CD8+ T cells responding to infection differentiate into short-lived effector cells destined to die or memory cells that provide long-lived protection. Omilusik et al. demonstrate that commitment to an effector cell fate is not necessarily terminal and that sustained transcriptional regulation is required to maintain subset-specific properties.
Abstract
CD8+ T cells responding to infection differentiate into a heterogeneous population composed of progeny that are short-lived and participate in the immediate, acute response and those that provide long-lasting host protection. Although it is appreciated that distinct functional and phenotypic CD8+ T cell subsets persist, it is unclear whether there is plasticity among subsets and what mechanisms maintain subset-specific differences. Here, we show that continued Id2 regulation of E-protein activity is required to maintain the KLRG1hi CD8+ T cell population after lymphocytic choriomeningitis virus infection. Induced deletion of Id2 phenotypically and transcriptionally transformed the KLRG1hi “terminal” effector/effector-memory CD8+ T cell population into a KLRG1lo memory-like population, promoting a gene-expression program that resembled that of central memory T cells. Our results question the idea that KLRG1hi CD8+ T cells are necessarily terminally programmed and suggest that sustained regulation is required to maintain distinct CD8+ T cell states.
Introduction
In response to acute infection, naive antigen-specific CD8+ T cells become activated, proliferate, and differentiate into a heterogeneous population of effector cells with the functional capacity to eliminate the pathogen. Many effector CD8+ T cells within this population are thought to be terminally fated to undergo apoptosis upon resolution of the infection. Others appear to be programmed for long-term survival and uniquely suited to protect the host upon reinfection (Chang et al., 2014). Considerable work in the field has focused on relating effector CD8+ T cell phenotype to cell fate. Two cell-surface receptors, killer cell lectin-like receptor G1 (KLRG1) and interleukin 7 receptor α (CD127), have been valuable in predicting the fates of CD8+ T cell populations at the peak of the effector response. During the effector phase of infection, CD8+ T cells expressing KLRG1 and low levels of CD127, called terminal effector (TE) cells, are often defined as terminally differentiated, have a shorter life span and exhibit minimal memory potential in adoptive transfer experiments. CD8+ T cells with low KLRG1 and high CD127 surface expression in the effector phase have been defined as memory-precursor (MP) T cells and show a greater propensity to survive after infection and exhibit increased stem-like properties such as self-renewal (Kaech et al., 2003; Joshi et al., 2007; Sarkar et al., 2008).
At memory time points, the relationship of the canonical markers, KLRG1 and CD127, to cell fate becomes less clear. Memory CD8+ T cells have been classified into subsets based on several criteria including location, effector function, capacity for self-renewal, and trafficking patterns. The best characterized distinction is that of effector memory (TEM) and central memory (TCM) T cells, based on CD62L and CCR7 expression (Sallusto et al., 1999). TEM cells that lack CD62L and CCR7 expression circulate through nonlymphoid tissues and the blood and are poised to provide immediate effector function but have limited proliferation potential upon recall (Mueller et al., 2013). TCM cells express CD62L and CCR7 and thus home to lymphoid tissues and provide a long-term, self-renewing pool of T cells (Mueller et al., 2013). Overlaying the KLRG1 and CD127 phenotypic characterization of T cells adds a level of complexity to defining memory T cell subsets. Although CD127 expression supports long-term survival of memory T cells, the classification of TEM and TCM has not explicitly included the expression of CD127 or exclusion of KLRG1. Within the TEM population, KLRG1 expression can be detected on a portion of cells (Masopust et al., 2006; Hikono et al., 2007; Phan et al., 2016; Kakaradov et al., 2017). This observation is consistent with TEM exhibiting more effector-like properties and being more terminally differentiated (Kaech and Cui, 2012); however, variable KLRG1 expression suggests the TEM population itself is heterogeneous. Furthermore, a sizeable population of CD8+ T cells defined as KLRG1hiCD127lo TE T cells at the effector stage survive after the infection has resolved and persist at memory time points, but the population continues to diminish relative to the KLRG1lo population, further supporting the idea that these cells are terminally fated (Olson et al., 2013).
Unique transcriptional programs have been described that drive the differentiation of CD8+ T cells during infection—with T-bet, Blimp-1, IRF4, Zeb2, and Id2 acting as critical regulators of the TE CD8+ T cell population and Tcf1, Eomes, Bcl6, Foxo1, Id3, and E proteins regulating the MP CD8+ T cell population (Kaech et al., 2003; Joshi et al., 2007; Zhou et al., 2010; Chang et al., 2014). Although it is clear that these transcriptional regulators are key for the generation of effector and memory CD8+ T cell populations, little is known about their roles in maintaining subset-specific gene-expression programs. When considering the transition of CD8+ effector T cells to memory populations, important questions arise: are effector CD8+ T cell populations unconditionally committed to their specified fate after infection resolution, or does plasticity exist and is active transcriptional regulation necessary to continually enforce subset specificity?
E-protein transcription factors (TFs) and their repressors, Id (inhibitor of DNA binding) proteins, have emerged as key regulators of effector and memory CD8+ T cell differentiation (Omilusik et al., 2013). E-protein activity increases upon T cell activation to induce a transcriptional network that promotes the formation of MP CD8+ T cells; and loss of E2A and HEB, two E-protein family members, results in increased frequencies of KLRG1hi effector CD8+ T cells after infection (D’Cruz et al., 2012). Id2 and Id3 are both thought to function by antagonizing E-protein activity, yet they differentially impact the formation of CD8+ effector and memory T cell populations after acute infection (Cannarile et al., 2006; Ji et al., 2011; Yang et al., 2011; Knell et al., 2013; Masson et al., 2013). Loss of Id2 in naive CD8+ T cells results in an effector CD8+ T cell population that is highly susceptible to apoptosis with impaired survival after several bacterial or viral infections (Cannarile et al., 2006; Knell et al., 2016). Importantly, Id2-deficient CD8+ T cells fail to form the KLRG1hi TE subset and instead adopt an MP phenotype (Cannarile et al., 2006; Knell et al., 2013; Masson et al., 2013). Conversely, CD8+ T cells lacking Id3 have defects in formation and survival of long-lived memory (Ji et al., 2011; Yang et al., 2011). Here, we consider the transcriptional networks that actively maintain CD8+ T cell commitment and the acquired phenotypic and functional subset-specific properties. In particular, we examine the role of Id2 and E-proteins in enforcing the TE CD8+ T cell fate after resolution of infection. We find that memory T cell fates are more plastic than anticipated and that sustained regulation of E-protein activity is necessary to maintain the effector or TE CD8+ T cell phenotype.
Results and discussion
Id2 is expressed in CD8+ T cells throughout the effector and into the memory phase after infection (Fig. S1 A; Cannarile et al., 2006). Id2 promotes survival and terminal differentiation of CD8+ T cells at the peak of several acute infections (Cannarile et al., 2006; Knell et al., 2013; Fig. S1, B–H). Effector and TE CD8+ T cells deficient for Id2 fail to accumulate after infection, and the lack of CD8+ T cells makes it difficult to attribute a functional role for Id2 in homeostasis of T cell populations at later time points. To circumvent this, we crossed mice with loxP-flanked Id2 alleles (Id2f/f; Niola et al., 2012) to Rosa26Cre-ERT2 mice (ERCre) to allow inducible deletion of Id2 upon tamoxifen treatment. These mice were further mated with P14 transgenic mice that express an H-2Db–restricted T cell antigen-receptor specific for lymphocytic choriomeningitis virus (LCMV) glycoprotein33–41. Treatment of naive Id2f/f-ERCre+ P14 mice with tamoxifen induced a near complete deletion of Id2 in CD8+ T cells when compared with Id2f/f-ERCre− P14 T cells that had been similarly treated (Fig. S1, B and C; mice called Id2WT or Id2KO after induced Id2 deletion with tamoxifen). Upon mixed transfer of P14 T cells into congenically distinct hosts, CD8+ T cells with induced deletion of Id2 responding to LCMV infection had an accumulation and differentiation defect on day 8 of infection compared with Id2WT CD8+ T cells consistent with our previously published results studying CD8+ T cells with germline Id2 deficiency after infection (Fig. S1, D and E; Cannarile et al., 2006; Knell et al., 2013). Further, when deletion was induced at the peak of infection (5–10 d), the Id2-deleted effector and TE CD8+ T cells rapidly declined within days compared with the Id2WT cells (Fig. S1, F–H).
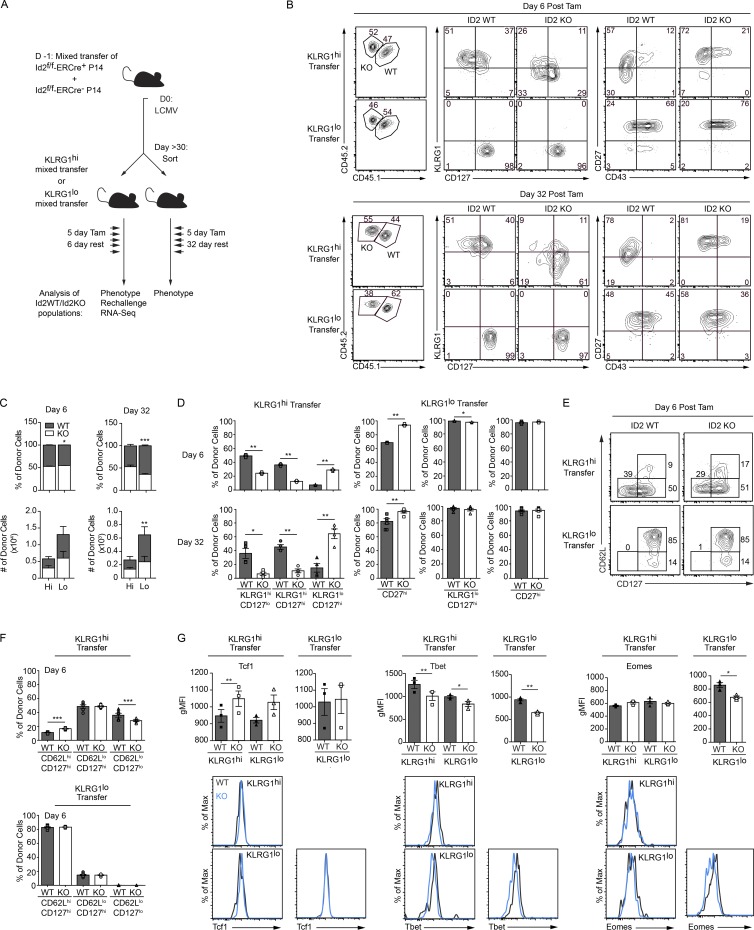
Having developed an efficient model in which to induce deletion of Id2, we sought to assess the role of Id2 in established memory CD8+ T cells. Untreated Id2f/f-ERCre+ and Id2f/f-ERCre− P14 CD8+ T cells were co-transferred at a 1:1 ratio into congenically distinct recipient mice 1 d before LCMV infection. After day 30 of infection, recipient mice were treated with tamoxifen to induce deletion of Id2 (Fig. 1 A; and Fig. S1, I and J). After tamoxifen treatment (15–16 d), Id2WT and Id2KO CD8+ T cells persisted in the blood, spleen, and lymph nodes (LNs) of the host at equivalent frequencies and numbers (Fig. 1, B and C; and Fig. S1 K). Strikingly, in this short duration of Id2 deficiency, the phenotype of the Id2KO CD8+ T cell population significantly changed compared with that of the Id2WT cells. Most prominently in the blood and spleen, the shorter-lived, more effector-like KLRG1hiCD127lo and KLRG1hiCD127hi Id2KO CD8+ T cell subsets almost completely disappeared, whereas the frequency of the more long-lived KLRG1loCD127hi Id2KO CD8+ T cells significantly increased (Fig. 1, B and D; Joshi et al., 2007; Sarkar et al., 2008). No differences in Ki67 were observed, indicating that the phenotypic changes occurred in the absence of proliferation (not depicted). Consistent with a conversion by the effector-like cells, the CD27loCD43lo CD8+ T cell population, a subset of long-lived effector T cells (Olson et al., 2013), was also lost upon Id2 deletion, whereas a proportional increase in the CD27hiCD43lo/hi CD8+ T cells was observed when compared with the Id2WT controls (Fig. 1, B and D). Furthermore, when analyzing the CD8+ T cells from the blood and spleen based on CD62L, a marker classically used to define circulating memory CD8+ T cells or TCM cells (Sallusto et al., 1999), we found that the frequency of the CD62LloCD127lo long-lived effector Id2KO CD8+ T cells declined compared with Id2WT CD8+ T cells. (Fig. 1, E and F). The loss of cells with a KLRG1hi phenotype in the Id2KO CD8+ T cell population was accompanied by increased expression of memory-associated TFs, Tcf1 and Foxo1 (Zhou et al., 2010; Hess Michelini et al., 2013), and a decrease in the effector-associated protein, T-bet (Joshi et al., 2007; Fig. 1 G). There were not significant differences in CD122 or CD127 levels for the Id2-deficient cells that had converted to a KLRG1lo phenotype compared with WT cells, indicating a similar potential for cytokine responsiveness (not depicted). We did note that at >20 d after induced Id2 deletion, the ratio of Id2KO to Id2WT donor cells began to fall, indicating that Id2 deficiency did eventually impair survival, in addition to profoundly affecting phenotype at earlier time points (Fig. S1 K). These results suggest that the effector differentiation state may be more actively controlled than previously appreciated and that continual Id2 transcriptional regulation is required to maintain the KLRG1hi effector/effector-memory CD8+ T cell differentiation.
Figure 1.
Late deletion of Id2 in antigen-specific CD8+ T cells results in conversion from a KLRG1hi to KLRG1lo population. (A) Schematic of experimental set-up. CD45.1 hosts were given a cotransfer of Id2f/f-ERCre− (CD45.1.2) and Id2f/f-ERCre+ (CD45.2) P14 CD8+ T cells then subsequently infected with LCMV. More than 30 d after infection, host mice were treated for 5 consecutive days with tamoxifen (Tam) to induce Id2 deletion. Id2f/f-ERCre− and Id2f/f-ERCre+ CD8+ T cells are called Id2WT or Id2KO, respectively, after Tam treatment. (B) Flow cytometry of transferred P14 CD8+ T cells from host peripheral blood lymphocytes (PBL) or spleen (Sp) and LN 15 or 16 d after the last Tam treatment, respectively. Frequency of Id2WT and Id2KO cells among P14 CD8+ T cells (left), KLRG1 and CD127 expression (middle), and CD27 and CD43 expression (right) are shown. Numbers in plots represent the percentage of cells. (C) Quantification of donor cell frequency and number or (D) frequency of populations from B. (E) Flow cytometry of cells from B for CD62L and CD127 expression. (F) Quantification of donor populations from E. (G) Expression of indicated proteins on KLRG1hi or KLRG1lo Id2WT or Id2KO donor populations. Data shown are representative of three (B and D–F) independent or cumulative of two or three (C and G) independent experiments; n = 3–5 mice per group. Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed paired Student’s t test).
The maintenance of Id2WT and Id2KO CD8+ T cells in a 1:1 ratio after Id2 deletion at early time points indicates that, in the absence of regulation of E-protein activity, effector-like Id2KO CD8+ T cells that have been presumed to be terminally fated dedifferentiate to resemble the long-lived KLRG1loCD127hiCD27hi T cell population. To understand if the loss of day 30 KLRG1hi effector/effector-memory Id2KO CD8+ T cells was due to altered cell fates, cell death, or increased KLRG1lo cell proliferation, we sorted, transferred, and induced Id2 deletion in CD8+ T cell subsets and followed them over time (Fig. 2 A). Untreated Id2f/f-ERCre+ and Id2f/f-ERCre− P14 CD8+ T cells were co-transferred into congenically distinct recipient mice that were infected the next day with LCMV. After day 30 of infection, KLRG1hiCD127lo and KLRG1loCD127hi P14 CD8+ T cells subsets that were Id2f/f-ERCre+ or Id2f/f-ERCre− were sorted from the spleen and LN of recipient mice. KLRG1hiCD127lo Id2f/f-ERCre+ and Id2f/f-ERCre− (KLRG1hi transfer) or KLRG1loCD127hi Id2f/f-ERCre+ and Id2f/f-ERCre− (KLRG1lo transfer) were mixed at a 1:1 ratio and transferred to new congenically distinct recipient mice, which were subsequently treated with tamoxifen to induce Id2 deletion (Fig. S2, A and B). The frequency and number of Id2WT and Id2KO CD8+ T cells in the spleens of recipient mice receiving the KLRG1hi CD8+ T cell transfer was not affected on day 6 or 32 after tamoxifen treatment (Fig. 2, B and C). However, by day 32, the proportion and number of Id2KO CD8+ T cells from the KLRG1lo transfer were significantly reduced compared with the Id2WT CD8+ T cells. The phenotype of the transferred KLRG1lo CD8+ T cells remained KLRG1loCD127hi and CD27hi after Id2 deletion, whereas the KLRG1hi Id2KO CD8+ T cells significantly down-regulated KLRG1 and up-regulated CD27, resembling a more long-lived T cell population (Fig. 2, B and D). The Id2WT cells in the KLRG1hi transfer partially up-regulated CD127, albeit not to the degree as the Id2KO CD8+ T cells, likely to support the long-term survival of the transferred cells. Notably, there is an increase in CD62L expression among KLRG1hi transferred cells after Id2 deletion (Fig. 2, E and F). The Id2KO KLRG1hi CD8+ T cells also significantly up-regulated Tcf1 expression and down-regulated T-bet expression consistent with reassignment from an effector-like phenotype to a memory-like phenotype (Fig. 2 G). The KLRG1lo transferred cells lost Eomes expression after Id2 deletion (Figs. 1 G and 2 F), suggesting that Id2 differentially affects the CD8+ T cell populations consistent with multiple roles—promoting survival in the KLRG1loCD127hi CD8+ T cells and enforcing differentiation in the KLRG1hiCD127lo CD8+ T cells. These results imply that differentiation to a KLRG1hi effector CD8+ T cell subset is a reversible process and that sustained Id2 regulation enforces the cell state of “terminal differentiation.”
Figure 2.
Id2 is necessary to maintain the terminal differentiation of antigen-specific CD8+ T cells. (A) Schematic of experimental set-up. CD45.1 hosts were given a cotransfer of Id2f/f-ERCre− (CD45.1.2) and Id2f/f-ERCre+ (CD45.2) P14 CD8+ T cells and then subsequently infected with LCMV. At >30 d after infection, KLRG1hiCD127lo and KLRG1loCD127hi, Id2f/f-ERCre−, and Id2f/f-ERCre+ P14 CD8+ T cells were sort purified. Id2f/f-ERCre− and Id2f/f-ERCre+ KLRG1hiCD127lo (KLRG1hi transfer) or Id2f/f-ERCre− and Id2f/f-ERCre+ KLRG1loCD127hi (KLRG1lo transfer) populations were co-transferred into naive CD45.1 hosts that were treated with tamoxifen (Tam). Id2f/f-ERCre− and Id2f/f-ERCre+ CD8+ T cells are called Id2WT or Id2KO, respectively, after Tam treatment. (B) Flow cytometry of transferred P14 CD8+ T cells from host spleen 6 d (top) or 32 d (bottom) after last Tam treatment. Frequency of Id2WT and Id2KO cells among P14 CD8+ T cells (left), KLRG1 and CD127 expression (middle), and CD27 and CD43 expression (right) for the KLRG1hi and KLRG1lo transfer are shown. Numbers in plots represent the frequency of cells in that quadrant. (C) Quantification of donor cell frequency and number from KLRG1hi (Hi) or KLRG1lo (Lo) transfers from B. (D) Quantification of donor populations from KLRG1hi (left) and KLRG1lo (right) transfer. (E) Flow cytometry of transferred P14 CD8+ T cells from host spleen 6 d after last Tam treatment. CD62L and CD127 expression for the KLRG1hi and KLRG1lo transfers is shown. (F) Quantification of donor populations from E. (G) Expression of indicated transcription factors on KLRG1hi or KLRG1lo Id2WT or Id2KO donor populations from hosts receiving KLRG1hi transferred (left) and KLRG1lo transferred (right) cells 6 d after last Tam treatment. Data shown are representative (B, E, and G) or cumulative (C, D, and F) of two independent experiments; n = 3–4 mice per group. Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed paired Student’s t test).
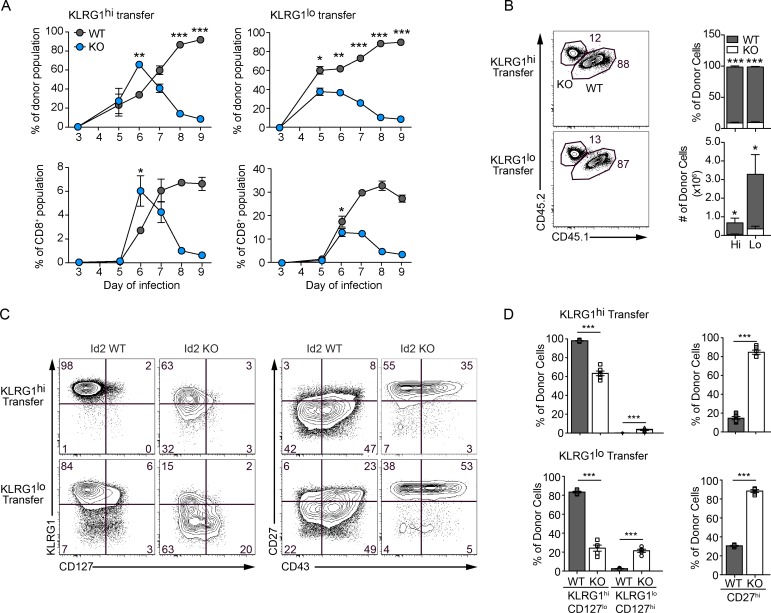
The loss of Id2 in long-lived KLRG1hi effector CD8+ T cells leaves them phenotypically resembling CD8+ T cells with more memory potential, so we sought to test their ability to recall after reinfection. After day 30 of LCMV infection, untreated KLRG1hiCD127lo and KLRG1loCD127hi Id2f/f-ERCre+ and Id2f/f-ERCre− P14 CD8+ T cells were sorted and co-transferred as above (Fig. 2 A), and 6 d after tamoxifen treatment to induce Id2 deletion, the host mice were rechallenged with LCMV. Following the kinetics of the CD8+ T cell response in the blood, we found that the Id2KO CD8+ T cells of the KLRG1hi transferred cells, which had largely lost KLRG1 expression, peaked 1 d sooner than the equivalent Id2WT CD8+ T cells after infection (Fig. 3 A). As previously observed in primary responses (Cannarile et al., 2006; Knell et al., 2013; Fig. S1), the CD8+ T cells deficient for Id2 failed to accumulate in the late effector stage of the secondary response, and after day 7 of reinfection the proportion of Id2KO CD8+ T cells of both the KLRG1hi and KLRG1lo transfer was largely diminished compared with the Id2WT CD8+ T cells. We examined the phenotype of the responding Id2WT and Id2KO CD8+ T cells in the spleen of recipients at day 9 after infection (Fig. 3, B–D). Although the Id2KO CD8+ T cells of the KLRG1lo transfer were significantly impaired in their ability to differentiate into KLRG1hiCD127lo effector cells much like in a primary response (Cannarile et al., 2006; Knell et al., 2013; Masson et al., 2013; Fig. S1), ∼60% of the Id2KO CD8+ T cells of the KLRG1hi transfer, despite being largely KLRG1lo at the time of rechallenge (Fig. 2, B and D), were able to re-express KLRG1. Yet, the Id2KO CD8+ T cells remained primarily CD27hi with increased expression of Tcf1 and Foxo1 and lower levels of T-bet compared with Id2WT CD8+ T cells (Fig. 3, C and D; and Fig. S2 C). The Id2KO CD8+ T cells likely persisted at a lower frequency at the late effector stage of rechallenge because of impaired survival demonstrated by reduced BCL2 (Fig. S2 C). Thus, KLRG1hi cells with induced Id2 deletion responded more rapidly to reinfection than their WT counterparts, showing characteristics consistent with a memory phenotype.
Figure 3.
Deletion of Id2 in KLRG1hi memory CD8+ T cells results in the faster initiation of a recall response. Id2f/f-ERCre− and Id2f/f-ERCre+ KLRG1hi or KLRG1lo P14 CD8+ T cells were co-transferred into naive CD45.1 hosts, and Id2 deletion was induced as described in Fig. 2 A. Host mice were infected with LCMV 7 d after the last tamoxifen treatment. Id2f/f-ERCre− and Id2f/f-ERCre+ CD8+ T cells are called Id2WT or Id2KO, respectively, following tamoxifen treatment. (A) The frequency of total Id2WT and Id2KO P14 donor cells (top) or Id2WT and Id2KO P14 cells of total CD8+ T cells (bottom) in PBL on indicated days after infection is shown. (B) Flow cytometry (left) and quantification (right) of the frequency of Id2WT and Id2KO P14 CD8+ T cells from host spleen 9 d after infection for KLRG1hi and KLRG1lo transfers. (C) Flow cytometry of KLRG1 and CD127 expression (left) and CD27 and CD43 expression (right) for KLRG1hi and KLRG1lo transfers are shown. (D) Quantification of donor populations from C after KLRG1hi (top) and KLRG1lo (bottom) transfer. Numbers in plots represent the percentage of cells. Data shown are cumulative (total donor frequency; A, top) or representative (total CD8+ population frequency; A, bottom), representative (B), or cumulative (C) of two independent experiments; n = 2–4 mice per group. Data are expressed as mean ± SEM. *, P ≤ 0.05; **, P < 0.01; ***, P < 0.001 (one-tailed paired [A and B] or two-tailed paired [D] Student’s t test).
To more thoroughly understand how Id2 affects the gene-expression profile of established KLRG1hi effector/effector-memory CD8+ T cell populations, we studied global transcriptional changes in CD8+ T cells that resulted from induced deletion of Id2. On day 26 of LCMV infection, KLRG1hiCD127lo and KLRG1loCD127hi Id2f/f-ERCre+ and Id2f/f-ERCre− P14 CD8+ T cells were sorted and co-transferred as in Fig. 2 A, and 6 d after tamoxifen treatment Id2WT and Id2KO T cell populations were sorted for RNA sequencing (RNA-Seq) analysis. Differential expression of genes associated with KLRG1hiCD127lo or KLRG1loCD127hi CD8+ T cell populations from 30 or more days of infection was confirmed by comparison of Id2WT CD8+ T cells from the KLRG1hi or KLRG1lo transfers (Fig. 4 A, left). We compared gene expression between the Id2WT and Id2KO CD8+ T cells from the KLRG1hi transfer and found that Id2KO KLRG1hi CD8+ T cells, in addition to down-regulating KLRG1 and T-bet protein expression, down-regulated mRNA of effector-associated genes (Prdm1, Tbx21, Zeb2, and Gzmb) and up-regulated mRNA of memory-associated genes (Id3, Ccr7, and Tcf7) upon Id2 deletion (Fig. 4 A, right). Principal component analysis confirmed that the induced deletion of Id2 resulted in the Id2KO KLRG1hi CD8+ T cells rapidly adopting a transcriptional profile that was distinct from WT KLRG1hi CD8+ T cells and more closely resembling MP or memory (TEM, TCM) cells (Fig. 4 B).
Figure 4.
Id2 regulation of E2A binding regulates expression of key effector CD8+ T cell genes. (A–D) Id2f/f-ERCre− (CD45.1.2) and Id2f/f-ERCre+ (CD45.2) KLRG1hi or KLRG1lo P14 CD8+ T cells were co-transferred into naive CD45.1 hosts, and Id2 deletion was induced as described in Fig. 2 A. Id2f/f-ERCre− and Id2f/f-ERCre+ CD8+ T cells are called Id2WT or Id2KO, respectively, after tamoxifen (Tam) treatment. On day 6 after the last Tam treatment, Id2WT and Id2KO cells were sorted for RNA-Seq. (A) Gene expression by cells from the Id2WT KLRG1hi transfer versus Id2WT KLRG1lo transfer (left) or from the Id2WT KLRG1hi transfer versus Id2KO KLRG1hi transfer (right) for genes differentially expressed by 1.5-fold or more, a coefficient of variation of ≤0.50, and an expression value of ≥10; colors indicate genes up-regulated 1.5-fold or more in the cells from the Id2WT KLRG1hi transfer relative to their expression in cells from the Id2WT KLRG1lo transfer (green) or vice versa (purple), or genes up-regulated in cells from the Id2WT KLRG1hi transfer relative to cells from the Id2KO KLRG1hi transfer (green) or vice versa (magenta). Labels in plots indicate genes of published relevance to CD8+ T cell differentiation and memory formation. (B) Principal component analysis of gene expression in Id2WT and Id2KO P14 CD8+ T cells from the KLRG1hi and KLRG1lo transfers, in TE and MP P14 CD8+ T cell populations at day 7 of LCMV infection, and in TCM, and TEM P14 CD8+ T cell populations at day 180 of LCMV infection. (C) Expression of TE- or MP-associated genes (top) or TEM- or TCM-associated genes (bottom) assessed in cells from the Id2KO KLRG1hi transfer versus cells from the Id2WT KLRG1hi transfer and plotted against p-value. Numbers in the corners indicate the total of those genes up-regulated in cells from the Id2KO KLRG1hi transfer (top left) or from the Id2WT KLRG1hi transfer (top right). P < 0.001 (χ2 test). (D) Relative expression of TE and MP genes (top) and TEM and TCM genes (bottom) from C that are putative E2A-target genes identified by ChIP-Seq (bar colors match dot colors in C). (E) Schematic of experimental set-up. CD45.1 host mice infected 1 d before received a cotransfer of Id2f/f-ERCre− and Id2f/f-ERCre+ P14 CD8+ T cells transduced with control shRNA targeting Cd19 or with shTcf3. At 15 d of infection, host mice were treated with tamoxifen (Tam) to induce Id2 deletion. (F) Flow cytometry of transferred cells 9 d after the last Tam treatment for KLRG1 and CD127 expression (left) and CD27 and CD43 expression (right). Numbers in the plots represent the percentage of cells. (G) Quantification of donor populations from highlighted populations in F. Data are representative of two (A–D) or three (F) and cumulative of three (G) independent experiments; n = 3–5 mice per group. Data are expressed as mean ± SEM. **, P < 0.01 (two-tailed paired Student’s t test).
To more carefully characterize the effect of induced Id2 deletion on genes that are effector or memory associated, we compared expression of genes differentially expressed between Id2WT and Id2KO CD8+ T cells from the KLRG1hi transfer to TE and MP gene signatures and TEM and TCM gene signatures previously established in our laboratory (Milner et al., 2017; Yu et al., 2017). It should be noted that TEM and TCM populations were isolated by sorting on the CD62Llo or CD62Lhi P14 CD8+ T cell populations and did not exclude KLRG1 to be consistent with other studies (Chang et al., 2014); therefore, it is likely that long-lived TE may be a component of our TEM signature. Id2KO CD8+ T cells lost expression of 79% of the TE signature genes and up-regulated expression of 69% of the MP-associated genes compared with the Id2WT CD8+ T cells (Fig. 4 C, top; P < 0.001). Similarly, Id2KO CD8+ T cells lost expression of 83% or up-regulated expression of 88% of TEM or TCM signature genes, respectively (Fig. 4 C, bottom; P < 0.001). These analyses indicate a substantial skewing away from an effector-like and toward a TCM CD8+ T cell transcriptional program by the so-called “terminally differentiated” KLRG1hiCD127lo Id2KO CD8+ T cells, rapidly following induction of Id2 deletion.
The dedifferentiation of long-lived KLRG1hi CD8+ T cells to memory-like KLRG1loCD127hi CD8+ T cells in the absence of continual Id2 regulation suggests that restrained E-protein activity is necessary to maintain a KLRG1hi effector CD8+ T cell phenotype. To confirm that E-protein activity, the presumed target of Id2, determines and maintains the fate of CD8+ T cells after infection, we analyzed genes differentially expressed between Id2WT or Id2KO KLRG1hi CD8+ T cells that are identified E2A targets (Leong et al., 2016) and are defined in the TE and MP or TEM and TCM gene signatures (Fig. 4 D). We found a substantial number of E2A-bound genes that were differentially up-regulated (Id3, Ccr7, Tcf7, and Cd27) and down-regulated (Prdm1, Ccl3, and Gzma) upon induced deletion of Id2. This suggests that Id2 mediation of E-protein activity plays a key role in enforcing effector CD8+ T cell terminal differentiation. It then follows that diminished levels of E2A would “rescue” the effector CD8+ T cell dedifferentiation observed in Id2-deficient cells. Therefore, we transduced Id2f/f-ERCre+ and Id2f/f-ERCre− P14 CD8+ T cells with a retroviral shRNA vector targeting the gene encoding E2A (shTcf3) or control shRNA (shCd19; Fig. 4 E and Fig. S3 A) as previously described (Shaw et al., 2016). Id2f/f-ERCre+ and Id2f/f-ERCre− P14 CD8+ T cells transduced with shTcf3 or shCd19 were mixed in a 1:1 ratio and co-transferred to congenically distinct recipients that had been infected 1 d earlier with LCMV. On day 15 of infection, recipient mice were treated with tamoxifen to induce deletion of Id2, and the phenotype of the transferred CD8+ T cells in recipient spleens was analyzed. As expected, the Id2KO CD8+ T cells expressing control shCd19 were unable to maintain a differentiated population of CD8+ T cells defined as KLRG1hiCD127lo and CD27loCD43lo. However, the Id2KO CD8+ T cells with diminished expression of Tcf3 were almost completely rescued with the frequency of KLRG1hiCD127lo and CD27loCD43lo populations nearly equivalent to Id2WT CD8+ T cell population (Fig. 4, F and G). Therefore, the loss of the effector-like CD8+ T cell population we observed upon Id2 deletion can be attributed largely to increased E-protein activity.
Loss of Id2 and enhanced E-protein activity disproportionally impact the KLRG1hi effector-like CD8+ T cell population at memory time points with a rapid and dramatic shift in phenotype and overall gene expression of KLRG1hi CD8+ T cells to a memory-like state, whereas the KLRG1lo CD8+ T cells showed moderately lower Eomes expression and impaired survival at later time points (by day 32). Although we noted a 1.9-fold increase in mRNA expression of Id2 in the KLRG1hiCD127lo compared with the KLRG1loCD127hi CD8+ T cells (Fig. 4 A, left) and higher Id2 reporter expression has been observed in TEM cells versus TCM cells after influenza infection (Masson et al., 2013), we did not detect differential Id2 reporter levels between KLRG1hiCD127lo and KLRG1loCD127hi CD8+ T cells or Id2 protein levels between TEM and TCM in our experiments (Fig. S3 B). This suggests that the increased dependency on Id2 observed in the KLRG1hi compared with KLRG1lo CD8+ T cells was not due to substantially differing expression levels. Using the assay for transposase-accessible chromatin–sequencing (ATAC-Seq) data for naive, TE, and MP or TCM and TEM CD8+ T cells assessed 0, 7, 30, or 180 d after LCMV infection, we analyzed chromatin accessibility of MP or TCM E-protein target genes that were differentially regulated upon Id2 deletion (Fig. 4 D). E-protein binding sites in Tcf7, Cd27, Cxcr5, Ccr7, and Id3, identified by using E2A chromatin immunoprecipitation–sequencing (ChIP-Seq; Leong et al., 2016), were accessible in the naive, TE, MP, TCM, and TEM populations analyzed (Fig. S3 C and not depicted). This suggests that other factors contribute to CD8+ T cell subset-specific E-protein regulation. For example, Zeb2, a transcription factor highly expressed in TE and TEM CD8+ T cells and found to be important for terminal differentiation (Dominguez et al., 2015; Omilusik et al., 2015; Kakaradov et al., 2017), can bind DNA at tandem, separated E-protein binding sites (Sekido et al., 1994; Remacle et al., 1999). We identified such a site within the Tcf7 gene (Fig. S3 C), suggesting Zeb2 may function in KLRG1hi effector-like CD8+ T cell populations to repress E-protein activity and thus the memory-gene expression program. Perhaps enhanced E-protein activity upon the loss of Id2 expression may outcompete Zeb2 for E-box sites, resulting in greater expression of Tcf7, which then could promote the TCM, long-lived memory phenotype.
Ultimately these results show that the concept of terminal differentiation may be flawed in the context of CD8+ memory T cell differentiation. KLRG1 may not be a definitive marker of terminal differentiation, and bona fide, terminally differentiated CD8+ T cells may die shortly after resolution of the infection, leaving a less-differentiated KLRG1hi population that persists at memory time points. Instead, we support the notion that CD8+ T cell differentiation is flexible and sustained inhibition of E-protein activity by Id2 allows the survival of an effector-like population that can reacquire at least a portion of the TCM, or long-lived memory gene-expression program, favoring the idea of plasticity of cell states over fixed cell fates among the heterogeneous, post-infection CD8+ T cell population.
Materials and methods
Mice
All mouse strains were bred and housed in specific pathogen–free conditions in accordance with the Institutional Animal Care and Use Guidelines of the University of California San Diego. Id2-YFP mice (Yang et al., 2011), Id2fl/fl mice (Niola et al., 2012), Rosa26Cre-ERT2 (ERCre) mice (Hess Michelini et al., 2013), P14 mice (with transgenic expression of H-2Db–restricted TCR specific for LCMV glycoprotein gp33), and CD45.1 and CD45.1.2 congenic mice were bred to obtain a fully C57BL/6J background. Both male and female mice were used throughout the study, with sex- and age-matched T cell donors and recipients.
Cell transfer, LCMV infection, and tamoxifen treatment
Congenically distinct Id2f/f-ERCre− and Id2f/f-ERCre+ P14 CD8+ T cells were mixed at a 1:1 ratio and adoptively transferred at 5 × 104 cells per CD45.1 recipient mouse. Mice were then infected with 2 × 105 PFU LCMV-Armstrong by intraperitoneal injection. At day 5 or >30 d after infection, Rosa26Cre-ERT2-mediated deletion of floxed alleles was induced by intraperitoneal injection for 5 consecutive days of 1 mg tamoxifen (Cayman Chemical Company) emulsified in 100 ml sunflower seed oil (Sigma-Aldrich). Alternatively, >30 d (or at 26 d for RNA-Seq analysis) after infection, KLRG1hiCD127lo and KLRG1loCD127hi populations from total P14 CD8+ T cells were sorted from recipient spleen and LN, mixed in a 1:1 ratio of KLRG1hiCD127lo Id2f/f- ERCre+/Id2f/f- ERCre− or KLRG1loCD127hi Id2f/f- ERCre+/Id2f/f- ERCre− and then transferred at 1.5 × 105 total cells per CD45.1 recipient mouse. The next day, tamoxifen was administered as above for five consecutive days to induce Id2 deletion. At 6 or 32 d after the last tamoxifen treatment, Id2WT and Id2KO populations were assessed by flow cytometry or sorted for RNA-Seq (as described in the RNA-Seq section), or recipients were rechallenged intraperitoneally with 2 × 105 PFU LCMV-Armstrong.
RT-PCR and qPCR
To confirm Id2 deletion or Tcf3 knockdown, total RNA was extracted by using Trizol (Life Technologies) from sorted Id2WT and Id2KO P14 CD8+ T cells. cDNA was synthesized by using Superscript II kit (Life Technologies) following the manufacturer's instructions. For qPCR, cDNA was quantitatively amplified by using Stratagene Brilliant II Syber Green master mix (Agilent Technologies). The abundance of transcripts was normalized to that of the housekeeping gene Hprt. The following primers were used: Id2 forward, 5′-TCCCTTCTGAGCTTATGTCG-3′ and Id2 reverse, 5′-GTCCATTCAACGTGTTCTCC-3′; Tcf3 forward, 5′-CATCCATGTCCTGCGAAGCCA-3′ and Tcf3 reverse, 5′-TTCTTGTCCTCTTCGGCGT-3′; Hprt forward, 5′-GGCCAGACTTTGTTGGATTT-3′, and Hprt reverse: 5′-CAACTTGCGCTCATCTTAGG-3′.
Flow cytometry and cell sorting
Single-cell suspensions were prepared from spleen, LN, or blood. The following antibodies were used for surface staining (all from eBioscience): CD8 (53-6.7), CD27 (LG-7F9), CD43 (1B11), CD45.1 (A20-1.7), CD45.2 (104), CD62L (MEL-14), CD122 (TM-b1), CD127 (A7R34), CXCR3 (CXCR3-173), and KLRG1 (2F1). Cells were incubated for 30 min at 4°C in PBS supplemented with 2% bovine growth serum and 0.1% sodium azide. Intracellular staining was performed by using the Foxp3 Transcription Factor Staining Buffer kit (eBioscience) and the following antibodies (all from eBioscience unless otherwise specified): BCL2 (3F11; BD PharMingen), Eomes (Dan11mag), Foxo1 (C29H4; Cell Signaling), Gzmb (GB12; Invitrogen), Id2 (ILCID2; Thermo Fisher Scientific), Tbet (eBio4B10), and Tcf1 (C63D9; Cell Signaling). Stained cells were analyzed by using LSRFortessa or LSRFortessa X-20 (BD) and FlowJo software (TreeStar). All sorting was performed on BD FACSAria or BD FACSAria Fusion instruments.
shRNA-mediated knockdown by retroviral transduction
DNA fragments encoding shRNA targeting mouse Tcf3 or Cd19 were subcloned into a custom retroviral vector containing ametrine as a reporter. Naive congenically distinct Id2f/f-ERcre+ and Id2f/f-ERcre− P14 CD8+ T cells were stimulated for 18 h in 6-well plates precoated with anti-CD3 and anti-CD28. After stimulation, cells were transduced by adding retroviral supernatants supplemented with 100 U/ml human IL-2 and 8 µg/ml polybrene, followed by centrifugation for 90 min at 2000 g at 37°C. After transduction, cells were incubated for 3 h at 37°C. Id2f/f- ERCre+/Id2f/f- ERCre− P14 CD8+ T cells were mixed 1:1 and 106 cells were transferred into day −1 LCMV-infected hosts, and remaining cells were cultured in vitro with 50 U/ml human IL-2 for 4 d to assess for knockdown efficiency by qPCR. On day 15 of infection, tamoxifen was administered as described in the tamoxifen treatment section for 5 consecutive days to induce Id2 deletion. At 9 d after the last tamoxifen treatment, Id2WT and Id2KO populations were assessed by flow cytometry.
RNA-Seq, ChIP-Seq, and ATAC-Seq
6 d after the last tamoxifen treatment, 103 Id2WT and Id2KO P14 cells from recipients receiving mixed transfers of KLRG1hiCD127lo or KLRG1loCD127hi transfer were sorted into Buffer TCL (Qiagen) with 1% 2-mercaptoethanol. RNA was isolated, and libraries were prepared following Immgen protocols (https://www.immgen.org/Protocols/11Cells.pdf). Libraries were analyzed on an Illumina HiSeq2500 sequencer. Samples were generated from two biological replicates, and ∼10 million paired-reads were generated per sample. Reads were mapped by using Tophat, and aligned reads in transcripts were counted with HTseq. Analysis was performed by using the GenePattern Multiplot Studio module. TE and MP (Yu et al., 2017) and TEM and TCM gene signatures (Milner et al., 2017) represent genes that have a 1.5-fold increased expression in that CD8+ T cell population. Principal component analysis used RNA-Seq data for TE and MP or TEM and TCM P14 CD8+ T cells from day 7 or 180 after LCMV infection. E2A Bio-Chip analysis was deposited by Leong et al. (2016) (GEO accession code GSE84974), and ATAC-Seq data from Immgen (http://rstats.immgen.org/Chromatin/chromatin.html) was analyzed in the University of California, Santa Cruz, genome browser.
Statistics
The one- or two-tailed paired Student's t test was used for comparisons between groups as stated in the figure legends. The χ2 test was used to determine significant enrichment of gene sets in the RNA-Seq data. Statistical analysis was performed by using GraphPad Prism software.
Online supplemental material
Fig. S1 shows analysis of CD8+ T cells responding to LCMV infection when Id2 deletion is induced in naive cells or at the peak of infection, as well as knockout efficiency when Id2 deletion is induced >30 d after infection. Fig. S2 shows the mix of Id2WT and Id2KO CD8+ T cells for KLRG1hi and KLRG1lo transfers and additional phenotyping after LCMV rechallenge. This relates to Figs. 2 and 3. Fig. S3 shows Tcf3 knockdown efficiency in Id2WT and Id2KO P14 CD8+ T cells transduced with retroviral shRNA constructs. Also, ATAC-Seq and E2A ChIP-Seq analysis of the Tcf7 loci for CD8+ T cell populations after LCMV infection is shown. This relates to Fig. 4.
Supplementary Material
Acknowledgments
We thank Toan C. Nguyen and the Flow Cytometry Core at the La Jolla Institute for Allergy and Immunology for their assistance with cell sorting. We also thank Dr. John Chang for helpful discussion and critical review of this manuscript.
This study was supported by the National Institutes of Health (1F31AG043222-01A1 to L.A. Shaw; AI067545 and A1072117 to A.W. Goldrath) and the Leukemia and Lymphoma Society (to K.D. Omilusik and A.W. Goldrath).
The authors declare no competing financial interests.
Author contributions: K.D. Omilusik designed and performed experiments, analyzed the data, and wrote the paper. M.S. Nadjsombati, L.A. Shaw, B. Yu, and J.J. Milner performed experiments and assisted with data analysis. A.W. Goldrath supervised the project, designed the experiments, analyzed the data, and wrote the paper.
References
- Cannarile M.A., Lind N.A., Rivera R., Sheridan A.D., Camfield K.A., Wu B.B., Cheung K.P., Ding Z., and Goldrath A.W.. 2006. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 7:1317–1325. 10.1038/ni1403 [DOI] [PubMed] [Google Scholar]
- Chang J.T., Wherry E.J., and Goldrath A.W.. 2014. Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 15:1104–1115. 10.1038/ni.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz L.M., Lind K.C., Wu B.B., Fujimoto J.K., and Goldrath A.W.. 2012. Loss of E protein transcription factors E2A and HEB delays memory-precursor formation during the CD8+ T-cell immune response. Eur. J. Immunol. 42:2031–2041. 10.1002/eji.201242497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C.X., Amezquita R.A., Guan T., Marshall H.D., Joshi N.S., Kleinstein S.H., and Kaech S.M.. 2015. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212:2041–2056. 10.1084/jem.20150186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess Michelini R., Doedens A.L., Goldrath A.W., and Hedrick S.M.. 2013. Differentiation of CD8 memory T cells depends on Foxo1. J. Exp. Med. 210:1189–1200. 10.1084/jem.20130392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikono H., Kohlmeier J.E., Takamura S., Wittmer S.T., Roberts A.D., and Woodland D.L.. 2007. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J. Exp. Med. 204:1625–1636. 10.1084/jem.20070322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Pos Z., Rao M., Klebanoff C.A., Yu Z., Sukumar M., Reger R.N., Palmer D.C., Borman Z.A., Muranski P., et al. . 2011. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat. Immunol. 12:1230–1237. 10.1038/ni.2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., and Cui W.. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12:749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., and Ahmed R.. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198. 10.1038/ni1009 [DOI] [PubMed] [Google Scholar]
- Kakaradov B., Arsenio J., Widjaja C.E., He Z., Aigner S., Metz P.J., Yu B., Wehrens E.J., Lopez J., Kim S.H., et al. . 2017. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 18:422–432. 10.1038/ni.3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knell J., Best J.A., Lind N.A., Yang E., D’Cruz L.M., and Goldrath A.W.. 2013. Id2 influences differentiation of killer cell lectin-like receptor G1(hi) short-lived CD8+ effector T cells. J. Immunol. 190:1501–1509. 10.4049/jimmunol.1200750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong Y.A., Chen Y., Ong H.S., Wu D., Man K., Deleage C., Minnich M., Meckiff B.J., Wei Y., Hou Z., et al. . 2016. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17:1187–1196. 10.1038/ni.3543 [DOI] [PubMed] [Google Scholar]
- Masopust D., Ha S.J., Vezys V., and Ahmed R.. 2006. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J. Immunol. 177:831–839. 10.4049/jimmunol.177.2.831 [DOI] [PubMed] [Google Scholar]
- Masson F., Minnich M., Olshansky M., Bilic I., Mount A.M., Kallies A., Speed T.P., Busslinger M., Nutt S.L., and Belz G.T.. 2013. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J. Immunol. 190:4585–4594. 10.4049/jimmunol.1300099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. . 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 552:253–257. 10.1038/nature24993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.N., Gebhardt T., Carbone F.R., and Heath W.R.. 2013. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 31:137–161. 10.1146/annurev-immunol-032712-095954 [DOI] [PubMed] [Google Scholar]
- Niola F., Zhao X., Singh D., Castano A., Sullivan R., Lauria M., Nam H.S., Zhuang Y., Benezra R., Di Bernardo D., et al. . 2012. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat. Cell Biol. 14:477–487. 10.1038/ncb2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J.A., McDonald-Hyman C., Jameson S.C., and Hamilton S.E.. 2013. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity. 38:1250–1260. 10.1016/j.immuni.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilusik K.D., Shaw L.A., and Goldrath A.W.. 2013. Remembering one’s ID/E-ntity: E/ID protein regulation of T cell memory. Curr. Opin. Immunol. 25:660–666. 10.1016/j.coi.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilusik K.D., Best J.A., Yu B., Goossens S., Weidemann A., Nguyen J.V., Seuntjens E., Stryjewska A., Zweier C., Roychoudhuri R., et al. . 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212:2027–2039. 10.1084/jem.20150194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A.T., Doedens A.L., Palazon A., Tyrakis P.A., Cheung K.P., Johnson R.S., and Goldrath A.W.. 2016. Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity. 45:1024–1037. 10.1016/j.immuni.2016.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle J.E., Kraft H., Lerchner W., Wuytens G., Collart C., Verschueren K., Smith J.C., and Huylebroeck D.. 1999. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 18:5073–5084. 10.1093/emboj/18.18.5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Förster R., Lipp M., and Lanzavecchia A.. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Sarkar S., Kalia V., Haining W.N., Konieczny B.T., Subramaniam S., and Ahmed R.. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205:625–640. 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R., Murai K., Funahashi J., Kamachi Y., Fujisawa-Sehara A., Nabeshima Y., and Kondoh H.. 1994. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol. Cell. Biol. 14:5692–5700. 10.1128/MCB.14.9.5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L.A., Bélanger S., Omilusik K.D., Cho S., Scott-Browne J.P., Nance J.P., Goulding J., Lasorella A., Lu L.F., Crotty S., and Goldrath A.W.. 2016. Id2 reinforces TH1 differentiation and inhibits E2A to repress TFH differentiation. Nat. Immunol. 17:834–843. 10.1038/ni.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.Y., Best J.A., Knell J., Yang E., Sheridan A.D., Jesionek A.K., Li H.S., Rivera R.R., Lind K.C., D’Cruz L.M., et al. . 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12:1221–1229. 10.1038/ni.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Zhang K., Milner J.J., Toma C., Chen R., Scott-Browne J.P., Pereira R.M., Crotty S., Chang J.T., Pipkin M.E., et al. . 2017. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18:573–582. 10.1038/ni.3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Yu S., Zhao D.M., Harty J.T., Badovinac V.P., and Xue H.H.. 2010. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 33:229–240. 10.1016/j.immuni.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.