Summary
While NME1 is well known for its ability to suppress metastasis of melanoma, the molecular mechanisms underlying this activity are not completely understood. Herein, we utilized a bioinformatics approach to systematically identify genes whose expression is correlated with the metastasis suppressor function of NME1. This was accomplished through a search for genes that were regulated by NME1, but not by NME1 variants lacking metastasis suppressor activity. This approach identified a number of novel genes, such as ALDOC, CXCL11, LRP1b and XAGE1 as well as known targets such as NETO2, which were collectively designated as an NME1-Regulated Metastasis Suppressor Signature (MSS). The MSS was associated with prolonged overall survival in a large cohort of melanoma patients in The Cancer Genome Atlas (TCGA). The median overall survival of melanoma patients with elevated expression of the MSS genes was greater than 5.6 years longer than patients with lower expression of the MSS genes. These data demonstrate that NMEl represents a powerful tool for identifying genes whose expression is associated with metastasis and survival of melanoma patients, suggesting their potential applications as prognostic markers and therapeutic targets in advanced forms of this lethal cancer.
Introduction
Melanoma is the most lethal form of skin cancer, with an enhanced ability to metastasize to distant organs via the hematogenous or lymphatic circulation (1). While recently-developed inhibitors of MAPK pathway components and immune checkpoint mediators represent the first meaningful progress in the treatment of advanced forms of melanoma (2, 3), most patients still develop resistance to these agents and succumb to metastatic disease. Improved prognostic markers and therapeutic targets are acutely needed to optimize therapy and achieve durable responses in patients with advanced melanoma.
Metastasis suppressor genes (MSGs) encode both proteins (4, 5) and miRNAs (6) with the unique ability to selectively inhibit metastatic potential of tumor cells, with little effect on growth of the primary tumor. As such, MSGs could represent powerful experimental tools for unraveling molecular mechanisms that specifically regulate the metastatic process. The NME1 gene (also known as NM23-H1 or NDPK-A) was the first MSG identified, by virtue of its diminished expression in a metastatic melanoma cell line of murine origin (7). Reduced NME1 expression has since been correlated with metastatic forms of melanoma (8) and many other cancers (9). In addition, forced expression of NME1 suppresses metastatic potential of numerous cancer cell lines both in cell culture and as xenografts in immunocompromised mice (10, 11). We recently showed that concomitant ablation of the NME1 and NME2 genes greatly enhances metastatic potential in a mouse model of ultraviolet light-induced melanoma (12), providing strong in vivo evidence of its metastasis suppressor function. Multiple reports suggest the NME1 protein regulates tumor cell motility via physical and functional interactions with a variety of signaling pathways (13). However, NME1 has also been reported to control RNA expression profiles in breast carcinoma (14) and melanoma cell lines (15), suggesting this activity could also mediate its metastasis suppressor function.
NME1 is a member of a 10-gene family (NME1-9 and RP2), with four of the encoded NME proteins exhibiting nucleoside diphosphate kinase (NDPK) activity (16). The NDPK activity is well-recognized for its role in balancing of intracellular pools of nucleotide diphosphates and triphosphates (17). NME1 is recruited to protein complexes within the cytoskeleton and other intracellular compartments, where the NDPK function may transfer NTPs directly to effector proteins through a process termed “substrate channeling” (18). A role for NDPK activity in the metastasis suppressor function of NME1 has yet to be firmly established, however, with at least one study suggesting it does not contribute (19). Nonetheless, relevance of NDPK is strongly suggested in other settings by the direct association of NME1/NDPK protein with a variety of cytoskeletal and membrane mediators of cell motility (13). In addition, NDPK activity has been shown to supply GTP to dynamin during membrane remodeling and endocytosis of cell surface receptors (20), a function which could well impact metastatic potential of cancer cells. Moreover, disruption of the NDPK active site by a site-directed point mutation of the catalytic histidine-118 residue (H118F) modestly impaired metastasis suppressor function of NME1 in the human melanoma cell line 1205Lu (11). NME1 also binds single-stranded DNA (21, 22), suggesting roles in transcription and DNA repair (23). Indeed, expression of NME1 and NME2 promotes nucleotide excision repair (NER) of UV-induced lesions (24), while NME1/2 deficiency renders mice prone to UV-induced melanoma in situ (25). Moreover, NME1 exhibits 3′-5′ exonuclease activity toward single-stranded DNA templates in vitro, consistent with roles in DNA proofreading and/or repair (26). Site-directed mutagenesis of NME1 and NME2 has identified a number of amino acid residues critical for enzymatic and metastasis suppressor functions (27–29). Our site-directed mutagenesis analysis of the NME1 molecule revealed amino acid residues E5 and K12 to be essential for both 3′–5′ exonuclease activity (i.e. DNA repair) and the metastasis suppressor function (11), a correlation which suggests NME1 may suppress metastasis-driving mutations in melanoma and other human cancers.
NME1 also regulates expression of genes that could serve as effectors of its metastasis suppressor activity. We posited that genes which were regulated by wild-type NME1, but not the suppressor-deficient mutants E5A and K12Q, would yield a signature enriched with genes that mediate metastasis suppressor activity. The approach involved forced expression of wild-type NME1 or one of the metastasis suppressor-deficient mutants in the human melanoma cell line, WM793, followed by assessment of their impacts on gene expression by microarray analysis. The approach identified multiple genes that alone and cumulatively are predictors of improved survival in patients with melanoma. This NME1-dependent metastasis suppressor signature (MSS) contains a number of genes with potential as prognostic markers and targets for therapy in advanced melanoma.
Materials and methods
Cell lines and culture conditions
The melanoma cell lines WM793 and WM278 were gifts of Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA, USA). WM793 and WM278 cells were cultured at 37ºC and 5% CO2 in Tu2% media, composed of the following: MCDB:Leibovitz-15 medium (4:1, v/v; Sigma-Aldrich, St. Louis, MO, USA) supplemented with 2 mM CaCl2, 2.5 μg/ml insulin and 2% fetal bovine serum (Life Technologies, Grand Island, NY, USA). MDA-MB-435s/M14 cells (referred to as M14 in this text) were obtained from R. Plattner (University of Kentucky) (30). 293t cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). M14 and 293t cells were maintained at 37ºC and 10% CO2 in DMEM supplemented with 10% fetal bovine serum, 4.5 g/l D-glucose, L-glutamine, and 110 mg/l sodium pyruvate. Wild-type and mutant variants of NME1 were prepared and characterized as described previously (11). Briefly, cDNA of wild-type or mutant variants of NME1 was cloned into mammalian expression vector pCI-EGFP (provided by S. Kraner, University of Kentucky), followed by transfection into WM793 or M14 cells. Stable transfectants were selected by two week exposure to G418, followed by enrichment for GFP-positive cells by cell sorting. Stable knockdown of NME1 expression in WM278 cells was achieved by lentiviral delivery of Mission shRNA (Sigma-Aldrich, St. Louis, MO, USA) TRCN0000010062 (shNME1#1) or Non-target shRNA pLK0.1 (SHC002, Sigma-Aldrich) as a negative control. Production of viral vectors and transduction were performed according to manufacturer’s instructions.
Experimental metastasis
Six-to-seven week old female nude mice (nu/nu, Harlan Laboratories, Indianapolis, IN) were injected intravenously with 2 × 106 cells in a total volume of 100 μl Hank’s Buffered Saline (HBSS, Invitrogen, Grand Island, NY). The mice were sacrificed three weeks later and the lungs were promptly removed, rinsed in PBS, and immersed in Bouin’s fixative (Polysciences, Warrington, PA) to preserve the tissue and enhance the visibility of potential metastatic lesions. The lesions were counted and their diameter was measured using a dissecting microscope.
RNA isolation and microarray analysis
Parental WM793 cells or WM793 stably expressing wild-type or mutant NME1 were seeded at a density of 1.5 × 105 per 100-mm plastic dish and grown for 3 days to 75–80% confluence in Tu2%. Total cellular RNA was harvested (RNeasy Extraction Kit, Qiagen, Valencia, CA, USA) from five independent dishes for each cell line and treatment group. Purified RNA samples were diluted to 1 mg/mL with RNAse-free deionized water and transferred to the University of Kentucky Microarray Core Facility for cDNA synthesis, hybridization and scanning using their established protocols (http://www.research.uky.edu/microarray/AffymetrixGeneChip.html). Genome-wide expression profiling was conducted using GeneChip® Human Exon 1.0 ST Arrays (Affymetrix, Santa Clara, CA, USA). Data analysis of the microarrays was conducted by Informatics Resources Center within the Institute for Genome Sciences at the University of Maryland, Baltimore (Baltimore, Maryland). Briefly, arrays were normalized by the Robust Multi-Array Average (RMA) method (31) implemented in R package. Differential expression analysis was performed with Linear Models of Microarrays (LIMMA) R package. A linear model was first fitted to expression data for each gene, and the empirical Bayes method was then used to assess differential expression between two wild-type NME1 cells and metastasis suppressor deficient cell lines (parent WM793, E5A or K12Q NME1 mutant lines). A P value cutoff of less than 0.01 was used to select significant probes. Microarray data from this study is deposited in the Gene Enrichment Omnibus under accession number GSE85978.
Enrichment analysis
Enrichment analysis was conducted using DAVID Bioinformatics Resources 6.8 Beta (32, 33) and QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). Exon array probe sets differentially expressed following NME1-wild type overexpression in WM793 cells (p < 0.05) were analyzed against the Gene Ontology Consortium (GO) database and Ingenuity Knowledge Base using all Human Exon 1.0 ST probe sets as background. Statistical significance was determined following multiple testing correction (i.e., Bonferroni and Benjamini-Hochberg).
Nanostring analysis of RNA expression
WM793, WM278 and M14 cells, and their respective stably-transfected variants, were plated at 1.5×105 per 100-mm plastic dish and grown for 3 days to 75–80% confluence. Cells were then serum-starved for 24 hours prior to harvesting total cellular RNA (RNeasy Extraction Kit, Qiagen, Valencia, CA, USA) from 3 independent dishes. For Nanostring transcriptome analysis a custom plate measuring 16 genes of interest was conducted using 250ng of total RNA (50ng/μl) per sample. Five additional housekeeping genes were measured and used for normalization. A complete list of gene targets and probe sequences can be found in Supplementary Table 1. Data analysis was conducted using the nSolver (v.2.5) user interface to operate the nCounter Advanced analysis module from Nanostring (Nanostring Technologies, Seattle, WA, USA). Genes of interested were only considered detectable if the mean of the normalized signal was greater than or equal to two standard deviations above the mean of the negative controls.
TCGA analyses
TCGA mRNA expression data, from Skin Cutaneous Melanoma (SKCM) samples, was obtained through GDAC FireHose (v01-28-2016). The results shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. The acquired dataset was run through the TCGA IlluminaHiSeq_RNASeqV2 pipeline. The mRNA expression data was imported into RStudio and was normalized using the voom function found in the LIMMA package (v1.0.136, RStudio Inc., Boston, MA, USA). The resulting normalized expression values were converted into z-scores and subsequently, expression profiles for specific genes of interest were generated and analyzed using heatmap.2 function, found in the gplots package.
Clinical information for TCGA SKCM patients was also obtained through FireHose version 01-28-2016 and imported into RStudio. In order to determine a relationship between gene expression and clinical outcome, SKCM patient identifiers from both clinical and z-score information were matched. Patients were grouped based on their gene expression profile and analyzed for alterations in both overall survival and recurrence-free survival (34). Kaplan-Meier survival analysis was completed through the survival package in R.
Results
NME1 regulates the profile of RNA expression in WM793 melanoma cells
To analyze the impact of NME1 on RNA expression profiles in melanoma, a cDNA containing the NME1 coding sequence without 5′- and 3′-untranslated sequences was expressed by stable transfection in the human melanoma cell line WM793. The parental WM793 line was derived from a vertical growth phase (VGP) melanoma and exhibits invasive characteristics in culture (35, 36), as well as very low endogenous expression of NME1 protein (11). For RNA expression analysis, total RNA was isolated from WM793 cells and the stably-transfected, WM793-derived line expressing NME1 protein at approximately 3–4× endogenous levels (11). Cells were subjected to 24 hours of serum starvation prior to RNA preparation to minimize induction of gene transcription by serum-associated growth factors. RNA samples were analyzed by microarray using the Affymetrix Human Exon 1.0 ST platform, which assesses expression for 21,014 gene products and variants.
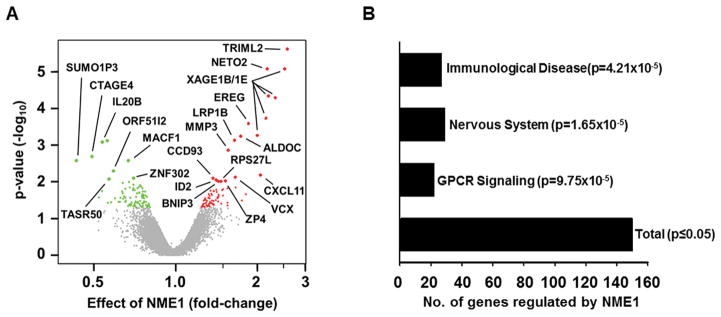
A comparison of expression profiles from the two WM793-derived cell lines identified a total of 153 RNAs whose expression was significantly altered (p < 0.05) by NME1 (Supplementary Table 2). Filtering of the data at higher stringency (p < 0.01) yielded 21 RNAs differentially regulated by NME1, 14 of which were upregulated and seven downregulated (Fig. 1A). Gene ontology analysis identified that half of the genes fell into three functional groupings that were significantly enriched in the list of NME1-regulated genes, namely immunological disease, nervous system development and function, and GPCR signaling (Fig. 1B). The remaining NME1-regulated genes that did not belong to these three ontological groups were associated with ontological groups that failed to achieve statistical significance in the analysis.
Figure 1. Microarray analysis of genes differentially expressed (D.E.) in WM793 cells after stable forced expression of NME1.
A) Volcano plot representation of RNA expression changes following forced NME1 expression in WM793 melanoma cells. Closed red circles depict transcripts upregulated by forced NME1 expression (p < 0.05), with green closed circles indicating downregulated transcripts. Transcripts not significantly regulated by NME1 shown in grey. Transcripts most robustly impacted by NME1 (p < 0.01) are shown as larger closed circles and are labeled with their corresponding gene ID. B) Shown is the number of D.E. genes significantly enriched in various pathways, as determined by gene ontology in DAVID and IPA analyses.
Impact of NME1 and point mutant variants on experimental metastasis of WM793 cells
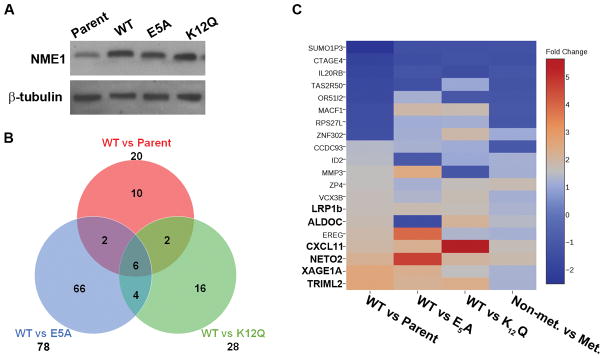
While NME1 regulates expression of gene networks in cancer-derived cell lines, the impacts of this activity on its metastasis suppressor function are not fully understood (15, 37). To identify specific genes and networks mediating metastasis suppressor function of NME1 in melanoma, a filtering strategy was devised to detect genes regulated by the wild-type NME1 protein, but not regulated by mutant variants of NME1 we have shown previously to lack metastasis suppressor activity. For the current study, a panel of WM793-derived cell lines was used that had been created by stable transfection to provide forced expression of wild-type NME1 and the NME1 point mutants glutamate-5 to alanine (E5A), lysine-12 to glutamine (K12Q), and histidine-118 to phenylalanine (H118F). The E5A variant is deficient in 3′–5′ exonuclease activity, variant K12Q is deficient in both the 3′–5′ exonuclease and NDPK activities, and H118F is deficient only in the NDPK activity. Of these point mutants, E5A and K12Q are deficient in metastasis suppressor activity in context of the human melanoma cell line 1205Lu (11). NME1 is rarely mutated in cancers, however, the use of point-mutations that fail to suppress metastasis allows for more streamlined examination of the metastasis suppressor functions of NME1 in experimental settings. The WM793 panel was considered for the current study in light of its higher levels of forced NME1 expression than obtained in the 1205Lu panel, with each NME1 mutant variant expressed at approximately 4-fold over endogenous NME1 levels (Fig. 2A) (11). Prior to RNA expression analysis, metastatic potential of the WM793 panel was measured by experimental metastasis assays, performed by injection of cells into the tail vein of athymic nude mice. Significant lung colonization by WM793 cells was seen in 60% of injected mice, while forced expression of wild-type NME1 strongly suppressed this activity as expected (8.7%; Table 1). In contrast, expression of the E5A or K12Q variants had no significant suppressor effect on lung colonizing activity (50% lung colony-positive for both), consistent with loss of metastasis suppressor activity, as previously described in 1205Lu cells (11). Also consistent with prior results in 1205Lu cells (11), expression of the NDPK-deficient variant H118F had no statistically significant effect on lung colonization in the WM793 line. Taken together, these analyses showed the WM793-derived cell lines expressing wildtype NME1 and the E5A and K12Q variants would provide a robust system for identifying genes whose expression tracks with the metastasis suppressor function of NME1.
Figure 2. Identification of NME1-regulated genes that track with metastasis suppressor activity of NME1.
A) Validation of wild-type (WT) or mutant NME1 over-expresssion in WM793 cells by immunoblot analysis. B) Venn diagram depicting the overlap of genes differentially expressed by WT versus metastasis suppressor deficient mutants of NME1 or parent cells, p=0.01. C) Heatmap showing the relative induction or repression of NME1-regulated genes in all comparisons of cells lines. The 6 genes common to all comparisons from (B) are shown in bold text.
Table 1.
Amino acid residues glutamate-5 and lysine-12 mediate ability of NME1 to suppress experimental metastasis potential of WM793 melanoma cells.
| Cell Lines1 | Experimental Metastasis | |||
|---|---|---|---|---|
| exogenous NME1 | NME1 enzymatic activity | % (Incidence) | Lesions/lung | |
| NDPK | 3′-5′ EXO | |||
| − | − | − | 60a (12/20) | 6a |
| wild-type | + | + | 8.7b (2/23) | 4a,b |
| E5A | + | − | 50a,c (5/10) | 1a,b,c |
| K12Q | − | − | 50a,c (5/10) | 2a,d |
| H118F | + | − | 25a,b,c (3/12) | 2a,b,e |
Experimental metastasis was measured in parental WM793 cells and lines with stable forced expression of the indicated wild type or mutant variants of NME1.
Values not showing a common superscript are significantly different (p < 0.05) as determined by Fisher’s exact test (incidence) and Student’s t-test (lesions/lung).
NDPK, nucleoside diphosphate kinase; EXO, exonuclease; E5A, glutamate-to-alanine substitution; K12Q, lysine-to-glutamine; H118F, histidine-to-phenylalanine.
+, in vitro enzymatic activity of recombinant NME1 variant > 95% of wild-type NME1.
−, in vitro enzymatic activity of recombinant NME1 variant < 5% of wild-type NME1.
Identification of a metastasis suppressor signature (MSS) of RNA expression in human melanoma cell lines
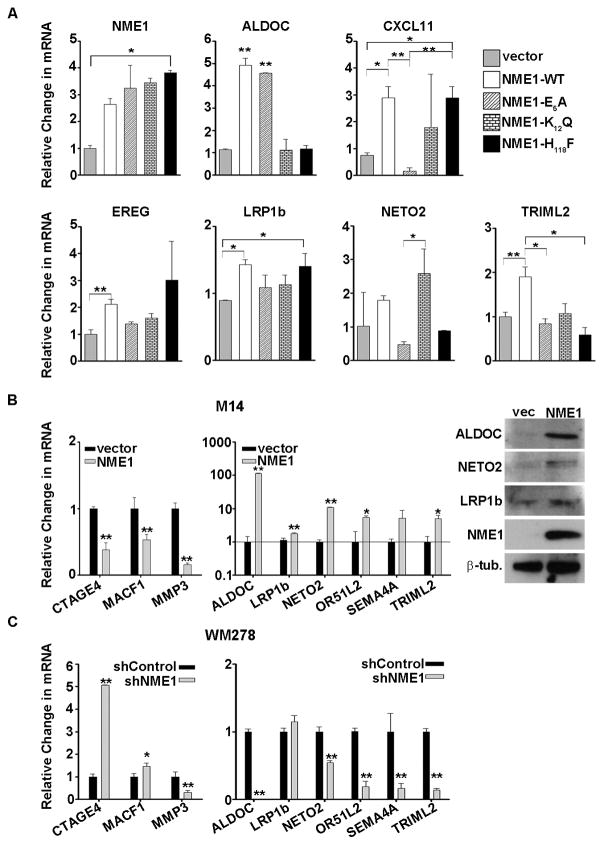
To better identify which of the 153 genes regulated by wild-type (WT) NME1 were associated with its metastasis suppressor activity, microarray analyses were also conducted in WM793 cells expressing the metastasis suppressor-deficient mutants of NME1 (E5A and K12Q). The marked loss of metastasis suppressor activity for the E5A and K12Q mutants made them ideal tools for identifying NME1-regulated genes mediating the suppressor function. It should be noted that the NME1 point mutations used in our filtering approach were generated in vitro for the purpose of dissecting structure-function relationships for the NME1 molecule and are not known to occur naturally within the NME1 sequence in melanoma or other forms of human cancer. The H118F-dependent gene expression profile was not included in our filtering approach for the MSS, as the H118F mutation did not significantly alter the metastasis suppressor activity of NME1 in vivo (Table 1). After three separate comparisons, (WT vs Parent, WT vs E5A, and WT vs K12Q) were compiled, six genes were consistently regulated by wild-type NME1 but not a metastasis suppressor-deficient mutant (Fig. 2B and 2C). This gene expression signature consisted of LDL receptor-related protein (LRP1b), C-X-C motif chemokine 11 (CXCL11), aldolase C (ALDOC), X antigen family member 1A (XAGE1A), neuropilin and tolloid-like 2 (NETO2) and tripartite motif family like 2 (TRIML2). For this report, these genes have been designated as the NME1-regulated metastasis suppressor signature (MSS). To validate results obtained by microarray analysis, Nanostring analysis was employed for quantitation of NME1 and the following NME1-regulated transcripts: ALDOC, CXCL11, EREG, LRP1b, NETO2 and TRIML2. In addition, we measured expression of MSS genes in H118F-expressing WM793 cells to identify any potential contributions of the NDPK function to their regulation. In agreement with microarray and immunoblot analyses, the Nanostring approach demonstrated a 3-to-4-fold induction of NME1 RNA in WM793 cell lines expressing wild-type NME1 and the variants E5A, K12Q and H118F (Fig. 3A). Consistent with the microarray analysis, Nanostring validated the induction of EREG, LRP1b and TRIML2 transcripts by wild-type NME1, and not the metastasis suppressor-deficient variants E5A and K12Q. Also similar to the microarray analyses, Nanostring results indicated that NME1-mediated induction of ALDOC mRNA was dependent on the NDPK function. Wild-type and E5A forms of NME1, which both retain full NDPK activity, induced ALDOC mRNA but the K12Q and H118F mutants did not. The Nanostring analysis yielded considerable variability with impacts of the K12Q mutant on expression of the CXCL11 and NETO2 transcripts limiting the ability to interpret if one or more enzymatic function of NME1 were critical for their expression. On the whole, however, the bulk of the microarray results were validated by the Nanostring approach.
Figure 3. NME1 regulated genes show variable dependence on NME1 enzymatic functions and cell line expression.
A) Nanostring mediated validation of target gene levels after stable expression of empty vector (gray bars), wildtype (WT), NDPK activity deficient NME1 mutants (K12Q and H118F), or 3′–5′ exonuclease deficient mutants (E5A and K12Q). B) The repression or induction of select NME1 target genes after stable expression of wildtype NME1 was further verified in M14 cells by Nanostring (left panel) and immunoblot analyses. C) The effect of NME1 knockdown on its target genes was measured by Nanostring analysis in WM278 cells after stable expression of a control or NME1 specific shRNA. *=p<.05, **=p<0.01.
Nanostring analysis was also used to measure levels of all 21 differentially-regulated genes after overexpression of wild-type NME1 in the M14 melanoma cell line. After normalization to several housekeeping genes, 9/21 of those genes were consistently altered in response to NME1, of which four were MSS genes. Three genes that showed repression by microarray analysis in WM793 also showed significant downregulation by wild-type NME1 in the M14 cells (Fig. 3B). Of the NME1-induced genes, the most robust regulation was that of ALDOC, which was induced over 100-fold (Fig. 3B). Other MSS transcripts strongly induced by NME1 in M14 cells were NETO2 (10-fold) and TRIML2 (5-fold), although LRP1b was only modestly upregulated in this cell line. Immunoblot analysis confirmed that NME1 overexpression induced steady-state concentrations of ALDOC, NETO2 and LRP1b proteins in M14 cells (Fig. 3B).
To further validate NME1-mediated regulation of these genes, the impact of NME1 knockdown was assessed was stably knocked down using lentiviral shRNA expression vectors (shNME1) in WM278 cells. This line was originally derived from a VGP melanoma and, in contrast with WM793 expresses relatively high amounts of NME1 protein. As predicted, loss of endogenous NME1 induced the expression of NME1-suppressed genes CTAGE4, MACF1 and MMP3 (Fig. 3C). Transcript levels of ALDOC, NETO2, OR51L2, SEMA4A and TRIML2 were strongly reduced by shNME1 in WM278 cells (Fig. 3C). Expression of the NME1-induced transcript encoding LRP1b was unaffected by the shNME1 treatment.
Interestingly, each of the MSS genes harbor functions and/or expression patterns suggesting relevance to melanoma phenotype and malignant progression. LRP1b is a compelling candidate as an effector of the metastasis suppressor function for NME1, with reports of potent invasion-suppressing activity across multiple cancer types including melanoma (38, 39). In addition, LRP1b is one of the top three most-frequently mutated genes in melanoma, consistent with a suppressor function (40). A closely related protein, LRP1, was recently shown to aid in suppressing melanoma metastasis, further suggesting that LRP1b is critical for NME1-mediated metastasis suppression (41). Chemokines such as CXCL11 are key players in tumor progression and metastasis, acting in both the primary tumor and metastatic niche. The actions of CXCL11 are pleotropic and context-dependent, due in part to autocrine and paracrine expression patterns of its cognate receptors CXCR3 and CXCR7 (42). Consistent with potential suppressor functions, CXCL11 stimulates recruitment of tumor-infiltrating lymphocytes in T cell lymphomas (43) and breast cancer (44), while its receptor CXCR3 is critical for response to anti-PD-1 therapy in preclinical studies (45). ALDOC is one of three aldolase isozymes (A–C), well-recognized for their roles in glycolysis but more recently implicated in novel glycolysis-independent functions in cell proliferation (46, 47) and motility/invasion (48). ALDOA interacts physically with ARNO, an ADP-ribosylation factor (Arf) and guanine exchange factor (GEF) implicated in cytoskeletal dynamics and membrane trafficking (49). Consistent with an anti-metastatic function, ALDOC expression is correlated with better prognosis in oral squamous cell carcinoma (50), while the isoenzyme ALDOA is upregulated by melanogenesis-induced cell quiescence in the metastatic melanoma cell line B16-F10 (51). Also relevant is the enriched expression of ALDOC protein in cells of neural crest lineage, from which melanocytes are derived (52, 53). XAGE1B is localized to nuclear speckles, suggesting a role in RNA processing and/or export, and its expression is strongly elevated in metastatic melanoma (54). NETO2 is structurally related to neuropilin, a VEGF receptor and coreceptor for semaphorins, the latter implicated in neural and vascular patterning. NETO2 overexpression has been observed in numerous cancers (55) and hemangiomas (56), and is downregulated in response to NME1 expression in the context of breast cancer (37). TRIML2 is a member of the XAGE subfamily/GAGE family of proteins that harbor E3 ubiquitin ligase activity (57), a function shown to regulate cancer metastasis (58, 59).
A subset of NME1-regulated genes is highly predictive of metastasis and reduced survival in human melanoma patients
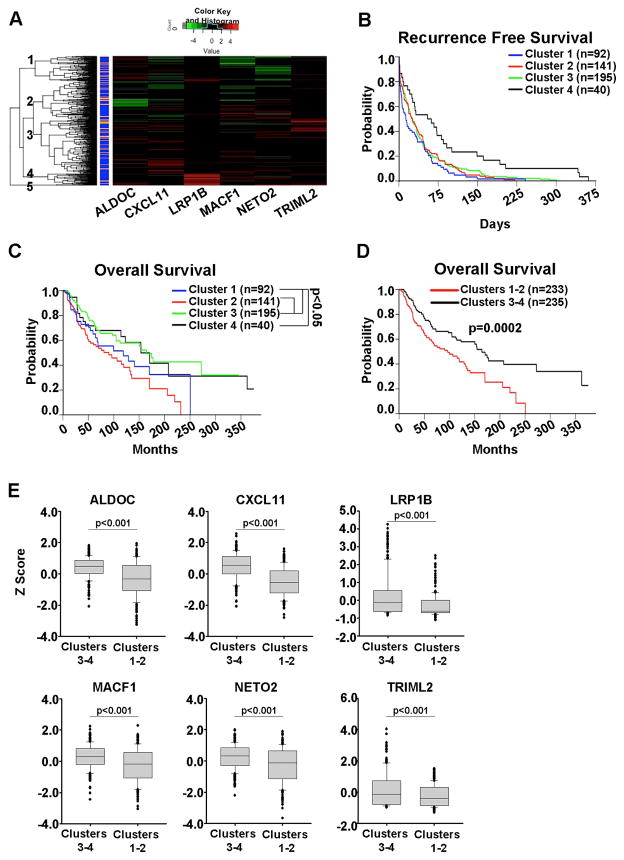
The ability of NME1 to suppress metastatic potential of melanoma cell lines suggests that genes under its regulatory control are associated with metastasis and survival in melanoma patients. To address this possibility, we analyzed expression of NME1-regulated genes within the skin cutaneous melanoma (SCKM) samples of The Cancer Genome Atlas (TCGA). Unsupervised hierarchical clustering was performed using the expression of 5 of the 6 core MSS genes (LRP1b, CXCL11, ALDOC, NETO2, and TRIML2). An additional target gene, MACF1, was included in the analysis because of the consistent significant changes observed upon manipulation of NME1 expression in multiple cell lines (Fig. 3B & 3C). The 468 patients within the SCKM TCGA separated into five general clusters based on expression of the 6 NME1-regulated genes (Fig. 4A). Cluster 5, however, only contained three patients and was excluded from further analysis. Few patients showed drastic changes in expression of any one individual gene (Fig. 4A) nor was there substantial predictive power for survival rates with any one gene (data not shown). When combined, however, the six gene signature was more powerful in predicting overall survival. No significant differences in recurrence-free survival were observed between the individual clusters (Fig. 4B). Despite somewhat closer clustering of groups 2 and 3 from the hierarchical clustering, the overall survival of patients in cluster 3 was significantly longer than both clusters 1 and 2 (Fig. 4C). Because there were no statistical differences in the overall survival probabilities between clusters 3 and 4, or between 1 and 2, the two pairs of sub-clusters were pooled for further analyses. Patients in pooled cluster 3–4 had a median survival time of 5.6 years longer than patients in cluster 1–2 (164.3 versus 96.3 months, p=0.0002) (Fig. 4D). Differences in survival times between the clusters was not due to uneven distribution of primary and metastatic patient samples as there was no significant difference in the total number of patients or percentage of metastatic or primary patients within the groups (Supplementary Table 4). When the expression of each of the 6 NME1-regulated genes was individually examined, all 6 were significantly (p<0.001) lower in cluster 1–2 compared to cluster 3–4 (Fig. 4E). Together, these data indicate that reduced expression of these six NME1-regulated genes is associated with poor survival.
Figure 4. Loss of NME1-induced genes identifies melanoma patients with poor survival.
A) Unsupervised heirarchical clustering based on the expression of NME1-regulated genes in the skin cutaneous melanoma (SKCM) portion of the TCGA identifies several clusters. Primary melanomas are designated in orange and metastatic samples in blue. Kaplan-Meier analysis of recurrence free- (B) and overall survival (C) of patients within each cluster. D) Kaplan-Meier analysis of overall survival after combining clusters 1 with 2, and cluster 3 with 4. E) RNA expression levels of indicated NME1 target genes within the clusters identified in (A). Stastical differences measured by ANOVA with Holm-Sidak post-hoc testing.
Expression of NME1-regulated mRNAs proved to be a stronger predictor of patient outcome than that of the NME1 transcript itself. In fact, after separating patients into the upper and lower quartiles for NME1 expression (Supplementary Fig. 1A), elevated NME1 mRNA expression was found to be associated with worse survival in the SCKM TCGA (Supplementary Fig. 1B). This counterintuitive observation could be explained by our recent demonstration that steady-state concentrations of NME1 mRNA are not the primary determinant of NME1 protein expression in metastatic cells in culture. Instead, NME1 protein is destabilized in metastatic cells via cathepsin-catalyzed degradation in the lysosomal compartment (60). Based on this observation, we predict that RNA/protein expression of the MSS genes would be more strongly correlated with intracellular concentrations of NME1 protein than NME1 mRNA. Unfortunately, the current lack of proteome data in the SCKN-TCGA collection precludes such an analysis. However, expression of 3/6 of the NME1-induced signature genes in the TCGA was inversely correlated with that of NME1 mRNA (Supplementary Fig. 1C). This could be explained by an induction of steady-state concentrations of NME1 mRNA levels in response to NME1 protein degradation; reduced expression of MSS genes could represent a response to NME1 protein downregulation, rather than to levels of the NME1 transcript.
Next, the “low NME1” and “high NME1” populations were individually subjected to hierarchical cluster analysis of recurrence-free survival, based on expression of the six-gene MSS. The analysis identified four clusters within each population (Supplementary Fig. 1D), with all clusters from the low NME1 group exhibiting longer recurrence-free survival than the clusters of the high NME1 group, as expected (Supplementary Fig. 1E, left; Supplementary Table 5). Interestingly, cluster 2 from the “high NME1” group exhibited significantly longer recurrence-free survival than that of cluster 4 of the same “high NME1” group (Supplementary Fig. 1E, right; Supplementary Table 5). No other differences in survival were observed between any clusters within the “high NME1” group or within the “low NME1” group. Expression of five of the six MSS genes was significantly different between clusters 2 and 4 (Supplementary Fig. 1F). Three MSS transcripts (ALDOC, CXCL11, LRP1B) were expressed at significantly higher levels in the longer-surviving patients of cluster 2 than in cluster 4. This positive correlation between expression of these MSS transcripts, which are induced by NME1 in melanoma cell lines, and extended survival in melanoma patients is consistent with roles as effectors of the metastasis suppressor function of NME1 in vivo. In addition, two other transcripts (MACF1 and NETO2) were expressed at significantly lower levels in cluster 2, suggesting they may be metastasis-driving in nature. Taken together, these analyses demonstrate that expression of the NME1-regulated MSS genes is a strong predictor of prolonged survival in melanoma patients.
Discussion
Rates of melanoma incidence are steadily increasing, as are death rates from melanoma patients not cured by tumor resection. Even recently-developed targeted therapies against the BRAF/MEK signaling axis and immune checkpoint mediators (e.g. CTLA-4, PD-1) have produced only modest improvements in overall survival for most patients with advanced melanoma (61). Off-target toxicities and acquired resistance to these agents remain impediments to durable responses. In the short term, clinical management of melanoma patients would be improved greatly by new molecular markers capable of identifying patients likely to progress to more aggressive forms. Such patients would represent the most appropriate candidates for undergoing therapy using these expensive and potentially toxic agents, and possibly for initiation of therapy at earlier stages of their disease. Unfortunately, no reliable markers exist for predicting the course of melanoma, in contrast with the molecular markers currently available for diagnosis and prognosis in breast carcinoma and other cancers (62). In the longer term, new therapeutic targets are needed if agents are to be developed with greater efficacy in blocking melanoma progression, and for eradicating metastatic disease once the cancer has progressed. Metastasis suppressor genes (MSGs) such as NME1 provide a unique opportunity to identify individual genes and gene networks that are either correlated with metastatic potential (e.g. prognostic indices), or are mediators of metastatic activity itself (e.g. therapeutic targets). The current study has identified gene networks whose expression is regulated in melanoma cells by the metastasis suppressor NME1, as well as genes that are predictive of better outcome and survival in a large cohort of human melanoma patients.
Ontological analysis identified immunological disease, nervous system development and function, and G protein-coupled receptors (GPCRs) as the three groupings most significantly impacted by forced expression of NME1 in WM793 melanoma cells (Supplementary Table 3). Regulation of genes involved in immune system function was robust (18.1% of significant genes) and is interesting in light of the critical role of immune cells in elimination of tumor cells in melanoma and other cancers (63), as well as the importance of immune cells in the metastatic niche (64). Of particular potential interest in this category is the presence of three chemokine/chemokine receptor genes (CXCL11, CCL8 and CCR2) and the interleukin-20 receptor beta (IL20RB). Genes identified in this functional grouping would seem worthy of further scrutiny as prognostic indicators for responsiveness to inhibitors of immune checkpoint molecules (e.g. PD-1 and CTLA-4), which are currently inadequate. The observed enrichment of genes involved in nervous system development and function is also intriguing in light of the neural crest origin of melanocytes, and the phenotypic similarities exhibited by neural crest stem cells and melanoma cells (65). Considering the close relationship proposed between stemness and metastatic potential in cancer cells (66), the genes identified in this category may represent rational drug targets for metastatic disease in melanoma and other cancers. GPCRs are becoming better recognized for their roles as key regulators of tumor and metastatic phenotypes, especially in melanoma (67). GPCRs are frequently mutated in melanoma and other cancers (68), exemplified by the identification of GRIN2A mutations in over 33% of human melanomas (69). GPCRs of particular interest were eleven olfactory receptor genes suppressed by NME1 in WM793 cells (Supplemental Table 3). Ectopic expression of olfactory receptor genes and proliferation-inhibitory activity has been described previously in melanocytes (70), an intriguing observation suggesting potential impact of NME1 on cellular proliferation and differentiation in melanoma.
Our study utilized NME1 variants that we showed previously to be deficient not only in metastasis suppressor activity, but also in NDPK and 3′-5′ exonuclease activities of the NME1 protein. While NME1 is rarely mutated in cancers, these variants represent powerful experimental tools to help differentiate the metastasis suppressor functions of NME1 from other potential housekeeping functions. In the current study as well as our prior one (11), metastasis suppressor function was disrupted significantly only in those variants lacking 3′–5′ exonuclease activity (E5A and K12Q), suggesting the possibility that this enzymatic function contributes in some way to regulation of the MSS genes. While a molecular mechanism underlying this regulatory function has yet to be described, the preference of the 3′–5′ exonuclease for single-stranded DNA substrates (22, 26) suggests a role in remodeling of regulatory elements in DNA that possess single-stranded character. Chromatin immunoprecipitation has demonstrated direct association of NME1 with DNA elements in a number of candidate target genes in another melanoma cell line (71), consistent with function as a canonical transcription factor, and many of those elements possess single-stranded character. Other transcriptional regulators with specificity for single-stranded motifs have been described, such as the single-stranded DNA binding factors that regulate transcription of the vascular smooth muscle actin promoter (72). Nevertheless, a transcriptional coregulatory function (i.e. non-DNA binding) has yet to be excluded for NME1, nor have post-transcriptional regulatory mechanisms.
Potential cancer-regulating functions have been ascribed to many of the NME1-regulated genes identified in our study, but the direction of regulation by NME1 was sometimes opposite of expected. For example, the NME1-upregulated genes EREG, CXCL11, NETO2 and XAGE1A have functional properties that are potentially cancer-driving in nature and their expression has been associated in some instances with cancer and malignant progression (43, 73, 74). These discrepancies could be explained by non-canonical functions in the setting of melanoma, tumor cell heterogeneity, or NME1-mediated regulation of expression of these proteins (e.g. translation initiation, protein stability) more concordant with their known cancer-relevant functions.
In a prior study conducted in the breast carcinoma cell line MDA-MB-435, microarray analysis also identified a large number of genes whose intracellular concentrations are regulated by NME1. Of particular interest were a number of RNA processing factors, such as GEMIN5, BOP1, ACIN1, PABP and HNRNPA2B (75), suggesting NME1 may regulate gene expression via impacts on RNA splicing, stability and other post-transcriptional events. Interestingly, only one gene regulated by NME1 in our melanoma-focused study, NETO2, was also shown to be regulated in the breast carcinoma cell analysis (14). Moreover, while NME1 upregulated expression of NETO2 RNA in both the WM793 and M14 melanoma cell lines, NETO2 was downregulated by NME1 in MDA-MDB-435 breast carcinoma cells. As discussed above, the discrepancy in regulation of NETO2 expression between melanoma and breast carcinoma-derived cell lines could be secondary to cell-specific differences in NETO2 function, tumor cell heterogeneity, and/or post-transcriptional processes. Also of note in our analysis was a prevalence of NME1-regulated cell surface receptors, consistent with prior studies conducted across a spectrum of cancer cell lines (75–77).
This study has identified a number of NME1-regulated transcripts that may have applications in diagnosis and prognosis for melanoma. Adapting this approach to other known MSGs would seem to hold potential for providing a wealth of novel prognostic markers and therapeutic targets for metastatic disease across a spectrum of human cancers. While expression of NME1 RNA has been shown to be associated with clinical outcome in melanoma (77), and the initial identification of NME1 as a metastasis suppressor evolved from an observation of reduced NME1 RNA expression in metastatic melanoma cells (7), expression of NME1 RNA itself has not proven to be a robust marker in melanoma or other cancers. This may well be due to the fact that reduced NME1 protein expression in metastatic melanoma cells is often not the result of reduced steady-state concentrations of the cognate NME1 RNA but instead caused by protein destabilization and lysosomal degradation (76). Our study provides proof-of-principle that NME1-regulated transcripts and their encoded proteins both individually and collectively hold promise as molecular markers for identifying melanoma subtypes, obtaining more reliable prognoses, and providing personalized medicine for optimal delivery of therapy. In addition, they represent promising molecular targets for development of novel therapies to manage melanoma in its advanced and lethal forms.
Supplementary Material
A) RNA expression levels of NME1 in the SCKM TCGA dataset samples sorted into the upper and lower quartiles of NME1 expression. B) Kaplan-Meier probabilities of recurrence free survival in “NME1 Low” and “NME1 High” groups. C) RNA expression levels of the indicated NME1 target genes in the “Low NME1” and “High NME1” groups. D) Pearson correlation based cluster analysis of the NME1-mediated gene signature in “NME1 Low” and “NME1 High” groups. E) Kaplan-Meier probabilities of recurrence free survival in all sub-clusters from the “High” and “Low” NME1 groups. For clarity, the only two subclusters with signfificant differences from the “High NME1” group, clusters 2 and 4, are also shown separately in the right panel. F) RNA expression levels of the indicated NME1 target genes in sub-clusters 2 and 4 from within the “High NME1” group. Significance was determined by Student’s t-test.
Acknowledgments
This work was supported by grants from the National Instititues of Health/National Cancer Institute (CA83237, CA159871 and CA159871-S1).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Shevde LA, Welch DR. Metastasis suppressor pathways--an evolving paradigm. Cancer Lett. 2003;198(1):1–20. doi: 10.1016/s0304-3835(03)00304-5. [DOI] [PubMed] [Google Scholar]
- 2.DePeralta DK, Boland GM. Melanoma: Advances in Targeted Therapy and Molecular Markers. Annals of surgical oncology. 2015;22(11):3451–8. doi: 10.1245/s10434-015-4702-1. [DOI] [PubMed] [Google Scholar]
- 3.Tsai KK, Daud AI. The Role of Anti-PD-1/PD-L1 Agents in Melanoma: Progress to Date. Drugs. 2015;75(6):563–75. doi: 10.1007/s40265-015-0376-z. [DOI] [PubMed] [Google Scholar]
- 4.Hurst DR, Welch DR. Metastasis Suppressor Genes: At the Interface between the Environment and Tumor Cell Growth. Int Rev Cel Mol Bio. 2011;286:107–80. doi: 10.1016/B978-0-12-385859-7.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9(4):253–64. doi: 10.1038/nrc2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayawardana K, Schramm SJ, Tembe V, et al. Identification, Review, and Systematic Cross-Validation of microRNA Prognostic Signatures in Metastatic Melanoma. J Invest Dermatol. 2016;136(1):245–54. doi: 10.1038/JID.2015.355. [DOI] [PubMed] [Google Scholar]
- 7.Steeg PS, Bevilacqua G, Kopper L, et al. Evidence for a novel gene associated with low tumor metastatic potential. Journal of the National Cancer Institute. 1988;80(3):200–4. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 8.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98(7):472–82. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 9.Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomembr. 2000;32(3):301–8. doi: 10.1023/a:1005597231776. [DOI] [PubMed] [Google Scholar]
- 10.MacDonald NJ, De La Rosa A, Steeg PS. The potential roles of nm23 in cancer metastasis and cellular differentiation. Eur J Cancer [A] 1995;31A:1096–100. doi: 10.1016/0959-8049(95)00152-9. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, McCorkle JR, Novak M, et al. Metastasis suppressor function of NM23-H1 requires its 3′;-5′ exonuclease activity. Int J Cancer. 2011;128:40–50. doi: 10.1002/ijc.25307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarrett SG, Novak M, Harris N, et al. NM23 deficiency promotes metastasis in a UV radiation-induced mouse model of human melanoma. Clin Exp Metastasis. 2013;30(1):25–36. doi: 10.1007/s10585-012-9495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snider NT, Altshuler PJ, Omary MB. Modulation of cytoskeletal dynamics by mammalian nucleoside diphosphate kinase (NDPK) proteins. Naunyn-Schmiedeberg’s archives of pharmacology. 2015;388(2):189–97. doi: 10.1007/s00210-014-1046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horak CE, Lee JH, Elkahloun AG, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer research. 2007;67(15):7238–46. doi: 10.1158/0008-5472.CAN-07-0962. [DOI] [PubMed] [Google Scholar]
- 15.McCorkle JR, Leonard MK, Kraner SD, et al. The metastasis suppressor NME1 regulates expression of genes linked to metastasis and patient outcome in melanoma and breast carcinoma. Cancer Genomics Proteomics. 2014;11(4):175–94. [PMC free article] [PubMed] [Google Scholar]
- 16.Boissan M, Dabernat S, Peuchant E, et al. The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Molecular and cellular biochemistry. 2009;329(1–2):51–62. doi: 10.1007/s11010-009-0120-7. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal RP, Robinson B, Parks RE. Nucleoside diphosphokinase from erythrocytes. Methods Enzymol. 1978;51:376–86. doi: 10.1016/s0076-6879(78)51051-3. [DOI] [PubMed] [Google Scholar]
- 18.Crawford RM, Treharne KJ, Arnaud-Dabernat S, et al. Understanding the molecular basis of the interaction between NDPK-A and AMPK alpha 1. Mol Cell Biol. 2006;26(15):5921–31. doi: 10.1128/MCB.00315-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Lee HY, Lee H. Inhibitory activity of nm23-H1 on invasion and colonization of human prostate carcinoma cells is not mediated by its NDP kinase activity. Cancer Lett JID - 7600053. 1999;145(1–2):93–9. doi: 10.1016/s0304-3835(99)00236-0. [DOI] [PubMed] [Google Scholar]
- 20.Boissan M, Montagnac G, Shen Q, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344(6191):1510–5. doi: 10.1126/science.1253768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Postel EH, Berberich SJ, Flint SJ, et al. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–80. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 22.Ma D, Xing Z, Liu B, et al. NM23-H1 cleaves and represses transcriptional activity of nuclease-hypersensitive elements in the PDGF-A promoter. J Biol Chem. 2002;277:1560–7. doi: 10.1074/jbc.M108359200. [DOI] [PubMed] [Google Scholar]
- 23.Postel E, Berberich SJ, Rooney JW, et al. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive elements. J Bioenerg Biomembr. 2000;32(3):277–84. doi: 10.1023/a:1005541114029. [DOI] [PubMed] [Google Scholar]
- 24.Yang M, Jarrett SG, Craven R, et al. YNK1, the yeast homolog of human metastasis suppressor NM23, is required for repair of UV radiation- and etoposide-induced DNA damage. Mutat Res. 2009;660:74–8. doi: 10.1016/j.mrfmmm.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarrett SG, Novak M, Dabernat S, et al. Metastasis suppressor NM23-H1 promotes repair of UV-induced DNA damage and suppresses UV-induced melanomagenesis. Cancer Res. 2012;72(1):133–43. doi: 10.1158/0008-5472.CAN-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma D, McCorkle JR, Kaetzel DM. The metastasis suppressor NM23-H1 possesses 3′-5′ exonuclease activity. The Journal of biological chemistry. 2004;279(17):18073–84. doi: 10.1074/jbc.M400185200. [DOI] [PubMed] [Google Scholar]
- 27.Postel EH, Abramczyk BA, Gursky SK, et al. Structure-based mutational and functional analysis identify human NM23-H2 as a multifunctional enzyme. Biochemistry JID - 0370623. 2002;41(20):6330–7. doi: 10.1021/bi025606+. [DOI] [PubMed] [Google Scholar]
- 28.Freije JM, Blay P, MacDonald NJ, et al. Site-directed mutation of Nm23-H1. Mutations lacking motility suppressive capacity upon transfection are deficient in histidine-dependent protein phosphotransferase pathways in vitro. J Biol Chem. 1997;272(9):5525–32. doi: 10.1074/jbc.272.9.5525. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald NJ, Freije JM, Stracke ML, et al. Site-directed mutagenesis of nm23-H1. Mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J Biol Chem JID - 2985121R. 1996;271(41):25107–16. doi: 10.1074/jbc.271.41.25107. [DOI] [PubMed] [Google Scholar]
- 30.Ganguly SS, Fiore LS, Sims JT, et al. c-Abl and Arg are activated in human primary melanomas, promote melanoma cell invasion via distinct pathways, and drive metastatic progression. Oncogene. 2012;31(14):1804–16. doi: 10.1038/onc.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parnell LD, Lindenbaum P, Shameer K, et al. BioStar: an online question & answer resource for the bioinformatics community. PLoS computational biology. 2011;7(10):e1002216. doi: 10.1371/journal.pcbi.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexaki VI, Javelaud D, Van Kempen LC, et al. GLI2-mediated melanoma invasion and metastasis. J Natl Cancer Inst. 2010;102(15):1148–59. doi: 10.1093/jnci/djq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steder M, Alla V, Meier C, et al. DNp73 exerts function in metastasis initiation by disconnecting the inhibitory role of EPLIN on IGF1R-AKT/STAT3 signaling. Cancer Cell. 2013;24(4):512–27. doi: 10.1016/j.ccr.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 37.Horak CE, Lee JH, Elkahloun AG, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67(15):7238–46. doi: 10.1158/0008-5472.CAN-07-0962. [DOI] [PubMed] [Google Scholar]
- 38.Sonoda I, Imoto I, Inoue J, et al. Frequent silencing of low density lipoprotein receptor-related protein 1B (LRP1B) expression by genetic and epigenetic mechanisms in esophageal squamous cell carcinoma. Cancer Res. 2004;64(11):3741–7. doi: 10.1158/0008-5472.CAN-04-0172. [DOI] [PubMed] [Google Scholar]
- 39.Prazeres H, Torres J, Rodrigues F, et al. Chromosomal, epigenetic and microRNA-mediated inactivation of LRP1B, a modulator of the extracellular environment of thyroid cancer cells. Oncogene. 2011;30(11):1302–17. doi: 10.1038/onc.2010.512. [DOI] [PubMed] [Google Scholar]
- 40.Nikolaev SI, Rimoldi D, Iseli C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2012;44(2):133–9. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 41.Pencheva N, Tran H, Buss C, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151(5):1068–82. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh AK, Arya RK, Trivedi AK, et al. Chemokine receptor trio: CXCR3, CXCR4 and CXCR7 crosstalk via CXCL11 and CXCL12. Cytokine Growth Factor Rev. 2013;24(1):41–9. doi: 10.1016/j.cytogfr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hensbergen PJ, Wijnands PG, Schreurs MW, et al. The CXCR3 targeting chemokine CXCL11 has potent antitumor activity in vivo involving attraction of CD8+ T lymphocytes but not inhibition of angiogenesis. Journal of immunotherapy. 2005;28(4):343–51. doi: 10.1097/01.cji.0000165355.26795.27. [DOI] [PubMed] [Google Scholar]
- 44.Chu Y, Yang X, Xu W, et al. In situ expression of IFN-gamma-inducible T cell alpha chemoattractant in breast cancer mounts an enhanced specific anti-tumor immunity which leads to tumor regression. Cancer Immunol Immunother. 2007;56(10):1539–49. doi: 10.1007/s00262-007-0296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chheda ZS, Sharma RK, Jala VR, et al. Chemoattractant Receptors BLT1 and CXCR3 Regulate Antitumor Immunity by Facilitating CD8+ T Cell Migration into Tumors. Journal of immunology. 2016 doi: 10.4049/jimmunol.1502376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mamczur P, Gamian A, Kolodziej J, et al. Nuclear localization of aldolase A correlates with cell proliferation. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbamcr.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Ritterson LC, Tolan DR. Targeting of several glycolytic enzymes using RNA interference reveals aldolase affects cancer cell proliferation through a non-glycolytic mechanism. J Biol Chem. 2012;287(51):42554–63. doi: 10.1074/jbc.M112.405969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritterson LC, Tolan DR. Aldolase sequesters WASP and affects WASP/Arp2/3-stimulated actin dynamics. J Cell Biochem. 2013;114(8):1928–39. doi: 10.1002/jcb.24538. [DOI] [PubMed] [Google Scholar]
- 49.Merkulova M, Hurtado-Lorenzo A, Hosokawa H, et al. Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am J Physiol Cell Physiol. 2011;300(6):C1442–C55. doi: 10.1152/ajpcell.00076.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li YJ, Huang TH, Hsiao M, et al. Suppression of fructose-bisphosphate aldolase C expression as a predictor of advanced oral squamous cell carcinoma. Head & neck. 2016;38(Suppl 1):E1075–85. doi: 10.1002/hed.24161. [DOI] [PubMed] [Google Scholar]
- 51.Cunha ES, Kawahara R, Kadowaki MK, et al. Melanogenesis stimulation in B16-F10 melanoma cells induces cell cycle alterations, increased ROS levels and a differential expression of proteins as revealed by proteomic analysis. Exp Cell Res. 2012;318(15):1913–25. doi: 10.1016/j.yexcr.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 52.Chambers D, Wilson LJ, Alfonsi F, et al. Rhombomere-specific analysis reveals the repertoire of genetic cues expressed across the developing hindbrain. Neural development. 2009;4:6. doi: 10.1186/1749-8104-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barriga EH, Maxwell PH, Reyes AE, et al. The hypoxia factor Hif-1alpha controls neural crest chemotaxis and epithelial to mesenchymal transition. The Journal of cell biology. 2013;201(5):759–76. doi: 10.1083/jcb.201212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zendman AJ, van Kraats AA, den Hollander AI, et al. Characterization of XAGE-1b, a short major transcript of cancer/testis-associated gene XAGE-1, induced in melanoma metastasis. Int J Cancer. 2002;97(2):195–204. doi: 10.1002/ijc.1584. [DOI] [PubMed] [Google Scholar]
- 55.Oparina NI, Sadritdinova AF, Snezhkina AV, et al. Increase in NETO2 gene expression is a potential molecular genetic marker in renal and lung cancers. Genetika. 2012;48(5):599–607. [PubMed] [Google Scholar]
- 56.Calicchio ML, Collins T, Kozakewich HP. Identification of signaling systems in proliferating and involuting phase infantile hemangiomas by genome-wide transcriptional profiling. Am J Pathol. 2009;174(5):1638–49. doi: 10.2353/ajpath.2009.080517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kung CP, Khaku S, Jennis M, et al. Identification of TRIML2, a novel p53 target, that enhances p53 SUMOylation and regulates the transactivation of proapoptotic genes. Molecular cancer research: MCR. 2015;13(2):250–62. doi: 10.1158/1541-7786.MCR-14-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paolino M, Choidas A, Wallner S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507(7493):508–12. doi: 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dadakhujaev S, Salazar-Arcila C, Netherton SJ, et al. A novel role for the SUMO E3 ligase PIAS1 in cancer metastasis. Oncoscience. 2014;1(3):229–40. doi: 10.18632/oncoscience.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiore LS, Ganguly SS, Sledziona J, et al. c-Abl and Arg induce cathepsin-mediated lysosomal degradation of the NM23-H1 metastasis suppressor in invasive cancer. Oncogene. 2014;33(36):4508–20. doi: 10.1038/onc.2013.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang T, Eldabaje R, Yang L. Current Status of Biological Therapies for the Treatment of Metastatic Melanoma. Anticancer Res. 2016;36(7):3229–41. [PubMed] [Google Scholar]
- 62.Mehta S, Shelling A, Muthukaruppan A, et al. Predictive and prognostic molecular markers for cancer medicine. Therapeutic advances in medical oncology. 2010;2(2):125–48. doi: 10.1177/1758834009360519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vinay DS, Ryan EP, Pawelec G, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Seminars in cancer biology. 2015;35(Suppl):S185–98. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 64.Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nature reviews Immunology. 2015;15(2):73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shakhova O. Neural crest stem cells in melanoma development. Current opinion in oncology. 2014;26(2):215–21. doi: 10.1097/CCO.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 66.Oskarsson T, Batlle E, Massague J. Metastatic stem cells: sources, niches, and vital pathways. Cell stem cell. 2014;14(3):306–21. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee HJ, Wall B, Chen S. G-protein-coupled receptors and melanoma. Pigment Cell Melanoma Res. 2008;21(4):415–28. doi: 10.1111/j.1755-148X.2008.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Hayre M, Vazquez-Prado J, Kufareva I, et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013;13(6):412–24. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei X, Walia V, Lin JC, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43(5):442–6. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gelis L, Jovancevic N, Veitinger S, et al. Functional Characterization of the Odorant Receptor 51E2 in Human Melanocytes. J Biol Chem. 2016 doi: 10.1074/jbc.M116.734517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cervoni L, Egistelli L, Eufemi M, et al. DNA sequences acting as binding sites for NM23/NDPK proteins in melanoma M14 cells. Journal of cellular biochemistry. 2006;98(2):421–8. doi: 10.1002/jcb.20808. [DOI] [PubMed] [Google Scholar]
- 72.Sun S, Stoflet ES, Cogan JG, et al. Negative regulation of the vascular smooth muscle alpha-actin gene in fibroblasts and myoblasts: disruption of enhancer function by sequence-specific single-stranded-DNA-binding proteins. Molecular and cellular biology. 1995;15(5):2429–36. doi: 10.1128/mcb.15.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, Iwanaga K, Choi KC, et al. Intratumoral epiregulin is a marker of advanced disease in non-small cell lung cancer patients and confers invasive properties on EGFR-mutant cells. Cancer Prev Res (Phila) 2008;1(3):201–7. doi: 10.1158/1940-6207.CAPR-08-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu K, Li SL, Gan YH, et al. Epiregulin promotes migration and invasion of salivary adenoid cystic carcinoma cell line SACC-83 through activation of ERK and Akt. Oral oncology. 2009;45(2):156–63. doi: 10.1016/j.oraloncology.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 75.Lee JH, Horak CE, Khanna C, et al. Alterations in Gemin5 expression contribute to alternative mRNA splicing patterns and tumor cell motility. Cancer research. 2008;68(3):639–44. doi: 10.1158/0008-5472.CAN-07-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fiore LS, Ganguly SS, Sledziona J, et al. c-Abl and Arg induce cathepsin-mediated lysosomal degradation of the NM23-H1 metastasis suppressor in invasive cancer. Oncogene. 2013 doi: 10.1038/onc.2013.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. Journal of the National Cancer Institute. 2006;98(7):472–82. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) RNA expression levels of NME1 in the SCKM TCGA dataset samples sorted into the upper and lower quartiles of NME1 expression. B) Kaplan-Meier probabilities of recurrence free survival in “NME1 Low” and “NME1 High” groups. C) RNA expression levels of the indicated NME1 target genes in the “Low NME1” and “High NME1” groups. D) Pearson correlation based cluster analysis of the NME1-mediated gene signature in “NME1 Low” and “NME1 High” groups. E) Kaplan-Meier probabilities of recurrence free survival in all sub-clusters from the “High” and “Low” NME1 groups. For clarity, the only two subclusters with signfificant differences from the “High NME1” group, clusters 2 and 4, are also shown separately in the right panel. F) RNA expression levels of the indicated NME1 target genes in sub-clusters 2 and 4 from within the “High NME1” group. Significance was determined by Student’s t-test.