. The opposite pattern was seen in LNA-miR-802–treated female liver: LNA-miR-802 upregulated (derepressed) 35 male- compared with only 4 female-biased mRNAs, and it downregulated 85 female- but only 2 male-biased mRNAs. These same patterns were seen for mRNAs that showed a weak sex bias (Fig. 5C). The strong enrichment of sex-biased genes among the targets of LNA-miR-1948 and LNA-miR-802 supports our hypothesis that these two miRNAs, by virtue of their sex-differential expression, contribute to the sex-biased expression of many liver-expressed mRNAs: the female-biased miR-802 represses male-biased mRNAs while upregulating other female-biased mRNAs in female liver; and the male-biased miR-1948 represses female-biased mRNAs while upregulating other male-biased mRNAs in male liver (Fig. 5D). Many of the LNA-responsive sex-biased genes showed a moderate sex-bias (less than twofold to threefold), and many of these genes showed expression changes of ∼twofold following LNA treatment (Supplemental Tables 3B and 3C).
Figure 5.
Functional analysis of miR-802-5p and miR-1948-5p. (A) Scheme: LNA sequences complementary to miR-802-5p and to miR-1948-5p were injected into female and male mice, respectively. LNA with a random sequence (LNA-negative) was used as control. The injected mice were euthanized 3 or 6 days after the first LNA injection on day 0, and liver RNA was analyzed by RNA-seq. (B) Venn diagrams showing the overlap of the mRNAs responsive to each LNA antimiR (239 LNA-miR-1948-responsive RNAs and 317 LNA-miR-802-responsive RNAs) (green) with the following two gene sets: the full set of 990 liver sex-biased genes (red) (Supplemental Table 3A), and the top 300 TargetScan-predicted targets of each miRNA (yellow). The LNA-responsive mRNAs were significantly enriched for sex-biased mRNAs, as indicated. (C) Table showing the numbers of sex-biased mRNAs, including weakly sex-biased mRNAs (see Materials and Methods) that were significantly upregulated or downregulated by each LNA antimiR at the indicated time point. Supplemental Table 3A shows the full lists of LNA-responsive mRNAs and their sex bias. (D) Stacked bar graphs of the data in panel C showing the strikingly distinct distributions of male- vs female-biased mRNAs that were upregulated vs downregulated by each LNA. Weakly sex-biased mRNAs responsive to each LNA were included, as shown. The sex bias of the upregulated vs downregulated mRNAs was significant at P < 0.001 (Fisher exact test). The sex bias of the distribution was also significant for LNA-miR-802-5p at day 3 (P = 0.0014), but not for LNA miR-1948-5p at day 6 (P = 0.21) (not shown).
Target genes of miR-1948 and miR-802
Examination of the full set of 239 miR-1948 target RNAs identified by LNA-miR-1948-5p (Supplemental Table 3B) revealed the presence of 20 Cyps P450, including 12 female-biased Cyps whose RNA was induced (i.e., derepressed) in the LNA-treated male liver. Further, the set of 317 miR-802-5p targets (Supplemental Table 3C) included 14 Cyps, of which 8 were female-biased RNAs repressed by LNA treatment. Interestingly, three of the female-biased Cyp RNAs were targeted by both miRNAs, but in opposite directions. Thus, RNAs for Cyp2a4, Cyp17a1, and Cyp3a41a were downregulated by miR-1948 in male liver and were induced by miR-802 in female liver, as judged by their responses to each LNA antimiR. Consistent with these findings, DAVID Functional Annotation Clustering identified Cyp P450 monooxygenase/endoplasmic reticulum as the strongest enriched cluster for both miRNA target gene sets (ES = 10.2 for miR-1948; ES = 7.0 for miR-802) (Supplemental Table 4). Immunity was the second strongest functional annotation cluster for the set of miR-1948 targets (28 genes, ES = 7.0) and included many interferon and other innate immune response genes. miRNA sequences were also targets of both sex-biased miRNAs (9 miRNAs were targets of miR-1948; 22 miRNAs were targets of miR-802) (Supplemental Table 3). This suggests that miR-1948 and miR-802 both belong to larger miRNA regulatory networks.
Discussion
We used small RNA-seq to identify 24 miRNAs that show sex-biased expression in mouse liver, and for two of these miRNAs, miR-1948 and miR-802, we investigated their hormonal regulation and their functional roles in young adult mouse liver. Our findings show that the male pattern of plasma GH pulse-stimulated liver STAT5 activity is essential for the sex-biased expression of both miR-1948 and miR-802, which are respectively activated and repressed in a GH/STAT5-dependent manner in male mouse liver. Further, LNA antimiR experiments demonstrated that miR-1948-5p and miR-802-5p, the major products of each miRNA locus, are each functional in regulating >100 downstream sex-biased mRNAs.
We initially identified miR-802 and miR-1948 as miRNAs of interest based on the sex-biased chromatin accessibility of the genomic regions harboring each miRNA gene, as determined by DNase hypersensitivity analysis (38). This analysis was based on the rationale that the presence of sex-biased open chromatin sites nearby a miRNA gene is likely to indicate sex-dependent regulation in cis by transcription factor(s) that bind at open chromatin and activate the miRNA promoter or a nearby enhancer in a sex-biased manner (46). miR-802 and miR-1948 were both confirmed by qPCR and by small RNA-seq to show significant sex bias in expression in the liver. Two other miRNA genes nearby sex-biased open chromatin, miR-21a and miR-455, were also confirmed to show sex-biased expression by RNA-seq. Analysis of liver miR-802 and miR-1948 levels in mouse models with alterations in GH signaling showed that the expression of both miRNAs in male liver is regulated by sex-dependent plasma GH stimulation and by the GH pulse-activated transcription factor STAT5. Thus, perturbation of the GH-stimulated liver STAT5 signaling axis by ablation of pituitary GH secretion (hypophysectomy) (15), by genetic deletion of STAT5a and STAT5b (STAT5-KO) (36), or by elimination of the GH-free periods between GH pulses [continuous GH infusion (16); and somatostatin-KO mice (49)] all led to downregulation of miR-1948 and derepression of miR-802 in male liver, but had no effect in female liver. These responses to treatments that alter hepatic GH signaling in male but not female liver are consistent with those seen for many but not all sex-biased mouse liver mRNAs (6).
We used several approaches to validate the authenticity of miR-1948 to rule out the possibility that it represents a spurious hairpin RNA, given its intronic origin, its low conservation in species other than rat, and the lack of prior characterization. Small RNA-seq analysis established that miR-1948-5p is >50-fold more abundant than its 3p arm counterpart, miR-1948-3p, indicating that the miRBase annotation that miR-1948-3p is the major form is incorrect for mouse liver. Small RNA-seq analysis of HEK293 cells overexpressing the miR-1948 hairpin together with ∼400 bp flanking sequence showed the same, highly asymmetrical processing of miR-1948, confirming the strong 5p arm bias seen in mouse liver. Specific interaction with mouse liver Argonaute was demonstrated for miR-1948 and for the conserved and better characterized miR-802 (52, 53), supporting the functionality of both miRNAs in mouse liver. These analyses illustrate the importance of directly validating a miRNA in a given experimental system, especially when the miRNA is not widely conserved. It is often presumed that such nonconserved miRNAs are nonfunctional. Our findings with miR-1948 challenge this idea, based on computational, biochemical, and functional analyses showing that miR-1948, although not conserved beyond the rat (Fig. 4A), does, in fact, play a functional role in regulating liver mRNA levels. Conceivably, human-specific miRNAs could play a functional role in human liver equivalent to that of miR-1948 in mouse liver.
GH regulates the liver transcriptome by both direct and indirect mechanisms. Sex-biased, GH-regulated genes with regulatory potential are candidate mediators of the indirect effects of GH on downstream sex-biased genes, and efforts to identify such indirect-acting factors have led to the discovery of GH/STAT5 target genes, including the sex-biased transcriptional repressors BCL6 (10, 46, 66) and CUX2 (17, 67). Our functional inhibition studies using LNAs complementary to miR-1948 and miR-802 identify these two GH-regulated miRNAs as sex-biased repressive factors that mediate a portion of the indirect effects of GH on downstream sex-biased mRNAs. Further, some of the effects of these miRNAs are likely to be indirect, as we observed both upregulation and downregulation of sex-biased RNAs when using LNAs to specifically inhibit each miRNA.
The question of miRNA function has been easy to answer for some miRNAs, such as miR-1 and miR-124 (68), which downregulate many target genes, and difficult for others, for which miRNA perturbation studies show no gross phenotypes or molecular effects (69, 70). This may reflect the built-in redundancy of complex miRNA networks, where each mRNA is regulated by multiple miRNAs (27), such that deletion or inhibition of one miRNA is compensated by the remaining miRNAs (71). Phenotypes associated with some miRNAs only become apparent when stress is applied to a biological system, when the ability of the system to maintain homeostasis is compromised (72). Thus, miRNAs may buffer the cell from transcriptional fluctuations, sharpen cell transitions, and generally confer robustness to biological systems (22, 73). Therefore, although functional inhibition of a sex-biased miRNA might induce clear-cut reciprocal effects on sex-biased target mRNAs, it could alternatively result in modest and perhaps variable changes in target mRNA levels for miRNAs that belong to a redundant miRNA network or play a subtle role in buffering transcriptional changes. To establish the functionality of miR-1948-5p and miR-802-5p, we used LNA-based antisense miRNA sequences to inhibit each sex-biased mRNA in vivo. We found that the set of mRNAs dysregulated by LNA targeting each sex-biased miRNA was significantly enriched for sex-biased mRNAs, and further, the sex-biased mRNAs responsive to each LNA showed a highly sex-biased distribution. Thus, in male liver, LNA-miR-1948-5p treatment upregulated (derepressed) female-biased mRNAs and downregulated male-biased mRNAs, whereas in female liver, LNA-miR-802-5p treatment upregulated male-biased mRNAs and downregulated female-biased mRNAs. It is uncertain why the sex-biased effects of LNA-miR-1948-5p were only apparent 3 days after LNA injection, whereas those of LNA-miR-802-5p were most striking after 6 days, but differences in the pharmacokinetics of each LNA could be a factor.
We identified comparatively few low P value LNA-responsive mRNAs, which could result from a number of factors. First, miRNAs primarily act by mRNA destabilization via poly-A shortening (74) and translational repression (75), and generally exert subtle repression of target mRNAs, typically less than twofold (21), as was seen in our studies. Unless such repressive effects are amplified by feedback and feed-forward loops, subtle changes in mRNA levels will be observed and may not be effectively captured by RNA-seq; even if detected, variation in the extent of repression between individual mice may result in high P values. Second, the impact of miRNA inhibition often is not pronounced unless stress is applied. Here, miRNA inhibition was performed in mice given LNAs under native conditions, where loss of a single miRNA may be compensated by built-in redundancies in the sex-biased gene miRNA regulatory network. Nevertheless, the substantial enrichment of the LNA anti-miRNA targets for sex-biased mRNAs, and the major differences in the directionality of responsiveness of male- vs female-biased mRNAs to both LNA-miR-1948 and LNA-miR-802 (Fig. 5) provide strong support for our conclusion that both sex-biased miRNAs are functional in regulating sex-biased mRNA levels in mouse liver. Although downregulation of mRNAs by miRNAs is most common, miRNAs are also known to stabilize certain mRNAs via direct and indirect mechanisms (76). The loss of such stabilization may explain why many female-biased mRNAs were downregulated by the miR-802 LNA antimiR in female liver, and, similarly, why many male-biased mRNAs were downregulated by the miR-1948 LNA antimiR in male liver.
The poor overlap between gene targets identified by LNA in vivo inhibition and the in silico predictions by TargetScan is not unexpected (45) and suggests that in vivo targets identified by inhibitory LNAs are not confined to mRNAs with 3′-UTR seed matches. This is consistent with the notion that many noncanonical mechanisms of miRNA targeting occur in animal cells. These include heterogeneity in miRNA 5′ processing (57), which generates different miRNA isoforms with different seed sequences, as seen in our analyses (imperfect seed pairing, noncanonical binding sites located in 5′ UTR, or exons that are outside of canonical 3′UTR, seedless interactions, indirect targets, and context-dependent targeting) (77–79). Further, some of the miRNA targets identified here could be indirect targets, as suggested by our finding that 22 other miRNA sequences were dysregulated by LNA-miR-802-5p and 9 other miRNAs in the case of LNA-miR-1948-5p. Several biochemical methods have been developed to identify miRNA-mRNA interactions directly (e.g., crosslinking, ligation, and sequencing of hybrids), in which miRNA-mRNA interactions in Argonaute pull-downs are ligated and can be detected by sequencing the resultant chimeric reads (80). Future studies may investigate the miRNA interactome in male and female liver to further characterize mRNA targets of miR-802-5p and miR-1948-5p, as well as those of the other sex-biased liver miRNAs that we identified (Table 2).
While this manuscript was under review, a study was published identifying miR-1948 as male-biased and miR-802 as female-biased in expression in adult mouse liver; however, no functional or hormonal regulatory studies were reported (81). miR-802-5p was much more abundant than miR-802-3p, consistent with our findings; however, miR-1948-5p and miR-1948-3p were similarly expressed, in contrast to the predominance of miR-1948-5p in our study. Conceivably, this discrepancy could result from age or strain differences; however, those details were not provided in (81).
In conclusion, miR-802 and miR-1948 are sex-biased, GH/STAT5-regulated miRNAs that are functional in young adult mouse liver. The targets of these sex-biased miRNAs include many sex-biased mRNAs, with substantial enrichment for Cyps P450 and other mRNAs important for microsomal/endoplasmic reticulum-based metabolism. Although the majority of GH-responsive genes are regulated at the transcriptional level through the action of GH-dependent transcription factors that bind DNA at chromatin sites in a sex-biased chromatin state (46), this study demonstrates that GH also exerts posttranscriptional control via sex-biased regulatory miRNAs, which may confer robustness to the complex network of sex-differentiated genes that underlie the widespread sex differences in liver metabolism and liver disease susceptibility (82).
Supplementary Material
Acknowledgments
Financial Support: This work was supported in part by National Institutes of Health Grant DK033765 and the Office of Research on Women's Health, National Institutes of Health (to D.J.W.).
Author Contributions: P.H. and D.J.W. conceived of the study jointly. P.H. designed and carried out all of the experimental work. Data analysis and writing of the manuscript was carried out jointly by P.H. and D.J.W. D.J.W. supervised the overall project and edited the manuscript.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- CPM
counts per million
- Cyp
cytochrome
- DHS
DNase hypersensitive site
- ES
enrichment score
- FDR
false discovery rate
- FPKM
fragments per read per kilobase
- GH
growth hormone
- IgG
immunoglobulin G
- IP
immunoprecipitation
- KO
knockout
- LNA
locked nucleic acid
- LNA-antimiR
microRNA inhibitory locked nucleic acid sequence
- miRNA
microRNA
- mRNA
messenger RNA
- nt
nucleotide
- PBS
phosphate-buffered saline
- qPCR
quantitative polymerase chain reaction
- RNA-seq
RNA sequencing
- RRID
Research Resource Identifier
- SDS
sodium dodecyl sulfate
- UTR
untranslated region
References
- 1.Waters MJ, Brooks AJ. Growth hormone and cell growth. Endocr Dev. 2012;23:86–95. [DOI] [PubMed] [Google Scholar]
- 2.Rotwein P. Mapping the growth hormone--Stat5b--IGF-I transcriptional circuit. Trends Endocrinol Metab. 2012;23(4):186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray PG, Higham CE, Clayton PE. 60 years of neuroendocrinology: the hypothalamo-GH axis: the past 60 years. J Endocrinol. 2015;226(2):T123–T140. [DOI] [PubMed] [Google Scholar]
- 4.Adams JM, Otero-Corchon V, Hammond GL, Veldhuis JD, Qi N, Low MJ. Somatostatin is essential for the sexual dimorphism of GH secretion, corticosteroid-binding globulin production, and corticosterone levels in mice. Endocrinology. 2015;156(3):1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansson JO, Edén S, Isaksson O. Sexual dimorphism in the control of growth hormone secretion. Endocr Rev. 1985;6(2):128–150. [DOI] [PubMed] [Google Scholar]
- 6.Waxman DJ, Holloway MG. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol. 2009;76(2):215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steyn FJ, Huang L, Ngo ST, Leong JW, Tan HY, Xie TY, Parlow AF, Veldhuis JD, Waters MJ, Chen C. Development of a method for the determination of pulsatile growth hormone secretion in mice. Endocrinology. 2011;152(8):3165–3171. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Bekaert AJ, Dupont J, Rouve S, Annesi-Maesano I, De Magalhaes Filho CD, Kappeler L, Holzenberger M. Exploring endocrine GH pattern in mice using rank plot analysis and random blood samples. J Endocrinol. 2011;208(2):119–129. [DOI] [PubMed] [Google Scholar]
- 9.Das RK, Banerjee S, Shapiro BH. Growth hormone: a newly identified developmental organizer. J Endocrinol. 2017;232(3):377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Laz EV, Waxman DJ. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol Cell Biol. 2012;32(4):880–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi HK, Waxman DJ. Plasma growth hormone pulse activation of hepatic JAK-STAT5 signaling: developmental regulation and role in male-specific liver gene expression. Endocrinology. 2000;141(9):3245–3255. [DOI] [PubMed] [Google Scholar]
- 12.Choi HK, Waxman DJ. Growth hormone, but not prolactin, maintains, low-level activation of STAT5a and STAT5b in female rat liver. Endocrinology. 1999;140(11):5126–5135. [DOI] [PubMed] [Google Scholar]
- 13.Waxman DJ, Ram PA, Park SH, Choi HK. Intermittent plasma growth hormone triggers tyrosine phosphorylation and nuclear translocation of a liver-expressed, Stat 5-related DNA binding protein. Proposed role as an intracellular regulator of male-specific liver gene transcription. J Biol Chem. 1995;270(22):13262–13270. [DOI] [PubMed] [Google Scholar]
- 14.Connerney J, Lau-Corona D, Rampersaud A, Waxman DJ. Activation of male liver chromatin accessibility and STAT5-dependent gene transcription by plasma growth hormone pulses. Endocrinology. 2017;158(5):1386–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wauthier V, Sugathan A, Meyer RD, Dombkowski AA, Waxman DJ. Intrinsic sex differences in the early growth hormone responsiveness of sex-specific genes in mouse liver. Mol Endocrinol. 2010;24:667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau-Corona D, Suvorov A, Waxman DJ. Feminization of male mouse liver by persistent growth hormone stimulation: Activation of sex-biased transcriptional networks and dynamic changes in chromatin states. Mol Cell Biol. 2017;37(19):e00301–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conforto TL, Zhang Y, Sherman J, Waxman DJ. Impact of CUX2 on the female mouse liver transcriptome: activation of female-biased genes and repression of male-biased genes. Mol Cell Biol. 2012;32(22):4611–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rinn JL, Rozowsky JS, Laurenzi IJ, Petersen PH, Zou K, Zhong W, Gerstein M, Snyder M. Major molecular differences between mammalian sexes are involved in drug metabolism and renal function. Dev Cell. 2004;6(6):791–800. [DOI] [PubMed] [Google Scholar]
- 19.Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20:1333–1351. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Klein K, Sugathan A, Nassery N, Dombkowski A, Zanger UM, Waxman DJ. Transcriptional profiling of human liver identifies sex-biased genes associated with polygenic dyslipidemia and coronary artery disease. PLoS One. 2011;6(8):e23506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5(5):396–400. [DOI] [PubMed] [Google Scholar]
- 25.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. [DOI] [PubMed] [Google Scholar]
- 27.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24(10):992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung L, Gustavsson C, Norstedt G, Tollet-Egnell P. Sex-different and growth hormone-regulated expression of microRNA in rat liver. BMC Mol Biol. 2009;10(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwekel JC, Vijay V, Han T, Moland CL, Desai VG, Fuscoe JC. Sex and age differences in the expression of liver microRNAs during the life span of F344 rats. Biol Sex Differ. 2017;8(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie X, Miao L, Yao J, Feng C, Li C, Gao M, Liu M, Gong L, Wang Y, Qi X, Ren J. Role of multiple microRNAs in the sexually dimorphic expression of Cyp2b9 in mouse liver. Drug Metab Dispos. 2013;41(10):1732–1737. [DOI] [PubMed] [Google Scholar]
- 33.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. [DOI] [PubMed] [Google Scholar]
- 34.Conforto TL, Waxman DJ. Sex-specific mouse liver gene expression: genome-wide analysis of developmental changes from pre-pubertal period to young adulthood. Biol Sex Differ. 2012;3(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holloway MG, Laz EV, Waxman DJ. Codependence of growth hormone-responsive, sexually dimorphic hepatic gene expression on signal transducer and activator of transcription 5b and hepatic nuclear factor 4alpha. Mol Endocrinol. 2006;20:647–660. [DOI] [PubMed] [Google Scholar]
- 36.Holloway MG, Cui Y, Laz EV, Hosui A, Hennighausen L, Waxman DJ. Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology. 2007;148(5):1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luque RM, Kineman RD. Gender-dependent role of endogenous somatostatin in regulating growth hormone-axis function in mice. Endocrinology. 2007;148(12):5998–6006. [DOI] [PubMed] [Google Scholar]
- 38.Ling G, Sugathan A, Mazor T, Fraenkel E, Waxman DJ. Unbiased, genome-wide in vivo mapping of transcriptional regulatory elements reveals sex differences in chromatin structure associated with sex-specific liver gene expression. Mol Cell Biol. 2010;30(23):5531–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan AR. BEDTools: the Swiss-Army Tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;. 47:11.12.11–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The ViennaRNA web services. Nucleic Acids Res. 2008;36:W70-W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green M, Loewenstein PM. Human adenoviruses: propagation, purification, quantification, and storage. Curr Protoc Microbiol. 2006;Chapter 14:Unit 14C.1. [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. [DOI] [PubMed] [Google Scholar]
- 45.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugathan A, Waxman DJ. Genome-wide analysis of chromatin states reveals distinct mechanisms of sex-dependent gene regulation in male and female mouse liver. Mol Cell Biol. 2013;33(18):3594–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther. 2015;15(2):142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desvignes T, Batzel P, Berezikov E, Eilbeck K, Eppig JT, McAndrews MS, Singer A, Postlethwait JH. miRNA nomenclature: a view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet. 2015;31(11):613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Low MJ, Otero-Corchon V, Parlow AF, Ramirez JL, Kumar U, Patel YC, Rubinstein M. Somatostatin is required for masculinization of growth hormone-regulated hepatic gene expression but not of somatic growth. J Clin Invest. 2001;107(12):1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5(4):395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruggieri A, Barbati C, Malorni W. Cellular and molecular mechanisms involved in hepatocellular carcinoma gender disparity. Int J Cancer. 2010;127(3):499–504. [DOI] [PubMed] [Google Scholar]
- 52.Kornfeld JW, Baitzel C, Könner AC, Nicholls HT, Vogt MC, Herrmanns K, Scheja L, Haumaitre C, Wolf AM, Knippschild U, Seibler J, Cereghini S, Heeren J, Stoffel M, Brüning JC. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494(7435):111–115. [DOI] [PubMed] [Google Scholar]
- 53.Wu X, Gong Z, Sun L, Ma L, Wang Q. MicroRNA-802 plays a tumour suppressive role in tongue squamous cell carcinoma through directly targeting MAP2K4. Cell Prolif. 2017;50(3):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hickey G, Hinrichs AS, Hubley R, Karolchik D, Learned K, Lee BT, Li CH, Miga KH, Nguyen N, Paten B, Raney BJ, Smit AF, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43(Database issue):D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17(12):1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H, Ye C, Ramirez D, Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS One. 2009;4(10):e7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4(2):76–84. [DOI] [PubMed] [Google Scholar]
- 59.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ekimler S, Sahin K. Computational methods for microRNA target prediction. Genes (Basel). 2014;5(3):671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akhtar MM, Micolucci L, Islam MS, Olivieri F, Procopio AD. Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 2016;44(1):24–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kauppinen S, Vester B, Wengel J. Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handb Exp Pharmacol. 2006(173):405–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Straarup EM, Fisker N, Hedtjärn M, Lindholm MW, Rosenbohm C, Aarup V, Hansen HF, Ørum H, Hansen JB, Koch T. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38(20):7100–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyer RD, Laz EV, Su T, Waxman DJ. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol Endocrinol. 2009;23:1914–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laz EV, Holloway MG, Chen CS, Waxman DJ. Characterization of three growth hormone-responsive transcription factors preferentially expressed in adult female liver. Endocrinology. 2007;148(7):3327–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. [DOI] [PubMed] [Google Scholar]
- 69.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3(12):e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19(R2):R169–R175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25(3):137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. [DOI] [PubMed] [Google Scholar]
- 73.Posadas DM, Carthew RW. MicroRNAs and their roles in developmental canalization. Curr Opin Genet Dev. 2014;27:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. [DOI] [PubMed] [Google Scholar]
- 76.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3(3):311–330. [DOI] [PubMed] [Google Scholar]
- 77.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. [DOI] [PubMed] [Google Scholar]
- 78.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69(8):3245–3248. [DOI] [PubMed] [Google Scholar]
- 79.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153(3):654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Helwak A, Tollervey D. Mapping the miRNA interactome by cross-linking ligation and sequencing of hybrids (CLASH). Nat Protoc. 2014;9(3):711–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Warnefors M, Mössinger K, Halbert J, Studer T, VandeBerg JL, Lindgren I, Fallahshahroudi A, Jensen P, Kaessmann H. Sex-biased microRNA expression in mammals and birds reveals underlying regulatory mechanisms and a role in dosage compensation. Genome Res. 2017;27(12):1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Humphries C. Sex differences: luck of the chromosomes. Nature. 2014;516(7529):S10–S11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.