The mitotic spindle checkpoint protects cells against chromosome missegregation. Petsalaki et al. show that Chmp4c, a protein in the ESCRT complex, has a surprising role in regulating spindle checkpoint signaling by promoting localization of the RZZ complex to unattached kinetochores.
Abstract
The mitotic spindle checkpoint delays anaphase onset in the presence of unattached kinetochores, and efficient checkpoint signaling requires kinetochore localization of the Rod–ZW10–Zwilch (RZZ) complex. In the present study, we show that human Chmp4c, a protein involved in membrane remodeling, localizes to kinetochores in prometaphase but is reduced in chromosomes aligned at the metaphase plate. Chmp4c promotes stable kinetochore–microtubule attachments and is required for proper mitotic progression, faithful chromosome alignment, and segregation. Depletion of Chmp4c diminishes localization of RZZ and Mad1-Mad2 checkpoint proteins to prometaphase kinetochores and impairs mitotic arrest when microtubules are depolymerized by nocodazole. Furthermore, Chmp4c binds to ZW10 through a small C-terminal region, and constitutive Chmp4c kinetochore targeting causes a ZW10-dependent checkpoint metaphase arrest. In addition, Chmp4c spindle functions do not require endosomal sorting complex required for transport–dependent membrane remodeling. These results show that Chmp4c regulates the mitotic spindle checkpoint by promoting localization of the RZZ complex to unattached kinetochores.
Introduction
The mitotic spindle checkpoint delays anaphase onset until all kinetochores are stably attached to microtubules emanating from opposing spindle poles (Lara-Gonzalez et al., 2012). This mechanism provides more time for the cell to correct erroneous kinetochore–microtubule attachments and avoid possible chromosome missegregation and aneuploidy. Conserved components of the spindle checkpoint pathway include the Mad (Mad1, Mad2, and BubR1) and the Bub (Bub1 and Bub3) proteins. Accumulation of Mad and Bub proteins on unattached kinetochores is essential to prevent activation of the anaphase-promoting complex/cyclosome and delay mitotic exit, but the molecular details are incompletely understood (Funabiki and Wynne, 2013; London and Biggins, 2014b).
Kinetochore localization of the Mad1-Mad2 heterotetramer is a major determinant of the spindle checkpoint activity (Maldonado and Kapoor, 2011; Kuijt et al., 2014). Bub1 interacts with Mad1 to activate the spindle checkpoint in budding yeast and Caenorhabditis elegans (London and Biggins, 2014a; Moyle et al., 2014). However, Mad1-Mad2 recruitment to kinetochores also requires the activity of the Rod–ZW10–Zwilch (RZZ) complex in metazoans (Buffin et al., 2005; Kops et al., 2005; Caldas et al., 2015; Silió et al., 2015). The RZZ complex is mainly present in metazoans (Karess, 2005; Vleugel et al., 2012), and cells lacking RZZ proteins are spindle checkpoint deficient (Basto et al., 2000; Chan et al., 2000; Williams et al., 2003).
Despite its important role in spindle checkpoint signaling, how RZZ is targeted to kinetochores remains unclear. The Knl1-Zwint1 protein complex was initially proposed to recruit RZZ to kinetochores, and RZZ localization may be regulated by Aurora B–dependent phosphorylation of Zwint1 (Wang et al., 2004; Kops et al., 2005; Kasuboski et al., 2011; Varma et al., 2013). Recently, it was shown that Knl1-Bub1 is also required for localization of the RZZ to kinetochores in HeLa cells independently of Zwint1 (Caldas et al., 2015). Furthermore, there is evidence of a Knl1- and Bub1-independent mechanism for RZZ and Mad1-Mad2 kinetochore targeting in HeLa and human retinal pigment epithelial cells suggesting additional factors are involved (Caldas et al., 2015; Silió et al., 2015). After chromosome biorientation at the metaphase plate, dynein transports RZZ and Mad1-Mad2 from kinetochores to the spindle poles via microtubules in a process called stripping, to silence the spindle checkpoint (Howell et al., 2001; Wojcik et al., 2001).
The endosomal sorting complex required for transport (ESCRT) machinery is required for biogenesis of multivesicular endosomes, viral budding, cytokinetic abscission, and sealing of the newly formed nuclear envelope during cell division (Hurley, 2015; Campsteijn et al., 2016). These functions require membrane remodeling and scission and involve assembly of cytosolic components of the ESCRT-III subcomplex into helical filaments that constrict and cut membrane invaginations in conjunction with the ATPase Vps4 that is essential for disassembly of ESCRT-III spirals and is also likely to provide energy input for membrane neck constriction (Adell et al., 2014). Chmp4c, an ESCRT-III component, is one of three human orthologues (Chmp4a, Chmp4b, and Chmp4c) of the yeast protein Snf7 that is involved in multivesicular body sorting (Hurley, 2015; Campsteijn et al., 2016). In late cytokinesis, Chmp4c is phosphorylated at S210, and this phosphorylation is required for proper Chmp4c localization to the midbody to delay abscission (Capalbo et al., 2012; Carlton et al., 2012). Furthermore, Chmp4c-recruitment to the midbody requires ALIX, another ESCRT protein that binds to Chmp4 components (McCullough et al., 2008; Christ et al., 2016). However, a role for Chmp4c independent of membrane-directed activities has not been previously reported. In the present study, we show that Chmp4c regulates the mitotic spindle checkpoint by promoting localization of the RZZ complex to unattached kinetochores.
Results
Chmp4c localizes to kinetochores in prometaphase
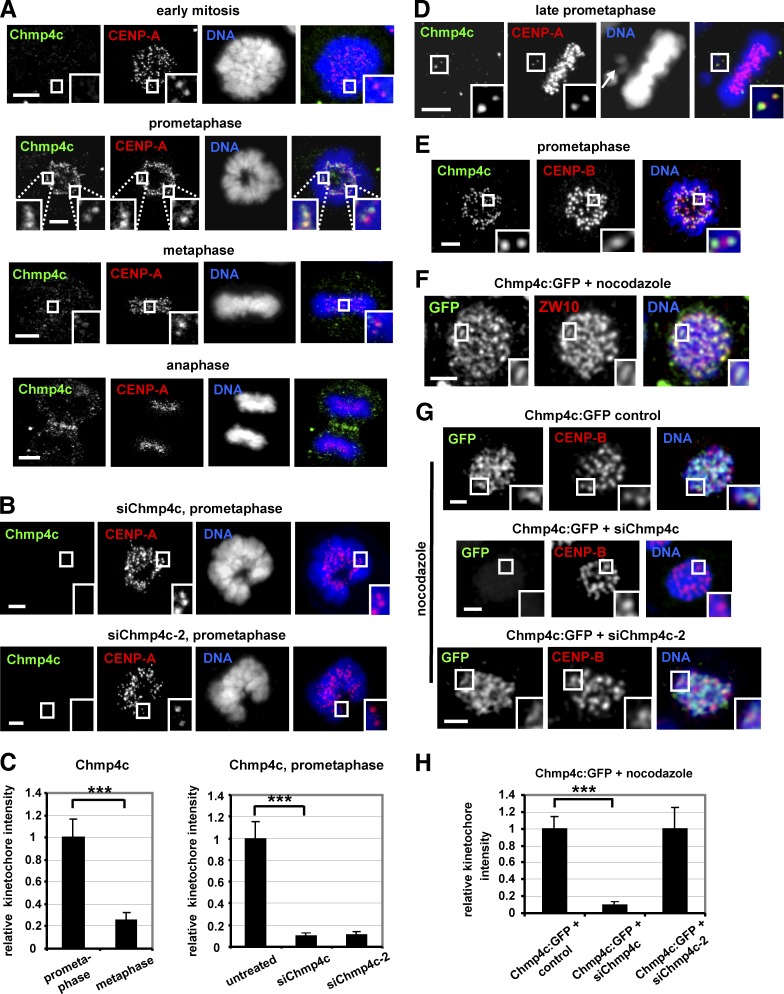
Chmp4c localizes to kinetochores in prometaphase and to the midzone in anaphase in human colon carcinoma BE cells (Fig. 1 A). However, Chmp4c kinetochore staining was undetectable in early mitosis, before condensed chromosomes form a ring-like morphology (Petsalaki and Zachos, 2014), and was diminished in chromosomes aligned at the metaphase plate compared with prometaphase cells or unaligned chromosomes (Fig. 1, A–D). The Chmp4c antibody used does not recognize Chmp4a or Chmp4b (Petsalaki and Zachos, 2016), and depletion of endogenous Chmp4c by two different siRNAs located in the open reading frame (siChmp4c) or the 3′-untranslated region (siChmp4c-2) reduced Chmp4c kinetochore staining compared with controls (Fig. 1, A–C; and Fig. S1 A), confirming the signal was specific. Also, Chmp4c was outside the centromere as revealed by staining for centromere protein (CENP)-B (Fig. 1 E). In cells treated with 3.32 µM nocodazole, which depolymerizes microtubules (Petsalaki and Zachos, 2014), the endogenous Chmp4c or Chmp4c fused to GFP (Chmp4c:GFP) colocalized with ZW10 and formed crescent-like shapes at kinetochores suggesting that Chmp4c is part of the kinetochore outer domain (Fig. 1, F and G; and Fig. S1 B; Hoffman et al., 2001). Furthermore, Chmp4c:GFP kinetochore staining was reduced after transfection with siChmp4c but not with siChmp4c-2 (Fig. 1, G and H). Also, Chmp4c staining at kinetochores was diminished in cells treated with 1 µM taxol, which stabilizes microtubules (Petsalaki and Zachos, 2014), compared with nocodazole-treated cells (Fig. S1, B and C). These results suggest that Chmp4c functions at prometaphase kinetochores in the absence of spindle poisons or cells treated with nocodazole but not taxol.
Figure 1.
Chmp4c localizes to kinetochores. (A–C) Cells were untreated or transfected with two different Chmp4c siRNAs (siChmp4c and siChmp4c-2). Relative green/red fluorescence intensity is shown, and values in prometaphase or untreated were set to one. (D) Example of a late-prometaphase cell exhibiting an unaligned chromosome (indicated by an arrow) and an acentric chromatin fragment. Chmp4c localizes to unaligned kinetochores (insets) but is diminished from kinetochores at the metaphase plate. (E) Localization of Chmp4c and CENP-B. (F) Localization of Chmp4c:GFP in cells treated with nocodazole for 4 h. (G and H) Localization of Chmp4c:GFP, resistant to degradation by siChmp4c-2, in cells transfected with negative siRNA (control), siChmp4c, or siChmp4c-2 and treated with nocodazole for 4 h. Relative green/red fluorescence intensity is shown, and values in the control were set to one. n > 200 kinetochores, 20 cells (C and H) from three independent experiments. Error bars show the SD. ***, P < 0.001 compared with the control. Statistical significant differences were determined by ANOVA and Student’s t test. Insets show 1.7× magnification of kinetochores. Bars, 5 µm.
Chmp4c-depletion induces chromosome alignment and segregation defects
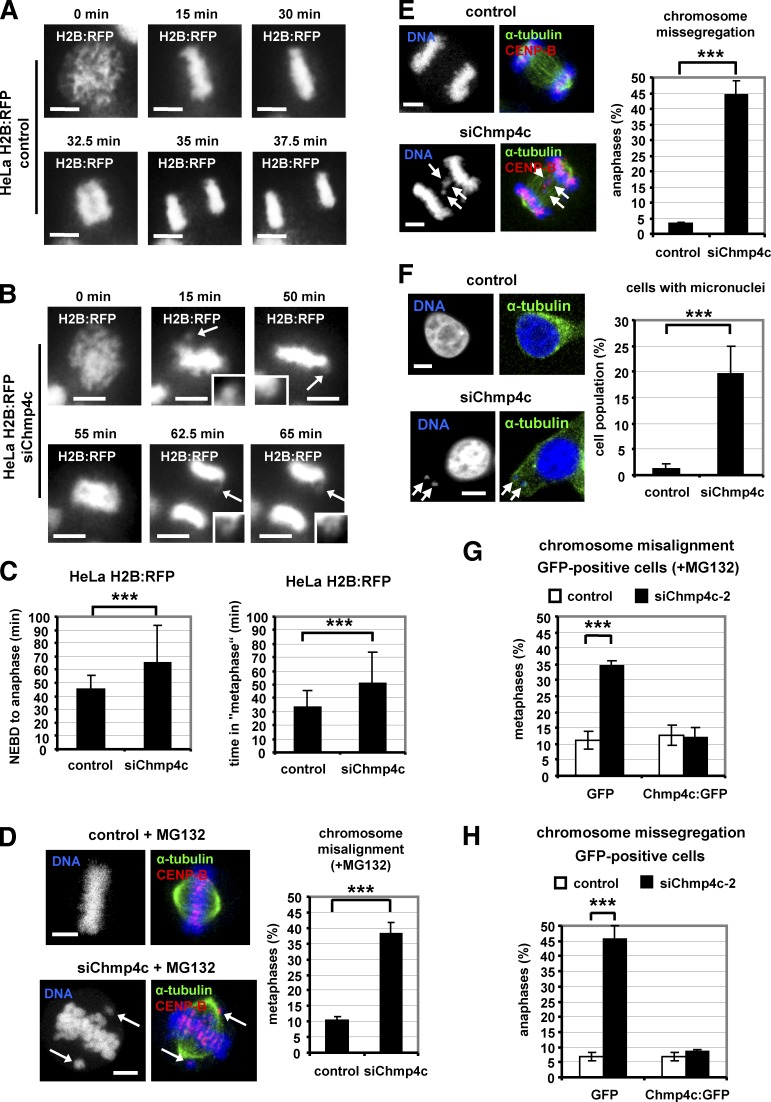
To investigate Chmp4c functions, HeLa cells stably expressing H2B:RFP were examined by live-cell imaging. Live-imaging with the use of fluorescence microscopy showed that 20 of 46 (43%) Chmp4c-deficient cells entered anaphase with misaligned chromosomes and divided with missegregated chromosomes compared with 1 of 32 (3%) controls in these movies (Fig. 2, A and B; and Videos 1 and 2). Also, live-imaging with the use of phase contrast showed that Chmp4c-depleted HeLa H2B:RFP cells entered anaphase in 65 ± 28 min (n = 173) after nuclear envelope breakdown (NEBD) compared with 45 ± 10 min in control cells (n = 137, ∼45% delay compared with controls), and most of the delay occurred in a metaphase-like state (Fig. 2 C; Fig. S1, D and E; and Videos 3 and 4). Similarly, Chmp4c-depleted BE cells entered anaphase in 70 ± 22 min (n = 53) after NEBD compared with 47 ± 12 min in controls (n = 52, ∼49% delay compared with controls; Fig. S1, F and G; and Videos 5 and 6). However, Chmp4c-deficient cells exhibited a similar rate of mitotic entry compared with controls (Fig. S1 H). These results show that Chmp4c-depletion delays anaphase onset.
Figure 2.
Chmp4c-depletion induces chromosome misalignment and missegregation. (A and B) Time-lapse fluorescence images of HeLa cells stably expressing H2B:RFP and transfected with negative (control) or Chmp4c siRNA (siChmp4c). Time is from the start of chromatin condensation. Arrows indicate misaligned or missegregated chromosomes (magnified in insets). Related to Videos 1 and 2. (C) Time from NEBD to anaphase onset or in a metaphase-like state (“metaphase”), calculated from time-lapse movies as in Fig. S1. Control: n = 137; siChmp4c: n = 173. (D) Chromosome misalignment. Cells were transfected as in A and treated with MG132 for 1 h. (E and F) Chromosome missegregation and frequency of interphase cells with micronuclei. Arrows indicate misaligned, missegregated chromosomes or micronuclei. n = 300 cells (D–F) from three independent experiments. (G and H) Cells expressing GFP or Chmp4c:GFP resistant to degradation by Chmp4c-2 siRNA (siChmp4c-2) were transfected with negative siRNA (control) or siChmp4c-2 in the presence of MG132 for 1 h (G) or without MG132 (H). n = 90 cells from three independent experiments. Error bars show the SD. ***, P < 0.001 compared with the control. Student’s t test was used. Bars, 5 µm.
In fixed samples, Chmp4c-deficient BE or HeLa cells treated with the proteasome inhibitor MG132 to block progression of mitosis beyond metaphase (Petsalaki and Zachos, 2014) exhibited increased chromosome misalignment compared with controls (Fig. 2 D and Fig. S2 A). Furthermore, Chmp4c-depletion was associated with increased chromosome missegregation and micronuclei formation compared with controls (Fig. 2, E and F; and Fig. S2 B). Expression of Chmp4c:GFP, which is resistant to degradation by siChmp4c-2 but not GFP alone, rescued chromosome alignment and segregation defects after depletion of the endogenous Chmp4c compared with controls (Fig. 2, G and H; Fig. S1 A; and Fig. S2 C). Chmp4c:GFP was expressed at approximate levels ninefold higher than the endogenous protein in control cells (Fig. S1 A) but exhibited correct localization to kinetochores (Fig. 1 G) and the midzone (Fig. S2 C) and correct localization and function at the midbody (Petsalaki and Zachos, 2016). These results show that Chmp4c is required for optimal mitotic progression, chromosome alignment, and segregation in human cells.
Chmp4c promotes stable kinetochore–microtubule attachments
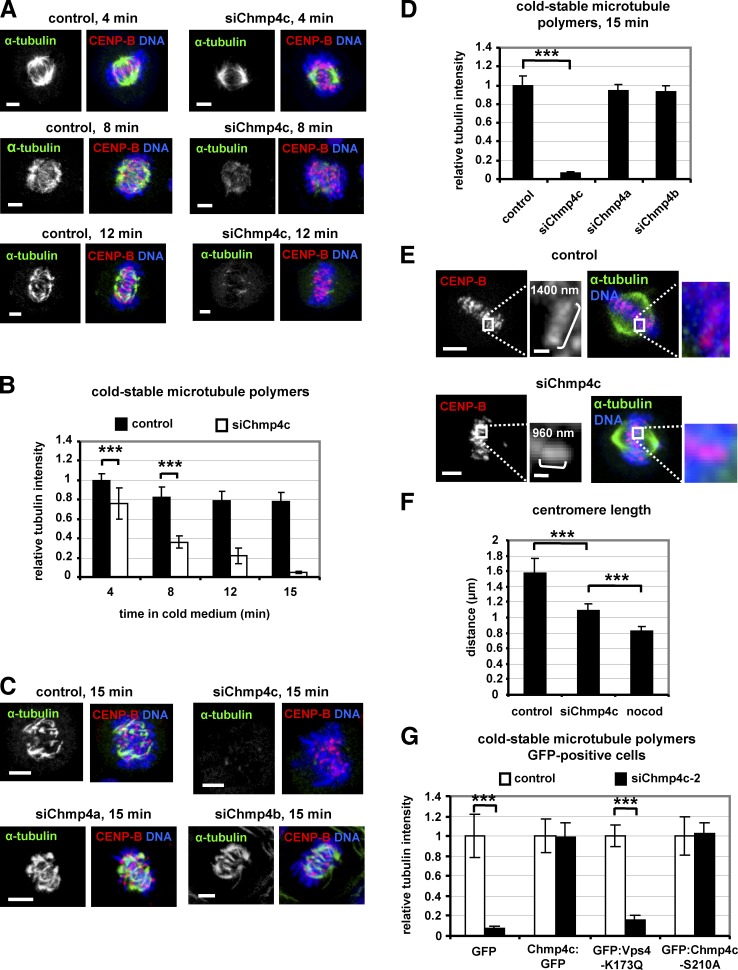
Metaphase-like delay and improper chromosome alignment may be the result of unstable kinetochore–microtubule attachments (Gaitanos et al., 2009). Chmp4c-deficient cells exhibited reduced cold-stable microtubule polymers (Sundin et al., 2011) compared with controls, suggesting that Chmp4c is required for robust kinetochore–microtubule attachments (Fig. 3, A and B; and Fig. S2 D). Please note that, depending on how long cells were incubated in cold medium, cold-stable microtubules could also be detected in Chmp4c-deficient cells but were always found less often than in controls. However, depletion of closely related ESCRT-III subunits Chmp4a or Chmp4b by siRNA did not reduce cold-stable microtubules compared with controls, and Chmp4a:GFP or Chmp4b:GFP was undetected at kinetochores in prometaphase cells, suggesting that the mitotic functions described earlier in this section are specific for the Chmp4c isoform (Fig. 3, C and D; and Fig. S2, E and F). Stable kinetochore–microtubule attachments promote strong pulling forces that cause spindle shortening and centromere stretching in metaphase (DeLuca et al., 2002; Emanuele and Stukenberg, 2007). Metaphase-like spindles were ∼20% longer from pole to pole in Chmp4c-deficient BE cells (P < 0.001) and 40% longer in Chmp4c-deficient HeLa cells compared with controls (Fig. S2 G). Furthermore, the average centromere length in Chmp4c-deficient BE cells was 1.09 µm compared with 1.58 µm in controls and 0.83-µm resting centromere distance in cells treated with a concentration of nocodazole (3.32 µM) that depolymerizes all kinetochore–microtubules (Fig. 3, E and F; Petsalaki and Zachos, 2014). These results suggest that pulling forces from kinetochore–microtubules are reduced in Chmp4c-deficient cells compared with controls. Expression of resistant WT Chmp4c:GFP or mutant GFP:Chmp4c-S210A, which is defective for the abscission checkpoint (Capalbo et al., 2012; Petsalaki and Zachos, 2016) but not GFP alone, rescued cold-stable microtubule polymers in GFP-positive cells after depletion of the endogenous Chmp4c by siChmp4c-2 compared with controls (Fig. 3 G and Fig. S3 A). Furthermore, Chmp4c was required for cold-stable microtubule polymers in GFP-positive cells expressing a dominant negative mutant Vps4 protein fused to GFP (GFP:Vps4-K173Q), which inhibits ESCRT-dependent membrane remodeling (Babst et al., 1998; Morita et al., 2007; Hurley, 2015) and abscission (Petsalaki and Zachos, 2016; Fig. 3 G and Fig. S3 B).
Figure 3.
Chmp4c-depletion reduces cold-stable microtubule polymers. (A–D) Cells transfected with negative siRNA (control), Chmp4c siRNA (siChmp4c), Chmp4a siRNA (siChmp4a), or Chmp4b siRNA (siChmp4b) were treated with ice-cold medium for the indicated times. (E) Centromere length in metaphase-like spindles. Insets show magnified centromeres, and values indicate the centromere length. Bars: (inset) 0.5 µm. (F) Quantification of centromere length from cells as in E or incubated with nocodazole (nocod) for 4 h. Control: n = 291 centromeres; 20 cells; siChmp4c: n = 697 centromeres, 26 cells; nocodazole: n = 200 centromeres, 20 cells from three independent experiments. (G) Cells expressing GFP, WT or S210A mutant Chmp4c:GFP resistant to degradation by siChmp4c-2, or GFP:Vps4-K173Q were treated with ice-cold medium for 15 min. Mean tubulin intensity is shown, and values in the controls were set to one. n = 90 cells (B, D, and G) from three independent experiments. Error bars show the SD. ***, P < 0.001 compared with the control. Statistically significant differences were determined by ANOVA and Student’s t test. Bars, 5 µm.
Chmp4c-S210 was not phosphorylated at kinetochores in prometaphase cells (Fig. S3 C). Also, GFP-positive cells expressing GFP:Chmp4c-S210A or GFP:Vps4-K173Q exhibited reduced frequency of cells at midbody stage compared with untreated or cells expressing WT Chmp4c:GFP (controls) despite a similar percentage of cells in prometaphase (Fig. S3 D). These results suggest that expression of Chmp4c-S210A or Vps4-K173Q does not prevent mitotic entry but causes cells to disassemble their midbodies more rapidly than controls, thus confirming that Vps4-K173Q and Chmp4c-S210A disrupt ESCRT functions under the experimental conditions used in our study. We propose that Chmp4c promotes stable kinetochore–microtubule attachments independently of its role in membrane remodeling.
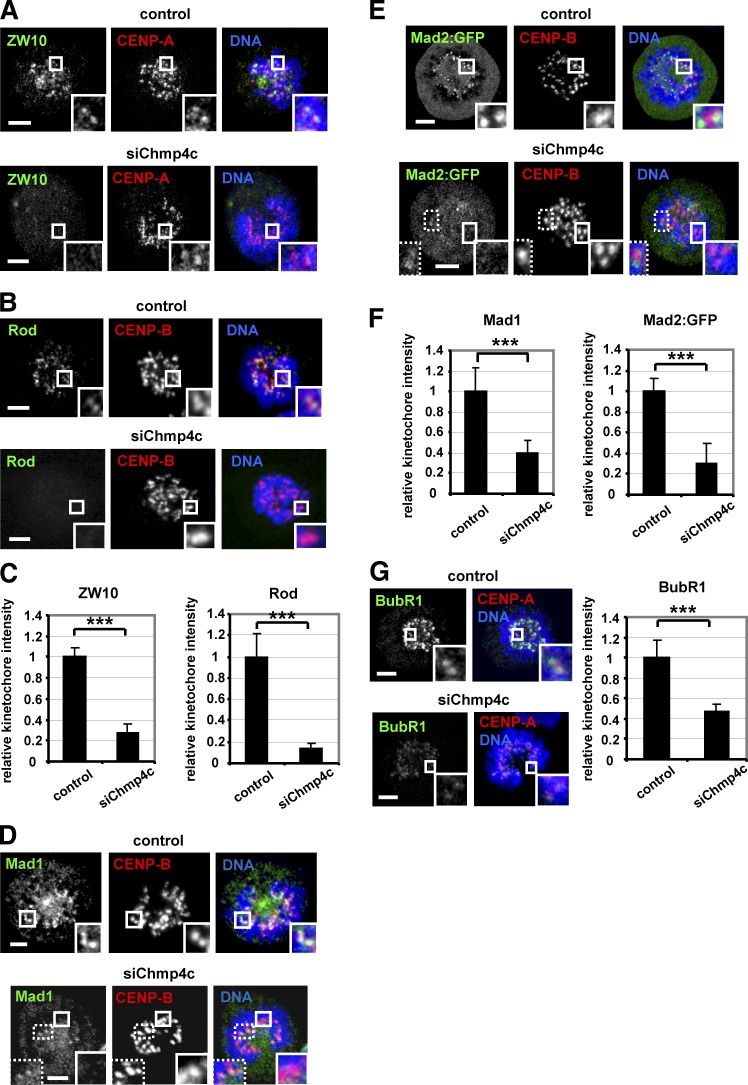
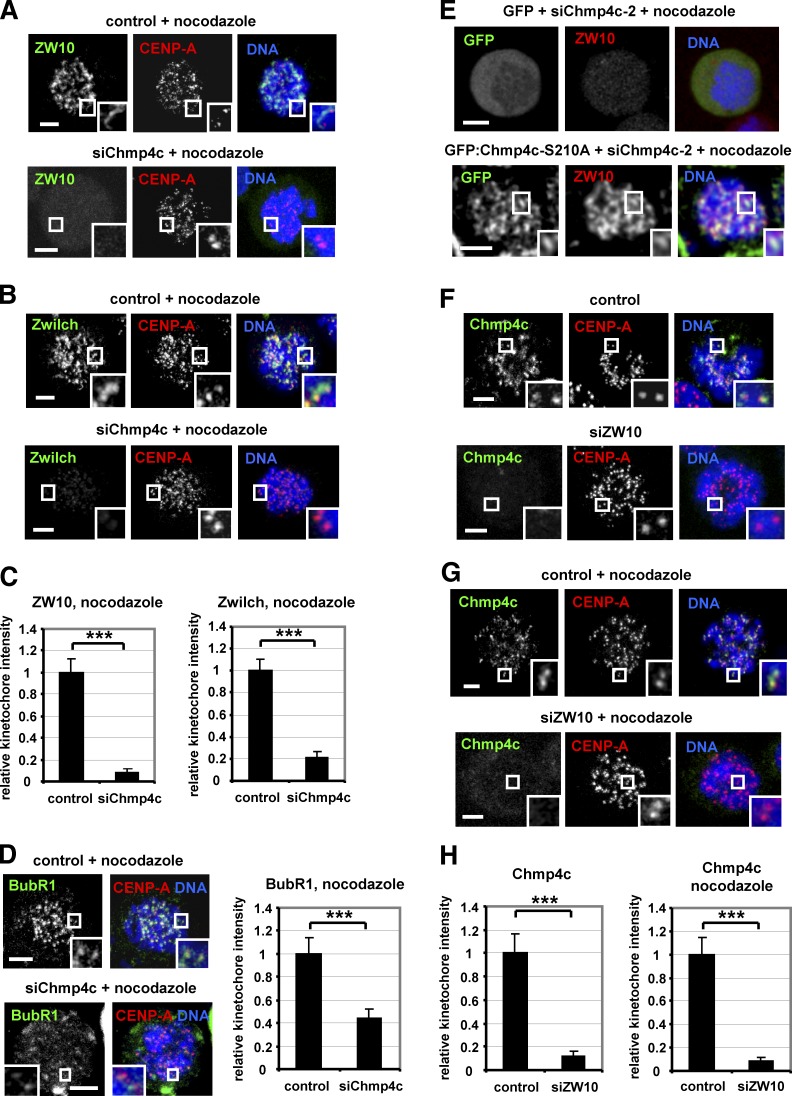
Chmp4c is required for optimal localization of spindle checkpoint proteins to kinetochores
Spindle checkpoint defects can promote chromosome missegregation. In the absence of spindle poisons, depletion of Chmp4c diminished localization of ZW10, Rod, Mad1, and Mad2:GFP to prometaphase kinetochores compared with control cells (Fig. 4, A–F). ZW10, Rod, Mad1, Mad2:GFP, BubR1, or Zwilch protein levels per se were not affected by Chmp4c-depletion (Fig. S3, E–G). Aurora B kinase promotes recruitment of ZW10 to kinetochores (Kasuboski et al., 2011). However, Chmp4c-deficient cells exhibited similar levels of total Aurora B at centromeres or phosphorylated Aurora B-S331 at kinetochores, a marker of Aurora B activation (Petsalaki et al., 2011), compared with controls (Fig. S4, A–C). Also, BubR1 kinetochore levels were reduced but remained detectable in Chmp4c-deficient compared with control cells (Fig. 4 G). These results suggest that Chmp4c is required for optimal spindle checkpoint signaling.
Figure 4.
Chmp4c is required for optimal localization of spindle checkpoint proteins to prometaphase kinetochores. Localization of ZW10 and Rod (A–C), Mad1 and Mad2:GFP (D–F), and BubR1 (G) in cells transfected with negative siRNA (control) or Chmp4c siRNA (siChmp4c). Relative green/red fluorescence intensity is shown, and values in the control were set to one. n > 200 kinetochores, 20 cells from three independent experiments. Error bars show the SD. ***, P < 0.001 compared with control. Student’s t test was used. In some Chmp4c-depleted cells, a few (typically two to six) prometaphase kinetochores exhibited detectable Mad1 or Mad2:GFP staining and are shown in insets with dotted lines (D and E). Insets show 1.7× magnification of kinetochores. Bars, 5 µm.
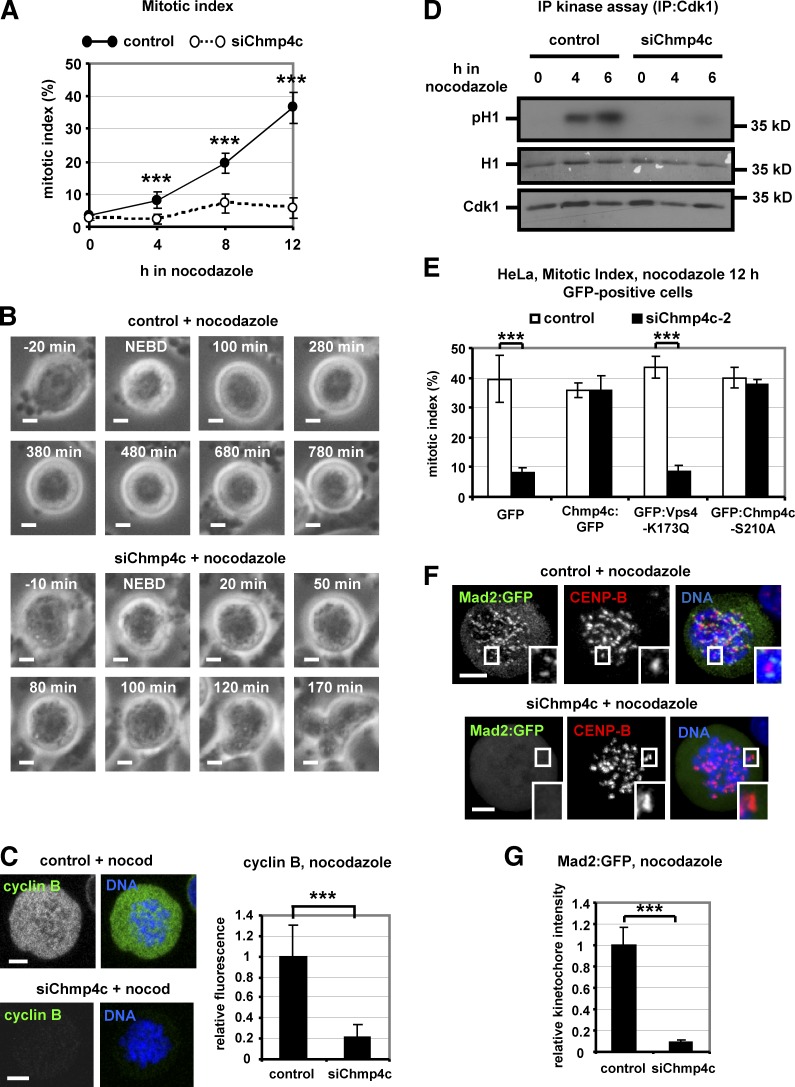
Chmp4c-deficient cells exit mitosis after microtubule depolymerization
To further investigate a role for Chmp4c in the spindle checkpoint, cells were incubated with 3.32 µM nocodazole to completely depolymerize kinetochore–microtubules (Petsalaki and Zachos, 2014). Chmp4c-depleted cells exhibited reduced mitotic index after prolonged nocodazole treatment compared with controls (Fig. 5 A and Fig. S4 D). Time-lapse analysis of BE cells for up to 12 h in nocodazole with the use of phase contrast showed that Chmp4c-depleted and control cells entered mitosis with similar kinetics as determined by NEBD and chromosome condensation (Fig. S4 E). However, all control cells that entered mitosis (n = 76) remained arrested in mitosis with condensed chromosomes for the duration of the experiment, whereas 76 of 85 (89%) mitotic Chmp4c-deficient cells decondensed their chromosomes after 109 ± 70 min (n = 76; Fig. 5 B and Videos 7 and 8). Also, Chmp4c-deficient and control cells entered mitosis with similar kinetics in the presence of 1 µM taxol, which stabilizes microtubules (Petsalaki and Zachos, 2014) and remained arrested in mitosis for the duration of the experiment, in agreement with our findings that Chmp4c does not localize to kinetochores in taxol-treated cells (Fig. S4 F). Consistent with mitotic exit in the presence of nocodazole, Chmp4c-deficient cells degraded cyclin B prematurely and failed to sustain high levels of Cdk1-associated kinase activity compared with controls (Fig. 5, C and D). Expression of resistant WT Chmp4c:GFP or mutant GFP:Chmp4c-S210A, which is defective for the abscission checkpoint, but not GFP alone, rescued mitotic accumulation in GFP-positive cells depleted of endogenous Chmp4c compared with controls (Fig. 5 E). Furthermore, Chmp4c was required for mitotic accumulation in cells expressing the GFP:Vps4-K173Q mutant protein that inhibits ESCRT-dependent membrane remodeling (Fig. 5 E; Babst et al., 1998; Morita et al., 2007). We propose that Chmp4c is required for mitotic arrest when kinetochores are unattached by nocodazole-treatment and that this function of Chmp4c is independent of its roles in membrane-directed activities.
Figure 5.
Chmp4c-deficient cells exit mitosis in nocodazole. (A) Mitotic index analysis in cells transfected with negative siRNA (control) or Chmp4c siRNA (siChmp4c) and treated with nocodazole. n = 300 cells per time point and per treatment, from three independent experiments. (B) Time-lapse analysis of BE cells transfected as in A and treated with nocodazole. Phase-contrast images of a control cell arrested with condensed chromatin (top) or a Chmp4c-deficient cell that exited mitosis, decondensed its chromatin and became multinucleated (bottom). Time is from NEBD. Related to Videos 7 and 8. (C) Cyclin B fluorescence in cells transfected as in A and treated with nocodazole (nocod) for 4 h. Mean cyclin B intensity is shown, and values in the control were set to one. n = 100 cells from three independent experiments. (D) Cdk1-associated histone H1 phosphorylation (pH1; top), ponceau staining of histone H1 (middle), and Western blot analysis of immunoprecipitated (IP) Cdk1 (bottom). (E) Mitotic index analysis. Cells expressing GFP, WT or S210A mutant Chmp4c:GFP resistant to degradation by Chmp4c-2 siRNA (siChmp4c-2), or GFP:Vps4-K173Q were transfected with negative siRNA (control) or siChmp4c-2 and treated with nocodazole for 12 h. n = 300 cells from three independent experiments. (F and G) Localization of Mad2:GFP in cells treated as in C. Relative green/red fluorescence intensity is shown, and values in the control were set to one. n > 200 kinetochores, 20 cells from three independent experiments. Error bars show the SD. ***, P < 0.001 compared with the control. Student’s t test was used. Insets in F show 1.7× magnification of kinetochores. Bars, 5 µm.
Chmp4c and ZW10 localize to kinetochores in an interdependent manner
Depletion of Chmp4c in cells treated with nocodazole and MG132 to inhibit mitotic exit diminished localization of Mad2:GFP, ZW10, and Zwilch to kinetochores compared with controls (Fig. 5, F and G; and Fig. 6, A–C). BubR1 was also reduced from kinetochores by ∼55% in Chmp4c-deficient cells compared with controls (Fig. 6 D). Expression of GFP:Chmp4c-S210A, resistant to degradation by siChmp4c-2, rescued localization of ZW10 to kinetochores in GFP-positive cells depleted of the endogenous Chmp4c compared with cells expressing GFP only (Fig. 6 E), further supporting that Chmp4c spindle checkpoint functions are independent of ESCRT membrane functions. However, Chmp4c-deficient or control cells exhibited similar levels of ZW10 at kinetochores in the presence of taxol (Fig. S4 G). Also, depletion of ZW10 impaired localization of Chmp4c to prometaphase kinetochores in the absence of spindle drugs or in the presence of nocodazole (Fig. 6, F–H; and Fig. S4 H), thus showing that Chmp4c and ZW10 promote each other’s presence at kinetochores.
Figure 6.
Chmp4c and ZW10 localize to kinetochores in an interdependent manner. (A–D) Localization of ZW10, Zwilch, and BubR1. Cells were transfected with negative siRNA (control) or Chmp4c siRNA (siChmp4c) and treated with nocodazole for 4 h. (E) Cells expressing GFP or GFP: Chmp4c-S210A resistant to degradation by Chmp4c-2 siRNA (siChmp4c-2) were transfected with siChmp4c-2 and treated with nocodazole for 4 h. 20 cells from two independent experiments were analyzed. (F–H) Chmp4c localization. Cells were transfected with negative siRNA (control) or ZW10 siRNA (siZW10) in the absence or presence of nocodazole for 4 h. Relative green/red fluorescence intensity is shown, and values in the control were set to one. n > 200 kinetochores, 20 cells (C, D, and H) from three independent experiments. Insets show 1.7× magnification of kinetochores. Error bars show the SD. ***, P < 0.001 compared with the control. Student’s t test was used. Bars, 5 µm.
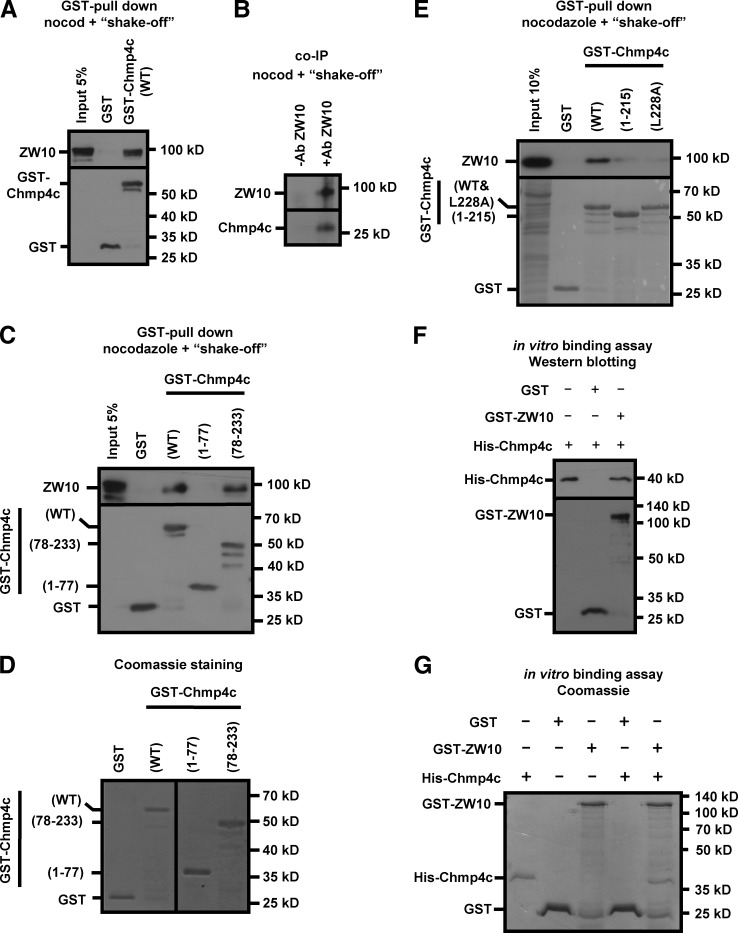
Chmp4c binds to ZW10
GST-Chmp4c associated with ZW10 in pull-down experiments (Fig. 7 A). Also, the endogenous Chmp4c coimmunoprecipitated with ZW10 in mitotic-enriched cell extracts (Fig. 7 B). Mapping of the ZW10 binding site on Chmp4c showed that the 78–233–amino acid region of human Chmp4c was required and sufficient for ZW10-binding (Fig. 7, C and D). The C-terminal sequence of Chmp4c (amino acids 216–233) contains an amphipathic helix that mediates interaction with the Chmp4c binding partner ALIX (McCullough et al., 2008). To investigate whether the ALIX-binding region of Chmp4c is required for binding to ZW10, we generated a GST-Chmp4c (1–215) truncation mutant lacking the ALIX-binding domain of Chmp4c or a GST-Chmp4c-L228A protein harboring mutation of Leucine-228, inside the Chmp4 recognition helix, to Alanine (McCullough et al., 2008). Both these proteins exhibited reduced binding to ZW10 in pull-down experiments compared with the WT GST-Chmp4c (Fig. 7 E). Also, purified His-tagged Chmp4c interacted with GST-ZW10 but not with GST in vitro, as determined by Western blotting or Coomassie staining of the gel in pull-down experiments (Fig. 7, F and G). Collectively, we propose that Chmp4c directly binds to ZW10 at kinetochores through a small C-terminal region containing the ALIX binding domain. In addition, Chmp4c associated with ZW10 in untreated cell extracts and colocalized with ZW10 in perinuclear compartments in interphase cells (Fig. S5, A and B; Civril et al., 2010).
Figure 7.
Chmp4c binds to ZW10. (A–C) GST-pull downs (A and C) and coimmunoprecipitation (coIP; B) from mitosis-enriched cells. ZW10, GST, or Chmp4c were detected by Western blotting. Ab, antibody; nocod, nocodazole. (D) Coomassie staining of GST proteins. (E) Western blot analysis (top) and ponceau staining (bottom) of GST pull-downs from mitosis-enriched cells. Amino acid numbers of Chmp4c fragments or mutations are shown in parentheses. (F) GST pull-downs with the use of purified proteins. 0.5 µg His-Chmp4c was incubated with 1 µg GST or GST-ZW10. His (top) or GST (bottom) was detected by Western blotting. His-Chmp4c input was 50%. (G) Coomassie-stained gel of GST pull-downs with the use of purified proteins. 1 µg His-Chmp4c was incubated with 2 µg GST or 1 µg GST-ZW10. His-Chmp4c input was 50% and GST or GST-ZW10 inputs were 100%. Ab, antibody; nocod, nocodazole.
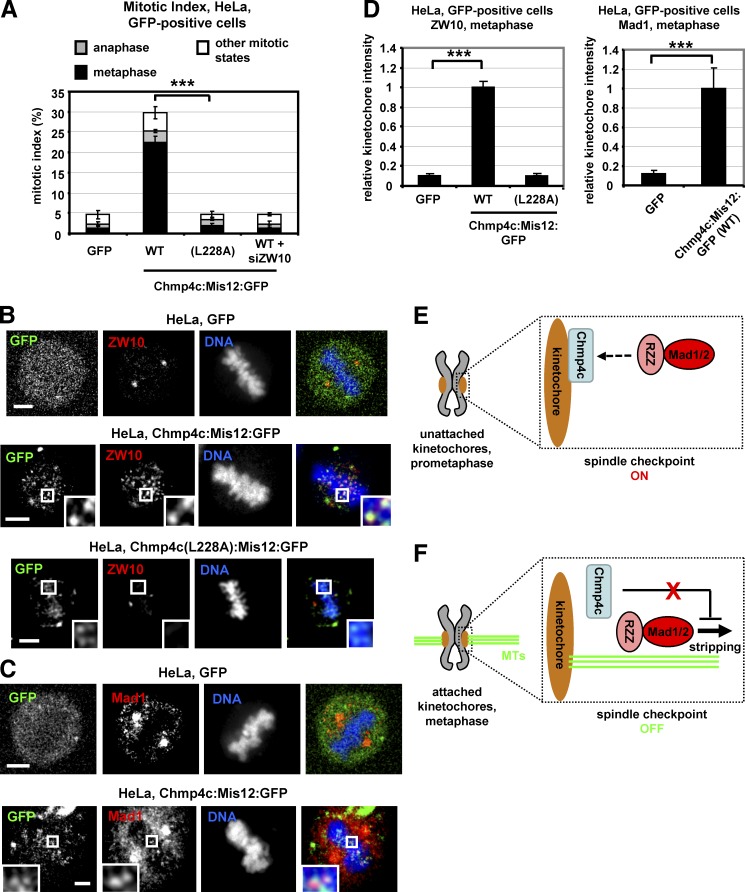
Constitutive Chmp4c kinetochore targeting induces a checkpoint metaphase arrest
Spindle checkpoint proteins, such as the RZZ complex and Mad1-Mad2, are reduced from kinetochores after chromosome alignment (Kops and Shah, 2012). We therefore investigated whether tethering Chmp4c to kinetochores by expressing the constitutively kinetochore localized Chmp4c:Mis12:GFP dissociates chromosome alignment from checkpoint signaling (Maldonado and Kapoor, 2011; Ballister et al., 2014). Expression of full-length (WT) Chmp4c fused to Mis12:GFP increased the mitotic index of GFP-positive cells compared with GFP-only controls, and ∼76% of Chmp4c:Mis12:GFP mitotic cells were in metaphase (Fig. 8 A). Also, in cells expressing Chmp4c:Mis12:GFP, localization of ZW10, Mad1, and Zwilch to kinetochores in the metaphase plate was increased compared with GFP-only, indicating a checkpoint arrest (Fig. 8, B–D; and Fig. S5 C). In contrast, cells expressing the mutant Chmp4c-L228A, which does not bind to ZW10, fused to Mis12:GFP exhibited reduced localization of ZW10 to metaphase plate kinetochores, and the mitotic index was significantly lower compared with cells expressing the WT Chmp4c:Mis12:GFP (Fig. 8, A, B, and D). Furthermore, depletion of ZW10 by siRNA prevented mitotic accumulation in cells expressing WT Chmp4c:Mis12:GFP (Fig. 8 A). These data show that forced localization of Chmp4c to kinetochores induces a metaphase arrest that is dependent on ZW10 kinetochore binding. This arrest is likely not due to unstable kinetochore–microtubule attachments because cold-stable microtubule bundles and chromosome alignment in cells expressing Chmp4c:Mis12:GFP were similar with GFP-controls (Fig. S5, D–G). We propose that Chmp4c kinetochore localization dictates spindle checkpoint activity.
Figure 8.
Expression of Chmp4c:Mis12:GFP induces accumulation of metaphase cells. (A) Cells were transfected with GFP, WT, or mutant (L228A) Chmp4c fused to Mis12:GFP in the absence or presence of ZW10 siRNA (siZW10) and analyzed for mitotic index and phenotypes at 24 h after transfection. GFP, tubulin, and DNA staining were used to determine mitotic index and fraction of cells in metaphase, anaphase, or all other mitotic states. n = 300 cells per condition from three independent experiments. (B–D) Localization of ZW10 or Mad1 at metaphase plate in cells treated as in A. Relative red fluorescence intensity is shown, and values in WT Chmp4c:Mis12:GFP were set to one. n > 100 kinetochores, 10 cells from three independent experiments. Error bars show the SD. ***, P < 0.001 compared with the control. Statistically significant differences were determined by ANOVA and Student’s t test. Insets in B and C show 1.7× magnification of kinetochores. Bars, 5 µm. (E and F) Model for the role of Chmp4c in the spindle checkpoint. MTs, microtubules; dashed arrow, RZZ binding to Chmp4c. See text for details.
Discussion
Chmp4c, a protein of the ESCRT complex, delays abscission in late cytokinesis; however, a role for Chmp4c in the spindle checkpoint has not been previously reported. Here, we show that human Chmp4c localizes to prometaphase kinetochores in the absence of spindle poisons or after treatment of cells with a concentration of nocodazole that completely depolymerizes spindle microtubules (Petsalaki and Zachos, 2014). However, Chmp4c is reduced from kinetochores in chromosomes aligned at the metaphase plate or in prometaphase cells treated with taxol, which stabilizes microtubules. Chmp4c is required for optimal chromosome alignment and segregation in the absence of spindle poisons and for mitotic arrest when kinetochores are unattached by nocodazole-treatment. Furthermore, Chmp4c binds to ZW10 in cell extracts and in vitro and promotes localization of the RZZ and Mad1-Mad2 complexes to prometaphase kinetochores. We propose that Chmp4c acts as a loading factor for the RZZ to unattached kinetochores. These results are consistent with recent findings that RZZ can localize to kinetochores independently of Knl1 and Bub1 in human cells (Caldas et al., 2015; Silió et al., 2015). However, Chmp4c is dispensable for ZW10 kinetochore localization and mitotic arrest in cells treated with taxol.
We also show that constitutive Chmp4c kinetochore targeting induces a ZW10-dependent checkpoint metaphase arrest suggesting that Chmp4c kinetochore localization prevents dynein-dependent removal of the RZZ and Mad1-Mad2 from attached kinetochores. Chmp4c could inhibit dynein function or make the attachment of RZZ/Mad1 to kinetochores so tight that it cannot be disrupted by dynein. Recently CENP-I, a member of the constitutive centromeric-associated network, was also reported to oppose dynein-mediated stripping by an unidentified mechanism (Matson and Stukenberg, 2014).
On the basis of those findings, we propose the following model for the role of Chmp4c in the spindle checkpoint: In prometaphase (Fig. 8 E), Chmp4c localizes to unattached kinetochores and promotes localization of the RZZ and Mad1-Mad2 to kinetochores to activate the spindle checkpoint. Upon formation of bipolar kinetochore–microtubule attachments in metaphase (Fig. 8 F), Chmp4c dissociates from kinetochores leading to dynein-dependent stripping of RZZ and Mad1-Mad2 and spindle checkpoint silencing.
Our model may appear at odds with delayed anaphase onset in Chmp4c-deficient cells in the absence of spindle drugs, which suggests an active checkpoint. Furthermore, how does RZZ localize to kinetochores in the absence of Chmp4c in cells treated with taxol (Famulski and Chan, 2007)? One possibility is that RZZ makes physical contacts with Knl1-Bub1 and/or Knl1-Zwint1 complexes independently of Chmp4c (Wang et al., 2004; Varma et al., 2013; Caldas et al., 2015; Zhang et al., 2015). Because Chmp4c is essential for RZZ kinetochore targeting when microtubules are completely depolymerized by nocodazole, we speculate that delivery of an additional factor, such as a protein or a posttranslational modification, by kinetochore–microtubules is required for the RZZ localization in the absence of spindle poisons or in taxol. For example, the Aurora B catalytic activity is enhanced by microtubule binding at kinetochores (Matson and Stukenberg, 2014). Thus, one possibility is that, in the presence of kinetochore–microtubules, activated Aurora B phosphorylates Zwint1 at kinetochores to promote RZZ kinetochore targeting (Kasuboski et al., 2011). Such alternative mechanisms may allow the spindle checkpoint to respond to more than one type of defect ensuring a higher level of chromosome segregation fidelity. Eventually however, Chmp4c-deficient cells fail to sustain the anaphase delay and enter anaphase with misaligned chromosomes.
Chmp4c is also required for stable kinetochore–microtubule attachments. Because RZZ regulates formation of Ndc80-mediated microtubule end-on attachments through an incompletely understood mechanism (Gassmann et al., 2008; Cheerambathur et al., 2013), one possibility is that misregulated RZZ localization results in destabilized kinetochore–microtubules in Chmp4c-deficient cells. Alternatively, Chmp4c may directly bind and stabilize kinetochore–microtubules or regulate the KMN protein network that mediates kinetochore–microtubule attachments (Varma and Salmon, 2012). Further experiments are required to explore these possibilities.
Chmp4c also interacts with ZW10 in interphase cells. In light of the role of ZW10 in vesicle trafficking, our results suggest that the Chmp4c-ZW10 interaction is also important for cellular functions outside the kinetochore and support a crosstalk between membrane trafficking and the mitotic spindle checkpoint (Hirose et al., 2004; Civril et al., 2010). Chmp4a and Chmp4b also harbor Alix-binding motifs in their C-terminal ends (McCullough et al., 2008). It is therefore possible that additional Chmp4 proteins interact with ZW10 in membrane compartments. In addition, it was recently shown that Chmp proteins participate in mitotic spindle disassembly and nuclear envelope resealing in telophase (Olmos et al., 2015; Vietri et al., 2015). Although a role for Chmp4c in these processes has not been established, animal cells may use Chmp proteins to help coordinate chromosome alignment and segregation with the late stages of cell division to ensure the ordered coupling of these mitotic processes. In conclusion, our study describes the identification of a novel kinetochore component that regulates spindle checkpoint signaling to ensure accurate chromosome segregation in human cells.
Materials and methods
Antibodies
Mouse monoclonal antibodies against GST (B-14), Rod (43-K), and CENP-B (F-4; Fig. 1 E and Fig. S4 G) and rabbit polyclonal antibodies against CENP-B (H-65), Cyclin B1 (H-433), His (H-15), and GFP (sc-8334) were obtained from Santa Cruz Biotechnology. Antibodies against Mad1 (ab5783, mouse monoclonal), BubR1 (ab4637, rabbit polyclonal, used for Western blotting in Fig. S3 G), and Chmp4c (ab155668, rabbit polyclonal) were obtained from Abcam. Mouse monoclonal antibodies against CENP-A (3–19; GTX13939) and Hec1 (9G3.23; GTX70268) were obtained from Genetex, and mouse monoclonal antibodies against α-tubulin (DM1A) and actin (AC-40) were obtained from Sigma-Aldrich. Anti-pS331 is a rabbit polyclonal antiserum raised against phosphorylated S331 of human Aurora-B (Petsalaki et al., 2011). A sheep polyclonal antibody against human BubR1 (used for immunofluorescence) was from S. Taylor (University of Manchester, Manchester, England, UK), and rabbit polyclonal antibodies against human ZW10 and Zwilch were from A. Musacchio (Max Planck Institute of Molecular Physiology, Dortmund, Germany). Anti–pChmp4c rabbit polyclonal antiserum against phosphorylated S210, S214, and S215 of human Chmp4c was a gift from P.P. D’Avino (University of Cambridge, Cambridge, England, UK; Capalbo et al., 2012).
Plasmids and cloning
Plasmids pCR3.1 GFP-EXN/chmp4c and pCR3.1 GFP-EXN/chmp4b encoding human Chmp4c or Chmp4b fused to GFP, respectively, were from P. Bieniasz and T. Zang (The Aaron Diamond AIDS Research Center, New York, NY; Jouvenet et al., 2011). pCMVGFP:Chmp4a encoding human Chmp4a fused to GFP was from P.P. D’Avino (Capalbo et al., 2012). Plasmid pEGFP-vps4-K173Q encoding human Vps4 harboring the Lysine 173 to Glutamine (K173Q) point mutation fused to EGFP into pEGFP-C1 vector (Clontech) was a gift from W. Sundquist (University of Utah, Salt Lake City, UT; Morita et al., 2007). To generate pGEX4T1/hChmp4c vectors encoding WT or truncated forms of GST-tagged human Chmp4c, Chmp4c cDNA sequences were amplified by PCR by using pCR3.1 GFP-EXN/chmp4c as the template and cloned into the pGEX4T1 vector (GE Healthcare) as BamHI–XhoI fragments. For pET28/hChmp4c vector encoding His-tagged human Chmp4c, full length Chmp4c cDNA was amplified by PCR by using pCR3.1 GFP-EXN/chmp4c as the template and cloned into the pET28a(+) vector (EMD Millipore) with XhoI–EcoRI. To generate the pcDNA4/GFP/Mis12/hChmp4c vector coding for N-terminal fusion of WT human Chmp4c to GFP:Mis12, the Chmp4c sequence was amplified by PCR and cloned into the pcDNA4/GFP/Mis12 vector (a gift from G. Kops, University Medical Centre Utrecht, Utrecht, Netherlands) as XhoI–EcoRI fragment. To generate plasmid pGEX4T1/hZW10 coding for full length GST-tagged human ZW10, the ZW10 cDNA was amplified by PCR by using pACEBac1/ZW10 (a gift from A. Musacchio) as the template and cloned into the pGEX4T1 vector with BamHI–SalI. All plasmids were completely sequenced.
siRNA sequences
Negative siRNAs (a pool of four different siRNAs: 5′-UAAGGCUAUGAAGAGAUAC-3′, 5′-AUGUAUUGGCCUGUAUUAG-3′, 5′-AUGAACGUGAAUUGCUCAA-3′, and 5′-UGGUUUACAUGUCGACUAA-3′) were from Thermo Scientific. Human Chmp4c located in the open reading frame (5′-GCUUGGGCUACCUAAACUA-3′), Chmp4c-2 located in the 3′-untranslated region (5′-GUAGAGGAGUCUUAUAUGA-3′), ZW10 (a pool of three different siRNAs: 5′-GCAUGGAGCUCACAAUACA-3′, 5′-CCAAGUAUUUGCACCUUUA-3′, and 5′-CAAGCCAGCUUGCAAGAAA-3′), Chmp4a (a pool of three different siRNAs: 5′-CAAACUGACGGGACAUUAU-3′, 5′-CUGAGUGGGUAUCCUGAUA-3′, and 5′-GAAGGAUCUUGCUACAGAA-3′), and Chmp4b (a pool of three different siRNAs: 5′-CAACAUGGACAUCGAUAAA-3′, 5′-CAGUCCCUCUACCAAAUGU-3′, and 5′-GCUGAAUCCUCAAUGGAAA-3′) siRNAs were obtained from Santa Cruz Biotechnology. Only the sense sequences of the siRNA duplexes are shown.
Mutagenesis
To generate Chmp4c-L228A (Leucine 228 to Alanine), the T682G and T683C point mutations were introduced to the human Chmp4c siRNA by using the Q5 site-directed mutagenesis kit (New England Biolabs). The pCR3.1 GFP-EXN/chmp4c was used to generate T628G point mutation, changing Chmp4c-S210 to alanine (Petsalaki and Zachos, 2016).
Protein purification
His-tagged human Chmp4c was expressed in BL21 (DE3) cells (Agilent Technologies) and purified by using the 6ΧHis purification kit (B-PER; Thermo Fisher Scientific). GST-tagged proteins were expressed in BL21 (DE3) cells and purified by using glutathione-agarose beads (Santa Cruz Biotechnology) or were eluted from beads by using the GST spintrap purification module (GE Healthcare).
Cell culture and treatments
Human colon carcinoma BE cells and cervical carcinoma HeLa cells were gifts from C. Marshall (Institute of Cancer Research, London, England, UK). BE cells are diploid and contain an oncogenic Kras-G13D mutation as well as the BRAF-G463V oncogenic mutation (Davies et al., 2002; Vial et al., 2003; Petsalaki and Zachos, 2014). Cells were grown in DMEM (Gibco) containing 10% FBS at 37°C in 5% CO2 and treated with 3.32 µM nocodazole (Applichem), 1 µM taxol (Applichem), or 10 µg/ml MG132 (474790; Calbiochem) as appropriate. Negative siRNA or siRNA duplexes designed to repress human Chmp4c, Chmp4a, Chmp4b, or ZW10 were transfected into cells 24 h before analysis or further drug treatment by using Lipofectamine 2000 (Invitrogen). For expression of GFP proteins, plasmids were transfected into cells in the absence or presence of appropriate siRNA duplexes 24 h before analysis or further drug-treatment by using Turbofect (Life Technologies).
Experiments were generally performed in BE cells unless otherwise stated. Transient expression of GFP-tagged proteins was more uniform in HeLa compared with BE cells; therefore, most rescue experiments were performed in HeLa cells as specified, and cells exhibiting relatively low GFP kinetochore levels were analyzed by fluorescence microscopy. All cell lines used exhibited consistent morphology and growth properties and were negative for mycoplasma contamination.
Enrichment of cells in mitosis (nocodazole and “shake-off”)
Cells were treated with 3.32 μΜ nocodazole for 16 h and mitotic cells isolated by “shake-off.” Microscopic examination had shown ∼70–80% of cells were in prometaphase after this treatment compared with 2–3% in untreated cells.
Time-lapse imaging
HeLa or BE cells were seeded onto Petri dishes with a 30-mm glass base (Greiner). An inverted fluorescence microscope (Observer D1; Zeiss) was used. Fluorescence images were taken by using a 63× Plan neofluar 0.75 NA Ph2 objective (Zeiss) and phase-contrast images with a 20× Plan neofluar 0.40 NA Ph2 objective (Zeiss). Imaging was performed in air, at 37°C in 5% CO2, by using a Zeiss AxioCam MRm camera and the Zeiss ZEN2 acquisition software.
Indirect immunofluorescence microscopy
For Chmp4c or Rod staining, cells were fixed in ice-cold methanol for 5 min at −20°C, washed twice with PBS at room temperature, and immunostained. For phospho-Aurora B-S331 or phospho-Chmp4c-S210, S214, and S215 staining (Petsalaki and Zachos, 2016), cells were extracted in prewarmed (37°C) Phem buffer (60 mM Pipes, 25 mM Hepes, pH 7.0, 10 mM EGTA, and 4 mM MgSO4) supplemented with 0.5% CHAPS and 100 nM microcystin (Sigma-Aldrich) for 5 min at room temperature, fixed with prewarmed (37°C) 4% paraformaldehyde in Phem buffer for 10 min at room temperature, permeabilized in 0.5% Triton X-100 in Phem buffer for 2 min at room temperature, washed twice with PBS, and immunostained as appropriate. For ZW10, Zwilch, or GFP staining, cells were fixed in 4% paraformaldehyde in cytoskeleton buffer (1.1 M Na2HPO4, 0.4 M KH2PO4, 137 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM EGTA, 5 mM Pipes, and 5 mM glucose, pH 6.1) for 5 min at 37°C, permeabilized in 0.5% Triton X-100 in cytoskeleton buffer, washed twice with PBS at room temperature, and immunostained.
FITC- or rhodamine-TRITC–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used as appropriate. DNA was stained with 10 µM TO-PRO-3 iodide (642/661 nm; Invitrogen), and cells were mounted in Vectashield medium (Vector Laboratories). Images were collected by using a laser-scanning spectral confocal microscope (TCS SP2; Leica), LCS Lite software (Leica), and a 63× Apochromat 1.40 NA oil objective. The low-fluorescence immersion oil (11513859; Leica) was used, and imaging was performed at room temperature. Average projections of image stacks were obtained by using the LCS Lite software.
To analyze fluorescence intensities, background readings were subtracted, and green/red fluorescence intensities were quantified by using LCS Lite. For cyclin B or tubulin fluorescence intensities (cold assays), background readings were subtracted, and cyclin B or tubulin fluorescence for each mitotic cell was quantified by analyzing an equal image area by using ImageJ (National Institutes of Health). For Fig. 8 D, ZW10 or Mad1 fluorescence intensity signals were quantified by using the LCS Lite ellipse tool by analyzing an equal image area around each kinetochore, and intensity values were normalized versus values obtained by analyzing an identical area within the cell adjacent to the kinetochores (Petsalaki and Zachos, 2016). Centromere or spindle length was measured with the use of the LCS Lite line tool.
Cold-induced microtubule depolymerization
To induce microtubule disassembly of non-kinetochore microtubules, cell media was replaced with ice-cold media, and cells were incubated on ice for 4–15 min as indicated. Subsequently, cells were fixed in 4% paraformaldehyde in cytoskeleton buffer, permeabilized in 0.5% Triton X-100, and stained for α-tubulin, and prometaphase cells were examined by fluorescence microscopy.
Mitotic and prometaphase indices
Cells were fixed in 4% paraformaldehyde in cytoskeleton buffer, permeabilized in 0.5% Triton X-100, stained, and examined for condensed chromatin (mitotic indices) or mitotic spindle formation (prometaphase indices) by fluorescence microscopy. For experiments with GFP-tagged proteins, the mitotic index was calculated as the percentage of mitotic green cells/total green cells.
Cdk1 immunoprecipitation and kinase assay
For Cdk1 immunoprecipitations, cells were sonicated three times for 10 s in ice-cold immunoprecipitation/kinase buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% Tween 20, 0.1 mM PMSF, 10 µg/ml leupeptin, 10 µg/ml aprotinin, 1 mM sodium fluoride, 10 mM sodium β-glycerophosphate, and 0.1 mM sodium vanadate) and incubated for another 30 min on ice. 1 mg cell lysate was incubated with 0.5 µg anti-Cdk1 antibody for 16 h followed by the addition of 10 µl protein A/G PLUS–agarose beads (Santa Cruz Biotechnology) for 1 h at 4°C. Samples were spun down and washed three times with immunoprecipitation/kinase buffer and three times with kinase buffer (50 mM Hepes, pH 7.5, 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT). Immunoprecipitated proteins on agarose beads were included in a 20-µl reaction in kinase buffer containing 1 µg histone H1 protein (Upstate), 50 µM ATP, and 2 µCi γ-ATP for 30 min at 30°C before analysis by SDS-PAGE. Radioactive labeling of the H1 substrate was determined by autoradiography.
Protein coimmunoprecipitation, GST pull-down and in vitro binding assays
Cells were lysed as described in the previous paragraph. For coimmunoprecipitations, 1 mg cell lysate was incubated with 1 µg antibody for 16 h followed by the addition of 10 µl protein A/G PLUS–agarose beads for 1 h at 4°C. For GST pull-downs or in vitro binding assays, 0.5 mg cell lysate or 0.5–1 µg purified His-Chmp4c, respectively, was incubated with 1–2 µg GST-protein on glutathione-agarose beads for 4 h, at 4°C. Samples were spun down and washed three times with immunoprecipitation/kinase buffer, and immunoprecipitated proteins on agarose beads were analyzed by SDS-PAGE and Western blotting.
Western blotting
Cells were lysed in ice-cold, whole-cell extract buffer (20 mM Hepes, 5 mM EDTA, 10 mM EGTA, 0.4 M KCl, 0.4% Triton X-100, 10% glycerol, 5 mM NaF, 1 mM DTT, 5 µg/ml leupeptin, 50 µg/ml PMSF, 1 mM benzamidine, 5 µg/ml aprotinin, and 1 mM Na3VO4) for 30 min on ice. Lysates were cleared by centrifugation at 15,000 g for 10 min, analyzed by SDS-PAGE, and transferred onto nitrocellulose membrane (Santa Cruz Biotechnology).
Densitometry
Densitometric analysis of bands was performed using ImageJ.
Statistical analysis
For kinetochore fluorescence or centromere length, several kinetochore pairs or centromeres per cell from a minimum of six cells per experiment from three independent experiments was analyzed for each treatment, and SD was calculated. For spindle length, ten cells per experiment from three independent experiments were analyzed for each treatment, and SD was calculated. For cyclin B or tubulin fluorescence, 30 cells per experiment from three independent experiments were analyzed for each treatment, and SD was calculated. For chromosome misalignment, SD from the mean from three independent experiments was calculated, and 30 cells per experiment and per treatment were analyzed. For chromosome missegregation and the micronuclei or mitotic index, SD from the mean from three independent experiments was calculated, and 100 cells per experiment and per treatment were analyzed. Statistically significant differences between two groups were calculated by using Student’s t test. Statistically significant differences between three or more groups (Fig. 1 C; Fig. 3, D and F; and Fig. 8, A and D) were determined by one-way ANOVA followed by Student’s t test between two groups. No statistical method was used to predetermine the sample size.
Online supplemental material
Fig. S1 shows that Chmp4c does not localize to kinetochores in taxol-treated cells and that Chmp4c-deficient cells exhibit delayed anaphase onset in the absence of spindle drugs compared with controls. Fig. S2 shows that Chmp4c-deficient HeLa cells exhibit increased chromosome misalignment and missegregation and reduced cold-stable microtubule polymers compared with controls; it also shows that Chmp4a and Chmp4b do not localize to kinetochores. Fig. S3 shows cold-stable microtubule polymers in cells expressing Chmp4c-S210A or Vps4-K173Q; it also shows that expression of Chmp4c-S210A or Vps4-K173Q reduces the midbody index compared with WT Chmp4c or untreated cells. Fig. S4 shows localization of Aurora B and phosphorylated Aurora B-S331 to centromeres or kinetochores in Chmp4c-deficient or control cells and shows mitotic entry, mitotic indices, and ZW10 localization in cells treated with taxol. Fig. S5 shows Chmp4c-ZW10 interaction in untreated cells; it also shows Zwilch localization, cold-stable microtubule polymers, and chromosome alignment in cells expressing Chmp4c:Mis12:GFP or GFP-only. Video 1 shows a control HeLa H2B:RFP cell exhibiting successful chromatin alignment in metaphase and segregation in anaphase. Video 2 shows a Chmp4c-depleted HeLa H2B:RFP cell entering anaphase with misaligned chromatin. Videos 3 and 4 show progression of a control and a Chmp4c-depleted HeLa H2B:RFP cell, respectively, from NEBD to anaphase. Videos 5 and 6 show progression of a control and a Chmp4c-depleted BE cell, respectively, from NEBD to anaphase. Video 7 shows a control BE cell arrested in mitosis in the presence of nocodazole. Video 8 shows a Chmp4c-deficient BE cell entering mitosis but decondensing its chromatin and becoming multinucleated in the presence of nocodazole.
Supplementary Material
Acknowledgments
We thank P. Bieniasz, P. D’Avino, J. DeLuca, G. Kops, A. Musacchio, W. Sundquist, S. Taylor, and T. Zang for providing reagents. We also thank A. Kyrkou for cloning GST-Chmp4c.
This work was supported by Worldwide Cancer Research (grant 15-0008) and by Fondation Santé (review cycle 2017). E. Petsalaki was supported by a postdoctoral fellowship from the Bodossaki Foundation and M. Dandoulaki by Worldwide Cancer Research.
The authors declare no competing financial interests.
Author contributions: E. Petsalaki, M. Dandoulaki, and G. Zachos performed experiments and analyzed the results. G. Zachos and E. Petsalaki designed the study and wrote the paper.
References
- Adell M.A.Y., Vogel G.F., Pakdel M., Müller M., Lindner H., Hess M.W., and Teis D.. 2014. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J. Cell Biol. 205:33–49. 10.1083/jcb.201310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E.J., and Emr S.D.. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister E.R., Riegman M., and Lampson M.A.. 2014. Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. J. Cell Biol. 204:901–908. 10.1083/jcb.201311113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Gomes R., and Karess R.E.. 2000. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat. Cell Biol. 2:939–943. 10.1038/35046592 [DOI] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., and Karess R.E.. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861. 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Caldas G.V., Lynch T.R., Anderson R., Afreen S., Varma D., and DeLuca J.G.. 2015. The RZZ complex requires the N-terminus of KNL1 to mediate optimal Mad1 kinetochore localization in human cells. Open Biol. 5:150160 10.1098/rsob.150160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campsteijn C., Vietri M., and Stenmark H.. 2016. Novel ESCRT functions in cell biology: Spiraling out of control? Curr. Opin. Cell Biol. 41:1–8. 10.1016/j.ceb.2016.03.008 [DOI] [PubMed] [Google Scholar]
- Capalbo L., Montembault E., Takeda T., Bassi Z.I., Glover D.M., and D’Avino P.P.. 2012. The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis. Open Biol. 2:120070 10.1098/rsob.120070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.G., Caballe A., Agromayor M., Kloc M., and Martin-Serrano J.. 2012. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 336:220–225. 10.1126/science.1217180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G.K.T., Jablonski S.A., Starr D.A., Goldberg M.L., and Yen T.J.. 2000. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2:944–947. 10.1038/35046598 [DOI] [PubMed] [Google Scholar]
- Cheerambathur D.K., Gassmann R., Cook B., Oegema K., and Desai A.. 2013. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 342:1239–1242. 10.1126/science.1246232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L., Wenzel E.M., Liestøl K., Raiborg C., Campsteijn C., and Stenmark H.. 2016. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 212:499–513. 10.1083/jcb.201507009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F., Wehenkel A., Giorgi F.M., Santaguida S., Di Fonzo A., Grigorean G., Ciccarelli F.D., and Musacchio A.. 2010. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 18:616–626. 10.1016/j.str.2010.02.014 [DOI] [PubMed] [Google Scholar]
- Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., et al. . 2002. Mutations of the BRAF gene in human cancer. Nature. 417:949–954. 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Moree B., Hickey J.M., Kilmartin J.V., and Salmon E.D.. 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159:549–555. 10.1083/jcb.200208159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., and Stukenberg P.T.. 2007. Xenopus Cep57 is a novel kinetochore component involved in microtubule attachment. Cell. 130:893–905. 10.1016/j.cell.2007.07.023 [DOI] [PubMed] [Google Scholar]
- Famulski J.K., and Chan G.K.. 2007. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr. Biol. 17:2143–2149. 10.1016/j.cub.2007.11.037 [DOI] [PubMed] [Google Scholar]
- Funabiki H., and Wynne D.J.. 2013. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma. 122:135–158. 10.1007/s00412-013-0401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos T.N., Santamaria A., Jeyaprakash A.A., Wang B., Conti E., and Nigg E.A.. 2009. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28:1442–1452. 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R. III, et al. . 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399. 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose H., Arasaki K., Dohmae N., Takio K., Hatsuzawa K., Nagahama M., Tani K., Yamamoto A., Tohyama M., and Tagaya M.. 2004. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23:1267–1278. 10.1038/sj.emboj.7600135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., and Salmon E.D.. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009. 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H. 2015. ESCRTs are everywhere. EMBO J. 34:2398–2407. 10.15252/embj.201592484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Zhadina M., Bieniasz P.D., and Simon S.M.. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13:394–401. 10.1038/ncb2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R. 2005. Rod-Zw10-Zwilch: A key player in the spindle checkpoint. Trends Cell Biol. 15:386–392. 10.1016/j.tcb.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Kasuboski J.M., Bader J.R., Vaughan P.S., Tauhata S.B., Winding M., Morrissey M.A., Joyce M.V., Boggess W., Vos L., Chan G.K., et al. . 2011. Zwint-1 is a novel Aurora B substrate required for the assembly of a dynein-binding platform on kinetochores. Mol. Biol. Cell. 22:3318–3330. 10.1091/mbc.E11-03-0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J., and Shah J.V.. 2012. Connecting up and clearing out: how kinetochore attachment silences the spindle assembly checkpoint. Chromosoma. 121:509–525. 10.1007/s00412-012-0378-5 [DOI] [PubMed] [Google Scholar]
- Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R. III, Tagaya M., and Cleveland D.W.. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60. 10.1083/jcb.200411118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt T.E.F., Omerzu M., Saurin A.T., and Kops G.J.P.L.. 2014. Conditional targeting of MAD1 to kinetochores is sufficient to reactivate the spindle assembly checkpoint in metaphase. Chromosoma. 123:471–480. 10.1007/s00412-014-0458-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Westhorpe F.G., and Taylor S.S.. 2012. The spindle assembly checkpoint. Curr. Biol. 22:R966–R980. 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- London N., and Biggins S.. 2014a Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 28:140–152. 10.1101/gad.233700.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., and Biggins S.. 2014b Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15:736–747. 10.1038/nrm3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado M., and Kapoor T.M.. 2011. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat. Cell Biol. 13:475–482. 10.1038/ncb2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson D.R., and Stukenberg P.T.. 2014. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J. Cell Biol. 205:541–554. 10.1083/jcb.201307137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Fisher R.D., Whitby F.G., Sundquist W.I., and Hill C.P.. 2008. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA. 105:7687–7691. 10.1073/pnas.0801567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.Y., Morham S.G., Gygi S.P., Rodesch C.K., and Sundquist W.I.. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227. 10.1038/sj.emboj.7601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle M.W., Kim T., Hattersley N., Espeut J., Cheerambathur D.K., Oegema K., and Desai A.. 2014. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J. Cell Biol. 204:647–657. 10.1083/jcb.201311015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y., Hodgson L., Mantell J., Verkade P., and Carlton J.G.. 2015. ESCRT-III controls nuclear envelope reformation. Nature. 522:236–239. 10.1038/nature14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki E., and Zachos G.. 2014. Chk2 prevents mitotic exit when the majority of kinetochores are unattached. J. Cell Biol. 205:339–356. 10.1083/jcb.201310071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki E., and Zachos G.. 2016. Clks 1, 2 and 4 prevent chromatin breakage by regulating the Aurora B-dependent abscission checkpoint. Nat. Commun. 7:11451 10.1038/ncomms11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki E., Akoumianaki T., Black E.J., Gillespie D.A., and Zachos G.. 2011. Phosphorylation at serine 331 is required for Aurora B activation. J. Cell Biol. 195:449–466. 10.1083/jcb.201104023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silió V., McAinsh A.D., and Millar J.B.. 2015. KNL1-Bubs and RZZ provide two separable pathways for checkpoint activation at human kinetochores. Dev. Cell. 35:600–613. 10.1016/j.devcel.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Sundin L.J., Guimaraes G.J., and Deluca J.G.. 2011. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol. Biol. Cell. 22:759–768. 10.1091/mbc.E10-08-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., and Salmon E.D.. 2012. The KMN protein network–chief conductors of the kinetochore orchestra. J. Cell Sci. 125:5927–5936. 10.1242/jcs.093724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Wan X., Cheerambathur D., Gassmann R., Suzuki A., Lawrimore J., Desai A., and Salmon E.D.. 2013. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J. Cell Biol. 202:735–746. 10.1083/jcb.201304197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial E., Sahai E., and Marshall C.J.. 2003. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 4:67–79. 10.1016/S1535-6108(03)00162-4 [DOI] [PubMed] [Google Scholar]
- Vietri M., Schink K.O., Campsteijn C., Wegner C.S., Schultz S.W., Christ L., Thoresen S.B., Brech A., Raiborg C., and Stenmark H.. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 522:231–235. 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- Vleugel M., Hoogendoorn E., Snel B., and Kops G.J.P.L.. 2012. Evolution and function of the mitotic checkpoint. Dev. Cell. 23:239–250. 10.1016/j.devcel.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Wang H., Hu X., Ding X., Dou Z., Yang Z., Shaw A.W., Teng M., Cleveland D.W., Goldberg M.L., Niu L., and Yao X.. 2004. Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279:54590–54598. 10.1074/jbc.M407588200 [DOI] [PubMed] [Google Scholar]
- Williams B.C., Li Z., Liu S., Williams E.V., Leung G., Yen T.J., and Goldberg M.L.. 2003. Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol. Biol. Cell. 14:1379–1391. 10.1091/mbc.E02-09-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaërou F., Karess R., and Hays T.. 2001. Kinetochore dynein: Its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007. 10.1038/ncb1101-1001 [DOI] [PubMed] [Google Scholar]
- Zhang G., Lischetti T., Hayward D.G., and Nilsson J.. 2015. Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat. Commun. 6:7162 10.1038/ncomms8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.