Abstract
The aim of this study was to investigate the predicting value of miR-146a/b for acute exacerbation chronic obstructive pulmonary disease (AECOPD) and COPD, and to explore their associations with inflammatory cytokines in AECOPD and stable COPD patients.
One hundred six AECOPD, 122 stable COPD patients, and 110 health volunteers with age and sex matched to total COPD patients (AECOPD and stable COPD) were enrolled. Blood samples were collected from all participants. Relative expression of miR-146a/b was determined by real-time polymerase chain reaction. Tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), interleukin-6 (IL-6), interleukin-8 (IL-8), leukotriene B4 (LTB-4) expression in serum from AECOPD and stable COPD patients were assessed using commercial ELISA kit.
Serum levels of miR-146a and miR-146b were down regulated in AECOPD patients compared with stable COPD patients and HCs. miR-146a and miR-146b are of good values for predicting the risk of AECOPD in HCs with AUC of 0.702 and 0.715. Additionally, miR-146a and miR-146b could distinguish AECOPD from stable COPD patients with AUC of 0.670 and 0.643. In AECOPD patients, levels of miR-146a in AECOPD patients were negatively associated with TNF-α, IL-6, IL-8, and LTE-4 expression. In stable COPD patients, miR-146a expressions were negatively correlated with TNF-α, IL-1β, IL-6, IL-8, and LTE-4 levels. And, the expressions of miR-146b in AECOPD patients were negatively associated with IL-1β and LTB-4 expression. While in stable COPD patients, miR-146b expressions were only negatively correlated with TNF-α level.
In conclusion, miR-146a and miR-146b were negatively correlated with inflammatory cytokines, and could be promising biomarkers for predicting the risk of AECOPD in stable COPD patients and healthy individuals.
Keywords: acute exacerbation chronic obstructive pulmonary disease, circulating, chronic obstructive pulmonary disease, inflammatory cytokines, miR-146a/b
1. Introduction
Characterized by almost irreversible obstruction of airflow and aggressive disease progression, chronic obstructive pulmonary disease (COPD) is driving increasing attention over decades due to its high mortality and fatal comorbidities.[1] Up to 2030, COPD will become the third cause of deaths worldwide, and the prevalence is still increasing in developing countries at present.[2] Unfortunately, COPD is over-diagnosed in the elderly and underdiagnosed in adults.[3,4] Although COPD has been described as a treatable disease and smoking cessation, β2 agonists as well as long-acting anticholinergic agents are effective to some extent, the treatment of COPD is still not satisfactory.[5]
MicroRNAs (miRNAs) are a growing family of small non-coding RNAs (19–25 nucleotides) that regulate gene expression through binding to the 3′ untranslated region (3′ UTR) of targeted messenger RNAs (mRNAs) to inhibit protein translation or degradation of mRNAs.[6] Dysregulation of miRNAs and their pathogenic roles in various diseases have been demonstrated by abundant studies. By altering gene expression, miRNAs regulate cellular activities such as proliferation, apoptosis, differentiation, and migration in multiple types of diseases including renal diseases, cardiovascular dysfunctions, lung diseases, and cancers.[7–10]
Destruction and inability of bronchioles and lung tissue, in which inflammatory disorder plays a critical role, are the base of pathophysiology in COPD.[11] T cells and T-helper-17 cells demonstrate strong associations with inflammatory cascade in COPD,[12,13] in which inflammatory cytokines are crucial in the early stage, such as tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and interleukin-8 (IL-8), after the release of which the inflammatory cells are concentrated in the site of inflammation to mediate the immune responses.[14,15] Therefore, our study aimed to investigate the predicting value of miR-146a/b for acute exacerbation chronic obstructive pulmonary disease (AECOPD) and stable COPD, and to explore their associations with inflammatory cytokines.
2. Materials and methods
2.1. Participants
One hundred six AECOPD patients and 122 stable COPD patients were consecutively enrolled at Department of Respiratory, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology from November 1, 2015 to July 31, 2016. The inclusion criteria of AECOPD patients were: age >40 years; diagnosed with COPD according to Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria; presentation of an acute exacerbation with at least 2 of the following major symptoms (increased dyspnoea, increased sputum purulence, increased sputum volume) or 1 major and 1 minor symptom (nasal discharge/congestion, wheeze, sore throat, cough) for at least 2 consecutive days. The exclusion criteria were: complicated with asthma, lung cancer, or other relevant lung diseases; history of severe infection, malignant tumors, autoimmune diseases. The inclusion criteria of stable COPD patients were: age >40 years; diagnosed with COPD according to GOLD criteria; without acute exacerbation in the last 6 months. The exclusion criteria were the same as AECOPD.
In the meantime, 110 health volunteers with age and sex matched to total COPD patients (AECOPD and stable COPD) underwent physical examinations in the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology were also recruited as health controls (HCs). Volunteers with infection, lung diseases, renal or hepatic dysfunction, and history of severe infection, malignant tumors, autoimmune diseases were excluded.
Written informed consents were obtained from all participants. This study was approved by the Ethics Committee of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology.
2.2. Sample collection
Peripheral blood was collected from AECOPD patients (at the day 1 on admission), stable COPD patients, and HCs. After standing at room temperature for 1 hour, peripheral blood was centrifuged at 1000 × g for 15 minutes at 4 °C, and the supernatant was subsequently acquired and further centrifuged at 16,000 × g for 10 minutes at 4 °C. The supernatant was then collected and used for further detection.
2.3. Real-time polymerase chain reaction
Total RNA in serum from all participants was extracted using TRIzol Reagent (TaKaRa, Japan), and then reversely transcribed with the PrimerScript Real-time reagent kit (TaKaRa, Japan). Quantitative analysis of miR-146a and miR-146b expression was determined using SYBR Premix Ex TaqTM II (TaKaRa, Japan). Levels of miR-146a/b were calculated utilizing the 2–ΔΔt method with U6 as the internal reference.
2.4. Enzyme-linked immuno sorbent assay (ELISA)
TNF-α, IL-1β, IL-6, IL-8, LTB-4 expression in serum from AECOPD and stable COPD patients were determined using commercial ELISA kit according to the manufacturer instructions (all from R&D Systems, USA)
2.5. Statistics
Statistical analysis was performed by SPSS 22.0 software (SPSS, Chicago, Illinois). Data were presented as mean ± standard deviation, median (25th-75th), or count (percentage). Comparison was determined by t test, Wilcoxon rank sum test, or Chi-square test. Receiver operating characteristic curve was performed to distinguish AECOPD patients and stable COPD patients from HCs, and to predict AECOPD risk from stable COPD patients. Spearman test was used to analyze the correlation of miR-146a/b expression with inflammatory cytokines. P value <.05 was considered significant.
3. Results
3.1. Baseline characteristics of participants
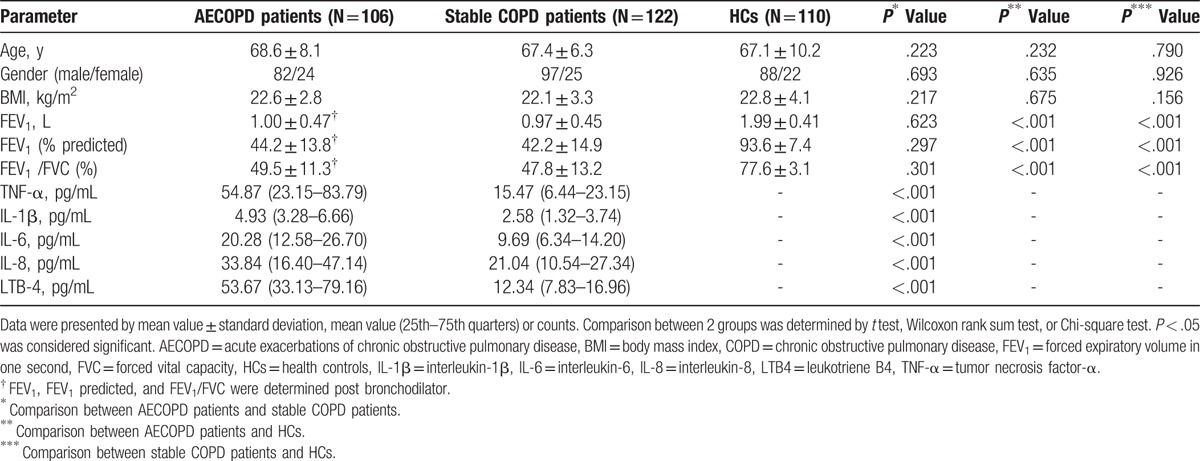
As listed in Table 1, mean age of AECOPD, stable COPD patients, and HCs were 68.6 ± 8.1 years, 67.4 ± 6.3 years, and 67.1 ± 10.2 years, respectively. There were 82 men and 24 women in AECOPD patients, 97 men and 25 women in COPD patients, and 88 men as well as 22 women in HCs. And body mass index (BMI) was 22.6 ± 2.8 kg/m2 in AECOPD group, 22.1 ± 3.3 kg/m2 in stable COPD group, and 22.8 ± 4.1 kg/m2 in HCs. No difference of demographic characteristics was found among groups. Mean levels of forced expiratory volume in one second (FEV1) in AECOPD patients (1.00 ± 0.47 L) post bronchodilator and COPD patients (0.97 ± 0.45 L) were obviously lower than that in HCs (all P < 0.001). In addition, FEV1 predicted and FEV1/forced vital capacity (FVC) of AECOPD patients were also obviously lower than that in HCs (all P < .001). Besides, TNF-α, IL-1β, IL-6, IL-8, and LTE-4 expression in AECOPD patients were markedly higher than that in stable COPD patients (all P < .001) (Table 1).
Table 1.
Baseline characteristics of AECOPD, stable COPD patients, and HCs.
3.2. Expression of miR-146a/b in AECOPD patients, stable COPD patients, and HCs
Comparisons of miR-146a/b levels in each group were shown in Fig. 1. miR-146a level was markedly down regulated in AECOPD patients (0.535 [0.258–0.759]) compared with stable COPD patients (0.874 [0.375–1.435], P < .001) and HCs (0.902 [0.445–1.388]) (P < .001), while no difference of miR-146a levels in stable COPD patients and HCs was found (P = .406) (Fig. 1A). Additionally, level of miR-146b also prominently reduced in AECOPD patients (0.669 [0.378–1.082]) compared with stable COPD (1.032 [0.485–1.764], P < .001) patients and HCs (1.232 [0.667–1.898]) (P < .001) (Fig. 1B). And expression of miR-146b in stable COPD patients and HCs were similar (P = 0.072).
Figure 1.
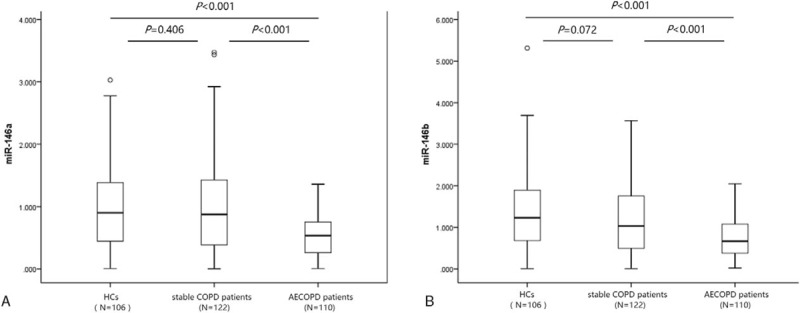
miR-146a/b expression in AECOPD patients, stable COPD patients, and HCs. (A) miR-146a; (B) miR-146b. Comparisons between groups were determined by t test. P < .05 was considered significant. AECOPD = acute exacerbation COPD, COPD = chronic obstructive pulmonary disease, HCs = health controls.
3.3. miR-146a/b expression in COPD and AECOPD patients with different disease severity
The relative expression of miR-146a in AECOPD patients was negatively associated with GOLD stages (P = 0.005) (Fig. 2A), while no difference of miR-146b relative expression among AECOPD patients with different GOLD stages were observed (P = .354) (Fig. 2B). In addition, as presented in Fig. 2C and D, the relative expression of miR-146a (P = .247) and miR-146b (P = .403) were similar in COPD patients with diversified GOLD stages.
Figure 2.
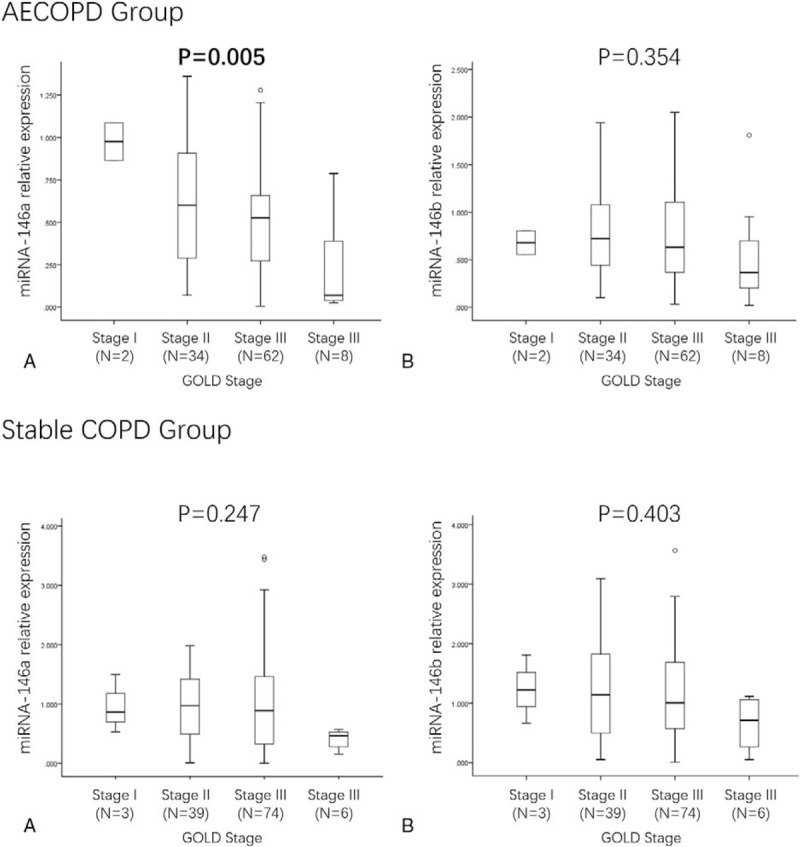
miR-146a/b expression in AECOPD and COPD patients with different stages. (A) miR-146a expression in AECOPD patients with different stages; (B) miR-146b expression in AECOPD patients with different stages; (C) miR-146a expression in COPD patients with different stages; (D) miR-146b expression in COPD patients with different stages. Comparison between groups was determined by Wilcoxon rank sum test. P < .05 was considered significant. AECOPD = acute exacerbation COPD, COPD = chronic obstructive pulmonary disease.
3.4. Predicting values of miR-146a/b for AECOPD, COPD and AECOPD status
As shown in Fig. 3A, miR-146a was of poor value for predicting the risk of COPD from HCs with AUC of 0.532 (95% CI: 0.457–0.606). While miR-146a could predict the risk of AECOPD from stable COPD patients and HCs with AUC of 0.670 (95% CI: 0.600–0.740) and 0.702 (95% CI: 0.632–0.771) (Fig. 3B, C). Additionally, the Youden indexes of miR-146a for predicting the risk of COPD in HCs, AECOPD in stable COPD patients, and AECOPD in HCs were 0.091, 0.388, and 0.365, respectively.
Figure 3.
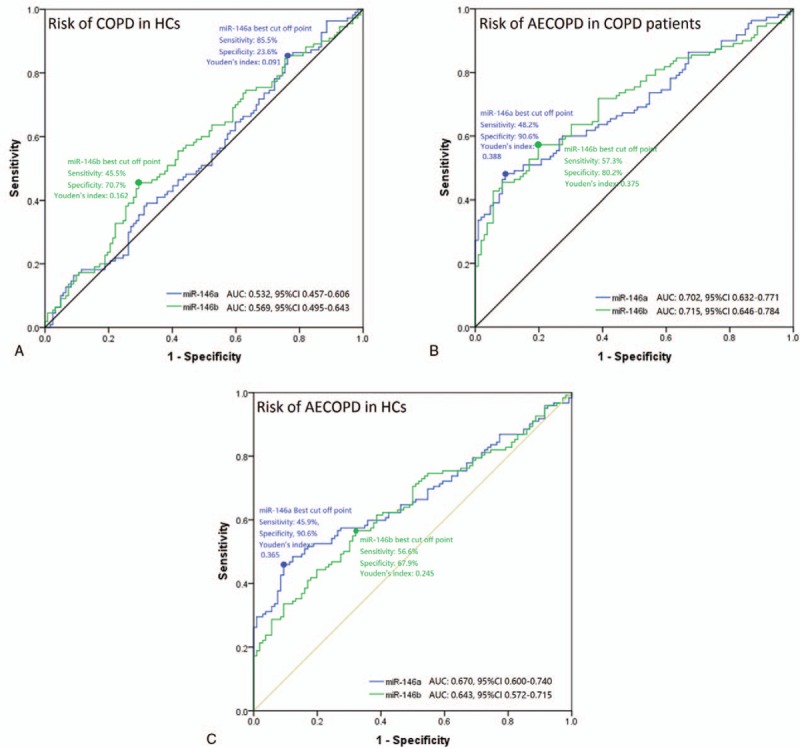
ROC curve analysis of miR-146a/b for predicting stable COPD and AECOPD status. (A) ROC curve analysis of miR-146a/b expression for predicting stable COPD risk from HCs. (B) ROC curve analysis of miR-146a/b expression for predicting AECOPD risk from HCs. (C) ROC curve analysis of miR-146a/b expression for predicting AECOPD risk from stable COPD status. AECOPD = acute exacerbation COPD, COPD = chronic obstructive pulmonary disease, HCs = health controls, ROC = receiver operating characteristic.
In addition, miR-146b also could distinguish AECOPD from stable COPD patients and HCs with AUC of 0.643 (95% CI: 0.572–0.715) and 0.715 (95% CI: 0.646–0.784) (Fig. 3B, C). However, poor predicting value for the risk of COPD from HCs of miR-146b was observed, and the AUC was 0.569 (95% CI: 0.495–0.643) (Fig. 3A). And the Youden indexes of miR-146b for diagnosing COPD in HCs, AECOPD in stable COPD patients and AECOPD in HCs were 0.162, 0.375, and 0.245.
3.5. Correlations of miR-146a/b with TNF-α, IL-1β, IL-6, IL-8 and LTE-4 in AECOPD patients and stable COPD patients
In AECOPD patients, levels of miR-146a in AECOPD patients were negatively associated with TNF-α (P < .001), IL-6 (P = .020), IL-8 (P = .036), and LTE-4 (P = .031) expression (Fig. 4 A, C–E). However, no correlation of miR-146a level with IL-1β expression was discovered (Fig. 4B). As for stable COPD patients, miR-146a levels were negatively correlated with TNF-α (P = .018), IL-1β (P = .029), IL-6 (P = .001), IL-8 (P = .036), and LTE-4 (P = .005) levels (Fig. 4F–J).
Figure 4.
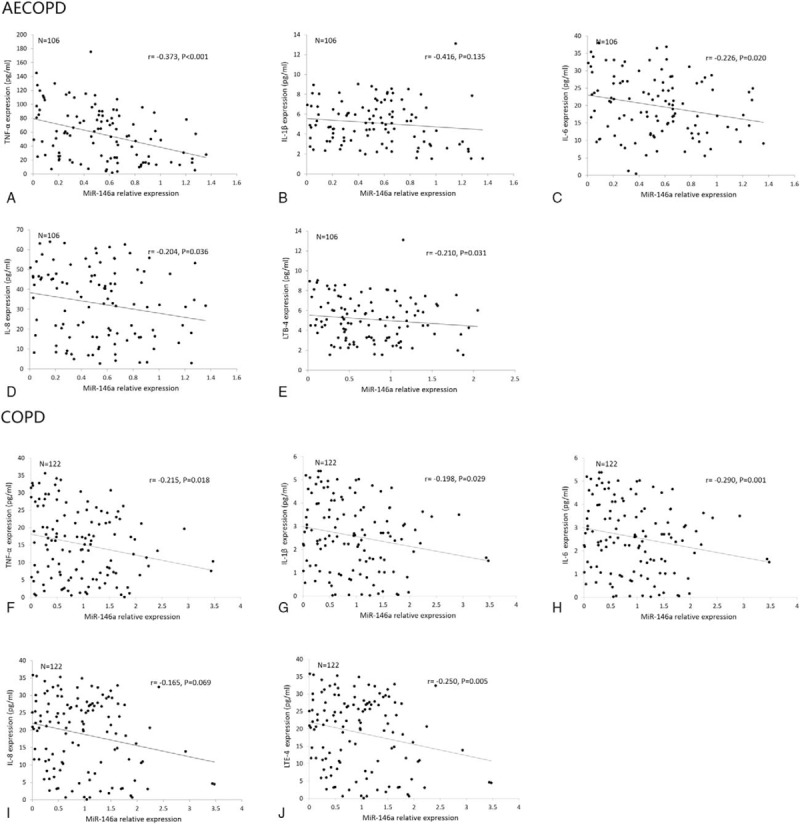
Correlation of miR-146a expression with TNF-α, IL-1β, IL-6, IL-8, LTB-4. (A–E) Correlations of miR-146a with TNF-α, IL-1β, IL-6, IL-8, LTB-4 in AECOPD patients. (F–J) Correlations of miR-146a with TNF-α, IL-1β, IL-6, IL-8, LTB-4 in stable COPD patients. AECOPD = acute exacerbation COPD, COPD = chronic obstructive pulmonary disease, IL-1β = interleukin-1β, IL-6 = interleukin-6, IL-8 = interleukin-8, LTB-4 = leukotriene B4; TNF-α = tumor necrosis factor-α.
As displayed in Fig. 5, levels of miR-146b in AECOPD patients were negatively associated with IL-1β (P = 0.045) and LTB-4 (P = 0.039) expression (Fig. 5B, E) but not with TNF-α (P = .389), IL-6 (P = .067), and IL-8 (P = 0.334) levels (Fig. 5A, C, D). Moreover, in stable COPD patients, miR-146b expression were only negatively correlated with TNF-α level (P = .002) (Fig. 5F).
Figure 5.
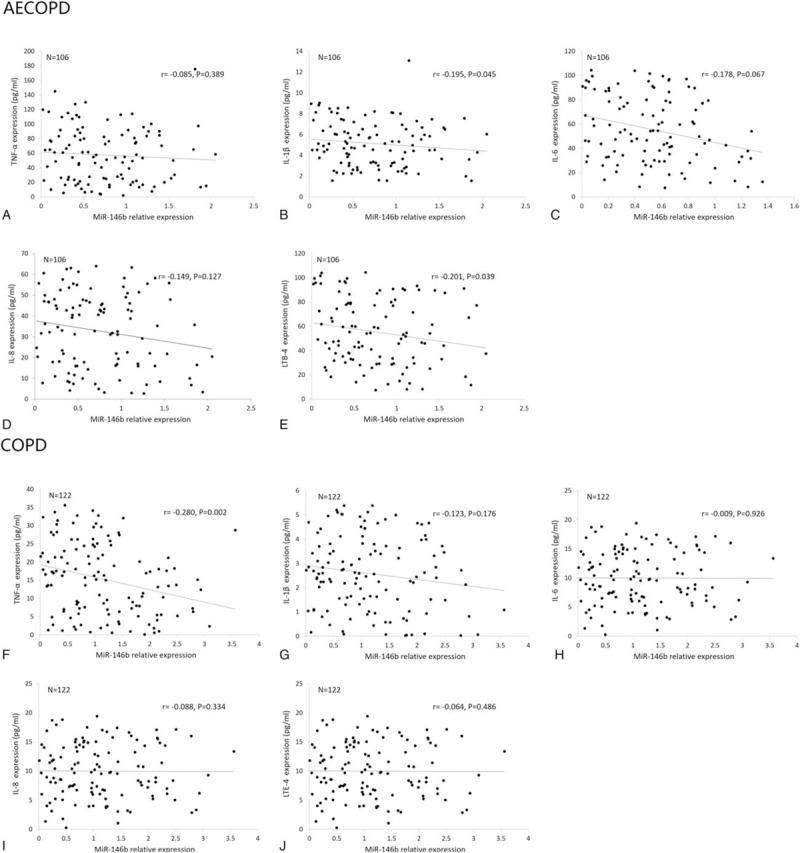
Correlation of miR-146b expression with TNF-α, IL-1β, IL-6, IL-8, LTB-4. (A–E) Correlations of miR-146b with TNF-α, IL-1β, IL-6, IL-8, LTB-4 in AECOPD patients. (F–J) Correlations of miR-146b with TNF-α, IL-1β, IL-6, IL-8, LTB-4 in stable COPD patients. Spearman test was used to analyze the correlations of miR-146a/b expression with inflammatory cytokines. P < .05 was considered significant. AECOPD = acute exacerbation COPD, COPD = chronic obstructive pulmonary disease, IL-1β = interleukin-1β, IL-6 = interleukin-6, IL-8 = interleukin-8, LTB-4 = leukotriene B4; TNF-α = tumor necrosis factor-α.
4. Discussion
In this study, we found: miR-146a and miR-146b were up regulated in AECOPD and COPD patients compared with HCs; miR-146a expression was negatively correlated with the disease severity of AECOPD; miR-146a and miR-146b could predict the risk of AECOPD in COPD patients and the risk of COPD in HCs with good AUCs; miR-146a expression was negatively associated with TNF-α, IL-6, IL-8, and LTE-4 expression in AECOPD patients and TNF-α, IL-1β, IL-6, IL-8, and LTE-4 levels in COPD patients, while miR-146b level was negatively associated with IL-1β and LTB-4 expression in AECOPD patients and TNF-α level in COPD patients.
Exacerbation of COPD patients is a critical issue of disease management, and data showed that the rate of exacerbations of individual patient increased with disease stages.[16] Infection is the most common reason of acute exacerbations, during which patients suffer from aggravating symptoms including cough, dyspnea, and purulent sputum.[2] During infections, inflammation is a process that cannot be ignored, and colonizing bacteria could even result in systemic inflammation.[17]
Dysregulated miRNAs were observed in numerous respiratory diseases, which consists of influenza A (H1N1), asthma, pulmonary fibrosis, pulmonary tuberculosis, and lung cancers.[7,18–21] Compared with smokers without airflow obstruction or healthy individuals, several miRNAs demonstrated dysregulations in COPD patients. And those differentially expressed miRNAs are discovered to be intimately involved in the pathogenesis of COPD.[22] miR-199a-5p is reported to be a crucial regulator of unfolded protein response in COPD patients.[23] And mouse model reveals that miR-223 level is negatively correlated with the expression of histone deacetylase 2 (HDAC2) in patients with COPD.[24] A study investigated the specific role of miRNAs in emphysematous lung destruction of COPD patients and illustrates that miR-638 promotes the maturity of emphysematous lung tissue and lung fibroblasts.[25]
miRNAs have been proved to be more abundantly expressed in tissues than in peripheral blood, and several studies demonstrate that the levels of miRNAs are dysregulated in lung tissue of patients with COPD and contribute to the pathological processes.[26,27] However, the diagnosis of COPD does not need histopathological confirmation, thus the lung tissue of COPD patients are usually not collected.[3] The detection of miRNA expression in circulation, namely the blood sample, is more easily to conduct, and it is widely reported that the miRNA could be detected in circulation of COPD patients.[22,28] Therefore, in our study, the blood samples were collected for the evaluation of miR-146a/b levels in COPD, AECOPD patients and HCs, and their correlations with inflammatory cytokines in COPD and AECOPD patients.
A down regulated miR-146a expression is illuminated to be associated with poor coronary collateral circulation in patients with coronary artery disease, which is an inflammation related disease, the results are partly in line with ours.[29] In the study of Cui et al,[30] a phenotype that has the functional polymorphism in the promoter region miR-146a is correlated with Alzheimer disease (AD) risk and the incidence of cognitive decline in AD patients, which is partly in consistent with our results. Accumulating evidence suggest that miR-146a, a nuclear factor kB (NF-kB) related miRNA, plays essential roles in innate immunological and inflammatory responses.[31] In atopic dermatitis (AD), miR-146 is also found to moderate chronic skin inflammatory through inhibiting NF-kB dependent innate immunology responses in keratinocytes.[32] Additionally, Zheng et al[33] discovered that miR-146a could suppress the NF-kB transcriptional activity and inflammatory factor synthesis, meanwhile it could also ameliorate chemotactic effect on macrophage through inhibiting TRAF6 activity. Recently, a study conducted on Tunisian population reveals a correlation of miR-146a and rheumatoid arthritis (RA), indicating that miR-146a might play a protective role in RA pathogenesis, which is mediated by plentiful pro-inflammatory cytokines through NF-kB pathways.[34] In addition, miR-146a expression in serum was illuminated to be negatively associated with the levels of TNF-α, IL-6, IL-8, and LTE-4 in AECOPD patients, and that miR-146a was negatively correlated with TNF-α, IL-6, IL-1β, IL-8, and LTE-4 levels in COPD patients in our study. The probable explanation of the correlation of miR-146a with those inflammatory cytokines may be that miR-146a is involved in the modulation of those cytokines expression. The investigations of the miR-146a mediating inflammatory cytokines in different cell types of COPD and AECOPD patients were few, however several studies illustrate that miR-146a is dysregulated in different types of cells in COPD patients. Such as, Jemal et al[1] observed a dysregulation of miR-146a in lung tissue of COPD patients and that miR-146a could target COX2 in fibroblasts.
And in the study of Dalbeth et al,[35] cell experiments on human monocytic THP-1 cells with MSU crystals shows that miR-146a inhibits gene expression of monosodium urate (MSU) crystal induced IL-1β, TNFα, monocyte chemoattractant protein-1 (MCP-1), and IL-8. miR-146a negatively mediates donor T cells through targeting TRAF6, and resulting in the reduction of TNF transcription in acute graft-versus-host disease (GVHD) mice models.[36] In the mice models with diabetes, it is observed that in miR-146a overexpressed transgenic mice the increased expression of IL-6, TNFα, and IL1β are prevented, suggesting miR-146a might play anti-inflammatory role by suppressing those inflammatory cytokines.[37] Although no study has revealed that miR-146a is involved in the regulation of LTE 4 expression, the negative association of miR-146a with TNF-α, IL-6, IL-1β, and IL-8 expression in our study might stem from that miR-146a could modulate the expression of those pro-inflammatory cytokines through multiple mechanisms.[35–37]
miR-146b, located on human chromosome 10 (10q24.32), is another member of miR-146 family that has been demonstrated to be a mediator of innate immune responses.[38] It is disclosed that miR-146b is increased in patients with osteoarthritis.[39] Despite that recent studies illustrate miR-146b might play a tumor suppressive role in human cancers through targeting pathways such as HuR/lincRNA-p21/β-catenin, its regulating role in inflammatory responses was relatively better established.[38,40] And miR-146–5p demonstrate a non-cell-autonomous tumor suppressor function via down regulation of IL-6, which is a pro-inflammatory cytokine and was also evaluated in our study.[41] And it is reported that over expression of miR-146a and miR-146b reduces secretion of inflammatory cytokines, including IL-12p70, IL-6, TNF-α, and IFN-γ via targeting TRAF6 and IRAK1 proteins.[42] It is also reported that miR-146b as well as miR-146a negatively regulates the levels of IL-6 and IL-8, which were illustrated to be senescence related inflammatory cytokines.[43] In this study, miR-146b was strikingly down regulated in AECOPD patients compared with COPD patients and HCs, which could be explained by the anti-inflammatory effect of miR-146b found by previous studies.[38,40–43] In addition, miR-146b level was negatively correlated with IL-1β and LTE-4 expression in AECOPD patients, and TNF-α level in COPD patients. A study shows that the TNF-α expression is over expressed in monocytes of obese patients after the suppression of miR-146b-5p by the antisense inhibitor.[44] Comer et al[45] illustrate that transfection of miR-146b mimics decreased the expressions of COX-2 and IL-β in human airway smooth muscles. However, miR-146b has not been found could regulate the LTE-4 expression.
Diagnostic value of miRNAs for AECOPD or COPD was not well investigated by previous studies, however, a prior study elucidated a good sensitivity and specificity of miR-21 and miR-181a for distinguishing the development of COPD.[28] In this present study, we found that miR-146a/b could predict the risk of AECOPD from stable COPD patients and HCs, additionally, the diagnostic value of miR-146a/b for predicting the risk of AECOPD in COPD patients was greater than that for predicting the risk of AECOPD in HCs, and the value of miR-146a/b for predicting the risk of COPD in HCs was the lowest. It might result from that the mechanisms of miR-146a/b mediating the levels of inflammatory cytokines reported by previous studies.[31,32,34–36,38,40–42,46–51]
Some limitations existed in this study: we did not investigate the expression of miR-146a/b in lung tissue, which was reported to be a place that also contains stable miR-146a/b beside peripheral blood, however, it is not ethical and appropriate to collect lung tissue from healthy individuals; detailed mechanisms of miR-146a/b modulating TNF-α, IL-1β, IL-6, IL-8, and LTE-4 were not investigated in our study; sample size of our study was relatively small.
5. Conclusion
In conclusion, our study revealed that miR-146a and miR-146b were negatively correlated with inflammatory cytokines, and could be promising biomarkers for predicting the risk of AECOPD in stable COPD patients and healthy individuals.
Footnotes
Abbreviations: 3′ UTR = 3′ untranslated region, AD = atopic dermatitis, AECOPD = acute exacerbation COPD, CI = confidence interval, COPD = chronic obstructive pulmonary disease, GVHD = graft-versus-host disease, HCs = health controls, HDAC2 = histone deacetylase 2, MacCM = macrophage-conditioned medium, MCP-1 = monocyte chemoattractant protein-1, miRNAs = microRNAs, MSU = monosodium urate, NF-kB= nuclear factor kB, RA = rheumatoid arthritis, SGBS = Simpson-Golabi-Behmel syndrome, TNF-α = tumor necrosis factor-α.
B-BC and Z-HL have contributed equally to this work.
Conflicts of Interest: The authors have no conflicts of interest to disclose.
References
- [1].Jemal A, Ward E, Hao Y, et al. Trends in the leading causes of death in the United States, 1970–2002. JAMA 2005;294:1255–9. [DOI] [PubMed] [Google Scholar]
- [2].Decramer M, Janssens W, Miravitlles M. Chronic obstructive pulmonary disease. Lancet 2012;379:1341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vollmer WM, Gislason T, Burney P, et al. Comparison of spirometry criteria for the diagnosis of COPD: results from the BOLD study. Eur Respir J 2009;34:588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hvidsten SC, Storesund L, Wentzel-Larsen T, et al. Prevalence and predictors of undiagnosed chronic obstructive pulmonary disease in a Norwegian adult general population. Clin Respir J 2010;4:13–21. [DOI] [PubMed] [Google Scholar]
- [5].Anthonisen NR, Connett JE, Murray RP. Smoking and lung function of Lung Health Study participants after 11 years. Am J Respir Crit Care Med 2002;166:675–9. [DOI] [PubMed] [Google Scholar]
- [6].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Booton R, Lindsay MA. Emerging role of MicroRNAs and long noncoding RNAs in respiratory disease. Chest 2014;146:193–204. [DOI] [PubMed] [Google Scholar]
- [8].Mansoori B, Mohammadi A, Shirjang S, et al. Micro-RNAs: the new potential biomarkers in cancer diagnosis, prognosis and cancer therapy. Cell Mol Biol (Noisy-Le-Grand) 2015;61:1–0. [PubMed] [Google Scholar]
- [9].Wang Z, Qin C, Zhang J, et al. MiR-122 promotes renal cancer cell proliferation by targeting Sprouty2. Tumour Biol 2017;39:1010428317691184. [DOI] [PubMed] [Google Scholar]
- [10].Riches K, Huntriss J, Keeble C, et al. Mapping the methylation status of the miR-145 promoter in saphenous vein smooth muscle cells from individuals with type 2 diabetes. Diab Vasc Dis Res 2017;14:122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cosio MG, Majo J, Cosio MG. Inflammation of the airways and lung parenchyma in COPD: role of T cells. Chest 2002;121:160S–5S. [DOI] [PubMed] [Google Scholar]
- [12].Alcorn JF, Crowe CR, Kolls JK. TH17 cells in asthma and COPD. Annu Rev Physiol 2010;72:495–516. [DOI] [PubMed] [Google Scholar]
- [13].Di Stefano A, Caramori G, Gnemmi I, et al. T helper type 17-related cytokine expression is increased in the bronchial mucosa of stable chronic obstructive pulmonary disease patients. Clin Exp Immunol 2009;157:316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zheng J, Shi Y, Xiong L, et al. The expression of IL-6, TNF-alpha, and MCP-1 in respiratory viral infection in acute exacerbations of chronic obstructive pulmonary disease. J Immunol Res 2017;2017:8539294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nadigel J, Audusseau S, Baglole CJ, et al. IL-8 production in response to cigarette smoke is decreased in epithelial cells from COPD patients. Pulm Pharmacol Ther 2013;26:596–602. [DOI] [PubMed] [Google Scholar]
- [16].Hoogendoorn M, Feenstra TL, Hoogenveen RT, et al. Association between lung function and exacerbation frequency in patients with COPD. Int J Chron Obstruct Pulmon Dis 2010;5:435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med 2008;359:2355–65. [DOI] [PubMed] [Google Scholar]
- [18].Song H, Wang Q, Guo Y, et al. Microarray analysis of microRNA expression in peripheral blood mononuclear cells of critically ill patients with influenza A (H1N1). BMC Infect Dis 2013;13:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Parker MW, Rossi D, Peterson M, et al. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Invest 2014;124:1622–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Singh Y, Kaul V, Mehra A, et al. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem 2013;288:5056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bjaanaes MM, Halvorsen AR, Solberg S, et al. Unique microRNA-profiles in EGFR-mutated lung adenocarcinomas. Int J Cancer 2014;135:1812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dang X, Qu X, Wang W, et al. Bioinformatic analysis of microRNA and mRNA Regulation in peripheral blood mononuclear cells of patients with chronic obstructive pulmonary disease. Respir Res 2017;18:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hassan T, Carroll TP, Buckley PG, et al. miR-199a-5p silencing regulates the unfolded protein response in chronic obstructive pulmonary disease and alpha1-antitrypsin deficiency. Am J Respir Crit Care Med 2014;189:263–73. [DOI] [PubMed] [Google Scholar]
- [24].Leuenberger C, Schuoler C, Bye H, et al. MicroRNA-223 controls the expression of histone deacetylase 2: a novel axis in COPD. J Mol Med (Berl) 2016;94:725–34. [DOI] [PubMed] [Google Scholar]
- [25].Christenson SA, Brandsma CA, Campbell JD, et al. miR-638 regulates gene expression networks associated with emphysematous lung destruction. Genome Med 2013;5:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shi L, Xin Q, Chai R, et al. Ectopic expressed miR-203 contributes to chronic obstructive pulmonary disease via targeting TAK1 and PIK3CA. Int J Clin Exp Pathol 2015;8:10662–70. [PMC free article] [PubMed] [Google Scholar]
- [27].Sato T, Liu X, Nelson A, et al. Reduced miR-146a increases prostaglandin E(2)in chronic obstructive pulmonary disease fibroblasts. Am J Respir Crit Care Med 2010;182:1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xie L, Wu M, Lin H, et al. An increased ratio of serum miR-21 to miR-181a levels is associated with the early pathogenic process of chronic obstructive pulmonary disease in asymptomatic heavy smokers. Mol Biosyst 2014;10:1072–81. [DOI] [PubMed] [Google Scholar]
- [29].Wang J, Yan Y, Song D, et al. Reduced plasma miR-146a is a predictor of poor coronary collateral circulation in patients with coronary artery disease. Biomed Res Int 2016;2016:4285942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cui L, Li Y, Ma G, et al. A functional polymorphism in the promoter region of microRNA-146a is associated with the risk of Alzheimer disease and the rate of cognitive decline in patients. PLoS One 2014;9:e89019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 2006;103:12481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rebane A, Runnel T, Aab A, et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J Allergy Clin Immunol 2014;134:836 e811–47 e811. [DOI] [PubMed] [Google Scholar]
- [33].Zheng CZ, Shu YB, Luo YL, et al. The role of miR-146a in modulating TRAF6-induced inflammation during lupus nephritis. Eur Rev Med Pharmacol Sci 2017;21:1041–8. [PubMed] [Google Scholar]
- [34].Hassine HB, Boumiza A, Sghiri R, et al. Micro RNA-146a but not IRAK1 is associated with rheumatoid arthritis in the Tunisian population. Genet Test Mol Biomarkers 2017;21:92–6. [DOI] [PubMed] [Google Scholar]
- [35].Dalbeth N, Pool B, Shaw OM, et al. Role of miR-146a in regulation of the acute inflammatory response to monosodium urate crystals. Ann Rheum Dis 2015;74:786–90. [DOI] [PubMed] [Google Scholar]
- [36].Stickel N, Prinz G, Pfeifer D, et al. MiR-146a regulates the TRAF6/TNF-axis in donor T cells during GVHD. Blood 2014;124:2586–95. [DOI] [PubMed] [Google Scholar]
- [37].Chen S, Feng B, Thomas AA, et al. miR-146a regulates glucose induced upregulation of inflammatory cytokines extracellular matrix proteins in the retina and kidney in diabetes. PLoS One 2017;12:e0173918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee HM, Kim TS, Jo EK. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep 2016;49:311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Budd E, de Andres MC, Sanchez-Elsner T, et al. MiR-146b is down-regulated during the chondrogenic differentiation of human bone marrow derived skeletal stem cells and up-regulated in osteoarthritis. Sci Rep 2017;7:46704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yang W, Yu H, Shen Y, et al. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/beta-catenin pathway. Oncotarget 2016;7:41505–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Al-Ansari MM, Aboussekhra A. miR-146b-5p mediates p16-dependent repression of IL-6 and suppresses paracrine procarcinogenic effects of breast stromal fibroblasts. Oncotarget 2015;6:30006–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Park H, Huang X, Lu C, et al. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J Biol Chem 2015;290:2831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bhaumik D, Scott GK, Schokrpur S, et al. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 2009;1:402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hulsmans M, Van Dooren E, Mathieu C, et al. Decrease of miR-146b-5p in monocytes during obesity is associated with loss of the anti-inflammatory but not insulin signaling action of adiponectin. PLoS One 2012;7:e32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Comer BS, Camoretti-Mercado B, Kogut PC, et al. MicroRNA-146a and microRNA-146b expression and anti-inflammatory function in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol 2014;307:L727–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Saba R, Sorensen DL, Booth SA. MicroRNA-146a: a dominant, negative regulator of the innate immune response. Front Immunol 2014;5:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Srivastava A, Nikamo P, Lohcharoenkal W, et al. MicroRNA-146a suppresses IL-17-mediated skin inflammation and is genetically associated with psoriasis. J Allergy Clin Immunol 2017;139:550–61. [DOI] [PubMed] [Google Scholar]
- [48].Song Y, Dou H, Li X, et al. Exosomal miR-146a contributes to the enhanced therapeutic efficacy of interleukin-1beta-primed mesenchymal stem cells against sepsis. Stem Cells 2017;35:1208–21. [DOI] [PubMed] [Google Scholar]
- [49].Luo X, Han M, Liu J, et al. Epithelial cell-derived micro RNA-146a generates interleukin-10-producing monocytes to inhibit nasal allergy. Sci Rep 2015;5:15937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Roos J, Enlund E, Funcke JB, et al. miR-146a-mediated suppression of the inflammatory response in human adipocytes. Sci Rep 2016;6:38339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jiang W, Liu J, Dai Y, et al. MiR-146b attenuates high-fat diet-induced non-alcoholic steatohepatitis in mice. J Gastroenterol Hepatol 2015;30:933–43. [DOI] [PubMed] [Google Scholar]


