Cell–cell adhesion and cell shape are regulated at adherens junctions during embryonic morphogenesis. Beati et al. show that the Drosophila LIM domain protein Smallish interacts with Bazooka, Canoe, and Src42A at adherens junctions. Loss-of-function and gain-of-function phenotypes reveal a function for Smallish in regulation of actomyosin contractility and cell shape.
Abstract
In epithelia, cells adhere to each other in a dynamic fashion, allowing the cells to change their shape and move along each other during morphogenesis. The regulation of adhesion occurs at the belt-shaped adherens junction, the zonula adherens (ZA). Formation of the ZA depends on components of the Par–atypical PKC (Par-aPKC) complex of polarity regulators. We have identified the Lin11, Isl-1, Mec-3 (LIM) protein Smallish (Smash), the orthologue of vertebrate LMO7, as a binding partner of Bazooka/Par-3 (Baz), a core component of the Par-aPKC complex. Smash also binds to Canoe/Afadin and the tyrosine kinase Src42A and localizes to the ZA in a planar polarized fashion. Animals lacking Smash show loss of planar cell polarity (PCP) in the embryonic epidermis and reduced cell bond tension, leading to severe defects during embryonic morphogenesis of epithelial tissues and organs. Overexpression of Smash causes apical constriction of epithelial cells. We propose that Smash is a key regulator of morphogenesis coordinating PCP and actomyosin contractility at the ZA.
Introduction
The regulation of cell–cell adhesion between epithelial cells is crucial for the control of morphogenetic movements during development (Haigo et al., 2003; Gumbiner, 2005; Lecuit and Yap, 2015). A major driving force for cell shape changes during morphogenesis is the contraction of the actomyosin network anchored at the belt-shaped adherens junction (AJ), the zonula adherens (ZA; Simões et al., 2014; Murrell et al., 2015; Siedlik and Nelson, 2015; Harris, 2017; Umetsu and Kuranaga, 2017). Links between the actomyosin network and the cell adhesion molecules of the ZA, the cadherins, are provided by actin-binding proteins that associate with the cytoplasmic tails of cadherins (Simões et al., 2010; Leckband and de Rooij, 2014; Takeichi, 2014). Among these linker proteins are α-catenin, vinculin, and afadin (Canoe [Cno] in Drosophila melanogaster; Sawyer et al., 2009).
The contractility of actomyosin is regulated via phosphorylation of the regulatory light chain of nonmuscle myosin II (Spaghetti squash [Sqh] in Drosophila) by Rho-associated coiled-coil containing kinase (Rho kinase [Rok] in Drosophila; Lilien and Balsamo, 2005; Julian and Olson, 2014) and its binding partner Shroom (Shrm; Hildebrand and Soriano, 1999). Overexpression of Shrm induces apical constriction of polarized epithelial cells (Haigo et al., 2003; Plageman et al., 2011). In Drosophila, Shrm binds to Rok and is required for its planar polarization during convergent extension movements of the epidermis (Simões et al., 2014). Among the phosphorylation targets of Rok is the polarity regulator Bazooka/Par-3 (Baz), which loses its planar polarization upon mutation or inhibition of Rok (Simões et al., 2010).
In Drosophila embryonic morphogenesis, Baz apparently has several key functions, as it is required for apical-basal polarity, planar cell polarity (PCP), and formation of the ZA in the neuroectodermal epithelium during germ band extension (Müller and Wieschaus, 1996; Bilder et al., 2003; Harris and Peifer, 2004; Zallen and Wieschaus, 2004). How these functions are coordinated at the molecular level is not well understood so far. In particular, very few factors are known that are not required for formation of the ZA as such, but that regulate adhesion and cortical tension at the ZA during epithelial morphogenesis.
Here we introduce Smash, a new ZA-associated Lin11, Isl-1, Mec-3 (LIM) domain protein in Drosophila that binds to Baz, to the Src family kinase Src42A, and to Cno. We show that Smash is planar polarized in the embryonic epidermis during germ band extension, being enriched at anterior–posterior (A/P) cell junctions between anterior and posterior cells, together with the key regulators of epithelial remodeling Sqh, Rok, and Cno and thus complementary to the enrichment of Baz at dorsal–ventral (D/V) junctions between dorsal and ventral cells (Zallen and Wieschaus, 2004; Simões et al., 2010). Embryos lacking Smash show defective PCP of Baz, Sqh, and Cno and fail to execute morphogenesis properly. By laser ablation experiments, we show that junctional tension in the larval epidermis is reduced in smash mutant animals. On the other hand, Smash overexpression causes apical constriction of epithelial cells. We propose that Smash mediates interactions between the polarity regulator Baz, the kinase Src42A, Cno, and the actomyosin network at the ZA to regulate cell shape and cortical tension during epithelial morphogenesis.
Results
The LIM protein Smash binds to PDZ domains of Baz
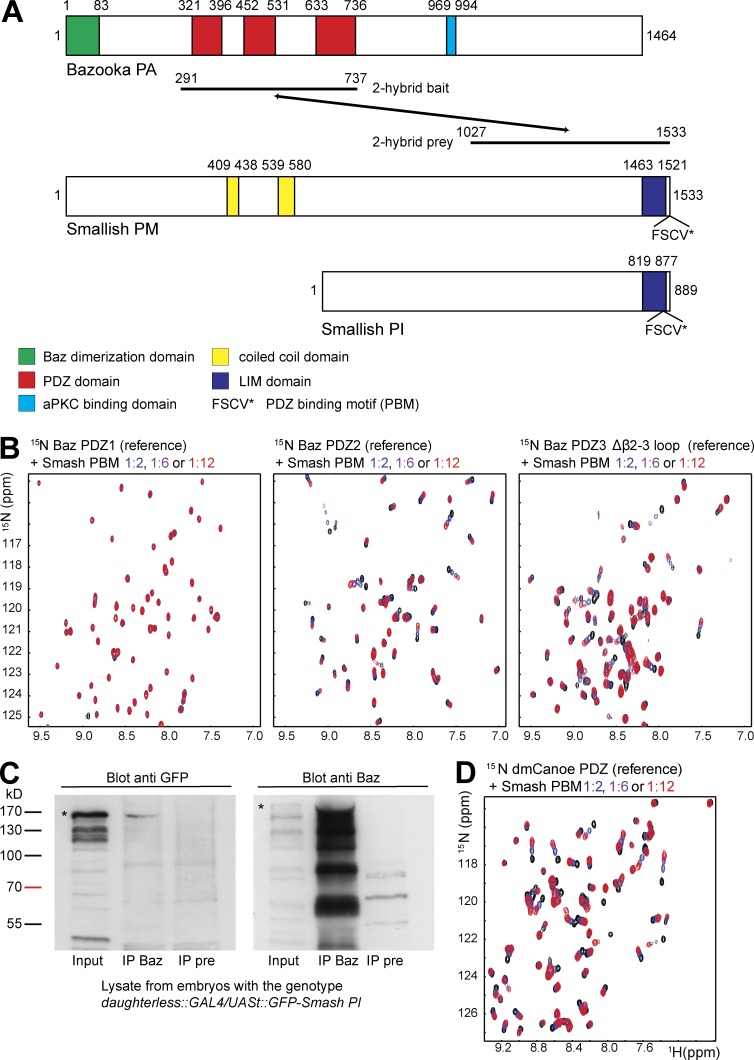
To identify binding partners of Baz involved in epithelial morphogenesis, we conducted a yeast two-hybrid screen using the three PDZ domains of Baz (aa 291–737) as bait (von Stein et al., 2005). One interacting clone encoded the C-terminal region (aa 1027–1533) of isoform PM of the predicted protein CG43427 (Fig. 1 A), which we named Smallish (Smash) because of its overexpression phenotype.
Figure 1.
Smash binds to Baz and Cno. (A) Domain structures of Baz and the Smash isoforms PM and PI. The region of Baz used as bait and the region of Smash isolated as prey in the yeast two-hybrid screen are indicated. Numbers correspond to amino acid residues in the respective proteins. (B) The PBM of Smash is recognized by the Baz PDZ2 and PDZ3 domains. Left: Overlay of a representative region of the 1H–15N correlation spectra of the Baz PDZ1 domain in the absence (black) and presence of a 2-fold (blue), 6-fold (purple), and 12-fold (red) stoichiometric excess of the Smash PBM peptide. Middle and right: Same as left, except the Baz PDZ2 (middle) and PDZ3 (right) domain. (C) GFP-Smash PI binds to Baz in Drosophila embryos. Lysates of embryos expressing GFP-Smash PI were subjected to IP with anti-Baz (IP Baz) or the preimmune serum of the same animal as control (IP pre). Western blots were probed with the indicated antibodies. Bands corresponding to full-length GFP-Smash PI and Baz are indicated by asterisks. (D) Overlay of a representative region of the 1H–15N correlation spectra of the Cno PDZ domain in the absence (black) and presence of a 2-fold (blue), 6-fold (purple), and 12-fold (red) stoichiometric excess of the Smash PBM peptide.
Smash PM is a 170-kD protein that is largely disordered except for two regions with predicted coiled-coil structure and a C-terminal module consisting of a LIM domain and a PDZ-binding motif (PBM; Fig. 1 A). The LIM-PBM module is conserved in invertebrates and homologous to vertebrate LMO7 proteins (Fig. S1 A). Several isoforms of Smash have been annotated (see http://flybase.org/reports/FBgn0263346.html). We have focused on the longest isoform Smash PM and the shorter isoform Smash PI, sharing the C-terminal LIM-PBM module (Figs. 1 A and S1 A).
The PBM of Smash interacts with the Baz PDZ2 and PDZ3 domains
PDZ domains recognize so-called PBMs, which are short, linear motifs most commonly located at the very C terminus of proteins (Songyang et al., 1997). Smash contains a class I PBM (FSCV) at the C terminus. We investigated by nuclear magnetic resonance (NMR) spectroscopy whether the Smash PBM can interact directly with the PDZ domains of Baz. NMR spectroscopy is an excellent tool to study intermolecular interactions (O’Connell et al., 2009; Wiesner and Sprangers, 2015), as the resonance frequencies (chemical shifts) are highly sensitive to the local chemical environment of the observed atomic nuclei. Therefore, addition of a ligand will result in chemical shift perturbations for the amino acids that constitute a binding pocket. For residues not involved in ligand binding or in case of a ligand that does not bind, the chemical shifts will remain unaltered.
To examine whether the Baz PDZ domains interact with the Smash PBM, we recorded 1H–15N correlation spectra of the individual 15N-labeled PDZ domains in the absence and presence of increasing amounts of unlabeled PBM peptide (Fig. 1 B). Although the spectrum of the Baz PDZ1 domain was not affected by the presence of the PBM even at a 12-fold stoichiometric excess (Fig. 1 B, left), we observed numerous significant changes in the spectra of the PDZ2 and PDZ3 domains upon stepwise addition of the Smash PBM (Fig. 1 B, middle and right). This demonstrates that Smash contains a PBM at its C terminus that can directly interact with Baz via the PDZ2 and PDZ3 domains. This finding was corroborated by coimmunoprecipitation (coIP) experiments using protein extracts from embryos overexpressing a GFP-Smash PI fusion protein (Fig. 1 C).
The Smash PBM binds to the PDZ domain of Canoe
A previous study on vertebrate LMO7 had revealed the binding of the C-terminal region of LMO7 to Afadin (Ooshio et al., 2004). To test whether the Smash PBM binds to the Cno/Afadin PDZ domain, we acquired 1H–15N correlation spectra of the 15N-labeled PDZ domain in the absence and presence of increasing amounts of unlabeled Smash PBM peptide. We found that addition of the Smash PBM to the Cno PDZ domain induced concentration-dependent chemical shift changes for a large number of residues (Fig. 1 D). This demonstrates that the Smash PBM interacts with the Cno PDZ domain.
Smash is a binding partner and phosphorylation substrate of Src42A
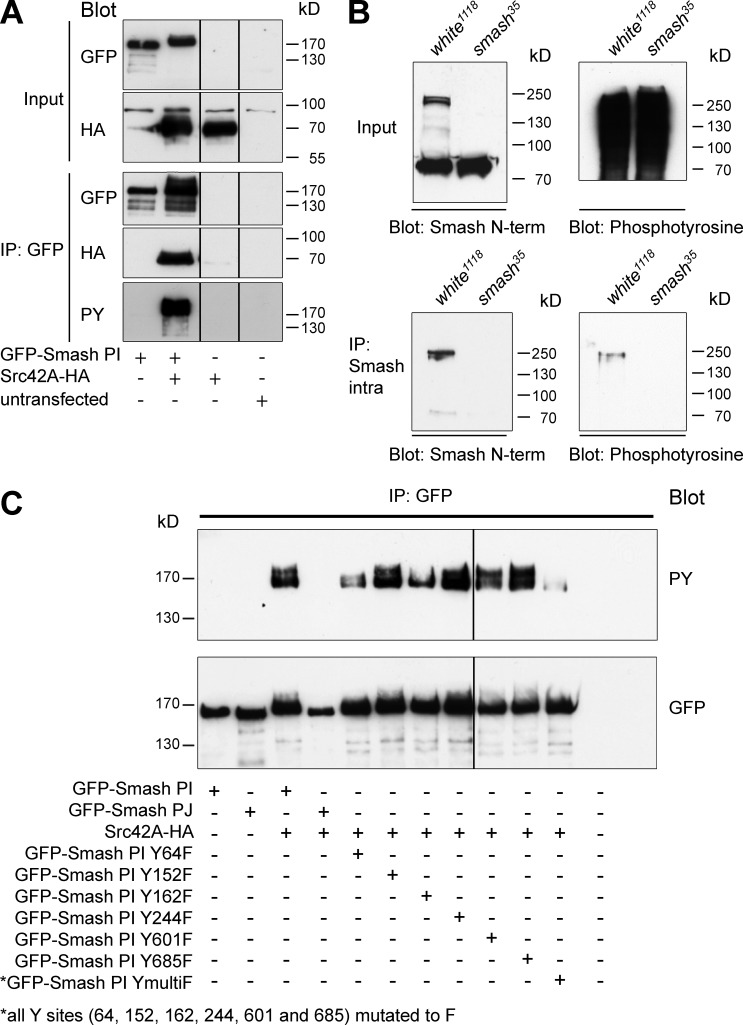
In a Drosophila genome-wide yeast two-hybrid screen, an interaction between Smash and Src42A, a well-known regulator of cell–cell adhesion and morphogenesis (Tateno et al., 2000; Takahashi et al., 2005; Murray et al., 2006; Shindo et al., 2008), was reported (Giot et al., 2003). We further investigated this interaction by coexpressing GFP-Smash PI with HA-tagged Src42A (Src42A-HA) in S2 cells. Full length Src42A-HA was pulled down by coIP with GFP-Smash PI (Fig. 2 A). Western blots of the IPs with anti-phosphotyrosine (anti-PY) showed a band corresponding in size to GFP-Smash PI (Fig. 2 A) that was absent in cells cotransfected with GFP-Smash PI and a mutant of Src42A lacking the kinase domain (Src42AΔΤK-HA; Fig. S1, B and C), indicating that Src42A directly phosphorylates GFP-Smash PI. To test whether endogenous Smash was phosphorylated in embryos, we pulled down Smash with anti-Smash-intra and probed the blot for PY. We detected a band corresponding in size to Smash that was absent in IPs of lysates from smash35 mutant embryos (Fig. 2 B), suggesting that Smash is Y-phosphorylated in vivo. We next mutated Y residues in Smash PI predicted by NetPhosK (http://www.cbs.dtu.dk/services/NetPhos-3.1/) as potential Src phosphorylation sites to F residues. However, none of the six GFP-Smash PI Y-to-F variants lacked Y phosphorylation (Fig. 2 C). Only mutation of all six Y residues of GFP-Smash PI to F (GFP-Smash PI YmultiF; see Fig. 8 A) caused a strong reduction in Y phosphorylation (Fig. 2 C). Interestingly, GFP-Smash PJ, lacking the LIM-PBM module (see Fig. 8 A), was not phosphorylated by Src42A (Fig. 2 C), pointing to a function for the LIM-PBM module in mediating the interaction between Smash and Src42A.
Figure 2.
Smash is a phosphorylation target of Src42A. (A) The indicated constructs were transfected into S2 cells. Lysates were either directly analyzed by Western blot (Input) or subjected to IP with anti-GFP, followed by Western blot with the indicated antibodies. HA, hemagglutinin epitope tag. (B) Endogenous Smash is tyrosine phosphorylated in vivo. Protein lysates of w1118 (control) or smash35 mutant embryos were directly analyzed by Western blot (Input) or subjected to IP with anti-Smash intra, followed by Western blot with the indicated antibodies. Note that in the IP a single band of ∼220 kD, which is absent in smash35 mutant embryos, is detected by anti-PY. This band corresponds in size to the band detected by anti-Smash N-term. The band of 80 kD visible in the input blot with anti-Smash N-term in both WT and smash35 mutant lysates is unspecific. (C) The indicated constructs were transfected into S2 cells. Lysates were subjected to IP with anti-GFP, and Western blots were probed with anti-PY and anti-GFP.
Figure 8.
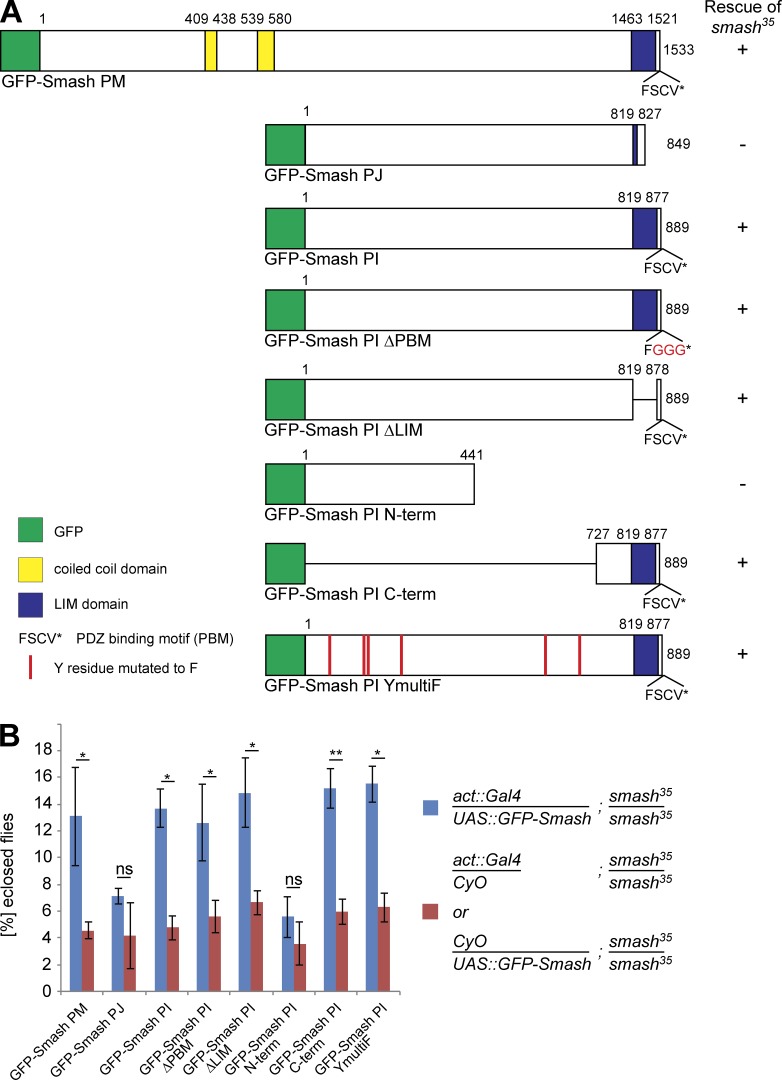
Overexpression of GFP-Smash PM and GFP-Smash PI rescues semilethality of smash35 mutant animals. (A) GFP-Smash constructs used for the rescue assays. + or – in the column to the right indicates whether semilethality of smash35 is rescued. The number of eclosed smash35 homozygous adults expressing the respective GFP-Smash construct under control of the ubiquitous act5C::Gal4 driver line was compared with the number of eclosed smash35 homozygous mutant adults from the same cross carrying only act5C::Gal4 or the respective UAS::GFP-Smash construct as negative control. Rescue was scored (+) when the eclosion rate of smash35 homozygous mutant adults expressing the rescue construct was significantly (P < 0.05) higher than in the negative control. (B) Quantification of the rescue assays. Blue bars, mean percentage of animals with the respective genotype ± SEM; red bars, mean percentage of animals with the respective genotype divided by 2 ± SEM; *, P < 0.05; **, P < 0.01; ns, not significant. GFP-Smash PM: P = 0.041, n = 663; GFP-Smash PJ: P = 0.143, n = 525; GFP-Smash PI: P = 0.014, n = 579; GFP-Smash PI ΔPBM: P = 0.045, n = 626; GFP-Smash PI ΔLIM: P = 0.033, n = 613; GFP-Smash PI N-term: P = 0.071, n = 875; GFP-Smash PI C-term: P = 0.009, n = 1072; GFP-Smash PI YmultiF: P = 0.021, n = 632.
Smash is expressed in ectodermal epithelia and muscles
To study the expression pattern and subcellular localization of Smash, we generated two antibodies against recombinant GST-Smash fusion proteins. Rabbit anti–Smash-intra is directed against aa 972–1278 of Smash-PM, whereas guinea pig anti–Smash-N-term is directed against aa 1–300 of Smash PM. Both antibodies specifically detect Smash, as demonstrated by the absence of signal in immunofluorescence stainings (see Fig. 5 E) and Western blots of protein extracts of smash35 mutant embryos (Fig. 2 B).
Figure 5.
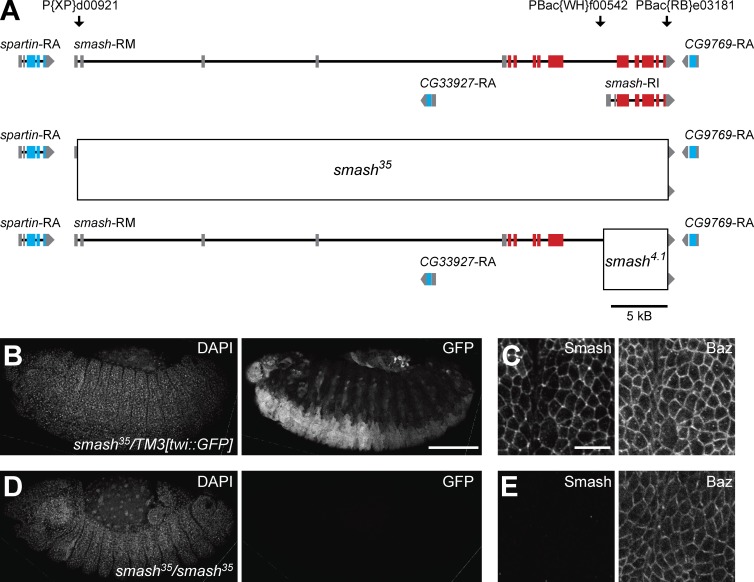
Generation of a null allele for smash. (A) Genomic structure of the smash locus. The exon-intron structures of the two smash transcripts smash-RM, encoding the Smash PM isoform, and smash-RI, encoding the Smash PI isoform, are indicated. Noncoding exons are gray, and coding exons are red. Exons of adjacent transcription units are shown in gray (noncoding) and blue (coding), respectively. The position of transposons used for the generation of defined deletions are indicated above the genomic map. The extent of the deletions in the smash35 (middle) and smash4.1 (bottom) alleles is indicated by boxes. (B and C) An embryo heterozygous for smash35 and the balancer chromosome TM3[twi::GFP] stained for DAPI, GFP (B), Smash, and Baz (C) shows expression of GFP in the mesoderm and Smash localized at the ZA in the epidermis. (D and E) A smash35 homozygous mutant embryo stained as in B and C lacks staining for GFP and Smash. C and E are higher-magnification views of the epidermis of the embryos shown in B and D, respectively. Anterior is to the left and dorsal is up. Bars: (B and D) 100 µm; (C and E) 10 µm.
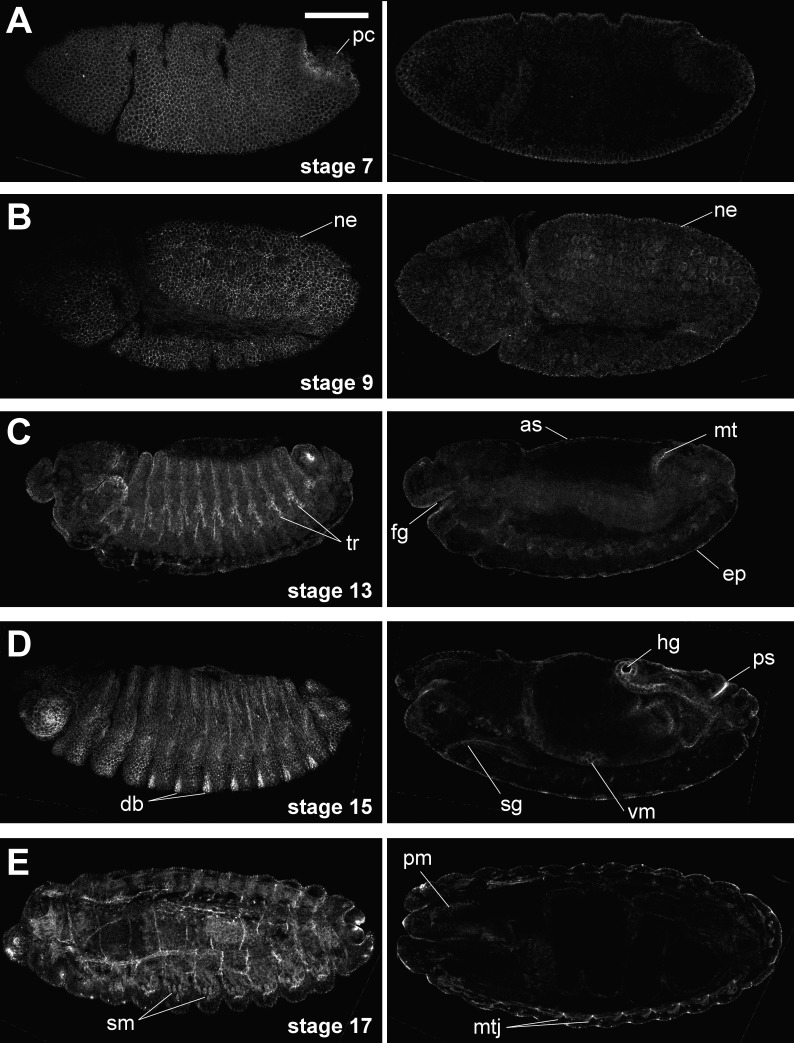
Smash was detectable from embryonic stage 5 onwards in all ectodermally derived epithelia, including the epidermis, fore- and hindgut, Malpighian tubules, salivary glands, amnioserosa, and tracheal tree (Fig. 3). No expression was detectable in neuroblasts and their descendants (Fig. S2 A). We also detected Smash expression in the somatic body wall muscles, the pharynx muscles, and the visceral muscles surrounding the midgut (Fig. 3, D [right] and E).
Figure 3.
Smash is expressed in ectodermal epithelia and muscles. (A–E) WT embryos of the indicated stages were stained with anti–Smash N-term and imaged by confocal microscopy. Left and right images in each row were taken from the same embryo. Images in the left column are superficial optical sections, and images in the right column are cross sections at a deeper focus level. pc, pole cells; ne, neuroectoderm; tr, tracheae; fg, foregut; as, amnioserosa; mt, Malpighian tubule; ep, epidermis; db, denticle belt; sg, salivary gland; vm, visceral musculature; hg, hind gut; ps, posterior spiracle; sm, somatic musculature; pm, pharynx musculature; mtj, myotendinous junction. Bar, 100 µm. Anterior is to the left; dorsal is up.
Smash is planar polarized at the ZA and enriched at tricellular junctions
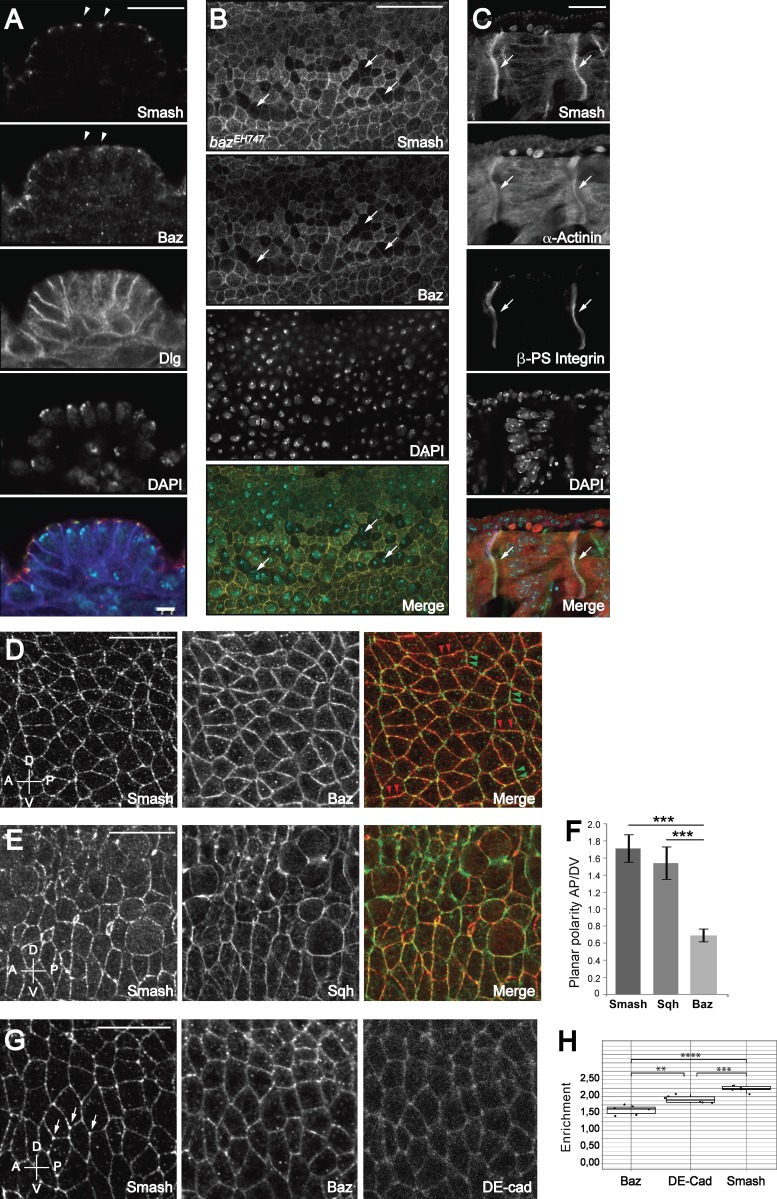
At the subcellular level, Smash was localized at the ZA in epithelia, where it colocalized with Baz but showed no overlap with the basolateral marker Discs-large (Dlg; Fig. 4 A and Fig. S2 A). Smash was also detected at several sites where Baz was absent, for instance at the leading edge of the dorsal epidermis during dorsal closure (Fig. S2, B and B′; Laplante and Nilson, 2011). Consistent with its binding to Baz, ZA localization of Smash was abolished in bazEH747 mutant cells in the embryonic epidermis (Fig. 4 B). ZA localization of Smash was completely abolished upon depolymerization of F-actin by treatment with cytochalasin D (Fig. S2, C–F). In the epidermis during germ band extension, Smash localization was planar polarized, showing a robust enrichment at A/P cell–cell contacts, where it colocalized with Sqh, Cno, and Rok (Fig. 4, D–F; and Fig. S3, C–H). We also noticed that Smash was significantly enriched at tricellular junctions in the embryonic epidermis at germ band extension (Fig. 4, G [left] and H).
Figure 4.
Subcellular localization of Smash in embryos. (A) Smash colocalizes with Baz at the ZA of the epidermis. Epidermis of an embryo at stage 13 stained for Smash (A, green in Merge), Baz (red in Merge), Dlg (blue in Merge), and DAPI (cyan in Merge). Arrowheads point to colocalization of Smash and Baz at the ZA. (B) In zygotic bazEH747 mutant embryos, maternal Baz is lost from individual cells at stage 11 (arrows). In these cells, Smash is also lost at the ZA (Smash and Merge panels). (C) Smash (C, green in Merge) is enriched at myotendinous junctions of an embryo at stage 16 and colocalizes there with α-actinin (red in Merge) and βPS integrin (blue in Merge). (D and E) Smash is planar polarized and enriched at A/P junctions. Embryos at stage 8 were stained for Smash (D, green in Merge) and Baz (D, middle and red in Merge) or for Smash (E, red in Merge) and Sqh-GFP (E, middle and green in Merge). In D (Merge panel), some A/P junctions are marked by green arrowheads and some D/V junctions are marked by red arrowheads. (F) Quantification of PCP of Smash, Sqh, and Baz in stage 8 embryos. ***, P < 0.001. n = 200 cell contacts analyzed for each protein. (G) Smash is enriched at tricellular junctions (G, arrows) to a higher extent than Baz (middle) and E-cadherin (right). (H) Quantification of the enrichment at tricellular junctions. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; n = 6 embryos. At least 10 cells were analyzed per embryo. In D–G, anterior is to the left and dorsal is up. Images in A–C and G are single optical sections taken with an Airyscan detector. Images in D and E are maximum-intensity projections of three optical sections 1 µm apart taken from the apical region of the epidermis with an Airyscan detector. Bars, 20 µm. Error bars in F and H show mean ± SEM.
In embryonic somatic muscles, Smash was strongly enriched at the contact sites between muscle fibers and epidermal tendon cells, the myotendinous junction. Here Smash colocalized with β-PS integrin and α-actinin (Fig. 4 C).
Generation of smash loss-of-function alleles
We generated two deletion alleles by recombination in trans of transposons with Flip recombinase target sites (Parks et al., 2004; Thibault et al., 2004). The allele smash35 was generated by recombination between the transposons P{XP}d00921 and PBac{RB}e03181 and thus removes the complete coding region of smash (Fig. 5 A). The allele smash4.1 was generated by recombination between the transposons PBac{WH}f00542 and PBac{RB}e03181 and deletes a 3′ portion of the coding region including the LIM-PBM module (Fig. 5 A). The deletions were verified by the absence of specific immunofluorescence signals in mutant embryos (Fig. 5 E) and the absence of a specific band corresponding to Smash in Western blots of lysates from mutant embryos (Fig. 2 B).
Embryos lacking maternal and zygotic smash expression show severe defects in epithelial morphogenesis
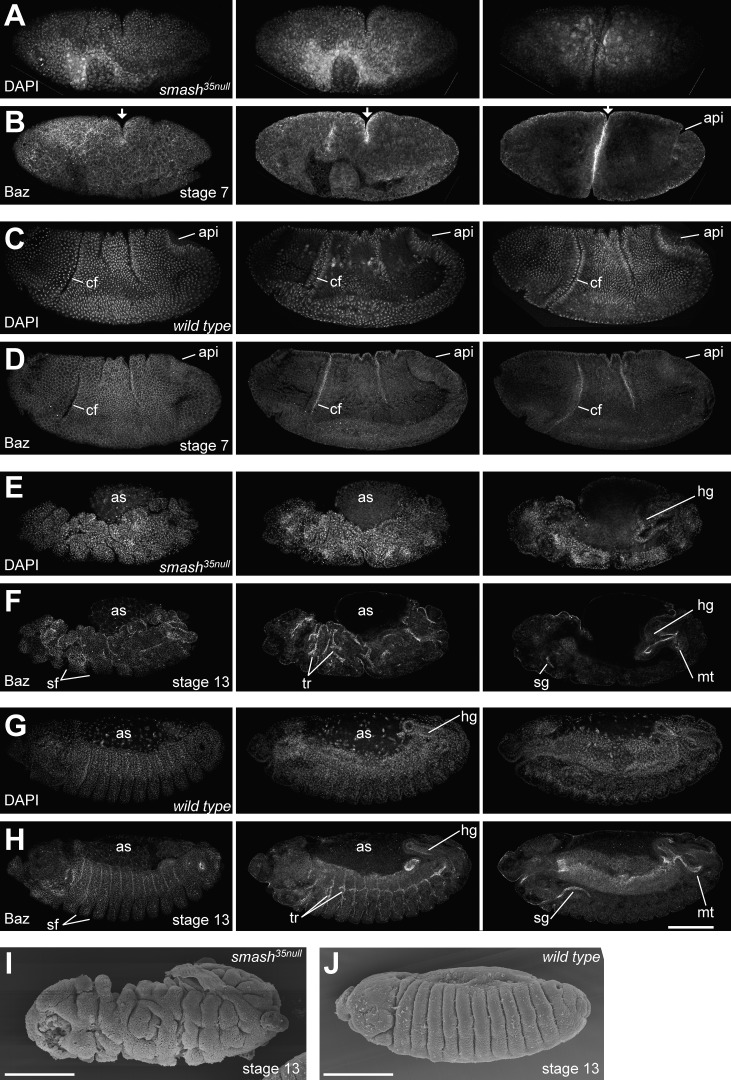
To uncover a functional requirement for smash during embryogenesis, we generated smash35 maternal and zygotic mutant embryos (smash35null embryos). The majority of smash35null embryos (68%, n = 50) showed dramatic defects in morphogenesis. This phenotype was characterized by the uncoordinated formation of furrows and invaginations (Fig. 6 and Videos 1 and 3). At gastrulation, many displayed an irregularly formed ventral furrow that frequently was twisted (Fig. 6, A and B; and Video 1). In addition, the cephalic furrow was often misplaced or missing, and additional furrows of varying depth formed in ectopic positions (Fig. 6, A and B; and Video 1). Concomitantly, germ band extension was delayed and proceeded eventually in an abnormal manner. At later developmental stages, other types of invaginations, furrows, and tubular organs formed in an abnormal, irregular manner, including the segmental furrows, the invaginations of fore- and hindgut, the salivary glands, and the tracheal tree (Fig. 6, E, F, and I; and Video 3). Altogether, the smash35 null embryos showed extremely aberrant morphogenesis compared with WT embryos (Fig. 6, C, D, G, H, and J; and Videos 2 and 4). We also pursued scanning EM on smash35null (Fig. 6 I) and WT embryos (Fig. 6 J), revealing the irregular shape of the epidermis upon loss of smash. Very similar phenotypes were observed in embryos with the maternal genotypes smash35/smash4.1 or smash4.1/smash4.1 (not depicted), confirming that morphogenesis defects were indeed caused by loss of smash function and not by a second site hit on the smash35 chromosome.
Figure 6.
Embryos lacking maternal and zygotic Smash show severe defects in morphogenesis. (A–H) smash35null embryos at stage 7 (A and B) and stage 13 (E and F) stained for DNA (DAPI) and Baz were compared with WT embryos at the corresponding stages (C, D, G, and H). Three different optical sections of the same embryo are shown for each stage. The left column shows the most superficial optical sections, whereas the middle and right columns show deeper optical sections to visualize internal organs. The mutant embryo in A and B lacks the cephalic furrow (cf) and instead has formed a deep ectopic furrow in the middle (arrows). It also fails to form a proper amnioproctodeal invagination (api). The mutant embryo in E and F has a very irregular shape, with deep clefts in its surface. Segmental furrows (sf) are irregular in shape, position, and depth. Morphogenesis of tubular organs such as hindgut (hg), Malpighian tubules (mt), salivary glands (sg), and tracheae (tr) is highly abnormal. The yolk covered by the amnioserosa (as) bulges out of the dorsal side of the embryo. (I and J) Scanning EM of smash35null (I) and WT (J) embryos at stage 13. Note the extremely irregular surface structure of the smash35null embryo in I. Bars, 100 µm. Anterior is to the left and dorsal is up. The full z-stacks of A–H are shown in Videos 1–4.
Smash is required for PCP of Baz, Sqh, and Cno in the embryonic epidermis
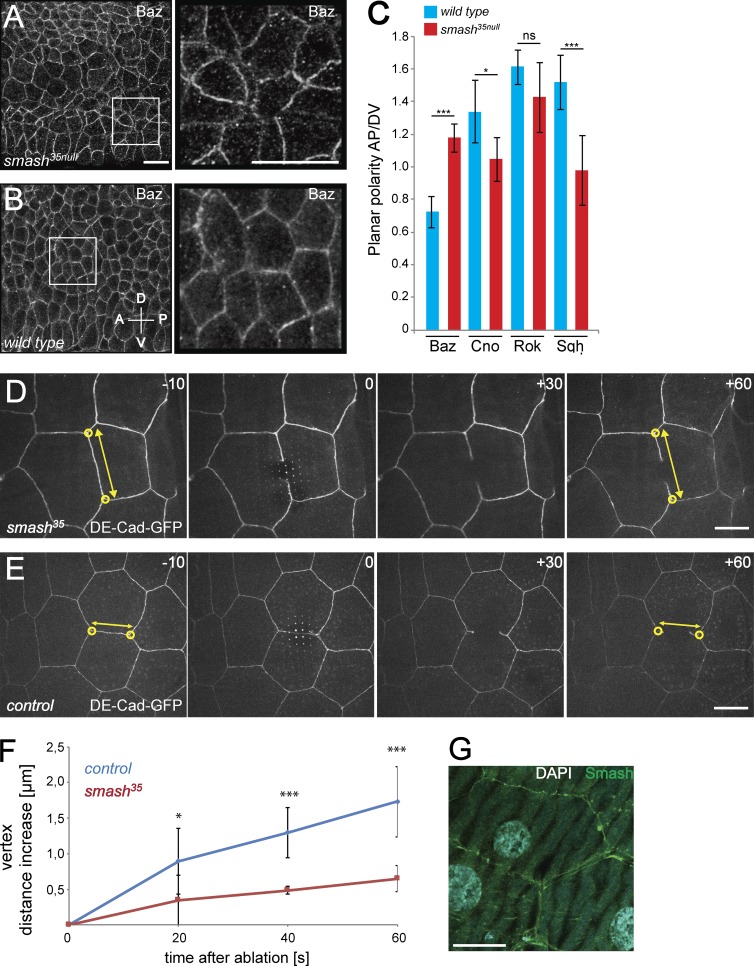
To understand the cellular basis of the smash loss-of-function phenotype in embryogenesis, we analyzed the subcellular localization of junction-associated cortical and transmembrane proteins with polarized localization in smash35null and WT embryos. Although apical-basal polarity of the neuroectodermal epithelium appeared normal in smash35null embryos (Fig. S3, A and B), PCP of Baz was abolished. Instead of being enriched at D/V junctions as in WT (Fig. 4, D [middle and right] and F; and Fig. 7, B and C), Baz showed a slight enrichment at A/P junctions in smash35null embryos (Fig. 7, A and C), indicating that Smash is required for preventing A/P enrichment of Baz. The subcellular localization of three additional proteins with planar polarized localization in the embryonic epidermis was analyzed in WT and smash35null embryos. Sqh-mCherry, Cno, and Rho kinase (Venus-Rok) were all enriched at A/P junctions in WT (Fig. S3, C, E, and G; and Fig. 7 C). In smash35null embryos, Sqh and Cno were evenly localized to A/P and D/V junctions, whereas Rok was unaffected and remained enriched at A/P junctions (Fig. S3, D, F, and H; and Fig. 7 C).
Figure 7.
PCP is altered and cell bond tension is reduced upon loss of smash function. (A and B) Smash35null embryos (A) and WT embryos (B) at stage 8 stained for Baz. Right: Higher-magnification views of the boxed areas in corresponding left panels. (C) Quantification of planar polarization of Baz, Cno, Rok, and Sqh in stage 8 embryos. ***, P < 0,001; *, P < 0.05; ns, not significant. For Baz, n = 160 cell contacts analyzed per genotype; for all other proteins, n = 200 cell contacts analyzed per genotype. (D and E) Live imaging of laser ablation of single cell bonds in the epidermis of smash35null (D) and control (E) third instar larvae. The ZA was marked with DE-cad-GFP. The time (seconds) relative to the time point of laser ablation (0) is given in each panel. The distance (double-headed yellow arrows) between vertices (yellow circles) of the ablated cell bond was measured over time. (F) Quantification of vertex distance increase over time in WT and smash35null larvae. Mean vertex displacement amplitude ± SEM: WT (20 s) 0.896 ± 0.462 µm, smash35 (20 s) 0.346 ± 0.353 µm; *, P = 0.0279; WT (40 s) 1.297 ± 0.352 µm, smash35 (40 s) 0.485 ± 0.055 µm; ***, P = 5.998 × 10−5; WT (60 s) 1.729 ± 0.490 µm, smash35 (60 s) 0.649 ± 0.182 µm; ***, P = 0.00014; mean vertex displacement speed ± SEM in first 60 s: WT 0.029 ± 0.007 µm/s, smash35 0.011 ± 0.003 µm/s; ***, P = 0.00014. P-values were determined using the two-sided unpaired t test. n = 7 representative videos were analyzed for each genotype. (G) Smash (green) localizes to the ZA of third instar larval epidermal cells. DNA was stained with DAPI (cyan). The image is a maximum-intensity projection of three adjacent optical sections at the level of the ZA taken with the Airyscan detector. Bars: (A and B) 10 µm; (D, E, and G) 20 µm. In A and B, anterior is to the left and dorsal is up. Error bars in C and F show mean ± SEM. See also Video 5.
Cell bond tension is reduced in animals lacking smash function
We noticed that cell junctions in the ventrolateral neuroectoderm of smash35null embryos at germ band extension had an irregular serpentine shape (Fig. 7 A) instead of being straight as in WT (Fig. 7 B), pointing to reduced cortical tension of actomyosin at the ZA. To directly investigate whether cell bond tension was altered in smash35null animals, we performed laser cutting experiments in the epidermis of living WT and smash35null larvae. With a pulsed UV laser, we cut the cortical actin belt at the level of the ZA marked by DE-Cad-GFP and followed the displacement of the corresponding cell vertices over time by live imaging (Fig. 7, D and E; and Video 5). Higher vertex displacement speed correlates with higher cell bond tension, as has been demonstrated before (Landsberg et al., 2009). We measured significantly reduced vertex displacement speed and amplitude in smash35null larvae compared with WT controls (Fig. 7 F), pointing to reduced cell bond tension in smash mutants. Consistent with the observed effects, Smash was expressed in the larval epidermis (Fig. 7 G).
Expression of GFP Smash rescues semilethality of smash35 mutant animals
Surprisingly, despite these strong phenotypes in the majority of mutant embryos, a fraction of smash35null embryos completed embryogenesis without major morphogenetic defects and developed to viable, fertile adults. However, the fitness of these escaper animals was strongly compromised. Homozygous mutant adults were weak and short-lived and frequently showed defects in wing eversion (not depicted). Lethality tests revealed that at 25°C, only 41.7% of homozygous smash35 mutant embryos eclosed as adults (Fig. S4 A). This number dropped further to 25% in the F2 generation (Fig. S4 B) and to less than 10% in the F2 generation of animals kept at 29°C (Fig. S4 C), pointing to a maternal effect of smash loss of function and reduced stress resistance of smash35 mutant animals.
To test whether the semilethality of smash35 homozygous mutant animals was indeed caused by loss of smash function and not by a linked second site mutation on the smash35 mutant chromosome, we performed rescue experiments. Semilethality of smash35 was completely rescued by chromosomal insertion of bacterial artificial chromosome CH321-21P3, which carries the complete genomic smash locus. Semilethality was also fully rescued by ubiquitous expression of full-length GFP-Smash PM or GFP-Smash PI, but not GFP-Smash PJ, which lacks the LIM-PBM module (Fig. 8). We pursued a structure–function analysis by generating a series of transgenic fly strains carrying deletion constructs of GFP-Smash PI (Fig. 8 A). A version of GFP-Smash PI lacking the N-terminal half but possessing the LIM-PBM module (GFP-Smash PI Cterm) rescued semilethality of the smash35 mutation, whereas a construct lacking the C-terminal half (GFP-Smash PI Nterm) failed to rescue (Fig. 8). Deletion versions of GFP-Smash PI lacking the LIM domain (GFP-Smash PI ΔLIM) or the PBM (GFP-Smash PI ΔPBM) both rescued, demonstrating that neither of these two domains alone is essential for rescue. Finally, a version of GFP-Smash PI lacking six potential phosphorylation sites for Src42A (GFP-Smash PI YmultiF) rescued the semilethality of smash35 (Fig. 8), demonstrating that phosphorylation of these sites by Src42A is dispensable for the function of Smash in this assay.
Overexpression of Smash causes morphogenetic defects in the embryonic epidermis
To uncover potential gain-of-function phenotypes, we overexpressed GFP-Smash PI and GFP-Smash PM with the UAS-Gal4 system (Brand and Perrimon, 1993) using tubulin::Gal4. Overexpression of GFP-Smash PI resulted in almost complete lethality in larval and pupal stages (Fig. S5 A). Very rare eclosing escaper flies were strongly reduced in size (Fig. S5 B), which was the reason we named the gene smallish.
Overexpression of GFP-Smash PM was lethal without any adult escapers. Almost 50% of embryos overexpressing GFP-Smash PM died before hatching as L1 (Fig. S5 C). The cuticles of ∼35% of embryos that died during embryogenesis exhibited anterior and dorsal holes (Fig. S5, E–I), indicating that GFP-Smash PM overexpression strongly interfered with epidermal development. Hatched larvae died before pupariation (Fig. S5 D).
Overexpression of GFP-Smash PM causes apical constriction of epithelial cells
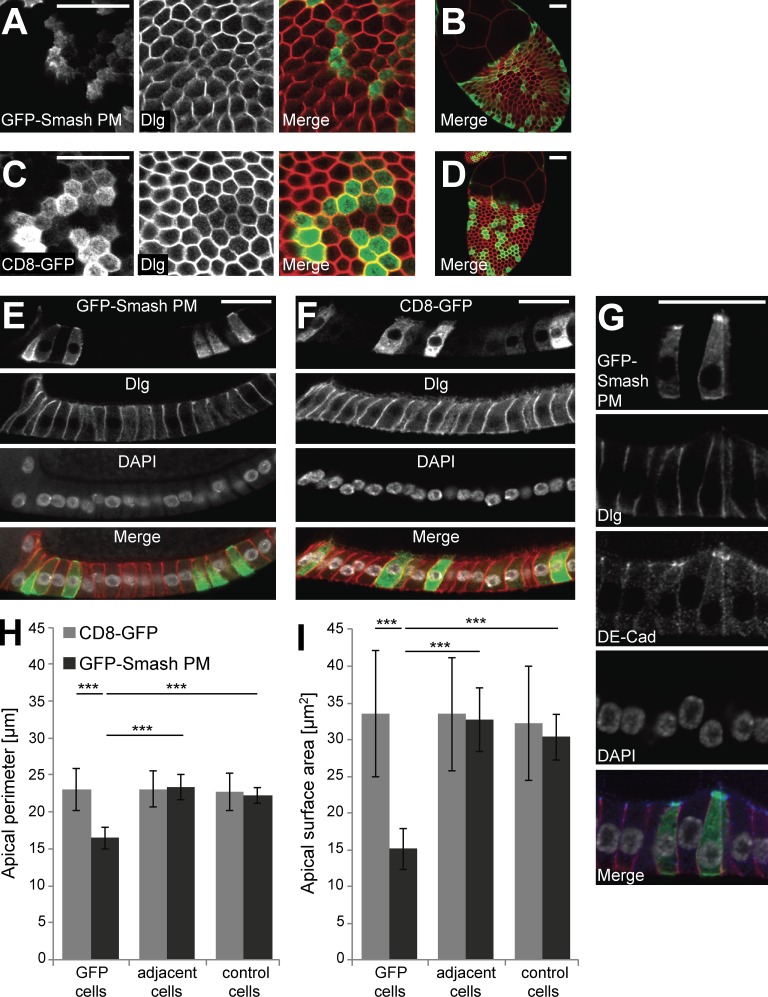
To further investigate the Smash gain-of-function phenotype at the cellular level, we overexpressed GFP-Smash PM in randomly induced clones in the follicular epithelium of adult ovaries. In egg chambers from stage 8 to 10A, overexpression of GFP-Smash PM induced apical constriction of follicular epithelial cells (Fig. 9). Compared with follicular epithelial cells overexpressing CD8-GFP as control (Fig. 9, C, D, F, H, and I), the apical perimeter of GFP-Smash PM–overexpressing cells was reduced by ∼30% and apical surface area was reduced by ∼55% (Fig. 9, A, B, E, and G–I). Cells immediately adjacent to GFP-Smash PM–overexpressing cells did not show any significant difference with respect to apical perimeter and apical surface area from cells further away from the clones, demonstrating that these effects were cell-autonomous (Fig. 9, H and I). We also observed a cell-autonomous increase in the staining intensity of ZA-associated DE-Cad in follicular epithelial cells overexpressing GFP-Smash PM (Fig. 9 G), which might be caused by a tighter clustering of DE-Cad in the ZA of apically constricted cells.
Figure 9.
Overexpression of GFP-Smash-PM causes apical constriction of follicular epithelial cells. (A) GFP-Smash PM (green in Merge) was overexpressed in the follicular epithelium of an egg chamber at stage 10A in clones. Dlg (red in Merge) marks cell outlines close to the apex of the cells. (B) Overview of the egg chamber shown in A. (C) Control clones overexpressing CD8-GFP. (D) Overview of the egg chamber shown in C. (E) Clones of GFP-Smash PM overexpressing cells shown from the side. Note the triangular shape of GFP-Smash PM–overexpressing cells. (F) Control clones overexpressing CD8-GFP shown from the side. Note the cylindrical shape of CD8-GFP–overexpressing cells that does not differ from adjacent cells. (G) Clones of GFP-Smash PM–overexpressing cells were stained for DE-Cad (blue in merged image). Bars, 20 µm. In E–G, apical is up. (H) Quantification of apical perimeter in follicular epithelial cells at stage 10A overexpressing GFP-Smash PM or CD8-GFP in small clones. Adjacent cells immediately touch GFP-positive cells, whereas control cells are more than one cell diameter away from GFP-positive cells. (I) Quantification of apical surface area in follicular epithelial cells at stage 10A overexpressing GFP-Smash PM or CD8-GFP in small clones. Definitions of cell categories as in H. ***, P < 0.001 by unpaired one-tailed t test. Error bars show mean ± SEM. n = 10 egg chambers.
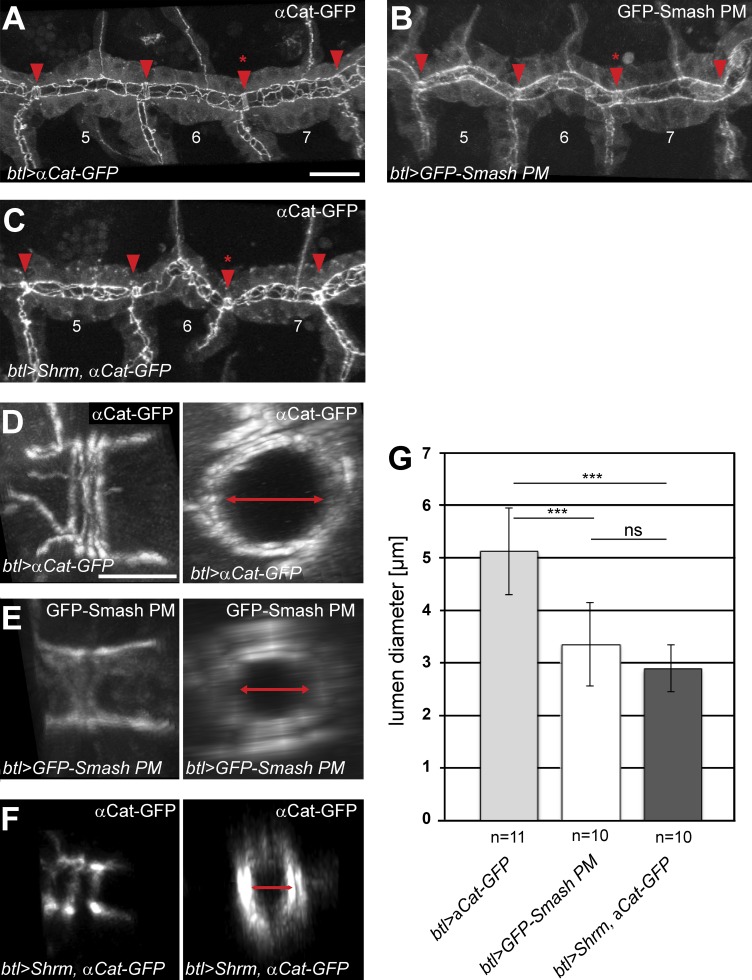
Next, we overexpressed GFP-Smash PM in the tracheal system using breathless::Gal4 to see whether gain-of-function phenotypes also occurred in tubular epithelial organs. Because the lumen of the main tracheal branches is formed by the apical surfaces of the tracheal cells, apical constriction should cause a reduction of the luminal diameter. Indeed, overexpression of GFP-Smash PM in the tracheae caused a statistically highly significant reduction in lumen diameter by ∼33%, measured at tracheal fusion points (Fig. 10, B, E, and G), compared with tracheal epithelial cells overexpressing α-catenin-GFP as control (Fig. 10, A, D, and G). Overexpression of Shrm, which was shown previously to induce apical constriction upon overexpression (Bolinger et al., 2010), caused a comparable reduction of tracheal lumen diameter (Fig. 10, C, F, and G).
Figure 10.
Overexpression of GFP-Smash PM or Shrm leads to reduced lumen diameter of the embryonic dorsal tracheal trunk. (A–F) αCatenin-GFP (A and D), GFP-Smash PM (B and E), or Shrm (C and F) together with αCatenin-GFP were overexpressed in the tracheal system of stage 16 embryos using breathless::Gal4 as driver. A–C are confocal images of GFP fluorescence in three adjacent segments of the dorsal tracheal trunk of living embryos of the indicated genotypes. Fusion points are indicated by red arrowheads. D–F are higher-magnification images of the AJs of ring-like fusion cells viewed from the side; right panels are the same junctions seen in cross section reconstructed from z-stacks. Lumen diameter in right panels is indicated by red double-headed arrows. (G) Quantification of lumen diameter measured at the fusion point between tracheal metameres 6 and 7 (red asterisks in A–C in the three genotypes indicated at the bottom). ***, P < 0,001; ns, not significant; Student’s t test. n = 11 for btl>αCat-GFP; n = 10 for btl>GFP-Smash PM and btl>Shrm, αCat-GFP. Bars: (A–C) 20 µm; (D–F) 5 µm. Error bars show mean ± SEM.
Discussion
Smash associates with multiple proteins at the ZA
We have shown that the LIM domain protein Smash, the Drosophila orthologue of vertebrate LMO7, localizes to the ZA and binds to the ZA-associated proteins Baz, Cno, and Src42A. The binding of Smash to PDZ domains 2 and 3 of Baz and to the single PDZ domain of Cno is mediated by its C-terminal PDZ binding motif. This finding implies that the binding of Smash to Baz and Cno is exclusive, unless Smash would form dimers or multimers, which appears likely because of the presence of two coiled-coil domains in Smash. If that were the case, then Smash would be able to link Baz and Cno in a large multiprotein complex at the ZA. For LMO7, binding to afadin, the homologue of Cno, and α-actinin and thus indirectly to actin was demonstrated (Ooshio et al., 2004; Yamada et al., 2004). Consistent with a direct or indirect interaction between Smash and F-actin, we found that junctional localization of Smash depends on F-actin and is abolished after depolymerization of F-actin after cytochalasin D treatment.
We confirmed the previously reported binding between Smash and Src42A (Giot et al., 2003), a well-known regulator of cell–cell adhesion at the ZA (Roura et al., 1999; Lilien and Balsamo, 2005; Takahashi et al., 2005; Wang et al., 2006). Src42A apparently phosphorylates multiple target sites in the Smash protein. However, a mutant version of GFP-Smash PI with mutation of six tyrosine residues that are potential phosphorylation sites for Src42A rescued the semilethality of smash35 animals, questioning the physiological relevance of Smash phosphorylation by Src42A.
The subcellular localization of Smash may be regulated by multiple binding partners
Although Smash in general colocalizes with Baz, Cno, and Src42A at the ZA of all ectodermal epithelia, the subcellular localization of these proteins is not identical. An example of different subcellular localization between Baz and Smash is the leading edge of the dorsal-most epidermal cells during dorsal closure. Smash localizes to the leading edge, whereas Baz is excluded from this region of the cells (Laplante and Nilson, 2011). A second example is the embryonic epidermis, where Baz is enriched at D/V junctions, whereas Smash, Sqh, Rok, and Cno are enriched at A/P junctions (Zallen and Wieschaus, 2004; Simões et al., 2010; Sawyer et al., 2011). These data indicate that the binding between Baz and Smash is not constitutive, and that additional binding partners including Cno, Src42A, and F-actin are likely to localize Smash at the ZA independent of its interaction with Baz. To test this experimentally is not trivial. Embryos lacking maternal and zygotic baz show very strong defects in epithelial tissue integrity very early in embryonic development, precluding the analysis of Smash localization during germ band extension (Müller and Wieschaus, 1996; Harris and Peifer, 2004). On the other hand, baz mutant embryos lacking only zygotic baz expression develop normally until late germ band extension because of the maternal load of Baz protein (Müller and Wieschaus, 1996; Zallen and Wieschaus, 2004). At later embryonic stages, many epidermal cells lose Baz protein, and in these cells, Smash is strongly reduced and lost from the ZA. However, this could be an indirect effect, as Baz is crucial for the integrity of the ZA in the embryonic ectoderm, and thus all ZA-associated proteins are mislocalized upon loss of Baz expression (Müller and Wieschaus, 1996; Harris and Peifer, 2004). Maternal expression of Src42A allows normal early embryonic development in the absence of zygotic Src42A expression (Takahashi et al., 2005; Sun et al., 2017). Removal of the maternal expression by the generation of germ line clones is technically difficult because the commonly used Flip recombinase–Flip recombinase target method cannot be used by reason of the chromosomal position of Src42A close to the centromere.
How does Smash regulate PCP?
Complementary planar polarization of proteins that directly interact with each other, as we show here for Smash and Baz, is not unprecedented. Like Smash, Rok is enriched at A/P junctions and can phosphorylate Baz, thus destabilizing Baz at A/P junctions (Simões et al., 2010, 2014). PCP of Baz, Cno, and Sqh was disturbed in smash35null embryos, whereas Rok was unaffected, establishing Smash as an essential component of the protein network regulating PCP in the embryonic epidermis. How could Smash function in the regulation of PCP? Because the smash loss-of-function phenotype is very similar to that of Rok with respect to PCP of Baz, Smash may be functioning as a cofactor for Rok, providing substrate specificity toward Baz. Alternatively, Smash could modulate the kinase activity or substrate specificity of Src42A, which has recently been implicated in the formation of basal rosettes during germ band extension and was shown to be planar polarized in the embryonic epidermis at this developmental stage (Sun et al., 2017). A function as a cofactor for a kinase as proposed here for Smash has been demonstrated for Baz, which binds to atypical PKC and provides substrate specificity toward Numb in Drosophila neuroblasts (Wirtz-Peitz et al., 2008).
Smash regulates embryonic morphogenesis and actomyosin contractility at the ZA
The majority of smash35null embryos showed severe morphogenesis defects. Although apical-basal polarity of epithelia and ZA integrity was unaffected, junctions were uneven in shape rather than straight as in WT, pointing to reduced cell bond tension. Laser ablation of cell bonds in the larval epidermis of WT and smash35null animals and the apical constriction gain-of-function phenotype observed upon overexpression of GFP-Smash PM support this hypothesis, raising the question as to the mechanism of Smash function in regulating actomyosin contractility. As speculated before, Smash could function by providing substrate specificity for Rok toward Sqh, thus inducing actomyosin contraction (Julian and Olson, 2014).
Structure–function analysis of Smash
The semilethality of smash35 mutant animals was fully rescued by expression of full-length GFP-Smash PM and PI, but not by GFP-Smash PJ, an isoform lacking the LIM-PBM module. These findings demonstrate the functional importance of the highly conserved LIM-PBM module and prove that semilethality was caused by loss of smash function and not by a second site mutation. The series of rescue experiments furthermore showed that a fragment of GFP-Smash PI containing the LIM-PBM module was sufficient for rescue of semilethality. Our finding that mutant versions of GFP-Smash PI lacking the LIM or PBM domains individually also rescued may be a result of the integration of Smash in a large multiprotein complex where many binding modules function in a partially redundant manner.
Smash shares many similarities with Shrm
The phenotypes described here for Smash show intriguing similarities to those of Shrm, an evolutionarily conserved actin-binding protein that induces apical constriction in Drosophila and vertebrate epithelial cells upon overexpression (Hildebrand and Soriano, 1999; Haigo et al., 2003; Bolinger et al., 2010; Simões et al., 2014). Smash and Shrm display a very similar expression pattern and subcellular localization during Drosophila embryogenesis (Bolinger et al., 2010; Simões et al., 2014). Both proteins localize to the ZA and are planar polarized in the epidermis during germ band extension with higher levels at A/P borders, and their junctional localization is F-actin dependent. Moreover, animals homozygous for a null mutation in Shrm are semilethal and show defects in junctional reorganization, PCP of Sqh, and tension of A/P junctions during germ band extension (Simões et al., 2014). Altogether, the numerous functional similarities of Smash and Shrm indicate that the two proteins may participate in a common molecular pathway.
Conclusions
Altogether, our work introduces Smash as a ZA-associated protein involved in the regulation of PCP and actomyosin-dependent apical constriction of epithelial cells. We propose that Smash is part of a molecular network containing, among others, the polarity regulators Baz and Cno, the regulator of cell–cell adhesion and morphogenesis Src42A, the regulatory subunit of nonmuscle myosin 2, Sqh, filamentous actin, and probably additional proteins that remain to be identified. Additional studies will be required to unravel the molecular mechanism of how Smash induces apical constriction and how it interacts with other known regulators of actomyosin contractility including Shrm, Rok, and Rho. It will be interesting to see whether LMO7 is a true functional homologue of Smash and how this relates to the function of LMO7 as a tumor suppressor for lung cancer in humans (Tanaka-Okamoto et al., 2009; Nakamura et al., 2011).
Materials and methods
Cloning of expression constructs for NMR binding assays
For NMR studies, gene fragments of the individual Baz PDZ domains were amplified by PCR from S2R cell cDNA or a synthetic gene fragment (Life Technologies) and cloned into a pET-M-41 vector (EMBL Heidelberg) containing an N-terminal His6-MBP expression tag followed by a TEV protease cleavage site. Of note, the loop connecting the β2 and β3 strands in the Baz PDZ3 domain contains an unstructured extension that is only present in Drosophila. Because this disordered region severely compromised the quality of the Baz PDZ3 NMR spectra, we removed this region from the PDZ3 construct. Deletion of the β2-β3 loop in the Baz PDZ3 domain was achieved using the QuikChange Site-Directed Mutagenesis protocol (Stratagene). The Cno PDZ domain (A0A0B4KF82, aa 1007–1116) was amplified by PCR from a synthetic gene fragment (Life Technologies) and cloned into a pET-M30 vector (EMBL Heidelberg) containing an N-terminal His6-GST expression tag followed by a TEV protease cleavage site. The Smash PBM peptide (DGIKFSCV) was cloned by QuikChange into a pRTDuet vector containing an N-terminal His6-GB1 (immunoglobulin binding domain B1 of streptococcal protein G) domain expression tag followed by a TEV protease cleavage site as template.
Expression and purification of recombinant proteins
The recombinant 15N-labeled Baz PDZ domains and the Cno PDZ domain were expressed as His6-MBP or His6-GST fusion constructs in Escherichia coli BL21-CodonPlus (DE3) RIL cells (Stratagene) in M9 minimal medium with 15NH4Cl as sole source of nitrogen. To facilitate PBM peptide production, the eight C-terminal residues of the Smash PBM were expressed in LB medium in fusion with a His6-tagged GB1 domain followed by a TEV protease cleavage site. All recombinant proteins for NMR studies were purified by Ni-affinity chromatography and size-exclusion chromatography. For the Baz PDZ domains as well as for the Cno PDZ domain, TEV protease was added after Ni-affinity chromatography to cleave the expression tag. All NMR constructs were buffer exchanged into NMR buffer (20 mM sodium phosphate, pH 6.5, 150 mM NaCl, and 1 mM DTT) for CSP experiments.
NMR spectroscopy
CSP studies were performed with 75–100-µM samples of 15N-labeled PDZ domain. 1H–15N-HSQC experiments were acquired at 20°C on a 600-MHz Bruker Avance-III spectrometer equipped with a room temperature probe head. NMR data were processed using the nmrPipe/nmrDraw software suite (Delaglio et al., 1995) and displayed with nmrView (http://www.onemoonscientific.com).
Cloning of epitope-tagged Smash and Src constructs
The coding sequence of Smash-PI was amplified from GH26442 (Drosophila Genomics Resource Center), whereas the coding sequences of Smash-PM, Smash-PJ, Src42A, and Src64B were amplified from embryonic cDNA by PCR. PCR-amplified coding sequences were cloned into the pENTR vector using the pENTR Directional TOPO Cloning kit (Invitrogen). N-terminally GFP-tagged Smash constructs were generated by recombination of the insert into appropriate Gateway destination vectors with pUASt (pTGW) or pUASp promotors (pPGW and pPGW-attB; Murphy Laboratory, Carnegie Institution of Washington, Baltimore, MD), respectively. C-terminally HA-tagged Src constructs were generated by recombination of the insert into the pPWH Gateway destination vector. Point mutations and deletions were introduced using the QuikChange II site-directed mutagenesis kit (Stratagene) and appropriate primers.
Western blotting
Primary antibodies were used for Western blotting according to standard procedures as follows: mouse anti-HA (1:2,000; 11-583-816-001; Roche), mouse anti–PY PT-66 (1:1,000; P3300; Sigma-Aldrich), rabbit anti-GFP (1:1,000; A11122; Invitrogen), mouse anti-GFP (1:1,000; Roche), rabbit anti-Baz (1:2,000; Wodarz et al., 1999), and guinea pig anti–Smash-N-term (1:500; this work).
Immunoprecipitation
Immunoprecipitation was performed as described previously (Wodarz, 2008). Embryos of an overnight collection of the appropriate genotype were dechorionated and lysed in lysis buffer (1% NP-40, 150 mM NaCl, and 50 mM Tris-HCl, pH 8.0) supplemented with protease inhibitors and phosphatase inhibitors (Roche). A total of 5 mg protein was preincubated with protein A–conjugated agarose. After centrifugation, 2 µl of the specific primary antibody or 2 µl of the corresponding preimmune serum as control were added to the respective lysates. Immunocomplexes were harvested using protein A–conjugated agarose (Roche), washed three times in lysis buffer, and boiled in 2× SDS sample buffer before SDS-PAGE and Western blotting. Lysates from S2 cells were processed accordingly by use of lysis buffer (1% Triton X-100, 150 mM NaCl, and 50 mM Tris-HCl) supplemented with protease inhibitors. GFP-tagged versions of Smash PI were immunoprecipitated with rabbit anti-GFP antibody (A11122; Invitrogen), without preincubation, and were directly subjected to SDS-PAGE and Western blotting.
Fly stocks and genetics
Flies were kept on standard medium at 25°C. white1118 was used as WT. The transposon insertion lines P{XP}d00921, PBac{WH}f00542, and PBac{RB}e03181 were obtained from the Exelixis Collection (Harvard University, Cambridge, MA) for the generation of smash mutant alleles. Fly stocks obtained from the Bloomington Drosophila stock center at the University of Indiana are indicated by the BL stock number in parentheses. Df(3R)ED5066 (BL 8092) is a deficiency completely removing the smash locus. tubulin::Gal4 (BL 5138), daughterless::Gal4 (BL 8641), act5C::Gal4 (BL 4414), and breathless::Gal4 (Förster and Luschnig, 2012) were used as driver lines. Clonal overexpression of GFP-Smash PM or CD8-GFP in the follicular epithelium was achieved by use of the hsFlp;;act5c < CD2 < Gal4 line (gift from S.L. Zipursky, University of California, Los Angeles, Los Angeles, CA; Pignoni and Zipursky, 1997). Clonal expression was initiated by a 10-min heat shock at 37°C in a water bath. The following mutant alleles were used: bazEH747 (Krahn et al., 2010; Shahab et al., 2015), smash35, and smash4.1 (this work). Overexpression of genes was achieved with the UAS-Gal4 system (Brand and Perrimon, 1993) using the following lines: UAS::α-Catenin-GFP (BL 58787); UAS::Shrm (gift from J.D. Hildebrand, University of Pittsburgh, Pittsburgh, PA; Bolinger et al., 2010); UAS::CD8-GFP (BL 32184); UASt::GFP-Smash PM, UASt::GFP-Smash PI, UASp::GFP-Smash PM, UASp::GFP-Smash PJ, UASp::GFP-Smash PI, UASp::GFP-Smash PI ΔPBM, UASp::GFP-Smash PI ΔLIM, UASp::GFP-Smash PI N-term, UASp::GFP-Smash PI C-term, and UASp::GFP-Smash PI YmultiF (this work). The subcellular localization of the regulatory subunit of nonmuscle myosin 2 (Sqh) was analyzed using the stocks y1, w1, cv1, sqhAX3; P{sqh-GFP.RLC} (BL 42234); and sqhAX3; sqh::Sqh-mCherry (Martin et al., 2009). The subcellular localization of Rok was analyzed using sqh::Venus-RokK116A (gift from J.A. Zallen, Memorial Sloan Kettering Cancer Center, New York, NY; Simões et al., 2010). During laser ablation experiments, AJs were imaged using endo::DE-Cad-GFP (gift from T. Lecuit, Institut de Biologie du Développement de Marseille, Marseille, France; Huang et al., 2009). Generation of transgenic flies by ΦC31-mediated targeted insertion at chromosomal position 22A was achieved by injection of attB-containing DNA constructs into embryos of the stock y1 M{vas-int.Dm}ZH-2A w[*]; M{3xP3-RFP.attP′}ZH-22A (BL 24481).
Generation of antibodies against Smash
Polyclonal antisera against Smash were generated by injection of purified GST fusion proteins comprising aa 1–300 (Smash N-term) into guinea pigs or aa 972–1278 (Smash intra) into rabbits (Eurogentec). Numbering refers to the full-length Smash PM isoform (GenBank accession no. ACL83464.2).
Immunohistochemistry and drug treatments
Embryos were fixed for 20 min in a 1:1 mixture of 4% formaldehyde in PBS and heptane. The vitelline membrane was removed by vigorous shaking in a 1:1 mixture of methanol and heptane, and embryos were washed in PBT (PBS containing 0.1% Tween-20) three times for 20 min. After blocking with PBT and 5% normal horse serum, embryos were incubated with primary antibodies in PBT and 5% normal horse serum. Primary antibodies used were rabbit anti-GFP (1:1,000; A11122; Invitrogen), mouse anti-GFP (1:1,000; A11120; Invitrogen), rabbit anti-Baz (1:1,000; Wodarz et al., 1999), rat anti–DE-cadherin DCAD2 (1:5; Developmental Studies Hybridoma Bank [DSHB]), mouse anti–Dlg 4F3 (1:20; DSHB), mouse anti–βPS integrin CF.6G11 (DSHB), rat anti–α-actinin MAC 276 (Babraham Biosciences Technologies), guinea pig anti–Smash N-term (1:500), and rabbit anti-Src42A (1:1,000; gift from K. Saigo, University of Tokyo, Tokyo, Japan; Takahashi et al., 2005). Embryos stained with rabbit anti–Smash intra (1:500) had to be fixed in boiling Triton salt solution instead of formaldehyde before removal of the vitelline membrane. DNA was stained with DAPI (Invitrogen). Secondary antibodies conjugated to Alexa Fluor 488, 567, and 647 were purchased from Invitrogen. F-actin staining was performed by use of Phalloidin conjugated to Alexa Fluor 555. After repeated washing in PBT, embryos were mounted in Mowiol 4-88 (Polysciences Europe). Cytochalasin D treatment of embryos was done as described (Harris and Peifer, 2005). Images were taken at room temperature on a confocal microscope (LSM 510 Meta; LSM 880 Airyscan; Zeiss) using 25× NA 0.8 Plan-Neofluar and 63× NA 1.40 Plan-Apochromat oil immersion objectives and processed using Zen black (Zeiss), Photoshop (Adobe Systems), and Illustrator (Adobe Systems).
Image analysis and quantification of planar polarity
Planar polarity was analyzed by measuring the mean intensity of AP cell edges (60°–90° relative to the AP axis) and the mean intensity of DV cell edges (0°–25° relative to the AP axis) at embryonic stage 8. Using ImageJ software (Schneider et al., 2012), we analyzed the mean pixel intensity and orientation for each junction. In each image, intensities were determined by 20 randomly chosen user-drawn lines for both edges parallel and perpendicular to the AP axis. A representative number of images was quantified for each experiment (n = 4 embryos for WT; n = 6 embryos for mutant). A mean value was obtained for each embryo. P-values were calculated using a two-sided unpaired t test. Error bars indicate SEM in all figures.
Quantification of Smash protein enrichment at tricellular junctions
For quantifying the enrichment of Smash accumulation at tricellular junctions, confocal z-stacks of embryonic epidermis stained for Smash, Baz, and DE-Cad were analyzed. For each confocal stack, a mean projection of the three slices with highest signals was generated. Tricellular junctions and bicellular junction regions, respectively, were manually marked in the Baz channel using circular regions of interest of the same size. Intensities for Smash, Baz, and DE-Cad were measured at tricellular junctions, at bicellular junction regions, and in the cell center (background) in six embryos for at least 10 cells per embryo. Enrichment at tricellular junctions was calculated by dividing the background-subtracted mean intensities at tricellular junctions by the background-subtracted mean intensities at bicellular junctions. Statistical significance was tested using a t test with Bonferroni correction for multiple testing.
Rescue experiments
For rescue experiments, we expressed GFP-tagged Smash constructs inserted at chromosomal position 22A in smash35 homozygous mutant animals using the UAS-Gal4 system. act5C::Gal4 was used as a ubiquitous driver line. Crosses for rescue experiments were set up according to the following scheme:
According to Mendelian laws, 11.11% of the progeny should have the following genotype if all animals except for homozygous balancer animals survive:
| (1) |
The animals with genotype 1 are those that are potentially rescued by expression of the respective GFP-Smash construct. The percentage of smash35 homozygous mutant animals carrying only the act5C::Gal4 driver or the respective UASp::GFP-Smash construct over the CyO balancer would be 22.22% according to Mendel, if all smash35 homozygous flies would survive, which is not the case. The corresponding two genotypes are as follows:
| (2) |
We counted the flies with the respective genotypes and determined the percentage relative to the total number of all surviving animals. We divided the percentage of homozygous smash35 flies not expressing the GFP-Smash construct (2) by 2 to allow comparison to the percentage of potentially rescued homozygous smash35 flies (1). If the percentage of animals with genotype 1 was significantly higher than the percentage of animals with genotype 2 divided by 2, we scored this as rescue of the semilethality of smash35 animals. Normal distribution was controlled using the Shapiro–Wilk test. P-values were calculated using a two-sided paired t test. Error bars indicate SEM.
Lethality assays
To determine lethality at different developmental stages, embryos of a defined genotype were deposited on an apple juice agar plate at a defined temperature (25°C or 29°C). After 24 h, hatched larvae were counted and transferred to a vial with fly food. The vial was incubated at the indicated temperature until all adult animals had eclosed. Pupae and eclosed animals were counted. Each experiment was done with n = 100 embryos in triplicate.
Generation of transgenic fly lines
Transgenic fly lines were generated by P-element–mediated transformation (Bachmann and Knust, 2008) and by ΦC31-mediated targeted insertion into chromosomal position 22A (Bischof et al., 2007). Transgenic flies for bacterial artificial chromosome CH321-21P3 were generated at Genetivision.
Laser ablation of cell bonds in L3 larvae
Immobilization of early L3 larvae were performed as described (Kakanj et al., 2016). The larvae had the genotype endo:DE-Cad-GFP, sqh::Sqh-mCherry (gift from T. Lecuit; Huang et al., 2009; Martin et al., 2009) or endo::DE-Cad-GFP, sqh::Sqh-mCherry; smash35/smash35. Only the green channel was imaged. Laser ablation of cell bonds was performed on an inverted spinning disk confocal microscope (Ultra-View VoX [PerkinElmer] or Inverse TiE [Nikon]) with a 60×/1.2 NA water-immersion objective equipped with 355-nm pulsed ultraviolet laser (DPSL-355/14; Rapp OptoElectronic, 14-mW mean power, 70-µJ per pulse). Laser ablation was induced at the plane of the AJs in the dorsal midline of abdominal segment A3, A4, or A5 with laser power of 0.25 µJ pulsed energy (measured after the objective). Laser ablation was conducted during time-lapse imaging. Larvae were imaged at ∼25°C on a spinning disk confocal microscope (Ultra-View VoX or Inverse TiE) with a single plan 60×/1.2 NA water-immersion objective and an attached CCD camera (C9100-50 CamLink; 1,000 × 1,000 pixels) controlled by Volocity software v.6.3. Images were taken every 0.5 s for 2–3 min, started ∼2 min before ablation, and finished ∼5 min after ablation. Images were processed using Fiji (National Institutes of Health). To analyze the vertex displacements of ablated cell bonds, we averaged the vertex distance increase from different ablation experiments in four time intervals of 20 s. First measurement point was defined as time of ablation. Standard errors were determined.
Cuticle preparations
A drop of yeast was placed on an apple juice agar plate with overnight embryo collection to remove hatching or surviving larvae. Embryos were allowed to develop another 24 h to exhibit their terminal phenotype. Embryos were dechorionated and placed within a drop of Hoyers mountant (100 µl Hoyers mountant mixed with 45 µl lactic acid) on a slide with a coverslip. Embryos were incubated at 65°C overnight. Images were taken with an AxioImager light microscope (Zeiss).
Online supplemental material
Fig. S1 is related to Figs. 1 and 2 and shows the alignment of the LIM-PBM module of different species and data on the interaction between GFP-Smash PI and Src42A-HA. Fig. S2 is related to Figs. 3 and 4 and shows details of the subcellular localization of Smash and the dependence of Smash localization on F-actin. Fig. S3 is related to Figs. 4 and 7 and shows the effect of smash loss-of-function on epithelial apical-basal polarity and PCP of Sqh, Cno, and Rok. Fig. S4 is related to Fig. 5 and shows lethality assays for smash35 and several controls. Fig. S5 is related to Figs. 9 and 10 and shows lethality assays and overexpression phenotypes for GFP-Smash PI and GFP-Smash PM. Videos 1–4 are related to Fig. 6 and show the full z-stacks of the confocal images in this figure. Video 5 is related to Fig. 7 and shows the live imaging of laser ablation experiments in the larval epidermis.
Supplementary Material
Acknowledgments
We thank J.A. Zallen, K. Saigo, T. Lecuit, S.L. Zipursky, and J.D. Hildebrand for fly stocks and reagents. We also thank the Bloomington Drosophila stock center at the University of Indiana and the Exelixis collection at Harvard University for sending fly stocks, the Developmental Studies Hybridoma Bank at the University of Iowa for sending hybridoma antibodies, and the Berkeley Drosophila genome project and the Drosophila Genomics Resource Center at the University of Indiana for sending DNAs. We thank F. Grawe and S. Köhler for help with scanning electron microscopy and M. Hollmann for quantification of Smash localization at tricellular junctions. We also thank E.A. Wimmer, R. Schuh, H.A. Müller, O. Bossinger, and members of our laboratories for comments and discussion.
Work in the laboratory of S. Luschnig was supported by the Swiss National Science Foundation and the “Cells-in-Motion” Cluster of Excellence (EXC 1003-CiM) at the University of Münster. F.A. Renschler was supported by a PhD fellowship of the International Max Planck Research Schools “From Molecules to Organisms.” Work in the laboratory of S. Wiesner was funded by the Max Planck Society. Work in the laboratory of A. Wodarz was funded by grants of the Deutsche Forschungsgemeinschaft (SFB 590, project A2; DFG FOR 1756, WO 989/9-1; Cluster of Excellence 171 “Nanoscale Microscopy and Molecular Physiology of the Brain”; and Cluster of Excellence 229 “Cellular Stress Responses in Aging-Associated Diseases”).
The authors declare no competing financial interests.
Author contributions: H. Beati, I. Peek, P. Hordowska, and M. Honemann-Capito performed the bulk of experimental work dealing with genetics, imaging, biochemistry, and molecular biology. J. Glashauser and S. Luschnig performed the experiments dealing with overexpression of Smash in tracheae. F.A. Renschler and S. Wiesner performed the NMR binding experiments. P. Kakanj performed the laser ablation experiments in the laboratory of M. Leptin. A. Ramrath performed the yeast two-hybrid screen. H. Beati, S. Luschnig, S. Wiesner, and A. Wodarz wrote the manuscript. A. Wodarz coordinated the project.
References
- Bachmann A., and Knust E.. 2008. The use of P-element transposons to generate transgenic flies. Methods Mol. Biol. 420:61–77. 10.1007/978-1-59745-583-1_4 [DOI] [PubMed] [Google Scholar]
- Bilder D., Schober M., and Perrimon N.. 2003. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 5:53–58. 10.1038/ncb897 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., and Basler K.. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 104:3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C., Zasadil L., Rizaldy R., and Hildebrand J.D.. 2010. Specific isoforms of Drosophila shroom define spatial requirements for the induction of apical constriction. Dev. Dyn. 239:2078–2093. 10.1002/dvdy.22326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H., and Perrimon N.. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., and Bax A.. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- Förster D., and Luschnig S.. 2012. Src42A-dependent polarized cell shape changes mediate epithelial tube elongation in Drosophila. Nat. Cell Biol. 14:526–534. 10.1038/ncb2456 [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., et al. 2003. A protein interaction map of Drosophila melanogaster. Science. 302:1727–1736. 10.1126/science.1090289 [DOI] [PubMed] [Google Scholar]
- Gumbiner B.M. 2005. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6:622–634. 10.1038/nrm1699 [DOI] [PubMed] [Google Scholar]
- Haigo S.L., Hildebrand J.D., Harland R.M., and Wallingford J.B.. 2003. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 13:2125–2137. 10.1016/j.cub.2003.11.054 [DOI] [PubMed] [Google Scholar]
- Harris T.J.C. 2017. Sculpting epithelia with planar polarized actomyosin networks: Principles from Drosophila. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2017.07.042. [DOI] [PubMed] [Google Scholar]
- Harris T.J.C., and Peifer M.. 2004. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 167:135–147. 10.1083/jcb.200406024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J.C., and Peifer M.. 2005. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J. Cell Biol. 170:813–823. 10.1083/jcb.200505127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J.D., and Soriano P.. 1999. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 99:485–497. 10.1016/S0092-8674(00)81537-8 [DOI] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A.M., and Hong Y.. 2009. Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA. 106:8284–8289. 10.1073/pnas.0900641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian L., and Olson M.F.. 2014. Rho-associated coiled-coil containing kinases (ROCK): Structure, regulation, and functions. Small GTPases. 5:e29846 10.4161/sgtp.29846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakanj P., Moussian B., Grönke S., Bustos V., Eming S.A., Partridge L., and Leptin M.. 2016. Insulin and TOR signal in parallel through FOXO and S6K to promote epithelial wound healing. Nat. Commun. 7:12972 10.1038/ncomms12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn M.P., Klopfenstein D.R., Fischer N., and Wodarz A.. 2010. Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 20:636–642. 10.1016/j.cub.2010.01.065 [DOI] [PubMed] [Google Scholar]
- Landsberg K.P., Farhadifar R., Ranft J., Umetsu D., Widmann T.J., Bittig T., Said A., Jülicher F., and Dahmann C.. 2009. Increased cell bond tension governs cell sorting at the Drosophila anteroposterior compartment boundary. Curr. Biol. 19:1950–1955. 10.1016/j.cub.2009.10.021 [DOI] [PubMed] [Google Scholar]
- Laplante C., and Nilson L.A.. 2011. Asymmetric distribution of Echinoid defines the epidermal leading edge during Drosophila dorsal closure. J. Cell Biol. 192:335–348. 10.1083/jcb.201009022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband D.E., and de Rooij J.. 2014. Cadherin adhesion and mechanotransduction. Annu. Rev. Cell Dev. Biol. 30:291–315. 10.1146/annurev-cellbio-100913-013212 [DOI] [PubMed] [Google Scholar]
- Lecuit T., and Yap A.S.. 2015. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 17:533–539. 10.1038/ncb3136 [DOI] [PubMed] [Google Scholar]
- Lilien J., and Balsamo J.. 2005. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of beta-catenin. Curr. Opin. Cell Biol. 17:459–465. 10.1016/j.ceb.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Martin A.C., Kaschube M., and Wieschaus E.F.. 2009. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 457:495–499. 10.1038/nature07522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.A., and Wieschaus E.. 1996. Armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J. Cell Biol. 134:149–163. 10.1083/jcb.134.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.J., Davidson C.M., Hayward N.M., and Brand A.H.. 2006. The Fes/Fer non-receptor tyrosine kinase cooperates with Src42A to regulate dorsal closure in Drosophila. Development. 133:3063–3073. 10.1242/dev.02467 [DOI] [PubMed] [Google Scholar]
- Murrell M., Oakes P.W., Lenz M., and Gardel M.L.. 2015. Forcing cells into shape: The mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol. 16:486–498. 10.1038/nrm4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Hori K., Tanaka-Okamoto M., Higashiyama M., Itoh Y., Inoue M., Morinaka S., and Miyoshi J.. 2011. Decreased expression of LMO7 and its clinicopathological significance in human lung adenocarcinoma. Exp. Ther. Med. 2:1053–1057. 10.3892/etm.2011.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M.R., Gamsjaeger R., and Mackay J.P.. 2009. The structural analysis of protein-protein interactions by NMR spectroscopy. Proteomics. 9:5224–5232. 10.1002/pmic.200900303 [DOI] [PubMed] [Google Scholar]
- Ooshio T., Irie K., Morimoto K., Fukuhara A., Imai T., and Takai Y.. 2004. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and alpha-actinin in epithelial cells. J. Biol. Chem. 279:31365–31373. 10.1074/jbc.M401957200 [DOI] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. 10.1038/ng1312 [DOI] [PubMed] [Google Scholar]
- Pignoni F., and Zipursky S.L.. 1997. Induction of Drosophila eye development by decapentaplegic. Development. 124:271–278. [DOI] [PubMed] [Google Scholar]
- Plageman T.F. Jr., Chauhan B.K., Yang C., Jaudon F., Shang X., Zheng Y., Lou M., Debant A., Hildebrand J.D., and Lang R.A.. 2011. A Trio-RhoA-Shroom3 pathway is required for apical constriction and epithelial invagination. Development. 138:5177–5188. 10.1242/dev.067868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roura S., Miravet S., Piedra J., García de Herreros A., and Duñach M.. 1999. Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J. Biol. Chem. 274:36734–36740. 10.1074/jbc.274.51.36734 [DOI] [PubMed] [Google Scholar]
- Sawyer J.K., Harris N.J., Slep K.C., Gaul U., and Peifer M.. 2009. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186:57–73. 10.1083/jcb.200904001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J.K., Choi W., Jung K.-C., He L., Harris N.J., and Peifer M.. 2011. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol. Biol. Cell. 22:2491–2508. 10.1091/mbc.E11-05-0411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahab J., Tiwari M.D., Honemann-Capito M., Krahn M.P., and Wodarz A.. 2015. Bazooka/PAR3 is dispensable for polarity in Drosophila follicular epithelial cells. Biol. Open. 4:528–541. 10.1242/bio.201410934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo M., Wada H., Kaido M., Tateno M., Aigaki T., Tsuda L., and Hayashi S.. 2008. Dual function of Src in the maintenance of adherens junctions during tracheal epithelial morphogenesis. Development. 135:1355–1364. 10.1242/dev.015982 [DOI] [PubMed] [Google Scholar]
- Siedlik M.J., and Nelson C.M.. 2015. Regulation of tissue morphodynamics: An important role for actomyosin contractility. Curr. Opin. Genet. Dev. 32:80–85. 10.1016/j.gde.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S. de M., Blankenship J.T., Weitz O., Farrell D.L., Tamada M., Fernandez-Gonzalez R., and Zallen J.A.. 2010. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell. 19:377–388. 10.1016/j.devcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S.M., Mainieri A., and Zallen J.A.. 2014. Rho GTPase and Shroom direct planar polarized actomyosin contractility during convergent extension. J. Cell Biol. 204:575–589. 10.1083/jcb.201307070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z., Fanning A.S., Fu C., Xu J., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., and Cantley L.C.. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 275:73–77. 10.1126/science.275.5296.73 [DOI] [PubMed] [Google Scholar]
- Sun Z., Amourda C., Shagirov M., Hara Y., Saunders T.E., and Toyama Y.. 2017. Basolateral protrusion and apical contraction cooperatively drive Drosophila germ-band extension. Nat. Cell Biol. 19:375–383. 10.1038/ncb3497 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Takahashi F., Ui-Tei K., Kojima T., and Saigo K.. 2005. Requirements of genetic interactions between Src42A, armadillo and shotgun, a gene encoding E-cadherin, for normal development in Drosophila. Development. 132:2547–2559. 10.1242/dev.01850 [DOI] [PubMed] [Google Scholar]
- Takeichi M. 2014. Dynamic contacts: Rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15:397–410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Tanaka-Okamoto M., Hori K., Ishizaki H., Hosoi A., Itoh Y., Wei M., Wanibuchi H., Mizoguchi A., Nakamura H., and Miyoshi J.. 2009. Increased susceptibility to spontaneous lung cancer in mice lacking LIM-domain only 7. Cancer Sci. 100:608–616. 10.1111/j.1349-7006.2009.01091.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno M., Nishida Y., and Adachi-Yamada T.. 2000. Regulation of JNK by Src during Drosophila development. Science. 287:324–327. 10.1126/science.287.5451.324 [DOI] [PubMed] [Google Scholar]
- Thibault S.T., Singer M.A., Miyazaki W.Y., Milash B., Dompe N.A., Singh C.M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H.L., et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36:283–287. 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- Umetsu D., and Kuranaga E.. 2017. Planar polarized contractile actomyosin networks in dynamic tissue morphogenesis. Curr. Opin. Genet. Dev. 45:90–96. 10.1016/j.gde.2017.03.012 [DOI] [PubMed] [Google Scholar]
- von Stein W., Ramrath A., Grimm A., Müller-Borg M., and Wodarz A.. 2005. Direct association of Bazooka/PAR-3 with the lipid phosphatase PTEN reveals a link between the PAR/aPKC complex and phosphoinositide signaling. Development. 132:1675–1686. 10.1242/dev.01720 [DOI] [PubMed] [Google Scholar]
- Wang Y., Du D., Fang L., Yang G., Zhang C., Zeng R., Ullrich A., Lottspeich F., and Chen Z.. 2006. Tyrosine phosphorylated Par3 regulates epithelial tight junction assembly promoted by EGFR signaling. EMBO J. 25:5058–5070. 10.1038/sj.emboj.7601384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner S., and Sprangers R.. 2015. Methyl groups as NMR probes for biomolecular interactions. Curr. Opin. Struct. Biol. 35:60–67. 10.1016/j.sbi.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., and Knoblich J.A.. 2008. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 135:161–173. 10.1016/j.cell.2008.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A. 2008. Extraction and immunoblotting of proteins from embryos. Methods Mol. Biol. 420:335–345. 10.1007/978-1-59745-583-1_21 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., and Knust E.. 1999. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 402:544–547. 10.1038/990128 [DOI] [PubMed] [Google Scholar]
- Yamada A., Irie K., Fukuhara A., Ooshio T., and Takai Y.. 2004. Requirement of the actin cytoskeleton for the association of nectins with other cell adhesion molecules at adherens and tight junctions in MDCK cells. Genes Cells. 9:843–855. 10.1111/j.1365-2443.2004.00768.x [DOI] [PubMed] [Google Scholar]
- Zallen J.A., and Wieschaus E.. 2004. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell. 6:343–355. 10.1016/S1534-5807(04)00060-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.