Abstract
In this study, we investigated the expression of microRNAs (miRNAs) in the skin samples from the Han and Uyghur populations in Xinjiang, China. The miRNA levels of the normal skin samples from 10 individuals of Uyghur or Han were tested by microarray and the expression differentiations were compared. Among the 3100 probes for microarray, a total of 247 miRNAs were differentially expressed in the Han versus Uyghur population, including 76 upregulated miRNAs and 171 downregulated miRNAs. The most significantly upregulated miRNAs were miR-141-3p, miR-1915-5p, kshv-miR-K12-2-5p, and miR-222-3p. And the most significantly downregulated miRNAs included miR-1207-3p and miR-625-3p. We have confirmed the upregulation of miR-141-3p and miR-1915-5p by qRT-PCR. There were no statistical correlations in the expression of miR-141-3p or miR-1915-5p with the age or gender within each group. Interestingly, the differentially expressed miRNAs were enriched in some cancer-related pathways, such as p53, mitogen-activated protein kinase, and WNT signal pathways. Collectively, these dysregulated expressions of the miRNAs may provide a better understanding of the differences in the incidence and mortality of skin-related carcinoma between the Uyghur and Han populations in Xinjiang.
Keywords: Han, microRNAs, skin tissue, Uyghur, Xinjiang
1. Introduction
MicroRNAs (miRNAs) are endogenous, small noncoding RNAs that can negatively regulate gene expression by interfering with the translation or stability of target genes.[1] They participate in the cellular proliferation, differentiation, and apoptosis and can also promote tumorigenesis by acting as oncogenes.[2] Among Caucasians, the cutaneous squamous cell carcinoma (SCC) is the 2nd most common cancer and is thought to be resulted from the accumulation of multiple genetic changes.[3] From 1992 to 2003, McDougall et al[4] had studied the incidence of cervical cancer in the United States and found that white Hispanics had the highest incidence rate of cervical squamous cell carcinoma (CSCC), while non-Hispanic whites had the lowest incidence rate, which concluded that the incidence of CSCC differed among races.
China is a country of diverse populations, and Xinjiang province is a multiethnic area, with rich genetic variation. In Xinjiang, Uyghur and Han are the main ethnic groups. The incidence of skin cancer is difference between Han and Uyghur population of Xinjiang. For example, the Kaposi sarcoma and malignant melanoma also have certain idiopathic and ethnic specificities and differences. In China, more than 90% of Kaposi sarcoma cases occur in Uyghur patients, and only sporadic cases occur in Han patients.[5] There is genetic heterogeneity in the occurrence of melanomas in the Uyghur and Han people of Xinjiang. The melanomas of Uyghur are mainly caused by chronic sunlight injury whereas the Han melanomas are still mainly the mucosal type [101].[6] Studying the miRNA profiles in different populations can facilitate understanding the mechanism causing the difference in skin cancer incidence in Xinjiang. In this study, the expression profiles of miRNAs in the skin samples from the Han and Uyghur populations in Xinjiang were investigated.
2. Materials and methods
2.1. Patients and specimen collection
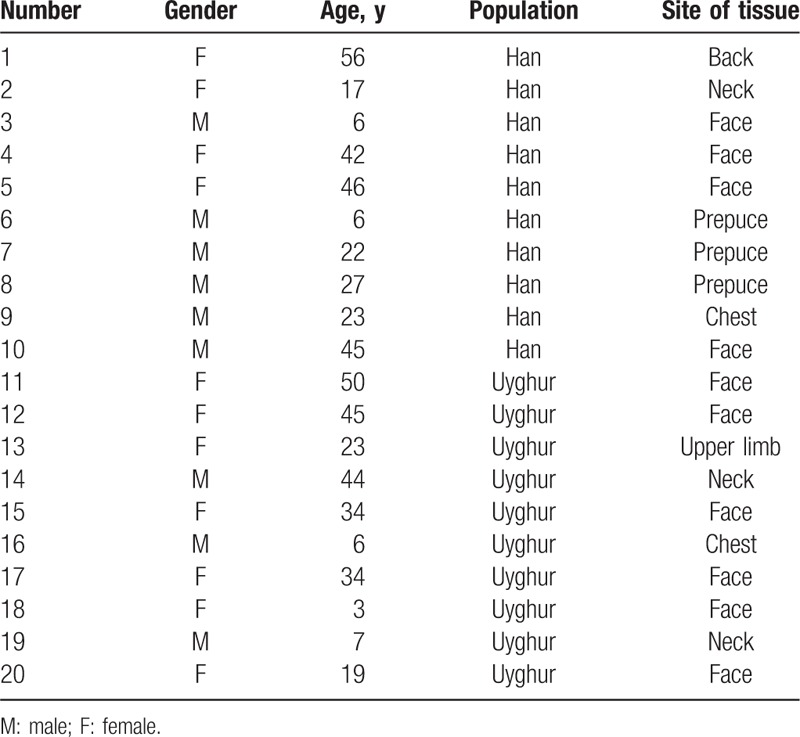
Twenty healthy people, including 10 Uyghur individuals and 10 Han individuals, were enrolled in this study (Table 1). Among the Uyghur subjects, there were 3 men and 7 women (average age, 26.5 ± 17.40 years), while the Han subjects included 6 men and 4 women (average age, 29 ± 17.40 years). There was no significant difference in the enrolled Uyghur and Han people at baseline level (such age, gender, or location of skin lesion). All the skin samples (n = 20) were obtained by surgical resection from January 2012 to September 2013 in the Department of Dermatology, People's Hospital of the Xinjiang Uyghur Autonomous Region.
Table 1.
The characteristics of the Han and Uyghur subjects.
The samples were flash frozen in liquid nitrogen and stored at −80 °C until the RNA extraction. The collected normal samples were further confirmed by histological examination. The protocol was approved by the Institutional Review Board of the People's Hospital of the Xinjiang Uyghur Autonomous Region, Urumqi, China. Parental consent was obtained for minors (younger than 18 years old), according to the ethics statement. And prior written consent for tissue donation (for research purposes) was obtained.
2.2. RNA extraction
Total RNAs were isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) and a miRNeasy mini kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The quality and quantity of isolated RNA were measured with a NanoDrop spectrophotometer (ND-1000, NanoDrop Technologies, Wilmington, DE) and the integrity of RNAs was determined by gel electrophoresis.
2.3. RNA labeling
The miRCURY Hy3/Hy5 power labeling kit (Exiqon, Skelstedet, Denmark) was used for miRNA labeling, according to the manufacturer's guidelines. Briefly, the RNAs were dissolved in 2 μL ddH2O, and then were mixed with 1 μL of CIP buffer and CIP. The mixture was then incubated for 30 minutes at 37 °C and was terminated by incubation for 5 minutes at 95 °C. Then, 3 μL of labeling buffer, 1.5 μL of fluorescent label (Hy3), 2 μL of dimethyl sulfoxide, and 2 μL of labeling enzyme were added to the mixture. The labeling reaction was incubated for 1 hour at 16 °C and was subsequently terminated by incubation at 65 °C for 15 minutes.
2.4. MicroRNA array
The microRNA array was performed according to the manual of the 7th generation miRCURYTM LNA Array kit (v18.0) (Exiqon, Skelstedet). The kit contains 3100 capture probes, representing all human, mouse, and rat miRNAs as well as all viral miRNAs related to these species. In addition, this array contains capture probes for 25 miRPlus human miRNAs. Briefly, the total RNA was isolated using TRIzol (Invitrogen) and miRNeasy mini kit (QIAGEN). After labeling, the hybridization buffer (25 μL) was added into the Hy3-labeled sample (25 μL). The mixture was denatured for 2 minutes at 95 °C, then incubated on ice for 2 minutes, and hybridized to the microarray for 16 to 20 hours at 56 °C in a 12-bay hybridization system (Hybridization System-Nimblegen Systems, Inc., Madison, WI). Then, the slides were scanned with the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA). The scanned images were imported into GenePix Pro 6.0 software (Axon Instruments) for grid alignment and data extraction. After normalization, the significantly differently expressed miRNAs were identified through volcano plot filtering.
2.5. RT-PCR
The isolated total RNAs were reverse transcribed with the miScript II Reverse-Transcription Kit (Qiagen, German) according to the manufacturer's instructions. The concentrations were measured using a Nanodrop spectrophotometer (ND-1000, NanoDrop Technologies). The expression levels of hsa-miR-141-3p, hsa-miR-1915-5p, and U6 were measured by RT-PCR with a miScript SYBR Green PCR kit (Qiagen, German) in a QiagenRoter-Gene Q (Qiagen). The primers used for the detection of hsa-miR-141_1, hsa-miR-1915-5p, and U6 were from the Hs_miR-141_1 miScript Primer Assay (MS00003501, Qiagen), hs_miR-1915-5p_1 miScript Primer Assay (MS00042238,Qiagen), and the Hs_RNU6 miScript Primer Assay (MS00033740, Qiagen). All reactions were performed in triplicate. The relative amount of miRNAs was normalized against U6 snRNA and the fold change for each miRNA was calculated by the 2-ΔΔCT method.
2.6. Target prediction
The predicted target genes and miRNA chromosomal locations were retrieved from the Microcosm database (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/ targets/v5/), TargetScan (http://www.targetscan.org), miRanda (http://www.microRNA.org), and GO analysis (http://www.geneontology.org). We only included miRNA targets that were present in more than 2 databases in order to exclude false positive results.
2.7. Statistical analysis
Statistical Program for the Social Sciences software 17.0 (SPSS Inc., Chicago, IL) was used for statistical analysis. All data are presented as the mean ± standard deviation. A one-way analysis of variance or t test was used to compare the miRNA expression level according to age and gender within Han or Uyghur groups. A P value < .05 was considered statistically significant.
3. Results
3.1. Expression profile of miRNAs in the normal skin samples in Uyghur and Han subjects from Xinjiang
To identify the differentially expressed miRNAs in the skin samples between Uyghur and Han subjects, miRNA microarray assay was performed. Only miRNAs that were altered by at least 2 folds in at least 2 of the samples were considered as significant candidates for differential miRNA expression. Among the 3100 human miRNA probes, we identified a total of 247 differentially regulated miRNAs (76 upregulated and 171 downregulated) in Han versus Uyghur subjects (Fig. 1). All raw and normalized miRNA expression data are publicly available in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE62354.
Figure 1.
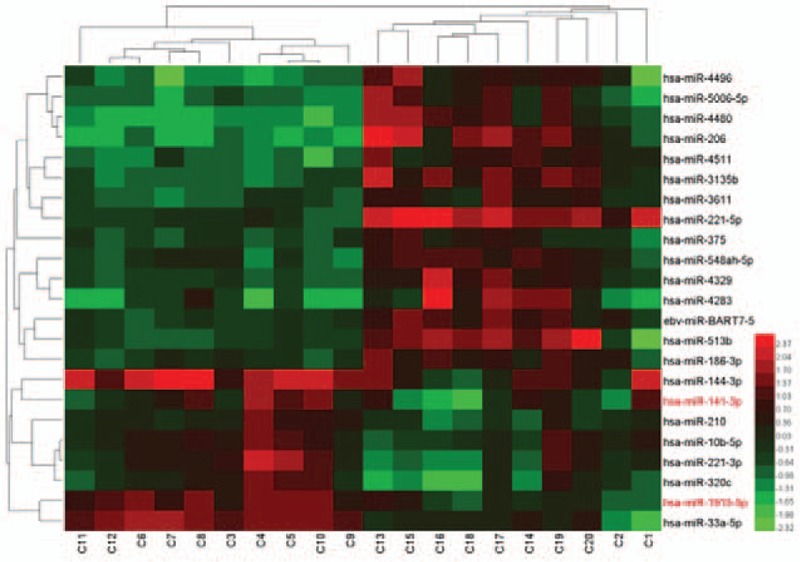
MicroRNAs (miRNAs) deregulated in the normal skin samples from Uyghur and Han individuals. A total of 20 normal skin samples were analyzed by a microRNA microarray with the 7th generation miRCURYTM LNA Array kit (v.18.0). Unsupervised hierarchical cluster analysis of miRNA expression in 20 normal skin tissues. Rows: miRNAs; columns: cases. The analysis demonstrated that the 2 chips in each case were consistent. For each miRNA, red represents higher expression, and blue represents lower expression (compared to the average). C1–C10 represents normal Han skin tissue. C11–C20 represents normal Uyghur skin tissue.
3.2. miRNA expression differentiations in the healthy Han and Uyghur skin subjects
In the Han population, 76 miRNAs were upregulated and 171 were downregulated compared with that of the Uyghur population (P < .05) (Table 2). The upregulated miRNAs that changed significantly included miR-1, miR-141-3p, miR-106b-5p, miR-1915-5p, miR-200c-3p, miR-221-3p, miR-222-3p, miR-3139, miR-4802-3p, miR-507, miR-541-3p, miR-876-3p, and kshv-miR-K12-2-5p. The downregulated miRNAs that changed most significantly included miR-1207-3p, miR-206, miR-2116-5p, miR-3667-5p, miR-374b-3p, miR-4423-5p, miR-4450, miR-4468, miR-542-3p, miR-548ao-3p, and miR-553. Among the upregulated miRNAs, the most significant upregulated were miR-141-3p, miR-1915-5p, kshv-miR-K12-2-5p, and miR-222-3p, whereas those among the downregulated miRNAs with the most significant downregulation included miR-1207-3p and miR-625-3p.
Table 2.
Selected differentially expressed microRNAs (miRNAs) in normal skin samples from Uyghur and Han populations.
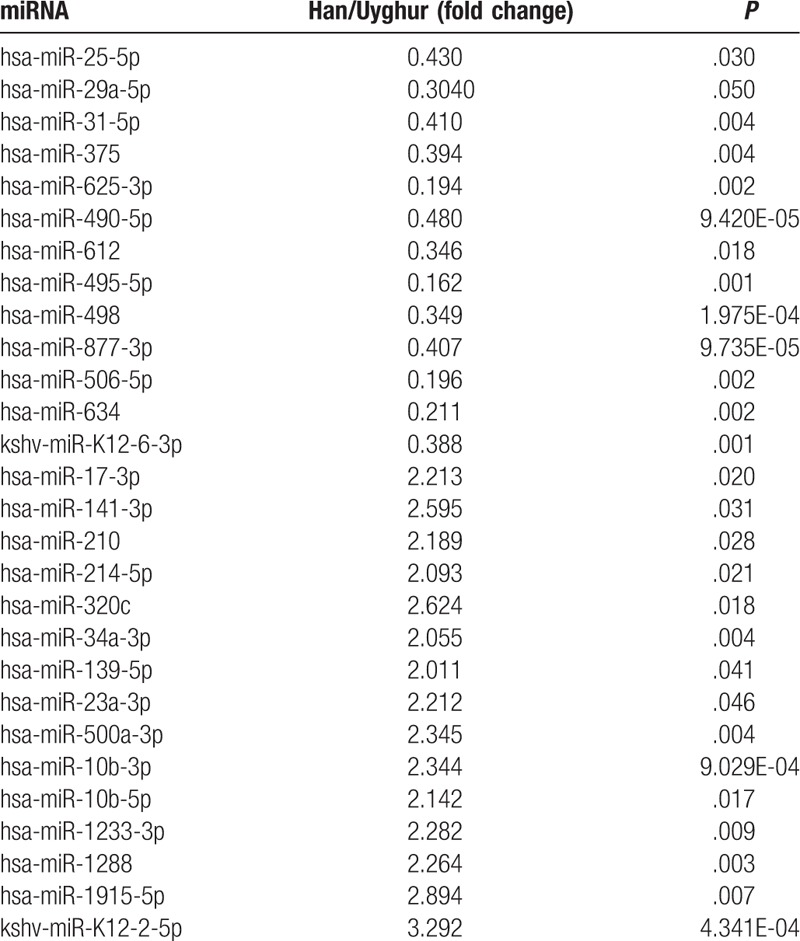
In the 76 upregulated miRNAs, 32 miRNAs have been reported in previous articles,[7–10] such as miR-139-5p, miR-23a-3p, miR-500a-3p, miR-10b-3p, miR-10b-5p, miR-1233-3p, etc. Six miRNAs, including miR-17-3p, miR-141-3p, miR-210, miR-214-5p, miR-320c, and miR-34a-3p may play a role in the pathogenesis of SCC. Kshv-miR-K12-2-5p is associated with Kaposi sarcoma and the other miRNAs partially contribute to the pathogenesis of hepatocellular carcinoma, breast cancer, kidney cancer, laryngeal cancer, colorectal cancer, gastric cancer, testis embryonic cell tumor as well as other cancer types.[11]
In the 171 downregulated miRNAs, 33 were described in previous reports, including miR-612, miR-495-5p, miR-498, miR-877-3p, miR-506-5p, miR-634, and so no. Five miRNAs, including miR-25-5p, miR-29a-5p, miR-31-5p, miR-375, and miR-625-3p have been reported to be associated with SCC.[12–15] The miRNA kshv-miR-K12-6-3p may play a role in the pathogenesis of Kaposi sarcoma and it also may participate in hepatocellular carcinoma, breast cancer, kidney cancer, laryngeal cancer, and rectal cancer.[16]
3.3. Validation of microarray results
To further confirm the microarray results, the RT-PCR method was used. The expression levels of miR-141-3p and miR-1915-5p were validated based on P-value and fold change, which showed that the levels of miR-141-3p and miR-1915-5p were 4.42 and 2.81 folds higher in tissue samples from Han subjects compared to Uyghur subjects, respectively. And the results obtained by RT-PCR were consistent with our microarray results (Fig. 2).
Figure 2.
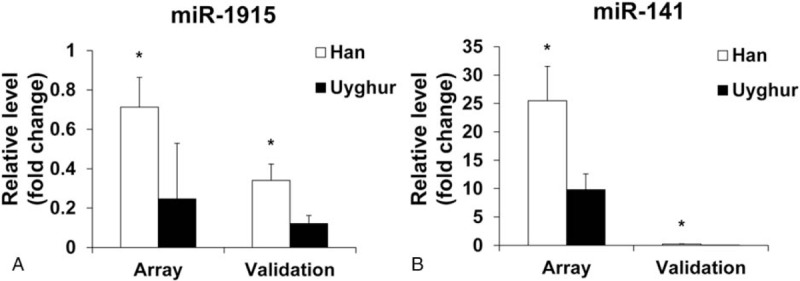
Validation of microarray results by RT-PCR. Expression patterns of miR-141-3p (A) and miR-1915-5p (B). The fold changes were compared. The left most 2 bars of each panel show miRNA levels determined by miRNA microarray assay on Han versus Uyghur tissues. The right most 2 bars show the level of the same miRNA validated by quantitative RT-PCR in the same tissues. Data are mean ± SEM. ∗P < .05 Han versus Uyghur tissues. miRNA = microRNA, RT-PCR = reverse-transcriptase polymerase chain reaction, SEM = standard error of the mean.
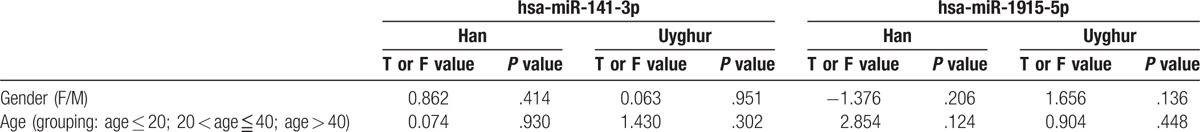
We also analyzed the correlations of miR-141-3p and miR-1915-5p expression with age (grouping: age ≤ 20; 20 < age ≤ 40; and age > 40) or gender among each population, respectively (Table 3). However, there was no statistically significant difference within-group in the expression of miR-141-3p and miR-1915-5p when stratifying subjects by age and gender (P > .05). In future studies, these results should be verified in a larger sample size.
Table 3.
The correlations of the 2 microRNAs (miRNAs) expression levels in Han and Uyghur populations.
3.4. Predicted targets of the identified miRNAs
There is strong evidence that the function of miRNAs mainly depend on their targets. Using the Gene Ontology and KEGG database, we identified that miR-141-3p and miR-1915-5p target genes include ZEB1, ZEB2, EIF4E, BAP1, CLOCK, CD133, and PAX2. The possible biological impacts of the differentially expressed miRNAs based on Gene Ontology are cell–cell signaling, cell proliferation, cell differentiation, and cell death. The KEGG pathways are enriched in some biological pathways, such as p53 signaling pathway, mitogen-activated protein kinase, WNT, and Chemokine signaling pathways (Fig. 3).
Figure 3.
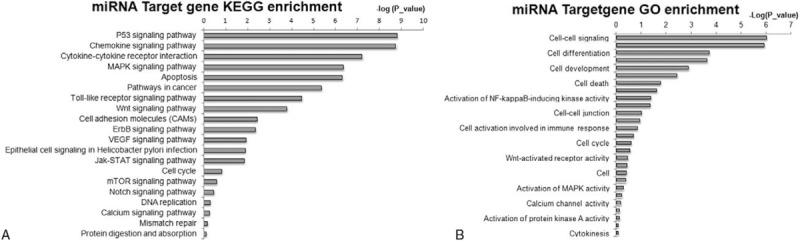
Pathway classifications of the predicted target genes regulated by differentially expressed microRNAs (miRNAs) in the normal skin samples from Uyghur and Han individuals. Pathway analysis was performed on genes predicted to be targets of differentially expressed miRNAs. (A) KEGG pathway classifications of predicted target genes; (B) GO enrichment of predicted target genes.
4. Discussion
Uyghur is the most populous minority ethnicity (10 million Uyghur people) and 99% of the total Uyghur population lives in Xinjiang. The Uyghur population has a high incidence of CSCC and esophageal squamous cell carcinoma. And, high incidence and mortality of melanoma and Kaposi sarcoma is also reported in this population.[17,18] Multiple studies have demonstrated that more than 1 miRNA is involved in the pathogenesis and therapy of cancers.[19–22] Therefore, we aimed to identify miRNAs expression differences between the Uyghur and Han populations.
By using the miRNA array kit, we detected the expression profiles in normal Uyghur and Han skin samples. And, we identified 247 differentially expressed miRNAs (171 miRNAs were upregulated in the Uyghur samples, and 76 miRNAs were upregulated in the Han samples). In further validation studies using RT-PCR, the expression levels of 2 selected miRNAs, hsa-miR-141-3p and hsa-miR-1915-5p, were in agreement with our array data. The identification of dysregulated miRNAs in normal Uyghur and Han skin samples has opened up new possibilities for the diagnosis of certain cancers. However, it should be noted that our understanding of these miRNAs is still limited.
Previous study has shown that miR-320 can regulate tumor angiogenesis of oral cancer by silencing Neuropilin 1.[23] miR-320 level is decreased in oral SCC cell lines and tumor tissues. In our study, although miR-320 was expressed at higher levels in Han subjects compared to Uyghur subjects, there was no significant difference between the 2 populations, which can be partially attributed to the small sample size. In future studies, we will increase the sample size to verify the miR-320 expression. Thus, our miRNA findings may contribute to the future development of antiangiogenic or anticancer drugs.
The miR-31 is an important miRNA that is capable of regulating tumor behavior.[24] Many studies have demonstrated that it displays differential expression and functionality in various tissue and cancers. For example, it reduced tumor metastases in breast cancer, but enhanced neoplasm invasion in colon cancer.[25,26] Many studies have found that miR-31 was expressed at high levels in most SCC samples.[27,28] In our study, miR-31-5p was significantly upregulated in the Uyghur population. Studies[29,30] have demonstrated that miRNAs have tissue specific expression. In laryngeal SCC, particularly in well differentiated and dysplasia tissue, miR-375 is downregulated and positively correlates with TNM stage and lymph node metastasis
Kaposi sarcoma is a chronic, progressive, malignant, pigmentary, vascular sarcoma caused by Human Herpes Virus-8. Blanco and coworkers[31] have found that the Human Herpes Virus-8 viral load increased significantly in infected Kaposi sarcoma tissue. Twelve miRNAs encode genes clustered in Kaposi sarcoma-associated herpesvirus (KSHV) ambush-related regions, which encode for the ambush-related nuclear antigen, LANA, v-cyclin, v-FLIP, and K12. It demonstrates that miR-K12-2, -4, -5, -6, -7, and -9 have obvious polymorphisms, and that the KSHV miRNA genes contribute to KSHV-related malignant carcinoma and pathogenesis after strict selection.[32] Skalsky et al[33] found that miR-155 has 100% seed sequence homology with miR-K12-11, which indicated that the host target genes regulated by has-miR-155 could also be regulated by miR-K12-11. In future studies, we should analyze the role of miR-155 in the pathogenesis of Kaposi sarcoma through its target genes.
As for the target genes of miR-141-3p, the E-cadherin transcriptional repressors contribute to epithelial-mesenchymal transition and cancer cell migration in cancer cell lines.[34] Another target gene of miR-141-3p, BAP1, inhibits PI3K activity, suppressing cell growth in both flies and humans.[35]CD133 and PAX2, the target genes of miR-1915-5p, regulate miRNA expression at both the transcriptional and posttranslational levels and are involved in the repair of adult renal progenitor cells.[36] The miR-1915-5p gene also represses BCL-2 expression in the apoptotic response to DNA damage by P53 induction.[37]
Studies[38] have demonstrated that KSHV-miR-K12-5 can promote viral immune escape. KSHV-miR-K12-3 and KSHV-miR-K12-7 participate in the body's humoral immune response by regulating the secretion of interleukin-6 and interleukin-10. KSHV-miR-K10 and KSHV-miR-K12 regulate kaposin proteins as viral miRNA.[32,39] We found unusual expression patterns of KSHV-miR-K12-2-5p and KSHV-miR-K12-6-3p in Uyghur and Han samples, respectively. Future studies should further analyze the roles of these miRNAs in viral immune escape and immune responses. The Xinjiang region has a high incidence of Kaposi sarcoma, however, no study has analyzed the relationship between viral miRNA and Kaposi sarcoma. Based on our initial miRNA screening, future studies can be performed to identify possible target genes in human tissue and blood, as well as in cells and animals for further verification.
In comparison to the miRNA expression profiles of Kaposi sarcoma in China, they reported that there were 15 miRNAs expressed in the Uyghur and Han population.[40] The miRNAs included 3 upregulated (kshv-miR-K12-2-5p, miR-23a-3p, and miR-221-3p, etc.) and 12 downregulated miRNAs (miR-1908, miR-4251, miR-483-3p, etc.). The miRNA expression levels of Kaposi sarcoma had obvious differences between the Han and Uyghur populations. Therefore, we speculate that the dysregulated expression of the above miRNAs may be markers of susceptibility to Kaposi sarcoma. Future works should focus on increasing the sample size. RNA pool from the female/male/young/aged skin/different body sites should be obtained to further confirm or improve the results of this study. Additionally, the basic clinical data of enrolled subjects were comparable at baseline level. RT-PCR results showed that there was no significant interindividual expression difference in miR-141-3p and miR-1915-5p. Thus, we speculate that there are no interindividual expression differences within the subpopulations.
In conclusion, we have investigated the expression of miRNAs in the skin samples from the Han and Uyghur populations. A total of 247 miRNAs were differentially expressed in the Han versus Uyghur population (76 upregulated and 171 downregulated). The upregulation of miR-141-3p and miR-1915-5p was confirmed by the RT-PCR. Our findings may provide a better understanding of the differences in the incidence and mortality of skin related carcinoma between the Uyghur and Han populations in Xinjiang.
Acknowledgments
The authors thank all of the families who willingly participated in this study. The authors also thank the support of the Clinical Dermatology Institute of the Xinjiang Uygur Autonomous Region and the National Clinical key specialty construction projects; and the Natural Science Foundation of China (Grant No. 81660454).
Footnotes
Abbreviations: CSCC = cervical squamous cell carcinoma, KSHV = Kaposi sarcoma-associated herpesvirus, miRNA = microRNA, SCC = squamous cell carcinoma.
XW and ZZ contributed equally to this work and should be considered first coauthors.
The authors have no conflicts of interest to disclose.
References
- [1].Hrustincova A, Votavova H, Dostalova Merkerova M. Circulating MicroRNAs: methodological aspects in detection of these biomarkers. Folia Biol (Praha) 2015;61:203–18. [DOI] [PubMed] [Google Scholar]
- [2].Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A 2008;105:5166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hameetman L, Commandeur S, Bavinck JN, et al. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer 2013;13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].McDougall JA, Madeleine MM, Daling JR, et al. Racial and ethnic disparities in cervical cancer incidence rates in the United States, 1992-2003. Cancer Causes Control 2007;18:1175–86. [DOI] [PubMed] [Google Scholar]
- [5].Wu XJ, Pu XM, Kang XJ, et al. One hundred and five Kaposi sarcoma patients: a clinical study in Xinjiang, Northwest of China. J Eur Acad Dermatol Venereol 2014;28:1545–52. [DOI] [PubMed] [Google Scholar]
- [6].Kang XJ, Shi XH, Chen WJ, et al. Analysis of KIT mutations and c-KIT expression in Chinese Uyghur and Han patients with melanoma. Clin Exp Dermatol 2016;41:81–7. [DOI] [PubMed] [Google Scholar]
- [7].Luo HN, Wang ZH, Sheng Y, et al. MiR-139 targets CXCR4 and inhibits the proliferation and metastasis of laryngeal squamous carcinoma cells. Med Oncol 2014;31:789. [DOI] [PubMed] [Google Scholar]
- [8].Bao L, Zhao J, Dai X, et al. Correlation between miR-23a and onset of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2014;38:318–30. [DOI] [PubMed] [Google Scholar]
- [9].Joerger M, Baty F, Fruh M, et al. Circulating microRNA profiling in patients with advanced non-squamous NSCLC receiving bevacizumab/erlotinib followed by platinum-based chemotherapy at progression (SAKK 19/05). Lung Cancer 2014;85:306–13. [DOI] [PubMed] [Google Scholar]
- [10].Xie Z, Chen G, Zhang X, et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS One 2013;8:e57502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Catrina Ene AM, Borze I, Guled M, et al. MicroRNA expression profiles in Kaposi's sarcoma. Pathol Oncol Res 2014;20:153–9. [DOI] [PubMed] [Google Scholar]
- [12].Tang J, Tao ZH, Wen D, et al. MiR-612 suppresses the stemness of liver cancer via Wnt/beta-catenin signaling. Biochem Biophys Res Commun 2014;447:210–5. [DOI] [PubMed] [Google Scholar]
- [13].Chu H, Chen X, Wang H, et al. MiR-495 regulates proliferation and migration in NSCLC by targeting MTA3. Tumour Biol 2014;35:3487–94. [DOI] [PubMed] [Google Scholar]
- [14].Kasiappan R, Shen Z, Tse AK, et al. 1,25-Dihydroxyvitamin D3 suppresses telomerase expression and human cancer growth through microRNA-498. J Biol Chem 2012;287:41297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhao Z, Ma X, Hsiao TH, et al. A high-content morphological screen identifies novel microRNAs that regulate neuroblastoma cell differentiation. Oncotarget 2014;5:2499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Morgan RW, Burnside J. Roles of avian herpesvirus microRNAs in infection, latency, and oncogenesis. Biochim Biophys Acta 2013;1809:654–9. [DOI] [PubMed] [Google Scholar]
- [17].Daniotti M, Ferrari A, Frigerio S, et al. Cutaneous melanoma in childhood and adolescence shows frequent loss of INK4A and gain of KIT. J Invest Dermatol 2009;129:1759–68. [DOI] [PubMed] [Google Scholar]
- [18].Zhang D, Pu X, Wu W, et al. Genotypic analysis on the ORF-K1 gene of human herpesvirus 8 from patients with Kaposi's sarcoma in Xinjiang, China. J Genet Genomics 2008;35:657–63. [DOI] [PubMed] [Google Scholar]
- [19].Huang SD, Yuan Y, Zhuang CW, et al. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol Cancer 2012;11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee JW, Choi CH, Choi JJ, et al. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res 2008;14:2535–42. [DOI] [PubMed] [Google Scholar]
- [21].Marshall V, Martro E, Labo N, et al. Kaposi sarcoma (KS)-associated herpesvirus microRNA sequence analysis and KS risk in a European AIDS-KS case control study. J Infect Dis 2010;202:1126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].de Rosa F, Fanini F, Guidoboni M, et al. MicroRNAs and dendritic cell-based vaccination in melanoma patients. Melanoma Res 2014;24:181–9. [DOI] [PubMed] [Google Scholar]
- [23].Wu YY, Chen YL, Jao YC, et al. Hong TM. miR-320 regulates tumor angiogenesis driven by vascular endothelial cells in oral cancer by silencing neuropilin 1. Angiogenesis 2014;17:247–60. [DOI] [PubMed] [Google Scholar]
- [24].Valastyan S. Weinberg RA. miR-31: a crucial overseer of tumor metastasis and other emerging roles. Cell Cycle 2010;9:2124–9. [DOI] [PubMed] [Google Scholar]
- [25].Valastyan S, Chang A, Benaich N, et al. Activation of miR-31 function in already-established metastases elicits metastatic regression. Genes Dev 2011;25:646–59. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [26].Cottonham CL, Kaneko S. Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem 2010;285:35293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tan X, Qin W, Zhang L, et al. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res 2011;17:6802–11. [DOI] [PubMed] [Google Scholar]
- [28].Liu CJ, Lin SC, Yang CC, et al. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck 2012;34:219–24. [DOI] [PubMed] [Google Scholar]
- [29].Yu X, Wu Y, Liu Y, et al. miR-21, miR-106b and miR-375 as novel potential biomarkers for laryngeal squamous cell carcinoma. Curr Pharm Biotechnol 2014;15:503–8. [DOI] [PubMed] [Google Scholar]
- [30].Sun X, Song Y, Tai X, et al. MicroRNA, expression and its detection in human supraglottic laryngeal squamous cell carcinoma. Biomed Rep 2013;1:743–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kouri V, Martinez PA, Blanco O, et al. Simultaneous quantification of human herpesvirus 8 DNA by real time PCR in different tissues of HIV infected cuban patients with Kaposi's sarcoma. Herpesviridae 2010;1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen M, Sun F, Han L, et al. Kaposi's sarcoma herpesvirus (KSHV) microRNA K12-1 functions as an oncogene by activating NF-kappaB/IL-6/STAT3 signaling. Oncotarget 2016;7:33363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Skalsky RL, Samols MA, Plaisance KB, et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol 2007;81:12836–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mahdavinezhad A, Mousavi-Bahar SH, Poorolajal J, et al. Evaluation of miR-141, miR-200c, miR-30b expression and clinicopathological features of bladder cancer. Int J Mol Cell Med 2015;4:32–9. [PMC free article] [PubMed] [Google Scholar]
- [35].Hyun S, Lee JH, Jin H, et al. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 2009;139:1096–108. [DOI] [PubMed] [Google Scholar]
- [36].Sallustio F, Serino G, Costantino V, et al. miR-1915 and miR-1225-5p regulate the expression of CD133, PAX2 and TLR2 in adult renal progenitor cells. PLoS One 2013;8:e68296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nakazawa K, Dashzeveg N, Yoshida K. Tumor suppressor p53 induces miR-1915 processing to inhibit Bcl-2 in the apoptotic response to DNA damage. FEBS J 2014;281:2937–44. [DOI] [PubMed] [Google Scholar]
- [38].Qin Z, Kearney P, Plaisance K, et al. Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukoc Biol 2010;87:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lei X, Zhu Y, Jones T, et al. A Kaposi's sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor beta pathway to promote cell survival. J Virol 2012;86:11698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wu XJ, Pu XM, Zhao ZF, et al. The expression profiles of microRNAs in Kaposi's sarcoma. Tumour Biol 2015;36:437–46. [DOI] [PubMed] [Google Scholar]


