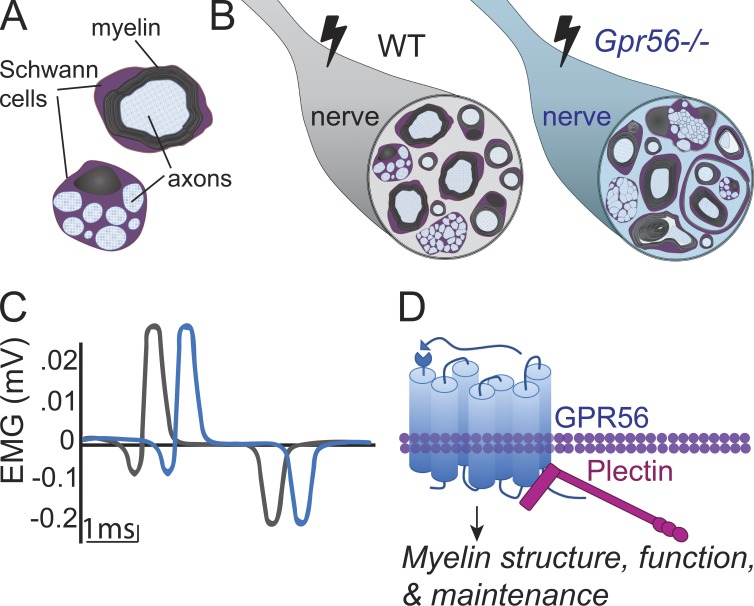
This study identifies GPR56/ADGRG1 as a conserved regulator of peripheral nervous system myelination and defines a novel interacting partner of GPR56: plectin. This study is directly relevant to human health because mutations in GPR56 and PLECTIN cause diseases with neuropathic symptoms.
Graphical Abstract
Abstract
Myelin is a multilamellar sheath generated by specialized glia called Schwann cells (SCs) in the peripheral nervous system (PNS), which serves to protect and insulate axons for rapid neuronal signaling. In zebrafish and rodent models, we identify GPR56/ADGRG1 as a conserved regulator of PNS development and health. We demonstrate that, during SC development, GPR56-dependent RhoA signaling promotes timely radial sorting of axons. In the mature PNS, GPR56 is localized to distinct SC cytoplasmic domains, is required to establish proper myelin thickness, and facilitates organization of the myelin sheath. Furthermore, we define plectin—a scaffolding protein previously linked to SC domain organization, myelin maintenance, and a series of disorders termed “plectinopathies”—as a novel interacting partner of GPR56. Finally, we show that Gpr56 mutants develop progressive neuropathy-like symptoms, suggesting an underlying mechanism for peripheral defects in some human patients with GPR56 mutations. In sum, we define Gpr56 as a new regulator in the development and maintenance of peripheral myelin.
Introduction
Myelin, the lipid-rich insulation that ensures rapid nerve conduction velocity, is generated by specialized glial cells: oligodendrocytes in the central nervous system (CNS) and Schwann cells (SCs) in the peripheral nervous system (PNS; Nave and Trapp, 2008). Disruption of the myelin sheath in patients with peripheral neuropathies (e.g., Charcot-Marie-Tooth disease [CMT]) causes severe symptoms, including pain and neuronal damage that can ultimately lead to paralysis (Harel and Lupski, 2014). Proper myelination is required for a high quality of life in humans, yet the molecular mechanisms governing myelinating glial cell development and myelination are poorly understood.
Recent works from our laboratory and others have identified the adhesion class of G protein-coupled receptors (aGPCRs) as key regulators of myelinating glial cell development (Monk et al., 2009, 2011; Glenn and Talbot, 2013; Mogha et al., 2013; Shin et al., 2013a; Liebscher et al., 2014; Ackerman et al., 2015; Giera et al., 2015; Petersen et al., 2015; Küffer et al., 2016; Salzman et al., 2016). aGPCRs represent the second-largest class of GPCRs in humans, yet remain largely understudied because of their genomic and structural complexity (Hamann et al., 2015). In brief, mature aGPCRs exist at the cell membrane as bipartite receptors consisting of an N-terminal fragment (NTF) facilitating adhesion and a C-terminal fragment (CTF) containing the seven-transmembrane pass signaling domain, and modulation of NTF–CTF interactions is associated with receptor activation (Hamann et al., 2015). Using a transcription activator-like effector nuclease–induced mutant allele of the aGPCR Gpr56/Adgrg1 in zebrafish (gpr56stl13/stl13), we previously determined that Gpr56 regulates proliferation of oligodendrocyte precursor cells (Ackerman et al., 2015). Analysis of published expression datasets revealed that Gpr56 is also robustly expressed in SCs (Miller et al., 2006; Weiss et al., 2016). Given the importance of Gpr126 (a closely related aGPCR) in regulating SC biology (Monk et al., 2009, 2011; Glenn and Talbot, 2013; Mogha et al., 2013; Liebscher et al., 2014; Petersen et al., 2015; Küffer et al., 2016) and the established role of GPR56 in oligodendrocyte precursor cells (Ackerman et al., 2015; Giera et al., 2015; Salzman et al., 2016), we sought to determine if GPR56 also regulates myelinating glial development in the PNS.
Here, we use zebrafish and rodent models to define the role of Gpr56 in SC development and myelination. Whereas loss of Gpr56 from the CNS results in hypomyelination because of a reduction in oligodendrocyte precursor cell proliferation and mature oligodendrocyte numbers, we find no apparent defect in SC proliferation or overall SC number in Gpr56 mutants. Instead, we demonstrate that GPR56 functions through RhoA in early SC development to regulate proper radial sorting of axons by SCs and that disrupted radial sorting in Gpr56 mutants likely results from altered cytoskeletal remodeling. Additionally, we find that loss of Gpr56 from mature nerve results in multiple myelin structural abnormalities, including increased myelin thickness, aberrant myelin domain organization, shortened internode length, and failed myelin maintenance. Finally, we identify plectin as a novel interacting partner of GPR56 in SCs. Plectin is a large cytoskeletal linker protein that causes multiple diseases (plectinopathies) when mutated in humans (Winter and Wiche, 2013). The most common plectinopathy is epidermolysis bullosa simplex with muscular dystrophy, a rare skin-blistering disorder that presents with late-onset muscular dystrophy, but mutations in PLECTIN have also been linked to defects in peripheral myelin ultrastructure (Bauer et al., 2001). We find that plectin physically interacts with GPR56, that plectin levels are up-regulated in the absence of Gpr56 (as expected for a receptor-ligand pair), and that Plectin mutant SCs phenocopy Gpr56 mutant SCs, suggesting that these proteins function together to regulate SC biology. Together, our studies define GPR56 as a novel and evolutionarily conserved regulator of peripheral nerve development and function with important implications for human health and disease.
Results
GPR56 regulates SC radial sorting via Gα12/13 and RhoA signaling
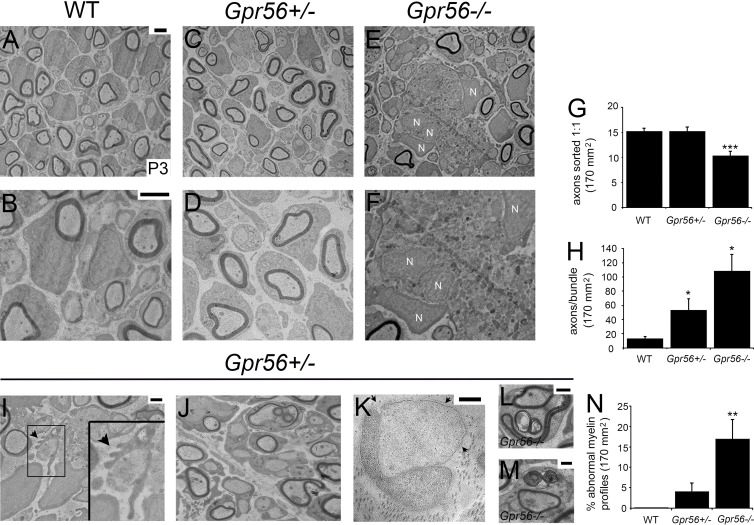
Radial sorting is the process by which immature SCs encompass bundles of naked axons and sort individual axonal segments into a 1:1 SC/axon segment relationship. The timing of radial sorting is essential; when radial sorting is impaired, both myelination and peripheral nerve maturation are delayed (Jessen and Mirsky, 2005; Feltri et al., 2016). Analysis of peripheral nerves from Gpr56 mouse mutants (Li et al., 2008) revealed radial sorting delays by transmission EM (TEM; Fig. 1). At postnatal day (P) 3 in Gpr56 mouse mutant sciatic nerves (SNs; n = 4 mutant mice), the number of axons sorted 1:1 with SCs was significantly reduced relative to WT (n = 3) and heterozygous (n = 3) siblings (Fig. 1 G; P < 0.0002, one-way ANOVA). Additionally, we observed abnormal cytoplasmic protrusions from sorting SCs (indicative of disrupted cytoskeletal rearrangements, Fig. 1, I and J), basal lamina trails reflecting aberrant SC process extension and retraction during radial sorting (Fig. 1 K), along with other myelin abnormalities in the SNs of Gpr56 mutant mice (Fig. 1, L–N; P < 0.002, one-way ANOVA). Deficient SC precursor proliferation can cause radial sorting delays (Feltri et al., 2016), and GPR56 is a known regulator of cell division (Wu et al., 2013; Bae et al., 2014; Ackerman et al., 2015; Giera et al., 2015); thus, we first tested if Gpr56 mutant SCs were less proliferative than WT. Interestingly, SC proliferation and total number were unaffected in Gpr56 mouse mutant SNs, as was overall peripheral (saphenous) nerve size (Fig. S1). Therefore, in contrast to its role in oligodendrocyte precursor cells, Gpr56 is dispensable for SC proliferation.
Figure 1.
GPR56 is required for efficient radial sorting. (A–F) TEM images of SN from (A and B) WT (n = 3), (C and D) Gpr56+/− (n = 3), and (E and F) Gpr56−/− mice (n = 4) on P3. SC precursor nuclei (N) surrounding and within axon bundles in Gpr56−/− nerves. Higher magnification images: B, D, and F. Bars, 2 µm. (G) Quantification of axons sorted 1:1 with SCs (***, P < 0.0002, one-way ANOVA). (H) Quantification of the number of axons per unsorted bundle (*, P < 0.04, one-way ANOVA). (I–M) TEM images of P3 SN Gpr56−/− (n = 4) animals. Bars: (I, J, L, and M) 1 µm; (I inset and K) 500 nm. (I) Large bundles of unsorted axons surrounded by SC processes are observed in a Gpr56−/− SN. Some SCs also exhibited abnormal cytoplasmic protrusions (arrowhead). Higher magnification of the boxed in region shown within the inset. In addition, (J, L, and M) myelin abnormalities (outfoldings) and (K) nearly naked axons associated with SCs and partially surrounded by SC basal lamina (arrows) were often observed. (N) Quantification of the percent of axons with myelin abnormalities revealed a significant increase in myelin defects in Gpr56−/− nerves compared with WT controls (**, P < 0.002, one-way ANOVA). (G, H, and N) Error bars represent SEM.
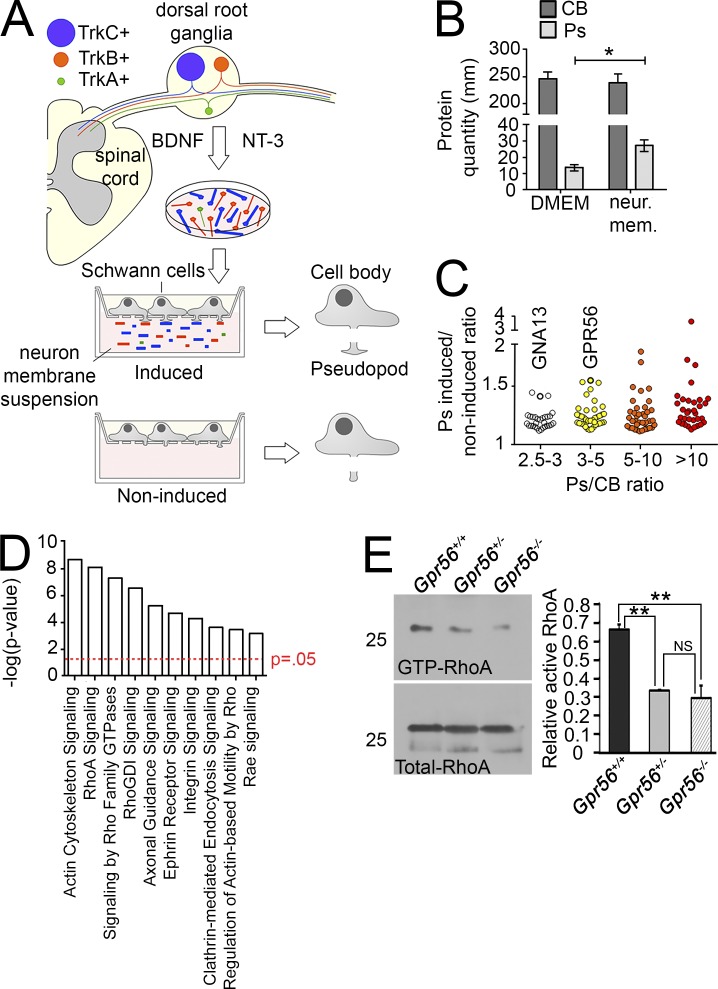
These findings suggest that GPR56 does not regulate radial sorting by affecting SC numbers. Given the cytoplasmic defects observed on P3 (Fig. 1, I and J), we hypothesized that GPR56 may function in controlling dynamics of immature SC protrusions and/or adhesion to axonal membranes. To test this, we used a modified Boyden chamber to mimic axon–SC interactions during radial sorting (Poitelon et al., 2015). In brief, purified rat SCs were seeded within the upper compartment of a Boyden chamber in which neuronal fragments were suspended below to induce SC “pseudopod” (mimicking cytoplasmic protrusion) growth. Pseudopods were then collected, processed, and analyzed by long-gradient nano reversed-phase liquid chromatography–mass spectrometry (LC-MS; Fig. 2, A and B; see Materials and methods for full details). Indeed, we determined that GPR56 is highly enriched in SC pseudopods (Fig. 2 C and Dataset 1), supporting the hypothesis that GPR56 autonomously regulates SC cytoplasmic protrusion dynamics during radial sorting.
Figure 2.
GPR56 and interacting proteins are enriched in pseudopods after induction by membranes of DRG neurons dependent on BDNF and NT-3. (A) Schematic representation of the pseudopod assay. DRG were dissected from E15.5 rats. Neurons were selected with BDNF and NT-3 growth factors. After purification, neurons were collected and homogenized. SCs were induced or not with neuronal membrane suspension. Pseudopods (Ps) were isolated from cell bodies (CBs), and both were analyzed by MS. (B) Quantification by bicinchoninic acid assay of the protein content of SC pseudopods and CBs after induction. Data are shown as mean ± SD; n (independent experiment) = 3. *, P < 0.05. (C) Distribution of proteins enriched by neuronal membranes. The abscissa axis indicates relative polarization of a protein between P and CB. The ordinate axis indicates relative enrichment of a protein by neuronal membranes in comparison to noninduced condition. Both GPR56 and GNA13 are found enriched in SC Ps after neuronal membranes induction. (D) Protein ontology analysis of the pathways most enriched in SC Ps. RhoA signaling is enriched by neuronal membranes. For more complete listings, see Dataset 1. Student’s t test (unpaired) was used to test for statistical significance. (E) GTP-RhoA pull-down assay revealed a significant decrease in the proportion of GTP-RhoA (active) to total RhoA in Gpr56−/− and Gpr56+/− animals relative to WT controls on P6 (**, P < 0.01, unpaired Student’s t test; data are shown as mean ± SEM; n = 4 nerves from n = 2 animals per technical replicate; n = 3 technical replicates).
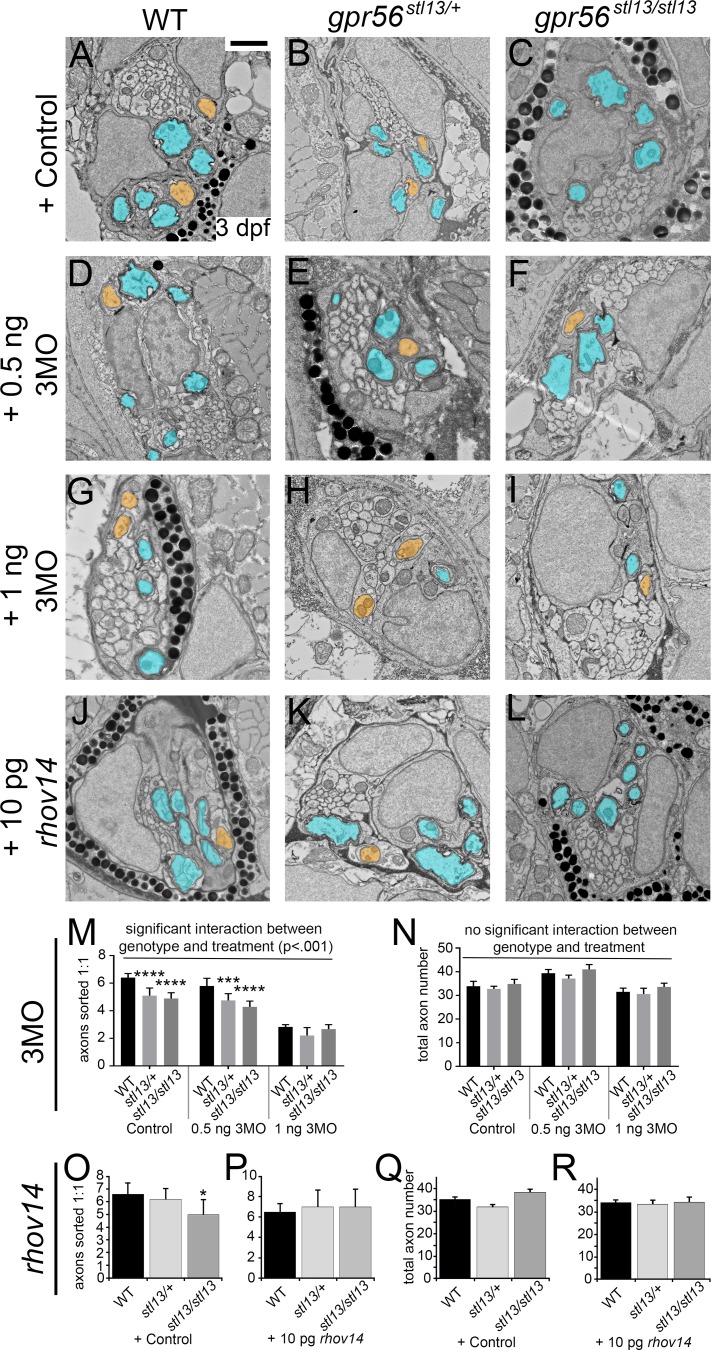
In the nervous system, GPR56 primarily signals through Gα12/13 and RhoA (Iguchi et al., 2008; Luo et al., 2011; Ackerman et al., 2015; Giera et al., 2015), and indeed, Gα13 (protein symbol GNA13) and RhoA signaling proteins are also enriched in SC pseudopods (Fig. 2, C and D; and Dataset 1). Further, we found a significant reduction in the relative concentration of active, GTP-bound RhoA in Gpr56 mutant nerves relative to WT sibling nerves on P6. We also detected a significant decrease in active, GTP-bound RhoA in heterozygous nerves relative to WT (Fig. 2 E, n = 12 nerves per genotype across three replicates), consistent with the observation that Gpr56+/− animals exhibit some mutant phenotypes onP3 (Fig. 1 H; P < 0.039, one-way ANOVA). To test whether GPR56 functions through Gα12/13 and RhoA to promote radial sorting, we used standard genetic manipulations to either enhance (via inhibition of Gα12/13) or suppress (via transient overexpression of constitutively active RhoA) radial sorting defects in gpr56stl13/stl13 mutant zebrafish (Lin et al., 2009; Ackerman et al., 2015). Strikingly, injection of antisense morpholinos that block translation of the three zebrafish transcripts encoding Gα12/13 (3MO) strongly enhanced radial sorting defects in the posterior lateral line nerve (pLLn) of gpr56stl13/stl13 mutants and induced radial sorting defects in heterozygote and WT siblings in a dose-dependent manner (P < 0.001, two-way ANOVA; Fig. 3, A–I and M). Conversely, transient overexpression of synthetic mRNA encoding constitutively active RhoA (rhov14) completely suppressed radial sorting defects in gpr56stl13/stl13 mutants, suggesting a positive role for RhoA in radial sorting via Gpr56 signaling (control WT vs. control gpr56stl13/stl13: P < 0.023, unpaired Student’s t test; rhov14 injected WT vs. rhov14 injected gpr56stl13/stl13: P < 0.58, unpaired Student’s t test; Phenol red: n = 3; WT: n = 3 stl13+/−, n = 4 stl13−/−; +rhov14: n = 3 animals per genotype; Fig. 3, J–L, O, and P). Total axon number was not affected by experimental conditions throughout (Fig. 3, N and Q–R). The importance of RhoA in maintenance of cell–cell contact points has been established (Parsons et al., 2010), suggesting that sorting delays in Gpr56 mutants may not result from impaired process extension but rather from an inability of SCs to maintain secure contacts with target axons during radial sorting. This hypothesis is consistent with the aforementioned signs of SC process retraction observed in P3 mouse mutant SNs (Fig. 1 K).
Figure 3.
Gpr56 signals through Gα12/13 and RhoA to promote radial sorting. (A–I) TEM images of the pLLn of WT, gpr56stl13/+, and gpr56stl13/stl13 larvae injected with (A–C) phenol red control (n = 6 animals per genotype), (D–F) 0.5 ng 3MOs targeting Gα12/13 signaling (3MO = gna12-MO, gna13a-MO, and gna13b-MO; n = 6 WT, n = 5 gpr56stl13/+, and n = 5 gpr56stl13/stl13 animals examined), or (G–I) 1 ng 3MO (n = 3 animals per genotype). (J–L) In a separate series of experiments, animals were injected with constitutively active rhov14 (rhov14: n = 3 animals per genotype; control: n = 3 each WT and gpr56stl13/+, n = 4 gpr56stl13/stl3). Sorted axons are pseudocolored orange. Myelinated axons are pseudocolored blue. (A–L) Bar, 1 µm. (M) Two-way ANOVA analysis revealed a significant interaction between genotype and treatment for 3MO experiments (****, P < 0.0001). Tukey’s multiple comparisons test was also performed and found a significant reduction in the number of sorted axons between control WT embryos and control heterozygous (****, P < 0.0001) as well as control mutant embryos (****, P < 0.0001). A significant decrease in sorted axon number was observed between 0.5-ng 3MO-treated WT and -treated heterozygous (***, P < 0.001) and -treated mutant embryos (****, P < 0.0001). No significant comparisons were found in the 1-ng 3MO condition. (N) Total axon number is unaffected by genotype or treatment (two-way ANOVA). (O) Quantification of the number of axons sorted 1:1 in phenol red controls (P < 0.03, unpaired Student’s t test) compared with (P) rhov14-injected embryos (no significant differences). (Q and R) Total axon number was unchanged by test condition (unpaired Student’s t test). Data reflect two independent experimental replicates. (M–R) Error bars represent SEM.
Myelin domain organization is impaired in Gpr56 mutants
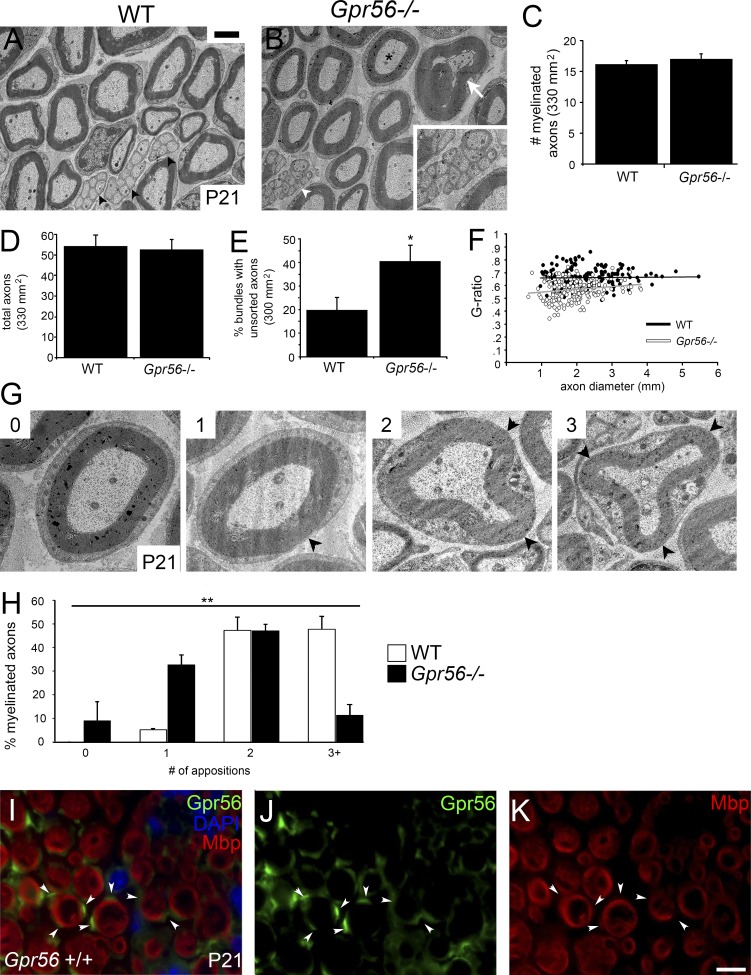
We’ve defined that GPR56 signaling through RhoA is required for timely radial sorting of axons during peripheral nerve development. Although this phenotype did not persist through adulthood (Fig. 4, A–D; n = 3 animals per genotype, P < 0.57, unpaired Student’s t test; Fig. S2, A–D), we noted several other pronounced alterations in mature peripheral nerve. First, we observed Remak bundle defects. Remak bundles are groups of small caliber axons sorted and individually encompassed by a single nonmyelinating SC. In Gpr56 mutants, Remak bundle morphology was impaired, such that interdigitation of SC processes between axons within a single bundle was incomplete (Fig. 4 E, P < 0.027, one-way ANOVA). In addition, we noted several deviations in mature myelin ultrastructure. Myelin thickness was significantly increased in Gpr56 mouse mutant SNs (Fig. 4 F; reflected in lower mean G-ratio, ∼180 fibers analyzed from three animals per genotype, P < 0.002, one-way ANOVA), and we noted myelin outfoldings commonly associated with hypermyelination (Fig. 4 B).
Figure 4.
GPR56 regulates mature myelin ultrastructure and domain organization. (A and B) TEM images of WT (n = 3) and Gpr56 mutant mouse (n = 3) SNs on P21. Bar, 2 µm. White arrow denotes myelin outfolding. Black arrowheads denote morphologically WT Remak bundles, which are incompletely formed in Gpr56 mutants (B, white arrowhead/inset). The asterisk marks an axon with no appositions. (C–E) Quantification of (C) total number of myelinated axons (P < 0.45, one-way ANOVA), (D) overall axon number (P < 0.81, one-way ANOVA), and (E) percentage of bundles containing axons that are not fully sheathed in Gpr56 mutants and WT sibling nerves (*, P < 0.03, one-way ANOVA). Error bars represent SEM. (F) Plots of G-ratios in mutants and WT controls (P < 0.002, one-way ANOVA). (G) TEM images of Gpr56−/− myelinated axons with 0, 1, 2, or 3 appositions. (H) Distribution of the percentage of myelinated axons (mean ± SD) per apposition number in WT (white bars) and Gpr56−/− SNs (black bars, **, P < 0.002, one-way ANOVA). (I–K) IHC showing GPR56 (green), DAPI (nuclear stain, blue), and MBP (marker of mature myelin) localization on P21 SNs. In WT SNs, GPR56 is detected in pockets (arrowheads) outside of MBP-positive rings but absent from mature axons. (J) GPR56 stain alone. (K) MBP stain alone. Bar, 5 µm.
In the PNS, myelin internodes possess two primary structural domains: Cajal bands (longitudinal channels of SC cytoplasm) and appositions (where the outermost layer of myelin contacts the SC plasma membrane; Sherman and Brophy, 2005). Strikingly, we also observed defects in the suborganization of peripheral myelin into Cajal bands and appositions in Gpr56 mouse mutant SCs (Fig. 4 G; 154 WT and 168 Gpr56−/− fibers from three animals per genotype, P < 0.002, one-way ANOVA), as well as SC cytoplasmic abnormalities in gpr56stl13/stl13 zebrafish mutant SCs (Fig. S2 B). The function of Cajal bands is poorly understood because few proteins have been linked to Cajal band formation (Court et al., 2004, 2009; Sherman et al., 2012; Walko et al., 2013). Of these proteins, Periaxin has been best studied; Periaxin stabilizes SC appositions, and Periaxin mutations cause neuropathy-like phenotypes in mouse and CMT disease in humans (Guilbot et al., 2001). Although Periaxin is well characterized for its role in CMT, mutations in PLECTIN (a cytolinker protein shown to regulate SC domain organization) are similarly associated with myelin maintenance defects in mice and neuropathy-like symptoms in humans (Walko et al., 2013; Winter and Wiche, 2013), demonstrating the importance of myelin domain architecture for sustained PNS health in humans. Interestingly, analysis by immunohistochemistry (IHC) revealed that in mature SNs, GPR56 is specifically localized to discrete cytoplasmic domains on the abaxonal SC membrane (Fig. 4, I–K) and, thus, poised to regulate Cajal band formation.
Altered expression of cytoskeletal genes in developing and mature Gpr56 mutant nerves
GPR56 signals via RhoA to regulate radial sorting (Figs. 1, 2, and 3), but the downstream signaling pathway(s) through which GPR56 regulates the development of mature myelin are not known. To determine what signaling pathway(s) may underlie the phenotypes observed in Gpr56 mutants during SC maturation in contrast to early SC development (radial sorting), we performed RNA sequencing (RNA-seq) on RNA isolated from SNs of P21 and P3 mutants and WT controls, respectively (n = 3 per genotype on P21; n = 4 per genotype on P3; Fig. S3). Importantly, we observed a significant decrease in the expression of Gpr56 at both developmental time points (Fig. S3 F). In accordance with the hypothesis that GPR56 normally functions in SC pseudopods to promote stable cell-cell contacts during radial sorting, and with the abnormal SC cytoplasmic extensions observed in Gpr56 mutant SCs on P3 (Fig. 1, I and J), we observed a significant increase in the expression of genes associated with dynamic actin filaments in mutant SN harvested on P3 during active radial sorting (Fig. S3 D). We also found a statistically significant enrichment (−log[p-value] <20) in gene ontology (GO) processes associated with actin dynamics, including (a) actin-myosin filament sliding, (b) actin-mediated cell contraction, (c) actin filament–based movement, and (d) actin filament–based process using g:Profiler (Dataset 2).
Interestingly, we also observed significant changes in expression of genes related to the cytoskeleton on P21, when Cajal band defects are pronounced in Gpr56 mutant nerves. Although we detected no differential expression of genes relating to the actin cytoskeleton, the expression of genes that promote microtubule assembly (Map2 and Ttll7) was significantly increased in mutant nerves relative to controls, whereas expression of genes that promote microtubule disassembly (e.g., Eml2) was significantly down-regulated (Fig. S3 E). We also found a statistically significant enrichment (-log[p-value] < 4) in the Cytoskeletal Remodeling GO pathway by g:Profiler (Dataset 2). The molecular mechanisms that guide Cajal band formation are poorly understood; however, microtubule disorganization is a common feature of other mutants that lack Cajal bands (Court et al., 2004). These findings suggest that precise microtubule organization is necessary for Cajal band formation and represents an important area of future research into the development of these understudied domains.
In addition to changes in the expression of genes involved in microtubule dynamics, we also observed a striking dysregulation of a cluster of genes that share the SREBP-1 binding motif. Although we observed a modest increase in expression of SREBP1 target genes on P3, by P21 we noted a significant decrease in the expression of genes that share the SREBP-1 motif. Loss of sterol regulatory element-binding factor-1c (Srebf1c) results in increased peripheral myelin thickness, Remak bundle defects, and peripheral neuropathy-like symptoms in mice (Cermenati et al., 2015). Finally, although the authors did not formally examine the role of Srebf1c in Cajal band formation, Srebf1c mutants did not present with overt Cajal band defects based on published TEM images of Srebf1c−/− peripheral nerve (Cermenati et al., 2015).
Our transcriptional profiling experiments (Fig. S3 and Dataset 2), combined with our genetic and biochemical analyses (Figs. 2 and 3) demonstrate that GPR56 signaling through Gα12/13 and RhoA is required to modify the actin skeleton for proper radial sorting during early SC development. These data further suggest that, during SC maturation, Gpr56 signals through SREBP1 pathway to regulate lipid biogenesis and Remak bundle biology and through an alternative mechanism to modify microtubule dynamics for proper Cajal band assembly.
Gpr56 is required for SC myelin maintenance
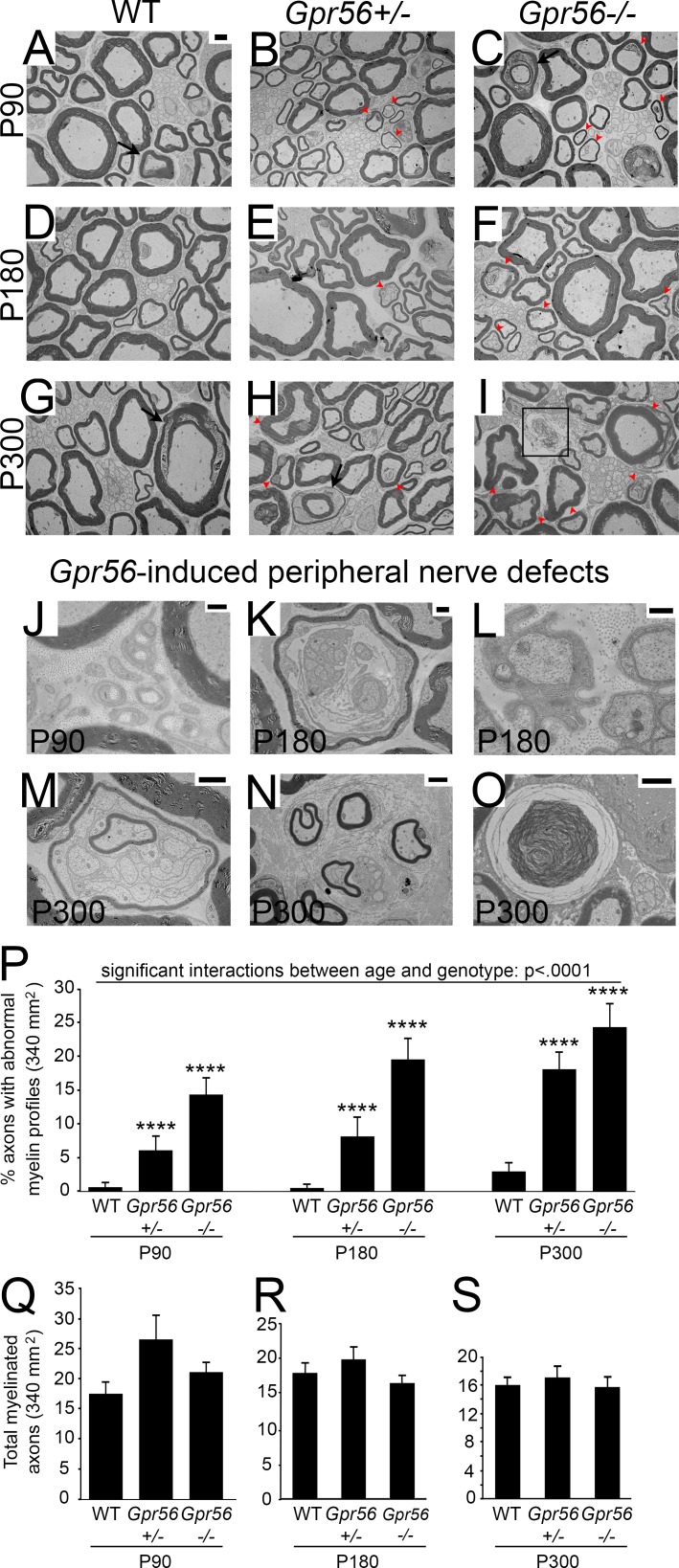
Defective organization of the myelin sheath precedes neuropathy-like symptoms in all mutants (e.g., Periaxin, Drp2, and Plectin) previously shown to have Cajal band defects (Court et al., 2004; Sherman et al., 2012; Walko et al., 2013). Gpr56 mutant mice present with Cajal band/apposition phenotypes, as well as Srebf1c-like hypermyelination and Remak SC defects (Cermenati et al., 2015); thus, we hypothesized that Gpr56 mutants would develop neuropathy-like symptoms with age as is observed in both Cajal band and Srebf1c mutants. Ultrastructural analyses of mouse SNs on P90, P180, and P300 (Fig. 5, A–I) revealed small yet significant increases in the percent of axons with abnormal myelin profiles in Gpr56 mutants and heterozygotes relative to WT siblings at each stage (n = 3 animals minimum per genotype/time point). Myelin abnormalities were composed of outfoldings and other defects: axon regeneration signatures (bands of Büngner), SCs myelinating other myelinating and Remak SCs, extensive cytoplasmic extensions, minifascicle formation, myelin debris (Fig. 5, J–O), and disrupted Schmidt-Lanterman incisures (Fig. S4, A and B). In addition, we observed a significant interaction between genotype and age with respect to myelin abnormalities (P < 0.0001, two-way ANOVA; Fig. 5 P), indicating that these phenotypes worsen with time. Axon number remained largely unchanged with age, suggesting that loss of Gpr56 does not induce widespread neuronal loss (Fig. 5, Q–S). Analysis of Gpr56 mouse mutant dorsal and ventral spinal roots, composed of purely sensory and purely motor neurons, respectively, revealed similar demyelinating signatures (Fig. S4, C–L).
Figure 5.
Gpr56 mutant mice show progressive accumulation of myelin abnormalities. (A–C) TEM images of SNs from (A) WT (n = 3), (B) Gpr56+/− (n = 3), and (C) Gpr56−/− (n = 4) mice on P90. (D–F) TEM images of SNs from (D) WT, (E) Gpr56+/−, and (F) Gpr56−/− mice on P180 (n = 3 animals per genotype). (G–I) TEM images of SNs from (G) WT, (H) Gpr56+/−, and (I) Gpr56−/− mice on P300 (n = 3 animals per genotype). Red arrowheads denote abnormal myelin profiles. Black arrows mark Schmidt-Lanterman incisures. Black box highlights myelin debris observed by P300. (A–I) Bar, 2 µm. (J–O) SC defects and pathologies observed in Gpr56 heterozygous and mutant peripheral nerves included bands of Büngner (J; bar, 500 nm), aberrant myelination of other SCs (K; bar, 2 µm), abnormal cytoplasmic protrusions (L; bar, 500 nm), myelinated Remak bundles (M; bar, 4 µm), minifascicles (N; bar, 2 µm), and myelin debris accumulation (O; bar, 2 µm). (P) Two-way ANOVA analysis revealed a significant interaction between age and genotype (****, P < 0.0001). Dunnett's multiple comparisons test also showed significant increases in the percent of axons with abnormal myelin profiles in Gpr56+/− and Gpr56−/− animals compared with WT at each time point (****, P < 0.0001). (Q–S) No significant differences between total myelinated axon number in WT and Gpr56−/− (P90: P < 0.15; P180: P < 0.44; P300: P < 0.89), nor between Gpr56+/− and Gpr56−/− (P90: P < 0.14; P180: P < 0.13; P300: P < 0.52) at any stage. Unpaired Student’s t test. (P–S) Error bars represent SEM.
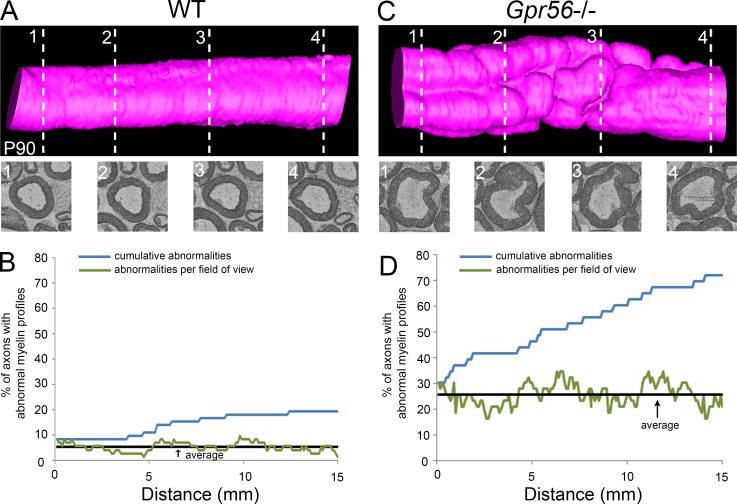
Given the importance of Gpr56 in the formation of Cajal bands, structures that span the entire internode and can reach nearly 1 mm in length in adult mice (Court et al., 2004), we hypothesized that standard TEM analysis may underestimate myelin abnormalities in Gpr56 mutants. Indeed, analysis of serial EM reconstructions of Gpr56 mouse mutant SN on P90 revealed a cumulative average of 68 ± 10% axons showing at least one myelin defect over 15 µm of internode, compared with 15 ± 6% in WT controls (Fig. 6, Fig. S5, and Videos 1, 2, 3, and 4).
Figure 6.
Gpr56 is required for myelin stability. (A–D) Serial scanning EM images (350, 100 nm apart) of WT and Gpr56−/− mouse SN on P90 (n = 3 animals per genotype) were acquired and reconstructed to generate the WT and Gpr56−/− models shown in A and C, respectively. Single sections taken from multiple locations along the nerve (1–4) are shown below each model (dashed lines show approximate locations). The percentage of axons with abnormal myelin profiles was calculated per section over a 15-µm distance for the WT (B) and Gpr56 mutant (D) reconstruction and is graphed to show the percentage of abnormalities per section (green line), the average percentage of abnormalities over all sections (black line) and the cumulative percentage of abnormalities (percentage of axons that show at least one abnormality over 15 µm [blue line]).
Gpr56 mutants display neuropathy-like symptoms with age
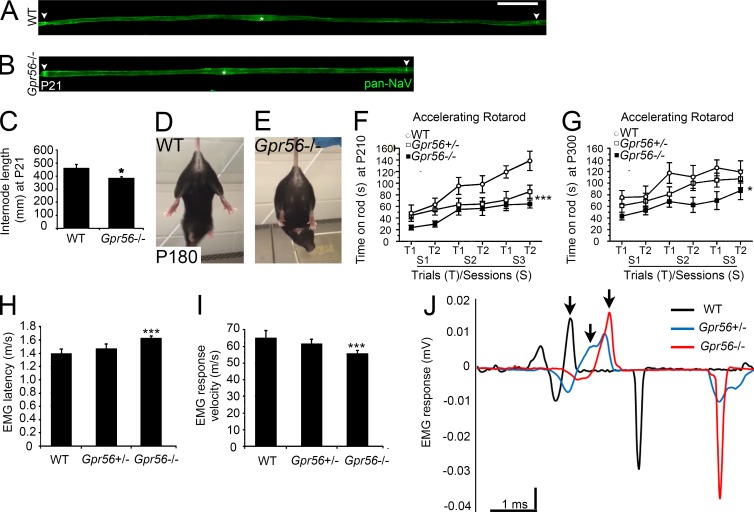
In addition to myelin loss, impaired Cajal band formation in Periaxin mutants is associated with shortened internodes, leading us to hypothesize that loss of Gpr56 would affect internode length (Court et al., 2004). Teased nerve fiber analysis revealed a significant reduction in internode length on P6 (not depicted) and on P21 in Gpr56 mutant mice (Fig. 7, A–C; P < 0.05, one-way ANOVA). Moreover, on P180, Gpr56 mutants displayed a hindlimb clasping phenotype (Fig. 7, D and E), and both Gpr56+/− (n = 16 on P180, n = 15 on P300) and Gpr56−/− mutants (n = 13 on P180, n = 12 on P300) performed poorly relative to WT controls (n = 11 on P180, n = 10 on P300) on the accelerated rotarod, indicative of impaired sensory-motor coordination and reduced grip strength (Fig. 7 F, P < 0.001; Fig. 7 G, P < 0.005; two-way ANOVA analysis). To test whether behavioral deficits observed in Gpr56 mutant and heterozygous mice reflected a reduction in nerve function, we performed electromyography (EMG) analyses in vivo and found a significant increase in EMG latency in Gpr56 mutants (n = 11) relative to controls (n = 5 WT, n = 12 Gpr56+/−) in animals aged P200–P300 (Fig. 7 H; P < 0.001, one-way ANOVA) with a corresponding reduction in response velocity (Fig. 7 I; P < 0.001, one-way ANOVA). See Fig. 7 J for example traces. Interestingly, although we observed no overt changes in Na+ channel structure by teased nerve analysis (Fig. 7, A and B), we detected a significant increase in the expression of genes related to K+ channel activity on P21 and a significant decrease in the expression of the Na+ channel clustering protein Ndrg4 (Fontenas et al., 2016) on both P3 and P21 (Fig. S3, C and D). GPR56 is also localized to sensory neurons (Fig. S4, M–O), and future work with cell type–specific knockout lines will be important for understanding the relative contributions of GPR56 in neurons versus SCs to peripheral nerve function.
Figure 7.
Gpr56 mutants display neuropathy-like symptoms. (A and B) IHC on teased nerve fibers from WT (n = 3) and Gpr56−/− (n = 4) mouse SNs on P21 stained for Pan-NaV to mark nodes of Ranvier (arrowheads). Bar, 50 µm. (C) Quantification of internode length on P21 (P < 0.05, one-way ANOVA). (D and E) Hindlimb clasping observed in Gpr56 mutants (n = 8/11) but not WT siblings on P180 (n = 1/11). (F and G) Quantification of time on rotarod to assess sensorimotor control on P180 (genotype effect: ***, P < 0.001, two-way ANOVA; n = 11 WT, n = 16 Gpr56+/−, n = 13 Gpr56−/−) and P300 (genotype effect, *, P < 0.02, two-way ANOVA; n = 10 WT, n = 15 Gpr56+/−, n = 12 Gpr56−/−). Error bars represent SEM. (H) Quantification of EMG latency (m/s) in animals aged P200–P300 (***, P < 0.001, one-way ANOVA). Error bars represent SD. (I) Quantification of EMG response velocity (m/s) in animals aged P200–P300 (***, P < 0.001, one-way ANOVA). Error bars represent SD. (J) Representative traces from EMG recordings from WT (black, n = 5), Gpr56+/− (blue, n = 12), and Gpr56−/− (red, n = 11) SNs. Arrows denote the point of the first positive deflection.
Plectin is a novel interacting partner of Gpr56 in peripheral nerve
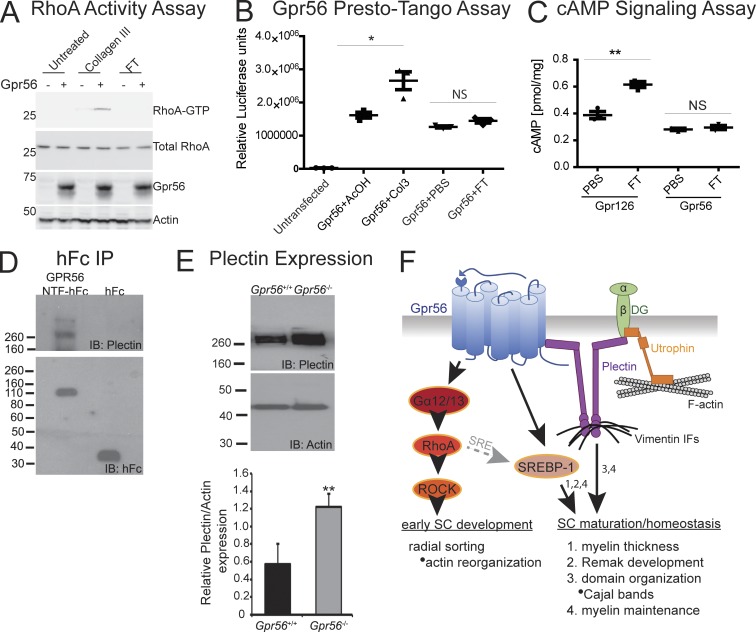
To further define how GPR56 regulates SC function, we next sought to determine which protein(s) interact with GPR56 to facilitate peripheral nerve development. We recently showed that cellular prion protein (Prnp/PrpC) is secreted from neurons and modulates GPR126 (an aGPCR closely related to GPR56) signaling and SC function. We used the previously published N-terminal flexible tail ([FT] residues 23–120) of PrPC (Küffer et al., 2016), which can activate GPR126 signaling in vitro and in vivo, to test whether PrPC can also activate Gpr56 signaling in vitro. We did not observe any significant increase in GPR56 signaling either through RhoA or through an SRE-mediated transcriptional response to FT compared with application of collagen III, a known ligand of GPR56 (Luo et al., 2011; Fig. 8, A and B; P < 0.03, unpaired Student’s t test). FT activates Gpr126 signaling through cAMP; to test whether GPR56 may signal through this alternative pathway in response to FT, we measured cAMP levels after FT addition to GPR126 or GPR56. As previously shown, FT robustly stimulates cAMP signaling through GPR126 (Küffer et al., 2016) but had no effect on GPR56 (Fig. 8 C; P < 0.0027, unpaired Student’s t test).
Figure 8.
Plectin and GPR56 physically and genetically interact in peripheral nerve. (A) GTP-RhoA assay to assess Gpr56 signaling in response to exogenous ligands. Addition of collagen III, a known activating ligand of Gpr56, to HEK 293T cells coexpressing Gpr56 result in elevation of GTP-RhoA levels over control (untransfected). Addition of prion protein (FT) does not induce Gpr56 signaling through RhoA. (B) Gpr56 Presto-Tango luciferase assay to assess Gpr56 signaling in HTLA cells. Addition of collagen III results in a significant elevation of luciferase compared with AcOH alone control (*, P < 0.05, unpaired Student’s t test). Addition of FT did not result in a significant elevation of luciferase relative to PBS controls (P = 0.0647, unpaired Student’s t test). (C) cAMP ELISA assay to assess cAMP signaling through Gpr56 and Gpr126 (positive control) after addition of FT. Addition of FT to HEK 293T cells expressing Gpr126 results in a significant elevation of cAMP (**, P < 0.01, unpaired Student’s t test) compared with PBS controls. Addition of FT to cells expressing Gpr56 does not induce cAMP signaling (P = 0.4575, unpaired Student’s t test). (A–C) Data are representative of three biologically independent experiments. Error bars represent SEM. (D) Co-IP to assess interactions between GPR56 and plectin in P5 WT SNs (n = 8 nerves from four animals per replicate; n = 3 replicates). Lane 1: plectin coimmunoprecipitates with GPR56 when the NTF of GPR56 fused to human IgG Fc (NTF-hFc) is used as a bait but not when pulling down with hFc alone (lane 2). (E) Analysis of plectin levels in P6 SNs. Plectin is significantly increased in Gpr56−/− nerves over sibling controls (**, P < 0.002, unpaired Student’s t test, n = 12 nerves from six animals; error bars represent SEM). (F) Graphic model: Gpr56 promotes function of developing and mature SCs. During early SC development, Gpr56 signals through Gα12/13 and RhoA to modulate the actin cytoskeleton for efficient radial sorting of axons. During SC maturation/homeostasis, Gpr56 functions upstream of SREBP1 to regulate myelin thickness and Remak SC biology, potentially through SRE signaling downstream of RhoA (gray dashed arrow). Concordantly, Gpr56 regulates mature myelin domain organization and myelin maintenance through direct interactions with the cytolinker protein plectin, which has been shown to physically interact with additional regulators of myelin domain organization (utrophin and β-dystroglycan). We therefore hypothesize that plectin serves as a docking center for cell signaling molecules that govern the partitioning of myelin into mature subdomains (Cajal bands and appositions) and helps generate these structures through its interactions with the cytoskeleton.
We next used an unbiased mass-spectrometry screen to determine what protein(s) physically interact with GPR56 (unpublished data). Interestingly, the top hit from this screen was plectin, a large cytolinker protein known to interact with and modulate cell surface signaling molecules through its coordinate reorganization of the actin and intermediate filament cytoskeletal networks (Winter and Wiche, 2013). PLECTIN is also mutated in a series of human diseases termed plectinopathies, including epidermolysis bullosa simplex with muscular dystrophy, a skin blistering disorder that also presents with neuropathy-like symptoms because of degeneration of intramuscular myelinated nerves (Bauer et al., 2001; Winter and Wiche, 2013). First, we verified that GPR56 and plectin physically interact in WT P5 SNs (n = 8 nerves from four animals) by coimmunoprecipitation (co-IP; Fig. 8 D). Interestingly, although we were able to detect binding of plectin to the CTF of GPR56 (not depicted), we observed robust binding of plectin to the extracellular NTF of GPR56 as well. Although this result most likely reflects binding of plectin to the larger GPR56 NTF-CTF complex, plectin has been shown to be secreted to the extracellular matrix in pathological contexts (Shin et al., 2013b). Future work will therefore be required to test whether plectin is secreted to serve as a direct activating ligand for GPR56.
We next tested whether loss of Gpr56 affects plectin expression in P6 SNs and found a significant increase in plectin levels in Gpr56 mutants (n = 7 replicates: 1 biological replicate = 2 pooled nerves from one animal) relative to WT (n = 8 replicates) and heterozygous (n = 5 replicates) siblings, as expected for a positive interacting partner of GPR56 (Fig. 8 E; P < 0.001, unpaired Student’s t test). Finally, plectin is strongly expressed in SCs and specifically localized to Cajal bands, and SC-specific KO of Plectin disrupts myelin domain organization and causes neuropathy-like symptoms with age (Walko et al., 2013). Thus, plectin is a novel binding partner for GPR56, and the presence of phenocopy coupled with both physical and genetic interactions (Fig. 8, D and E) strongly suggest that GPR56 and plectin function together to promote SC function and long-term health of peripheral nerves.
Discussion
GPR56 in myelinating glial cell development
In the CNS, oligodendrocytes form the myelin sheath that both protects and insulates axons for rapid action potential propagation (Jessen and Mirsky, 2005). We previously defined GPR56 as a novel regulator of oligodendrocyte development (Ackerman et al., 2015; Giera et al., 2015; Salzman et al., 2016). In the CNS, GPR56 regulates the timing of the transition between oligodendrocyte precursor cell proliferation and differentiation. In the absence of GPR56, oligodendrocyte precursor cells prematurely exit the cell cycle resulting in fewer numbers of myelinating oligodendrocytes; however, the myelinating oligodendrocytes that are produced generate phenotypically normal myelin (Ackerman et al., 2015; Giera et al., 2015). In the PNS, SCs ensure long-term health of peripheral axons (Jessen and Mirsky, 2005). In contrast to the role of GPR56 in regulating the numbers of myelinating glia in the CNS, GPR56 does not affect SC proliferation or overall numbers. Instead, we observe novel and distinct functions of GPR56 in SC development (radial sorting) and in SC myelination (thickness, organization, and maintenance), and our data suggest defects in cytoskeletal dynamics as underlying causes of both early and late functions of GPR56 in peripheral nerve.
RhoA-dependent regulation of radial sorting by GPR56
During early SC development, we find that GPR56 signaling is required for timely sorting of axons. Many proteins have been shown to function during radial sorting, including small Rho-GTPases (Nodari et al., 2007; Pereira et al., 2009; Feltri et al., 2016). RhoA is robustly expressed during radial sorting (Taylor et al., 2003; Melendez-Vasquez et al., 2004; Nodari et al., 2007) and specifically enriched in SC pseudopods. The function of RhoA in SCs has not been thoroughly examined, however, given the established role of the related Rho GTPase Rac in radial sorting (Nodari et al., 2007; Pereira et al., 2009; Feltri et al., 2016). Rac positively regulates the formation/protrusion of lamellipodia-like structures from SCs, whereas RhoA activation typically inhibits process extension (Pereira et al., 2009). Indeed, up-regulation of Rho kinase (ROCK; a major downstream effector of RhoA signaling) in ILK SC-specific KO mice results in severe radial sorting delays because of impaired process extension (Pereira et al., 2009). In contrast, we find that reduced RhoA signaling in Gpr56 mutants likewise impairs radial sorting, which can be restored upon transient overexpression of constitutively active RhoA.
These results indicate that RhoA levels must be very tightly regulated during SC development, because both increased signaling in ILK mutants impairs SC process extension, and decreased signaling in Gpr56 mutants delays radial sorting theoretically through impaired SC adhesion to axonal membranes. In accordance with this hypothesis, a previous study demonstrated that inhibition of ROCK in vitro results in aberrant actin fiber organization and reduced SC adhesion because of loss of stress fibers (Melendez-Vasquez et al., 2004). ROCK is a serine–threonine kinase that regulates actin organization through phosphorylation of proteins that modulate cytoskeletal dynamics (Amano et al., 1996). Myosin II light chain is a direct target of ROCK, functions to stabilize and increase the contractility of actin-myosin filaments (Tan et al., 1992), and is required for ROCK-dependent reorganization of actin fibers in SCs (Melendez-Vasquez et al., 2004). RNA-seq of Gpr56 mutant nerves during radial sorting (P3) revealed a significant dysregulation of genes that modify the actin and actomyosin cytoskeleton, including Myosin light chain 1 (Myl1, also known as Mhc1/Mhc3), Myosin Light Chain, Phosphorylatable, Fast Skeletal Muscle (Mylpf, also known as Mlc2b), and Mybpc1, a myosin binding protein. These findings support the hypothesis that GPR56 signals through RhoA and ROCK to modify the actin and actomyosin cytoskeleton to establish firm SC-axon contacts during radial sorting (Fig. 8 F, working model).
Gpr56 regulates peripheral myelin structure and maintenance
Although radial sorting recovers in Gpr56 mutants with time, we show that loss of GPR56 leads to multiple defects in mature SCs. First, we observed a significant increase in myelin thickness despite the delay in myelination onset. RNA-seq analysis of mutant nerves on P21 (postradial sorting defects) revealed a significant decrease in the expression of a cluster of SREBP1 target genes. SREBP1 is a transcription factor that regulates membrane lipid composition, and deletion of the primary isoform of SREBP1 (Srebp1c) results in hypermyelination, incomplete separation of axons by Remak SCs, and neuropathy-like symptoms with age (Cermenati et al., 2015). Although the signaling pathways upstream of SREBP1 activity are poorly defined, analysis of the promoter region of human and mouse SREBP1 revealed SRE binding sites (Eberlé et al., 2004). GPR56-dependent RhoA signaling is known to function upstream of SRE to regulate neural precursor migration (Iguchi et al., 2008), suggesting that GPR56 may also signal through this cascade to regulate SREBP1 expression and in turn, myelin thickness and Remak SC dynamics (Fig. 8 F, working model).
In addition to changes in myelin thickness and Remak SC biology, we also observed a striking defect in mature myelin domain organization in Gpr56 mutants—a significant reduction in Cajal band number. Mature myelin is longitudinally organized into two primary structural domains: Cajal bands and appositions. Cajal bands are cytoplasmic channels separated from one another by regions of completely compact SC membrane called appositions. The molecular mechanisms that drive Cajal band development are poorly understood, in part because few proteins have been linked to Cajal band development. Mutations in the large intracellular cytolinker protein PLECTIN in humans are linked to peripheral nerve defects (Winter and Wiche, 2013), and a recent study in mice defined plectin as a key regulator of myelin domain organization (Walko et al., 2013). In SCs, plectin is specifically localized to Cajal bands where it interacts with β-dystroglycan and utrophin to restructure vimentin intermediate filaments in the cytoskeleton (Walko et al., 2013). Interestingly, β-dystroglycan is also a member of the Periaxin–DRP2–dystroglycan complex that stabilizes appositions for proper formation of Cajal bands (Sherman and Brophy, 2005). Here, we define plectin as a novel interacting protein of GPR56: GPR56 and plectin physically interact, and loss of Gpr56 results in up-regulation of plectin, as expected if plectin mediates GPR56 downstream signaling.
Plectin has now been shown to interact with proteins that are specifically enriched in Cajal bands (utrophin and GPR56) as well β-dystroglycan, which is present in both Cajal bands and in appositions as part of the Periaxin–DRP2–dystroglycan complex. Furthermore, plectin is known to function as a scaffold for receptors that mediate intracellular signaling (Castañón et al., 2013). Together, these data suggest that that in myelinating SCs, plectin could serve as a docking center for both structural (Periaxin, DRP2, and utrophin) and signaling (GPR56 and β-dystroglycan) molecules required for efficient compartmentalization and long-term integrity of the myelin sheath (Fig. 8 F, working model). Although the downstream signaling pathways that promote peripheral myelin organization remain elusive, our RNA-seq analysis on P21 (when changes in Cajal bands are present in Gpr56 mutant nerves) revealed significant dysregulation of genes that regulate microtubule dynamics. Disorganization of the microtubule cytoskeleton is present in Periaxin mutants, which also present with Cajal band defects (Court et al., 2004), supporting the hypothesis that reorganization of microtubules is required for mature myelin domain assembly.
Neuropathy-like symptoms in GPR56 mutants: links to human health
Several of the developmental defects observed in Gpr56 mutants (e.g., hypermyelination and reduced Cajal bands) are known to precede disruptions in myelin integrity (Court et al., 2004, 2009; Sherman et al., 2012; Walko et al., 2013; Cermenati et al., 2015), and indeed, myelin maintenance is also defective in Gpr56 mutants relative to controls. We demonstrate that in Gpr56−/− mouse adults, impaired myelin stability coupled with shortened internode length results in behavioral deficits consistent with the recorded EMG latency delay in Gpr56 mutants. Similar phenotypes are often noted in mouse models of peripheral neuropathy and in human neuropathy patients (Martini, 1999), suggesting that mutations in GPR56 could cause neuropathy-like symptoms in humans. Mutations in GPR56 cause several neurological disorders affecting the CNS, including bilateral perisylvian polymicrogyria (BPP or BPPR; OMIM #615752). BPP is a cortical brain malformation resulting in severe neurological impairments, including epilepsy and oromotor apraxia, and a minority of BPP patients also present with arthrogryposis multiplex congenital and exhibit EMG signatures consistent with PNS pathology (Poduri et al., 2010). Our work defines GPR56 as a novel, evolutionarily conserved regulator of SC development and myelin maintenance and suggests a mechanism underlying PNS defects observed in BPP patients.
Materials and methods
Zebrafish
Stocks and rearing conditions
Zebrafish (Danio rerio) were maintained in the Washington University Zebrafish Consortium facility. All experiments were performed in compliance with Washington University's institutional animal care and use committee. Embryos were collected from pairwise matings and reared at 28.5°C in egg water (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, and 0.33 mM MgSO4). Embryos and larvae were staged in days postfertilization (dpf) by using morphological markers (Kimmel et al., 1995). The gpr56stl13/stl13 line was genotyped as described previously (Ackerman et al., 2015).
3MO and synthetic mRNA injection
3MOs targeting gna12, gna13a, and gna13b (3MO) are previously published (Lin et al., 2009; Ackerman et al., 2015) and were provided by F. Lin (University of Iowa, Iowa City, IA). 3MOs were combined and diluted in sterile water supplemented 1:10 with phenol red dye and then injected at a final concentration of 0.5 ng each in a total volume of 1 nl or 1 ng each in a total volume of 2 nl. To control for adverse side effects from the injection process, we also injected control siblings with an equal volume of phenol red diluted 1:10 in water (phenol red–injected controls). Constitutively active Rho (rhov14) was provided by L. Solnica-Krezel (Washington University, St. Louis, MO) and was transcribed and injected as described previously (Lin et al., 2009; Ackerman et al., 2015).
TEM
For zebrafish larvae, TEM was performed as described (Czopka and Lyons, 2011) by using a PELCO BioWave Pro with SteadyTempTM Digital Plus water-recirculating system. Microwave-assisted fixation was used for all steps before Epon treatment. In brief, embryos were fixed in modified Karnovsky’s fix (4% paraformaldehyde, 2% glutaraldehyde, and 0.1 M sodium cacodylate, pH 7.4) and left overnight at 4°C. After fixation, embryos were postfixed in 2% osmium tetroxide in 0.1M sodium cacodylate and 0.1M imidazole, pH 7.4, washed, and treated with saturated uranyl acetate. Embryos were then dehydrated by using increasing concentrations of ethanol followed by 100% acetone and then infiltrated overnight in an acetone-Epon mixture. Embryos were then transferred to 100% Epon, embedded in molds, and baked for at least 48 h at 65°C. Fish were examined at approximately the same body segment level to control for variability along the anterior/posterior axis. For preparation of pLLns from 6-mo-old adult fish, we anesthetized animals in an ice-water bath and severed the hindbrain before dissecting a portion of the anterior pLLn, and fixation of adult tissue occurred as described previously (Monk et al., 2009). In brief, adult tissues were fixed via microwave-assisted fixation in modified Karnovsky’s fix as earlier in this paragraph and then postfixed in 2% osmium tetroxide in 0.1M sodium cacodylate, washed, and dehydrated in increasing concentrations of ethanol followed by 100% propylene oxide. The tissues were then submerged in a 2:1 mixture of propolyene oxide/EPON for 1 h, followed by a 1:1 mixture overnight. The next day, the tissues were transferred to 100% Epon before embedding and baking at 65°C. Thin sections (70 nm) for all samples were mounted on mesh grids and stained with 8% uranyl acetate followed by Sato’s lead stain. We viewed sections on a Jeol (JEM-1400) electron microscope, and images were acquired with an AMT V601 digital camera. All images were processed and analyzed by using ImageJ and Adobe Photoshop software. At 3 dpf, quantifications were performed on the entire nerve. For rhov14 transient overexpression experiments, we examined five nerves from three WT, five nerves from three gpr56stl13/+, and seven nerves from four gpr56stl13/stl13 larvae for phenol red controls; and six nerves from three WT, six nerves from three gpr56stl13/+, and five nerves from three gpr56stl13/stl13 larvae injected with 10 pg rhov14 synthetic mRNA. For 3MO experiments, we examined 10 nerves from 6 WT, 10 nerves from 6 gpr56stl13/+, and 9 nerves from 6 gpr56stl13/stl13 larvae for phenol red control treatment. For larvae treated with 0.5 ng 3MO, we examined 10 nerves from 6 WT, 8 nerves from 5 gpr56stl13/+, and 7 nerves from 5 gpr56stl13/stl13 animals. For larvae treated with 1 ng 3MO, we examined six nerves from three WT, five nerves from three gpr56stl13/+, and six nerves from n = 3 gpr56stl13/stl13 animals. For larval studies at 5 dpf, we examined seven nerves from four WT and six nerves from three gpr56stl13/stl13 animals. For adult studies, three nerves from three individuals at 6 mo of age for WT and gpr56stl13/+, and four nerves from three gpr56stl13/stl13 animals were analyzed.
Mice
Lines and rearing conditions
All mouse (Mus musculus) experiments were performed in compliance with Washington University's Institutional Animal Care and Use Committee as well as the Animal Care and Use Committee at Boston Children’s Hospital. The Gpr56 knockout mice were obtained pure on C57BL/6 (n = 11 generations) from the Mutant Mouse Regional Resource Centers (032342-UCD), maintained on a C57BL/6 background, and genotyped as described previously (Li et al., 2008).
SC cultures and preparation of neuronal membranes
Primary rat SCs were prepared from SN on P3 as described (Feltri et al., 1992). SCs were maintained in SC medium: DMEM (Gibco), 1% FCS, 2 mM l-glutamine, 2 µM Forskolin (Calbiochem), and 2 ng/ml recombinant human NRG1-β 1 (R&D) and passaged not more than three times.
Neuronal membranes were prepared from dorsal root ganglia (DRG) of rat embryos at 15 gestational days. In brief, 20 DRGs were plated on 9.5-cm2 dishes in C media: MEM (Gibco), 10% FBS, 4 g l-1 d-glucose, 2 mM l-glutamine, 25 ng/ml BDNF (PeproTech), and 10 ng/ml NT-3 (PeproTech). After 12 h, cells were switched for 2 wk in NB medium (Gibco), 1X B27 supplement (Gibco), 4 g/l d-glucose, 2 mM l-glutamine, 25 ng/ml BDNF (PeproTech), and 10 ng/ml NT-3 (PeproTech). To obtain neuron-only cultures, the cells were treated with FUDR (10 µM floxuridine and 10 µM uridine) in NB medium for three cycles (2 d with FUDR, 2 d without FUDR). Neurons were collected by scraping the network with forceps and homogenized in PBS with a Dounce homogenizer. The homogenized solution was centrifuged at 300 g for 20 min to remove debris and collagen. The supernatant was centrifuged at 30,000 g for 1 h. The pellets were resuspended in DMEM (67 µl for each 9.5-cm2 dish) and stored at −80°C for 1 mo. To test their activity, membranes were centrifuged onto serum-starved, rat primary SCs, incubated at 37°C for an additional 20 min, lysed, and analyzed by Western blotting for Akt activation.
Pseudopod assay
Pseudopod assay was performed as described previously (Poitelon et al., 2015). In brief, starved SCs were plated onto the upper compartment of the Boyden chamber insert (3414; Corning). Cells were maintained in DMEM, 2 mM l-glutamine and 0.5% BSA for 4 h at 37°C. Then pseudopods were allowed to grow for 2 h at 37°C by replacing bottom-chamber DMEM with neuronal membrane suspension. After induction, inserts were washed twice in PBS, fixed with ice-cold methanol for 30 min on ice, and washed again in PBS. For each filter, either the cell bodies on the upper surface or the pseudopods on the undersurface were removed with a cotton swab.
Sample preparation for shotgun proteomics for SC pseudopod assay
Microporous filters with either pseudopods or cell bodies were manually cut into tiny pieces and suspended in 200 µl of cold lysis buffer (50 mM Tris-FA, 150 mM NaCl, 0.5% sodium deoxycholate, 2% SDS, 2% IGEPAL CA-630 [Sigma-Aldrich], pH 8.0) supplemented with protease/phosphatase inhibitor cocktail tablets (Roche Applied Science). The samples were sonicated until the liquid turned clear and were then placed on ice for 30 min, followed by a 30-min centrifugation at 20,000 g under 4°C. Supernatant from all samples was carefully transferred to Eppendorf tubes, and protein concentrations were measured by bicinchoninic acid assay kit (Pierce Biotechnology, Inc.).
For each extraction, 100 µg of protein was subjected to a surfactant-aided precipitation/on-pellet digestion procedure (An et al., 2015) to generate tryptic peptides for LC-MS analysis. Proteins first undergo reduction and alkylation by 3 mM tris(2-carboxyl)phosphine and 20 mM iodoacetamide. Both steps were performed under 37°C for 30 min in darkness, with constant oscillation in an Eppendorf Thermomixer (Eppendorf). Proteins were then precipitated by addition of eight volumes of chilled acetone, and the samples were incubated under −20°C overnight. Precipitated proteins were collected by centrifugation at 20,000 g under 4°C for 30 min, and the supernatant portion was removed. This step allowed the elimination of detergents and unwanted contaminants (e.g., phospholipids), which will compromise the analytical quality of the LC-MS system. Protein pellets were rinsed with 500 µl of chilled acetone/water mixture (85:15, vol/vol percentage), air-dried for 5 min, and resuspended in 80 µl Tris-FA. Trypsin from porcine pancreas (Sigma-Aldrich) at a concentration of 0.25 µg/μl was added to the samples in two steps, at a final enzyme/substrate ratio of 1:20 (wt/wt percentage). The samples were incubated under 37°C overnight (18–20 h) with constant oscillation to achieve complete proteolytic cleavage. The digestion step was terminated by addition of 1 µl formic acid, after which the samples were centrifuged at 20,000 g under 4°C for 30 min and transferred to LC vials.
Long-gradient nano reversed-phase LC-MS for SC pseudopod assay
The nano RPLC system used in the present study consists of a Spark Endurance autosampler (Emmen) and an ultrahigh-pressure Eksigent Nano-2D Ultra capillary/nano LC system. A nano LC/nanospray setup featuring low-void-volume and high-chromatographic reproducibility was used so that comprehensive separation of complex peptide samples could be achieved. Mobile phase A was 0.1% formic acid in 2% acetonitrile, whereas mobile phase B was 0.1% formic acid in 88% acetonitrile. Peptides were first loaded onto a large inner-diameter trap (300 µm inner diameter × 1 cm, packed with Zorbax 3M C18 material) with 1% B at 10 µl min−1, and the trap was washed for 3 min before bringing in line with the nano LC flow path. A series of optimized gradients (250 nl/min) was used to back-flush the trapped peptides onto the nano LC column (75 µm inner diameter × 100 cm, packed with Pepmap 3M C18 material), which was heated homogenously to 52°C to improve chromatographic resolution and reproducibility. The 7-h gradient profile used for extensive peptide separation was listed as follows: 3–8% B over 15 min, 8–24% B over 215 min, 24–38% B over 115 min, 38–63% B over 55 min, 63–97% B in 5 min, and finally isocratic at 97% B for 15 min.
An LTQ Oribtrap XL mass spectrometer (Thermo Fisher Scientific) was used for peptide detection. Reproducibility of ionization efficiency was maintained via rinsing and conditioning of the nanospray needle (by dripping 50% methanol after every three runs). The instrument was carefully calibrated for mass accuracy and Fourier transform transmission. One complete scan cycle consisted of one MS1 survey scan (m/z 310−2,000) at a resolution of 60,000 and seven subsequent MS2 scans by collision-induced dissociation activation mode to fragment the top seven most-abundant precursors in the survey scan. The automatic gain control target was set to 8 × 106, which allowed highly sensitive detection with little compromise to mass accuracy and resolution. Dynamic exclusion was enabled with the following settings: repeat count = 1, repeat duration = 30 s, exclusion list size = 500, and exclusion duration = 40 s. The activation time was 30 ms with an isolation width of 3 D for ion trap mass spectrometry; the normalized activation energy was 35%, and the activation q was 0.25. All 20 samples were randomly analyzed.
Protein identification and quantification for SC pseudopod assay
LC-MS.raw files were matched to a combined Swiss-Prot Mus musculus and Rattus norvegicus protein database because of the incomprehensiveness of the current rat protein database. Files were matched by SEQUEST by using a target-decoy search strategy, in which a concatenated protein database with both forward and reverse sequences was used (Elias et al., 2005). The searching parameters include 15 ppm for precursor ion mass, 1.0 D tolerance for fragment ion mass, 2 for maximal missed cleavages, carbamidomethylation of cysteine as fixed modification, and oxidation of methionine as dynamic modification. Peptide inference, protein grouping, and false-discovery rate control was accomplished by Scaffold (v4.3.2; Proteome Software Inc.; Searle, 2010). Corresponding parameters include for minimal peptide length, 25 ppm for parent mass tolerance; 0.1 for DeltaCn; 2.1, 2.2, 2.6, 3.5 (pseudopods); and 1.1, 1.4, 1.7, 2.5 (cell bodies) for Xcorr thresholds for singly, doubly, triply, and quadruply charged ions.
Chromatographic alignment and global intensity-based MS1 feature detection/extraction were accomplished by SIEVE (v2.1.377; Lopez et al., 2011). The major steps we took are as follows. (a) Alignment of LC-MS runs via the use of ChromAlign algorithm. The quality of LC-MS runs was monitored and benchmarked by the alignment scores and base-peak ion current intensity among all runs. (b) Determination of quantitative “frames” based on m/z and retention time in the aligned collective dataset. The reliability of quantification was assured by retaining only frames with high-quality area-under-the-curve data (e.g., signal/noise ratio >10). (c) Calculation of peptide ion intensities for each frame. The resulting .sdb files were incorporated with spectrum reports exported from Scaffold to associated MS2 fragmentation scans with each frame by using customized R scripts. Another in-house-developed R package was used for the normalization of ion current intensities for each protein (by quantile ion intensities among individual runs) and the aggregation of sun intensities from frame levels to protein levels. The calculation of protein-abundance ratio, as well as the evaluation of statistical significance with Student’s t test (unpaired) between induced and noninduced (i.e., control) groups, was conducted to determine proteins with significant changes in abundance. Detailed information about the quantitative proteomics workflow is described in several pieces of our previous works (Tu et al., 2014a,b; An et al., 2015; Shen et al., 2015).
Bioinformatics for SC pseudopod assay
The pathway and signaling networks were analyzed by using Ingenuity Pathways Analysis software (Ingenuity Systems). Lists of proteins with relative enrichment in pseudopods by neuronal membrane were associated with canonical pathways in Ingenuity Pathways Analysis. The probability of association between the dataset and the canonical pathway proteins was measured by using Fisher’s exact test (Dataset 1). Networks of proteins were algorithmically generated based on their connectivity.
RT-PCR
Standard RT-PCR was performed on cDNAs derived from purified mouse DRG and purified rat SCs (gifts from Y. Sasaki, J. Milbrandt laboratory, Washington University, St. Louis, MO). For mouse Gpr56, the following primer pair was used to amplify a 577-bp fragment that spans exons 5–10: 5′-CAGTTACAGAGCCTGGAGTC-3′ and 5′-TGTGAGTGGCTTCTACCTCT-3′. For rat Gpr56, the following primer pair was used to amplify a 573-bp fragment that spans exons 9–12: 5′-GAGGGCTGTGAGACTGTTAG-3′ and 5′-TAGAGGGGTAGATGACATGG-3′. The following primers were used to amplify a 702-bp fragment from exons 3–6 of mouse and exons 2–6 of rat Gapdh: 5′-GCCATCAACGACCCCTTCATT-3′ and 5′-CGCCTGCTTCACCACCTTCTT-3′.
TEM
For standard EM analyses, tissues were drop-fixed in modified Karnovsky’s fix and processed as described previously (Monk et al., 2011). Semithin sections (200 nm) were stained with toluidine blue and imaged with an AxioCam MRm on a light microscope (Zeiss AxioImager M2). Thin sections (70 nm) were treated as described for zebrafish analyses. For examination of dorsal and ventral roots, two roots each were analyzed from two WT and three Gpr56−/− animals on P180. The number of animals examined at time point for SN analyses is as follows: P3, six independent regions (4,000×) of nerve from three WT, three Gpr56+/−, and four Gpr56−/−; P14, four independent regions (2,000×) of nerve from three WT, three Gpr56+/−, and three Gpr56−/−; P21, four independent regions (2,000×) of nerve from three WT and three Gpr56−/−; P90, three independent regions (2,000×) of nerve from three WT, three Gpr56+/−, and four Gpr56−/−; P180, three independent regions (2,000×) of nerve from three WT, three Gpr56+/−, and three Gpr56−/−; and P300, three independent regions (2,000×) of nerve from three WT, three Gpr56+/−, and three Gpr56−/−.
For all experiments, myelinated fibers showing myelin outfoldings, infoldings, or demyelinating profiles were considered abnormal.
Antibodies for IHC
We used the following primary antibodies: mouse anti–Gpr56-H11 (1:1,000; Jeong et al., 2012), rabbit anti–Ki67 (1:400; Abcam; Mogha et al., 2013), rabbit anti–TUJ1 (1:1,000; Covance; Mogha et al., 2013), rat anti–MBP (1:10; AbD Serotec; Mogha et al., 2013), and mouse anti–pan sodium channel (IgG1, 1:400; Sigma-Aldrich; Saito et al., 2003). Fluorescently conjugated secondary antibodies were applied at a concentration of 1:1,000 (Invitrogen). Rhodamine-phalloidin (1:100; Invitrogen; Terada et al., 2012) was used for staining of Schmidt-Lanterman incisures.
IHC
For tissue sections, nerves were harvested and drop-fixed in 4% paraformaldehyde overnight, transferred into 30% sucrose, and then embedded in optimal cutting temperature compound. IHC was done as described previously on 7–10-µm-thick sections (Jeong et al., 2012). Stained sections were imaged with an AxioCam MRm on a light microscope (Zeiss AxioImager M2), and analyses were performed in ImageJ software. SNs and DRG from three WT were used for expression analyses. SNs from three WT and two Gpr56+/− mice (controls) and three Gpr56−/− mice were used for quantification of cell proliferation on P3. For teased nerve preparations, nerves were harvested and fixed for 30 min in 4% paraformaldehyde before teasing on glass slides. Samples were then permeabilized for 15 min in acetone at −20°C before antibody staining.
RNA sequencing
Mouse SNs were harvested on P3 and P21 (n = 4 WT and n = 4 Gpr56−/− on P3; n = 3 WT and n = 3 Gpr56−/− on P21), flash frozen in liquid nitrogen, and stored at −80°C. Total RNA was extracted by using an RNeasy Mini kit (74104; Qiagen). First, nerves were allowed to thaw on ice for at least 5 min in Buffer RLT containing β-mercaptoethanol. During this time, small spring-loaded surgical scissors were used to cut the nerves into smaller pieces while the nerves were still in the liquid. Scissors were cleaned with 100% ethanol between each sample. Once thawed, nerves were homogenized by using a TissueRuptor for ∼30–60 s followed by vortexing briefly, then passing the sample through a syringe and successively smaller needles (22 gauge and then 28 gauge) at least 10 times each or until large clumps of tissue were no longer visible. We then followed manufacturer instructions for the RNeasy Mini kit to obtain total RNA for each animal. RNA was eluted in 30 μl elution buffer and then submitted on dry ice to the Genome Technology Access Center at Washington University in St. Louis, MO, for processing and RNA-seq. Libraries were generated by using Clontech Library Prep kits, and 12 samples were run on a single lane of Illumina 3000, 1 × 50 bp. Approximately 20–40 million mapped reads were generated per sample. All reads were mapped with STAR (Dobin et al., 2013).
R/Bioconductor was used for subsequent analysis. Differential expression analysis was performed by using the packages, DESeq (Anders and Huber, 2010), and edgeR (Robinson et al., 2010) in R (http://www.R-project.org). The original p-values were corrected via false-discovery rate for multiple testing errors. In addition to a false-discovery rate of <0.05, we considered genes differentially expressed if the adjusted p-value was <0.05 and fold change >1.4× (log2[fold change] >0.5). Individual P3 and P21 volcano plots and comparison between the two developmental stages were generated on normalized gene expression by using R. GO analysis was performed with MetaCore and g:Profiler.
Morphometry
G-ratios were calculated from a total of ∼180 myelinated fibers per genotype (∼60 fibers each from three mice per genotype) by taking the ratio of axon diameter to fiber diameter. Measurements were obtained by using ImageJ software. Internodal lengths were similarly measured from IHC images in ImageJ software. For internode analyses, a total of a total of 21 WT and 64 Gpr56−/− fibers were measured per genotype on P21 (n = 3 mice per genotype). For apposition quantifications from electron micrographs, a total of 154 WT and 168 Gpr56−/− myelinated fibers were analyzed on P21 (n = 3 mice per genotype). For analysis of Schmidt-Lanterman incisures on P300, a total of 104 WT fibers and 93 Gpr56−/− fibers were analyzed (n = 3 mice per genotype).
Serial scanning EM and reconstruction
Nerves from P90 mice (three nerves from three WT and three Gpr56−/− animals) were harvested and drop-fixed in modified Karnovsky’s fixative overnight. Nerves were then washed into 0.1 M sodium cacodylate before staining, embedding, and imaging at Renovo Neural Inc. Approximately 400 serial sections were imaged (100 nm each in thickness) per nerve, and data were analyzed and reconstructed by using IMOD software (Kremer et al., 1996). Videos depicting data and models from serial EM analyses (Videos 1, 2, 3, and 4) were generated in ImageJ software.
Motor function
Motor function tests were performed in the Animal Behavior Core Lab at Washington University School of Medicine. The accelerated rotarod was used to assess sensorimotor function and grip strength and was performed on a cohort of 11 WT, 16 Gpr56+/−, and 13 Gpr56−/− mice on P180, and the same cohort was used for testing on P300 (n = 10 WT, n = 15 Gpr56+/−, and n = 12 Gpr56−/−) as described previously (Idol et al., 2014).
Electrophysiology
Animals (WT, n = 5; Gpr56+/−, n = 12; Gpr56−/−, n = 11) were anesthetized by using 4% isoflurane/96% oxygen (induction) and 2% isoflurane/98% oxygen (maintenance) administered by inhalation. After preparation of the sterile field, the right SN was isolated from the sciatic notch to below the point of trifurcation, distal to the popliteal artery. Cathodic, monophasic electrical impulses (duration = 50 μs, frequency = single, amplitude = 1 mA) were generated by a single-channel isolated pulse stimulator (model 2100; A-M Systems Inc.) and delivered to the SN proximal to the implanted device via bipolar platinum/iridium microwire electrodes (Medwire). Resulting EMGs were then differentially recorded in the tibialis anterior muscle by using intramuscular microwire electrodes (4 mil; California Fine Wire). Measured signals were band-pass filtered (low pass = 1 Hz, high pass = 5 kHz, notch = 60 Hz) and amplified (gain = 1,000–10,000×) by using a two-channel microelectrode AC amplifier (model 1800; A-M Systems, Inc.) before being recorded on a desktop PC (Dell Computer Corp.) equipped with a data acquisition board (DT3003/PGL; Data Translations). EMG traces were digitally recorded while peak latency and maximal amplitude were calculated by using custom Matlab software (The MathWorks, Inc.).
Western blot and GTP-Rho pull-down assay
For analysis of RhoA activity in mouse nerves, P6 litters consisting of Gpr56+/+, Gpr56+/−, and Gpr56−/− mice were used in each of three separate experiments (we combined four nerves from the same genotype for one pull-down and did three separate experiments, with a total of 12 for each genotype group). P6 SNs were dissected under a Leica stereo microscope (MZ 6; Leica Pte Ltd.); four nerves from same the genotype were combined in one group, powdered on liquid nitrogen, then lysed in ice-cold RIPA buffer (1% Nonidet P-40, 50 mM Tris, pH 7.6, 120 mM NaCl, and 1 mM EDTA) containing protease inhibitor cocktail set 1 (Calbiochem). The lysates were cleared of insoluble materials by centrifugation at 16,000 g for 10 min at 4°C. The protein concentration was determined by a Bio-Rad protein assay method (Bio-Rad) according to the manufacturer's protocol, and equal amounts of protein were used for each pull-down assay. The GTP-Rho pull-down assay was performed as previously described (Luo et al., 2011). In short, tissues were pulverized on liquid nitrogen, lysed in 300 µl of ice-cold RIPA buffer containing protease inhibitors with a cell disruptor for 10 min and homogenized with a 26-gauge syringe needle. Equal amounts of total protein were incubated with 60 µg GST-RBD beads (Cytoskeleton) at 4°C for 90 min. The beads were washed twice with lysis buffer and once with Tris-buffered saline buffer. Bound Rho proteins were eluted by Laemmli sample buffer and detected by Western blot by using mouse monoclonal anti–RhoA antibody (ARH04, 1:1000; Cytoskeleton). For analysis of GPR56-dependent RhoA signaling in vitro, HEK 293T cells were plated in 6-well plates (500,000 cells/well) and transfected with mouse Gpr56 (Luo et al., 2014) or not. 24 h posttransfection, cells were subjected to serum starvation for 36 h followed by treatment with human collagen III (84 nM, CC054, positive control; Sigma-Aldrich) or full-length recombinant FT (res. 23–110, 10 µM) and as a control with either 0.5 M acetic acid or PBS, respectively, for 5 min. Cells were lysed in lysis buffer (PBS + 1% Triton X-100 + 1× protease inhibitors) followed by pull-down of RhoA-GTP (Luo et al., 2011). We also blotted for GPR56 (ab172361, concentration of 1 µg/ml; Abcam) to assess the success of transfection and actin (MAB1501, 200 ng/ml antibody and c concentration; Chemicon) for a loading control. Data are representative of three biologically independent experiments.
Gpr56 PRESTO-Tango assay
HTLA cells (a HEK293 cell line stably expressing a tTA-dependent luciferase reporter and a β-arrestin2-TEV fusion gene) were provided by the laboratory of B. Roth (University of Noth Carolina, Chapel Hill, NC; Kroeze et al., 2015). Cells were plated in six-well plates (500,000 cells/well) and transfected with either Gpr56 Tango plasmid (Addgene) or as a control with pCDNA3 plasmid. 24 h posttransfection, cells were subjected to serum starvation for further 24 h followed by treatment with human collagen III (150 nM; CC054; Sigma-Aldrich) or full-length recombinant FT (res. 23–110, 10 µM), and as a control, with either 0.5 M acetic acid or PBS, respectively, overnight. Cells were washed with PBS and processed with Bright-Glo luciferase system (E2610; Promega) according to manufacturer’s protocol. 200 µl of Bright-Glo solution, diluted in the assay buffer, was added to 25 µg of cell lysate, and followed by 20 min of incubation, luciferase activity was measured (as a function of chemiluminescence). Data are representative of three biologically independent experiments.
cAMP assay
cAMP assay was done with a colorimetric competitive immunoassay (Enzo Life Sciences) as described previously (Küffer et al., 2016). In brief, HEK 293T cells were transfected with either human GPR126 (cloned into pCDNA3.1/V5-His-TOPO vector) or mouse Gpr56 (cloned into pCDNA3.1 vector) and 48 h posttransfection were treated with either PBS or full-length recombinant FT (10 µM) and then processed for quantification of cAMP. Data are representative of three biologically independent experiments.
Co-IP and immunoblotting
Co-IP and immunoblotting were done as previously described (Luo et al., 2011). In brief, P5 mouse SNs (n = 8 nerves from four animals for each co-IP) were lysed in RIPA buffer as described for the GTP-RhoA pull-down assay. After extraction at 4°C with rocking, insoluble material was removed by centrifugation, and lysates were precleared for 1 h at 4°C with protein G-Sepharose (101241; Invitrogen). The lysate was equally divided into two; half of the lysate was used for GPR56 IP, and the other half was used for the control IP. Protein G beads were then used to pull-down the GPR56NTF-hFc protein complex. Immunocomplexes were subjected to SDS-PAGE and Western blot by using pan-plectin antibody (MAB5674; Sigma-Aldrich), and rabbit anti–human IgG Fc antibody (31142; Thermo Fisher Scientific) following standard protocols.
Plectin expression assay
P6 mouse SNs (n = 12 nerves from six animals per genotype) were collected and lysed as described for the GTP-RhoA pull-down assay. Plectin levels were assessed by Western blot by using a pan-plectin antibody (MAB5674; Sigma-Aldrich). Two nerves were pooled from each animal to make up a single replicate.
Statistical analyses
Analyses were performed blinded to genotype and treatment. All data are presented as the mean ± SEM unless otherwise noted. To represent significance, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. A minimum of two technical replicates was performed for each described experiment. Although we did not perform statistical tests to predetermine sample size, our samples sizes are similar to previously published studies in the field. Statistical analyses were performed by using one-way ANOVA, two-way ANOVA, or unpaired Student’s t test with two tails with unequal variance when required conditions were met. A departmental statistician was consulted to confirm the appropriateness of each statistical analysis performed.
Online supplemental material
Fig. S1 shows IHC analysis of cellular proliferation and the total cell number in P3 mutant and control mouse SNs as well as an assessment of overall saphenous nerve size by toluidine blue staining, which can be affected by cell number. Fig. S2 shows TEM images assessing pLLn integrity and myelin ultrastructure in gpr56stl13/stl13 zebrafish mutants during development and in adulthood to show conservation with mouse phenotypes. Fig. S3 shows RNA-seq analysis of the transcriptome of Gpr56 mutant and WT control nerves on P3 and P21. Fig. S4 Shows further structural analysis of Gpr56 mutant nerves: Schmidt Lanterman incisure quantification by IHC, toluidine blue, and TEM images of Gpr56−/− dorsal and ventral roots compared to control roots on P180. Analysis of Gpr56 expression in neurons versus SCs is shown. Fig. S5 shows additional TEM reconstructions to supplement observations in Fig. 6. Video 1 shows serial reconstruction of a myelinated fiber from a WT mouse SN on P90 generated from 350 successive scanning EM images (100 nm thick). Video 2 shows serial scanning EM images (210 sections shown, 100 nm thick) from a WT mouse SN on P90. Video 3 shows serial reconstruction of a myelinated fiber from a Gpr56−/− mutant mouse SN on P90 generated from 350 successive scanning EM images (100 nm thick). Video 4 shows serial scanning EM images (210 sections shown, 100 nm thick) from a Gpr56−/− mutant mouse SN on P90. Dataset 1 is a summary of all proteins significantly upregulated in SC pseudopods after induction and analysis of the major cellular pathways found to be enriched. Dataset 2 is a summary of gProfiler results for significantly enriched or depleted pathways in Gpr56 mutant nerves based on RNA-seq analyses on P3 and P21. Raw data files can be accessed from Zenodo (https://zenodo.org/record/1154250).
Supplementary Material
Acknowledgments
We thank members of the Monk laboratory for helpful discussions. We thank Fang Lin for 3MOs, Lila Solnica-Krezel for the rhov14 construct, Jeffrey Milbrandt for cDNAs, and Robyn Roth for assistance with TEM. We thank Charleen Johnson and Zachary Spence for husbandry assistance, Bo Zhang for statistical consultations, and Marcus Mahar for his aid in RNA-seq analysis. Finally, we thank Gerhard Wiche for sharing his expertise on plectin biology.
This work was supported by a predoctoral fellowship from the National Institutes of Health to S.D. Ackerman (F31 NS087801), to B.L. Harty (F31 NS094004), to M. D’Rozario (F32 NS100461), to M.L. Feltri (R01NS045630), and to W.Z. Ray (K23 NS084932); by a research grant from the Muscular Dystrophy Association to X. Piao and K.R. Monk (293295); and by grants to K.R. Monk from the National Institutes of Health (R01 NS079445) and the Edward J. Mallinckrodt, Jr. Foundation. A. Aguzzi is the recipient of an Advanced Grant of the European Research Council, a European Union Seventh Framework Programme grant (NEURINOX), the Swiss National Science Foundation, the Clinical Research Priority Programs Small RNAs and Human Hemato-Lymphatic Diseases, SystemsX.ch (PrionX and SynucleiX), and the Novartis Foundation. A.K.K. Lakkaraju was supported by a grant from the Synapsis Foundation.
The authors declare no competing financial interests.
Author contributions: Y. Poitelon and M.L. Feltri designed the SC pseudopods assay, which was executed by Y. Poitelon, J. Li, and J. Qu. A.K.K. Lakkaraju and A. Aguzzi designed the PrP experiments, which were performed by A.K.K. Lakkaraju. W.Z. Ray and M.R. MacEwan designed the electrophysiology experiments, which were performed by M.R. MacEwan and P. Gamble. The remaining experiments were designed by S.D. Ackerman, X. Piao, and K.R. Monk and executed by S.D. Ackerman, R. Luo, A. Mogha, M. D’Rozario, N.E. Sanchez, and B.L. Harty. S.D. Ackerman and K.R. Monk wrote the manuscript, which was read and edited by all authors.
References
- Ackerman, S.D., Garcia C., Piao X., Gutmann D.H., and Monk K.R.. 2015. The adhesion GPCR Gpr56 regulates oligodendrocyte development via interactions with Gα12/13 and RhoA. Nat. Commun. 6:6122. 10.1038/ncomms7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano, M., Ito M., Kimura K., Fukata Y., Chihara K., Nakano T., Matsuura Y., and Kaibuchi K.. 1996. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271:20246–20249. 10.1074/jbc.271.34.20246 [DOI] [PubMed] [Google Scholar]
- An, B., Zhang M., Johnson R.W., and Qu J.. 2015. Surfactant-aided precipitation/on-pellet-digestion (SOD) procedure provides robust and rapid sample preparation for reproducible, accurate and sensitive LC/MS quantification of therapeutic protein in plasma and tissues. Anal. Chem. 87:4023–4029. 10.1021/acs.analchem.5b00350 [DOI] [PubMed] [Google Scholar]
- Anders, S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, B.I., Tietjen I., Atabay K.D., Evrony G.D., Johnson M.B., Asare E., Wang P.P., Murayama A.Y., Im K., Lisgo S.N., et al. 2014. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science. 343:764–768. 10.1126/science.1244392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, J.W., Rouan F., Kofler B., Rezniczek G.A., Kornacker I., Muss W., Hametner R., Klausegger A., Huber A., Pohla-Gubo G., et al. 2001. A compound heterozygous one amino-acid insertion/nonsense mutation in the plectin gene causes epidermolysis bullosa simplex with plectin deficiency. Am. J. Pathol. 158:617–625. 10.1016/S0002-9440(10)64003-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañón, M.J., Walko G., Winter L., and Wiche G.. 2013. Plectin-intermediate filament partnership in skin, skeletal muscle, and peripheral nerve. Histochem. Cell Biol. 140:33–53. 10.1007/s00418-013-1102-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermenati, G., Audano M., Giatti S., Carozzi V., Porretta-Serapiglia C., Pettinato E., Ferri C., D’Antonio M., De Fabiani E., Crestani M., et al. 2015. Lack of sterol regulatory element binding factor-1c imposes glial Fatty Acid utilization leading to peripheral neuropathy. Cell Metab. 21:571–583. 10.1016/j.cmet.2015.02.016 [DOI] [PubMed] [Google Scholar]
- Court, F.A., Sherman D.L., Pratt T., Garry E.M., Ribchester R.R., Cottrell D.F., Fleetwood-Walker S.M., and Brophy P.J.. 2004. Restricted growth of Schwann cells lacking Cajal bands slows conduction in myelinated nerves. Nature. 431:191–195. 10.1038/nature02841 [DOI] [PubMed] [Google Scholar]
- Court, F.A., Hewitt J.E., Davies K., Patton B.L., Uncini A., Wrabetz L., and Feltri M.L.. 2009. A laminin-2, dystroglycan, utrophin axis is required for compartmentalization and elongation of myelin segments. J. Neurosci. 29:3908–3919. 10.1523/JNEUROSCI.5672-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka, T., and Lyons D.A.. 2011. Dissecting mechanisms of myelinated axon formation using zebrafish. Methods Cell Biol. 105:25–62. 10.1016/B978-0-12-381320-6.00002-3 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlé, D., Hegarty B., Bossard P., Ferré P., and Foufelle F.. 2004. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 86:839–848. 10.1016/j.biochi.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Elias, J.E., Haas W., Faherty B.K., and Gygi S.P.. 2005. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods. 2:667–675. 10.1038/nmeth785 [DOI] [PubMed] [Google Scholar]
- Feltri, M.L., Scherer S.S., Wrabetz L., Kamholz J., and Shy M.E.. 1992. Mitogen-expanded Schwann cells retain the capacity to myelinate regenerating axons after transplantation into rat sciatic nerve. Proc. Natl. Acad. Sci. USA. 89:8827–8831. 10.1073/pnas.89.18.8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltri, M.L., Poitelon Y., and Previtali S.C.. 2016. How Schwann cells sort axons: New concepts. Neuroscientist. 22:252–265. 10.1177/1073858415572361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenas, L., De Santis F., Di Donato V., Degerny C., Chambraud B., Del Bene F., and Tawk M.. 2016. Neuronal Ndrg4 is essential for nodes of Ranvier organization in zebrafish. PLoS Genet. 12:e1006459. 10.1371/journal.pgen.1006459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giera, S., Deng Y., Luo R., Ackerman S.D., Mogha A., Monk K.R., Ying Y., Jeong S.J., Makinodan M., Bialas A.R., et al. 2015. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat. Commun. 6:6121. 10.1038/ncomms7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn, T.D., and Talbot W.S.. 2013. Analysis of Gpr126 function defines distinct mechanisms controlling the initiation and maturation of myelin. Development. 140:3167–3175. 10.1242/dev.093401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbot, A., Williams A., Ravisé N., Verny C., Brice A., Sherman D.L., Brophy P.J., LeGuern E., Delague V., Bareil C., et al. 2001. A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum. Mol. Genet. 10:415–421. 10.1093/hmg/10.4.415 [DOI] [PubMed] [Google Scholar]
- Hamann, J., Aust G., Araç D., Engel F.B., Formstone C., Fredriksson R., Hall R.A., Harty B.L., Kirchhoff C., Knapp B., et al. 2015. International union of basic and clinical pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 67:338–367. 10.1124/pr.114.009647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel, T., and Lupski J.R.. 2014. Charcot-Marie-Tooth disease and pathways to molecular based therapies. Clin. Genet. 86:422–431. 10.1111/cge.12393 [DOI] [PubMed] [Google Scholar]
- Idol, R.A., Wozniak D.F., Fujiwara H., Yuede C.M., Ory D.S., Kornfeld S., and Vogel P.. 2014. Neurologic abnormalities in mouse models of the lysosomal storage disorders mucolipidosis II and mucolipidosis III γ. PLoS One. 9:e109768. 10.1371/journal.pone.0109768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi, T., Sakata K., Yoshizaki K., Tago K., Mizuno N., and Itoh H.. 2008. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G α 12/13 and Rho pathway. J. Biol. Chem. 283:14469–14478. 10.1074/jbc.M708919200 [DOI] [PubMed] [Google Scholar]
- Jeong, S.J., Luo R., Li S., Strokes N., and Piao X.. 2012. Characterization of G protein-coupled receptor 56 protein expression in the mouse developing neocortex. J. Comp. Neurol. 520:2930–2940. 10.1002/cne.23076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen, K.R., and Mirsky R.. 2005. The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 6:671–682. 10.1038/nrn1746 [DOI] [PubMed] [Google Scholar]
- Kimmel, C.B., Ballard W.W., Kimmel S.R., Ullmann B., and Schilling T.F.. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kroeze, W.K., Sassano M.F., Huang X.P., Lansu K., McCorvy J.D., Giguère P.M., Sciaky N., and Roth B.L.. 2015. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22:362–369. 10.1038/nsmb.3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küffer, A., Lakkaraju A.K.K., Mogha A., Petersen S.C., Airich K., Doucerain C., Marpakwar R., Bakirci P., Senatore A., Monnard A., et al. 2016. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature. 536:464–468. 10.1038/nature19312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Jin Z., Koirala S., Bu L., Xu L., Hynes R.O., Walsh C.A., Corfas G., and Piao X.. 2008. GPR56 regulates pial basement membrane integrity and cortical lamination. J. Neurosci. 28:5817–5826. 10.1523/JNEUROSCI.0853-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebscher, I., Schön J., Petersen S.C., Fischer L., Auerbach N., Demberg L.M., Mogha A., Cöster M., Simon K.U., Rothemund S., et al. 2014. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Reports. 9:2018–2026. 10.1016/j.celrep.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F., Chen S., Sepich D.S., Panizzi J.R., Clendenon S.G., Marrs J.A., Hamm H.E., and Solnica-Krezel L.. 2009. Galpha12/13 regulate epiboly by inhibiting E-cadherin activity and modulating the actin cytoskeleton. J. Cell Biol. 184:909–921. 10.1083/jcb.200805148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, M.F., Kuppusamy R., Sarracino D.A., Prakash A., Athanas M., Krastins B., Rezai T., Sutton J.N., Peterman S., and Nicolaides K.. 2011. Mass spectrometric discovery and selective reaction monitoring (SRM) of putative protein biomarker candidates in first trimester Trisomy 21 maternal serum. J. Proteome Res. 10:133–142. 10.1021/pr100153j [DOI] [PubMed] [Google Scholar]
- Luo, R., Jeong S.J., Jin Z., Strokes N., Li S., and Piao X.. 2011. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. USA. 108:12925–12930. 10.1073/pnas.1104821108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, R., Jeong S.J., Yang A., Wen M., Saslowsky D.E., Lencer W.I., Araç D., and Piao X.. 2014. Mechanism for adhesion G protein-coupled receptor GPR56-mediated RhoA activation induced by collagen III stimulation. PLoS One. 9:e100043. 10.1371/journal.pone.0100043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini, R. 1999. P0-deficient knockout mice as tools to understand pathomechanisms in Charcot-Marie-Tooth 1B and P0-related Déjérine-Sottas syndrome. Ann. N. Y. Acad. Sci. 883:273–280. 10.1111/j.1749-6632.1999.tb08589.x [DOI] [PubMed] [Google Scholar]
- Melendez-Vasquez, C.V., Einheber S., and Salzer J.L.. 2004. Rho kinase regulates schwann cell myelination and formation of associated axonal domains. J. Neurosci. 24:3953–3963. 10.1523/JNEUROSCI.4920-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.J., Rangwala F., Williams J., Ackerman P., Kong S., Jegga A.G., Kaiser S., Aronow B.J., Frahm S., Kluwe L., et al. 2006. Large-scale molecular comparison of human schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res. 66:2584–2591. 10.1158/0008-5472.CAN-05-3330 [DOI] [PubMed] [Google Scholar]
- Mogha, A., Benesh A.E., Patra C., Engel F.B., Schöneberg T., Liebscher I., and Monk K.R.. 2013. Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J. Neurosci. 33:17976–17985. 10.1523/JNEUROSCI.1809-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk, K.R., Naylor S.G., Glenn T.D., Mercurio S., Perlin J.R., Dominguez C., Moens C.B., and Talbot W.S.. 2009. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 325:1402–1405. 10.1126/science.1173474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk, K.R., Oshima K., Jörs S., Heller S., and Talbot W.S.. 2011. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development. 138:2673–2680. 10.1242/dev.062224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave, K.A., and Trapp B.D.. 2008. Axon-glial signaling and the glial support of axon function. Annu. Rev. Neurosci. 31:535–561. 10.1146/annurev.neuro.30.051606.094309 [DOI] [PubMed] [Google Scholar]
- Nodari, A., Zambroni D., Quattrini A., Court F.A., D’Urso A., Recchia A., Tybulewicz V.L., Wrabetz L., and Feltri M.L.. 2007. β1 integrin activates Rac1 in Schwann cells to generate radial lamellae during axonal sorting and myelination. J. Cell Biol. 177:1063–1075. 10.1083/jcb.200610014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, J.T., Horwitz A.R., and Schwartz M.A.. 2010. Cell adhesion: Integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11:633–643. 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, J.A., Benninger Y., Baumann R., Gonçalves A.F., Ozçelik M., Thurnherr T., Tricaud N., Meijer D., Fässler R., Suter U., and Relvas J.B.. 2009. Integrin-linked kinase is required for radial sorting of axons and Schwann cell remyelination in the peripheral nervous system. J. Cell Biol. 185:147–161. 10.1083/jcb.200809008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, S.C., Luo R., Liebscher I., Giera S., Jeong S.J., Mogha A., Ghidinelli M., Feltri M.L., Schöneberg T., Piao X., and Monk K.R.. 2015. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron. 85:755–769. 10.1016/j.neuron.2014.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri, A., Chitsazzadeh V., D’Arrigo S., Fedrizzi E., Pantaleoni C., Riva D., Busse C., Küster H., Duplessis A., Gaitanis J., et al. 2010. The syndrome of perisylvian polymicrogyria with congenital arthrogryposis. Brain Dev. 32:550–555. 10.1016/j.braindev.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitelon, Y., Bogni S., Matafora V., Della-Flora Nunes G., Hurley E., Ghidinelli M., Katzenellenbogen B.S., Taveggia C., Silvestri N., Bachi A., et al. 2015. Spatial mapping of juxtacrine axo-glial interactions identifies novel molecules in peripheral myelination. Nat. Commun. 6:8303. 10.1038/ncomms9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, F., Moore S.A., Barresi R., Henry M.D., Messing A., Ross-Barta S.E., Cohn R.D., Williamson R.A., Sluka K.A., Sherman D.L., et al. 2003. Unique role of dystroglycan in peripheral nerve myelination, nodal structure, and sodium channel stabilization. Neuron. 38:747–758. 10.1016/S0896-6273(03)00301-5 [DOI] [PubMed] [Google Scholar]
- Salzman, G.S., Ackerman S.D., Ding C., Koide A., Leon K., Luo R., Stoveken H.M., Fernandez C.G., Tall G.G., Piao X., et al. 2016. Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron. 91:1292–1304. 10.1016/j.neuron.2016.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, B.C. 2010. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 10:1265–1269. 10.1002/pmic.200900437 [DOI] [PubMed] [Google Scholar]
- Shen, X., Hu Q., Li J., Wang J., and Qu J.. 2015. Experimental null method to guide the development of technical procedures and to control false-positive discovery in quantitative proteomics. J. Proteome Res. 14:4147–4157. 10.1021/acs.jproteome.5b00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, D.L., and Brophy P.J.. 2005. Mechanisms of axon ensheathment and myelin growth. Nat. Rev. Neurosci. 6:683–690. 10.1038/nrn1743 [DOI] [PubMed] [Google Scholar]
- Sherman, D.L., Wu L.M., Grove M., Gillespie C.S., and Brophy P.J.. 2012. Drp2 and periaxin form Cajal bands with dystroglycan but have distinct roles in Schwann cell growth. J. Neurosci. 32:9419–9428. 10.1523/JNEUROSCI.1220-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, D., Lin S.T., Fu Y.H., and Ptácek L.J.. 2013a. Very large G protein-coupled receptor 1 regulates myelin-associated glycoprotein via Gαs/Gαq-mediated protein kinases A/C. Proc. Natl. Acad. Sci. USA. 110:19101–19106. 10.1073/pnas.1318501110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S.J., Smith J.A., Rezniczek G.A., Pan S., Chen R., Brentnall T.A., Wiche G., and Kelly K.A.. 2013b. Unexpected gain of function for the scaffolding protein plectin due to mislocalization in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 110:19414–19419. 10.1073/pnas.1309720110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J.L., Ravid S., and Spudich J.A.. 1992. Control of nonmuscle myosins by phosphorylation. Annu. Rev. Biochem. 61:721–759. 10.1146/annurev.bi.61.070192.003445 [DOI] [PubMed] [Google Scholar]
- Taylor, A.R., Geden S.E., and Fernandez-Valle C.. 2003. Formation of a beta1 integrin signaling complex in Schwann cells is independent of rho. Glia. 41:94–104. 10.1002/glia.10170 [DOI] [PubMed] [Google Scholar]
- Terada, N., Saitoh Y., Ohno N., Komada M., Saitoh S., Peles E., and Ohno S.. 2012. Essential function of protein 4.1G in targeting of membrane protein palmitoylated 6 into Schmidt-Lanterman incisures in myelinated nerves. Mol. Cell. Biol. 32:199–205. 10.1128/MCB.05945-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, C., Li J., Sheng Q., Zhang M., and Qu J.. 2014a. Systematic assessment of survey scan and MS2-based abundance strategies for label-free quantitative proteomics using high-resolution MS data. J. Proteome Res. 13:2069–2079. 10.1021/pr401206m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, C., Sheng Q., Li J., Shen X., Zhang M., Shyr Y., and Qu J.. 2014b. ICan: An optimized ion-current-based quantification procedure with enhanced quantitative accuracy and sensitivity in biomarker discovery. J. Proteome Res. 13:5888–5897. 10.1021/pr5008224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walko, G., Wögenstein K.L., Winter L., Fischer I., Feltri M.L., and Wiche G.. 2013. Stabilization of the dystroglycan complex in Cajal bands of myelinating Schwann cells through plectin-mediated anchorage to vimentin filaments. Glia. 61:1274–1287. 10.1002/glia.22514 [DOI] [PubMed] [Google Scholar]
- Weiss, T., Taschner-Mandl S., Bileck A., Slany A., Kromp F., Rifatbegovic F., Frech C., Windhager R., Kitzinger H., Tzou C.H., et al. 2016. Proteomics and transcriptomics of peripheral nerve tissue and cells unravel new aspects of the human Schwann cell repair phenotype. Glia. 64:2133–2153. 10.1002/glia.23045 [DOI] [PubMed] [Google Scholar]
- Winter, L., and Wiche G.. 2013. The many faces of plectin and plectinopathies: pathology and mechanisms. Acta Neuropathol. 125:77–93. 10.1007/s00401-012-1026-0 [DOI] [PubMed] [Google Scholar]
- Wu, M.P., Doyle J.R., Barry B., Beauvais A., Rozkalne A., Piao X., Lawlor M.W., Kopin A.S., Walsh C.A., and Gussoni E.. 2013. G-protein coupled receptor 56 promotes myoblast fusion through serum response factor- and nuclear factor of activated T-cell-mediated signalling but is not essential for muscle development in vivo. FEBS J. 280:6097–6113. 10.1111/febs.12529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.