The R47H variant of the microglia gene TREM2 has been linked to a significantly higher risk of Alzheimer’s disease. In this study, Song et al. generate human TREM2-expressing mice and demonstrate that R47H leads to a decreased microglia number and activation as well as a decreased presence of soluble TREM2 on neurons and plaques in a mouse model of Alzheimer’s disease.
Abstract
Alzheimer’s disease (AD) is a neurodegenerative disease that causes late-onset dementia. The R47H variant of the microglial receptor TREM2 triples AD risk in genome-wide association studies. In mouse AD models, TREM2-deficient microglia fail to proliferate and cluster around the amyloid-β plaques characteristic of AD. In vitro, the common variant (CV) of TREM2 binds anionic lipids, whereas R47H mutation impairs binding. However, in vivo, the identity of TREM2 ligands and effect of the R47H variant remain unknown. We generated transgenic mice expressing human CV or R47H TREM2 and lacking endogenous TREM2 in the 5XFAD AD model. Only the CV transgene restored amyloid-β–induced microgliosis and microglial activation, indicating that R47H impairs TREM2 function in vivo. Remarkably, soluble TREM2 was found on neurons and plaques in CV- but not R47H-expressing 5XFAD brains, although in vitro CV and R47H were shed similarly via Adam17 proteolytic activity. These results demonstrate that TREM2 interacts with neurons and plaques duing amyloid-β accumulation and R47H impairs this interaction.
Introduction
Alzheimer’s disease (AD) is the most common form of late-onset dementia and currently lacks approved therapies to halt or reverse disease progression. Although the pathogenesis of sporadic AD is still uncertain, it is thought to involve production and extracellular aggregation of amyloid β (Aβ) peptides and accumulation of intracellular cytotoxic tau aggregates, which result in neuron and synapse loss and ultimately cognitive impairment (Querfurth and LaFerla, 2010). For decades, it has been known that astrocytes and microglia proliferate, associate with plaques, and assume an activated morphology during disease (Itagaki et al., 1989). Imaging studies using tracers for the inflammatory marker TSPO have also identified AD-associated increases in signal (Kreisl et al., 2013; Fan et al., 2017). Very recently, a population of AD-associated microglia with a distinct transcriptional profile was identified by single-cell RNA seq (Keren-Shaul et al., 2017). A more direct link between microglia and AD was identified by a series of GWAS studies in the past decade linking several microglial genes to AD risk, including CD33 (Griciuc et al., 2013), TREM2 (Guerreiro et al., 2013; Jonsson et al., 2013), ABCA7 (Steinberg et al., 2015), and PLCG2 (Sims et al., 2017), supporting that microglia and/or other brain myeloid cells play a role in disease progression. However, it remains controversial whether this role is protective or harmful overall (Meyer-Luehmann and Prinz, 2015). Microglia can act as a double-edged sword by clearing Aβ peptides and aggregates via phagocytosis or enzyme secretion while simultaneously releasing inflammatory mediators that can exacerbate disease (Querfurth and LaFerla, 2010; Heneka et al., 2015).
Of the various microglia-expressed risk variants, the R47H mutation in TREM2 (Guerreiro et al., 2013; Jonsson et al., 2013) stands out as having an effect size similar to the well-characterized ApoE ε4 allele (Corder et al., 1993; Rebeck et al., 1993). TREM2 is an immunoreceptor expressed exclusively in the myeloid lineage, especially microglia (Bouchon et al., 2001). In vitro, TREM2 binds anionic lipids (Wang et al., 2015), lipidated ApoE and ApoJ (Atagi et al., 2015; Bailey et al., 2015), high-density and low-density lipoproteins (Yeh et al., 2016; Song et al., 2017), and apoptotic cells (Hsieh et al., 2009), but in vivo ligands are not known. Engagement of TREM2 by ligands activates downstream protein tyrosine phosphorylation through its adaptor DAP12 (Xing et al., 2015), stimulating mTOR signaling, energetic and anabolic metabolism (Ulland et al., 2017), proliferation, and survival (Otero et al., 2009) while suppressing TLR-induced inflammatory cytokine production (Hamerman et al., 2006; Turnbull et al., 2006). TREM2 is also shed from the cell surface by metalloproteinases such as Adam10 and Adam17 and released as a soluble protein (Wunderlich et al., 2013; Kleinberger et al., 2014; Feuerbach et al., 2017), which has been proposed to mediate cell survival and inflammation (Wu et al., 2015; Zhong et al., 2017).
In Aβ-driven AD mouse models, TREM2 deficiency impairs microglial activation and association with Aβ plaques with variable effects on total Aβ levels (Ulrich et al., 2014, 2017; Jay et al., 2015, 2017; Painter et al., 2015; Wang et al., 2015; Yeh et al., 2017). Given the strong, consistent association of R47H with increased AD risk, it is of utmost importance to understand how this variant affects TREM2 function in vivo to effectively target TREM2 for AD therapy. Loss of function would indicate a protective role and favor TREM2-enhancing therapies, whereas gain of function would indicate a harmful role and favor TREM2 inhibition. In vitro studies seem to suggest loss of function. We and others have shown decreased ligand binding of the R47H variant. Another rare variant weakly associated with AD, R62H, also showed reduced ligand binding (Yeh et al., 2016; Song et al., 2017). Accordingly, R47 and R62 map to a basic surface that likely participates in binding anionic ligands (Kober et al., 2016). Furthermore, postmortem specimens from AD patients bearing R47H have decreased microglial response to plaques (Korvatska et al., 2015; Yuan et al., 2016). However, the in vivo effect of the R47H polymorphism has not been studied in a controlled experimental setting. Moreover, the human and mouse genes share only ∼77% sequence identity, meaning that nonconserved regions could potentially modulate the repertoire of ligands, overall binding affinity, and the effects of variants. To address these issues, we generated two transgenic mouse lines by introducing either common variant (CV) or R47H human TREM2 (hTREM2) into mouse TREM2 (mTREM2)–deficient mice and crossed them with the 5XFAD mouse model of AD (Oakley et al., 2006). We found that CV but not R47H can augment plaque-associated microgliosis and enhance microglial activation. These findings demonstrate that R47H is a partial loss-of-function variant that affects microglia activation and proliferation in response to Aβ plaques. Importantly, we found a widespread presence of soluble TREM2 shed from microglia in situ on neuron cell bodies, on Aβ plaques, and throughout brain tissue in mice that express CV but not in those expressing R47H. In vitro, various activators of myeloid cells, such as LPS, TNFα, and IFNγ, induced Adam17-mediated cleavage of both CV and R47H TREM2 from the cell surface, suggesting that R47H does not directly impact shedding of soluble TREM2. These results show for the first time that TREM2 broadly interacts with neurons and plaques in vivo, whereas the R47H variant impairs this interaction.
Results and discussion
CV and R47H hTREM2 show a similar pattern and level of expression in myeloid cells of transgenic mice
To understand the in vivo properties of hTREM2 and the R47H polymorphism, we generated several BAC transgenic mouse lines expressing either the hTREM2 CV or the R47H polymorphism (Fig. 1 A). We first assessed cell surface expression of hTREM2 in each of these lines to ensure that both CV and R47H transgenic mice generated using this strategy express hTREM2 on the cell surface in a pattern paralleling that of endogenous TREM2, i.e., on myeloid cell populations previously shown to express TREM2. We selected two lines—one carrying CV and one carrying R47H—that expressed similar levels of hTREM2 on the cell surface. Thioglycollate-elicited peritoneal macrophages were collected on day 3 after injection and stained for hTREM2, revealing expression of both transgenes at similar levels in these lines (Fig. S1 A). Bone marrow–derived macrophages (BMDMs) were stained for hTREM2 after 2 d of culture, showing hTREM2 specifically on F4/80+-differentiated cells, again at similar levels in both transgenic lines (Fig. S1 B). We next bred these transgenic lines to mTrem2−/− (KO) mice to obtain CV-KO and R47H-KO transgenic mice lacking endogenous TREM2. Previously, we demonstrated that TREM2-deficient microglia are sensitized to growth factor withdrawal because of a defect in mTOR signaling (Ulland et al., 2017). CV-KO BMDMs were more viable than KO BMDMs after 48 h of CSF1 withdrawal, as measured by propidium iodide staining (Fig. S1 C). This rescue of cell viability was heightened by addition of human HDL, a previously identified ligand for TREM2 (Yeh et al., 2016; Song et al., 2017). R47H-KO BMDMs demonstrated an intermediate phenotype with and without HDL (Fig. S1 C). These data indicate not only that the CV and R47H transgenes are expressed in the appropriate cell types but also that they function properly in mouse cells.
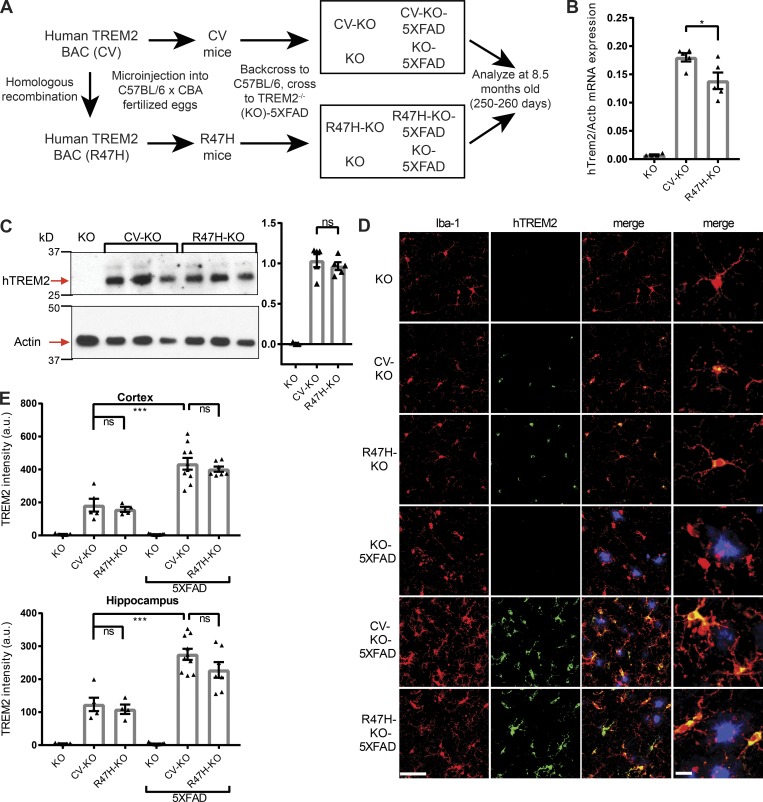
Figure 1.
hTREM2 is expressed at comparable levels in microglia of CV and R47H transgenic mice. (A) Diagram showing generation of 5XFAD and non-5XFAD humanized TREM2 mice. (B) hTREM2 mRNA expression in KO, CV-KO, and R47H-KO whole cortical tissue. (C) Immunoblot of hTREM2 and actin in the PBS-insoluble fraction of hippocampal tissue homogenates shows similar relative hTREM2 protein abundance between CV-KO and R47H-KO by densitometric analysis normalized to actin. (D) Confocal microscopy of cortex of CV-KO, R47H-KO, or KO 5XFAD or non-5XFAD mice shows colocalization of hTREM2 C terminus (green) and microglial marker Iba-1 (red); methoxy-X04 staining for plaques is shown in blue. Bars, 50 µm for first three columns, 10 µm for last column. (E) Quantification of staining intensity of hTREM2 in the cortex and hippocampus. Staining intensity is similar between CV and R47H and increased in 5XFAD compared with non-5XFAD. *, P < 0.05; ***, P < 0.001 by one-way ANOVA with Holm-Sidak multiple comparisons testing. Data are presented as mean ± SEM.
To study the differences between CV and R47H in the brain during AD, we then crossed these mice to 5XFAD mice, in which amyloid pathology is driven by overexpression of mutant human amyloid precursor protein and presenilin-1 (Oakley et al., 2006), and compared these CV+ mTrem2−/− 5XFAD (CV-KO-5XFAD) and R47H+ mTrem2−/− 5XFAD (R47H-KO-5XFAD) mice to mTrem2−/− 5XFAD (KO-5XFAD) and non-5XFAD mice at 8.5 mo of age (Fig. 1 A). To verify comparable hTREM2 expression in the brain, we measured hTREM2 DNA, mRNA, and protein levels in brain homogenates. Despite notable differences in CV and R47H transgene copy numbers (Fig. S1 D), only a 20% difference was observed between CV and R47H mRNAs in transgenic lines (Fig. 1 B). Moreover, quantification of hTREM2 protein expression in hippocampal tissue homogenates of CV and R47H transgenes by immunoblotting showed specific bands for hTREM2 that were similar in intensity by densitometric analysis (Fig. 1 C). To confirm microglia-specific expression, we performed immunofluorescence microscopy of brain sections using an antibody against the C terminus of hTREM2. hTREM2 colocalized exclusively with microglia marker Iba-1 in both non-5XFAD and 5XFAD groups (Fig. 1 D), and quantification of staining intensity yielded similar levels on a per-voxel basis in CV-KO and R47H-KO, which were increased in CV-KO-5XFAD and R47H-KO-5XFAD to similar extents (Fig. 1 E).
Alternatively spliced isoforms of hTREM2 putatively encoding soluble TREM2 could also be detected, and their expression tracked with the main hTREM2 isoform in non-5XFAD (Fig. S1 E) and 5XFAD (Fig. S1 F) brains. The hTREM2 locus also contains hTREML1 and hTREML2 genes, which were present in the integrated BAC (Fig. S1 D). hTREML1 was not expressed in the brain (Fig. S1 G); hTREML2 showed a low level of expression that was similar between transgenes and between 5XFAD and non-5XFAD brains (Fig. S1 H). Therefore, TREML2 expression is unlikely to confound differences in brain innate immune responses between genotypes. Because of a lack of standardized reagents for comparing mTREM2 and hTREM2 levels, we were unable to determine whether CV and R47H are expressed at similar levels as endogenous mTREM2. It is theoretically possible that hTREM2 variants are both overexpressed in our model and that this leads to unnoticed nonphysiological effects. However, the overall similar pattern and level of expression of both transgenes in microglia and other myeloid cells, on both the mRNA and protein levels, suggest that CV and R47H can be compared functionally in vivo.
R47H polymorphism impairs microgliosis
To understand the effect of the R47H variant on microglia in situ, we first examined microglial numbers and microglia–plaque interaction, which have previously been shown to be altered by TREM2 deficiency. As quantified by Iba-1 immunoreactivity, Aβ-driven microgliosis throughout the cortex and hippocampus was enhanced by both CV and R47H transgenes compared with KO (Fig. 2 A), with CV having a larger effect. RT quantitative PCR (qPCR) for the constitutively expressed microglia marker Tyrobp (Dap12 protein) in whole cortical tissue confirmed these differences in microglial mass (Fig. 2 B). To examine microgliosis specifically around plaques, we examined Iba-1+ microglia and methoxy-X04+ plaques using a semiautomated workflow in Imaris and Matlab. We identified the positions of microglial nuclei and plaques within confocal images (Fig. 2 C) and determined the density of microglial nuclei within 0–15 µm and 0–30 µm spherical shells surrounding plaques in the cortex (Fig. 2 D) and hippocampus (Fig. 2 E). As expected, density was greater in the more plaque-restricted 0–15 µm shell than the wider 0–30 µm shell, indicating that microglia are indeed concentrated near plaques. Significantly higher plaque-associated microglial density was evident in CV-KO-5XFAD mice than in KO-5XFAD mice, whereas similar density was observed in R47H-KO-5XFAD mice and KO-5XFAD mice.
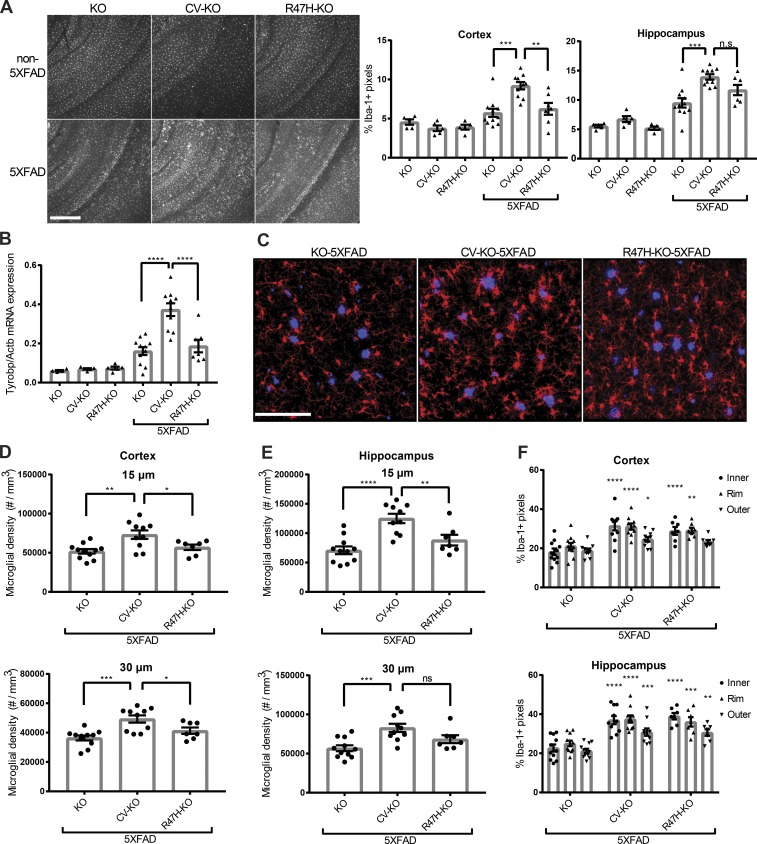
Figure 2.
CV-KO-5XFAD mice have increased numbers of plaque-associated microglia compared with R47H-KO-5XFAD or KO-5XFAD mice. (A) Representative fluorescence images and quantification of Iba-1 area coverage from the cortex and hippocampus of 8.5-mo-old KO-5XFAD, CV-KO-5XFAD, or R47H-KO-5XFAD mice, and littermate non-5XFAD mice, showing higher overall microglia staining in CV-KO-5XFAD. Bar, 300 µm. (B) mRNA expression of constitutively expressed microglia marker Tyrobp in whole cortical tissue shows increase in microglia mass in KO-5XFAD and R47H-KO-5XFAD compared with non-5XFAD, with a further increase in CV-KO-5XFAD. (C) Representative confocal images of 5XFAD cortices, showing plaques (blue, methoxy-X04) and plaque-associated microglia (red, Iba-1). Bar, 100 µm. (D and E) Density of microglia within a 15-µm or 30-µm shell around plaque surfaces within the cortex (D) or hippocampus (E), showing higher microglia density in CV-KO-5XFAD compared with R47H-KO-5XFAD and KO-5XFAD. (F) Coverage of plaques by microglia processes inside the plaque (inner), at the plaque rim (rim), or in a ribbon outside the rim (outer) in the cortex (top) or subiculum (bottom). Both CV-KO-5XFAD and R47H-KO-5XFAD have increased coverage of plaques compared with KO-5XFAD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, by one-way ANOVA (A–E) or two-way ANOVA (F) with Holm-Sidak multiple comparisons testing. Statistical significance in F is referenced to respective “inner,” “rim,” or “outer” values in KO-5XFAD. Data are presented as mean ± SEM.
In addition to the number of microglia around plaques, we measured the extent of direct contact of Iba-1+ processes with Aβ plaques, which has been postulated as a mechanism of limiting plaque toxicity (Wang et al., 2016; Yuan et al., 2016). We measured the fractional microglial coverage of plaque “inner,” “rim,” and “outer” pixels (Fig. 2 F). Both CV-KO-5XFAD and R47H-KO-5XFAD mice had more microglial coverage of plaques than KO-5XFAD mice, but did not differ significantly from each other. Because microglia clustering around plaques has been proposed to promote Aβ clearance, we also measured Aβ levels in the cortex and hippocampus of 5XFAD mice. Flash-frozen tissue was homogenized sequentially in PBS and guanidine solutions to obtain PBS-soluble and -insoluble fractions. Aβ1-40 and Aβ1-42 levels were measured in both fractions by ELISA. In CV-KO-5XFAD mice, compared with other groups, we observed a modest reduction of PBS-soluble but not PBS-insoluble Aβ1-40 and Aβ1-42 in the hippocampus (Fig. S2 A). Aβ levels were similar between genotypes in the cortex (Fig. S2 B). As a complementary measure of insoluble Aβ, total fibrillar plaque area in the cortex, subiculum, and hippocampus (excluding subiculum) was determined by fluorescence microscopy of methoxy-X04 staining, showing no differences between genotypes (Fig. S2 C). This contrasts with our previous study of TREM2 deficiency in the 5XFAD model that showed a difference in all these Aβ species (Wang et al., 2015). However, this is not totally unexpected, given seeming dependency on time point and gene dose in other studies of TREM2 in AD mouse models (Ulrich et al., 2014; Jay et al., 2015, 2017). Detection of an impact of R47H variant on microglial clearance of Aβ in our mouse model of AD may require more time, consistent with the slower course of human AD. All in all, these data provide strong evidence for a partial loss-of-function effect of R47H in terms of microglia number, which is more consistent and prompt than its effect on Aβ or on microglia–plaque contact.
R47H polymorphism reduces microglial activation
To more clearly understand the effect of the R47H variant on microglia, we performed RT-qPCR analysis of whole cortex for microglia activation–related transcripts (Fig. 3 A). The genes Spp1, Gpnmb, and Cst7, encoding osteopontin, osteoactivin, and cystatin F, respectively, are among the most up-regulated in AD models (Orre et al., 2014; Wang et al., 2015) and are indicative of a unique microglial transcriptional profile associated with accumulation of Aβ plaques, part of which is TREM2-dependent (Keren-Shaul et al., 2017). We noticed that Cst7 was up-regulated in all 5XFAD mice (Fig. 3 A) and showed larger differences between genotypes than the constitutive marker Tyrobp (Fig. 2 B), whereas activation markers Spp1 and Gpnmb were much more up-regulated in CV-KO-5XFAD than in other genotypes (Fig. 3 A). Thus, although expression of Cst7 is indicative of microglial activation, expression of Spp1 and Gpnmb more directly reflects TREM2 signaling and is highly sensitive to the R47H variant. Next, we determined RNA levels of the proinflammatory cytokines Tnf, Il6, and Il1b. On a whole-tissue level, Il1b expression was higher in CV-KO-5XFAD mice, but neither Tnf nor Il6 transcripts differed among genotypes (Fig. 3 A).
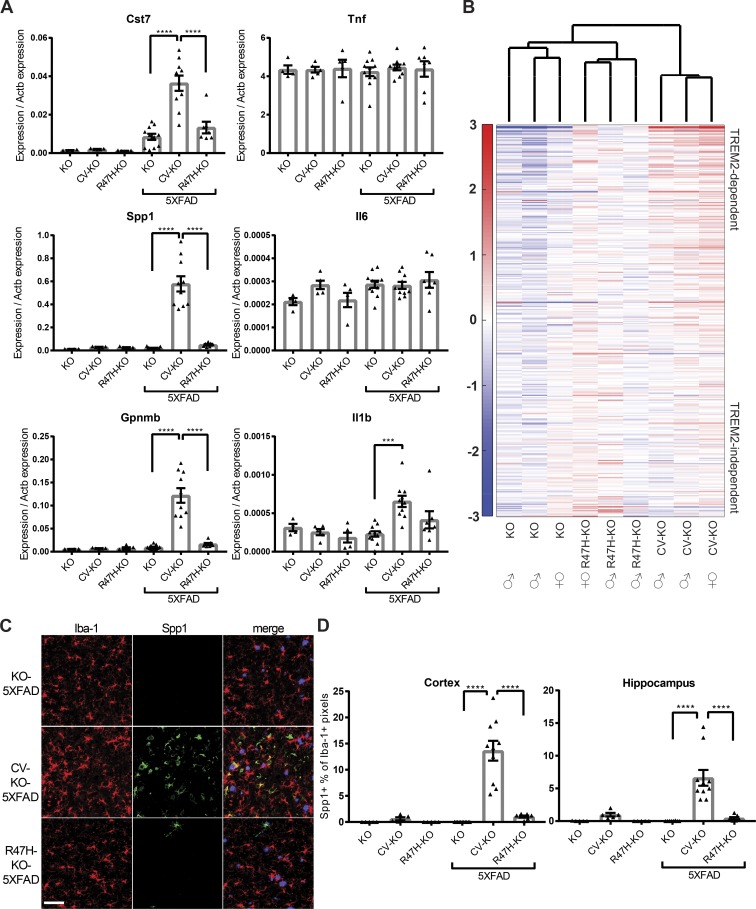
Figure 3.
CV-KO-5XFAD mice have increased microglia activation compared with R47H-KO-5XFAD and KO-5XFAD mice. (A) RT-qPCR on whole cortical tissue for neurodegeneration-associated microglial activation markers Cst7, Spp1, and Gpnmb, as well as classical inflammatory cytokines Tnf, Il6, and Il1b, showing dramatically higher microglial activation markers but not inflammatory cytokine transcripts in CV-KO-5XFAD compared with other groups. (B) Microarray analysis of activation markers in sorted microglia shows that KO-5XFAD and R47H-KO-5XFAD cluster together and separately from CV-KO-5XFAD. Gene names and values are given in Table S1. Some variability within CV-KO-5XFAD, R47H-KO-5XFAD, and KO-5XFAD groups was also observed, with females in all groups having higher activation markers. Color scale represents log2 deviation from the row mean. (C) Spp1 protein is detected by confocal microscopy in CV-KO-5XFAD brains and largely absent from R47H-KO-5XFAD and KO-5XFAD brains. Bar, 50 µm. (D) The percent of Iba-1+ (microglia) pixels that were also Spp1+ was quantified in cortex and hippocampus for all 5XFAD and non-5XFAD groups. ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA with Holm-Sidak multiple comparisons testing. Data are presented as mean ± SEM.
To acquire a global snapshot of transcriptional differences between microglia in different conditions, we performed microarray analysis of sorted microglia from brains of 8.5-mo-old 5XFAD animals. A gene list of microglial activation markers was compiled from our previous data using the same platform (Wang et al., 2015) by selecting genes up-regulated at least twofold between 5XFAD and non-5XFAD microglia and ordering them based on their up-regulation in TREM2-deficient microglia. Genes that were more highly up-regulated in TREM2-sufficient compared with TREM2-deficient microglia were deemed TREM2-dependent, and genes with similar up-regulation were deemed TREM2-independent (Table S1). Unsupervised clustering of samples based on this microglia activation signature grouped KO-5XFAD and R47H-KO-5XFAD together, distinct from CV-KO-5XFAD, and revealed a stepwise increase in activation from KO-5XFAD to R47H-KO-5XFAD to CV-KO-5XFAD (Fig. 3 B). Furthermore, differences between the groups were more pronounced among TREM2-dependent than TREM2-independent genes. We verified that Spp1 was up-regulated on the protein level in a TREM2-dependent manner by confocal microscopy (Fig. 3 C). A large proportion of CV-KO-5XFAD microglia in cortex and hippocampus was positive for Spp1 staining, whereas few positive microglia were observed in R47H-KO-5XFAD and virtually none in KO-5XFAD (Fig. 3 D). We conclude that CV mediates activation of microglia, which is partially affected by R47H, and Spp1 is a sensitive marker of TREM2-dependent activation on the mRNA and protein levels. The effects of R47H on microglia activation are seemingly more dramatic than its effect on microglia numbers, perhaps suggesting that microglia activation plays a key role in AD progression.
Brains of CV but not R47H mice show plaque- and neuron-coincident soluble TREM2
Given recent studies on TREM2 polarization during Aβ accumulation (Yuan et al., 2016), we sought to shed light on localization of CV and R47H hTREM2. As shown in Fig. 1, hTREM2 is exclusively detected in Iba-1+ microglia by confocal microscopy using an antibody directed against its intracellular C terminus. However, brain sections stained with an antibody against hTREM2 extracellular domain (ECD) showed diffuse distribution of hTREM2 outside Iba-1+ voxels (Fig. 4 A) and hTREM2 C terminus+ voxels (Fig. 4 B) in CV-KO-5XFAD mice, suggesting the presence of soluble TREM2. The intensity of hTREM2 ECD staining was quantified both within Iba-1+ voxels and within Iba-1− voxels in the cortex (Fig. 4 C) and hippocampus (Fig. 4 D). Although hTREM2 ECD intensity within microglia remained relatively constant between non-5XFAD and 5XFAD images, hTREM2 intensity outside microglia dramatically increased in CV-KO-5XFAD images and slightly increased in R47H-KO-5XFAD images. This nonmicroglial staining was absent in KO-5XFAD.
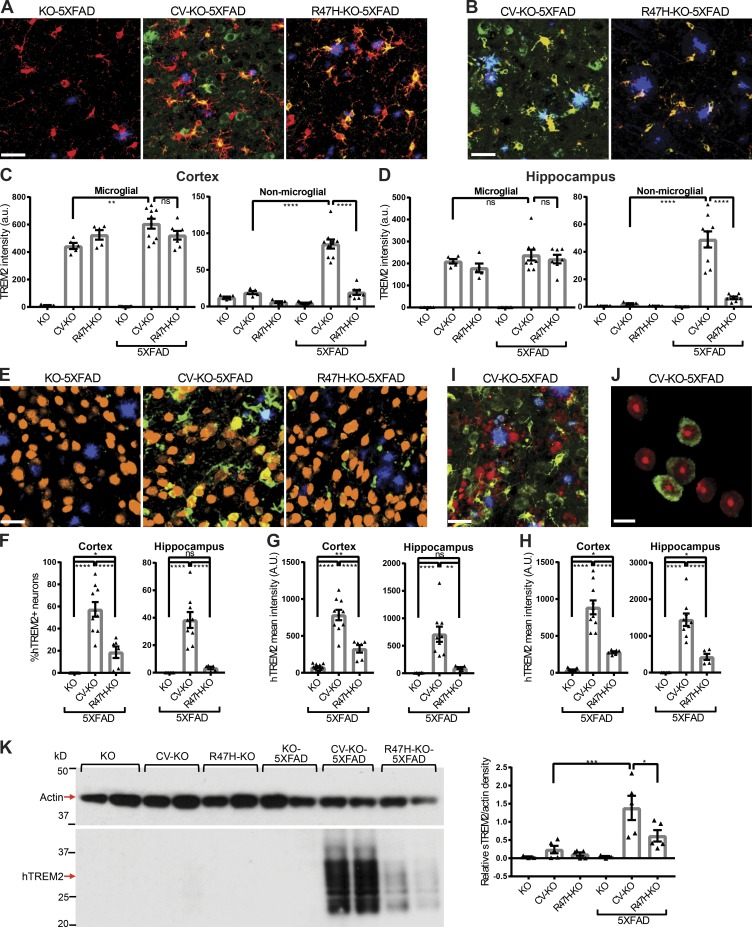
Figure 4.
CV-KO-5XFAD mice show unique localization of soluble hTREM2 to neurons, plaques, and other nonmicroglial spaces. (A) Representative confocal images showing plaques (methoxy-X04, blue), microglia (Iba-1, red), and hTREM2 ECD (green) in the cortex. Bar, 30 µm. (B) Representative confocal images showing plaques (blue), hTREM2 C terminus (red), and hTREM2 ECD (green). In CV-KO-5XFAD brains, hTREM2 ECD is present in voxels that lack either Iba-1 or hTREM2 C terminus staining, whereas in R47H-KO-5XFAD, hTREM2 ECD largely colocalizes with Iba-1 and hTREM2 C terminus. Bar, 30 µm. (C and D) Mean intensity of hTREM2 ECD staining was quantified in Iba-1–positive (microglial) voxels and Iba-1–negative (nonmicroglial) voxels in (C) cortex and (D) hippocampus, showing similar expression levels in microglia but a significant increase outside of microglia in CV-KO-5XFAD brains only. (E) Representative confocal images showing plaques (methoxy-X04, blue), neuronal soma (NeuN, orange), and hTREM2 (green). Bar, 30 µm. (F) Frequency of hTREM2+ neurons and (G) mean hTREM2 intensity in neuronal soma was quantified in cortex and hippocampus. (H) Mean intensity of hTREM2 staining was quantified in methoxy-X04+ plaques in cortex and hippocampus. hTREM2 ECD staining on plaques and neuronal soma was significantly higher in CV-KO-5XFAD compared with R47H-KO-5XFAD. (I) Representative confocal image of plaques (blue), APP+ dystrophic neurites (red), and hTREM2 ECD (green), showing a relative lack of soluble TREM2 on dystrophic neurites. Bar, 30 µm. (J) Representative confocal slice showing nuclei (ToPro-3, red) and hTREM2 ECD (green) within NeuN+ neuronal soma. hTREM2+ neurons do not show nuclear abnormalities characteristic of apoptosis. Bar, 10 µm. (K) Immunoblot of the PBS-soluble fraction of hippocampal homogenates for hTREM2, showing a hTREM2 smear at lower molecular weight than full-length hTREM2, indicating soluble TREM2. Densitometric analysis normalized to actin was performed, showing that CV-KO-5XFAD mice had more soluble TREM2 than CV-KO and R47H-KO-5XFAD mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA with Holm-Sidak multiple comparisons testing. Data are presented as mean ± SEM.
A particularly intense hTREM2 ECD staining was observed on a fraction of nonmicroglial cells and Aβ plaques. Costaining for NeuN and hTREM2 ECD demonstrated that the hTREM2 ECD+ cells are neurons (Fig. 4 E). Quantification of hTREM2 intensity specifically in NeuN+ neuronal soma showed that CV-KO-5XFAD mice had a higher frequency of hTREM2+ neurons (Fig. 4 F) and higher hTREM2 intensity in neuronal soma (Fig. 4 G) than R47H-KO-5XFAD mice in both cortex and hippocampus. Similarly, hTREM2 intensity in Aβ plaques was higher in CV-KO-5XFAD mice than R47H-KO-5XFAD mice in both cortex and hippocampus (Fig. 4 H). Although TREM2 has been reported to bind damage-associated phospholipids such as phosphatidylserine (Wang et al., 2015), hTREM2 ECD did not colocalize to APP+ dystrophic neurites (Fig. 4 I), and nuclei of hTREM2+ neurons did not show morphological abnormalities such as pyknosis or fragmentation (Fig. 4 J). To verify that the unique distribution of hTREM2 ECD reflected soluble TREM2, we performed immunoblots on the detergent-free PBS-soluble fraction of hippocampal tissue homogenates for TREM2 (Fig. 4 K). A low-molecular-weight hTREM2 smear was detected, with the lower range of the smear corresponding to the predicted soluble hTREM2 size of 20 kD. Densitometry confirmed higher soluble TREM2 levels in CV-KO-5XFAD compared with R47H-KO-5XFAD.
We were able to take advantage of reagents targeting both the extracellular and the intracellular domains of human TREM2 to show for the first time that soluble TREM2 production markedly increases during Aβ accumulation in a mouse model and coincides with neurons and plaques in vivo, suggesting binding. Because soluble and full-length TREM2 share nearly the entire ECD, this staining pattern provides insight into the distribution of hTREM2 ligands in the brain in situ. This unexpected distribution of hTREM2 in tissues of CV-KO-5XFAD mice also has several implications for the role of TREM2 in AD. Although the 5XFAD model does feature neuronal apoptotic cell death (Eimer and Vassar, 2013) and TREM2 has been reported to recognize apoptotic neurons (Takahashi et al., 2005; Hsieh et al., 2009), the sheer number and normal nuclear morphology of hTREM2+ neurons suggest that soluble TREM2 staining of neurons reflects cellular events distinct from apoptosis. Whether soluble TREM2 can affect signaling in neurons remains to be determined, but a handful of studies have indicated a prosurvival and/or proinflammatory effect of soluble TREM2 on myeloid cells (Wu et al., 2015; Zhong et al., 2017). Moreover, the presence of soluble TREM2 on plaques may indicate binding to plaque-associated phospholipids or directly to Aβ peptides and may affect plaque structure or provide signals to nearby cells. Importantly, this process also appears to be strongly affected by the R47H variant in vivo, which raises the possibility that shedding of soluble TREM2 may be causally linked to disease progression.
CV and R47H are equally shed through activation of Adam17
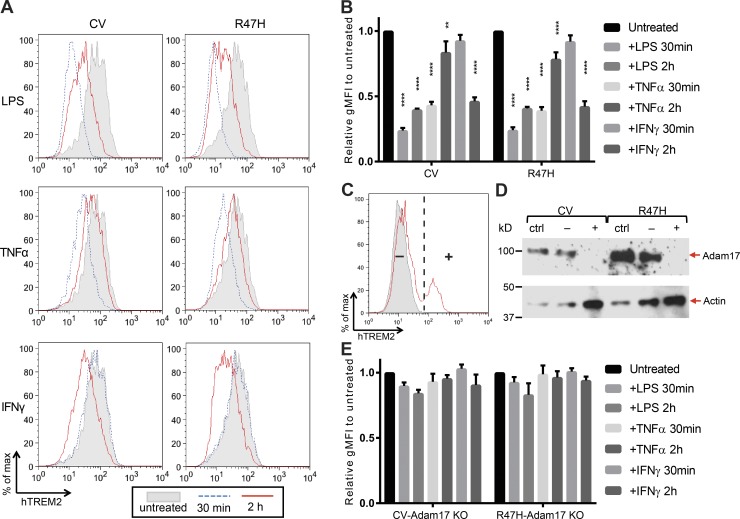
We sought to shed light on the differences in soluble TREM2 observed between genotypes. To that end, we generated RAW264.7 macrophage-like cell lines that lack endogenous TREM2 and overexpress either CV or R47H. Previous work had indicated that diverse immunostimulatory molecules such as LPS, TNFα, and IFNγ could lead to down-regulation of cell-surface TREM2 (Bouchon et al., 2001; Turnbull et al., 2006). We found that in our system, surface expression of both CV and R47H, as measured by flow cytometry, could indeed be down-regulated by these molecules within 2 h of stimulation (Fig. 5, A and B). Although LPS and TNFα led to a rapid down-regulation of surface hTREM2 at 30 min and partial recovery by 2 h, IFNγ had no detectable effect at 30 min but decreased surface hTREM2 by 2 h. To determine whether these changes were a result of cleavage, we lentivirally transduced cells with Cas9 and a guide RNA against Adam17, which was previously shown to cleave TREM2 (Kleinberger et al., 2014; Feuerbach et al., 2017). Unlike nontransduced cells, a fraction of transduced cells retained TREM2 surface expression upon LPS treatment (Fig. 5 C); this seemingly cleavage-resistant fraction (+) was purified from the remainder of cleavage-sensitive transduced cells (−) by FACS, and both populations were analyzed by immunoblot for Adam17 protein levels (Fig. 5 D). The (−) population showed only a slight decrease in Adam17 protein relative to nontransduced cells, possibly reflecting the presence of heterozygous Adam17 knockout, whereas the (+) population showed virtually no residual Adam17 expression and was considered Adam17 KO. LPS, TNFα, and IFNγ treatment all failed to decrease TREM2 surface expression in CV-Adam17 KO and R47H-Adam17 KO (Fig. 5 E). As soluble TREM2 is shed constitutively by RAW264.7 cells, we were unable to consistently detect an increase in the concentration of soluble TREM2 in the culture supernatant (not depicted). Thus, the change in cell surface TREM2 more sensitively reflected Adam17-induced TREM2 cleavage upon cytokine stimulation. Other studies have suggested that Adam10 is the main sheddase of TREM2 (Schlepckow et al., 2017; Thornton et al., 2017). At least in our system, Adam10 in the absence of Adam17 is not sufficient for cytokine-induced cleavage. These results suggest that various myeloid cell signaling pathways that may be activated in neuropathology can induce Adam17-dependent release of soluble TREM2 and that R47H polymorphism does not directly impact this process.
Figure 5.
Various immunostimulatory signals induce similar CV and R47H hTREM2 cleavage from the cell surface. (A) Representative flow cytometry plots demonstrating that both CV and R47H hTREM2 are lost from the cell surface of transduced RAW264.7 cells upon treatment with LPS, TNFα, or IFNγ. (B) Quantification of gMFI of LPS, TNFα, and IFNγ treated cells relative to untreated cells. Shown is mean ± SEM for three independent experiments. (C) Adam17 was knocked out in CV- and R47H-expressing RAW264.7 cells using lentivirally delivered Cas9 and guide RNA. LPS-stimulated cells were stained for hTREM2. Nontransduced cells (ctrl; gray shaded) uniformly lost hTREM2 surface expression. Transduced cells (solid red line) had two populations, one that retained hTREM2 surface expression (+) and one that lost hTREM2 surface expression (−), and these two populations were sorted by FACS. (D) Immunoblot for Adam17 demonstrates that the CV and R47H (+) populations have highly efficient knockout of Adam17 protein expression relative to either ctrl or (−) populations. (E) Experiments conducted as in A and B using Adam17 KO cells show that the effects of LPS, TNFα, and IFNγ on hTREM2 surface expression are ablated in Adam17-deficient cells. Shown is mean ± SEM for three independent experiments. **, P < 0.01; ****, P < 0.0001 by one-way ANOVA with Holm-Sidak multiple comparisons testing.
Taking these in vitro results into consideration, the blunted increase in soluble TREM2 in R47H-KO mice during AD progression may be a result of several nonexclusive mechanisms: (a) R47H hTREM2 itself may not be bound and retained by ligands in the brain as well as CV hTREM2, which is consistent with in vitro data showing decreased ligand binding of R47H; (b) shedding of soluble TREM2 may be reduced because of impaired activation of microglia and Adam17, although in vitro R47H polymorphism does not impact the Adam17-mediated proteolysis event directly; (c) reduced microglial activation may also impact the induction of alternatively spliced forms of TREM2 proposed to encode soluble TREM2; and (d) the total amount of TREM2 available for generating soluble TREM2 may be decreased because of global reduction of microglial mass.
Conclusions
All in all, our results provide conclusive evidence for an in vivo reduction of function effect of R47H, and thus a protective role of TREM2 in human disease. Our data also suggest that microglial activation in AD leads to widespread shedding of soluble TREM2, which then interacts with plaques and neurons, and that this phenomenon is also affected by R47H polymorphism. Further studies are required to elucidate the functional impact of soluble TREM2 in AD, as well as additional differences between CV and R47H, e.g., whether R47H affects the biochemical composition of Aβ and/or the degree of Aβ oligomerization and amyloid maturation. Studies examining other significant TREM2 polymorphisms may provide additional insights into microglia function during AD. However, given that R47H polymorphism leads to clear defects in microglia number and activation state and defects in soluble TREM2 activity, it is likely that proper TREM2 function, and perhaps downstream microgliosis, microglial activation, and soluble TREM2 production, plays a protective role in AD.
Materials and methods
Mice
Trem2−/− 5XFAD mice were generated as previously described (Wang et al., 2015). The CTD-2210D2 bacterial artificial chromosome (BAC) clone was purchased from Thermo Fisher Scientific, and the R47H polymorphism was introduced by recombination using the Quick and Easy BAC Modification kit (Gene Bridges). The entire BAC was injected into the nucleus of fertilized eggs from C57BL/6 × CBA mice, and CV and R47H founders were obtained. The two lines selected for further work were subsequently backcrossed onto the C57BL/6 background for at least four generations (some done in the process of crossing to Trem2−/− and 5XFAD mice, both of which are on a C57BL/6 background). All mice were bred and housed in specific pathogen-free conditions. The Institutional Animal Care and Use Committee at Washington University in St. Louis approved all protocols used in this study. Sample size for animals was chosen based on previous studies. No animals were excluded from analysis. Animals were sacrificed between the ages of 245 and 260 d (8.5 mo), with each experimental group containing both transgenic and nontransgenic mice. Investigators were not blinded to experimental groups, but all samples and data were processed in a high-throughput or automated fashion.
Cell culture and biochemical assays
To obtain thioglycollate-elicited peritoneal macrophages, mice were injected intraperitoneally with 1 ml of 3% thioglycollate medium (prepared in water and autoclaved; Sigma-Aldrich), and cells were harvested by peritoneal lavage after 3 d. Cells were subsequently stained for flow cytometry.
To prepare BMDMs, femurs and tibias were removed and flushed with PBS. Cells were counted and plated at 2 × 106 cells/100 mm Petri dish in RPMI supplemented with GlutaMAX, penicillin/streptomycin, nonessential amino acids, pyruvate, and 10% heat-inactivated FBS and 10% L-cell conditioned medium as a source of CSF1. Cells were cultured for 4–6 d before use. Human HDL (Millipore) was supplemented at a concentration of 50 µg/ml.
RAW264.7 cells were cultured on nontissue culture–treated 12-well plates in RPMI supplemented with GlutaMAX, penicillin/streptomycin, pyruvate, and 5% FBS. RAW264.7 cells were transduced with Cas9 overexpression lentivirus and transfected with plasmid-expressing guide RNA against mouse TREM2. TREM2-negative cells were sorted by FACS. lentiCas9-Blast and lentiGuide-Puro were gifts from F. Zhang (Broad Institute, Cambridge, MA; plasmids 52962 and 52963, respectively, from Addgene). These TREM2-deficient RAW264.7 cells were retrovirally transduced with either CV or R47H driven under the CMV promoter, and cells were sorted for hTREM2 expression by FACS. Cells were stimulated for 30 min or 2 h with 10 ng/ml LPS (K12; InvivoGen), 20 ng/ml recombinant TNFα (PeproTech), or 20 ng/ml recombinant IFNγ (PeproTech). For sorting Adam17-KO populations, cell were stimulated for 30 min with 10 ng/ml LPS and then stained for hTREM2.
Preparation of brain samples
Mice were anesthetized with ketamine/xylazine and perfused with ice-cold PBS containing 1 U/ml of heparin. Right brain hemispheres were fixed in 4% PFA overnight at 4°C, rinsed in PBS, and incubated overnight at 4°C in 30% sucrose before freezing in a 2:1 mixture of 30% sucrose and optimal cutting temperature compound. Serial 40 µm coronal sections were cut on a cryo-sliding microtome. Cortices and hippocampi of the left-brain hemispheres were carefully dissected out and flash frozen for biochemical analysis. A portion of cortex was preserved in RNAlater (Ambion) until all samples were collected.
Confocal microscopy
Floating sections were blocked with 3% BSA and 0.25% Triton X-100 in PBS, and then stained with anti–Iba-1 (rabbit polyclonal, 1:5,000; Wako; or goat polyclonal, 1:1,000; Abcam), anti–human TREM2 ECD (goat polyclonal, 1:500; R&D Systems), anti–human TREM2 C terminus (D814C rabbit mAb, 1:500; Cell Signaling), anti-Spp1 (goat polyclonal, 1:500; R&D Systems), anti-APP (22C11 mouse mAb, 1:1,000; Millipore) and/or anti-NeuN (D3S3I rabbit mAb, 1:500; Cell Signaling) overnight at 4°C followed by staining with anti–rabbit IgG DyLight 549 (1:2,000; Vector), anti–goat IgG Alexa Fluor 488 (1:2,000; Abcam), anti–rabbit IgG Alexa Fluor 647 (goat polyclonal, 1:1,000; Invitrogen), anti–goat IgG-biotin (donkey polyclonal, 1:2,000; Invitrogen), streptavidin Alexa Fluor 647 (1:2,000; Invitrogen), methoxy-X04 (3 µg/ml; Tocris), and/or TO-PRO-3 iodide (300 nM; Thermo Fisher Scientific) for 1 h at room temperature. All antibodies were used in blocking buffer, and between all incubations, sections were washed for 10 min in PBS three times. Images were collected using a Nikon A1Rsi+ confocal microscope. Three-dimensional image segmentation of microglia, plaques, and neurons, and extraction of parameters were performed in Imaris 8.1 (Bitplane), and further processing was performed using automated scripts in Matlab (Mathworks). For detailed image analysis procedures, see Supplemental Methods.
Flow cytometry
Staining for human TREM2 was performed using in-house–generated unconjugated 21E10 antibody (15 µg/ml) followed by PE-conjugated goat (Fab′)2 anti–mouse IgG1 (1:100; SouthernBiotech). Staining for myeloid cells was performed using BV421–anti-CD45 (30-F11, 1:200; BioLegend), FITC–anti-CD11b (M1/70, 1:200; eBioscience), and APC–anti-F4/80 (BM8, 1:100; eBioscience). Samples were incubated with primary antibodies in blocking buffer (anti–mouse CD16/32) for 30 min on ice, washed two times with FACS buffer (2% BCS, 0.01% NaN3 in PBS), and incubated with secondary antibody in FACS buffer for 30 min on ice. Samples were read using a FACSCalibur (BD), and data were analyzed in FlowJo.
Immunoblot
Samples were prepared from PBS-soluble or PBS-insoluble guanidine-soluble fractions of brain homogenates (as described in Aβ ELISA), or from cell lysates, by adding 1× NuPAGE LDS sample buffer (Invitrogen) and 10% β-mercaptoethanol and boiling for 5 min. Samples were run on NuPAGE 4–12% Bis-Tris precast gels (Invitrogen), transferred to nitrocellulose membranes, and blocked with 5% milk in PBS + 0.05% Tween-20 (PBS-T) for 1 h at room temperature. Membranes were incubated with in-house–generated 29E3 anti–human TREM2 antibody (mouse IgG1, 4 µg/ml), anti-TACE/ADAM17 (rabbit polyclonal, 1:1,000; Novus), or anti–pan-actin (rabbit polyclonal, 1:4,000; Cell Signaling,) in 0.5% milk/PBS-T overnight at 4°C, and incubated in HRP-conjugated anti–rabbit Ig (goat polyclonal, 1:1,000; SouthernBiotech) or HRP-conjugated anti–mouse IgG1 (goat polyclonal, 1:1,000; SouthernBiotech) at room temperature for 1 h, with three washes of PBS-T between steps. Blots were detected by SuperSignal West Pico Chemiluminescent Substrate or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) with autoradiographic film. Developed films were scanned using an Epson Perfection V370 photo scanner and analyzed using Image Studio Lite (LI-COR).
Copy number determination, RT-qPCR, and microarray analysis
For copy number analysis, genomic DNA was isolated from the cerebellum of 5-mo-old mice using the QIAamp DNA Mini kit and amplified by real-time qPCR using primer pairs against two genomic reference loci (Lin et al., 2010) as well as three loci within the BAC (Table 1). Relative copy numbers were determined by ΔΔCt, normalizing to the mean of both reference loci.
Table 1. qPCR primers.
| Name | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| Genomic locus | ||
| RPP38 (ref) | TCATTGGCTTAAAATGTGT | TTTATTTTAAGGGGTTGTAATG |
| RC3H2 (ref) | ACTAAGTCAAGGCTACTGTG | TTCTGGTGTCAGTATGGAAG |
| hTREM2 | ATCCCTGCCCAGTCCACCCTTGATGGCT | CCATCCTTCTCCTCCTGGCCTGCATCTTT |
| hTREML1 | GCGTACGTTTCTCACAGACC | CCCACTCCTCAAGAACCCAT |
| hTREML2 | TTCACCCCAGGACTCATCAC | GGCATGGAGGGTAGTCTGTT |
| Gene | ||
| Actb | GGAGGGGGTTGAGGTGTT | TGTGCACTTTTATTGGTCTCAAG |
| Tyrobp | GAGTGACACTTTCCCAAGATGC | CCTTGACCTCGGGAGACCA |
| hTREM2 iso1 | GCATCTCCAGGAGCCTCTTG | TGAGAAAGATGCAGGCCAGG |
| hTREM2 iso2 | CGGGATGCTGGAGATCTCTG | TCTCAGCCCTGGAGATGCT |
| hTREM2 iso3 | CATCTCCAGGCCATCTCAAG | AGGAGGAGAAGGATGGAAGT |
| hTREML1 | CTGGCGAGTATGGCTGCAT | TGCCAATCTTATGGGTCTCTTCT |
| hTREML2 | CCTATAAGGGCTACAAAAACCGC | CCCATCAAGGGGTACAGGA |
| Spp1 | CACTCCAATCGTCCCTACAGT | CTGGAAACTCCTAGACTTTGACC |
| Gpnmb | CATTCCCATCTCGAAGGTGAAA | AAATGGCAGAGTCGTTGAGGA |
| Cst7 | GGAGCTGTACTTGCCGAGC | CATGGGTGTCAGAAGTTAGGC |
| Tnf | GCCTCTTCTCATTCCTGCTTGT | TGATCTGAGTGTGAGGGTCTGG |
| Il1b | GAAATGCCACCTTTTGACAGTG | TGGATGCTCTCATCAGGACAG |
| Il6 | GCCAGAGTCCTTCAGAGAGATACA | CTTGGTCCTTAGCCACTCCTTC |
Total RNA was isolated by tissue homogenization in TRIzol reagent (Invitrogen), and single-strand cDNA was synthesized with qScript cDNA Supermix (Quantabio). Real-time qPCR was performed using iTaq SYBR Green Supermix (Bio-Rad) and the LightCycler 96 detection system (Roche) or StepOnePlus (Applied Biosystems). Primers were chosen from PrimerBank or custom-designed (Wang et al., 2012). Human TREM2 isoform three-primer sequences were from a previous publication (Jin et al., 2014). The list of primers is in Table 1.
For microarray analysis of sorted microglia, perfused brains were homogenized using the Neural Dissociation kit (T) (Miltenyi) per the manufacturer’s instructions. The homogenized tissue was run through a 70-µm cell strainer and resuspended and centrifuged in 30% Percoll (GE). The resulting pellet was stained for CD11b and CD45, and CD11b+CD45+ cells were sorted using a FACSAriaII (BD). RNA was extracted from sorted cells using the RNeasy Micro Plus kit (Qiagen) per the manufacturer’s instructions and submitted for microarray analysis at the Genome Technology Access Center at Washington University in St. Louis. Samples were amplified using the Nugen Ovation Pico SL and run on the Affymetrix Gene 1.0 ST platform.
Image analysis
For measurement of Iba-1 area coverage, coronal sections from −1.5 mm to −3 mm from bregma were stained for Iba-1 as described in Confocal microscopy and imaged by epifluorescence microscopy at low magnification. A region encompassing portions of cortex, hippocampus, and midbrain was drawn in ImageJ (National Institutes of Health), omitting edges of the section and/or image to limit artifacts, and percent area coverage was determined by batch processing in ImageJ, setting as a threshold a fixed intensity greater than the mean for the selected region, i.e., 30 arbitrary units greater than mean. A similar procedure was performed for quantification of fibrillar plaque coverage, except that the threshold was set to twice the mean of the selected region.
For measurement of microglial density around plaques, plaque volume and center of mass were determined using the Surfaces function of Imaris on methoxy-X04 image data. Microglial nuclei were identified using the Spots function of Imaris for colocalized Iba-1 and ToPro-3 nuclear staining. To calculate the number of microglial nuclei within a given distance from plaque surfaces, each plaque was modeled as an idealized sphere with the same volume and center of mass. Pairwise Euclidean distances were calculated between each microglial nucleus and each plaque, taking into account the idealized plaque radius; e.g., for a microglia-plaque pair separated by 40 µm center-to-center with an equivalent plaque radius of 10 µm, the distance would be calculated as 30 µm from the plaque surface. For each plaque, the number of microglia within the specified distance was enumerated. Microglia were allowed to be counted for multiple plaques. Next, the occupiable volume for microglia was calculated by finding the volume of the sphere (or spherical segment) that falls within the specified distance. As an example, for a plaque of radius 10 µm and a cutoff of 30 µm, the volume of a 40-µm sphere is determined. If either the top or bottom of this sphere (or both) extends beyond the top or bottom of the z-stack, respectively, the volume is calculated using the formula for spherical segments instead of the formula for a sphere. Finally, the number of microglia is divided by the occupiable volume to obtain the density, and these values are averaged for all the plaques for a given animal.
For measurement of plaque coverage by microglia, three sequential slices were z-projected by maximum intensity separately for methoxy-X04 and Iba-1 images. Plaques were traced by setting two intensity thresholds, a floor for nonplaque and a ceiling for definite plaque pixels. For pixels in between thresholds, a Laplacian of Gaussian edge detection algorithm was implemented in Matlab, followed by dilation of edges, filling in of small holes, and fine erosion of the final shape to achieve a smooth border. Only contiguous shapes above a certain size were retained. To label “inner,” “rim,” and “outer” pixels, a combination of image erosion and image dilation was performed on plaque traces. After slightly eroding plaque traces, pixels within this smaller area were deemed “inner.” Next, plaque traces were slightly dilated, and additional pixels included in this larger area were deemed “rim.” Finally, plaque traces were further dilated, and these additional pixels were deemed “outer.” A similar tracing process was used for microglia. Finally, the percent coverage of plaque pixels, of “inner,” “rim,” or “outer” pixels, was calculated as the percent of those pixels that were also labeled as microglia pixels. The remainder of the z-stack was divided into three-slice projections and processed the same way.
For measurement of Spp1+ pixels within Iba-1+ pixels, z-stacks were projected by maximum intensity projection, and thresholds for both Spp1 and Iba-1 were determined by visual inspection and used for all images. For each z-projected image, the percentages of Iba-1+ pixels and Spp1+ Iba-1+ pixels were calculated, and the ratio was calculated.
For measurement of percent of positive neurons and the mean intensity of plaques and neurons with regard to hTREM2 staining, first the Surfaces function in Imaris was used on methoxy-X04 and NeuN images, respectively, to label plaques and neuron cell bodies. From Imaris, the mean intensity of the hTREM2 channel within each object was exported to Microsoft Excel. A threshold of mean hTREM2 intensity was set to determine the percent positively staining neurons, and the means across all relevant objects was simply averaged to obtain the final mean intensity.
To display hTREM2 and nuclear staining only in neuronal cell bodies, the Surfaces function in Imaris was used to mask any NeuN-negative voxels in both channels.
Aβ ELISA
Cortical tissue was homogenized first in PBS containing protease inhibitor (Roche) at 0.5× concentration. Remaining insoluble material was pelleted. Supernatant was collected as the PBS-soluble fraction, and the pellet was resuspended in 5.5 M guanidine and 50 mM Tris, pH 8.0, buffer and further homogenized and incubated on a rotator for 3 h at room temperature. The samples were centrifuged again to pellet insoluble material, and the supernatant was collected as the PBS-insoluble guanidine fraction.
2G3 mAb (Aβ1-40) or 21F12 mAb (Aβ1-42) was coated onto immunosorbent ELISA plates (Thermo Fisher Scientific) overnight at 4°C. Samples were diluted and incubated on plates overnight at 4°C. Sample protein content was also measured by DC protein assay (BioRad). Plates were subsequently incubated with biotinylated 3D6 mAb (both Aβ1-40 and Aβ1-42), followed by HRP-conjugated streptavidin (SouthernBiotech) for 1 h each. Between incubation steps, plates were washed three times with PBS-T. Plates were developed with O-phenylenediamine dihydrochloride substrate (Sigma-Aldrich).
Statistics
Data in figures are presented as mean ± SEM. Unless otherwise stated, statistical analysis was performed using Prism (GraphPad). Statistical analysis to compare the mean values for multiple groups was performed using a one-way or two-way ANOVA, as appropriate, with Holm-Sidak multiple comparisons testing.
Data availability
All relevant data and computer code are available from the authors. Microarray data are accessible through GEO Series accession number GSE108595.
Online supplemental material
Fig. S1 provides additional characterization of the transgenic mice. Fig. S2 contains measurements of amyloid accumulation. Table S1 contains the list of activation genes and expression values shown in Fig. 3 B.
Supplementary Material
Acknowledgments
Microarray analysis was performed through the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine, partially supported by a National Cancer Institute Cancer Center Support Grant (P30 CA91842) to the Siteman Cancer Center and by a Institute of Clinical and Translational Sciences/Clinical and Translational Science Awards grant from the National Center for Research Resources (UL1TR000448), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research. Confocal microscopy was performed through the use of the Washington University Center for Cellular Imaging supported by the Washington University School of Medicine in St. Louis, the Children’s Discovery Institute of Washington University, and St. Louis Children’s Hospital (grant CDI-CORE-2015-505), and the National Institute for Neurological Disorders and Stroke (grant NS086741). Generation of transgenic mice was performed with the help of the Mouse Genetics Core at Washington University School of Medicine with partial support from NIH grants P30DK052574 and P30DK020579. We thank Dr. Richard Cho and Cell Signaling Technology for generously providing anti–human TREM2 C terminus antibody. We thank Drs. Jason Ulrich and David Holtzman for providing helpful discussion and reagents.
T.K. Ulland was supported by NIH grant 5T32CA009547-30. This work was also supported by NIH grant RF1 AG05148501 and a Cure Alzheimer’s Fund grant to M. Colonna.
The authors declare no competing financial interests.
Author contributions: W.M. Song, S. Joshita, Y. Zhou, and T.K. Ulland designed and performed experiments. S. Joshita generated CV and R47H mouse lines. S. Gilfillan generated and maintained KO-5XFAD, CV-KO-5XFAD, and R47H-KO-5XFAD mice. M. Colonna designed and supervised the project. W.M. Song and M. Colonna wrote the manuscript.
References
- Atagi Y., Liu C.C., Painter M.M., Chen X.F., Verbeeck C., Zheng H., Li X., Rademakers R., Kang S.S., Xu H., et al. 2015. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). J. Biol. Chem. 290:26043–26050. 10.1074/jbc.M115.679043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.C., DeVaux L.B., and Farzan M.. 2015. The Triggering Receptor Expressed on Myeloid Cells 2 Binds Apolipoprotein E. J. Biol. Chem. 290:26033–26042. 10.1074/jbc.M115.677286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon A., Hernández-Munain C., Cella M., and Colonna M.. 2001. A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J. Exp. Med. 194:1111–1122. 10.1084/jem.194.8.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., and Pericak-Vance M.A.. 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 261:921–923. 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- Eimer W.A., and Vassar R.. 2013. Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Aβ42 accumulation and Caspase-3 activation. Mol. Neurodegener. 8:2 10.1186/1750-1326-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Brooks D.J., Okello A., and Edison P.. 2017. An early and late peak in microglial activation in Alzheimer’s disease trajectory. Brain. 140:792–803. 10.1093/brain/aww349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerbach D., Schindler P., Barske C., Joller S., Beng-Louka E., Worringer K.A., Kommineni S., Kaykas A., Ho D.J., Ye C., et al. 2017. ADAM17 is the main sheddase for the generation of human triggering receptor expressed in myeloid cells (hTREM2) ectodomain and cleaves TREM2 after Histidine 157. Neurosci. Lett. 660:109–114. 10.1016/j.neulet.2017.09.034 [DOI] [PubMed] [Google Scholar]
- Griciuc A., Serrano-Pozo A., Parrado A.R., Lesinski A.N., Asselin C.N., Mullin K., Hooli B., Choi S.H., Hyman B.T., and Tanzi R.E.. 2013. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 78:631–643. 10.1016/j.neuron.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S., Younkin S., et al. Alzheimer Genetic Analysis Group . 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368:117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamerman J.A., Jarjoura J.R., Humphrey M.B., Nakamura M.C., Seaman W.E., and Lanier L.L.. 2006. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J. Immunol. 177:2051–2055. 10.4049/jimmunol.177.4.2051 [DOI] [PubMed] [Google Scholar]
- Heneka M.T., Carson M.J., El Khoury J., Landreth G.E., Brosseron F., Feinstein D.L., Jacobs A.H., Wyss-Coray T., Vitorica J., Ransohoff R.M., et al. 2015. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14:388–405. 10.1016/S1474-4422(15)70016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Koike M., Spusta S.C., Niemi E.C., Yenari M., Nakamura M.C., and Seaman W.E.. 2009. A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J. Neurochem. 109:1144–1156. 10.1111/j.1471-4159.2009.06042.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itagaki S., McGeer P.L., Akiyama H., Zhu S., and Selkoe D.. 1989. Relationship of microglia and astrocytes to amyloid deposits of Alzheimer disease. J. Neuroimmunol. 24:173–182. 10.1016/0165-5728(89)90115-X [DOI] [PubMed] [Google Scholar]
- Jay T.R., Miller C.M., Cheng P.J., Graham L.C., Bemiller S., Broihier M.L., Xu G., Margevicius D., Karlo J.C., Sousa G.L., et al. 2015. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212:287–295. 10.1084/jem.20142322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay T.R., Hirsch A.M., Broihier M.L., Miller C.M., Neilson L.E., Ransohoff R.M., Lamb B.T., and Landreth G.E.. 2017. Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J. Neurosci. 37:637–647. 10.1523/JNEUROSCI.2110-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.C., Benitez B.A., Karch C.M., Cooper B., Skorupa T., Carrell D., Norton J.B., Hsu S., Harari O., Cai Y., et al. 2014. Coding variants in TREM2 increase risk for Alzheimer’s disease. Hum. Mol. Genet. 23:5838–5846. 10.1093/hmg/ddu277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J., et al. 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368:107–116. 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kleinberger G., Yamanishi Y., Suárez-Calvet M., Czirr E., Lohmann E., Cuyvers E., Struyfs H., Pettkus N., Wenninger-Weinzierl A., Mazaheri F., et al. 2014. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 6:243ra86 10.1126/scitranslmed.3009093 [DOI] [PubMed] [Google Scholar]
- Kober D.L., Alexander-Brett J.M., Karch C.M., Cruchaga C., Colonna M., Holtzman M.J., and Brett T.J.. 2016. Neurodegenerative disease mutations in TREM2 reveal a functional surface and distinct loss-of-function mechanisms. eLife. 5:707–720. 10.7554/eLife.20391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korvatska O., Leverenz J.B., Jayadev S., McMillan P., Kurtz I., Guo X., Rumbaugh M., Matsushita M., Girirajan S., Dorschner M.O., et al. 2015. R47H Variant of TREM2 Associated With Alzheimer Disease in a Large Late-Onset Family: Clinical, Genetic, and Neuropathological Study. JAMA Neurol. 72:920–927. 10.1001/jamaneurol.2015.0979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl W.C., Lyoo C.H., McGwier M., Snow J., Jenko K.J., Kimura N., Corona W., Morse C.L., Zoghbi S.S., Pike V.W., et al. Biomarkers Consortium PET Radioligand Project Team . 2013. In vivo radioligand binding to translocator protein correlates with severity of Alzheimer’s disease. Brain. 136:2228–2238. 10.1093/brain/awt145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.-T., Tseng L.H., Kamiyama H., Kamiyama M., Lim P., Hidalgo M., Wheelan S., and Eshleman J.. 2010. Quantifying the relative amount of mouse and human DNA in cancer xenografts using species-specific variation in gene length. Biotechniques. 48:211–218. 10.2144/000113363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M., and Prinz M.. 2015. Myeloid cells in Alzheimer’s disease: culprits, victims or innocent bystanders? Trends Neurosci. 38:659–668. 10.1016/j.tins.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Oakley H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. 2006. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26:10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre M., Kamphuis W., Osborn L.M., Jansen A.H.P., Kooijman L., Bossers K., and Hol E.M.. 2014. Isolation of glia from Alzheimer’s mice reveals inflammation and dysfunction. Neurobiol. Aging. 35:2746–2760. 10.1016/j.neurobiolaging.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Otero K., Turnbull I.R., Poliani P.L., Vermi W., Cerutti E., Aoshi T., Tassi I., Takai T., Stanley S.L., Miller M., et al. 2009. Macrophage colony-stimulating factor induces the proliferation and survival of macrophages via a pathway involving DAP12 and β-catenin. Nat. Immunol. 10:734–743. 10.1038/ni.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter M.M., Atagi Y., Liu C.C., Rademakers R., Xu H., Fryer J.D., and Bu G.. 2015. TREM2 in CNS homeostasis and neurodegenerative disease. Mol. Neurodegener. 10:43 10.1186/s13024-015-0040-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth H.W., and LaFerla F.M.. 2010. Alzheimer’s disease. N. Engl. J. Med. 362:329–344. 10.1056/NEJMra0909142 [DOI] [PubMed] [Google Scholar]
- Rebeck G.W., Reiter J.S., Strickland D.K., and Hyman B.T.. 1993. Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron. 11:575–580. 10.1016/0896-6273(93)90070-8 [DOI] [PubMed] [Google Scholar]
- Schlepckow K., Kleinberger G., Fukumori A., Feederle R., Lichtenthaler S.F., Steiner H., and Haass C.. 2017. An Alzheimer-associated TREM2 variant occurs at the ADAM cleavage site and affects shedding and phagocytic function. EMBO Mol. Med. 9:1356–1365. 10.15252/emmm.201707672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R., van der Lee S.J., Naj A.C., Bellenguez C., Badarinarayan N., Jakobsdottir J., Kunkle B.W., Boland A., Raybould R., Bis J.C., et al. GERAD/PERADES, CHARGE, ADGC, EADI . 2017. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49:1373–1384. 10.1038/ng.3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Hooli B., Mullin K., Jin S.C., Cella M., Ulland T.K., Wang Y., Tanzi R.E., and Colonna M.. 2017. Alzheimer’s disease-associated TREM2 variants exhibit either decreased or increased ligand-dependent activation. Alzheimers Dement. 13:381–387. 10.1016/j.jalz.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S., Stefansson H., Jonsson T., Johannsdottir H., Ingason A., Helgason H., Sulem P., Magnusson O.T., Gudjonsson S.A., Unnsteinsdottir U., et al. DemGene . 2015. Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat. Genet. 47:445–447. 10.1038/ng.3246 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Rochford C.D.P., and Neumann H.. 2005. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 201:647–657. 10.1084/jem.20041611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton P., Sevalle J., Deery M.J., Fraser G., Zhou Y., Ståhl S., Franssen E.H., Dodd R.B., Qamar S., Gomez Perez-Nievas B., et al. 2017. TREM2 shedding by cleavage at the H157-S158 bond is accelerated for the Alzheimer’s disease-associated H157Y variant. EMBO Mol. Med. 9:1366–1378. 10.15252/emmm.201707673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull I.R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., and Colonna M.. 2006. Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 177:3520–3524. 10.4049/jimmunol.177.6.3520 [DOI] [PubMed] [Google Scholar]
- Ulland T.K., Song W.M., Huang S.C., Ulrich J.D., Sergushichev A., Beatty W.L., Loboda A.A., Zhou Y., Cairns N.J., Kambal A., et al. 2017. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell. 170:649–663.e13. 10.1016/j.cell.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich J.D., Finn M.B., Wang Y., Shen A., Mahan T.E., Jiang H., Stewart F.R., Piccio L., Colonna M., and Holtzman D.M.. 2014. Altered microglial response to Aβ plaques in APPPS1-21 mice heterozygous for TREM2. Mol. Neurodegener. 9:20 10.1186/1750-1326-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich J.D., Ulland T.K., Colonna M., and Holtzman D.M.. 2017. Elucidating the Role of TREM2 in Alzheimer’s Disease. Neuron. 94:237–248. 10.1016/j.neuron.2017.02.042 [DOI] [PubMed] [Google Scholar]
- Wang X., Spandidos A., Wang H., and Seed B.. 2012. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40(Database issue, D1):D1144–D1149. 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cella M., Mallinson K., Ulrich J.D., Young K.L., Robinette M.L., Gilfillan S., Krishnan G.M., Sudhakar S., Zinselmeyer B.H., et al. 2015. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 160:1061–1071. 10.1016/j.cell.2015.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ulland T.K., Ulrich J.D., Song W., Tzaferis J.A., Hole J.T., Yuan P., Mahan T.E., Shi Y., Gilfillan S., et al. 2016. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213:667–675. 10.1084/jem.20151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Byers D.E., Jin X., Agapov E., Alexander-Brett J., Patel A.C., Cella M., Gilfilan S., Colonna M., Kober D.L., et al. 2015. TREM-2 promotes macrophage survival and lung disease after respiratory viral infection. J. Exp. Med. 212:681–697. 10.1084/jem.20141732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich P., Glebov K., Kemmerling N., Tien N.T., Neumann H., and Walter J.. 2013. Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and γ-secretase-dependent intramembranous cleavage. J. Biol. Chem. 288:33027–33036. 10.1074/jbc.M113.517540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., Titus A.R., and Humphrey M.B.. 2015. The TREM2-DAP12 signaling pathway in Nasu-Hakola disease: a molecular genetics perspective. Res. Rep. Biochem. 5:89–100. 10.2147/RRBC.S58057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh F.L., Wang Y., Tom I., Gonzalez L.C., and Sheng M.. 2016. TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron. 91:328–340. 10.1016/j.neuron.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Yeh F.L., Hansen D.V., and Sheng M.. 2017. TREM2, Microglia, and Neurodegenerative Diseases. Trends Mol. Med. 23:512–533. 10.1016/j.molmed.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Yuan P., Condello C., Keene C.D., Wang Y., Bird T.D., Paul S.M., Luo W., Colonna M., Baddeley D., and Grutzendler J.. 2016. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron. 90:724–739. 10.1016/j.neuron.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Chen X.F., Wang T., Wang Z., Liao C., Wang Z., Huang R., Wang D., Li X., Wu L., et al. 2017. Soluble TREM2 induces inflammatory responses and enhances microglial survival. J. Exp. Med. 214:597–607. 10.1084/jem.20160844 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data and computer code are available from the authors. Microarray data are accessible through GEO Series accession number GSE108595.