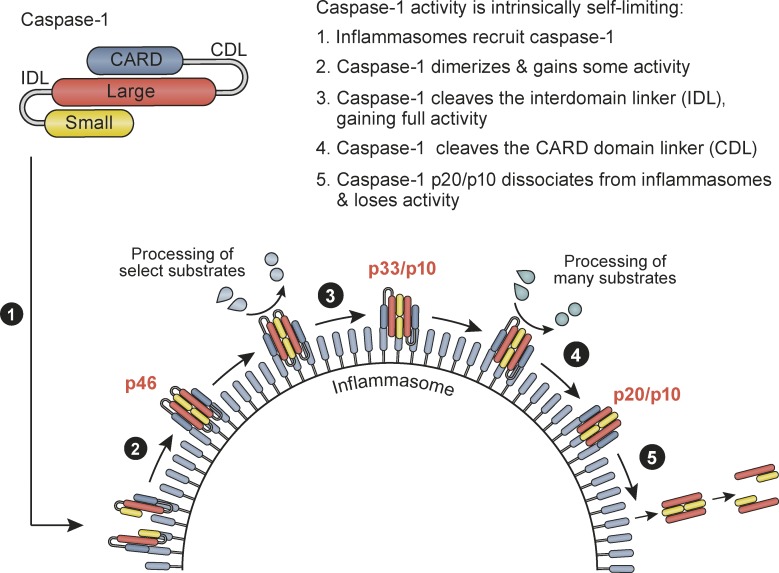
The inflammasome generates caspase-1 p20/p10, presumed to be the active protease. Boucher et al. demonstrate that the inflammasome contains an active caspase-1 species, p33/p10, and functions as a holoenzyme. Further caspase-1 self-processing generates and releases p20/p10 to terminate protease activity.
Abstract
Host-protective caspase-1 activity must be tightly regulated to prevent pathology, but mechanisms controlling the duration of cellular caspase-1 activity are unknown. Caspase-1 is activated on inflammasomes, signaling platforms that facilitate caspase-1 dimerization and autoprocessing. Previous studies with recombinant protein identified a caspase-1 tetramer composed of two p20 and two p10 subunits (p20/p10) as an active species. In this study, we report that in the cell, the dominant species of active caspase-1 dimers elicited by inflammasomes are in fact full-length p46 and a transient species, p33/p10. Further p33/p10 autoprocessing occurs with kinetics specified by inflammasome size and cell type, and this releases p20/p10 from the inflammasome, whereupon the tetramer becomes unstable in cells and protease activity is terminated. The inflammasome–caspase-1 complex thus functions as a holoenzyme that directs the location of caspase-1 activity but also incorporates an intrinsic self-limiting mechanism that ensures timely caspase-1 deactivation. This intrinsic mechanism of inflammasome signal shutdown offers a molecular basis for the transient nature, and coordinated timing, of inflammasome-dependent inflammatory responses.
Graphical Abstract
Introduction
Inflammasomes are signaling hubs that assemble in response to cell stress or microbial infection and provide an activation platform for the zymogen protease, caspase-1. Upon activation, caspase-1 triggers the maturation and secretion of potent proinflammatory mediators (IL-1β and IL-18) and induces cell lysis (pyroptosis), culminating in the activation of the immune system and antimicrobial defense (Schroder and Tschopp, 2010). Other proinflammatory signaling pathways are tightly controlled, with signal initiation eliciting negative feedback mechanisms to shut down inflammatory signaling within a set time window (Liew et al., 2005). Isolated studies have suggested that caspase-1 enzymatic function can be suppressed via cysteine oxidation by reactive oxygen species (Meissner et al., 2008) or prevented by primate-specific caspase activation and recruitment domain (CARD)-only proteins (Schroder and Tschopp, 2010). General mechanisms, including signal feedback loops, that control the duration of caspase-1 activity in inflammasome-signaling cells are unknown.
Results
Active caspase-1 is predominantly a transient species, p33/p10
Full-length caspase-1 is recruited to inflammasomes via its N-terminal CARD that interacts with a CARD domain presented by these signaling hubs (e.g., that of polymerized ASC). Caspase-1 recruitment to the hub enables its activation (Schroder and Tschopp, 2010), likely by increasing the local concentration of caspase-1 to facilitate the dimerization of caspase-1 monomers (Datta et al., 2013). Indeed, caspase-1 dimerization enables its protease activity (Fig. S1, A and B). A CARD domain linker (CDL) separates the CARD of caspase-1 from its C-terminal catalytic domain, which is composed of large (p20) and small (p10) subunits, separated by an interdomain linker (IDL; Fig. 1 A). Caspase-1 can undergo self-processing at multiple sites within the two linker domains (Broz et al., 2010; Fig. 1 A), to potentially generate a variety of dimeric caspase-1 species (Fig. 1 B). In macrophages, cytokine processing by caspase-1 requires caspase-1 cleavage at the IDL (p20↓p10; Broz et al., 2010) and is temporally associated with cleavage of the CDL (CARD↓p20; Mariathasan et al., 2006; Broz et al., 2010). Because inflammasome activation elicits caspase-1 p20 and p10 cleavage fragments, and recombinant p20/p10 is catalytically active (Fig. S1 C; Ramage et al., 1995; Walsh et al., 2011; Datta et al., 2013), it is widely assumed that the active species of caspase-1 in cells is a tetramer composed of two p20 and two p10 subunits (p20/p10; Thornberry et al., 1992). However, the active species of cellular caspase-1 has not been defined experimentally.
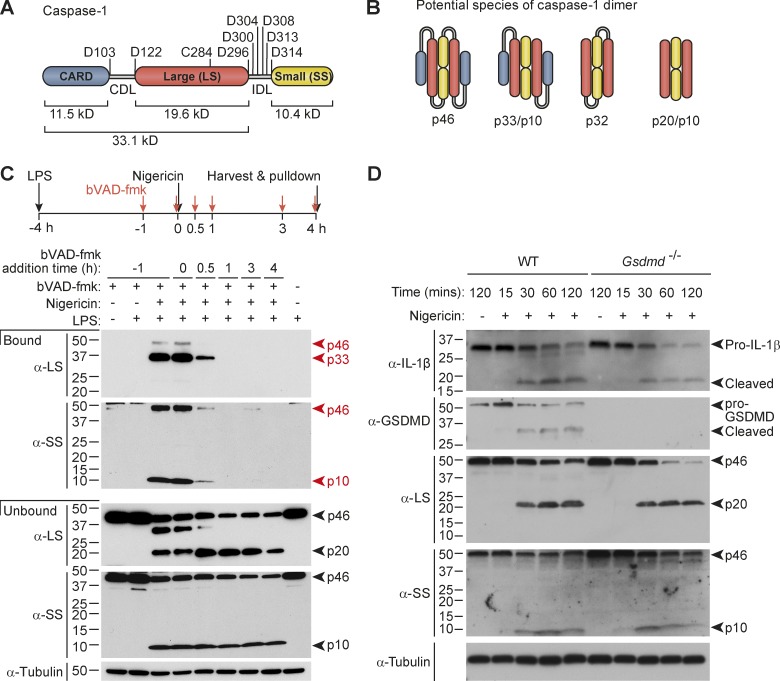
Figure 1.
Active caspase-1 is predominantly a transient p33/p10 species in nigericin-stimulated macrophages. (A) Representation of potential self-processing sites within the CDL and IDL of mouse caspase-1, relative to the catalytic cysteine (C284). (B) Possible species of dimeric caspase-1 generated by CDL and/or IDL cleavage. (C) Pull-down of active caspase-1 from mouse macrophages, using the bVAD-fmk caspase activity probe. Macrophages were left untreated or primed with LPS for 4 h, and then stimulated with nigericin for a further 4 h before addition of 1% IGEPAL into the well, to lyse cells directly in their culture medium. bVAD-fmk was applied to cells 1 h before (−1 h), during (0 h), or after (0.5, 1, 3, 4 h) nigericin addition. Streptavidin-coated beads pulled down active caspase-1 bound to the biotin-labeled activity probe in mixed lysates/supernatants. Streptavidin-bound and -unbound fractions were analyzed by Western blot using antibodies directed against the caspase-1 large and small subunits (LS, SS). (D) LPS-primed WT and Gsdmd−/− macrophages were stimulated with nigericin and harvested at various time points until 2 h after nigericin stimulation. Cell supernatants were precipitated and resuspended in cell extracts, and the kinetics of caspase-1 substrate cleavage (pro-IL-1β cleavage to p17, pro-GSDMD to GSDMD p30) was examined by Western blot. All data are representative of at least three independent experiments.
We examined the active species of caspase-1 in inflammasome-activated cells, first using an activity probe that covalently binds the catalytic cysteine of active caspases and inhibits further protease activity. We used a stimulation protocol that activates the well-characterized NLRP3-ASC-caspase-1 inflammasome, in which mouse bone marrow macrophages are first primed with LPS for 4 h to induce the expression of NLRP3 and pro–IL-1β, and are then exposed to the bacterial toxin, nigericin, to trigger NLRP3 inflammasome assembly (Mariathasan et al., 2006). A cell-permeable pan-caspase activity probe, biotin-VAD(OMe)-fmk (bVAD-fmk; Faleiro et al., 1997), was applied to cells at various times before and after nigericin stimulation, to trap caspases in their active form and suppress further autoprocessing. Streptavidin pulled down active caspase-1 species, which were then identified by Western blot using antibodies raised against the caspase-1 small (p10) and large (p20) subunits (α-SS, α-LS; Fig. 1 C). Strikingly, the major caspase-1 fragments pulled down by bVAD-fmk/streptavidin were p10 and p33, a cleavage product generated by caspase-1 IDL but not CDL processing (Fig. 1, A–C). By contrast, the bVAD-fmk activity probe did not detectably pull down p20, an anticipated component of active caspase-1. Full-length caspase-1 (p46) was pulled down as a minor species, in keeping with the accepted activation model for initiator caspases in which caspase dimerization is the first step for the protein to acquire basal enzymatic function such as the ability to self-cleave (Boatright et al., 2003). Surprisingly, when the caspase activity probe was applied to cells 30 min after nigericin, its ability to pull down p33/p10 diminished, and bVAD-fmk addition thereafter failed to capture any active caspase-1 species (Fig. 1 C), indicating that p33/p10 is a transient species. bVAD-fmk addition at early time points (≤30 min after nigericin stimulation) that allow it to capture p33/p10 suppressed the generation of p20 (Fig. 1 C). Given that bVAD-fmk binds to caspases only in their active forms and inhibits their further processing, this indicates that early bVAD-fmk application trapped the major active species, caspase-1 p33/p10, and prevented its subsequent processing to p20/p10, which could not be captured by later addition of bVAD-fmk. This suggests that contrary to general assumption, p20/p10 is not the major active caspase-1 species generated by cellular inflammasome signaling, and may instead be an inactive caspase-1 species. The observation that caspase-1 is rapidly deactivated in cells was confirmed using the YVAD-afc fluorescent caspase-1 substrate to monitor protease activity over time in lysates of nigericin-stimulated, Gsdmd-deficient macrophages unable to undergo rapid pyroptotic cell death. Caspase-1–like activity was restricted to cell lysates in which protease activity declined over time, and caspase-1–like activity was not observed in the cell culture media of these samples (Fig. S1 C). If inflammasome assembly triggers caspase-1 activation and then subsequent deactivation, then this suggests that caspase-1–dependent substrate processing occurs within a set time window. Indeed, pro–IL-1β and pro–gasdermin-D (GSDMD), were predominantly cleaved in a burst between 15 and 30 min after nigericin stimulation (Figs. 1 D and S1 D), coinciding with the duration of caspase-1 activity (Fig. 1 C). Pyroptotic pores or cell rupture were not primarily responsible for curtailed substrate processing, because Gsdmd deficiency, or the addition of the osmoprotectant glycine, delayed cell lysis (Fig. S1 E) but did not extend the kinetics of caspase-1–dependent substrate processing (Figs. 1 D and S1 D).
p33/p10 self-cleavage generates p20/p10 and deactivates caspase-1
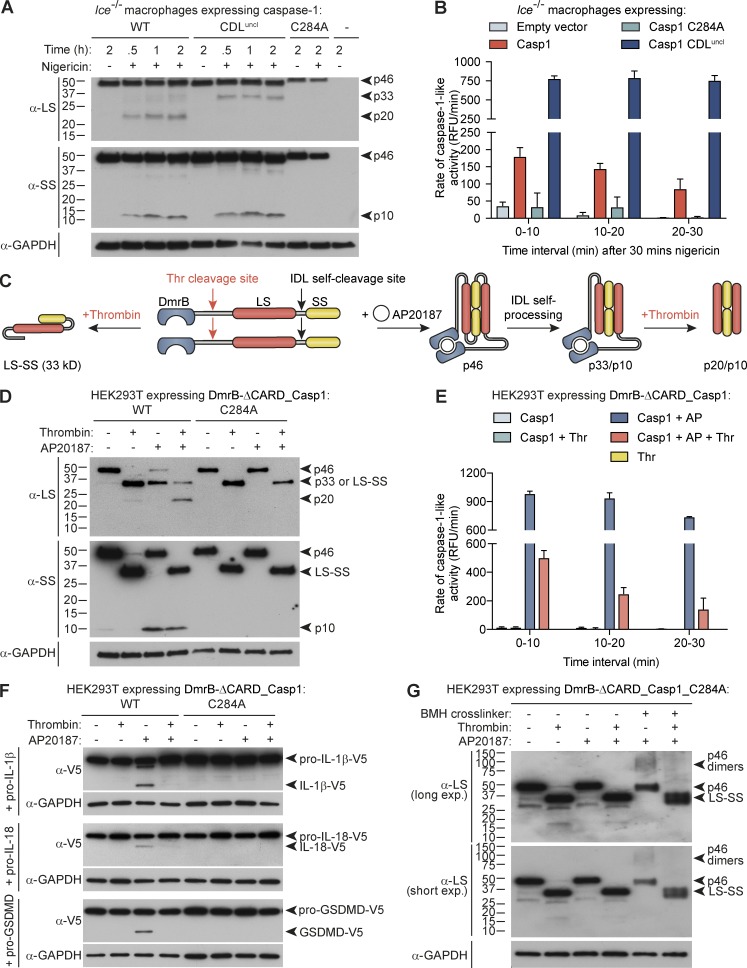
The aforementioned data are at odds with the common belief, based on activity of recombinant p20/p10, that caspase-1 self-processing within the CDL is required for caspase-1–dependent cytokine processing. To definitively establish that CDL processing is not required for cellular caspase-1 activity, and in fact deactivates caspase-1, we first retrovirally reconstituted caspase-1 expression in Casp1−/−/Casp11null/null (Ice−/−) macrophages. We ectopically expressed a caspase-1 mutant in which the CDL self-cleavage sites are mutated to prevent their processing (CDL-uncleavable caspase-1 compound mutant in which preferred [D] and suboptimal [E] cleavage sites are substituted for alanine, E102A/D103A/E121A/D122A, CDLuncl; Fig. 2 A), alongside WT caspase-1 and its catalytic cysteine mutant (C284A), and performed activity assays after cells were stimulated for 30 min with nigericin. Similar to endogenous caspase-1 (Fig. 1 C), ectopically expressed WT caspase-1 lost activity over time (Figs. 2 B and S2 A). Mutation of the CDL cleavage sites to prevent self-processing substantially increased caspase-1 activity at 30 min after nigericin stimulation, and unlike WT caspase-1, the activity of caspase-1 CDLuncl was remarkably stable over the next 30 min (Figs. 2 B and S2 A). To further confirm that CDL cleavage inactivates cellular caspase-1 protease activity, we also used the reverse approach, in which CDL cleavage was controlled in trans. For this, we replaced caspase-1 CDL self-cleavage sites with a thrombin cleavage site, within an engineered construct that enables caspase-1 dimerization to be induced by a dimerizer drug (AP20187; Fig. 2 C). Here, recombinant thrombin cleaved dimerized caspase-1 at the CDL (Fig. 2 D), and this triggered dimeric caspase-1 to rapidly lose activity over time (Figs. 2 E and S2 B), akin to endogenous and ectopic WT caspase-1 in macrophages (Figs. 1 C and 2 B). Dimerized caspase-1 cleaved its natural substrates pro-GSDMD, pro–IL-1β, and pro–IL-18, but this was blocked when the caspase-1 CDL was cleaved by thrombin (Fig. 2 F). Thus, CDL cleavage is not required for caspase-1 activity, but rather inactivates cellular caspase-1 protease function.
Figure 2.
Caspase-1 processing at the CDL destabilizes caspase-1 dimers and terminates protease activity. Bone marrow progenitors from Ice−/− (Casp1−/−/Casp11null/null) mice were retrovirally reconstituted for mouse caspase-1 (WT, C284 catalytic mutant, or CDL-uncleavable mutant) during their differentiation to macrophages. (A) Differentiated macrophages were then left untreated or LPS-treated for 6 h, or were LPS-primed for 4 h before stimulation with nigericin for 0.5, 1, or 2 h. Cell extracts and cell-free supernatants were mixed and analyzed by immunoblot. (B) Differentiated, transduced macrophages were LPS-primed for 4 h, before stimulation with nigericin for 30 min. Cell culture medium was then replaced with caspase assay buffer containing the caspase-1 fluorogenic substrate, YVAD-afc, and caspase-1–like activity was monitored over the next 30 min. (C) An engineered caspase-1 system allowed controlled caspase-1 dimerization by AP20187 (AP) and CDL cleavage by thrombin (thr). 1 µM AP20187 was added to HEK293T-expressing caspase-1 constructs (WT vs. catalytic mutant, C284A) for 10 min. Cell culture medium was replaced for caspase activity buffer supplemented with digitonin to lyse cells for activity assays (± 20 U/ml thrombin). (D) Immunoblot analysis of caspase-1 cleavage by thrombin after incubation for 1 h. (E) Caspase activity assay, in which thrombin was added to cell lysates at the same time as the YVAD-afc fluorogenic substrate. (F) Cleavage assay in which thrombin was added to caspase-1–expressing cell lysates at the same time as natural substrates (lysates from HEK293T ectopically expressing V5-tagged substrates) and incubated for 0.5 h. (G) Thrombin was added to caspase-1_C284A–expressing cell lysates and incubated for 1 h. Bismaleimidohexane was then added to lysates for 1 h to cross-link caspase-1 dimers. Graphs are mean + SD of triplicate wells from a single experiment. All data are representative of at least two (G) or three (A–F) independent experiments.
Our observation that cellular caspase-1 CDL cleavage to generate p20/p10 renders the protease inactive was surprising, given that recombinant preparations of caspase-1 p20/p10 exhibit protease activity (Fig. S1 C; Ramage et al., 1995; Walsh et al., 2011; Datta et al., 2013). In vitro expression of ΔCARD-caspase-1 yields high protein concentration, triggering spontaneous caspase-1 dimerization and IDL cleavage to generate an active p20/p10 protease (Fig. S1 C). However, recombinant caspase-1 (p20/p10) is reported to rapidly lose its quaternary structure and become inactive when diluted to 10 nM (Walsh et al., 2011), a concentration close to total (active plus inactive) cellular caspase-1 levels (Walsh et al., 2011), which are substantially lower than the dissociation constant of the caspase-1 p20/p10 tetramer (KD ∼110 µM; Datta et al., 2013). Indeed, we confirm here that diluting recombinant p20/p10 results in reduced protease activity per nM caspase-1 (Fig. S2 C). In all, these biochemical characteristics of caspase-1 predict that cellular full-length caspase-1 monomers will only dimerize, gain activity, and self-cleave if they are recruited to a complex (i.e., the inflammasome) that allows a high local concentration of monomers. They further predict that the quaternary structure of cellular caspase-1 dimers will only be stable while they remain bound to their activation complex. Given that caspase-1 interacts with inflammasome signaling hubs via its CARD, we hypothesized that CDL cleavage to remove the CARD releases the p20/p10 tetramer from the signaling hub and in so doing renders the tetramer unstable, terminating activity. Indeed, when cellular dimers of the caspase-1 catalytic mutant were cleaved at the CDL by thrombin, this destabilized their dimeric structure (Fig. 2 G) that is required for protease activity (Fig. S1 B). Thus, active caspase-1 p33/p10 self-cleaves at the CDL to generate p20/p10, and in so doing inactivates protease function by destabilizing caspase-1 dimer structure.
The inflammasome is the site of caspase-1 activity
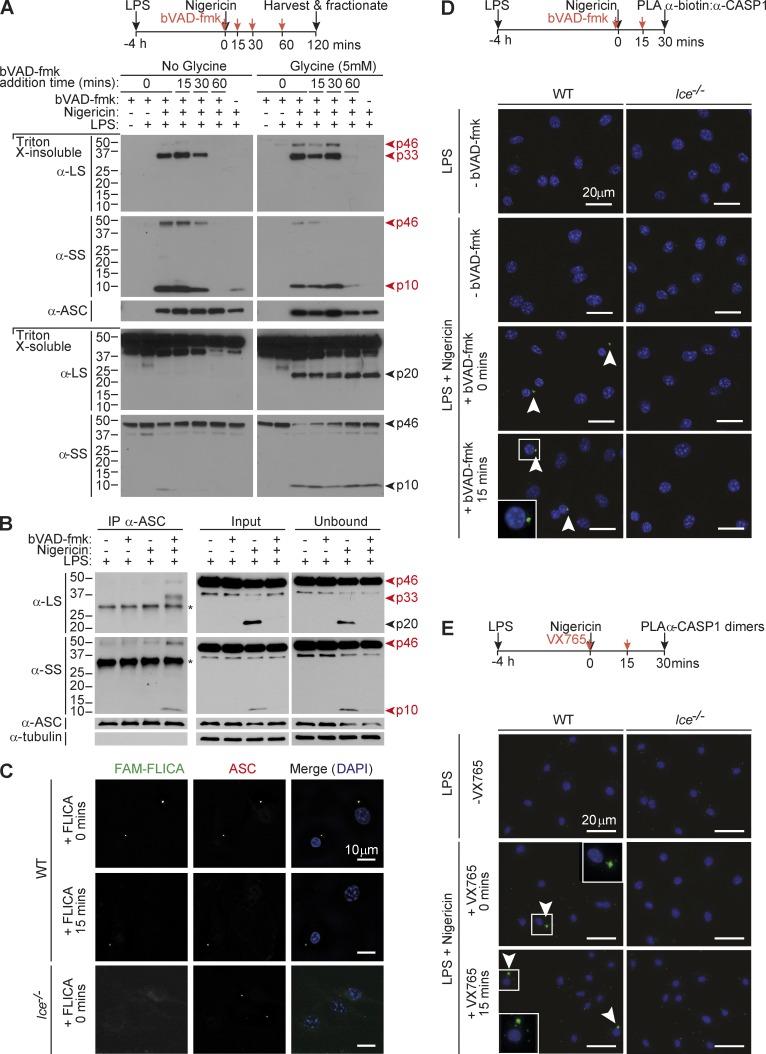
We thus hypothesized that inflammasomes would be the major site of caspase-1 activity (p33/p10 species) and CDL cleavage would trigger the release of p20/p10 from the signaling hub, leading to dissociation of p20/p10 tetramers and caspase-1 deactivation. To examine whether p33, and not p20, was bound to the inflammasome, we added bVAD-fmk to cells at various times after nigericin stimulation to trap active caspase-1 species on the complex and prevent their further self-cleavage. The cell lysates were then fractionated to isolate polymerized ASC (the ASC speck, found in the Triton X-100–insoluble fraction), or ASC was immunoprecipitated, to capture ASC-bound caspase-1 species. Indeed when bVAD-fmk was applied to cells within 30 min of nigericin treatment, caspase-1 p33/p10, but not p20/p10, was captured in the ASC speck–containing fraction (Fig. 3 A). bVAD-fmk application 1 h after nigericin stimulation failed to trap any caspase-1 species on the ASC speck (Fig. 3 A). When cell rupture was delayed by glycine, p20 was retained within the cell but did not cofractionate with the Triton X-100–insoluble ASC speck (Fig. 3 A). In glycine-untreated cells, caspase-1 p20 was released into the cell culture medium, where it was inactive (Fig. S3 A). Likewise, p33/p10 was the dominant species coimmunoprecipitated with ASC in bVAD-fmk–treated macrophages (Fig. 3 B) and macrophages exposed to the caspase-1 inhibitor VX765 (Fig. S3 B), whereas p20 was not coimmunoprecipitated, despite being a prominent fragment in the unbound and input fractions of stimulated cells.
Figure 3.
Caspase-1 activity localizes to the inflammasome. (A) Mouse macrophages were either left untreated or LPS-primed for 4 h before nigericin stimulation for a further 2 h. bVAD-fmk was applied to cells at various times after nigericin (0, 15, 30, 60 min). Cell supernatants were removed and subjected to streptavidin pull-down, which failed to detect active caspase-1 species in this fraction (see Fig. S3 A for pull-down analysis of cell culture medium). Cells were harvested and fractionated. The ASC speck-containing (Triton X-100–insoluble) fraction and the Triton X-100–soluble fraction were analyzed for caspase-1 species by Western blot. Cells were treated with 5 mM glycine 30 min before nigericin to delay cell rupture (right). (B) Macrophages were LPS-primed (4 h) and stimulated with nigericin for 30 min (with and without bVAD-fmk application immediately before nigericin). ASC was immunoprecipitated, and coimmunoprecipitated caspase-1 species were examined by immunoblot. *, Nonspecific band. (C) Macrophages were LPS-primed (4 h) and stimulated with nigericin for 30 min. The FAM-FLICA caspase-1 activity probe was added at the indicated times after nigericin stimulation to localize caspase-1 activity relative to ASC. (D and E) LPS-primed WT or Casp1-deficient Ice−/− macrophages were stimulated with nigericin. Caspase inhibitors (bVAD-fmk caspase activity probe, VX765) were applied at the indicated times after nigericin stimulation, with (D) or without (E) 5 mM glycine in the cell culture media. 30 min after nigericin stimulation, cells were fixed and PLA was performed to detect active caspase-1 (α-biotin/α-caspase-1 LS; (D) or caspase-1 dimers (α-caspase-1 LS:PLUS/α-caspase-1 LS:MINUS; E). DAPI labeled the nucleus. All data are representative of at least three (A–D) or two (E) independent experiments. Arrowheads indicate foci of caspase-1 activity or dimers.
To examine the subcellular localization of endogenous active caspase-1 species, the FAM-FLICA caspase-1 probe or the bVAD-fmk caspase probe was applied to nigericin-treated macrophages, and active and total caspase-1 were localized relative to ASC. Both activity probes localized active caspase-1 to the ASC speck, and total caspase-1 was also enriched on the ASC speck in stimulated cells (Figs. 3 C and S4). We also performed proximity ligation assay (PLA) to verify that active caspase-1 was confined to the inflammasome, first by using antibodies against caspase-1 and biotin to detect bVAD-fmk–labeled caspase-1, in stimulated macrophages. The resultant PLA signal localized to a single focus characteristic of the ASC speck (Fig. 3 D). PLA was also used to detect dimeric caspase-1 species, by conjugating the same caspase-1 monoclonal antibody to plus and minus probes and detecting the PLA between these antibodies. Dimeric caspase-1 in inflammasome-stimulated cells likewise localized to a single speck-like focus (Fig. 3 E). Unlike endogenous and ectopic WT caspase-1, which did not coimmunoprecipitate with ASC unless cells were exposed to a caspase-1 inhibitor before nigericin (Figs. 3B and S3 B), ectopically expressed, CDL-uncleavable caspase-1 (p33) remained in the ASC speck fraction in both inhibitor-treated and -untreated cells (Fig. S5 A), indicating that p33–ASC interaction is stabilized by preventing CDL cleavage. The activity of CDL-uncleavable caspase-1 also colocalized with the ASC speck (Fig. S5 B). In all, our data indicate that caspase-1 is predominantly active when it is bound to a signaling hub such as polymeric ASC and show that CDL self-cleavage releases the p20/p10 protease from the hub, leading to tetramer dissociation. This explains why caspase-1 dimers and caspase-1 activity are localized to the ASC speck (Fig. 3, D and E) and cellular p20 is not pulled down with the bVAD-fmk activity probe (Figs. 1 C and S3 A). Thus, the inflammasome–caspase-1 complex functions as a holoenzyme that incorporates an intrinsic self-limiting mechanism, in which caspase-1 CDL self-cleavage governs the duration of holoenzyme activity.
Inflammasome size specifies active caspase-1 species and deactivation kinetics
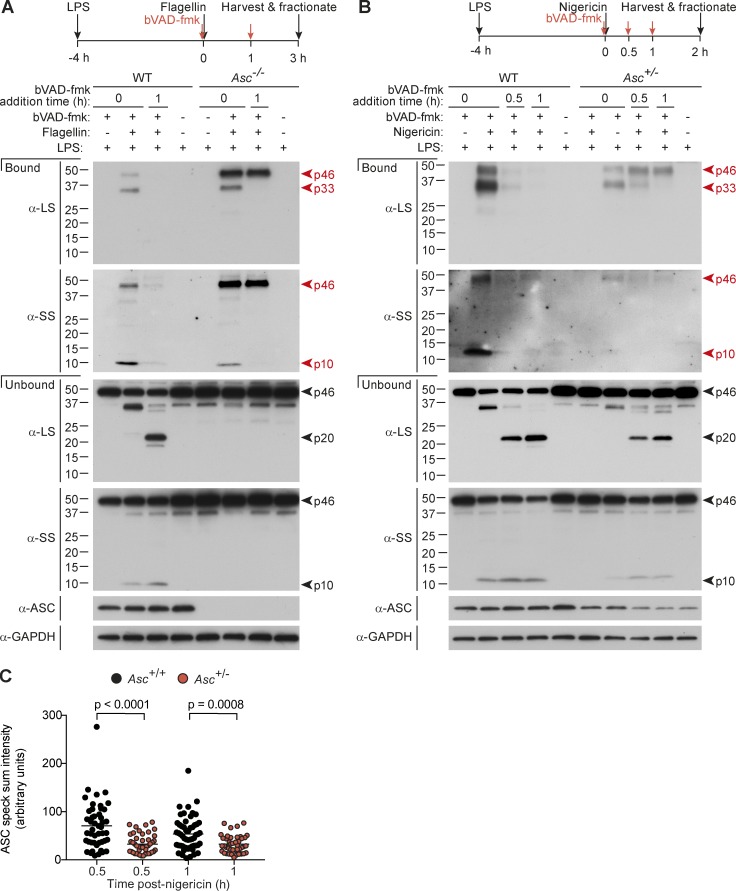
We next explored the signaling determinants that control caspase-1 activation and deactivation by CDL cleavage. Many inflammasomes (e.g., NLRP3, AIM2) obligatorily require ASC for signaling to caspase-1, but some inflammasomes can elicit specific caspase-1 signals in Asc-deficient cells. For example, the NLRC4 inflammasome directly recruits caspase-1 to trigger pyroptosis without apparent caspase-1 autoprocessing to p20 in Asc-deficient macrophages, whereas in WT cells, NLRC4 signaling triggers ASC polymerization, cytokine processing, and full caspase-1 processing to p20 and p10 (Broz et al., 2010). This suggests that efficient caspase-1 autocleavage to p20 requires the ASC speck, a conclusion also reached by studies with ASC point mutants that cannot polymerize or facilitate caspase-1 cleavage to p20 but retain the ability to activate caspase-1 for pyroptosis (Dick et al., 2016). A fully uncleavable caspase-1 mutant (i.e., uncleavable at both the IDL and CDL) activated on the NLRC4 or AIM2 inflammasomes induced pyroptosis but not cytokine processing (Broz et al., 2010), suggesting that dimeric full-length caspase-1 is an active, stable species in specific scenarios (and indeed, we see caspase-1 p46 as a minor active species in Fig. 1 C), but full-length dimeric caspase-1 is unable to efficiently cleave pro-IL-1β. To definitively establish whether the nature of the inflammasome affects the species of active caspase-1 generated (p33/p10 vs. p46), we transfected WT versus Asc−/− macrophages with flagellin to activate the NLRC4 inflammasome and compared the active caspase-1 species generated on the NLRC4-ASC-caspase-1 speck versus the smaller NLRC4-caspase-1 signaling hub. As for NLRP3, NLRC4 activation in WT cells generated p46, p33, and p10 as major active caspase-1 fragments, and active caspase-1 species could not be pulled down when bVAD-fmk was applied 1 h after flagellin (Fig. 4 A). In contrast, in Asc−/− macrophages, although p33/p10 was an active species, the dominant active species was p46, which retained substantial activity 1 h after flagellin (Fig. 4 A), indicating extended and stable activity kinetics. This suggests that p46 is unable to efficiently self-cleave on this hub to generate p33/p10 or terminate protease activity (p20/p10). These data indicate that either (1) ASC-caspase-1 interactions enable caspase-1 self-processing and deactivation in a manner not supported by other caspase-1 signaling hubs such as NLRC4-caspase-1, or (2) the size or density of the signaling hub (e.g., large, compact ASC speck versus small NLRC4 oligomers) is critical for the efficient generation of p33/p10 and the inactivation of this species by CDL cleavage. To generate an intermediate ASC signaling platform, we activated NLRP3 in Asc+/− macrophages. Both p46 and p33/p10 were dominant active caspase-1 species in Asc+/− macrophages, and p46 and p33 activity was more stable over time in Asc+/− macrophages compared with WT macrophages (Fig. 4 B). As anticipated, specks from Asc+/− macrophages had half the ASC content of WT specks (Fig. 4 C). This suggests that the number of available binding sites for caspase-1 (i.e., the size of the inflammasome) is a critical factor specifying the nature of caspase-1 species that are generated, and their activity duration.
Figure 4.
Inflammasome size specifies active caspase-1 species and activity duration. (A–C) LPS-primed macrophages were transfected with flagellin (A) or stimulated with nigericin (B and C). bVAD-fmk was applied at the indicated time points (A and B). (A and B) Pull-down of active caspase-1 from macrophages. 1% IGEPAL was added to macrophages at 3 h after flagellin and 2 h after nigericin stimulation, to lyse cells directly in their culture medium. Streptavidin-coated beads pulled down active caspase-1 bound to the biotin-labeled activity probe in mixed lysates/supernatants. (C) ASC speck intensity of Asc+/+ and Asc+/− macrophages (4 h LPS + nigericin, with VX765 added 1 h before nigericin to prevent cell death), quantified from microscopy images. Each circle is one speck (n = 40–55), and significance between samples was assessed by nonparametric, unpaired Mann–Whitney test. All data are representative of at least three (A and B) or two (C) independent experiments.
Cell type specifies caspase-1 deactivation kinetics
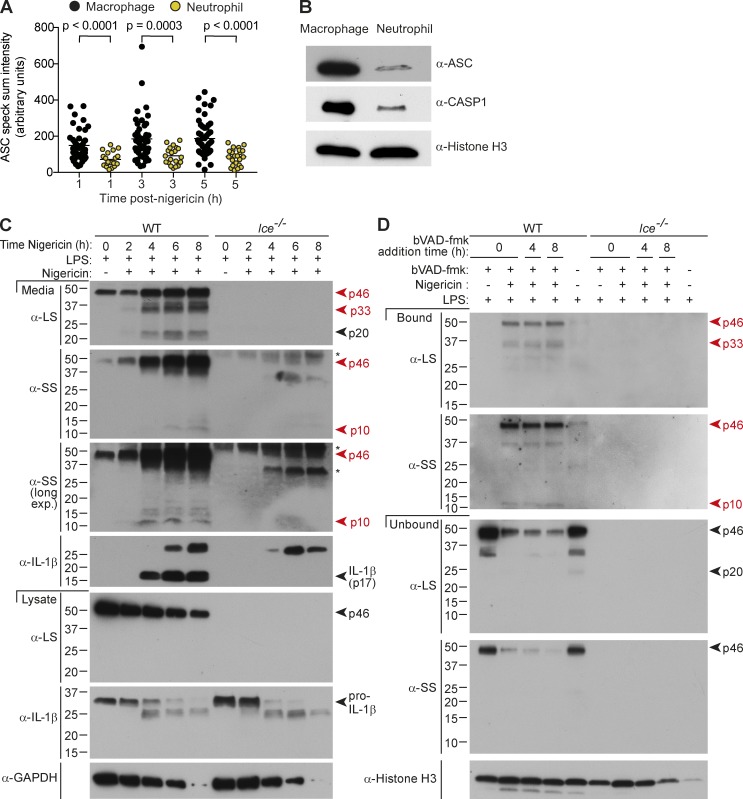
We recently reported that neutrophils exhibit prolonged kinetics of IL-1β production compared with macrophages (Chen et al., 2014), and we note that neutrophils assemble a smaller ASC speck than macrophages (Fig. 5 A) because they express substantially less ASC per cell (Fig. 5 B). Given these findings, coupled with the extended duration of caspase-1 activity in Asc+/− macrophages (Fig. 4 B), we speculated that caspase-1 activity may be prolonged in neutrophils compared with macrophages. Unlike nigericin-stimulated macrophages in which mature IL-1β (p17) was generated in a burst (Fig. 1 D), levels of secreted IL-1β p17 continued to rise up to 6 h after nigericin stimulation in neutrophils (Fig. 5 C), and p46, p33, and p10 were major and sustained cleavage fragments pulled down by bVAD-fmk (Fig. 5 D). Further, bVAD-fmk application up to 8 h after nigericin stimulation successfully trapped the active p46 and p33/p10 species (Fig. 5 D), confirming that neutrophil caspase-1 activity duration is indeed extended relative to macrophages. Thus, caspase-1 CDL self-cleavage within the inflammasome holoenzyme also tailors the duration of caspase-1 activity to the identity of the responding cell. ASC speck size may not be the only factor controlling cell type–specific caspase-1 activity kinetics; we also noted that caspase-1 is less abundant in neutrophils than macrophages on a per-cell basis (Fig. 5 B), which is also likely to affect its ability to dimerize and autoprocess on the inflammasome.
Figure 5.
Cell type specifies caspase-1 activity duration. (A–D) LPS-primed (A and B) macrophages or (A–D) neutrophils were left untreated or stimulated with nigericin. (A) ASC speck intensity of Ice−/− macrophages and neutrophils (4 h LPS + nigericin), quantified from microscopy images. Each circle is one speck (n = 20–55), and significance between samples was assessed by nonparametric, unpaired Mann–Whitney test. (B) Immunoblot for inflammasome component expression in whole-cell extracts (4 h LPS). (C) Cell lysates and cell-free supernatants were harvested at various times after nigericin stimulation (0, 2, 4, 8 h), and analyzed by Western blot. *, Nonspecific band. (D) bVAD-fmk was applied at the indicated time points. Supernatant was removed, and cells were lysed 8 h after nigericin stimulation, and active caspase-1 was pulled down using streptavidin beads. Streptavidin-bound and -unbound fractions from mixed supernatants/extracts were analyzed by Western blot. All data are representative of at least three (A, B, C [WT], D [WT]) or two (C [Ice−/−], D [Ice−/−]) independent experiments.
Discussion
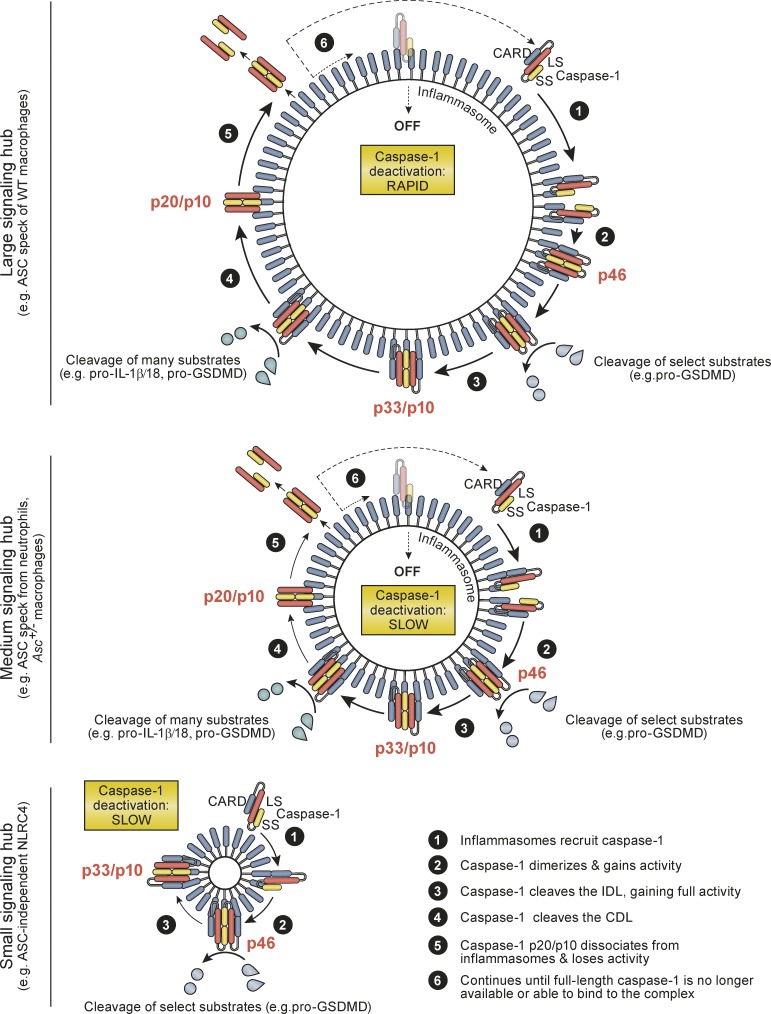
Proinflammatory signaling pathways must be tightly controlled, because failure to shut down inflammatory signaling leads to inflammatory diseases. Since the discovery of inflammasomes in 2002, inflammasome research has focused on activation mechanisms and pathway association with a plethora of human diseases, but surprisingly little is known of how cells and tissues deactivate inflammasomes and their elicited caspases to shut down inflammatory signaling. Given that in vivo inflammasome activity is transient in acute inflammation (e.g., infection models), mechanisms for signal inhibition must exist to allow for inflammation resolution and the return to homeostasis. The existence of such negative regulatory mechanisms is also indicated by a universal principle of signal transduction, in which signal initiation elicits negative feedback mechanisms to curtail cell signaling in a timely manner. Here we have characterized the molecular mechanisms of caspase-1 activation and deactivation by inflammasomes and report an elegant self-limiting signaling mechanism by which caspase-1 activation is intrinsically coupled to its timely deactivation. Our data show that caspase-1 clustering on inflammasomes generates active caspase-1 p46 dimers that self-process at the IDL to generate a second active species, p33/p10, which remains bound to the inflammasome. The activity of caspase-1 p33/p10 is then halted by a second self-cleavage event, which removes the CARD domain, releasing caspase-1 (p20/p10) from the inflammasome (Fig. 6). Because free caspase-1 p20/p10 is unstable at the concentrations found in cells, caspase-1 release from the inflammasome triggers a loss of dimer structure and terminates protease activity. A molecular timer also governs caspase-9 deactivation during apoptosome signaling, but via a distinct mechanism involving caspase-9 displacement from the apoptosome (Malladi et al., 2009), rather than cleavage to release the enzymatic subunits from the activation complex as we describe here for caspase-1. This mechanism of caspase-1 protease activation intrinsically coupled to deactivation by CDL cleavage is thus unique among caspase regulatory mechanisms reported to date.
Figure 6.
Summary model for mechanisms by which inflammasomes direct the active caspase-1 species and duration of caspase-1 activity. See Discussion for details.
Caspase-1 activity duration is also specified by activation platform size, which differs between distinct inflammasome complexes and cell types, with large signaling hubs turning over protease activity more quickly (Fig. 6). Cellular caspase-1 activity duration is likely specified by cell type and inflammasome size at three levels: (1) the number of available caspase-1–binding sites on the inflammasome; (2) the kinetics of CDL cleavage-driven caspase-1 p20/p10 release from inflammasomes; and (3) the number of successive rounds of caspase-1 binding and release from inflammasomes, which will be dictated by cellular concentrations of full-length caspase-1 relative to available binding sites on the inflammasome. Individual cell types, and individual inflammasome complexes, may thereby tailor the kinetics of caspase-1 activity duration to modulate the chronicity of inflammasome signaling. Future studies should identify cell types with extended caspase-1 activity duration, such as neutrophils identified here. Prolonged signaling by such cells may be critical for inflammasome-driven chronic inflammatory diseases in humans.
Unlike apoptotic caspases that are either initiator or executioner caspases, caspase-1 is both an initiator and executioner caspase. Compared with other caspases, recombinant caspase-1 tested in vitro is promiscuous in its substrate repertoire, but in cells, the caspase-1 substrate repertoire is relatively small (Agard et al., 2010; Walsh et al., 2011). Our discovery that the inflammasome–caspase-1 complex functions as a holoenzyme has interesting implications for caspase-1 substrate selection. Because cellular caspase-1 activity is confined to the inflammasome, caspase-1 substrates may be additionally specified by their capacity for recruitment to the inflammasome.
In summary, we report the first definition of active cellular caspase-1 species as p46 and a transient species, p33/p10, and demonstrate that the inflammasome is not merely an inert platform that facilitates caspase-1 activation. The inflammasome–caspase-1 complex constitutes an active holoenzyme, as caspase-1 requires association with the complex to retain its activity. Assembly of the holoenzyme and resultant protease activity then enables caspase-1 CDL cleavage to generate p20/p10, with kinetics specified by cell type and inflammasome size. p20/p10 has been presumed to be an active species of caspase-1 in inflammasome-signaling cells, but here we show that CDL cleavage to generate p20/p10 in fact destabilizes caspase-1 structure and thus terminates caspase-1 signaling. Our data provide a mechanism for self-limiting inflammasome signaling that explains inflammatory resolution after immune challenge and reveals the molecular basis for the coordinated timing of caspase-1 cellular programs.
Materials and methods
Mice
Mice not generated on the C57BL/6 background were backcrossed at least 10 times to C57BL/6, and all experiments were performed using age- and sex-matched animals. C57BL/6, Ice−/− (Casp1−/−/Casp11null/null; Kuida et al., 1995) Asc−/− and Asc+/− (Mariathasan et al., 2004), and Gsdmd−/− (Kayagaki et al., 2015) mice were housed in specific pathogen-free facilities at the University of Queensland. The University of Queensland’s animal ethics committee approved all experimental protocols.
Preparation of mouse macrophages and neutrophils
Bone marrow–derived macrophages were differentiated as previously described (Schroder et al., 2012a) and stimulated on day 7 of differentiation. Bone marrow neutrophils were MACS purified using an α-Ly6G–FITC antibody (1A8; BioLegend) and α-FITC beads as previously described (Chen et al., 2014). MACS-purified neutrophils achieved >98% purity, as assessed by flow cytometry.
Retroviral transduction
The coding sequence of mouse caspase-1 was cloned into a replication-defective mouse stem cell retroviral construct (pMSCV). Caspase-1 mutants were generated using overlapping PCR site-directed mutagenesis. The retroviral packing cell line, PlatE, was Lipofectamine-transfected with pMSCV vectors and incubated at 32°C for 2 d for virus production (Schroder et al., 2012b). Cell-free retrovirus was prepared and used to spinfect Ice−/− bone marrow cells on day 2 of their CSF-1–directed differentiation into macrophages. Equal numbers of transduced (GFP+) differentiated macrophages were seeded for each construct for inflammasome assays.
Inflammasome and pyroptosis assays
Differentiated mouse bone marrow–derived macrophages were plated at a density of 106 cells/ml in full media (RPMI-1640, 10% FBS, 1× Glutamax, and 150 ng/ml recombinant human CSF-1 [endotoxin-free, produced in insect cells by the UQ Protein Expression Facility]). Bone marrow neutrophils were used for experiments directly after purification and were plated at a density of 107 cells/ml in opti-MEM (Life Technologies) supplemented with 1.2 µg/ml aprotinin (Sigma-Aldrich). Cells were primed with ultrapure Escherichia coli K12 LPS (100 ng/ml) for 4 h, before exposure to 5 µM nigericin (Sigma-Aldrich) or transfected with flagellin (0.4% Lipofectamine LTX + 150 ng/ml of ultrapure flagellin; InvivoGen) for the indicated times. For most applications, macrophage cell culture medium was replaced with opti-MEM (supplemented with 150 ng/ml human CSF-1) before nigericin stimulation. IL-1β in cell-free supernatants was analyzed by ELISA (eBioscience). Cellular cytotoxicity was quantified using the Cytox96 nonradioactive cytotoxicity assay (Promega) and displayed as a percentage of total cellular LDH (100% cell lysis control). Western blots of cell extracts and methanol/chloroform-precipitated supernatants were performed using standard procedures (Gross, 2012). Antibodies for immunoblot were against IL-1β (polyclonal goat; R&D Systems), caspase-1 p20 (LS; Casper-1; Adipogen), caspase-1 p10 (SS; M-20; Santa Cruz Biotechnology; or casper-2; Adipogen), ASC ([N-15]-R; Santa Cruz Biotechnology), GSDMD (126–138; Sigma-Aldrich), α-tubulin (B5-1-2; Sigma-Aldrich), and GADPH (polyclonal mouse; BioScientific).
Active caspase pulldown
bVAD-fmk (Faleiro et al., 1997) was applied to cells to label and trap active caspases during inflammasome activation. Mouse bone marrow–derived macrophages (106) or neutrophils (3 × 106) were primed and stimulated as described. 10 µM bVAD-fmk (Santa Cruz) was applied to cells at various times before or after the inflammasome agonist, as indicated. For bVAD-fmk pull-down of entire cell output (lysate plus supernatant), cells were lysed directly in opti-MEM with 1% IGEPAL CA630 and incubated on ice for 5 min in the presence of protease inhibitors (cOmplete mini protease inhibitor cocktail; Roche) and benzonase. Paramagnetic streptavidin beads (Promega) were added to cell lysates and incubated with gentle rotation at 4°C for 3 h or overnight. The streptavidin-unbound fraction was methanol/chloroform-precipitated (Gross, 2012), and beads were washed three times with buffers with increasing salt concentrations (150–500 mM NaCl, 50 mM Hepes, pH 8.0, and 1% IGEPAL CA630). Beads were then pelleted and resuspended in NuPage LDS sample buffer (Thermo Fisher Scientific) supplemented with 10 mM DTT and boiled for 10 min to release bVAD-fmk–trapped proteins from the beads. The mixture was centrifuged for 1 min at 6000 g, and the soluble (streptavidin-bound) fraction was separated by gel electrophoresis alongside the unbound fraction.
ASC speck isolation
106 macrophages per condition were treated with LPS, nigericin, and bVAD-fmk as indicated; cell supernatant was removed for streptavidin pull-down assay; and the cells were lysed on ice for 5 min in lysis buffer (50 mM Hepes, pH 8.0, 0.5% Triton X-100, and Roche cOmplete mini protease inhibitor cocktail). Cell lysates were then centrifuged for 15 min at 6,000 g. The resulting supernatant (Triton X-100–soluble fraction) was precipitated (Gross, 2012), and the pellet was once again washed with lysis buffer and dried. The Triton X-100–soluble and –insoluble fractions were then resuspended in NuPage LDS sample buffer. Cell fractions were analyzed by immunoblot.
ASC immunoprecipitation
5 × 106 macrophages per condition were primed with LPS for 4 h and then stimulated with nigericin for 30 min. DMSO vehicle control, bVAD-fmk (10 µM), or VX765 (10 µM) were applied 5 min before nigericin. Cells were then lysed in buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 0.5% IGEPAL, 1 mM ATP, protease inhibitors (cOmplete Mini; Roche), and benzonase. Clarified lysates were incubated with α-ASC antibody (N15, 0.4 µg/ml; Santa Cruz Biotechnology) with Dynabeads for 2 h rotating at 4°C. Immunoprecipitated samples, input (8%), and unbound fractions were analyzed by immunoblot.
Preparation of recombinant caspase-1
Mouse caspase-1 (ΔCARD) was expressed as a GST-DmrB fusion protein with a C-terminal His8 tag in E. coli BL21 Gold. Expression was induced with 200 µM IPTG for 8 h at 18°C, and ΔCARD_caspase-1 was purified using Ni Sepharose 6 Fast Flow resin as described (Boucher et al., 2014). Active caspase-1 was quantified via active site titration using zVAD-fmk (Boucher et al., 2014). For experiments in which different concentrations of recombinant p20/p10 were assayed for activity, recombinant caspase-1 p20/p10 was first diluted to 1 µM in high-salt buffer (50 mM Hepes, pH 7.4, 50 mM NaCl, 1 M sodium citrate, 0.01% CHAPS, and 10 mM DTT) for 30 min at 37°C and then further diluted in caspase activity buffer (200 mM NaCl, 50 mM Hepes, pH 8.0, 50 mM KCl, 200 µM Ac-YVAD-afc, and 10 mM DTT) for 5 min before caspase-1–like activity on the substrate Ac-YVAD-afc was quantified.
Caspase-1 dimerizer system
ΔCARD_Caspase-1 (residues 91–402 of the full-length mouse protein) and ΔCARD_Caspase-1_C284A were cloned as N-terminal DmrB fusions into pEF6. A ΔCARD_Caspase-1 variant was engineered to prevent self-processing but allow cleavage by thrombin, in which residues 99–103 (including self-processing sites E102/D103) were mutated to a thrombin cleavage site (LVPRG), and residues E121 and D122 were substituted with alanine to prevent self-processing. HEK293T were transfected with 500 ng DmrB-ΔCARD_caspase-1 plasmids using Lipofectamine 2000 for 18 h. 4 × 105 cells were then seeded per well in 24-well plates. 4 h later, cells were exposed to 1 µM AP20187 for 10 min to mediate DmrB dimerization. Cell culture medium was then replaced with caspase assay buffer (200 mM NaCl, 50 mM Hepes, pH 8.0, 50 mM KCl, and 100 µg/ml digitonin). For the substrate cleavage assay, the buffer was supplemented with 10 mM DTT; cell extract from HEK293T expressing pro–IL-1β-V5, pro–IL-18-V5, or pro-GSDMD-V5; and 20 U/ml thrombin (Sigma-Aldrich), where appropriate, to cleave the engineered caspase-1 protein. The reaction was incubated for 0.5 h. For the cross-linking experiment, caspase assay buffer was supplemented with 20 U/ml thrombin (Sigma-Aldrich). 1 h later, 2 mM bismaleimidohexane (Thermo Fisher Scientific) was added for 1 h at RT.
Caspase-1–like activity assay
Cells were seeded in opaque, flat-bottom 96-well plates (Costar) before cell stimulation, at 105 cells (untransduced macrophages, transfected HEK293T) or 4 × 104 GFP+ cells (transduced macrophages) per well. Macrophages were LPS-primed and stimulated with nigericin as described, and transfected HEK293T were treated with 1 µM DmrB dimerizer drug, AP20187. 30 min after nigericin stimulation (macrophages) or 10 min after AP20187 treatment (HEK293T), the cell culture supernatant was removed and replaced with buffer (200 mM NaCl, 50 mM Hepes, pH 8.0, 50 mM KCl, 100 µM Ac-YVAD-afc, 100 µg/ml digitonin, and 10 mM DTT). Fluorogenic caspase activity was measured using an M1000 Tecan spectrofluorometer at 37°C with 400-nm excitation and 505-nm emission, at set time intervals.
Immunofluorescence microscopy and PLA
Macrophages were seeded at 1–1.5 × 105 on glass coverslips in full medium in 24-well plates. Cells were LPS-primed for 4 h in opti-MEM, and then stimulated with nigericin. 10 µM bVAD-fmk, VX765, or FAM-FLICA (used as per manufacturer’s instructions; ImmunoChemistry) was added to the cells as indicated. After 30 min of nigericin stimulation, coverslips were washed twice with PBS (or 1× Apoptosis Wash Buffer supplied with Caspase-1 assay Kit for FAM-FLICA) and incubated in PBS containing 4% paraformaldehyde for 15 min.
For immunofluorescence microscopy, cells were stained with the following antibodies: mouse anti–caspase-1 p20 (LS, Casper-1, 1:100; Adipogen), mouse anti–caspase-1 p10 (SS, Casper-2, 1:100; Adipogen), rabbit anti-ASC (N15, 1:300; Santa Cruz Biotechnology), and goat anti-biotin (ab6643, 1:100; Abcam). Alexa Fluor 488– and 594–conjugated secondary antibodies were purchased from Molecular Probes (Invitrogen). Coverslips were mounted in ProLong Gold Reagent (Life Technologies) and imaged using a DeltaVision Olympus IX80 inverted wide-field deconvolution microscope equipped with Olympus Plan-Apochromat ×60/1.35 oil lens and a Lumencor 7 line LED lamp. Images were captured using a Photometrics Coolsnap HQ2 camera. Images were deconvolved using 10 cycles of iterative deconvolution using SofWorx. Imaging analysis of all data were performed using ImageJ software (NIH). ASC speck sum intensity between cell types and genotypes were calculated as area × mean intensity using the particle analysis function in ImageJ.
For PLA, coverslips were washed twice with PBS, before Duolink In Situ PLA was performed using Duolink In Situ Detection Reagents Orange (Sigma-Aldrich), according to the manufacturer’s instructions. Primary antibodies were mouse anti–caspase-1 p20 (LS, Casper-1, 1:100; Adipogen) and goat anti-biotin (ab6643, 1:500; Abcam), used in conjunction with anti-goat minus and anti-mouse plus Duolink PLA probes. Alternatively, Duolink plus and minus probes were each conjugated to the mouse anti–caspase-1 p20 (LS, Casper-1, 1:100; Adipogen) monoclonal antibody (using Duolink In Situ Probemaker; Sigma-Aldrich), and PLA detected the signal between these antibodies. Coverslips were mounted with ProLong Gold antifade with DAPI. Confocal images were acquired on a Zeiss LSM 710 laser-scanning microscope (Plan-Apochromatic 40×/1.3 oil objective) driven by Zen software (ZEN 2009; Zeiss). Images were also acquired on an Olympus BX51 fluorescence microscope with an Olympus DP70 CCD camera (Plan-Apochromatic 60×/1.35 oil objective). Images were processed and analyzed using ImageJ.
Online supplemental material
Fig. S1 displays additional data related to Fig. 1, showing that cellular caspase-1 activity is induced by dimerization and terminated in a manner independent of GSDMD or frank cell lysis. Fig. S2 shows additional data related to Fig. 2, and demonstrates that caspase-1 cleavage at the CDL in cis or trans inactivates proteolytic activity, and dilution of recombinant caspase-1 p20/p10 triggers its loss of relative activity. Fig. S3 displays additional data related to Fig. 3, and demonstrates that caspase-1 is active only on the inflammasome, and not when released into cell culture medium. In keeping with this, p33/10 but not p20/p10 is bound to ASC. Fig. S4 displays additional data related to Fig. 3, showing that active caspase-1 colocalizes with the ASC speck. Fig. S5 displays additional data related to Fig. 3, and demonstrates that a CDL-uncleavable mutant of caspase-1 localizes to the ASC speck.
Supplementary Material
Acknowledgments
We thank Kylie Georgas for her assistance with figure preparation, Dr. Vishva Dixit for reagents, and Drs. Nicholas Condon and Nicholas Hamilton for assistance with image analysis.
This work was supported by a project grant from the Australian Research Council to K. Schroder (DP160102702). D. Boucher was supported by postdoctoral fellowships from the Fonds de Recherche du Québec - Santé and the University of Queensland. R.C. Coll was supported by a postdoctoral research-industry fellowship from the University of Queensland. K.W. Chen was supported by a postgraduate scholarship from the ANZ Trustee Medical Research Program. K.J. Stacey was supported by a National Health and Medical Research Council Senior Research Fellowship (1059729). K. Schroder was supported by an Australian Research Council Future Fellowship (FT130100361) and a National Health and Medical Research Council of Australia Career Development Fellowship (1141131). Imaging was performed in the Australian Cancer Research Foundation–funded IMB Cancer Biology Imaging Facility.
K. Schroder holds a full-time Associate Professorial Research Fellow appointment at the University of Queensland and conducts research focused on inflammation and inflammasomes. K. Schroder and R.C. Coll are co-inventors on patent applications for NLRP3 inhibitors, which have been licensed to Inflazome Ltd., a company headquartered in Dublin, Ireland. Inflazome is developing drugs that target the NLRP3 inflammasome to address unmet clinical needs in inflammatory disease. The authors have no further conflicts of interest to declare.
Author contributions: D. Boucher, M. Monteleone, R.C. Coll, K.W. Chen, C.M. Ross, J.L. Teo, G.A. Gomez, C.L. Holley, and D. Bierschenk designed and performed experiments. K.J. Stacey, A.S. Yap, and J.S. Bezbradica assisted in specific areas of research conceptualization. D. Boucher and K. Schroder designed the research. K. Schroder supervised the study and wrote the manuscript, with input from all authors.
References
- Agard N.J., Maltby D., and Wells J.A.. 2010. Inflammatory stimuli regulate caspase substrate profiles. Mol. Cell. Proteomics. 9:880–893. 10.1074/mcp.M900528-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright K.M., Renatus M., Scott F.L., Sperandio S., Shin H., Pedersen I.M., Ricci J.E., Edris W.A., Sutherlin D.P., Green D.R., and Salvesen G.S.. 2003. A unified model for apical caspase activation. Mol. Cell. 11:529–541. 10.1016/S1097-2765(03)00051-0 [DOI] [PubMed] [Google Scholar]
- Boucher D., Duclos C., and Denault J.B.. 2014. General in vitro caspase assay procedures. Methods Mol. Biol. 1133:3–39. 10.1007/978-1-4939-0357-3_1 [DOI] [PubMed] [Google Scholar]
- Broz P., von Moltke J., Jones J.W., Vance R.E., and Monack D.M.. 2010. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 8:471–483. 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.W., Groß C.J., Sotomayor F.V., Stacey K.J., Tschopp J., Sweet M.J., and Schroder K.. 2014. The neutrophil NLRC4 inflammasome selectively promotes IL-1β maturation without pyroptosis during acute Salmonella challenge. Cell Reports. 8:570–582. 10.1016/j.celrep.2014.06.028 [DOI] [PubMed] [Google Scholar]
- Datta D., McClendon C.L., Jacobson M.P., and Wells J.A.. 2013. Substrate and inhibitor-induced dimerization and cooperativity in caspase-1 but not caspase-3. J. Biol. Chem. 288:9971–9981. 10.1074/jbc.M112.426460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick M.S., Sborgi L., Rühl S., Hiller S., and Broz P.. 2016. ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat. Commun. 7:11929 10.1038/ncomms11929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faleiro L., Kobayashi R., Fearnhead H., and Lazebnik Y.. 1997. Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. EMBO J. 16:2271–2281. 10.1093/emboj/16.9.2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross O. 2012. Measuring the inflammasome. Methods Mol. Biol. 844:199–222. 10.1007/978-1-61779-527-5_15 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526:666–671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Kuida K., Lippke J.A., Ku G., Harding M.W., Livingston D.J., Su M.S., and Flavell R.A.. 1995. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 267:2000–2003. 10.1126/science.7535475 [DOI] [PubMed] [Google Scholar]
- Liew F.Y., Xu D., Brint E.K., and O’Neill L.A.. 2005. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5:446–458. 10.1038/nri1630 [DOI] [PubMed] [Google Scholar]
- Malladi S., Challa-Malladi M., Fearnhead H.O., and Bratton S.B.. 2009. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 28:1916–1925. 10.1038/emboj.2009.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D.M., Vucic D., French D.M., Lee W.P., Roose-Girma M., Erickson S., and Dixit V.M.. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218. 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Mariathasan S., Weiss D.S., Newton K., McBride J., O’Rourke K., Roose-Girma M., Lee W.P., Weinrauch Y., Monack D.M., and Dixit V.M.. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 440:228–232. 10.1038/nature04515 [DOI] [PubMed] [Google Scholar]
- Meissner F., Molawi K., and Zychlinsky A.. 2008. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat. Immunol. 9:866–872. 10.1038/ni.1633 [DOI] [PubMed] [Google Scholar]
- Ramage P., Cheneval D., Chvei M., Graff P., Hemmig R., Heng R., Kocher H.P., Mackenzie A., Memmert K., Revesz L., et al. 1995. Expression, refolding, and autocatalytic proteolytic processing of the interleukin-1 beta-converting enzyme precursor. J. Biol. Chem. 270:9378–9383. 10.1074/jbc.270.16.9378 [DOI] [PubMed] [Google Scholar]
- Schroder K., and Tschopp J.. 2010. The inflammasomes. Cell. 140:821–832. 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- Schroder K., Irvine K.M., Taylor M.S., Bokil N.J., Le Cao K.A., Masterman K.A., Labzin L.I., Semple C.A., Kapetanovic R., Fairbairn L., et al. 2012a Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proc. Natl. Acad. Sci. USA. 109:E944–E953. 10.1073/pnas.1110156109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Sagulenko V., Zamoshnikova A., Richards A.A., Cridland J.A., Irvine K.M., Stacey K.J., and Sweet M.J.. 2012b Acute lipopolysaccharide priming boosts inflammasome activation independently of inflammasome sensor induction. Immunobiology. 217:1325–1329. 10.1016/j.imbio.2012.07.020 [DOI] [PubMed] [Google Scholar]
- Thornberry N.A., Bull H.G., Calaycay J.R., Chapman K.T., Howard A.D., Kostura M.J., Miller D.K., Molineaux S.M., Weidner J.R., Aunins J., et al. 1992. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 356:768–774. 10.1038/356768a0 [DOI] [PubMed] [Google Scholar]
- Walsh J.G., Logue S.E., Lüthi A.U., and Martin S.J.. 2011. Caspase-1 promiscuity is counterbalanced by rapid inactivation of processed enzyme. J. Biol. Chem. 286:32513–32524. 10.1074/jbc.M111.225862 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.