Interactions between actin nucleators and the exocyst in yeast and mammals control membrane remodeling. van Gisbergen et al. now describe For1F, a fusion of an exocyst subunit (Sec10) and an actin nucleation factor (formin), retained in the moss lineage for more than 170 million years, which provides unique insight into the regulation of exocytosis by actin.
Abstract
Exocytosis, facilitated by the exocyst, is fundamentally important for remodeling cell walls and membranes. Here, we analyzed For1F, a novel gene that encodes a fusion of an exocyst subunit (Sec10) and an actin nucleation factor (formin). We showed that the fusion occurred early in moss evolution and has been retained for more than 170 million years. In Physcomitrella patens, For1F is essential, and the expressed protein is a fusion of Sec10 and formin. Reduction of For1F or actin filaments inhibits exocytosis, and For1F dynamically associates with Sec6, another exocyst subunit, in an actin-dependent manner. Complementation experiments demonstrate that constitutive expression of either half of the gene or the paralogous Sec10b rescues loss of For1F, suggesting that fusion of the two domains is not essential, consistent with findings in yeast, where formin and the exocyst are linked noncovalently. Although not essential, the fusion may have had selective advantages and provides a unique opportunity to probe actin regulation of exocytosis.
Introduction
Vesicle trafficking, particularly exocytosis and endocytosis, is fundamental for cell growth. Exocytosis delivers new membrane and external material, whereas endocytosis recycles excess membrane and catabolized secretory products. In eukaryotes that generate an extracellular matrix encasing the cell, exocytosis is a critical determinant of cell shape and polarity. Plant cells are an excellent example as they are surrounded by a cell wall that physically constrains the cell. Construction and modifications of this wall by delivery of building material and remodeling enzymes from within the cell eventually shape the cell.
During exocytosis, the highly conserved exocyst complex tethers secretory vesicles to the plasma membrane before vesicle fusion occurs. The exocyst is comprised of eight subunits and is essential for plant growth and development (Synek et al., 2006; Hála et al., 2008). Although animals and fungi tend to have one copy of each subunit, plants have evolved multigene families encoding for some of the subunits (Elias et al., 2003; Synek et al., 2006; Cvrčková et al., 2012). The most extreme example is the EXO70 subunit, which has greatly expanded in plants (Cvrčková et al., 2012). Based on these expansions, it has been hypothesized that plants have multiple exocyst complexes to execute specific exocytic pathways leading to distinct cell wall patterning (Cvrčková et al., 2012). Thus, it is not surprising that the exocyst complex in plants has been implicated in a variety of processes, including autophagy (Kulich et al., 2013), cell polarity (Cole et al., 2005; Wen et al., 2005; Synek et al., 2006), cell plate formation (Fendrych et al., 2010; Wu et al., 2013), and pathogen resistance (Stegmann et al., 2013).
Because of its anatomical simplicity relative to seed plants, the moss Physcomitrella patens provides an opportunity to study the function of the exocyst complex in cell morphogenesis. Mosses have few cell types that comprise simple tissues, usually only a single cell layer thick. Consistent with this, P. patens has 13 copies of EXO70, in contrast to the 23 found in Arabidopsis and 47 in rice (Cvrčková et al., 2012). In analyzing the complement of P. patens exocyst subunits, we were particularly intrigued by one of the three copies of Sec10. This copy is actually predicted to encode a domain in a larger protein that contains both an N-terminal Sec10 domain and a C-terminal formin domain.
Formins are nucleators of actin filaments, and are essential for multiple actin-based processes. In plants, formins have been implicated in cell division and polarized growth (Ingouff et al., 2005; Vidali et al., 2009b; van Gisbergen et al., 2012; van Gisbergen and Bezanilla, 2013), as well as pathogen defense responses (de Almeida Engler et al., 2004; Favery et al., 2004). Although actin and exocytosis have been linked via noncovalent interactions between actin nucleators and the exocyst in fission yeast (Jourdain et al., 2012) and animals (Zuo et al., 2006), the fusion of a component of the exocyst subunit to an actin filament–nucleating factor within a single protein provides a unique opportunity to study the relationship between actin and exocytosis in a eukaryotic system where cell shape is a direct readout of exocytic activity.
Results
Formin 1F is comprised of two domains: An N-terminal Sec10 and a C-terminal class I formin
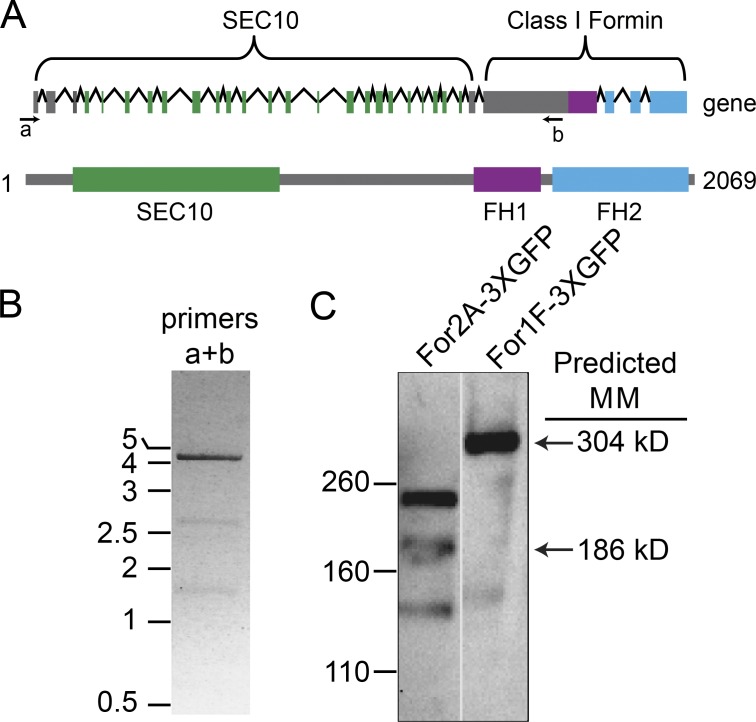
The Formin 1F (For1F) gene is predicted to encode a protein with a novel domain configuration, comprised of an N-terminal exocyst subunit and a C-terminal actin filament–nucleating factor (Zimmer et al., 2013). This gene structure is remarkable because the 5′ end of the gene has similar numbers of coding exons as the other two moss Sec10 genes, and the 3′ end of the gene has a similar genomic structure to the other five moss class I formins (Fig. 1 A). Additionally, similar to all other formins in the P. patens genome, For1F has a large exon that in part encodes the formin homology (FH) 1 domain. The predicted amino acid sequence of the Sec10 domain of For1F is 64% similar to Arabidopsis thaliana Sec10 and 70% and 53% similar to P. patens Sec10b and Sec10a, respectively. This sequence similarity suggests that the For1F Sec10 domain is a functional Sec10 subunit, as opposed to a divergent relative no longer capable of interacting with the exocyst.
Figure 1.
For1F encodes for a protein comprised of an N-terminal Sec10 domain and a C-terminal formin domain. (A) For1F gene model and protein schematic. Top: In the gene model, boxes indicate exons and lines are introns. For both the gene model and the protein prediction, green represents sequences with similarity to Sec10; purple, FH1; blue, FH2; and gray boxes, no known sequence similarity. Numbers indicate amino acid positions. Arrows labeled a and b under the gene model are primers used to perform RT-PCR. (B) RT-PCR of the For1F transcript amplified with primers a and b. Numbers indicate size in kb. (C) Immunoblots performed with an antibody to GFP of cell extracts from a line with For2A (Vidali et al., 2009b) or For1F tagged endogenously with sequences encoding for three tandem GFP molecules. Numbers to the left indicate size in kilodaltons.
Because this domain configuration has not been observed in any other species to date (Grunt et al., 2008), we hypothesized that the predicted gene may have been improperly assembled. We analyzed mRNA transcribed from this region to determine whether the region comprises one or two genes. Because it is extremely difficult for reverse transcription to transcribe through the highly GC-rich region encoding for the polyproline-rich FH1 domain, we used a reverse primer just upstream of this region to generate the cDNA template (Fig. 1 A). Using a forward primer in the first predicted exon of the region encoding for the Sec10 domain together with the reverse primer upstream of the FH1 domain (Fig. 1 A), we amplified a product that spans the Sec10 domain and the formin domain, consistent with the presence of a single gene (Fig. 1 B).
Although a continuous mRNA is consistent with a single gene, it is possible that alternative splice variants are generated and only separate proteins are translated. To determine the size of the protein that is generated from this locus, we tagged the 3′ end of the genomic locus in frame with sequences encoding for three tandem copies of monomeric EGFP (hereafter referred to as GFP; Vidali et al., 2009b). In lines that were properly integrated, we extracted proteins and probed immunoblots with a GFP antibody. We found that For1F-3XmEGFP (hereafter referred to as For1F-GFP) migrates around 300 kD, closely corresponding to the predicted size of a protein containing the N-terminal Sec10, the C-terminal formin, and three tandem copies of GFP (Fig. 1 C), demonstrating that this locus encodes a single protein. The presence of Sec10, a subunit of the exocyst complex, and formin, an actin-interacting module, in one protein is intriguing and suggests that in P. patens there is a direct link between exocytosis and the actin cytoskeleton.
Fusion of Sec10 and a formin is present throughout diverse mosses
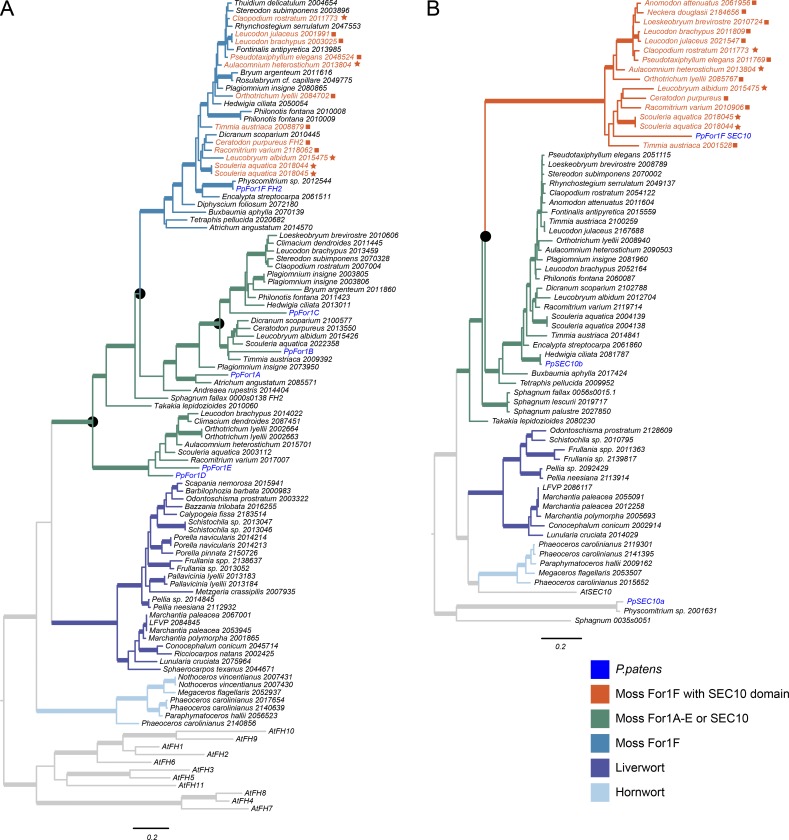
To gain insight into the evolution of the unique domain configuration in For1F, we used class I formin FH2 domain sequences to retrieve bryophyte sequences from the 1KP (Matasci et al., 2014) and Ceratodon purpureus (Szövényi et al., 2015) transcriptome databases. We also searched the Sphagnum fallax genome (Shaw et al., 2016). Using these sequences, we performed phylogenetic analysis under the maximum likelihood (ML) information criterion using the Jones, Taylor, and Thornton model of protein evolution. We found that all moss class I formins fall into a well-supported clade (bootstrap support = 100%), entirely separate from other bryophyte class I formin sequences (liverwort and hornwort class I formins; Fig. 2 A). In the mosses, an early gene duplication gave rise to two main gene clades: one containing For1D and For1E, and a sister clade containing the remaining class I formins. A later gene duplication within the sister clade gave rise to two more gene lineages: one containing only For1F and another containing For1A, For1B, and For1C (Fig. 2 A).
Figure 2.
Phylogenetic analysis of FH2 and Sec10 domains. ML phylogenies of bryophyte amino acid sequences homologous to either a class I formin FH2 domain (A) or a SEC10 domain (B). Thickened lines indicate strong ML bootstrap support (≥85%). Black circles indicate most likely gene duplication events. Stars next to gene names indicate a For1F FH2 domain and a SEC10 domain on the same transcript assembly. Squares indicate species with SEC10 transcript assemblies with truncated For1F domains, and either separate or absent FH2 domain assemblies.
We found that 28 moss species, spanning all major moss lineages except the early diverging Takakiopsida and Sphagnopsida, have a gene encoding the For1F FH2 domain. We were unable to identify gene sequences with homology to For1F from the genome of S. fallax (Szövényi et al., 2015), or from the transcriptomes of Sphagnum palustre, Sphagnum lescurii (Sphagnopsida), or Takakia lepidozioides (Takakiopsida). These data indicate that the gene duplication that gave rise to For1F occurred very early in moss history, likely more than 200 million years ago (Hedges et al., 2015). We found four moss species apart from P. patens that have a Sec10 domain on a transcript that extends all the way to a For1F FH2 domain (Fig. 2, stars). We found an additional seven species that had a For1F transcript that includes an FH2 domain, and a separate transcript assembly that includes both a Sec10 domain clearly homologous to the P. patens For1F Sec10 domain, and a truncated formin sequence (no FH2 domain; Fig. 2, squares). In the case of C. purpureus, the two identified transcripts map immediately next to each other on the genome (unpublished data), suggesting that they are encoded by a single locus as in P. patens. Last, we found three species with transcripts that encode a Sec10 domain and a truncated formin sequence, but with no matching FH2 transcript in the 1KP database (Matasci et al., 2014). We inferred a Sec10/formin fusion in all 14 of these distantly related species. Because the Sec10 domain is found only associated with FH2 domains that group with For1F, it suggests that this fusion happened early on, after the divergence of Takakiopsida and Sphagnopsida from the rest of the mosses.
To see if the fusion evolved independently from the other Sec10 proteins, we performed a similar analysis looking at the phylogeny of the Sec10 domains (Fig. 2 B). Besides the For1F Sec10 domain, P. patens has two additional Sec10 genes, Sec10a and Sec10b. Sec10a and its homologues that we retrieved only from Physcomitrium species and S. fallax are in a separate clade to the other two Sec10 genes (bootstrap support = 100%). Single Sec10 homologues from Takakia lepidozioides and Sphagnum species are sisters to a clade of Sec10 genes that includes both Sec10b and the For1F Sec10 domain. This indicates that sometime after the divergence of Takakiopsida and Sphagnopsida, there was a gene duplication that led to Sec10b and a monophyletic clade of Sec10s that are fused to a For1F formin domain (Fig. 2 B). Although the timing of the Sec10 duplication is less clear than it is for For1F, both Sec10 gene clades have sequences retrieved from taxa likely separated by more than 170 million years of evolution (Hedges et al., 2015). Thus, the Sec10–formin fusion is not unique to P. patens but is found in many mosses, and likely happened early on in moss history.
For1F is essential for plant viability
Vidali et al. (2009b) previously silenced For1F using an RNAi construct targeting a small region of the FH2 domain, which resulted in plants 30% smaller than control RNAi plants. However, targeting this region of the For1F gene results in only a 75% reduction of the transcript, not complete elimination (Vidali et al., 2009b). To increase the silencing efficiency, we generated several constructs containing different regions of the gene. We found that an RNAi construct containing 305 bp of the coding sequence of the Sec10 region of For1F recapitulated the data in Vidali et al. (2009b). In contrast, transformation of RNAi constructs containing the first 3,908 bp of the coding sequence of For1F, which encompasses the entire Sec10 domain, or the 5′ and 3′ untranslated regions yielded only 2–14 silenced plants per transformation. This yield is 35–250 times lower than the control constructs, which yield a mean of 490 silenced plants. These data suggest that silencing For1F is incompatible with life. In support of this, we were unable to generate a knockout line using homologous recombination, suggesting that For1F is essential for viability.
To determine if silencing the exocyst complex results in a similar loss-of-function phenotype as silencing For1F, we generated an RNAi construct targeting Sec6, which is the only subunit encoded by a single-copy gene in moss. Because Sec6 is a single-copy gene and is a core subunit of the complex, it should be part of every exocyst complex. Silencing Sec6 by targeting the entire coding sequence did not yield any surviving plants, suggesting that the exocyst is essential for viability. We also silenced the other two Sec10 genes in P. patens (Fig. S1). Silencing Sec10a did not affect plant size in 7-d-old plants. However, silencing Sec10b resulted in 50% smaller plants. Silencing both Sec10a and Sec10b simultaneously did not have an additional effect on plant size consistent with the lack of a phenotype observed when Sec10a was silenced alone (Fig. S1). These data suggest that Sec10b, not Sec10a, is required in young plants. However, silencing of Sec10b was not lethal, suggesting that either the silencing was not efficient or that Sec10b is redundant with the For1F Sec10 domain.
For1F is involved in exocytosis
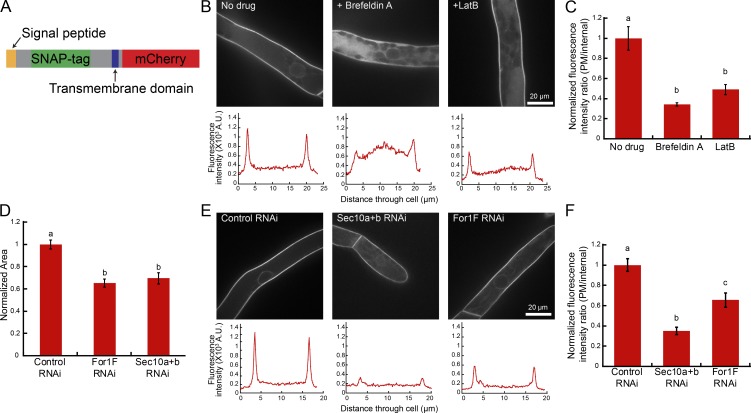
To determine whether For1F is required for efficient exocytosis, we developed a quantitative exocytosis assay in which we measured secretion of a fluorescently labeled transmembrane protein. We generated a chimeric transmembrane protein comprised of the signal peptide from For1B (Vidali et al., 2009b) fused to a SNAP-tag followed by the transmembrane domain of For1B fused to mCherry (SNAP-TM-mCherry; Fig. 3 A). We stably transformed the SNAP-TM-mCherry construct into a line that also expresses GFP-β-glucuronidase (GUS) fused to a nuclear localization signal. This line (hereafter referred to as SNAP-TM-mCherry) is similar to wild type (Fig. S2). The nuclear GFP-GUS protein in this line serves as a visual marker allowing identification of actively silencing plants in our transient RNAi assay. In brief, all RNAi constructs contain inverted repeats of GUS sequences fused to inverted repeats of sequences targeting the gene of interest (Bezanilla et al., 2005). Constructs were transformed into protoplasts expressing GFP-GUS and allowed to regenerate for 1 wk, and then actively silencing plants were identified by the absence of nuclear GFP-GUS fluorescence.
Figure 3.
Exocytosis assay demonstrates that For1F, Sec10b, and actin contribute to secretion. (A) Schematic of SNAP-TM-mCherry, the chimeric transmembrane protein used in the secretion assay. See also Fig. S2. (B) Representative images of cells in a 7-d-old SNAP-TM-mCherry plant regenerated from protoplasts with indicated treatments. Below the images are representative line traces of the fluorescence intensity from a cell transect. (C) Quantification of the ratio of plasma membrane (PM) to internal fluorescence intensity. Error bars represent SEM (control, 7; BFA, 7; LatB, 6), and letters above the bars indicate statistical groups with α < 0.05 from an ANOVA analysis. (D) Quantification of the area of 7-d-old plants expressing the indicated RNAi constructs. All data are normalized to the control. Error bars represent SEM (n = 30 plants for each condition), and letters above the bars indicate statistical groups with α < 0.05 from an ANOVA analysis. See also Fig. S1. (E) Representative images of cells from 7-d-old SNAP-TM-mCherry plants silenced with indicated RNAi constructs. Below the images are representative line traces of the fluorescence intensity from a cell transect. (F) Quantification of the ratio of plasma membrane to internal fluorescence intensity. Error bars represent SEM (control RNAi, 6; Sec10a+b-RNAi, 4; For1F-RNAi, 8), and letters above the bars indicate statistical groups with α < 0.05 from an ANOVA analysis.
In the SNAP-TM-mCherry line, the mCherry resides on the cytosolic side of the membrane, clearly labeling the plasma membrane of protonemal cells (Fig. 3, B and E). Because membrane protein delivery depends on the exocytic pathway, interrupting exocytosis should result in lower fluorescence at the plasma membrane and higher internal fluorescence. To identify defects in exocytosis, we took a ratio of the fluorescence intensity at the plasma membrane and the fluorescence intensity inside the cell. As a proof of concept, we transformed plants with a control RNAi construct that only targets the nuclear GFP-GUS and regenerated silenced plants for 4 d. At day 4 we treated plants for 24 h with the exocytosis-inhibiting drug brefeldin A (BFA; Fig. 3 B). To eliminate variations in the intensity measurements between experiments, all data were normalized to the control of that day (Fig. 3 C). BFA treatment dramatically increases the internal signal, presumably because of accumulation of the transmembrane protein in the ER, demonstrating that this assay can quantitatively measure exocytosis. Treatment with latrunculin B (LatB), a drug that depolymerizes the actin cytoskeleton, resulted in about a 50% decrease in the plasma membrane to internal fluorescence ratio, demonstrating that actin is required for efficient exocytosis in P. patens.
To address whether For1F silencing also decreases the intensity ratio, we used the weak For1F silencing construct (Vidali et al., 2009b) that results in plants 37% smaller than control RNAi plants (Fig. 3 D). In addition, we silenced Sec10a and Sec10b, which should reduce exocyst function and also results in smaller plants similar in size to For1F RNAi plants (Fig. 3 D and Fig. S1). We found that the plasma membrane SNAP-TM-mCherry intensity was reduced upon simultaneous silencing of Sec10a and Sec10b, indicating that very little SNAP-TM-mCherry reaches the plasma membrane (Fig. 3 E). Similarly, cells silencing For1F exhibited a reduction in peak values (Fig. 3 F). Collectively, these results demonstrate that efficient exocytosis contributes to plant size, requiring both exocyst and For1F function.
For1F dynamically associates with the Sec6, a component of the exocyst complex
To analyze the intracellular localization of For1F, we took advantage of the For1F-GFP line that was used to demonstrate that For1F is a fusion of Sec10 and formin domains. Using quantitative growth assays, we found that For1F-GFP grows similarly to wild type (Fig. S2), suggesting that For1F-GFP is functional. To test if For1F associates with the exocyst complex, we tagged Sec6 in the For1F-GFP line. We inserted sequences encoding for three tandem mRuby2 molecules in frame with the 3′ end of Sec6. Tagging Sec6 also did not affect plant size (Fig. S2), suggesting that Sec6-3XmRuby2 (hereafter referred to as Sec6-mRuby) is functional. Because Sec6 is the only component of the exocyst complex that is encoded by a single gene copy, every exocyst complex should be tagged with mRuby2.
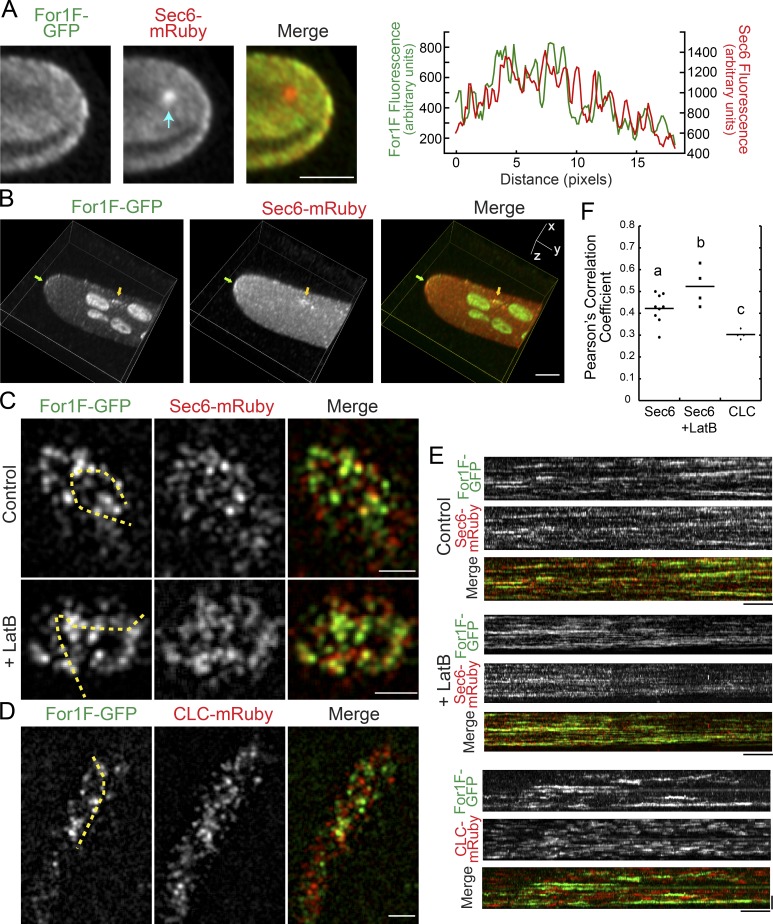
In protonemal filaments, we found that For1F-GFP and Sec6-mRuby localize to the cytoplasm and three distinct membranous areas in the cell: both For1F and Sec6 were weakly enriched at the cell apex (Fig. 4, A and B), localized to dynamic punctae at the cell cortex (Fig. 4 C, Fig. S4 A, and Videos 1 and 2), and decorated the extending cell plate (Fig. 5 and Video 3). We found that For1F and Sec6 partially overlap. For example, at the cell apex, regions that contained high levels of Sec6 often overlapped with regions enriched in For1F, as demonstrated by the line trace in Fig. 4 A. We also observed foci containing both For1F and Sec6 form at the plasma membrane (Fig. 4 B, arrow).
Figure 4.
For1F-GFP dynamically associates with Sec6-mRuby. (A) Laser scanning confocal image of a single focal plane at the apex of a protonemal cell expressing endogenously tagged For1F-GFP and Sec6-mRuby. See also Fig. S2. Arrow indicates intracellular Sec6-mRuby accumulation. Bar, 5 µm. Graph shows the fluorescence intensity from a line trace at the plasma membrane of the cell. (B) Volume view of a z-stack acquired on a laser scanning confocal microscope. Images were deconvolved with NIS-elements (five iterations, type Richardson-Lucy.) For1F-GFP (green) and Sec6-mRuby (red) concentrate at the cell tip (green arrow) and on other regions of the plasma membrane (yellow arrow). Bar, 5 µm. (C) VAEM images of the cortex of cells expressing endogenously tagged For1F-GFP and Sec6-mRuby in the presence (control) or absence (LatB) of actin filaments. Bar, 2 µm. Also see Videos 1 and 4. (D) VAEM images of the cortex of cells expressing For1F-GFP and CLC-mRuby. Bar, 2 µm. Also see Video 6. (E) Kymographs generated from the yellow dashed lines shown in C and D. Bars: (horizontal) 1 min; (vertical) 2 µm. (F) PCCs from kymographs generated by drawing a line through the brightest Sec6-mRuby or CLC-mRuby particles present in the first frame of a 10-min time-lapse acquisition (see Fig. S4). Letters indicate statistical groups with α < 0.05 from a Fisher’s least significant difference ANOVA test.
Figure 5.
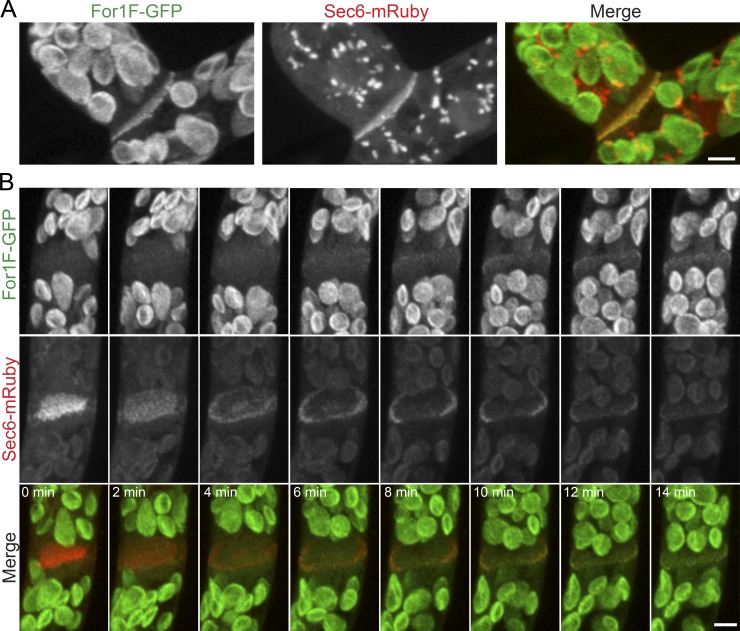
Both For1F-GFP and Sec6-mRuby localize to the developing cell plate during cell division. (A) Images are maximum projections of z-stacks acquired with a laser scanning confocal of a late phragmoplast in a branching cell expressing For1F-GFP and Sec6-mRuby. Sec6-mRuby localizes to prominent intracellular structures, which is particularly evident in branching cells. (B) Images are maximum projections of z-stacks acquired with a laser scanning confocal microscope from a time-lapse image series of a dividing cell. Sec6-mRuby localizes to the phragmoplast throughout cell division, whereas For1F-GFP localizes when the phragmoplast reaches the mother cell wall. Large globular structures in the green channel in A and B are chloroplasts, which autofluoresce under these imaging conditions. Bars, 5 µm. Also see Video 3.
To characterize the cortical population, we used variable-angle epifluorescence microscopy (VAEM) to simultaneously image For1F-GFP and Sec6-mRuby. We found that For1F-GFP and Sec6-mRuby partially overlapped at the cell cortex, similar to what was observed at the cell apex (Fig. 4 C) and to what has been observed for exocyst complex components in A. thaliana (Fendrych et al., 2013). Both For1F and Sec6 cortical particles did not translocate along the membrane, but rather appeared and disappeared from the membrane, which is readily apparent in the kymographs in Fig. 4 E. Interestingly, For1F-GFP particles resided on the membrane for 78 ± 62 s and were often populated with Sec6. However, the Sec6 particles appeared to bind and unbind from the same site on the scale of seconds (Fig. 4 E and Fig. S3). To investigate particle behavior with increased time resolution, we acquired 30-s videos with nearly continuous acquisition. With this time resolution, it was apparent that Sec6 rapidly dissociated from puncta containing both For1F and Sec6, with For1F remaining on the plasma membrane throughout the 30-s-long time-lapse acquisition (Fig. S3 and Video 2). We also observed bright Sec6 puncta accumulate For1F and then dissociate from the cortex (Fig. S3, arrow). In the absence of actin, both For1F and Sec6 particles were more stable (Fig. 4 C, Fig. S3, and Videos 4 and 5).
To investigate the nature of the For1F cortical puncta, we generated a line in the For1F-GFP background that expresses a clathrin light chain (CLC), a marker of endocytic activity (Ito et al., 2012), fused to mRuby2 at its C terminus. Similar to For1F-GFP, CLC-mRuby2 (hereafter referred to as CLC-mRuby) labels cortical dots (Fig. 4 D and Video 6). However, in contrast to dual imaging of For1F and Sec6, For1F puncta mostly do not overlap with CLC puncta. To assess the amount of overlap between For1F-GFP and CLC-mRuby versus Sec6-mRuby, we analyzed the dynamic behavior of individual CLC or Sec6-mRuby particles. We identified CLC or Sec6-mRuby particles and then generated kymographs to analyze their behavior through time (Fig. 4 E and Fig. S3). CLC-mRuby also binds and unbinds without translocating along the cortex, but does not often overlap with For1F and has a significantly shorter residence time (28 ± 13 s) as compared with For1F (78 ± 62 s). We measured Pearson’s correlation coefficient (PCC) of the two signals in the kymographs (Materials and methods and Fig. S4) to determine the degree of overlap with For1F-GFP. We found that For1F and CLC are largely nonoverlapping with a low PCC (Fig. 4 F). The PCC increased when analyzing For1F-GFP and Sec6-mRuby, and is even larger for For1F-GFP and Sec6-mRuby in the absence of actin filaments (Fig. 4 F). These data are consistent with the finding that dissociation of Sec6 from For1F sites requires dynamic actin and suggest that the plasma membrane has distinct domains for endocytosis and exocytosis.
During cell division in both branching and apical cells, we observed Sec6-mRuby and For1F at the developing cell plate (Fig. 5). In addition to the cell plate, we found that Sec6 localizes to intracellular structures (Fig. 4 A, arrow), which are particularly prominent in subapical branching cells (Fig. 5 A). Although both Sec6 and For1F labeled the cell plate, we did observe that Sec6-mRuby marked the developing cell plate at very early stages, before it expanded to reach the cell cortex (Fig. 5 B and Video 3). In contrast, For1F-GFP was detectable only once the expanding cell plate had reached the cortex. These data suggest that different exocyst complexes may form during cell division. Early during cell plate formation, the exocyst may not associate with For1F, suggesting that association with actin may not be required at this stage. However during later stages, when the cell plate must join with the mother cell wall, exocyst complexes linked directly to actin remodeling may predominate.
Actin influences cortical For1F dynamics, and active areas of filament remodeling are associated with For1F
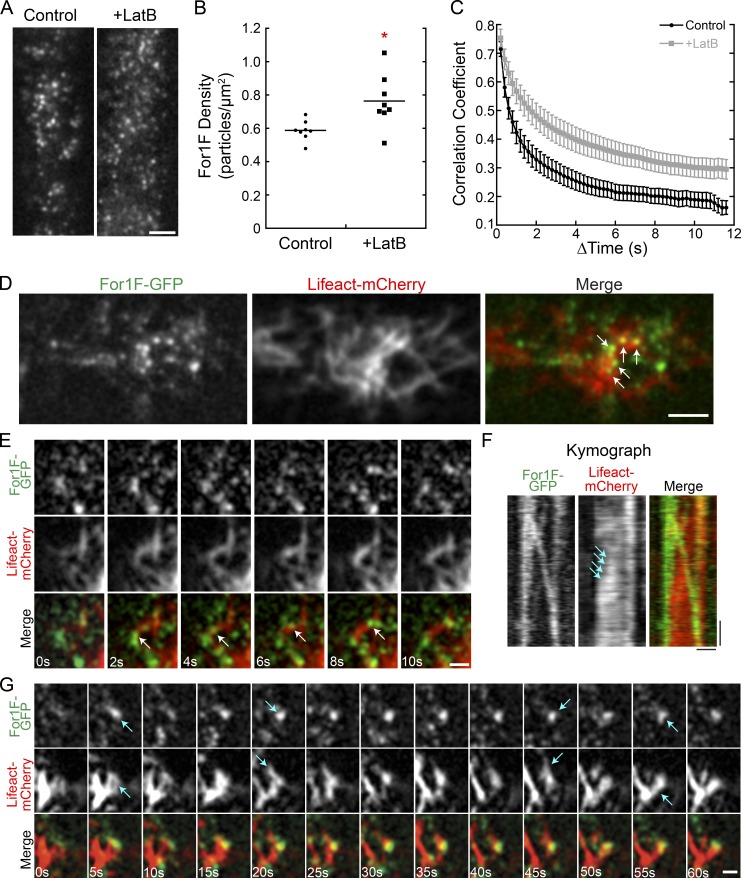
In the absence of actin filaments, we observed that For1F and Sec6 particles remain associated longer at the cell cortex (Fig. 4, C, E, and F; and Videos 4 and 5). This suggests that actin filaments may play a role in dissociating For1F/Sec6 complexes and removing them from the membrane. To quantify how actin influences cortical For1F behavior, we imaged For1F-GFP in the presence and absence of actin filaments (Fig. 6 A and Video 7). We identified all For1F particles in a field, enabling us to determine the particle density. We found that cortical For1F particles were denser in the absence of actin (Fig. 6, A and B), suggesting that For1F targeting to the cell cortex is independent of actin, but removal requires actin filaments. Additionally, time-lapse imaging of For1F-GFP revealed that For1F particles were more dynamic in the presence of actin (Video 7). We quantified global changes in cortical For1F particle distribution by calculating the correlation coefficient of the intensity of the For1F-GFP signal at all pixel locations between time points. This analysis (Vidali et al., 2010) examines the degree of change in the images of For1F-GFP. A smaller change between images results in a shallower decay of the correlation coefficient and indicates decreased dynamics. We found that the correlation coefficient decayed more slowly in the absence of actin, indicating that For1F particles are less dynamic (Fig. 6 C).
Figure 6.
Cortical For1F-GFP dynamics are dampened in the absence of actin, and cortical For1F-GFP associates with areas of active actin remodeling. (A) VAEM images of the cell cortex in a line expressing endogenously tagged For1F-GFP show that in the absence of actin, more For1F remains at the cell cortex. Bar, 2 µm. Also see Video 7. (B) Quantification of For1F-GFP density in the presence and absence of actin filaments. The mean density from 150 frames of a time-lapse acquisition was measured for eight cells for each condition. The asterisk indicates statistical significance with α < 0.05 from an ANOVA analysis. (C) Quantification of cortical For1F-GFP dynamics in the presence or absence of actin filaments. The correlation coefficient between two images was calculated at all possible temporal spacings (time interval). Error bars represent SEM (n = 10 cells). (D) Representative VAEM image from a time-lapse acquisition of the cortex of a cell expressing Lifeact-mCherry and endogenously tagged For1F-GFP. Arrows indicate For1F-GFP at actin filament junctions. Bar, 5 µm. (E) Example where For1F-GFP travels linearly along an existing actin filament (arrows). Bar, 1 µm. Also see Video 8. (F) Kymograph of For1F-GFP linear trajectory from E. Arrows indicate increase in Lifeact-mCherry signal as the particle traverses along a preexisting actin filament. Bars: (horizontal) 1 µm; (vertical) 2 s. (G) Example of a relatively stationary For1F-GFP associated with polymerizing and depolymerizing actin filaments, indicated by arrows. Bar, 1 µm. Also see Video 9.
To determine the kind of actin activity that For1F may associate with, we analyzed the localization of For1F-GFP and actin simultaneously. We expressed LifeAct-mCherry, a validated marker for imaging actin in living protonemal cells (Vidali et al., 2009a; Wu and Bezanilla, 2014), in the For1F-GFP line. We found that a portion of the cortical For1F dots localizes to actin filaments (Fig. 6 D). Often we observed For1F particles at actin filament junctions (Fig. 6 D, arrows). Occasionally, we observed For1F particles move in linear trajectories along a preexisting actin filament (Fig. 6 E, arrows; and Video 8). Kymograph analysis of the trajectory in Fig. 6 E reveals that a For1F particle splits off from an existing particle, and as it moves, the actin signal in its wake increases (Fig. 6 F, arrows), which suggests that this particular particle may be generating an actin filament. However, more often, we observed that relatively stationary cortical For1F particles associated with focal points of polymerizing and depolymerizing filaments (Fig. 6 G, arrows; and Video 9). Together these data suggest that For1F associates with active areas of actin filament remodeling.
The Sec10 and formin domains independently rescue viability
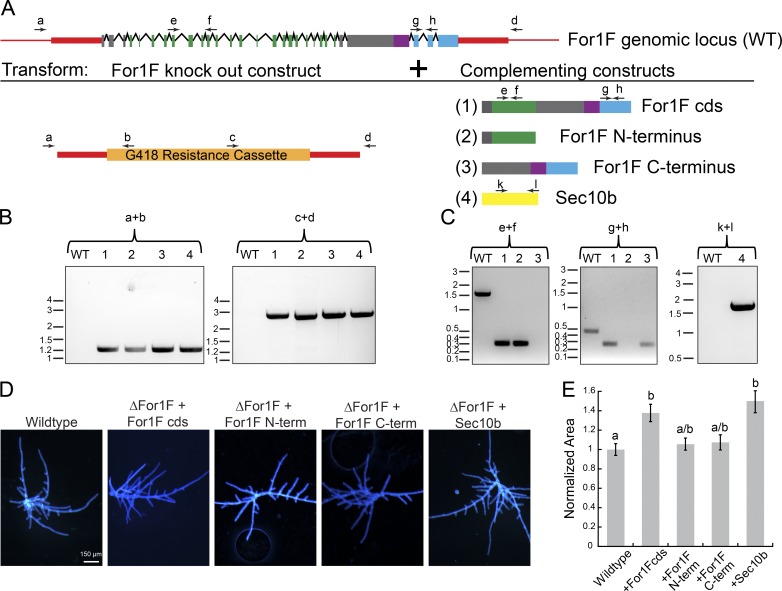
To investigate whether both the Sec10 and formin domains are required for For1F function, we performed a complementation experiment. We reasoned that because For1F is essential, it would only be possible to delete the For1F locus in the presence of a complementing construct. Therefore, we simultaneously transformed a For1F knockout construct with a construct driving expression of full length, the N terminus (Sec10 domain) or the C terminus (formin domain) of For1F from a constitutive promoter (Fig. 7 A). The expression construct was targeted to integrate into a noncoding locus that is aphenotypic when disrupted (Schaefer and Zrÿd, 1997). Resulting lines were genotyped for correct disruption of the For1F locus (Fig. 7 B) and the presence of the expression construct (Fig. 7 C). The untagged full-length protein rescued the disruption of the For1F locus (Fig. 7 D), producing on average larger plants than wild type (Fig. 7 E). Paradoxically, expression of either the N terminus (Sec10 domain) or the C terminus (formin domain) also rescued and quantitatively resembled wild-type plants (Fig. 7, D and E). These data suggest that that the fusion is not essential for viability and protonemal growth. We hypothesize that either domain is sufficient, because in addition to For1F, P. patens has two more Sec10 proteins, and five more class I formins. These additional proteins might transiently interact with the constitutively expressed portions of For1F in the rescue experiments, reconstituting a protein that functions similar to For1F. If this is the case, then it suggests that constitutive expression of another Sec10 could also rescue disruption of the For1F locus. To test this, we disrupted the For1F locus and simultaneously expressed Sec10b (Fig. 7 A). Resulting lines were genotyped for proper disruption of the For1F locus (Fig. 7 B) and the presence of the expression construct (Fig. 7 C). Consistent with our hypothesis, we found that plants expressing Sec10b in the absence of For1F resembled wild type (Fig. 7 D). Interestingly, constitutive expression of Sec10b rescued to the same extent as expressing full-length For1F (Fig. 7 E). Collectively, these data suggest that both Sec10 and formin activities are required, but need not be on the same polypeptide.
Figure 7.
Fusion of the Sec10 and formin domains is not essential for For1F function in protonemal growth. (A) Schematic representation of the For1F genomic locus and the complementation experiments in which the For1F knockout construct was simultaneously transformed with one of the four indicated expression constructs. Red regions indicate genomic noncoding sequence of the For1F locus, and red boxes were sequences incorporated into the knockout construct. For both the For1F gene model and the expressed constructs, green represents sequences with similarity to Sec10; purple, FH1; blue, FH2; and gray boxes, no known sequence similarity. Arrows labeled with letters above the gene model and expression constructs are primers used to perform genotyping. (B) Genotyping for proper integration of the knockout construct into the For1F genomic locus at both 5′ (primers a + b) and 3′ (primers c + d) ends. Wild type (WT) does not produce a PCR product because the wild-type locus cannot anneal to primers b and c, which are specific to the antibiotic resistance cassette. Size in kb is indicated to the left of each gel. (C) Genotyping for the presence of the expression construct. Primers e + f amplify a 0.305-kb region of the coding sequence (cds) of For1F in the Sec10 region. The equivalent genomic region is 1.506 kb. Primers g + h amplify a 0.288-kb region of the For1F cds in the formin region. The equivalent genomic region is 0.471 kb. As expected, wild type only amplifies the genomic fragment in each case, whereas lines that contain the rescue constructs lack the genomic fragment and only amplify the cds fragment. Primers k + l amplify a 1.567-kb region of the Sec10b cds. No amplification for the genomic amplicon is observed in wild type or the Sec10b rescued line because the genomic amplicon is 5.116 kb and did not amplify with the short extension time used to amplify the cds amplicon. Size in kb is indicated to the left of each gel. (D) Images of 7-d-old plants regenerated from protoplasts. Plants were stained with calcofluor. Bar, 150 µm. (E) Plant area was quantified by measuring the area of calcofluor fluorescence. All data are normalized to the wild-type control. Error bars represent SEM (wild type, n = 169; ΔFor1F+For1Fcds, n = 50; ΔFor1F+For1FNterm, n = 48; ΔFor1F+For1FCterm, n = 50; ΔFor1F+Sec10b, n = 50), and letters above the bars indicate statistical groups with α < 0.05 from ANOVA.
Discussion
Here we validate the For1F gene model, demonstrating that the encoded protein is a fusion of two conserved proteins: Sec10, a subunit of the exocyst complex, and a class I formin. We show that For1F is an essential gene that contributes to exocytosis, dynamically associates with the exocyst complex, and localizes to sites of active actin remodeling in vivo.
Consistent with participation in exocytosis, we found that For1F localizes to areas with high membrane turnover. Using imaging, we distinguished between exocytic and endocytic membrane activity and found that cortical For1F does not overlap with endocytic particles labeled with CLC-mRuby. Rather, we found that For1F dynamically associates with Sec6. In fact, we observed an interesting behavior at the membrane where Sec6 particles rapidly associated and dissociated from stable For1F particles. However, in the absence of actin filaments, cortical For1F and Sec6 particles remained associated with For1F for longer and their colocalization increased, suggesting that For1F and Sec6 may be removed from the membrane by an actin-dependent process. For1F may mediate this actin-dependent removal from the membrane, as we found that For1F associates with areas of active actin remodeling. Although we observed very few actin elongation events associated with For1F, it is possible that For1F generates actin filaments into the cytosol, away from the VAEM imaging plane, especially because For1F dynamics are dependent on actin filaments.
The dynamic association between For1F and Sec6 at the cell cortex shows that For1F and the exocyst complex, visualized by Sec6, function and reside at common exocytic sites. Interestingly, partial overlap was also observed for bona fide exocyst subunit components in A. thaliana (Fendrych et al., 2013). Thus, if For1F is part of the exocyst complex, then our data suggest that the exocyst complex in P. patens rapidly assembles and disassembles in cells, which has been observed in Neurospora crassa (Riquelme et al., 2014) and suggested in A. thaliana (Synek et al., 2017). During cell division, Sec6 localizes early, whereas For1F is detectable only in late phragmoplasts. This may be additional evidence for the existence of distinct exocyst complexes: one that functions early, lacking For1F and one that contains For1F and functions late when the phragmoplast reaches the mother cell cortex. The late phragmoplast may need to have For1F to drive actin-mediated removal of the exocyst as the cell plate matures and joins the mother cell wall. Future studies analyzing the localization and dynamics of additional exocyst components will be necessary to study the dynamics of exocyst complex formation at the cell cortex and during cell division.
To date, a fusion of Sec10 and formin has not been identified in other species (Grunt et al., 2008). Indeed, the Simple Modular Architecture Research Tool (Letunic et al., 2015; Letunic and Bork, 2018), which searches for cooccurring domains in public databases, only identified the fusion in P. patens. However, by searching the 1KP (Matasci et al., 2014) and C. purpureus (Szövényi et al., 2015) transcriptomes, and the S. fallax genome (Shaw et al., 2016), we identified several sequences from distantly related mosses that contain both a Sec10 and formin on the same transcript. Using phylogenetic analyses, we showed that class I formins in mosses have undergone several gene duplications that are specific to the moss lineage, with For1F falling squarely into its own clade (bootstrap support = 100%). All transcript sequences that contained both a Sec10 and a formin domain resided specifically within the For1F clade. By performing phylogenetic analyses of Sec10 genes, we found that similarly there were moss-specific gene duplications of Sec10. Interestingly, the Sec10 sequences that are fused to a formin domain also reside in a single, well-supported clade (bootstrap support = 100%), and were found in fewer transcriptomes than other Sec10 homologue sequences. This underrepresentation is likely a result of the inherent 3′ bias in the transcriptomic dataset used to identify the sequences, coupled with the difficulty in reverse transcribing through the highly GC-rich sequence encoding for the FH1 domain, located 3′ to the Sec10 region. Together these data indicate that deep in moss history, after the divergence of the Takakiopsida and the Sphagnopsida, a gene fusion event occurred that generated the For1F gene lineage.
To gain insight into whether the fusion of these two proteins is required for For1F function, we performed a complementation experiment where we simultaneously knocked out the For1F gene and constitutively expressed rescuing constructs. Surprisingly we discovered that the fusion is not essential for viability or protonemal growth, because we could rescue by constitutively expressing either half of For1F or a completely different Sec10 gene. Given that P. patens has two additional Sec10s and five additional class I formins, these paralogues expressed at endogenous levels might transiently interact with the constitutively expressed portions of Sec10 or formin. These transient interactions would reconstitute a protein that functions similar to For1F, suggesting that Sec10 interacts with a class I formin but does not need to be on the same polypeptide for full function, which is consistent with findings that the exocyst subunit Sec3 interacts noncovalently with the formin For3 in fission yeast (Jourdain et al., 2012). Although the fusion of Sec10 and the class I formin is not essential for viability, it may have evolved as a mechanism to more tightly control the expression levels of these proteins that link exocyst function to actin polymerization. In other lineages, these two processes are likely still linked noncovalently, as in fission yeast, but the proteins have remained as independent genes, and other mechanisms may be used to regulate their expression levels. Alternatively, there may be specific developmental transitions in mosses we have not yet uncovered that require the fusion of the two domains. Either way, this fusion gene in P. patens offers a fantastic opportunity to explore the linkage between actin and exocytosis that likely extends to many eukaryotic systems.
Materials and methods
Construct design and generation
cDNA
To generate cDNA for constructs or to identify the 5′ end of For1F, RNA was first isolated from 1-wk-old moss protonemal tissue using the RNeasy plant mini kit (QIAGEN) according to the manufacturer’s recommendations. cDNA was generated using either oligo(dT) for total cDNA, or a gene-specific primer for specific gene fragments as detailed in the RNAi constructs and Expression constructs sections, and SuperScript III Reverse transcription (Invitrogen) following the manufacturer’s protocol. All primers used are referenced by number and listed in Table S1.
Locus-tagging constructs
A four-way in vitro recombination cloning system (Invitrogen) was used to tag the For1F and Sec6 loci. Approximately 1,000 bp at the end of each gene, excluding the stop codon, were amplified with primers in Table S1 to create the 5′ targeting arm, and ∼1,000 bp downstream of the stop codon were amplified with primers in Table S1 to create the 3′ targeting arm. PCR products were transferred into pDONR vectors using a Gateway BP clonase (Invitrogen) reaction following the manufacturer’s instructions. A vector encoding the fluorescent protein (pENT-L5L4-3xmEGFP [For1F locus] or pENT-L5L4-3XmRuby2 [Sec6 locus]) and another vector containing an antibiotic resistance cassette (pENT-R4R3-HYG [For1F locus] or pENT-R4R3-G418 [Sec6 locus]) were also used in the reaction. All four fragments were cloned into the pGEM-gate destination vector using the Gateway LR clonase II plus enzyme. The resulting constructs (For1F-3xGFP and Sec6-3XmRuby) contain the end of the gene (5′ targeting arm), followed by triple GFP/mRuby, an antibiotic selection cassette and the region downstream of the gene (3′ targeting arm). Restriction sites were also engineered into the construct to enable linearization for moss transformation.
Knockout construct
A three-way in vitro recombination cloning system (Invitrogen) was used to generate the For1F knockout construct. Approximately 1,000 bp upstream of the For1F gene was amplified with primers in Table S1 to create the 5′ targeting arm, and ∼1,000 bp downstream of the stop codon was amplified with primers in Table S1 to create the 3′ targeting arm. PCR products were transferred into pDONR vectors using a Gateway BP clonase (Invitrogen) reaction, generating entry clones. The entry clones were combined with a vector containing an antibiotic resistance cassette (pENT-R4R3-G418) and the pGEM-gate destination vector using Gateway LR clonase II plus to generate the knockout construct. Restriction sites were engineered into the construct to enable linearization for moss transformation.
RNAi constructs
RNAi constructs were generated by amplifying a region of sequence from the target gene, generating an entry clone from that amplicon using pENTR/D-topo (Invitrogen), and then performing a Gateway LR clonase recombination reaction to move the target gene sequence into the final RNAi vector, pUGGi (Bezanilla et al., 2005). The following description details generation of the amplicons that were used to make the entry clones.
Two RNAi constructs targeting different regions of the For1F Sec10 domain were generated. The small RNAi construct used primers 1065 and 1039 to amplify a 305-bp region of the For1F cDNA in the Sec10 region. The cDNA template for this amplicon was generated using primer 981. The larger RNAi construct encompassing the first 3,908 bp of For1F was amplified from cDNA generated with primer 1304. We also generated an RNAi construct targeting the For1F 5′ and 3′ untranslated regions. The UTRs were amplified with primers 1458 + 1459 and 1308 + 1327. Both amplicons were cloned into pENTR/D-topo. The 3′UTR region was removed from the backbone vector with a NotI digestion and then cloned into pENT-5′UTR linearized with NotI.
The Sec6 RNAi construct was generated from a PCR amplification of the Sec6 coding sequence with primers 1591 and 1592 off total cDNA. The Sec10ab RNAi construct was generated by amplifying Sec10a and Sec10b off total cDNA with primers 1566 + 1567 and 1568 + 1569, respectively. PCR products were digested with EcoRI, ligated, and then amplified with primers 1566 and 1569. This amplicon, which is a fusion of the two fragments, was then cloned into pENTR/D-topo.
Expression constructs
To generate the SNAP-TM-mCherry construct, we amplified a region of the For1B cDNA that encodes for the first 130 amino acids from total cDNA using primers 1046 and 1047 and cloned this into pENTR/D-Topo. This region encodes the signal peptide (aa 1–30) and the transmembrane domain (aa 142–162). Between sequences encoding for the signal peptide and the transmembrane domain, we used megaprimer PCR (Barik, 1997) to insert sequences encoding for a SNAP tag using primers 1048 and 1045. After the sequence encoding for the transmembrane domain, we used megaprimer PCR (Barik, 1997) to insert sequences encoding mCherry, thereby generating pENT-SNAP-TM-mCherry. Using a Gateway LR clonase-mediated recombination with pTH-UBIgate (Vidali et al., 2007), we generated the expression construct SNAP-TM-mCherry.
Because of the presence of a highly GC-rich region, which could not be reverse transcribed, it was necessary to reconstruct the full-length For1F cDNA as follows. The large exon containing the GC-rich region was amplified from genomic DNA and cloned into pENTR/D-topo to generate pENT-FH1. Next, the region 3′ of the large exon was amplified from total cDNA and cloned into pENTR/D-topo to generate pENT-FH2. To isolate the 5′ region of the For1F cDNA, we first generated cDNA using primer 1304, just upstream of the GC-rich region. Using this cDNA as a template, the Sec10 region was amplified by PCR and cloned into pENTR/D-Topo to generate pENT-Sec10. pENT-FH1 and pENT-FH2 were digested with BanI and AscI. The fragment released from the pENT-FH2 digest was ligated into pENT-FH1. BanI cuts pENT-FH2 twice, resulting in a small BanI fragment that was ligated in subsequently and screened for orientation. This resulted in pENT-FH1FH2. pENT-FH1FH2 and pENT-Sec10 were then digested with NdeI and AscI, and the released fragment of pENT-FH1FH2 was ligated into pENT-Sec10, generating pENT-For1F. Gateway LR clonase-mediated recombination with pTH-UBIgate generated pTH-UBI-For1F.
To generate a construct encoding for the N-terminal region of For1F, we amplified the 5′ region of For1F using pENT-For1F as template with primers 1495 and 1496 incorporating attB1 and attB5r sites. Using a Gateway BP clonase reaction, this amplicon was transferred to pDONR-P1P5r to generate pENT-Nterm. The construct encoding for the C-terminal region of For1F was done in a few steps. First, a 3′ region of the gene was amplified off pENT-For1F with primers 1518 and 1519 to generate a fragment with no stop codon. This was transferred into pDONRP1P5r with a BP reaction generating pENT-CtermNOSTOP. The 3′ end of the gene was amplified off pENT-For1F using primers 1517 and 1306, and transferred into pENTR/D-topo, generating pENT-Cterm. To remove the stop codon and replace the attL2 site in pENT-Cterm with the attR5 site, an EcoRV fragment containing this region was replaced with an EcoRV fragment from pENT-CtermNOSTOP, generating pENT1-Cterm. pENT1-Cterm and pENT-Nterm were then combined with pENT-3XFlag and pTH-UBIgate using Gateway LR clonase II plus to generate pTHUBI-NtermFlag (For1F Nterm) and pTHUBI-CtermFlag (For1F Cterm).
To clone full-length Sec10b, the Sec10b cDNA was amplified from total cDNA with primers 1762 and 1763 and transferred into pENTR/D-topo. A Gateway LR clonase recombination reaction with pTH-UBIgate was done to generate pTH-UBI-Sec10b.
To generate CLC-mRuby–expressing construct, the CLC gene (Pp3c7_10150) was amplified from cDNA isolated from 7-d-old P. patens with primers 2303 and 2302 (Table S1). The PCR fragment was cloned into pDONR-P1P5r with Gateway BP clonase to generate the entry clone CLC-L1R5. The entry clones mRuby2-L5L2 and CLC-L1R5 were cloned into the destination vector pTZUbi-gate with LR clonase II plus to generate pTZUbi-CLC-mRuby2.
Tissue culture and protoplast transformation
Moss tissue culture and protoplast transformations were done as described in Vidali et al. (2007). Isolation of stable transformants was performed as described in Wu et al. (2011). Plant area measurements were taken 6 to 8 d after transformation by imaging chlorophyll autofluorescence as described in Vidali et al. (2007). Quantification of plant area by calcofluor fluorescence was performed as previously described (Wu et al., 2011).
Analysis of the For1F transcript and protein
RT-PCR of For1F
Reverse transcription was performed with primer 982 on RNA isolated from 7-d-old tissue, and PCR was performed with primers 1303 and 1304.
Immunoblotting
Immunoblots of cell extracts were performed using a polyclonal GFP antibody (van Gisbergen et al., 2012). About 100 mg of 7-d-old tissue was ground to a powder using a mortar and pestle in liquid nitrogen and resuspended in 0.9 ml of extraction buffer (50 mM Hepes-NaOH, pH 7.5, 150 mM NaCl, 10 mM EDTA, 1% casein, 1% Triton X-100, 2 mM DTT, 1X Protease Arrest cocktail [G-Biosciences], 20 µg/ml leupeptin, 2 mM phenylmethanesulfonyl fluoride, and 2.5% SDS). Extracts were spun for 5 min at 4°C. 50–100 µl of the supernatant was methanol-precipitated, resolubilized with SDS loading buffer, separated on a 6% SDS-PAGE gel, and transferred to nitrocellulose. Nonspecific sites were blocked with 5% milk in TBS-T (125 mM NaCl, 25 mM Tris-HCl, pH 8, and 0.05% Tween-20). Blots were incubated in GFP primary antibody at a 1:5,000 dilution in 1% BSA TBS-T overnight at 4°C, washed three times in TBS-T, and incubated in secondary antibody (goat anti–rabbit) at a 1:100,000 dilution in TBS-T for 1 h at room temperature. After three washes in TBS-T, the membranes were incubated in 800 µl of SuperSignal West Femto Chemiluminescent Substrate kit (Thermo Fisher Scientific) solution for 5 min. Chemiluminescence was detected using a gel documentation system (BioRad).
Phylogenetic analysis
FH2 and SEC10 DNA sequences were translated into protein and aligned using MAFFT (Katoh and Standley, 2013), as implemented in Geneious (Kearse et al., 2012). The resulting alignments were trimmed to only include regions of reliable homology. Regions of alignment uncertainty were removed using Gblocks (Talavera and Castresana, 2007). Model selection was performed using the Akaike Information Criterion (Akaike, 1974), as implemented on the Datamonkey webserver (Pond and Frost, 2005). Phylogenetic analysis was performed under the ML information criterion using RAxML (Stamatakis, 2014), as implemented on the CIPRES web portal (Miller, M.A., W. Pfeiffer, and T. Schwartz. 2010. Gateway Computing Environments Workshop.). To assess statistical support for phylogenetic relationships, 100 ML rapid bootstrap replicates were performed. Topologies shown represent the most likely trees recovered from each analysis, with thickened branches representing ML bootstrap support ≥85%. Final alignments and trees will be deposited at the dryad data depository.
Exocytosis assay
We stably transformed the SNAP-TM-mCherry construct into a line expressing a nuclear localized GFP-GUS protein. For drug treatment experiments, this line was transformed with a control RNAi plasmid (Bezanilla et al., 2005) that targets GUS. 4 d after transformation, plants were moved to selection, and on day 6, plants were treated with 0.5% DMSO (control), 15 µM BFA, or 25 µM LatB for 24 h. RNAi experiments were performed similarly but without drug treatments. Plants were imaged 7 d after transformation with a spinning disc confocal. Regenerating plants were mounted on slides as described in Vidali et al. (2009a). Slides were imaged on an inverted microscope (model Ti-E; Nikon Instruments Inc.) equipped with a spinning disk head (model CSU-X1; Yokogawa) and a 512 × 512 electron multiplying charge-coupled device camera (iXON; Andor Technology). Images were collected with a 1.4 NA 60× oil immersion objective (Nikon Instruments Inc.) at room temperature. Laser power and exposure times were kept consistent within experiments. Actively silencing plants were identified by the absence of nuclear GFP. No more than one cell per plant was imaged at the medial focal plane.
The SNAP-TM-mCherry signal was quantified by drawing a line scan across the cell through an area that is not nuclear or highly vacuolated in ImageJ. The line scan was plotted, and mean peak values and the mean between peak values were calculated. All values were normalized to the control sample of the same day.
Laser-scanning confocal microscopy
Protonemal tissue was either mounted on a glass slide as previously described in Wu and Bezanilla (2014) or regenerated in custom-made microfluidic imaging chambers (Bascom et al., 2016) and then imaged with a laser-scanning confocal microscope as previously described in Wu and Bezanilla (2014). The image acquisition process was controlled by NIS-Elements AR software (Nikon Instruments Inc.), and images were further processed (including background subtraction and enhanced contrast with standard settings) and quantified with ImageJ.
VAEM
5- to 6-d-old plants regenerated from protoplasts were mounted on slides as previously described in Wu and Bezanilla (2014). Imaging was performed on a Ti-E inverted microscope (Nikon Instruments Inc.) equipped with a mirror-based T-FL-TIRF illuminator (Nikon Instruments Inc.) as described in Wu and Bezanilla (2014).
To quantify the extent of overlap between For1F-GFP and Sec6-mRuby or CLC-mRuby, kymographs were generated from a line segment that transected the majority of the bright cortical Sec6-mRuby or CLC-mRuby particles observed in the first frame of a time-lapse acquisition. PCCs were calculated on the kymographs using the coloc2 plug-in in Fiji. For1F-GFP particle density was quantified using the Track Mate plug-in in Fiji (Tinevez et al., 2017). Correlation coefficient analysis was performed in MatLab (Mathworks) as previously described in Vidali et al. (2010).
Online supplemental material
Fig. S1 shows that silencing Sec10b results in a growth defect, whereas silencing Sec10a has no effect on growth. Fig. S2 shows that For1F-GFP, Sec6-mRuby, and SNAP-TM-mCherry grow similar to wild type. Fig. S3 shows that rapid dissociation of Sec6-mRuby from For1F-GFP cortical spots depends on actin. Fig. S4 describes the method used to analyze the dynamic association of For1F-GFP and Sec6mRuby or CLC-mRuby. Video 1 shows a 10-min time-lapse acquisition of simultaneous VAEM imaging of cortical For1F-GFP and Sec6-mRuby from Fig. 4 C. Video 2 shows a 30-s time-lapse acquisition of simultaneous VAEM imaging of cortical For1F-GFP and Sec6-mRuby from Fig. S3. Video 3 shows For1F-GFP and Sec6-mCherry localization during cell division. Images are maximum projections of a confocal z-stack. Video 4 shows a 10-min time-lapse acquisition of simultaneous VAEM imaging of cortical For1F-GFP and Sec6-mRuby in the presence of LatB from Fig. 4 C. Video 5 shows a 30-s time-lapse acquisition of simultaneous VAEM imaging of cortical For1F-GFP and Sec6-mRuby in the presence of LatB from Fig. S3. Video 6 shows a 10-min time-lapse acquisition of simultaneous VAEM imaging of cortical For1F-GFP and CLC-mRuby from Fig. 4 D. Video 7 shows time-lapse VAEM imaging of cortical For1F-GFP from Fig. 6 A. Video 8 shows time-lapse acquisition of simultaneous VAEM imaging of cortical For1F-GFP and Lifeact-mCherry from Fig. 6 E. Video 9 shows time-lapse acquisition of simultaneous VAEM imaging of cortical For1F-GFP and Lifeact-mCherry from Fig. 6 G. Table S1 lists the primers used in this study.
Supplementary Material
Acknowledgments
We would like to thank C.J. Bascom for technical support.
This work was supported by grants to M. Bezanilla from the National Science Foundation (grant MCB-1330171) and the David and Lucille Packard Foundation, and by grants from the Marine Biologicial Laboratory including the Laura and Arthur Colwin Endowed Summer Research Fellowship, the Burr and Susie Steinbach Fellowship Funds, and the Nikon Fellowship. P.A.C. van Gisbergen was supported by the Gilgut Fellowship from the Plant Biology Graduate Program at the University of Massachusetts Amherst.
The authors declare no competing financial interests.
Author contributions: P.A.C. van Gisbergen and M. Bezanilla conceptualized the project. Methodology was by P.A.C. van Gisbergen, S.-Z. Wu, M.E. Bartlett, and M. Bezanilla. Investigation was by P.A.C. van Gisbergen, S.-Z. Wu, M. Chang, K.A. Pattavina, M.E. Bartlett, and M. Bezanilla. M. Bezanilla wrote the original draft. P.A.C. van Gisbergen, S.-Z. Wu, M.E. Bartlett, and M. Bezanilla reviewed and edited the paper. M. Bezanilla acquired funding and supervised the project.
References
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19:716–723. 10.1109/TAC.1974.1100705 [DOI] [Google Scholar]
- Barik S. 1997. Mutagenesis and gene fusion by megaprimer PCR. Methods Mol. Biol. 67:173–182. [DOI] [PubMed] [Google Scholar]
- Bascom C.S. Jr., Wu S.-Z., Nelson K., Oakey J., and Bezanilla M.. 2016. Long-Term Growth of Moss in Microfluidic Devices Enables Subcellular Studies in Development. Plant Physiol. 172:28–37. 10.1104/pp.16.00879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M., Perroud P.-F., Pan A., Klueh P., and Quatrano R.S.. 2005. An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol (Stuttg). 7:251–257. 10.1055/s-2005-837597 [DOI] [PubMed] [Google Scholar]
- Cole R.A., Synek L., Žárský V., and Fowler J.E.. 2005. SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol. 138:2005–2018. 10.1104/pp.105.062273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrčková F., Grunt M., Bezvoda R., Hála M., Kulich I., Rawat A., and Zárský V.. 2012. Evolution of the land plant exocyst complexes. Front. Plant Sci. 3:159 10.3389/fpls.2012.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler J., Van Poucke K., Karimi M., De Groodt R., Gheysen G., Engler G., and Gheysen G.. 2004. Dynamic cytoskeleton rearrangements in giant cells and syncytia of nematode-infected roots. Plant J. 38:12–26. 10.1111/j.1365-313X.2004.02019.x [DOI] [PubMed] [Google Scholar]
- Elias M., Drdova E., Ziak D., Bavlnka B., Hala M., Cvrckova F., Soukupova H., and Zarsky V.. 2003. The exocyst complex in plants. Cell Biol. Int. 27:199–201. 10.1016/S1065-6995(02)00349-9 [DOI] [PubMed] [Google Scholar]
- Favery B., Chelysheva L.A., Lebris M., Jammes F., Marmagne A., De Almeida-Engler J., Lecomte P., Vaury C., Arkowitz R.A., and Abad P.. 2004. Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell. 16:2529–2540. 10.1105/tpc.104.024372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M., Synek L., Pecenková T., Toupalová H., Cole R., Drdová E., Nebesárová J., Sedinová M., Hála M., Fowler J.E., and Zársky V.. 2010. The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell. 22:3053–3065. 10.1105/tpc.110.074351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M., Synek L., Pecenková T., Drdová E.J., Sekeres J., de Rycke R., Nowack M.K., and Zársky V.. 2013. Visualization of the exocyst complex dynamics at the plasma membrane of Arabidopsis thaliana. Mol. Biol. Cell. 24:510–520. 10.1091/mbc.E12-06-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunt M., Zárský V., and Cvrcková F.. 2008. Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 8:115 10.1186/1471-2148-8-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hála M., Cole R., Synek L., Drdová E., Pecenková T., Nordheim A., Lamkemeyer T., Madlung J., Hochholdinger F., Fowler J.E., and Zárský V.. 2008. An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell. 20:1330–1345. 10.1105/tpc.108.059105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S.B., Marin J., Suleski M., Paymer M., and Kumar S.. 2015. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32:835–845. 10.1093/molbev/msv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J.N., Guérin C., Robert H., Sørensen M.B., Van Damme D., Geelen D., Blanchoin L., and Berger F.. 2005. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7:374–380. 10.1038/ncb1238 [DOI] [PubMed] [Google Scholar]
- Ito E., Fujimoto M., Ebine K., Uemura T., Ueda T., and Nakano A.. 2012. Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J. 69:204–216. 10.1111/j.1365-313X.2011.04782.x [DOI] [PubMed] [Google Scholar]
- Jourdain I., Dooley H.C., and Toda T.. 2012. Fission yeast sec3 bridges the exocyst complex to the actin cytoskeleton. Traffic. 13:1481–1495. 10.1111/j.1600-0854.2012.01408.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., and Standley D.M.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. . 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulich I., Pečenková T., Sekereš J., Smetana O., Fendrych M., Foissner I., Höftberger M., and Zárský V.. 2013. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic. 14:1155–1165. [DOI] [PubMed] [Google Scholar]
- Letunic I., and Bork P.. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., and Bork P.. 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43(D1):D257–D260. 10.1093/nar/gku949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N., Hung L.-H., Yan Z., Carpenter E.J., Wickett N.J., Mirarab S., Nguyen N., Warnow T., Ayyampalayam S., Barker M., et al. . 2014. Data access for the 1,000 Plants (1KP) project. Gigascience. 3:17 10.1186/2047-217X-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S.L.K., and Frost S.D.W.. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 21:2531–2533. 10.1093/bioinformatics/bti320 [DOI] [PubMed] [Google Scholar]
- Riquelme M., Bredeweg E.L., Callejas-Negrete O., Roberson R.W., Ludwig S., Beltrán-Aguilar A., Seiler S., Novick P., and Freitag M.. 2014. The Neurospora crassa exocyst complex tethers Spitzenkörper vesicles to the apical plasma membrane during polarized growth. Mol. Biol. Cell. 25:1312–1326. 10.1091/mbc.E13-06-0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D.G., and Zrÿd J.P.. 1997. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11:1195–1206. 10.1046/j.1365-313X.1997.11061195.x [DOI] [PubMed] [Google Scholar]
- Shaw A.J., Schmutz J., Devos N., Shu S., Carrell A.A., and Weston D.J.. 2016. The sphagnum genome project: A new model for ecological and evolutionary genomics. In Advances in Botanical Research. Academic Press, Cambridge, MA: 167–187. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R.G., Westphal L., Rosahl S., McDowell J.M., and Trujillo M.. 2013. The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal. Behav. 8:e27421 10.4161/psb.27421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek L., Schlager N., Eliás M., Quentin M., Hauser M.-T., and Zárský V.. 2006. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 48:54–72. 10.1111/j.1365-313X.2006.02854.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek L., Vukašinović N., Kulich I., Hála M., Aldorfová K., Fendrych M., and Žárský V.. 2017. EXO70C2 Is a Key Regulatory Factor for Optimal Tip Growth of Pollen. Plant Physiol. 174:223–240. 10.1104/pp.16.01282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szövényi P., Perroud P.-F., Symeonidi A., Stevenson S., Quatrano R.S., Rensing S.A., Cuming A.C., and McDaniel S.F.. 2015. De novo assembly and comparative analysis of the Ceratodon purpureus transcriptome. Mol. Ecol. Resour. 15:203–215. 10.1111/1755-0998.12284 [DOI] [PubMed] [Google Scholar]
- Talavera G., and Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Tinevez J.-Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., and Eliceiri K.W.. 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115:80–90. [DOI] [PubMed] [Google Scholar]
- van Gisbergen P.A.C., and Bezanilla M.. 2013. Plant formins: membrane anchors for actin polymerization. Trends Cell Biol. 23:227–233. 10.1016/j.tcb.2012.12.001 [DOI] [PubMed] [Google Scholar]
- van Gisbergen P.A.C., Li M., Wu S.-Z., and Bezanilla M.. 2012. Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J. Cell Biol. 198:235–250. 10.1083/jcb.201112085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Augustine R.C., Kleinman K.P., and Bezanilla M.. 2007. Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell. 19:3705–3722. 10.1105/tpc.107.053413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., and Bezanilla M.. 2009a Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS One. 4:e5744 10.1371/journal.pone.0005744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., van Gisbergen P.A.C., Guérin C., Franco P., Li M., Burkart G.M., Augustine R.C., Blanchoin L., and Bezanilla M.. 2009b Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA. 106:13341–13346. 10.1073/pnas.0901170106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Burkart G.M., Augustine R.C., Kerdavid E., Tüzel E., and Bezanilla M.. 2010. Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell. 22:1868–1882. 10.1105/tpc.109.073288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T.-J., Hochholdinger F., Sauer M., Bruce W., and Schnable P.S.. 2005. The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol. 138:1637–1643. 10.1104/pp.105.062174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.-Z., and Bezanilla M.. 2014. Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. eLife. 3:e03498 10.7554/eLife.03498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Tan X., Wu C., Cao K., Li Y., and Bao Y.. 2013. Regulation of cytokinesis by exocyst subunit SEC6 and KEULE in Arabidopsis thaliana. Mol. Plant. 6:1863–1876. 10.1093/mp/sst082 [DOI] [PubMed] [Google Scholar]
- Wu S.-Z., Ritchie J.A., Pan A.-H., Quatrano R.S., and Bezanilla M.. 2011. Myosin VIII regulates protonemal patterning and developmental timing in the moss Physcomitrella patens. Mol. Plant. 4:909–921. 10.1093/mp/ssr068 [DOI] [PubMed] [Google Scholar]
- Zimmer A.D., Lang D., Buchta K., Rombauts S., Nishiyama T., Hasebe M., Van de Peer Y., Rensing S.A., and Reski R.. 2013. Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics. 14:498 10.1186/1471-2164-14-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X., Zhang J., Zhang Y., Hsu S.-C., Zhou D., and Guo W.. 2006. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat. Cell Biol. 8:1383–1388. 10.1038/ncb1505 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.