Abstract
Pharmacogenetics is the genetic basis of pharmacokinetics, genetic testing, and clinical management in diseases. Evaluation about genetic alterations of drug metabolizing enzymes in human genome contributes toward understanding the interindividual and interethnic variability for clinical response to potential toxicants. CYP2E1 gene encodes a drug-metabolizing enzyme that metabolizes mostly small, polar molecules, including toxic laboratory chemicals. The aim of this study was to investigate CYP2E1 polymorphisms and gene profile in a Chinese Uygur population. Frequencies for the CYP2E1 mutated alleles and genotypes were screened in 100 unrelated random healthy Uygur volunteers. PCR and direct sequencing revealed a total of 32 polymorphisms, of which 5 novel mutations were presented. Rs 943975 was the most common single nucleotide polymorphism (SNP). The allele frequencies of CYP2E1∗1A, ∗4, ∗7A, and ∗7C were 65.5, 2, 19.5, and 13%, respectively. The most common genotype combinations were CYP2C19∗1A/∗1A (43%) and ∗1A/∗7C (24%). Functional prediction for 2 nonsynonymous mutations G173S and V179I was performed using MutationTaster, sorting intolerant from tolerant, and PolyPhen-2. The observations of the present study give rise to useful information on CYP2E1 polymorphisms in Chinese Uygur individuals. The results suggest important clinical implications for the use of medications metabolized by CYP2E1 among Uygurs.
Keywords: allele frequencies, CYP2E1, genetic polymorphism, nonsynonymous mutations, Uygur populations
1. Introduction
Polymorphisms in coding genes of drug-metabolizing enzymes lead to alterations of the metabolism function, affect drug response, and contribute to susceptibility to cancers. Cytochrome P450 (CYP) supergene family, which comprises 57 functional genes and 58 pseudogenes, is divided into 18 families. These genes encode enzymes capable of catalyzing oxidative metabolism of most drugs, toxic chemicals, and other lipophilic xenobiotics.[1]
Interindividual variations in DNA sequence and the distribution of the common variant alleles vary among different ethnic populations.[2] Genetic variations of CYPs related to variability in drug pharmacokinetics within and between individuals.[3] So far, extensive polymorphism occurs in 1, 2, and 3 CYP-family members and their potential clinical implications in drug therapy in different population have been identified in quantity.[4–6] These data provide indisputable evidence to predict relative susceptibility of an individual or populations to the adverse effects and relative sensitivity to drugs.[7]
Human cytochrome P450, family 2, subfamily E, polypeptide 1 (CYP2E1), is located on chromosome 10q.24 consisting of 9 exons and 8 introns. CYP2E1 gene produces an important I phase metabolic enzyme constitutively expressed in the liver as well as many other tissues. The most significant role of CYP2E1 is previously deemed to its corresponding acceleration of in ethanol-induced hepatotoxicity reaction. Subsequently, CYP2E1 is of interest because of its ability to metabolize and activate a wide array of small toxicological substrates, to more hydrophobic compounds and drugs, including potential carcinogens.[8–10]
Several studies have evaluated associations of CYP2E1 polymorphism with drug metabolizing enzyme genotypes. But there are still not enough data for functionally polymorphism of CYP2E1 in different ethnic groups. Here, we aimed to investigate the CYP2E1 genotype profile in a random Uygur population. The present result makes it possible to achieve optimal quality use of medicines in CYP2E1-related drug treatment.
2. Materials and methods
2.1. Study population
The study protocol was approved by the Ethics Committee of the Xizang Minzu University and was performed in accordance with the Declaration of Helsinki.
A total of 100 unrelated healthy volunteers including 50 males and 50 females (aged 19–52 years) were recruited from the Xizang Minzu University between October 2010 and December 2011. Written informed consent was obtained from each volunteer. All of the collected subjects were Chinese Uygur residents living in the Xinjiang Autonomous Region of China. They had at least 3 generations of Uygur paternal ancestry without any known ancestry from other ethnicities. Individuals with any type of medical illness, organ transplant, drug or alcohol addiction, and pregnant females were excluded from the study.
2.2. DNA extraction and polymerase chain reaction (PCR)
Blood samples (5 mL) were collected from the volunteers in an ethylenediaminetetraacetic acid tube (Jiangsu Kangjie Medical Devices Co, Ltd, Jiangyan, China) and stored at −80°C. Total genomic DNA was isolated using the Whole Blood Genomic DNA Purification Kit (GoldMag Ltd, Xi’an, China) following standard procedures according to the manufacturer's instructions. Primer pairs for amplification and sequencing of the promoter region, exons, introns, and the 3′-untranslated region (UTR) of CYP2E1 gene were listed in Table 1. The PCR was carried out in a final volume of 10 μL containing 5 μL HotStar Taq Master Mix (Qiagen China Co, Ltd, Shanghai, China), 20 ng template genomic DNA, 0.25 μM of each primer (Sangon, Shanghai, China) and 3 μL deionized water. The PCR program was used as follows: 1 initial denaturation step at 95°C for 15 minutes, followed by 35 denaturation cycles of 30 seconds at 94°C, 30 seconds of annealing at 55°C to 65°C and 1 minute of extension at 72°C, followed by a final elongation cycle at 72°C for 10 minutes.
Table 1.
Primers for CYP2E1 gene amplification and sequencing.

The PCR product was purified using TIANgel Midi Purification Kit (TIANGEN, Beijing, China) referring to the manufacturer's protocol. After purification, the PCR products were sequenced using ABI BigDye Terminator Cycle Sequencing Kit (version 3.1, Applied Biosystems, Thermo Fisher Scientific, Inc, Waltham, MA) on an ABI Prism3100 sequencer (Applied Biosystems, Thermo Fisher Scientific Inc).
2.3. Variants discovery and data analysis
We used SPSS 17.0 statistical packages (SPSS, Chicago, IL) to perform statistical calculations. The generated output and mutations were manually inspected by comparison to the wild-type nucleotide sequence of CYP2E1 (NG_008383.1, National Center for Biotechnology Information database) using Finch TV and Blast online software (https://blast.ncbi.nlm.nih.gov/Blast.cgi). All the positions of CYP2E1 polymorphism loci were assigned with a reference cluster ID (rs number) from NCBI Database of Single Nucleotide Polymorphisms (SNP) build, when available. The CYP allele nomenclature is quoted based on the Human Cytochrome P450 Allele Nomenclature Committee tables (http://www.cypalleles.ki.se/).
Then we compared the CYP2E1 allele frequency with 1000 Genome population frequencies (www.hapmap.org).[11]χ2 test was used. P < .05 was considered to indicate a statistically significant difference. Hardy–Weinberg equilibrium (HWE) for each genetic variant and pairwise linkage disequilibrium (LD) between loci pairs were analyzed by Haploview 4.1 (http://broad.mit.edu/mpg/haploview). Haplotypes were constructed from the selected tag SNPs and haplotype frequencies were derived for the study population. Haplotype blocks were defined based on the Gabriel definition (D′> 0.9; minimum allele frequency> 5%).[12]
2.4. Protein function prediction
The online tools MutationTaster (http://www.mutationtaster.org/), sorting intolerant from tolerant (SIFT) (http://blocks.fhcrc.org/sift/SIFT.html), and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph/) were used to predict the protein function of nonsynonymous SNPs in CYP2E1 exon regions.
MutationTaster evaluates disease-causing potential of sequence alterations denoted by 4 types: disease_causing_automatic, disease_causing, polymorphism, and polymorphism_automatic, which we coded as “A,” “D,” “N,” and “P,” respectively.
“A” and “D” categories are regarded as deleterious.[13] The SIFT output was then divided into 4 categories based on the previous reported reference: tolerant (0.201–1.00), borderline (0.101–0.20), potentially intolerant (0.051–0.10), and intolerant (0.00–0.05). PolyPhen-2 predicts the effect of an amino acid substitution and the results were divided into 3 categories: benign (0.00–0.15), possibly damaging (0.15–0.85), and probably damaging (0.85–1.00) with scores ranging from 0 to 1.[14,15]
3. Result
3.1. Genetic variants
In the present study, direct sequencing revealed the frequencies of CYP2E1 gene variations in this Uygur population (Table 2). A total of 32 different polymorphisms were detected. Among them, polymorphism 1361C>T, located in CYP2E1 intron 2 nearing exon 1, has the highest frequency of 99%. 11610T>A and 11615A>G in the UTR region both had a frequency of 59% and −333A>T in gene promotor accounted for 53%.
Table 2.
Frequency distribution of CYP2E1 genetic variants in the present study.
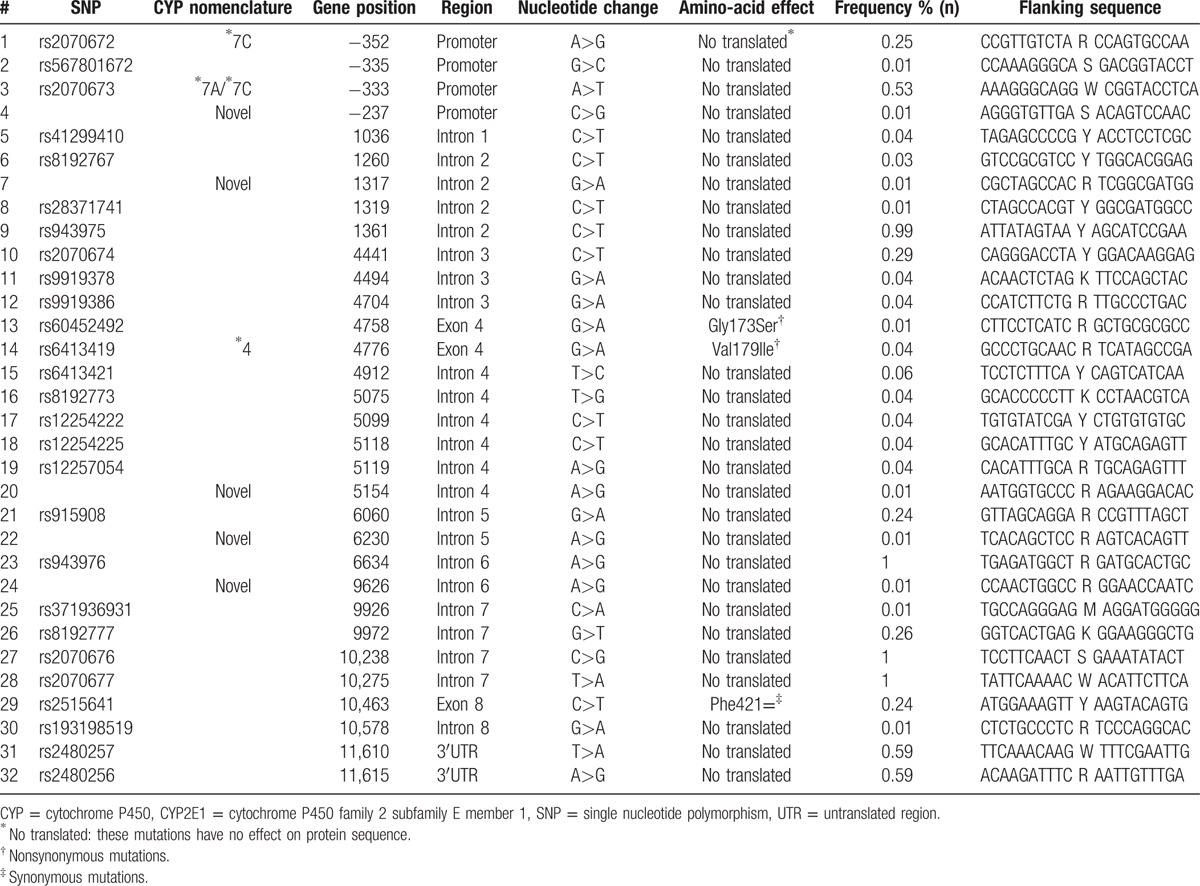
Within exon 8, a synonymous mutation 10463C>T (Phe421=), which had 24% frequency, was identified. In gene exon 4, polymorphisms 4776G>A and 4758G>A caused amino acid alterations Val179Ile (4%) and Gly173Ser (1%) respectively. Besides, 5 variants −237C>G, 1317G>A, 5154A>G, 6230A>G, and 9626A>G were first identified and had not previously been reported in the NCBI database or the Human Cytochrome P450 Allele Nomenclature Committee tables. The 5 novel variants were all located in no coding region with the same frequency of 1%.
3.2. Allele and genotype frequencies
All the identified frequencies of allele and genotype were in accordance with the Hardy–Weinberg equilibrium (P>.05) and presented in Table 3. Altogether we detected 4 alleles and 6 genotypes in the current population. The wild-type allele CYP2E1∗1A had the highest frequency of 65.5%. The CYP2E1∗7A allele had 19.50% frequency, followed by the CYP2E1∗7C with 13.00%. The frequency of CYP2E1∗4 was rare in the Chinese Uygur population (2%), which leads to a residue change but had no influence on enzyme activity putatively.
Table 3.
Allele and genotype frequencies of CYP2E1 variants in the 100 Chinese Uygur subjects.
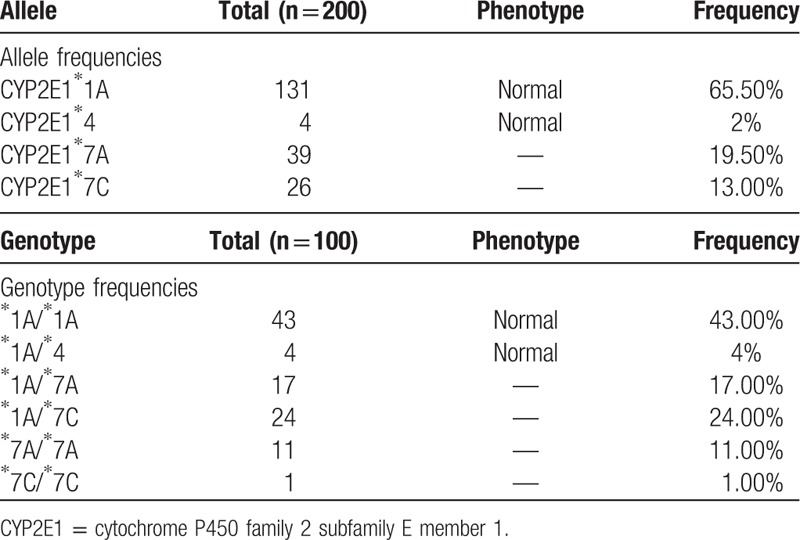
In relation to the genotypes, 43 cases (43%) exhibited ∗1A/∗1A wide-type homozygote, which had the highest genotype frequency and normal enzyme activity. There are 24% individuals who accompanied with heterozygote genotype ∗1A/∗7C and 17% individuals with ∗1A/∗7A. The frequency of homozygote genotype ∗7A/∗7A was 11%. In addition, ∗1A/∗4 genotype was relatively rare (4%). Only 1 individual displayed ∗7C/∗7C homozygous variant genotype based on identified polymorphisms.
The CYP2E1 allele frequencies were further compared with 1000 Genome population frequencies. CYP2E1 alleles that have been mostly reported were listed in Table 4.
Table 4.
CYP2E1 allele frequencies in 1000 Genome population.

We did not discover CYP2E1∗1B, CYP2E1∗2, CYP2E1∗3, CYP2E1∗5B, CYP2E1∗6, and CYP2E1∗7B alleles in the current Uygur group. CYP2E1∗7A (rs2070673) occurs at a frequency of 45.16% in Chinese Dai, 56.8% in Chinese Han (Beijing), and 57.62% in Chinese Han (South). The frequency of allele ∗7C (rs2070672) in the 3 populations is 38.71%, 22.82%, and 22.38%, respectively.
3.3. Linkage disequilibrium analysis
Haploview software was then applied to evaluate the linkage disequilibrium between the identified SNPs according to the coefficient of linkage disequilibrium D′ values. SNPs with lower frequencies have little power to detect LD and D values of each pair of SNPs represent the degree of linkage. The consequence affirmed 2 LD blocks across CYP2E1: −352A>G (rs2070672), −333G>C (rs567801672) consisted of the small block 1; 9972G>T (rs8192777), 10238C>G (rs2070676), 10275T>A (rs2070677), 10463C>T (rs2515641), 11610T>A (rs2480257), and 11615A>G (rs2480256) consisted of the big block 2, which spans 1 kb region. These SNPs were obviously linked with high D′ (D′≥0.96) and tightly correlated (Fig. 1).
Figure 1.

LD analysis of CYP2E1 in the Chinese Uygur population. LD is indicated by bright red (very strong: LOD ≥ 2, D′ = 1), light red (LOD ≥ 2, D′ < 1) and blue (LOD < 2, D′ = 1) for intermediate LD, and white (none: LOD < 2, D′ < 1). CYP2E1 = cytochrome P450 family 2 subfamily E member 1, LD = linkage disequilibrium.
3.4. Protein function prediction of nonsynonymous mutation
By amino acid sequence alignment, 2 mutations 4758G>A (Gly173Ser, G173S) and 4776G>A (Val179Ile, V179I) were nonsynonymous SNPs. Functional prediction of the 2 amino acid substitutions was performed. Each variant was given a score based on the impact of its mutation on protein function.
MutationTaster showed that alteration G173S was disease causing while V179N was classified into polymorphism. Degree of protein damage analyzed by PolyPhen-2 was positive correlation with score value in HumVar. As shown in Fig. 2, our report indicated that G173S and V179I are both probably damaging in terms of both HumDiv and HumVar models (score>0.990). SIFT score ranged from 0 to 1 and less than 0.05 was considered deleterious. However, the results presented by SIFT were not consistent with PolyPhen-2. SIFT prediction of G173S was scored 0.96, which meant the alteration was tolerant. For V179I, the SIFT score was lower (score = 0.23). It was near the borderline (0.101–0.20) but still predicted as being tolerant.
Figure 2.
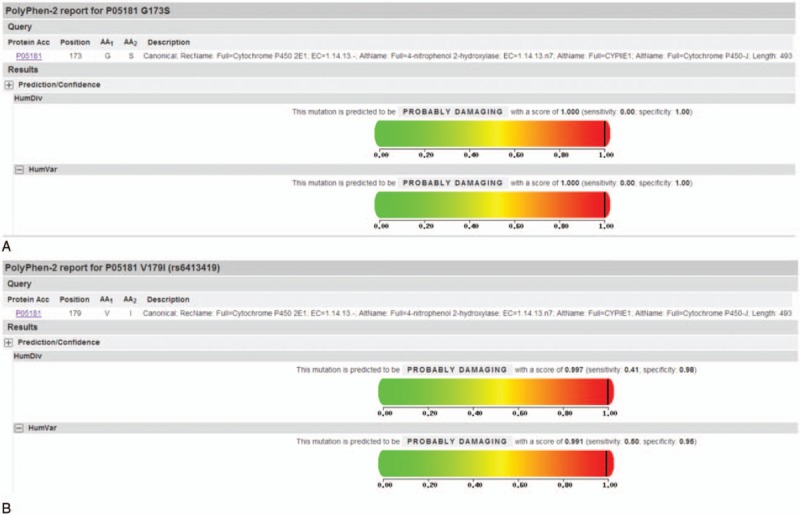
Protein prediction of synonymous mutation G173S and V179I using the PloyPhen-2 software. A, Prediction of the mutation G173S. B, Prediction of the mutation V179I.
4. Discussion
The current study expounds CYP2E1 genetic variants including the distribution of SNPs, the frequency of allele, and genotype in Chinese Uygur subjects for the first time.
By direct PCR and sequencing, 32 polymorphisms were revealed. Most SNPs seated in no translated region. In exons areas, 2 nonsynonymous mutations (G173S, V179I) and 1 synonymous mutation (Phe421=) were detected. Moreover, 1361C>T in intron2 was the most common polymorphism in the present subjects. Five SNPs (−237C>A, −1317G>A, 5154A>G, 6230A>G, 9620A>G) in the no-coding region were never reported before.
So far, more than 100 SNPs have been found in CYP2E1 but only several might alter the enzyme activity.[16] According to the racial comparison, the CYP2E1 gene shows genetic polymorphisms that vary markedly in their frequency among different ethnic populations. Among them, linkage disequilibrium of rs2031920 and rs3813867 leads to the CYP2E1∗5B haplotype and was associated with restriction enzyme sites RsaI and PstI. SNP rs6413432 relate to restriction enzyme site Dra I and form CYP2E1∗6 haplotype. The 3 genetic polymorphisms have been largely reported to alter transcription of the CYP2E1 gene and affected outcomes of cancers such as gastric cancer, breast cancer, and non-small cell lung cancer.[17,18] rs6413420 was detected to affect the gene transcription and led to an activation of CYP2E1. rs72559710, results in CYP2E1∗2 haplotype, causes an R76H amino acid exchange, lowers enzyme synthesis and catalytic activity. The additional functional SNP, rs55897648 (CYP2E1∗3), causes V389I substitution, but has no effect on enzyme activity.[19]
Based on the published data about CYP2E1 allele distribution in Asian, European, and American, CYP2E1∗5B, CYP2E1∗6, and CYP2E1∗7 showed frequent occurrence.[20–23] In particular, CYP2E1∗5B has been universally reported in various ethnic groups in quantity. Wang et al[24] determined the genotype and allele distribution patterns of CYP2E1 polymorphisms among 103 healthy participants in the mainland Chinese Uygur. The mutant allele frequencies were 12.1%, 18.8% in CYP2E∗5B and CYP2E∗6, respectively. CYP2E1∗4 (rs6413419) were not determined.[24] But CYP2E1∗5B and CYP2E1∗6 alleles were not discovered among our current Uygur subjects. Another haplotype, CYP2E1∗4, has 2% frequency and results in an amino acid substitution, Val179Ile. Besides, rs2070673 and rs2070672 both exhibited lower frequency compared with rs2070673, rs2070672 when comparing to Chinese Dai, Chinese Han (Beijing) in Chinese Han (South). Rs2070673 may be a genetic biomarker of susceptibility to benzene toxicity, based on the fact that the enzyme activity and mRNA expression of “TT” genotype were significantly upregulated on the phenol treatment B lymphocyte cell lines derived from the Han Chinese population.[25] The evident genetic variants in the coding or noncoding regions of CYP2E1 among different populations may be explained by the different lifestyles, culture, and history. The interethnic difference suggests that disease susceptibilities or efficacy and toxicity of many medications may differ in the diverse ethnic populations.
Through the bioinformation analysis, the functional prediction by different tools on mutations G173S and V179I exhibits a great inconsistency. PolyPhen-2 predicted that G173S was probably damaging and MutationTaster predicted it was disease causing. But SIFT score of the substitution was 0.96, which can be judged as tolerated. In respect for V179I, PolyPhen-2 prediction result is also probably damaging, but SIFT predicted it to be tolerated with the score of 0.23, which is consistent with the MutationTaster result: polymorphism to some extent. The inconsistency of functional prediction may be due to the fact that different algorithms are based on different training data. Each has its own strength and weakness. According to related analysis, the accuracy of SIFT and PolyPhen-2 prediction typically reaches 63% and 75% with false positive rates as high as 19% and 9%, while MutationTaster may have a higher true positive rate.[26,27] The previous study by Fairbrother et al[28] has indicated that no significant alternation was observed in enzyme activity for chlorzoxazone hydroxylation between CYP2E1 V179I mutant and wild-type. It is probably due to the structural similarities of the 2 amino acid residues and the fact that position 179 is on the periphery of the whole protein.[29] Based on these studies we presented V179I have no influence on enzyme activity. But for G173S, no evidence demonstrated its effect on normal enzyme function of CYP2E1. From the above, functional prediction has become an indispensable step to detect the influence of genetic variation. But the credible effects of genetic variants identified in our study need further experimental data to support.
In summary, 32 polymorphisms were detected on the CYP2E1 gene, including 5 novel mutations (−237C>G, 1317G>A, 5154A>G, 6230A>G, 9626A>G), 2 nonsynonymous mutations (G173S, V179I), and 1 synonymous mutation (Phe421=). The basic determination in polymorphisms of CYP2E1 in the Uygur population may contribute to individualized medicine and provide advantage for dose adjustment of certain drug therapies for the sake of reduction in adverse effects and even death.
Acknowledgments
The author are grateful to the healthy individuals, the clinicians, and hospital staff for their participation and contribution.
Footnotes
Abbreviations: CYP = cytochrome P450, CYP2E1 = cytochrome P450 family 2 subfamily E member 1, EDTA = ethylenediaminetetraacetic acid, HWE = Hardy–Weinberg equilibrium, LD = linkage disequilibrium, PCR = polymerase chain reaction, SIFT = sorting intolerant from tolerant, SNP = single nucleotide polymorphism, UTR = untranslated region.
This study was supported by Science and Technology Agency Project of Xizang (Tibet) Autonomous (No. 2015ZR-13-11), National Natural Science Foundation of China (No. 31760312) and National Key Research and Development Plan of China (No. 2017YFC1201303).
The authors have no conflicts of interest to disclose.
References
- [1].Uehara S, Uno Y, Inoue T, et al. Novel marmoset cytochrome P450 2C19 in livers efficiently metabolizes human P450 2C9 and 2C19 substrates, s-warfarin, tolbutamide, flurbiprofen, and omeprazole. Drug Metab Dispos 2015;43:1408–16. [DOI] [PubMed] [Google Scholar]
- [2].Jin T, Yang H, Zhang J, et al. Polymorphisms and phenotypic analysis of cytochrome P450 3A4 in the Uygur population in northwest China. Int J Clin Exp Pathol 2015;8: 7083–91.2. [PMC free article] [PubMed] [Google Scholar]
- [3].Palma BB, Moutinho D, Urban P, et al. Cytochrome P450 expression system for high-throughput real-time detection of genotoxicity: application to the study of human CYP1A2 variants. Mutat Res Genet Toxicol Environ Mutagen 2016;806:24–33. [DOI] [PubMed] [Google Scholar]
- [4].Jin T, Zhang X, Geng T, et al. Genotype-phenotype analysis of CYP2C19 in the Tibetan population and its potential clinical implications in drug therapy. Mol Med Rep 2016;13:2117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang C, Cheng Y, Dai R, et al. Polymorphisms of drug-metabolizing enzyme CYP2J2 in Wa population from Yunnan Province of China. Int J Clin Exp Pathol 2016;9:4260–8. [Google Scholar]
- [6].Shi X. Genetic analysis of drug-metabolizing enzyme CYP3A5 polymorphisms in Tibetans in China. Int J Clin Exp Pathol 2016;9:3717–25. [Google Scholar]
- [7].Arici M, Özhan G. The genetic profiles of CYP1A1, CYP1A2 and CYP2E1 enzymes as susceptibility factor in xenobiotic toxicity in Turkish population. Saudi Pharm J 2017;25:294–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev 1997;77:517–44. [DOI] [PubMed] [Google Scholar]
- [9].Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med 2008;44:723–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ingelman-Sundberg M, Ronis MJ, Lindros KO, et al. Ethanol-inducible cytochrome P4502E1: regulation, enzymology and molecular biology. Alcohol Alcohol Suppl 1994;2:131–9. [PubMed] [Google Scholar]
- [11].Abecasis GR, Altshuler D, Auton A, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–9. [DOI] [PubMed] [Google Scholar]
- [13].Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions †. Hum Mutat 2011;32:894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ding Y, Xu D, Zhang X, et al. Genetic polymorphisms and phenotypic analysis of drug-metabolizing enzyme CYP2C19 in a Li Chinese population. Int J Clin Exp Pathol 2015;8:13201–8. [PMC free article] [PubMed] [Google Scholar]
- [15].Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lu Y, Zhu X, Zhang C, et al. Role of CYP2E1 polymorphisms in breast cancer: a systematic review and meta-analysis. Cancer Cell Int 2017;17:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ada AO, Bilgen S, Karacaoglan V, et al. Association between the TP53 and CYP2E1∗5B gene polymorphisms and non-small cell lung cancer. Arh Hig Rada Toksikol 2016;67:311–6. [DOI] [PubMed] [Google Scholar]
- [18].Feng J, Pan X, Yu J, et al. Functional PstI/RsaI polymorphism in CYP2E1 is associated with the development, progression and poor outcome of gastric cancer. PLoS One 2012;7:e44478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shahriary GM, Galehdari H, Jalali A, et al. CYP2E1∗5B, CYP2E1∗6, CYP2E1∗7B, CYP2E1∗2, and CYP2E1∗3 allele frequencies in Iranian populations. Asian Pac J Cancer Prev 2012;13:6505–10. [DOI] [PubMed] [Google Scholar]
- [20].Persson I, Johansson I, Bergling H, et al. Genetic polymorphism of cytochrome P4502E1 in a Swedish population. Relationship to incidence of lung cancer. FEBS Lett 1993;319:207–11. [DOI] [PubMed] [Google Scholar]
- [21].Soya SS, Padmaja N, Adithan C. Genetic polymorphisms of CYP2E1 and GSTP1 in a South Indian population—comparison with North Indians, Caucasians and Chinese. Asian Pac J Cancer Prev 2005;6:315–9. [PubMed] [Google Scholar]
- [22].Yang B, O’Reilly DA, Demaine AG, et al. Study of polymorphisms in the CYP2E1 gene in patients with alcoholic pancreatitis. Alcohol 2001;23:91–7. [DOI] [PubMed] [Google Scholar]
- [23].Liu S, Park JY, Schantz SP, et al. Elucidation of CYP2E1 5′ regulatory RsaI/Pstl allelic variants and their role in risk for oral cancer. Oral Oncol 2001;37:437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang SM, Zhu AP, Li D, et al. Frequencies of genotypes and alleles of the functional SNPs in CYP2C19 and CYP2E1 in mainland Chinese Kazakh, Uygur and Han populations. J Hum Genet 2009;54:372–5. [DOI] [PubMed] [Google Scholar]
- [25].Zhang J, Yin L, Liang G, et al. Detection of CYP2E1, a genetic biomarker of susceptibility to benzene metabolism toxicity in immortal human lymphocytes derived from the Han Chinese Population. Biomed Environ Sci 2011;24:300–9. [DOI] [PubMed] [Google Scholar]
- [26].Wu J, Li Y, Jiang R. Integrating multiple genomic data to predict disease-causing nonsynonymous single nucleotide variants in exome sequencing studies. PLoS Genet 2014;10:e1004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lai SWS, Lopes RM, Doherty E, et al. Analysis of BRCA gene missense mutations. 2016;2:91–108. [Google Scholar]
- [28].Fairbrother KS, Grove J, de Waziers I, et al. Detection and characterization of novel polymorphisms in the CYP2E1 gene. Pharmacogenetics 1998;8:543–52. [DOI] [PubMed] [Google Scholar]
- [29].Pratt-Hyatt M, Lin HL, Hollenberg PF. Mechanism-based inactivation of human CYP2E1 by diethyldithocarbamate. Drug Metab Dispos 2010;38:2286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]


