Loncarek and Bettencourt-Dias review molecular mechanisms of centriole biogenesis amongst different organisms and cell types.
Abstract
The centriole is a multifunctional structure that organizes centrosomes and cilia and is important for cell signaling, cell cycle progression, polarity, and motility. Defects in centriole number and structure are associated with human diseases including cancer and ciliopathies. Discovery of the centriole dates back to the 19th century. However, recent advances in genetic and biochemical tools, development of high-resolution microscopy, and identification of centriole components have accelerated our understanding of its assembly, function, evolution, and its role in human disease. The centriole is an evolutionarily conserved structure built from highly conserved proteins and is present in all branches of the eukaryotic tree of life. However, centriole number, size, and organization varies among different organisms and even cell types within a single organism, reflecting its cell type–specialized functions. In this review, we provide an overview of our current understanding of centriole biogenesis and how variations around the same theme generate alternatives for centriole formation and function.
Introduction on centriole structure and function
Centrioles are microtubule (MT)-based structures that form centrosomes and cilia and have diverse functions in our cells, such as cell polarity, signaling, cell proliferation, and motility. The centrosome is an important MT-nucleating and signaling center of the cell (Arquint et al., 2014; Conduit et al., 2015). The animal centrosome is composed by two centrioles surrounded by a protein-rich material, the pericentriolar matrix (PCM). In mitosis, two centrosomes organize the poles of the mitotic spindle, which defines its bipolarity, and mediate spindle positioning through the interactions of their astral MTs with the cell cortex (Roubinet and Cabernard, 2014; Ramkumar and Baum, 2016). In interphase, centrosomes and their anchored MTs regulate the positioning of many molecules and structures such as nuclei and the Golgi along with the stability of cellular junctions and adhesions, helping to define cell shape and polarity (Akhmanova et al., 2009; Etienne-Manneville, 2013; Gavilan et al., 2015). When fully mature, centrioles form a structure termed the basal body that is essential to nucleate a cilium (Fig. 1 A). Cilia functions include cell motility, movement of fluids, and specialized sensory functions such as a response to light. In addition, most cilia, whether motile or immotile, are signaling entities. It is likely that the centrosome plays many other undescribed roles as several novel functions were recently described, such as positioning of the cilium (Mazo et al., 2016), promoting polarized secretion at the immune synapse (Stinchcombe et al., 2015), locally regulating actin nucleation (Farina et al., 2016; Obino et al., 2016), and concentrating protein translation and degradation machinery (Hehnly et al., 2012; Amato et al., 2014; Vertii et al., 2016).
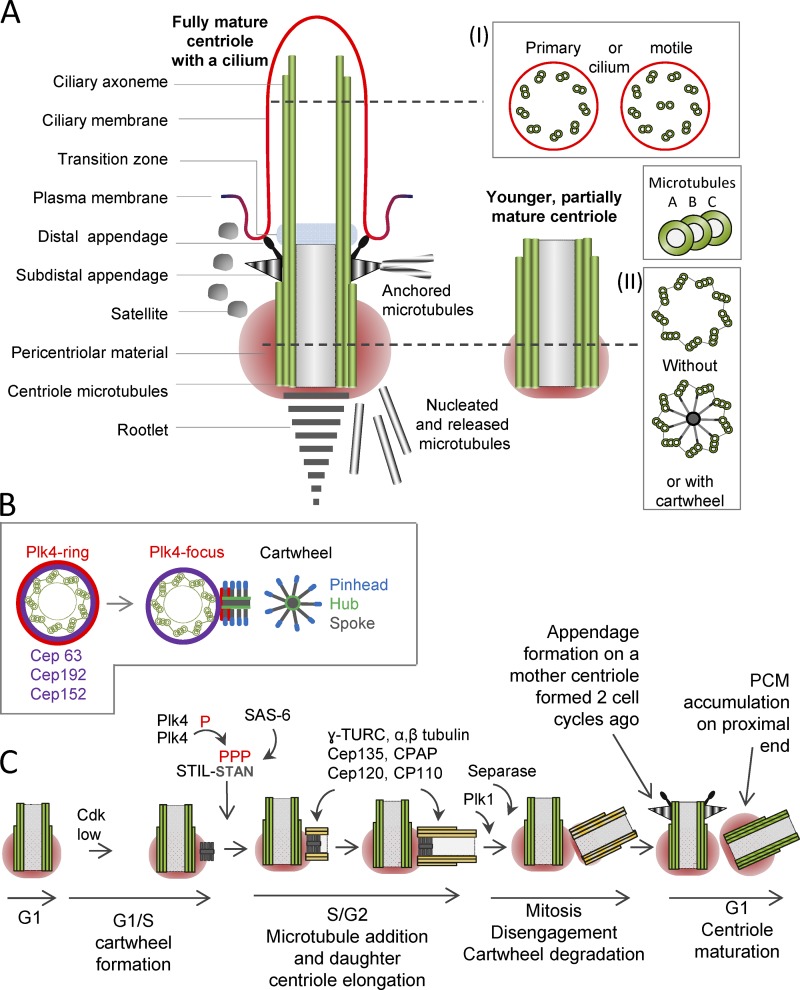
Figure 1.
Centrosome structure and centriole duplication cycle in vertebrates. (A) Image shows longitudinal sections of two unduplicated centrioles during G1. Both centrioles are made of nine MT triplets organized in a ninefold radial symmetry. A fully mature centriole is decorated with DAs and SDAs, a fibrous rootlet. It also nucleates a ciliary axoneme at its distal end and organizes the transition zone on the axoneme’s proximal end. Satellites surround the mature centriole in many vertebrate species, whereas its proximal end is embedded in the PCM. The MTs that are nucleated in the PCM are either being released to other parts of the cell or are anchored on SDAs. Younger centrioles lack appendages and have less abundant PCM. Panel I indicates a cross section of an axoneme in primary and motile cilia. The axoneme in a motile cilium has as a pair of central MTs, although motility can also exist without the MT central pair if the MT doublets have dynein arms. Panel II indicates a cross section through the proximal parts of the centrioles with or without cartwheel. (B) Initiation of centriole formation. The mother centriole is shown in a cross section. During G1, Plk4, an initiator of centriole formation, is organized in a ringlike pattern around the mother centriole along with centrosomal proteins Cep63, Cep152, and Cep192, which aid in Plk4 recruitment. Centriole initiation begins at the G1/S transition by focusing Plk4 to the site of the future daughter centriole and by forming a ninefold-symmetrical cartwheel, a structure composed of a central hub and nine radially organized spokes and pinheads. (C) Canonical centriole duplication cycle. For simplicity, only one longitudinally sectioned centriole is depicted. With the degradation of cyclin B, a conserved cascade of centrosomal proteins initiates daughter centriole formation. Plk4 binds to and phosphorylates STIL on its STAN domain and allows its association with SAS-6. These three proteins form the cartwheel perpendicularly to the proximal wall of the mother centriole. Cdk2 promotes centriole elongation and prevents reduplication. Other proteins assemble into the cartwheel and help the formation of the daughter MT wall. Daughter centriole MTs elongate during S and G2 phases of the cell cycle. Mother and daughter centrioles stay associated until the end of mitosis. Each sister G1 cell inherits one mother and one daughter centriole. During G1, the daughter centriole acquires its own PCM and the ability to duplicate. The mother centriole, formed two cell cycles ago, reaches its final maturation and acquires proximal appendages and DAs. In vertebrates, the cartwheel is lost from maturing daughters in early mitosis. For more details, see main text (section Building a centriole).
Centrioles may drastically vary in size among different organisms and within different cell types of the same organism; however, all centrioles share a ninefold symmetry and a proximal–distal polarity (Fig. 1 A). Centriole diameter is ∼200–220 nm, whereas length ranges from 150 (Drosophila melanogaster embryo) to 500 nm (human somatic cells) to >1,000 nm in the sperm of Drosophila and some other organisms. Most centrioles have a wall of nine triplet MTs: a complete inner A tubule and two partial B and C tubules, although in certain organisms, the centriole wall has MT doublets or singlets (Carvalho-Santos et al., 2011). Centriole symmetry is defined during early stages of its formation by a ninefold symmetrical cartwheel (Fig. 1 A; Gönczy, 2012), a scaffolding structure present in the proximal lumen of young and sometimes older (mature) centrioles.
The proximal end of centrioles organize PCM critical for the centrosome to nucleate MTs (Fig. 1 A; Conduit et al., 2015). PCM size depends on the size of the centriole, the amount of available components, and the activity of regulatory kinases such as Polo-like kinase 1 (PLK1; for a model of PCM accumulation see Woodruff et al., 2014; Zwicker et al., 2014; Conduit et al., 2015). The PCM was originally observed by electron microscopy as an electron-dense amorphous material surrounding centrioles with γ-tubulin ring complexes that nucleate MTs. Recent superresolution microscopy of PCM components revealed that the PCM is not an amorphous material but a well-defined supramolecular complex (Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012; Lukinavičius et al., 2013). During interphase, some PCM components such as pericentrin extend radially from the centriole, with the carboxy terminus pericentrin-AKAP450 centrosomal targeting (PACT) domain adjacent to the centriole wall and the amino terminus extending outward into the PCM. Many other components are organized in toroids around the centriole. In mitosis, the PCM is more abundant but less structured. Mitotic kinases such as PLK1 phosphorylate critical components of the PCM such as SPD5 in Caenorhabditis elegans (Laos et al., 2015; Wueseke et al., 2016; Woodruff et al., 2017) and CNN/CDK5RAP2 in Drosophila, leading to the recruitment of MT polymerizers and stabilizers that concentrate tubulin, thus promoting MT nucleation (Conduit et al., 2010, 2014a,b; Feng et al., 2017; Woodruff et al., 2017).
At their distal end, fully mature centrioles form nine distal appendages (DAs; Fig. 1 A), and in vertebrates, a variable number of more robust sub-DAs (SDAs). DAs are required for the formation of cilia as they interact with vesicles to form a vesicular cap, which fuses with the cytoplasmic membrane of the cell during ciliogenesis (Sorokin, 1962; Tanos et al., 2013; Lu et al., 2015). DAs are equivalent to fibers observed at the distal end of basal bodies in motile cilia (Ishikawa et al., 2005; Tanos et al., 2013; Tateishi et al., 2013). During interphase, SDAs anchor MTs and position the centriole and primary cilium (Piel et al., 2000; Delgehyr et al., 2005; Tateishi et al., 2013; Mazo et al., 2016). SDAs are molecularly equivalent to the basal feet, a structure that associates with the basal bodies of motile cilia and help their positioning (Kunimoto et al., 2012; Tateishi et al., 2013). In cycling cells, SDAs are transiently lost from centrioles before mitosis and are rebuilt in G1, although the physiological significance of such dynamics is not understood. On their proximal end, centrioles often have another fibrous appendage-like striated structure called a rootlet, which varies in size depending on the cell type. Rootlets link centrioles and basal bodies of various animal cilia and green algae flagella to the cell body (Lechtreck and Melkonian, 1991) and contribute to their long-term stability and/or function (Yang et al., 2005; Mohan et al., 2013; Chen et al., 2015).
Additional structures found in the vicinity of vertebrate centrosomes are centriolar satellites (Fig. 1 A). Satellites are poorly understood nonmembrane particles (70–100 nm), which contain components such as OFD1 and PCM1 (Hori and Toda, 2017). Deregulation of satellite components perturbs spindle assembly, centriole duplication, and the initial steps of ciliogenesis, suggesting that satellites regulate centrosome–cilia complexes by their sequestration and/or trafficking (Delgehyr et al., 2005; Kurtulmus et al., 2016).
Recent work has begun to elucidate the molecular underpinnings of different modes of centriole biogenesis in different organisms and cell types. In this review, we focus on our current understanding of molecular processes that drive centriole formation in different cellular contexts, aiming at discussing common frameworks and diverse aspects that underlie each.
Building a centriole
In cycling somatic cells, centrioles assemble in association with preexisting (mother) centrioles in the canonical centriole duplication process. However, centrioles can also form without detectable precursors by the so-called de novo pathway or around specialized structures known as deuterosomes. Those different modes of centriole formation can occur independently in time and space or simultaneously (Bettencourt-Dias and Glover, 2007). These modes use some specific, but mostly conserved, molecules to build centrioles (Meunier and Azimzadeh, 2016). Cells have molecular mechanisms that adjust the number of centrioles to the requirement of a given cell. For instance, somatic cycling cells strictly restrict the number of mature centrioles able to form cilia, whereas multiciliated cells (MCCs) produce and mature hundreds of centrioles (Meunier and Azimzadeh, 2016). In this section, we discuss the common steps in centriole assembly and describe several examples where these steps take a different turn to satisfy specific physiological requirements.
Initiation of a centriole
Initiation of centriole formation is driven by the sequential action of a highly conserved set of proteins. In humans, three proteins, Plk4 (Sak or PLK4 in Drosophila and ZYG-1 in C. elegans), SCL-interrupting locus protein (STIL; Ana-2 in Drosophila and SAS-5 in C. elegans), and SAS-6, comprise a seed for centriole initiation (Fig. 1 C; Arquint and Nigg, 2016). Plk4, a serine/threonine kinase, is the master driver of centriole duplication (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Kleylein-Sohn et al., 2007). Plk4 phosphorylates STIL, its key binding partner, on multiple sites (Moyer et al., 2015; Kraatz et al., 2016), allowing STIL binding with SAS-6 and initiation of centriole formation. SAS-6 is a key structural component of a cartwheel, the structure that institutes a ninefold symmetry of the centriole (Leidel et al., 2005; Nakazawa et al., 2007; Kitagawa et al., 2011; van Breugel et al., 2011; Keller et al., 2014). This was recently demonstrated by the ability of recombinant SAS-6 and its binding partner Bld10 to self-organize into a ninefold symmetrical cartwheel structure in vitro (Guichard et al., 2017).
Initial binding of Plk4 to centrioles in human cells is mediated by Cep63, Cep192, and Cep152, which encircle the proximal ends of mother centrioles (Fig. 1 B; Dzhindzhev et al., 2010; Brown et al., 2013; Kim et al., 2013; Lukinavičius et al., 2013; Sonnen et al., 2013). In flies, recruitment of SAK/Plk4 requires only the Cep152 orthologue Asterless, and in worms, the recruitment of ZYG-1 requires only the Cep192 orthologue SPD-2 (Delattre et al., 2006; Pelletier et al., 2006; Dzhindzhev et al., 2010). After cartwheel formation, centrosomal p4.1-associated protein (CPAP; Sas-4 in Drosophila) aids formation of the centriole MT wall (Pelletier et al., 2006; Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009). In some species such as humans, nine MT singlets are first assembled, followed by addition of the second and third set of MTs (Guichard et al., 2010). It is not known what regulates addition of the second and the third MT set. Additional proteins such as Cep135 (Bld10 in Drosophila) and Cep120 associate with CPAP, facilitate the formation and stabilization of the centriole MT wall, and drive centriole elongation (Fig. 1 C; Comartin et al., 2013; Lin et al., 2013a,b).
Elongation of a centriole
Centriole size varies in different species and in different tissues within a species. Unlike cytosolic MTs, centriole MTs grow at a very slow rate and are extremely stable (Kochanski and Borisy, 1990). In recent years, a variety of work has shown that there are very specialized MT-associated proteins (MAPS) that regulate the slower but steady elongation of centriolar MTs (Sharma et al., 2016; Zheng et al., 2016). Overexpression of several centrosomal proteins such as CPAP or Cep120 leads to the elongation of centriole tubules in some mammalian cells (Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009; Lin et al., 2013b). CPAP associates directly with centriole tubules via its N terminus, mediating centriole growth and stabilization (Sharma et al., 2016; Zheng et al., 2016). Proteins such as hPOC5 (Azimzadeh et al., 2009), Asterless (Galletta et al., 2016), Cep295 (Chang et al., 2016), Centrobin (Gudi et al., 2015), and SPICE1 (Comartin et al., 2013) also positively regulate centriole elongation, although the mechanisms are unknown (Fig. 1 C). In addition, proteins that cap distal centriole ends, such as CP110 and Cep97, restrict centriole elongation in vertebrate but not invertebrate cells (Spektor et al., 2007; Schmidt et al., 2009; Franz et al., 2013). In flies, the MT-depolymerizing kinesin-13 KLP10A/Kin13 has an important role in restricting elongation through MT depolymerization (Delgehyr et al., 2012). In conclusion, how centriole elongation is regulated and how this process varies across tissues and species is still a fundamentally unanswered question.
Maturation of a centriole and acquisition of appendages
New centrioles must be stabilized to become competent for duplication in a process termed “centriole-to-centrosome conversion” (Fig. 1 C). This involves the acquisition of ANA1/CEP295 for the recruitment of critical factors for duplication, such as Asl/CEP152 (Lončarek et al., 2010; Wang et al., 2011; Izquierdo et al., 2014; Kong et al., 2014; Fu et al., 2016). Proteins such as centrobin and Cep120, which are recruited to daughter centrioles in their first cell cycle, are gradually lost during the conversion (Mahjoub et al., 2010; Januschke et al., 2013).
To become competent for ciliogenesis and MT anchoring, centrioles must additionally mature on their distal ends and build DAs and SDAs (Fig. 1 C). The assembly of SDAs is initiated by recruitment of ODF2 (Ishikawa et al., 2005; Huang et al., 2017) and followed by the recruitment of various proteins that are poorly characterized, including CCDC120, CCDC68, and trichoplein. hNinein and CEP170 are part of the SDA, and both bind MTs. Other proteins that localize near SDAs and function in anchoring MTs to mother centrioles, include those comprising the dynein–dynactin complex (containing p50/dynamitin, p150Glued, and p24), EB1, and Kif3a (Huang et al., 2017). The formation of DAs requires the recruitment of OFD1 and C2cd3 (Ye et al., 2014) at the centriole, followed by the recruitment of Cep83, which then recruits other DA components, including Cep89, SCLT1, FBF1, and Cep164 (Tanos et al., 2013; Thauvin-Robinet et al., 2014). All identified DA proteins are essential for appendage and cilia formation. Functional studies indicate that DA proteins are essential for centriole docking to the membrane, recruitment of intraflagellar transport complex, and axoneme extension (Schmidt et al., 2012; Joo et al., 2013; Tanos et al., 2013; Ye et al., 2014; Sánchez and Dynlacht, 2016).
Maintenance of a centriole
Centrioles and basal bodies are very stable structures, with fluorescence recovery after photobleaching of α-tubulin at the basal body in Tetrahymena thermophila showing little turnover (Pearson et al., 2009). Moreover, upon nematode egg fertilization, the sperm centriole that is inherited after fertilization can be traced at least until the 550-cell stage (Balestra et al., 2015). Finally, in vertebrate cells, centriole MTs show also little turnover and are resistant to drug- and cold-induced MT depolymerization (Kochanski and Borisy, 1990), and they resist MT-generated forces and MT instability in mitosis (Belmont et al., 1990).
Several factors have been shown to contribute to centriole stability. The cartwheel is important for the maintenance of immature centrioles (Izquierdo et al., 2014). In certain species, δ- and ε-tubulins found on the centriole contribute to the formation and/or stability of the triplet MTs (Winey and O’Toole, 2014). Ultrastructural analysis revealed linkers between the A and B tubules and between the A and C tubules that may help to stabilize the centriole (Winey and O’Toole, 2014). In addition, centriole MTs are modified by detyrosination, acetylation, and glutamylation and contain Δ2-tubulin, all of which can contribute to their stability directly or through association with other proteins (Janke, 2014). Injections of antibodies against the more characterized PTM, glutamylated tubulin (Bobinnec et al., 1998), and interference with a molecular linker between glutamylated tubulin and the PCM were suggested to lead to centriole disassembly (Madarampalli et al., 2015), indicating that those modifications are important for centriole stability.
However, centrioles can be eliminated from cells at certain development stages. In 1933, Huettner and Rabinowitz (1933) showed that centrosomes are eliminated in Drosophila eggs, and the same was later shown in some terminally differentiated cells such as skeletal muscle (Tassin et al., 1985; Srsen et al., 2009). It is not yet known how they disassemble, but it was recently shown in the female germline in Drosophila that loss of Polo kinase leads to the loss of PCM and centriole elimination, whereas artificial tethering of Polo to centrosomes prevented centriole loss in the oocyte (Pimenta-Marques et al., 2016). This strongly suggests that centrioles require active maintenance through a PCM-mediated mechanism and that cells have strategies for their elimination.
Regulation of centriole number
Canonical centriole duplication
In proliferating vertebrate cells, centriole formation follows the canonical pathway whereby new centrioles assemble in association with mother centrioles (Vorobjev and Chentsov YuS, 1982). There are two unduplicated centrioles during G1 (Fig. 1 A). At the beginning of S phase, the proximal end of each (mother) centriole provides an environment for the initiation of one perpendicularly oriented daughter centriole, also known as procentriole (Fig. 1, B and C). Initially, a cartwheel forms near the wall of the mother centriole, marking the site of procentriole assembly. Next, there is addition of centriole MTs, which continue to elongate during interphase. In mitosis, each duplicated centriole forms one pole of a mitotic spindle and segregates into one of the sister cells. The mother and daughter centrioles separate from each other and form two centrosomes at mitotic exit by assembling an independent PCM around them. By synchronizing the duplication and segregation of their two centrioles with the DNA cycle, somatic cycling cells maintain two centrosomes over generations. However, for this strategy to succeed, the mother centriole must duplicate only once per cell cycle and form only one procentriole.
Control of procentriole number. Control of the number of procentrioles formed in one duplication cycle requires fine-tuned cellular levels of centriole initiating factors such as Plk4, STIL, and SAS-6, because their stoichiometry controls the litter size. Overexpression of Plk4, and to a lesser extent STIL and SAS-6, leads to simultaneous formation of multiple daughter centrioles per mother centriole (Habedanck et al., 2005; Kleylein-Sohn et al., 2007; Guderian et al., 2010). Plk4 levels are regulated at a transcriptional level but, more importantly, via protein turnover. Plk4 degradation depends on its homodimerization and autophosphorylation of the residues critical for the binding of the SCFb-TrCP E3 ubiquitin ligase, which mediates its degradation (Cunha-Ferreira et al., 2009; Rogers et al., 2009; Guderian et al., 2010; Moyer et al., 2015). During G1, Plk4 is localized in a Cep63/Cep152/Cep192-dependent manner in a ringlike pattern around centrioles (Fig. 1 B). Interestingly, although Cep63, Cep152, and Cep192 remain localized in a ring throughout the cell cycle, Plk4 localization is reduced to a discrete site marking the position of the future daughter centriole after procentriole initiation (Sonnen et al., 2012; Kim et al., 2013). Asymmetric dotlike localization of Plk4 persists until the end of the cell cycle when the two centrioles separate (Sonnen et al., 2012; Kim et al., 2013). It is unclear how Plk4 achieves an asymmetric high local concentration, escaping its own destruction, and what mechanisms prevent the accumulation of Plk4 around the mother centriole once the procentriole is initiated. In this respect, Plk4 association with STIL was suggested to protect Plk4 from degradation (Dzhindzhev et al., 2014; Ohta et al., 2014; Arquint et al., 2015; Klebba et al., 2015).
Association between mother and daughter centriole. Centrosomes have an intrinsic mechanism to inhibit overduplication of a mother centriole once it is associated (engaged) with a procentriole (Tsou and Stearns, 2006a). This centrosome-intrinsic block to reduplication was first proposed by Wong and Stearns (2003) after a series of cell fusion experiments. Fusion of the cells containing either duplicated or unduplicated centrioles with cells in different cell cycle stages has demonstrated that when exposed to cytoplasm permissive for centriole duplication, only unduplicated mother centrioles initiate duplication, whereas the mother centrioles already engaged to daughter centrioles do not. Laser ablation of the daughter centrioles in S phase–arrested HeLa cells promoted a new duplication cycle on the mother centrioles, showing that the block of reduplication originates from the daughter (Loncarek et al., 2008).
So, how does engagement prevent reduplication? Fong et al. (2016) disrupted cartwheel maintenance by chemical and genetic manipulation of Plk4 and STIL to show that the loss of STIL from the cartwheel triggers a new reduplication cycle on the mother centriole. Under experimental conditions such as those causing cell cycle arrest that allow mother centriole reduplication during interphase, distancing of daughter centrioles from the wall of mother centrioles leads to reduplication even without the dissolution of the cartwheel (Shukla et al., 2015). So, it is possible that the mother centriole senses the vicinity of the Plk4–STIL complex, which operates in trans and inhibits the initiation of additional centrioles.
How centrioles maintain or lose association is also not clear. One explanation of the association between mother and daughter centriole is the existence of a physical link established between the two centrioles during duplication and which persists until it is severed during centriole disengagement. Candidate structures were seen using cryotomography on isolated centrioles (Guichard et al., 2010) and in Drosophila spermatocytes (Stevens et al., 2010).
In recent years, a series of investigations have aimed to unravel the components responsible for centriole engagement. Separase is the proteolytic enzyme that cleaves cohesin connecting two sister chromatids during mitosis and is indispensable for disengagement of isolated human centrioles in Xenopus laevis extracts in vitro (Tsou and Stearns, 2006b). However, homologous inactivation of both separase genes in human HCT116 cells slowed, but did not prevent, disengagement (Tsou et al., 2009). Similarly, defects in the centriole cycle have not been reported in a separase-knockout mouse (Kumada et al., 2006). In C. elegans embryos, separase depletion affected centriole disengagement and duplication of the sperm-derived centrioles at the meiosis–mitosis transition but not during mitotic centriole duplication cycles (Cabral et al., 2013). Thus, separase may not be universally needed for disengagement. The data describing the role of cohesin in engagement are contradictory as well. Inhibition of cohesin cleavage did not prevent centriole disengagement in HCT116 cells (Tsou et al., 2009), whereas a different study conducted in both U2OS cells and isolated centrioles found that it did (Schöckel et al., 2011). Cohesin cleavage was not sufficient to drive centriole disengagement in Drosophila (Oliveira and Nasmyth, 2013), but the depletion of a short form of Shugoshin, a protector of cohesin integrity, was sufficient for centriole disengagement in mouse fibroblasts (Wang et al., 2008).
Mitotic Plk1 has also emerged as a necessary protein for centriole disengagement in human mitotic (Tsou et al., 2009; Kim et al., 2015) and interphase-arrested cells (Lončarek et al., 2010; Kong et al., 2014). Overactive Plk1 in human cells leads to premature centriole disengagement and reduplication in a CPAP-dependent manner (Shukla et al., 2015). In mitosis, Plk1 may also promote centriole disengagement via Plk1-dependent phosphorylation and subsequent degradation of the PCM protein Pericentrin (Lee and Rhee, 2012; Matsuo et al., 2012; Seo et al., 2015). Centrioles also prematurely disengage after various types of genotoxic stress, a process dependent on Plk1 (Inanç et al., 2010) and/or the ubiquitin proteasome system (Douthwright and Sluder, 2014).
In conclusion, the existing data point to multiple possible mechanisms leading to the loss of centriole association, which may result in uncoupling of the centriole with the cell cycle. These mechanisms may be cell type– or cell cycle–specific. Many studies exploring centriole cycle rely on biochemical methods or on conventional low-resolution immunofluorescence analyses and therefore lack an in-depth ultrastructural centriole analysis needed for unambiguous interpretation of the data. Increased usage of superresolution imaging techniques together with more direct and immediate techniques to perturb gene function in centriole studies will resolve some of these conundrums and help us understand the nuances of centriole cycle regulation among various cells and cell cycle conditions.
Centrosome association and segregation. Preservation of mother–daughter centriole association during interphase is also critical for proper segregation of duplicated centrioles during cell division. After mitosis, separated mother and daughter centrioles each assemble a centrosome. During early G1, the younger centriole moves extensively in the cytoplasm, whereas the older centriole stays near the cell center. As the younger centriole matures and duplicates, its movement becomes attenuated and in most cell types, both centrosomes are adjacent to each other near the physical center of the cell in S and G2 phases (Piel et al., 2000). Some research suggests that the two mother centrioles are tethered by a proteinacious filament composed of several coiled-coil proteins including rootletin, LRRC45, and CEP68 (Flanagan et al., 2017). These proteins dock to the proximal side of mother centrioles via NEK2-associated protein 1 (C-NAP1), CEP135, and centlein. In the G2 phase of the cell cycle, Plk1 activates NEK2A, which phosphorylates the linker filaments, allowing their dissolution. Together with the activity of motor proteins such as kinesin Eg5, this leads to the separation of two centrosomes at the onset of mitosis (Agircan et al., 2014).
Regulation of centriole cycle by cyclin-dependent kinases (Cdks). Cdk1 and Cdk2 regulate the centriole cycle in several embryonic and somatic systems (Hinchcliffe and Sluder, 2002). Cdk2 activity was shown to be necessary for centriole duplication and/or reduplication in a variety of systems, comprising Xenopus embryos (Lacey et al., 1999), Xenopus egg extracts (Hinchcliffe et al., 1999), sea urchin zygotes (Schnackenberg et al., 2008), and CHO cells (Matsumoto et al., 1999; Meraldi et al., 1999) as well as for HPV-16 E7-dependent centriole reduplication in mouse fibroblasts (Duensing et al., 2006). Cdk2 and Cdk4 were proposed to promote centriole reduplication in mouse fibroblasts by hyperphosphorylating the centrosomal protein nucleophosmin, a putative inhibitor of centriole duplication (Okuda et al., 2000; Adon et al., 2010). However, in mammalian somatic cells, Cdk2 is not essential for cell cycle progression (Berthet et al., 2003) or for the centriole cycle (Duensing et al., 2006). In Cdk2-null cells, centrioles still duplicate; thus, if Cdk2 drives centriole duplication, its role can be overtaken by a redundant Cdk/cyclin combination. Contrary to its positive role in centriole formation, Cdk2 was proposed to play an inhibitory role in centriole reduplication via phosphorylation of Cep76 and inhibition of premature Plk1 activation (Barbelanne et al., 2016). Cdk1 has been proposed to inhibit premature initiation of centriole formation in mitosis via its inhibitory binding and phosphorylation of STIL, which prevents both its presence at the centrosome and association with Plk4 (Arquint and Nigg, 2014; Zitouni et al., 2016). In fast-dividing Drosophila embryos, Cdk1 is important for phosphorylation and priming of Sas-4 for Polo recruitment to the young centrioles and for their timely conversion to centrosomes (Novak et al., 2016). A systematic approach will be required to unravel which steps of the centriole cycle are responsive to the activity of various Cdks.
MCCs and deuterosomes
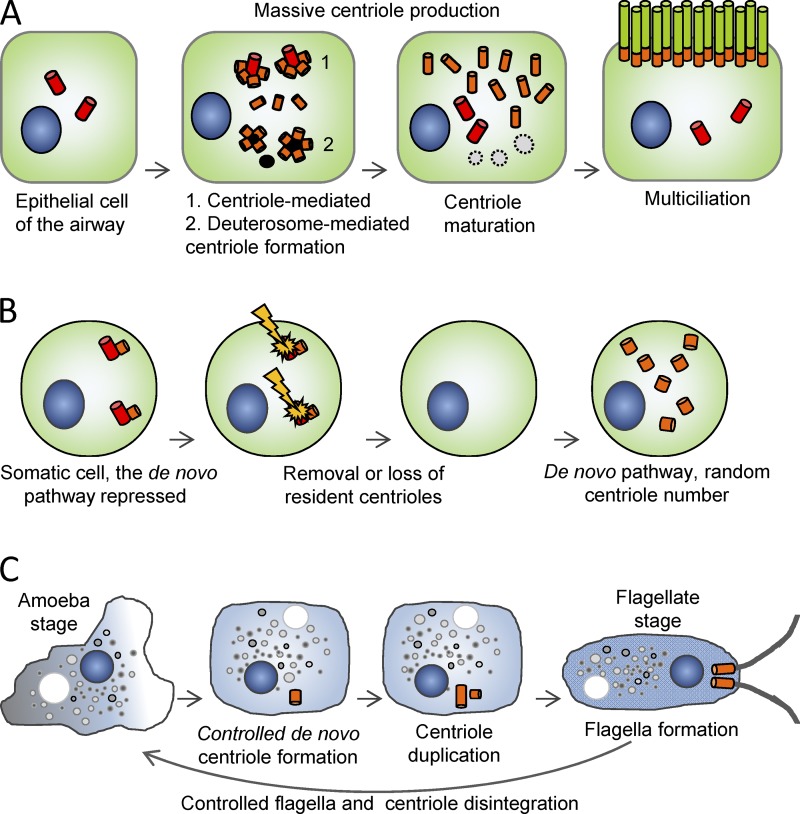
Somatic cycling cells must restrict the number of mature centrioles to two. However, in tissues such as respiratory airways, oviduct, and brain ependymal, some cells undergo differentiation into MCCs. MCCs generate hundreds of centrioles that are converted into basal bodies and produce hundreds of motile cilia (Fig. 2 A). Cilia beat at the cell surface and promote a fluid flow that is vital for numerous tissue-specific processes. Some centrioles in MCCs form in association with the preexisting two centrioles, which duplicate with the increased litter size. However, for massive centriole production, MCCs develop specialized electron-dense structures called deuterosomes, which nucleate centrioles in their vicinity. The descriptions of deuterosome-mediated centriole formation go back to the 1960s (Sorokin, 1968), but the cascade of molecular events that trigger deuterosome formation and support a massive centriole production have only recently emerged (Meunier and Azimzadeh, 2016).
Figure 2.
Noncanonical modes of centriole formation. (A) MCC of the airway. Multiciliation requires a rapid production of hundreds of centrioles. Some form around preexisting centrioles in a rosettelike arrangement (1), but most centrioles are formed around spherical proteinacious structures called deuterosomes (2). Centrioles are then released from deuterosomes or centrioles, mature, associate with the cell surface, and form motile cilia. (B) Somatic cycling cells form new centrioles by canonical duplication (Fig. 1). But if resident centrioles are removed (by a laser beam, microsurgery, or lost as a result of missegregation during cytokinesis), centrioles can form de novo, without any visible precursors. However, the control over centriole numbers is usually lost. (C) Centriole formation in the free-living freshwater protist Naegleria. Naegleria reproduce and divide without centrioles when they are in their amoeba state. Exposed to nutritional and environmental challenges, Naegleria rapidly form the first centriole de novo, followed by their duplication and the formation of two swimming flagella. Centrioles and flagella are disintegrated when the organism reverts to its amoeba stage. See references in main text (section De novo centriole formation).
In many tissues, the process of multiciliation starts with down-regulation of the Notch signaling pathway followed by differentiation of MCCs. This event promotes the expression of two regulatory genes, GEMC1 and multicilin (MCIDAS), triggering the differentiation program of MCCs (Stubbs et al., 2012; Amato et al., 2014; Terré et al., 2016). A complex containing MCIDAS, E2F4/5, and Dp1 then in turn modulates the expression of selected cell cycle regulators and transcriptionally up-regulates hundreds of genes, including those used for canonical centriole duplication (Vladar and Stearns, 2007; Hoh et al., 2012). Massive centriole formation is also somehow promoted by the transcription factor Myb (Tan et al., 2013). In addition, downstream from MCIDAS is cyclin O, expression of which is up-regulated in MCCs and required for proper deuterosome structure, subsequent centriole maturation, and centriole docking to the membrane (Funk et al., 2015). Maturation of young centrioles into basal bodies and ciliogenesis is under the control of the RFX/FOXJ1 signaling network (Choksi et al., 2014; Jackson and Attardi, 2016).
Transcriptional up-regulation of conserved centriole duplication genes explains the formation of multiple daughter centrioles around existing parental centrioles in MCCs. To form deuterosomes, MCCs need a specific deuterosome-forming factor Deup1 (or CCDC67), a paralog of the centrosomal protein Cep63 (Zhao et al., 2013). Like Cep63, Deup1 associates with Cep152, but unlike Cep63, which localizes specifically to parental centrioles, Deup1 associates with deuterosomes. Cep152 recruitment to the deuterosomes requires CCDC78 (Klos Dehring et al., 2013). This is then followed by PLK4 recruitment and the rest of the centriole assembly pathway used by the canonical centriole duplication pathway (Klos Dehring et al., 2013; Zhao et al., 2013). How these centrioles mature in these terminally differentiated cells is not known.
MCCs from mouse ependyma form <100 cilia, a small number compared with other multiciliated epithelia, which form 200–300 cilia. It was suggested that in these cells, deuterosomes form only in association with the young centriole and not freely in the cytoplasm (Al Jord et al., 2014). This finding differs from the early studies of MCC formation in fetal rat lungs (Sorokin, 1968), where deuterosomes were found associated with the fibrous material adjacent to the Golgi. In agreement, deuterosomes in MCCs of trachea were not seeded from preexisting centrioles (Zhao et al., 2013). It is noteworthy that MCCs in airways form a larger number of cilia (200–300). Finally, MCCs of olfactory neurons only moderately up-regulate centriole numbers (Jenkins et al., 2009). It is not clear whether in these cells, centriole amplification uses deuterosomes or only the centrosome-mediated pathway (Cuschieri and Bannister, 1975). It is possible that the presence of deuterosomes, their origin, and their overall number is related to the total number of cilia in MCCs. In support of this possibility, species that generate a smaller number of cilia in their MCCs (for instance, zebrafish) or that do not have MCCs (for instance, invertebrates) do not have Deup1.
Multiciliation also occurs in some primitive plants such as mosses, where numerous centrioles can be produced by a structure called the blepharoplast (Mizukami and Gall, 1966; Vaughn and Renzaglia, 2006). The blepharoplast is a hollow sphere with initially radially arranged short centrioles along its wall. As the sphere enlarges during spermatid differentiation, centrioles elongate until the blepharoplast finally breaks up and releases individual centrioles. The mechanisms leading to the formation of this intriguing structure are slowly emerging (Klink and Wolniak, 2003; Wolniak et al., 2015).
De novo centriole formation
Finally, centrioles can form de novo without any visible precursors. In cycling somatic vertebrate cells, the de novo pathway is silenced by resident centrosomes. It can be activated after resident centrioles are destroyed by a laser beam or removed by microsurgery (Khodjakov et al., 2002; La Terra et al., 2005; Uetake et al., 2007). The de novo pathway in somatic human cells leads to the formation of a random number of centrioles, which are scattered in the cytoplasm (Fig. 2 B). The de novo pathway uses a common Plk4/STIL/SAS-6/CPAP molecular cascade for centriole biogenesis, raising the question of how cells with resident centrioles normally suppress random centriole formation in their cytoplasm (Rodrigues-Martins et al., 2007). A study by Lopes et al. (2015) offered insight into this question. They demonstrated that the activation of Plk4 requires dimerization and trans-phosphorylation of its Thr172 residues, which occurs only at higher Plk4 concentrations. Plk4 is naturally a low abundant protein, and its cytosolic levels are not sufficient for Plk4 activation. Resident centrioles serving as Plk4 concentrators allow Plk4 to achieve the levels necessary for its self-activation, assuring that the new centrioles form only in association with existing centrosomes. In support of this idea, a moderate Plk4 overexpression leads to the formation of additional daughters in association with existing centrioles, whereas higher Plk4 overexpression leads to de novo centriole formation (Kleylein-Sohn et al., 2007; Lopes et al., 2015). Similarly, inhibition of Plk4 degradation leads to the formation of de novo centrioles in the cytoplasm (Wang et al., 2011).
The de novo centriole pathway is naturally widely used by a variety of organisms. During parthenogenesis (development without fertilization) in some insects, centrioles are lost during oogenesis but are formed after oocyte activation (Miki-Noumura, 1977; Szöllosi and Ozil, 1991; Riparbelli and Callaini, 2003; Manandhar et al., 2005). Centrioles form de novo after parthenogenetic activation of sea urchin eggs (Kato and Sugiyama, 1971; Kallenbach and Mazia, 1982; Kallenbach, 1983) or in early embryogenesis in species where both paternal and maternal gametes lose centrioles during gametogenesis (Szollosi et al., 1972; Schatten, 1994; Courtois and Hiiragi, 2012a). A mouse zygote is a classic illustration as it does not initially contain centrioles and successfully divides without them. Then, during the blastomere stage, centrioles form and continue to duplicate by the canonical pathway thereafter (Gueth-Hallonet et al., 1993; Courtois et al., 2012b). Furthermore, centrioles form de novo in centriole-less cells of a Chlamydomonas reinhardtii mutant defective in centriole segregation (Marshall et al., 2001). De novo formation of centrioles has been originally documented in the amoeboflagellate Naegleria gruberi (Fulton and Dingle, 1971). Naegleria is a free-living freshwater protist, which reproduces and divides without centrioles when it is in its amoeba state. However, if exposed to various nutritional and environmental changes Naegleria undergoes a rapid (within 1 h) metamorphosis into a swimming flagellate (Fig. 2 C), forming two centrioles and two flagella. In this system, the first centriole forms de novo immediately followed by its duplication (Fritz-Laylin et al., 2016).
Although these examples of naturally occurring de novo centriole formation illustrate that de novo centriole formation can be controlled to yield centrioles in the proper number, the molecular mechanisms have remained enigmatic.
Centriole and centrosome requirement
The requirement for centrosomes, centrioles, and cilia is species- and tissue type–specific. Higher plants, yeasts, and amoebas do not have centrioles, canonical centrosomes, or cilia. Lower plants like mosses, ferns, cycads, and ginkgo make centrioles only in the cells which yield flagellate sperms as referred to previously (Renzaglia et al., 2000). All animals studied to date build centrioles, centrosomes, and cilia except planarians (flatworms), which have no centrosomes but have centrioles and motile cilia in MCCs.
Recent studies have demonstrated that the loss of centrioles or centrosomes is variably tolerated. For instance, somatic cells of fruit flies appear more tolerant to centrosome loss than somatic vertebrate cells. Without centrosomes, divisions in the fly occur slower and with defects, but compensatory cell proliferation can still make up for the lost cells (Basto et al., 2006; Rodrigues-Martins et al., 2008; Poulton et al., 2014). Nevertheless, flies without centrioles are uncoordinated and sterile because of the absence of sensory cilia and sperm and ultimately die. Vertebrates seem to have pathways sensitive to errors caused by the lack of centrosomes. In mouse and human somatic cells, centrosome loss triggers 53BP1 and USP28-mediated pathways, which activate p53 and block cell proliferation (Fong et al., 2016; Lambrus et al., 2016; Meitinger et al., 2016). Which types of errors are detected by 53BP1 and USP28 pathways in acentrosomal cells are not known (Lambrus and Holland, 2017). Divisions in zebrafish and C. elegans embryos as well as fly embryos and spermatocytes, however, seem sensitive to the absence of centriole-mediated bipolar spindle assembly (O’Connell et al., 2001; Yabe et al., 2007; Rodrigues-Martins et al., 2008).
In vertebrates, centrosome number and structure defects can lead to disease. Numerical and structural centriole aberrations can cause chromosome attachment and segregation errors (Ganem et al., 2009; Silkworth et al., 2009) and alter the behavior of interphase cells by increasing their invading and migrating capacity (Godinho and Pellman, 2014; Godinho et al., 2014). Centrosome numerical and structural defects can also perturb the formation and the function of cilia. Already in the 19th century, the German biologist Theodor Boveri suggested a link between centrosome amplifications and cancer. This remained a hypothesis until recently, when it became possible to manipulate the centrosome number in flies and mice to demonstrate that amplified centrosomes can both trigger and accelerate tumor development (Basto et al., 2008; Coelho et al., 2015; Kulukian et al., 2015; Vitre et al., 2015; Serçin et al., 2016; Levine et al., 2017). Centrosome deregulation caused by mutations in centrosome protein coding genes can also cause microcephaly, a neurological condition resulting in smaller-than-normal brain size (Bettencourt-Dias et al., 2011; Chavali et al., 2014). It has been proposed that microcephaly results from a decreased pool of neural precursors as a result of abnormal asymmetric divisions in the neural stem cells and/or abnormal mitotic divisions that lead to aneuploidy, death, and p53-mediated cell cycle arrest. Microcephaly can also be induced by Zika virus. Recent work shows that the Zika virus leads to centrosome perturbations, abnormal spindle positioning, and premature differentiation of neural progenitors (Gabriel et al., 2017; Wolf et al., 2017). Infection with other viruses such as oncogene human papilloma viruses increases centrosome number (Korzeniewski et al., 2011). The causality in those relationships is yet to be determined.
Conclusion
It is gradually becoming clear that different modes of centriole formation observed across different organisms and different cell types of the same organism are controlled variations of the same centriole assembly blueprint. However, to fully understand centriole functions and how centrioles contribute to human diseases, it will be critical to understand the nuances of the molecular pathways that operate in physiological cellular contexts. Until then, because of the diversity in their number, structure, and function, centrioles will rightfully remain a central enigma in cell biology.
Acknowledgments
We thank members of the J. Loncarek and M. Bettencourt-Dias laboratory for suggestions to the review.
Work in the laboratory of M. Bettencourt-Dias is funded by the Gulbenkian Foundation, a European Research Council Consolidator Grant (CoG683528), and a Fundaçao para a Ciencia e Tecnologia grant (PTDC/BIM-ONC/6858/2014). Research in J. Loncarek’s laboratory was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and the Center for Cancer Research.
The authors declare no competing financial interests.
References
- Adon A.M., Zeng X., Harrison M.K., Sannem S., Kiyokawa H., Kaldis P., and Saavedra H.I.. 2010. Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells. Mol. Cell. Biol. 30:694–710. 10.1128/MCB.00253-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agircan F.G., Schiebel E., and Mardin B.R.. 2014. Separate to operate: control of centrosome positioning and separation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130459 10.1098/rstb.2013.0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A., Stehbens S.J., and Yap A.S.. 2009. Touch, grasp, deliver and control: functional cross-talk between microtubules and cell adhesions. Traffic. 10:268–274. 10.1111/j.1600-0854.2008.00869.x [DOI] [PubMed] [Google Scholar]
- Al Jord A., Lemaître A.I., Delgehyr N., Faucourt M., Spassky N., and Meunier A.. 2014. Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature. 516:104–107. [DOI] [PubMed] [Google Scholar]
- Amato R., Morleo M., Giaquinto L., di Bernardo D., and Franco B.. 2014. A network-based approach to dissect the cilia/centrosome complex interactome. BMC Genomics. 15:658 10.1186/1471-2164-15-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C., and Nigg E.A.. 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 24:351–360. 10.1016/j.cub.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Arquint C., and Nigg E.A.. 2016. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem. Soc. Trans. 44:1253–1263. 10.1042/BST20160116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C., Gabryjonczyk A.M., and Nigg E.A.. 2014. Centrosomes as signalling centres. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130464 10.1098/rstb.2013.0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C., Gabryjonczyk A.M., Imseng S., Böhm R., Sauer E., Hiller S., Nigg E.A., and Maier T.. 2015. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. eLife. 4:e07888 10.7554/eLife.07888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Hergert P., Delouvée A., Euteneuer U., Formstecher E., Khodjakov A., and Bornens M.. 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185:101–114. 10.1083/jcb.200808082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra F.R., von Tobel L., and Gönczy P.. 2015. Paternally contributed centrioles exhibit exceptional persistence in C. elegans embryos. Cell Res. 25:642–644. 10.1038/cr.2015.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbelanne M., Chiu A., Qian J., and Tsang W.Y.. 2016. Opposing post-translational modifications regulate Cep76 function to suppress centriole amplification. Oncogene. 35:5377–5387. 10.1038/onc.2016.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., and Raff J.W.. 2006. Flies without centrioles. Cell. 125:1375–1386. 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., and Raff J.W.. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042. 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L.D., Hyman A.A., Sawin K.E., and Mitchison T.J.. 1990. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 62:579–589. 10.1016/0092-8674(90)90022-7 [DOI] [PubMed] [Google Scholar]
- Berthet C., Aleem E., Coppola V., Tessarollo L., and Kaldis P.. 2003. Cdk2 knockout mice are viable. Curr. Biol. 13:1775–1785. 10.1016/j.cub.2003.09.024 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., and Glover D.M.. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463. 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., and Glover D.M.. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Hildebrandt F., Pellman D., Woods G., and Godinho S.A.. 2011. Centrosomes and cilia in human disease. Trends Genet. 27:307–315. 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov A., Mir L.M., Rieder C.L., Eddé B., and Bornens M.. 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143:1575–1589. 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.J., Marjanović M., Lüders J., Stracker T.H., and Costanzo V.. 2013. Cep63 and cep152 cooperate to ensure centriole duplication. PLoS One. 8:e69986 10.1371/journal.pone.0069986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral G., Sans S.S., Cowan C.R., and Dammermann A.. 2013. Multiple mechanisms contribute to centriole separation in C. elegans. Curr. Biol. 23:1380–1387. 10.1016/j.cub.2013.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J.B., and Bettencourt-Dias M.. 2011. Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194:165–175. 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-W., Hsu W.-B., Tsai J.-J., Tang C.-J.C., and Tang T.K.. 2016. CEP295 interacts with microtubules and is required for centriole elongation. J. Cell Sci. 129:2501–2513. 10.1242/jcs.186338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali P.L., Pütz M., and Gergely F.. 2014. Small organelle, big responsibility: the role of centrosomes in development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130468 10.1098/rstb.2013.0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.V., Kao L.R., Jana S.C., Sivan-Loukianova E., Mendonça S., Cabrera O.A., Singh P., Cabernard C., Eberl D.F., Bettencourt-Dias M., and Megraw T.L.. 2015. Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila. J. Cell Biol. 211:435–453. 10.1083/jcb.201502032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi S.P., Lauter G., Swoboda P., and Roy S.. 2014. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 141:1427–1441. 10.1242/dev.074666 [DOI] [PubMed] [Google Scholar]
- Coelho P.A., Bury L., Shahbazi M.N., Liakath-Ali K., Tate P.H., Wormald S., Hindley C.J., Huch M., Archer J., Skarnes W.C., et al. 2015. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 5:150209 10.1098/rsob.150209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comartin D., Gupta G.D., Fussner E., Coyaud É., Hasegan M., Archinti M., Cheung S.W.T., Pinchev D., Lawo S., Raught B., et al. 2013. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 23:1360–1366. 10.1016/j.cub.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Conduit P.T., Brunk K., Dobbelaere J., Dix C.I., Lucas E.P., and Raff J.W.. 2010. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 20:2178–2186. 10.1016/j.cub.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Conduit P.T., Feng Z., Richens J.H., Baumbach J., Wainman A., Bakshi S.D., Dobbelaere J., Johnson S., Lea S.M., and Raff J.W.. 2014a The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell. 28:659–669. 10.1016/j.devcel.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P.T., Richens J.H., Wainman A., Holder J., Vicente C.C., Pratt M.B., Dix C.I., Novak Z.A., Dobbie I.M., Schermelleh L., and Raff J.W.. 2014b A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife. 3:e03399 10.7554/eLife.03399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P.T., Wainman A., and Raff J.W.. 2015. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16:611–624. 10.1038/nrm4062 [DOI] [PubMed] [Google Scholar]
- Courtois A., and Hiiragi T.. 2012a Gradual meiosis-to-mitosis transition in the early mouse embryo. Mouse Development: From Oocyte to Stem Cells. 55:107–114. [DOI] [PubMed] [Google Scholar]
- Courtois A., Schuh M., Ellenberg J., and Hiiragi T.. 2012b The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol. 198:357–370. 10.1083/jcb.201202135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19:43–49. 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- Cuschieri A., and Bannister L.H.. 1975. The development of the olfactory mucosa in the mouse: electron microscopy. J. Anat. 119:471–498. [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Canard C., and Gönczy P.. 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16:1844–1849. 10.1016/j.cub.2006.07.059 [DOI] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J., and Bornens M.. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118:1565–1575. 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- Delgehyr N., Rangone H., Fu J., Mao G., Tom B., Riparbelli M.G., Callaini G., and Glover D.M.. 2012. Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Curr. Biol. 22:502–509. 10.1016/j.cub.2012.01.046 [DOI] [PubMed] [Google Scholar]
- Douthwright S., and Sluder G.. 2014. Link between DNA damage and centriole disengagement/reduplication in untransformed human cells. J. Cell. Physiol. 229:1427–1436. 10.1002/jcp.24579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing A., Liu Y., Tseng M., Malumbres M., Barbacid M., and Duensing S.. 2006. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene. 25:2943–2949. 10.1038/sj.onc.1209310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718. 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J., Debski J., Dadlez M., and Glover D.M.. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24:2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2013. Microtubules in cell migration. Annu. Rev. Cell Dev. Biol. 29:471–499. 10.1146/annurev-cellbio-101011-155711 [DOI] [PubMed] [Google Scholar]
- Farina F., Gaillard J., Guérin C., Couté Y., Sillibourne J., Blanchoin L., and Théry M.. 2016. The centrosome is an actin-organizing centre. Nat. Cell Biol. 18:65–75. 10.1038/ncb3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Caballe A., Wainman A., Johnson S., Haensele A.F.M., Cottee M.A., Conduit P.T., Lea S.M., and Raff J.W.. 2017. Structural Basis for Mitotic Centrosome Assembly in Flies. Cell. 169:1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan A.M., Stavenschi E., Basavaraju S., Gaboriau D., Hoey D.A., and Morrison C.G.. 2017. Centriole splitting caused by loss of the centrosomal linker protein C-NAP1 reduces centriolar satellite density and impedes centrosome amplification. Mol. Biol. Cell. 28:736–745. 10.1091/mbc.E16-05-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong C.S., Mazo G., Das T., Goodman J., Kim M., O’Rourke B.P., Izquierdo D., and Tsou M.F.. 2016. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. eLife. 5:e16270 10.7554/eLife.16270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A., Roque H., Saurya S., Dobbelaere J., and Raff J.W.. 2013. CP110 exhibits novel regulatory activities during centriole assembly in Drosophila. J. Cell Biol. 203:785–799. 10.1083/jcb.201305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin L.K., Levy Y.Y., Levitan E., Chen S., Cande W.Z., Lai E.Y., and Fulton C.. 2016. Rapid centriole assembly in Naegleria reveals conserved roles for both de novo and mentored assembly. Cytoskeleton (Hoboken). 73:109–116. 10.1002/cm.21284 [DOI] [PubMed] [Google Scholar]
- Fu J., Lipinszki Z., Rangone H., Min M., Mykura C., Chao-Chu J., Schneider S., Dzhindzhev N.S., Gottardo M., Riparbelli M.G., et al. 2016. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol. 18:87–99. 10.1038/ncb3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton C., and Dingle A.D.. 1971. Basal bodies, but not centrioles, in Naegleria. J. Cell Biol. 51:826–836. 10.1083/jcb.51.3.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk M.C., Bera A.N., Menchen T., Kuales G., Thriene K., Lienkamp S.S., Dengjel J., Omran H., Frank M., and Arnold S.J.. 2015. Cyclin O (Ccno) functions during deuterosome-mediated centriole amplification of multiciliated cells. EMBO J. 34:1078–1089. 10.15252/embj.201490805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E., Ramani A., Karow U., Gottardo M., Natarajan K., Gooi L.M., Goranci-Buzhala G., Krut O., Peters F., Nikolic M., et al. 2017. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell. 20:397–406. [DOI] [PubMed] [Google Scholar]
- Galletta B.J., Jacobs K.C., Fagerstrom C.J., and Rusan N.M.. 2016. Asterless is required for centriole length control and sperm development. J. Cell Biol. 213:435–450. 10.1083/jcb.201501120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavilan M.P., Arjona M., Zurbano A., Formstecher E., Martinez-Morales J.R., Bornens M., and Rios R.M.. 2015. Alpha-catenin-dependent recruitment of the centrosomal protein CAP350 to adherens junctions allows epithelial cells to acquire a columnar shape. PLoS Biol. 13:e1002087 10.1371/journal.pbio.1002087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho S.A., and Pellman D.. 2014. Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130467 10.1098/rstb.2013.0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho S.A., Picone R., Burute M., Dagher R., Su Y., Leung C.T., Polyak K., Brugge J.S., Théry M., and Pellman D.. 2014. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 510:167–171. 10.1038/nature13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435. 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Guderian G., Westendorf J., Uldschmid A., and Nigg E.A.. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J. Cell Sci. 123:2163–2169. 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Gudi R., Haycraft C.J., Bell P.D., Li Z., and Vasu C.. 2015. Centrobin-mediated regulation of the centrosomal protein 4.1-associated protein (CPAP) level limits centriole length during elongation stage. J. Biol. Chem. 290:6890–6902. 10.1074/jbc.M114.603423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueth-Hallonet C., Antony C., Aghion J., Santa-Maria A., Lajoie-Mazenc I., Wright M., and Maro B.. 1993. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J. Cell Sci. 105:157–166. [DOI] [PubMed] [Google Scholar]
- Guichard P., Chrétien D., Marco S., and Tassin A.M.. 2010. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 29:1565–1572. 10.1038/emboj.2010.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P., Hamel V., Le Guennec M., Banterle N., Iacovache I., Nemčíková V., Flückiger I., Goldie K.N., Stahlberg H., Lévy D., et al. 2017. Cell-free reconstitution reveals centriole cartwheel assembly mechanisms. Nat. Commun. 8:14813 10.1038/ncomms14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., and Nigg E.A.. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hehnly H., Chen C.T., Powers C.M., Liu H.L., and Doxsey S.. 2012. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr. Biol. 22:1944–1950. 10.1016/j.cub.2012.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe E.H., and Sluder G.. 2002. Two for two: Cdk2 and its role in centrosome doubling. Oncogene. 21:6154–6160. 10.1038/sj.onc.1205826 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Li C., Thompson E.A., Maller J.L., and Sluder G.. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 283:851–854. 10.1126/science.283.5403.851 [DOI] [PubMed] [Google Scholar]
- Hoh R.A., Stowe T.R., Turk E., and Stearns T.. 2012. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One. 7:e52166 10.1371/journal.pone.0052166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A., and Toda T.. 2017. Regulation of centriolar satellite integrity and its physiology. Cell. Mol. Life Sci. 74:213–229. 10.1007/s00018-016-2315-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Xia Y., Zhang D., Wang S., Bao Y., He R., Teng J., and Chen J.. 2017. Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68. Nat. Commun. 8:15057 10.1038/ncomms15057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner A.F., and Rabinowitz M.. 1933. Demonstration of the central body in the living cell. Science. 78:367–368. 10.1126/science.78.2025.367-a [DOI] [PubMed] [Google Scholar]
- Inanç B., Dodson H., and Morrison C.G.. 2010. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol. Biol. Cell. 21:3866–3877. 10.1091/mbc.E10-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., and Tsukita S.. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524. 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Izquierdo D., Wang W.J., Uryu K., and Tsou M.F.. 2014. Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Reports. 8:957–965. 10.1016/j.celrep.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P.K., and Attardi L.D.. 2016. p73 and FoxJ1: Programming Multiciliated Epithelia. Trends Cell Biol. 26:239–240. 10.1016/j.tcb.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C. 2014. The tubulin code: molecular components, readout mechanisms, and functions. J. Cell Biol. 206:461–472. 10.1083/jcb.201406055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J., and Gonzalez C.. 2013. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15:241–248. 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- Jenkins P.M., McEwen D.P., and Martens J.R.. 2009. Olfactory cilia: linking sensory cilia function and human disease. Chem. Senses. 34:451–464. 10.1093/chemse/bjp020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo K., Kim C.G., Lee M.-S., Moon H.-Y., Lee S.-H., Kim M.J., Kweon H.-S., Park W.-Y., Kim C.-H., Gleeson J.G., and Kim J.. 2013. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc. Natl. Acad. Sci. USA. 110:5987–5992. 10.1073/pnas.1220927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach R.J.R. 1983. The induction of de novo centrioles in sea urchin eggs: a possible common mechanism for centriolar activation among parthenogenetic procedures. Eur. J. Cell Biol. 30:159–166. [PubMed] [Google Scholar]
- Kallenbach R.J.R., and Mazia D.. 1982. Origin and maturation of centrioles in association with the nuclear envelope in hypertonic-stressed sea urchin eggs. Eur. J. Cell Biol. 28:68–76. [PubMed] [Google Scholar]
- Kato K.H.K., and Sugiyama M.. 1971. On the de novo formation of the centriole in the activated sea urchin egg. Dev. Growth Differ. 13:359–366. 10.1111/j.1440-169X.1971.00359.x [DOI] [PubMed] [Google Scholar]
- Keller D., Orpinell M., Olivier N., Wachsmuth M., Mahen R., Wyss R., Hachet V., Ellenberg J., Manley S., and Gönczy P.. 2014. Mechanisms of HsSAS-6 assembly promoting centriole formation in human cells. J. Cell Biol. 204:697–712. 10.1083/jcb.201307049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L., Sluder G., Cassels G., Sibon O., and Wang C.L.. 2002. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 158:1171–1181. 10.1083/jcb.200205102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee K., and Rhee K.. 2015. PLK1 regulation of PCNT cleavage ensures fidelity of centriole separation during mitotic exit. Nat. Commun. 6:10076 10.1038/ncomms10076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Park J.E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M.C., Flückiger I., Gönczy P., and Steinmetz M.O.. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell. 144:364–375. 10.1016/j.cell.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba J.E., Buster D.W., McLamarrah T.A., Rusan N.M., and Rogers G.C.. 2015. Autoinhibition and relief mechanism for Polo-like kinase 4. Proc. Natl. Acad. Sci. USA. 112:E657–E666. 10.1073/pnas.1417967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., and Nigg E.A.. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202. 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Klink V.P., and Wolniak S.M.. 2003. Changes in the abundance and distribution of conserved centrosomal, cytoskeletal and ciliary proteins during spermiogenesis in Marsilea vestita. Cell Motil. Cytoskeleton. 56:57–73. 10.1002/cm.10134 [DOI] [PubMed] [Google Scholar]
- Klos Dehring D.A., Vladar E.K., Werner M.E., Mitchell J.W., Hwang P., and Mitchell B.J.. 2013. Deuterosome-mediated centriole biogenesis. Dev. Cell. 27:103–112. 10.1016/j.devcel.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanski R.S., and Borisy G.G.. 1990. Mode of centriole duplication and distribution. J. Cell Biol. 110:1599–1605. 10.1083/jcb.110.5.1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B.F., Mogensen M.M., Spektor A., Dynlacht B.D., Khodjakov A., and Gönczy P.. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19:1012–1018. 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Farmer V., Shukla A., James J., Gruskin R., Kiriyama S., and Loncarek J.. 2014. Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. J. Cell Biol. 206:855–865. 10.1083/jcb.201407087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski N., Treat B., and Duensing S.. 2011. The HPV-16 E7 oncoprotein induces centriole multiplication through deregulation of Polo-like kinase 4 expression. Mol. Cancer. 10:61 10.1186/1476-4598-10-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraatz S., Guichard P., Obbineni J.M., Olieric N., Hatzopoulos G.N., Hilbert M., Sen I., Missimer J., Gonczy P., and Steinmetz M.O.. 2016. The Human Centriolar Protein CEP135 Contains a Two-Stranded Coiled-Coil Domain Critical for Microtubule Binding. Structure. 24:1358–1371. 10.1016/j.str.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Kulukian A., Holland A.J., Vitre B., Naik S., Cleveland D.W., and Fuchs E.. 2015. Epidermal development, growth control, and homeostasis in the face of centrosome amplification. Proc. Natl. Acad. Sci. USA. 112:E6311–E6320. 10.1073/pnas.1518376112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumada K., Yao R., Kawaguchi T., Karasawa M., Hoshikawa Y., Ichikawa K., Sugitani Y., Imoto I., Inazawa J., Sugawara M., et al. 2006. The selective continued linkage of centromeres from mitosis to interphase in the absence of mammalian separase. J. Cell Biol. 172:835–846. 10.1083/jcb.200511126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto K., Yamazaki Y., Nishida T., Shinohara K., Ishikawa H., Hasegawa T., Okanoue T., Hamada H., Noda T., Tamura A., et al. 2012. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 148:189–200. 10.1016/j.cell.2011.10.052 [DOI] [PubMed] [Google Scholar]
- Kurtulmus B., Wang W., Ruppert T., Neuner A., Cerikan B., Viol L., Dueñas-Sánchez R., Gruss O.J., and Pereira G.. 2016. WDR8 is a centriolar satellite and centriole-associated protein that promotes ciliary vesicle docking during ciliogenesis. J. Cell Sci. 129:621–636. 10.1242/jcs.179713 [DOI] [PubMed] [Google Scholar]
- Lacey K.R., Jackson P.K., and Stearns T.. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA. 96:2817–2822. 10.1073/pnas.96.6.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus B.G., and Holland A.J.. 2017. A New Mode of Mitotic Surveillance. Trends Cell Biol. 27:314–321. 10.1016/j.tcb.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus B.G., Daggubati V., Uetake Y., Scott P.M., Clutario K.M., Sluder G., and Holland A.J.. 2016. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J. Cell Biol. 214:143–153. 10.1083/jcb.201604054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laos T., Cabral G., and Dammermann A.. 2015. Isotropic incorporation of SPD-5 underlies centrosome assembly in C. elegans. Curr. Biol. 25:R648–R649. 10.1016/j.cub.2015.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra S., English C.N., Hergert P., McEwen B.F., Sluder G., and Khodjakov A.. 2005. The de novo centriole assembly pathway in HeLa cells. J. Cell Biol. 168:713–722. 10.1083/jcb.200411126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S., Hasegan M., Gupta G.D., and Pelletier L.. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158. 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Lechtreck K.F., and Melkonian M.. 1991. Striated microtubule-associated fibers: identification of assemblin, a novel 34-kD protein that forms paracrystals of 2-nm filaments in vitro. J. Cell Biol. 115:705–716. 10.1083/jcb.115.3.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., and Rhee K.. 2012. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle. 11:2476–2485. 10.4161/cc.20878 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., and Gönczy P.. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125. 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Levine M.S., Bakker B., Boeckx B., Moyett J., Lu J., Vitre B., Spierings D.C., Lansdorp P.M., Cleveland D.W., Lambrechts D., et al. 2017. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev. Cell. 40:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-C., Chang C.-W., Hsu W.-B., Tang C.-J.C., Lin Y.-N., Chou E.-J., Wu C.-T., and Tang T.K.. 2013a Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 32:1141–1154. 10.1038/emboj.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.N., Wu C.T., Lin Y.C., Hsu W.B., Tang C.J., Chang C.W., and Tang T.K.. 2013b CEP120 interacts with CPAP and positively regulates centriole elongation. J. Cell Biol. 202:211–219. 10.1083/jcb.201212060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J., Hergert P., Magidson V., and Khodjakov A.. 2008. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 10:322–328. 10.1038/ncb1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lončarek J., Hergert P., and Khodjakov A.. 2010. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr. Biol. 20:1277–1282. 10.1016/j.cub.2010.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C.A., Jana S.C., Cunha-Ferreira I., Zitouni S., Bento I., Duarte P., Gilberto S., Freixo F., Guerrero A., Francia M., et al. 2015. PLK4 trans-Autoactivation Controls Centriole Biogenesis in Space. Dev. Cell. 35:222–235. 10.1016/j.devcel.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Lu Q., Insinna C., Ott C., Stauffer J., Pintado P.A., Rahajeng J., Baxa U., Walia V., Cuenca A., Hwang Y.S., et al. 2015. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17:228–240. 10.1038/ncb3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius G., Lavogina D., Orpinell M., Umezawa K., Reymond L., Garin N., Gönczy P., and Johnsson K.. 2013. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr. Biol. 23:265–270. 10.1016/j.cub.2012.12.030 [DOI] [PubMed] [Google Scholar]
- Madarampalli B., Yuan Y., Liu D., Lengel K., Xu Y., Li G., Yang J., Liu X., Lu Z., and Liu D.X.. 2015. ATF5 Connects the Pericentriolar Materials to the Proximal End of the Mother Centriole. Cell. 162:580–592. 10.1016/j.cell.2015.06.055 [DOI] [PubMed] [Google Scholar]
- Mahjoub M.R., Xie Z., and Stearns T.. 2010. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J. Cell Biol. 191:331–346. 10.1083/jcb.201003009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar G., Schatten H., and Sutovsky P.. 2005. Centrosome reduction during gametogenesis and its significance. Biol. Reprod. 72:2–13. 10.1095/biolreprod.104.031245 [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Vucica Y., and Rosenbaum J.L.. 2001. Kinetics and regulation of de novo centriole assembly: Implications for the mechanism of centriole duplication. Curr. Biol. 11:308–317. 10.1016/S0960-9822(01)00094-X [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Hayashi K., and Nishida E.. 1999. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol. 9:429–432. 10.1016/S0960-9822(99)80191-2 [DOI] [PubMed] [Google Scholar]
- Matsuo K., Ohsumi K., Iwabuchi M., Kawamata T., Ono Y., and Takahashi M.. 2012. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr. Biol. 22:915–921. 10.1016/j.cub.2012.03.048 [DOI] [PubMed] [Google Scholar]
- Mazo G., Soplop N., Wang W.J., Uryu K., and Tsou M.B.. 2016. Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia. Dev. Cell. 39:424–437. 10.1016/j.devcel.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F., Anzola J.V., Kaulich M., Richardson A., Stender J.D., Benner C., Glass C.K., Dowdy S.F., Desai A., Shiau A.K., and Oegema K.. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 214:155–166. 10.1083/jcb.201604081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., and Agard D.A.. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168. 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A.M., Bartek J., and Nigg E.A.. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88–93. 10.1038/10054 [DOI] [PubMed] [Google Scholar]
- Meunier A., and Azimzadeh J.. 2016. Multiciliated Cells in Animals. Cold Spring Harb. Perspect. Biol. 8:a028233 10.1101/cshperspect.a028233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki-Noumura T. 1977. Studies on the de novo formation of centrioles: aster formation in the activated eggs of sea urchin. J. Cell Sci. 24:203–216. [DOI] [PubMed] [Google Scholar]
- Mizukami I., and Gall J.. 1966. Centriole replication. II. Sperm formation in the fern, Marsilea, and the cycad, Zamia. J. Cell Biol. 29:97–111. 10.1083/jcb.29.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S., Timbers T.A., Kennedy J., Blacque O.E., and Leroux M.R.. 2013. Striated rootlet and nonfilamentous forms of rootletin maintain ciliary function. Curr. Biol. 23:2016–2022. 10.1016/j.cub.2013.08.033 [DOI] [PubMed] [Google Scholar]
- Moyer T.C., Clutario K.M., Lambrus B.G., Daggubati V., and Holland A.J.. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209:863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., and Hirono M.. 2007. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17:2169–2174. 10.1016/j.cub.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Novak Z.A., Wainman A., Gartenmann L., and Raff J.W.. 2016. Cdk1 Phosphorylates Drosophila Sas-4 to Recruit Polo to Daughter Centrioles and Convert Them to Centrosomes. Dev. Cell. 37:545–557. 10.1016/j.devcel.2016.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell K.F., Caron C., Kopish K.R., Hurd D.D., Kemphues K.J., Li Y., and White J.G.. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558. 10.1016/S0092-8674(01)00338-5 [DOI] [PubMed] [Google Scholar]
- Obino D., Farina F., Malbec O., Sáez P.J., Maurin M., Gaillard J., Dingli F., Loew D., Gautreau A., Yuseff M.I., et al. 2016. Actin nucleation at the centrosome controls lymphocyte polarity. Nat. Commun. 7:10969 10.1038/ncomms10969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., and Kitagawa D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5:5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda M., Horn H.F., Tarapore P., Tokuyama Y., Smulian A.G., Chan P.K., Knudsen E.S., Hofmann I.A., Snyder J.D., Bove K.E., and Fukasawa K.. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 103:127–140. 10.1016/S0092-8674(00)00093-3 [DOI] [PubMed] [Google Scholar]
- Oliveira R.A., and Nasmyth K.. 2013. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Curr. Biol. 23:R601–R603. 10.1016/j.cub.2013.04.003 [DOI] [PubMed] [Google Scholar]
- Pearson C.G., Giddings T.H. Jr., and Winey M.. 2009. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol. Biol. Cell. 20:904–914. 10.1091/mbc.E08-08-0835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., O’Toole E., Schwager A., Hyman A.A., and Müller-Reichert T.. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., and Bornens M.. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330. 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta-Marques A., Bento I., Lopes C.A., Duarte P., Jana S.C., and Bettencourt-Dias M.. 2016. A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science. 353:aaf4866 10.1126/science.aaf4866 [DOI] [PubMed] [Google Scholar]
- Poulton J.S., Cuningham J.C., and Peifer M.. 2014. Acentrosomal Drosophila epithelial cells exhibit abnormal cell division, leading to cell death and compensatory proliferation. Dev. Cell. 30:731–745. 10.1016/j.devcel.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkumar N., and Baum B.. 2016. Coupling changes in cell shape to chromosome segregation. Nat. Rev. Mol. Cell Biol. 17:511–521. 10.1038/nrm.2016.75 [DOI] [PubMed] [Google Scholar]
- Renzaglia K.S., Nickrent D.L., and Garbary D.J.. Duff RJT . 2000. Vegetative and reproductive innovations of early land plants: implications for a unified phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:769–793. 10.1098/rstb.2000.0615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli M.G., and Callaini G.. 2003. Drosophila parthenogenesis: a model for de novo centrosome assembly. Dev. Biol. 260:298–313. 10.1016/S0012-1606(03)00243-4 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2007. Revisiting the role of the mother centriole in centriole biogenesis. Science. 316:1046–1050. 10.1126/science.1142950 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2008. From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 7:11–16. 10.4161/cc.7.1.5226 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Roberts D.M., Peifer M., and Rogers S.L.. 2009. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239. 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubinet C., and Cabernard C.. 2014. Control of asymmetric cell division. Curr. Opin. Cell Biol. 31:84–91. 10.1016/j.ceb.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Sánchez I., and Dynlacht B.D.. 2016. Cilium assembly and disassembly. Nat. Cell Biol. 18:711–717. 10.1038/ncb3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten G. 1994. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev. Biol. 165:299–335. 10.1006/dbio.1994.1256 [DOI] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., and Pereira G.. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101. 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., and Nigg E.A.. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Schnackenberg B.J., Marzluff W.F., and Sluder G.. 2008. Cyclin E in centrosome duplication and reduplication in sea urchin zygotes. J. Cell. Physiol. 217:626–631. 10.1002/jcp.21531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöckel L., Möckel M., Mayer B., Boos D., and Stemmann O.. 2011. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol. 13:966–972. 10.1038/ncb2280 [DOI] [PubMed] [Google Scholar]
- Seo M.Y., Jang W., and Rhee K.. 2015. Integrity of the Pericentriolar Material Is Essential for Maintaining Centriole Association during M Phase. PLoS One. 10:e0138905 10.1371/journal.pone.0138905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serçin Ö., Larsimont J.C., Karambelas A.E., Marthiens V., Moers V., Boeckx B., Le Mercier M., Lambrechts D., Basto R., and Blanpain C.. 2016. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat. Cell Biol. 18:100–110. 10.1038/ncb3270 [DOI] [PubMed] [Google Scholar]
- Sharma A., Aher A., Dynes N.J., Frey D., Katrukha E.A., Jaussi R., Grigoriev I., Croisier M., Kammerer R.A., Akhmanova A., et al. 2016. Centriolar CPAP/SAS-4 Imparts Slow Processive Microtubule Growth. Dev. Cell. 37:362–376. 10.1016/j.devcel.2016.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Kong D., Sharma M., Magidson V., and Loncarek J.. 2015. Plk1 relieves centriole block to reduplication by promoting daughter centriole maturation. Nat. Commun. 6:8077 10.1038/ncomms9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Scholl L.M., and Cimini D.. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 4:e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Schermelleh L., Leonhardt H., and Nigg E.A.. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Gabryjonczyk A.M., Anselm E., Stierhof Y.D., and Nigg E.A.. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126:3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15:363–377. 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S.P. 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3:207–230. [DOI] [PubMed] [Google Scholar]
- Spektor A., Tsang W.Y., Khoo D., and Dynlacht B.D.. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell. 130:678–690. 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Srsen V., Fant X., Heald R., Rabouille C., and Merdes A.. 2009. Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 10:28 10.1186/1471-2121-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.R., Roque H., and Raff J.W.. 2010. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell. 19:913–919. 10.1016/j.devcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe J.C., Randzavola L.O., Angus K.L., Mantell J.M., Verkade P., and Griffiths G.M.. 2015. Mother Centriole Distal Appendages Mediate Centrosome Docking at the Immunological Synapse and Reveal Mechanistic Parallels with Ciliogenesis. Curr. Biol. 25:3239–3244. 10.1016/j.cub.2015.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs J.L., Vladar E.K., Axelrod J.D., and Kintner C.. 2012. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 14:140–147. 10.1038/ncb2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi D., Calarco P., and Donahue R.P.. 1972. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11:521–541. [DOI] [PubMed] [Google Scholar]
- Szöllosi D., and Ozil J.P.. 1991. De novo formation of centrioles in parthenogenetically activated, diploidized rabbit embryos. Biol. Cell. 72:61–66. 10.1016/0248-4900(91)90079-3 [DOI] [PubMed] [Google Scholar]
- Tan F.E., Vladar E.K., Ma L., Fuentealba L.C., Hoh R., Espinoza F.H., Axelrod J.D., Alvarez-Buylla A., Stearns T., Kintner C., and Krasnow M.A.. 2013. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development. 140:4277–4286. 10.1242/dev.094102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.J., Fu R.H., Wu K.S., Hsu W.B., and Tang T.K.. 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11:825–831. 10.1038/ncb1889 [DOI] [PubMed] [Google Scholar]
- Tanos B.E., Yang H.J., Soni R., Wang W.J., Macaluso F.P., Asara J.M., and Tsou M.F.. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27:163–168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A.M., Maro B., and Bornens M.. 1985. Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 100:35–46. 10.1083/jcb.100.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K., Yamazaki Y., Nishida T., Watanabe S., Kunimoto K., Ishikawa H., and Tsukita S.. 2013. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J. Cell Biol. 203:417–425. 10.1083/jcb.201303071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terré B., Piergiovanni G., Segura-Bayona S., Gil-Gómez G., Youssef S.A., Attolini C.S., Wilsch-Bräuninger M., Jung C., Rojas A.M., Marjanović M., et al. 2016. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 35:942–960. 10.15252/embj.201592821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauvin-Robinet C., Lee J.S., Lopez E., Herranz-Pérez V., Shida T., Franco B., Jego L., Ye F., Pasquier L., Loget P., et al. 2014. The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nat. Genet. 46:905–911. 10.1038/ng.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M.F., and Stearns T.. 2006a Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 18:74–78. 10.1016/j.ceb.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., and Stearns T.. 2006b Mechanism limiting centrosome duplication to once per cell cycle. Nature. 442:947–951. 10.1038/nature04985 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Wang W.J., George K.A., Uryu K., Stearns T., and Jallepalli P.V.. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 17:344–354. 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Loncarek J., Nordberg J.J., English C.N., La Terra S., Khodjakov A., and Sluder G.. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176:173–182. 10.1083/jcb.200607073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M., Hirono M., Andreeva A., Yanagisawa H.A., Yamaguchi S., Nakazawa Y., Morgner N., Petrovich M., Ebong I.O., Robinson C.V., et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science. 331:1196–1199. 10.1126/science.1199325 [DOI] [PubMed] [Google Scholar]
- Vaughn K.C., and Renzaglia K.S.. 2006. Structural and immunocytochemical characterization of the Ginkgo biloba L. sperm motility apparatus. Protoplasma. 227:165–173. 10.1007/s00709-005-0141-3 [DOI] [PubMed] [Google Scholar]
- Vertii A., Hehnly H., and Doxsey S.. 2016. The Centrosome, a Multitalented Renaissance Organelle. Cold Spring Harb. Perspect. Biol. 8:a025049 10.1101/cshperspect.a025049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B., Holland A.J., Kulukian A., Shoshani O., Hirai M., Wang Y., Maldonado M., Cho T., Boubaker J., Swing D.A., et al. 2015. Chronic centrosome amplification without tumorigenesis. Proc. Natl. Acad. Sci. USA. 112:E6321–E6330. 10.1073/pnas.1519388112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E.K., and Stearns T.. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178:31–42. 10.1083/jcb.200703064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.A., and Chentsov YuS. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949. 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.J., Soni R.K., Uryu K., and Tsou M.F.. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193:727–739. 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang Y., Duan Q., Jiang N., Huang Y., Darzynkiewicz Z., and Dai W.. 2008. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev. Cell. 14:331–341. 10.1016/j.devcel.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., and O’Toole E.. 2014. Centriole structure. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130457 10.1098/rstb.2013.0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf B., Diop F., Ferraris P., Wichit S., Busso C., Missé D., and Gönczy P.. 2017. Zika virus causes supernumerary foci with centriolar proteins and impaired spindle positioning. Open Biol. 7:160231 10.1098/rsob.160231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolniak S.M., Boothby T.C., and van der Weele C.M.. 2015. Posttranscriptional control over rapid development and ciliogenesis in Marsilea. Methods Cell Biol. 127:403–444. 10.1016/bs.mcb.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Wong C., and Stearns T.. 2003. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat. Cell Biol. 5:539–544. 10.1038/ncb993 [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Wueseke O., and Hyman A.A.. 2014. Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130459 10.1098/rstb.2013.0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., and Hyman A.A.. 2017. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 169:1066–1077. [DOI] [PubMed] [Google Scholar]
- Wueseke O., Zwicker D., Schwager A., Wong Y.L., Oegema K., Jülicher F., Hyman A.A., and Woodruff J.B.. 2016. Polo-like kinase phosphorylation determines Caenorhabditis elegans centrosome size and density by biasing SPD-5 toward an assembly-competent conformation. Biol. Open. 5:1431–1440. 10.1242/bio.020990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T., Ge X., and Pelegri F.. 2007. The zebrafish maternal-effect gene cellular atoll encodes the centriolar component sas-6 and defects in its paternal function promote whole genome duplication. Dev. Biol. 312:44–60. 10.1016/j.ydbio.2007.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Gao J., Adamian M., Wen X.H., Pawlyk B., Zhang L., Sanderson M.J., Zuo J., Makino C.L., and Li T.. 2005. The ciliary rootlet maintains long-term stability of sensory cilia. Mol. Cell. Biol. 25:4129–4137. 10.1128/MCB.25.10.4129-4137.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Zeng H., Ning G., Reiter J.F., and Liu A.. 2014. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. USA. 111:2164–2169. 10.1073/pnas.1318737111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhu L., Zhu Y., Cao J., Li S., Huang Q., Xu T., Huang X., Yan X., and Zhu X.. 2013. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat. Cell Biol. 15:1434–1444. 10.1038/ncb2880 [DOI] [PubMed] [Google Scholar]
- Zheng X., Ramani A., Soni K., Gottardo M., Zheng S., Ming Gooi L., Li W., Feng S., Mariappan A., Wason A., et al. 2016. Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat. Commun. 7:11874 10.1038/ncomms11874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitouni S., Francia M.E., Leal F., Montenegro Gouveia S., Nabais C., Duarte P., Gilberto S., Brito D., Moyer T., Kandels-Lewis S., et al. 2016. CDK1 Prevents Unscheduled PLK4-STIL Complex Assembly in Centriole Biogenesis. Curr. Biol. 26:1127–1137. 10.1016/j.cub.2016.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker D., Decker M., Jaensch S., Hyman A.A., and Jülicher F.. 2014. Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc. Natl. Acad. Sci. USA. 111:E2636–E2645. 10.1073/pnas.1404855111 [DOI] [PMC free article] [PubMed] [Google Scholar]