Abstract
CYP725A4 is a P450 enzyme from Taxus cuspidata that catalyzes the formation of taxadiene-5α-ol (T5α-ol) from taxadiene in paclitaxel biosynthesis. Past attempts expressing CYP725A4 in heterologous hosts reported the formation of 5(12)-oxa-3(11)-cyclotaxane (OCT) and/or 5(11)-oxa-3(11)-cyclotaxane (iso-OCT) instead of, or in addition to, T5α-ol. Here we report that T5α-ol is produced as a minor product by Escherichia coli expressing both taxadiene synthase (TS) and CYP725A4. The major products were OCT and iso-OCT, while trace amounts of unidentified monooxygenated taxanes were also detected by gas chromatography-mass spectrometry. Since OCT and iso-OCT had not been found in nature, we tested the hypothesis that protein-protein interaction of CYP725A4 with redox partners, such as cytochrome P450 reductase (CPR) and cytochrome b5, may affect the products formed by CYP725A4, possibly favoring the formation of T5α-ol over OCT and iso-OCT. Our results show that coexpression of CYP725A4 with CPR from different organisms did not change the relative ratios of OCT, iso-OCT, and T5α-ol, while cytochrome b5 decreased overall levels of the products formed. Although unsuccessful in finding conditions that promote T5α-ol formation over other products, we used our results to clarify conflicting claims in the literature and discuss other possible approaches to produce paclitaxel via metabolic and enzyme engineering.
Keywords: Cytochrome b5, Diterpene, Metabolic engineering, P450 reductase, Paclitaxel, Taxane
1. Introduction
Paclitaxel (brand name Taxol™) is a well-known antimitotic drug originally obtained from the bark of yew trees (Taxus spp.) and widely used in the treatment of breast, ovarian and lung cancer, among others [1, 2]. The production of this drug (and its semisynthetic derivative docetaxel) relies on manufacturing processes that use paclitaxel precursors from cultivated yews as starting materials [3], or the induced biosynthesis of paclitaxel and its precursors in yew cell cultures [4]. Despite these well-established sources, shortages of paclitaxel and docetaxel (also used for a wide range of cancers) still occur, because of the need for new alternative methods to produce these anticancer drugs. One alternative is the use of metabolically engineered organisms that express biosynthetic genes from yew, which enable the biotechnological production of paclitaxel and its precursors.
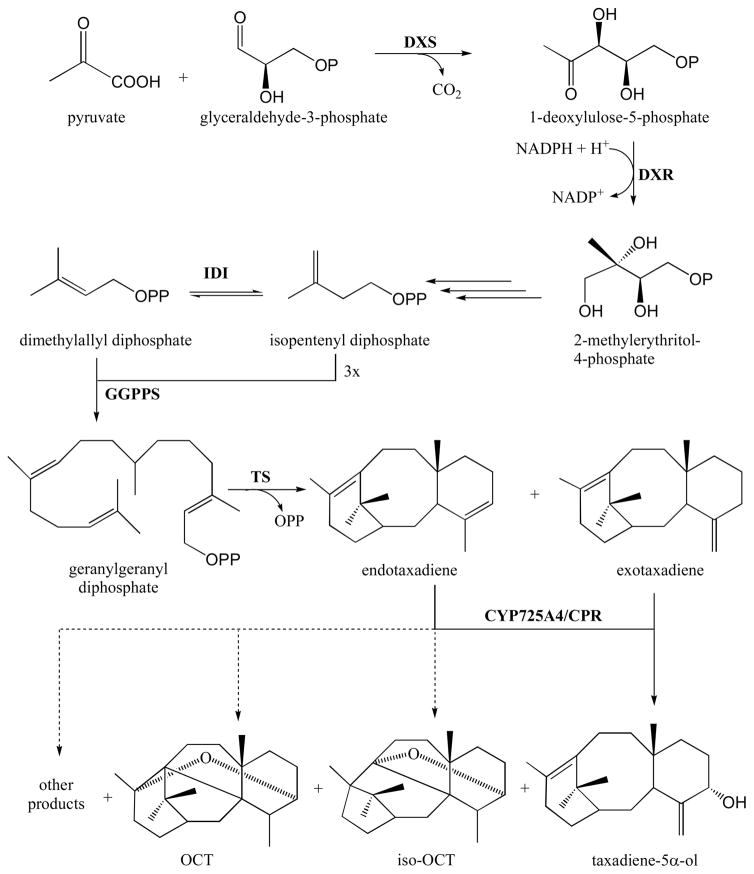
To produce paclitaxel, yew cells rely on the methylerythritol phosphate (MEP) pathway to generate isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) from pyruvate and glyceraldehyde-3-phosphate (Fig. 1). The two universal isoprenoid precursors IPP and DMAPP are used by geranylgeranyl diphosphate synthase (GGPPS) to form geranylgeranyl diphosphate (GGPP) [5], which is the substrate of taxadiene synthase (TS), the first enzyme in the paclitaxel pathway [6]. When expressed in E. coli, TS converts GGPP into two isomers of taxadiene, namely taxa-4(5),11(12)-diene (endotaxadiene) as the major product (87–94%), and taxa-4(20),11(12)-diene (exotaxadiene) as a minor product (5–13%) [7, 8]. This is consistent with the proposed role of endotaxadiene as the native metabolic intermediate in paclitaxel biosynthesis [9], although exotaxadiene can also serve as a substrate for the next enzyme in the pathway, CYP725A4 [10].
Fig. 1.
Biosynthesis of paclitaxel precursors and side products. The MEP pathway enzymes include 1-deoxy-D-xylulose-5-phosphate synthase (DXS), 1-deoxy-D-xylulose reductase (DXR), and isopentenyl diphosphate isomerase (IDI). Geranylgeranyl diphosphate synthase (GGPPS) and taxadiene synthase (TS) convert MEP pathway products into endotaxadiene and exotaxadiene. CYP725A4 and cytochrome P450 reductase (CPR) convert endotaxadiene and exotaxadiene into taxadiene-5α-ol, which is a paclitaxel precursor. Dashed lines show converison of endotaxadiene into OCT, iso-OCT and other side products, which do not lead to paclitaxel formation.
CYP725A4 is a cytochrome P450 enzyme believed to be responsible for the formation of taxadiene-5α-ol (T5α-ol) in the biosynthetic pathway to paclitaxel in yew [10, 11]. Like most plant P450s, CYP725A4 depends on a cytochrome P450 reductase (CPR) for its activity [12]. Some CPRs and other P450 interacting proteins like cytochrome b5 (Cb5) have been shown to modulate the activities of P450s [13, 14]. Endotaxadiene is presumed to be the natural substrate of CYP725A4 (at least in planta) since it is the main product of TS [7–9]. However, when heterologously expressed in E. coli, CYP725A4 has been shown to convert endotaxadiene mainly to 5(12)-oxa-3(11)-cyclotaxane (OCT) and 5(11)-oxa-3(11)-cyclotaxane (iso-OCT) (Fig. 1) [15–17] in vivo, neither of which have been reported to occur naturally in yew, nor serve as precursors to paclitaxel.
Following the first hydroxylation step catalyzed by CYP725A4, several more reactions are required to form paclitaxel [18]. These include other P450-catalyzed hydroxylations, acylation reactions (one of which requires assembly of a phenylalanine-derived acyl side chain), oxidation of a C-9 hydroxyl to a ketone, and oxetane ring formation [19, 20]. The specific order in which these reactions occur is not completely understood, but most of the genes responsible for these reactions have already been isolated and characterized [20]. However, only the first two paclitaxel biosynthetic genes (TS and CYP725A4) have been successfully used to produce paclitaxel precursors (i.e. taxadienes and T5α-ol), while CYP725A4 mainly generates undesired non-paclitaxel forming side products (i.e. OCT and iso-OCT) [15–17]. Thus, it still remains a challenge to find and optimize heterologous expression systems that can effectively use TS and CYP725A4 to produce high levels of T5α-ol [16, 21].
In this communication, we report the results of our own attempts to produce T5α-ol in E. coli by expressing TS and CYP725A4, along with upstream isoprenoid pathway genes, GGPPS, and P450 interacting proteins. Similar experiments have already been done previously by other groups [15–17, 22], but we deem it important to publish our independently generated data, especially considering the variable and confusing results reported in the literature. Our aim is to investigate the effect of redox partners on product formation and bring attention to the limitations of using TS and CYP725A4 in producing paclitaxel precursors (at least in E. coli), which may not have been adequately addressed in previous publications, and stimulate more interdisciplinary studies into this bottleneck in paclitaxel metabolic engineering.
2. Materials and Methods
2.1. Reagents, plasmids, and primers
Antibiotics and isopropyl-β-D-1-thiogalactopyranoside (IPTG) were purchased from Gold Biotechnology (St. Louis, MO, USA). Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, Iowa, USA). Reagents for PCR were obtained from Clontech (Madison, WI, USA). Restriction enzymes and all other chemicals were purchased from Thermo Fisher Scientific (Waltham, MA, USA) unless otherwise specified. Plasmids and primers used in this study are listed in Tables 1 and 2, respectively.
Table 1.
Plasmids used in this study
| Vector | Promoter | Antibiotic markera | Gene/enzyme inserts | Replicon | Source/ Reference |
|---|---|---|---|---|---|
| pENTR™/SD/D-TOPO® | T7 | Kan | None | pUC | Invitrogen |
| pIRS | T7 | Spec | dxs, idi, dxr | CloDF13 | [23] |
| pGG_DEST | T7 | Cam | GGPPS | P15A | [23] |
| pGG_coTS79 | T7 | Cam | GGPPS, TS | P15A | This study |
| pCW′_2A6_hNPR | tac | Amp | CYP2A6, hNPR | M13 | [24] |
| pCW′_coCYP725A4/coTCPR (fused) | tac | Amp | CYP725A4, TCPR | M13 | This study |
| pCW′_coCYP725A4_hNPR (bicistronic) | tac | Amp | CYP725A4, hNPR | M13 | This study |
| pCW′_NcoI_coCYP725A4_hNPR (bicistronic) | tac | Amp | CYP725A4, hNPR | M13 | This study |
| pCW′_coCYP725A4_TCPR (bicistronic) | tac | Amp | CYP725A4, TCPR | M13 | This study |
| pCW′_coCYP725A4_coCYP725A4/coTCPR (bicistronic/fused) | tac | Amp | CYP725A4, CYP725A4, TCPR | M13 | This study |
| pCW′_coTxCb5_coCYP725A4/coTCPR (bicistronic/fused) | tac | Amp | TxCb5, CYP725A4, TCPR | M13 | This study |
Kan= kanamycin; Spec = spectinomycin; Cam = chloramphenicol; Amp = ampicillin
Table 2.
Primers used in this study
| Primers | Sequence (5′-3′) |
|---|---|
| coTSdN79_F1cacc | CACCATGGTCGATGATATTCCGC |
| coTS_R1 stp | GCTCACACCTGAATCGGGTC |
| pCWb_coT5H_NdeI_F1 | GCTTAGGAGGTCATATGGCTCTGCTGCTGGCAGTG |
| pCWb_coTCPR_HindIII_R1 stp | TATCATCGATAAGCTTACCAGATGTCACGCAGATAGCG |
| pCWb_coT5H_XbaI_R1 | AAAATTATTTCTAGACTACGGACGCGGGAACAGTTT |
| pCWb_SD_NcoI_mut | AATAATTTTGTTTAACTTTAAGAAGGAGATATAACCATGGCTGACTCCCACGTG |
| GC_pCWb_SD_NcoI_mut | CACGTGGGAGTCAGCCATGGTTATATCTCCTTCTTAAAGTTAAACAAAATTATT |
| pCWb_coTCPR_NcoI_F1gs | GGAGATATAACCATGGGCTCTACCGGTAGTCGTCG |
| pCWb_coT5H_NcoI_F1 | GGAGATATAACCATGGCTCTGCTGCTGGCT |
| pCWb_TxCb5.6_NdeI_F1 | GCTTAGGAGGTCATATGGAAGGTACTAAAGTTTTCACCCTGG |
| pCWb_TxCb5.6_XbaI_R1stp | AAAATTATTTCTAGATTAGCTGCTCGGTTGGCTC |
2.2. Construction of recombinant plasmids
A codon optimized truncated taxadiene synthase (coTS79) was synthesized by GenScript (Piscataway, NJ, USA), and amplified by PCR using primers coTSdN79_F1cacc and coTS_R1 stp (Table 2), with an Advantage® HD Polymerase Mix (Clontech), according to the manufacturer’s instructions (Three-Step PCR Method). The PCR product (~ 2.5 kb) was cloned into pENTR™/SD/D-TOPO® vector using a pENTR™ Directional TOPO® Cloning Kit (Invitrogen, Waltham, MA, USA) and transferred to a destination vector pGG_DEST using LR clonase II (Invitrogen) according to the manufacturer’s instructions to generate pGG_coTS79 expression plasmid (Fig. S1 in Supplementary Material). Vector pGG_DEST is derived from the pACYCDuet plasmid [23], and harbors the gene GGPPS from Abies grandis under the control of T7 promoter.
A fusion protein consisting of CYP725A4 and TCPR (designated as coCYP725A4/coTCPR) was codon optimized for expression in bacteria, synthesized by GenScript, and then cloned into pCWori expression vector as follows. Two restriction sites, NdeI and HindIII, were introduced into the synthetic coCYP725A4/coTCPR construct by PCR as above, using primers pCWb_coT5H_NdeI_F1, and pCWb_coTCPR_HindIII_R1 stp (Table 2). The resulting PCR product (~ 3.5 kb) was treated with DpnI (New England Biolabs, USA) at 37°C for 1 h and purified using Nucleospin Extract II (Clontech) according to the manufacturer’s instructions, generating NdeI-coCYP725A4/coTCPR-HindIII DNA fragment. DpnI digestion was performed to destroy the original plasmid DNA used as template in PCR. To prepare the vector, the plasmid pCW′_2A6_hNPR was digested with NdeI and HindIII (FastDigest™, Thermo Scientific) at 37°C for 15 min (which removed CYP2A6 and hNPR). The resulting linearized vector was purified using agarose gel electrophoresis, and NdeI-coCYP725A4/coTCPR-HindIII was ligated to it using an In-Fusion® cloning kit (Clontech) to generate the expression plasmid pCW′_coCYP725A4/coTCPR (Fig. S2 in Supplementary Material).
pCW′_2A6_hNPR had two multiple cloning sites which were occupied by a human P450 gene (CYP2A6) and a human P450 reductase gene (hNPR). Each of these genes was preceded by a ribosome binding site, while the expression of the whole bicistronic transcript containing both genes was under the control of a single tandem tac promoter [24]. CYP2A6 was replaced with CYP725A4 with a stop codon to make a bicistronic construct that expressed CYP725A4 and hNPR as two separate proteins from the same transcript. hNPR was then replaced with TCPR to make a bicistronic construct that expressed CYP725A4 with its native P450 reductase. hNPR was also replaced with CYP725A4/TCPR fusion protein to express a separate CYP725A4 that can potentially interact with TCPR that is already linked to another CYP725A4. CYP725A4 was then replaced with Taxus x media Cb5 (TxCb5) to coexpress a native Cb5 with the CYP725A4/TCPR fusion protein.
To replace CYP2A6 with CYP725A4, NdeI and XbaI restriction sites were introduced at the 5′ and 3′ ends of coCYP725A4 by PCR as above using pCWb_coT5H_NdeI_F1 and pCWb_coT5H_XbaI_R1 as primers (Table 2) and synthetic coCYP725A4/coTCPR as template. The reverse primer pCWb_coT5H_XbaI_R1 added a TAG stop codon at the 3′ end of coCYP725A4. The PCR product (~1.5 kb) was treated with DpnI and purified using Nucleospin Extract II as above to generate the NdeI-coCYP725A4-XbaI DNA fragment. The vector pCW′_2A6_hNPR was linearized with NdeI and XbaI, removing CYP2A6 (~1.5 kb) which was then replaced with NdeI-coCYP725A4-XbaI using an In-Fusion® cloning kit (Fig. S3 in Supplementary Material).
To replace hNPR with TCPR, NcoI restriction site was introduced at the 5′ end of the hNPR gene in pCW′_coCYP725A4_hNPR using a QuikChange® Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA), with pCWb_SD_NcoI_mut and GC_pCWb_SD_NcoI_mut as primers (Table 2). These mutagenic primers contain the NcoI site, which would enable the removal of the hNPR gene using NcoI and HindIII. PCR was started at 95°C for 2 min, followed by 18 cycles of 95°C for 20 sec, 60°C for 10 sec and 68°C for 4 min, with a 5 min extension at 68°C at the end of the final cycle. The mutated plasmid was transformed into XL-Gold ultracompetent E. coli cells (Agilent Technologies) and selected on LB plate (10 g/L tryptone, 5 g/L yeast extract, 10 g/L sodium chloride and 12 g/L agar) containing 100 mg/L ampicillin. A colony was picked from the plate and grown overnight at 37 °C in LB medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L sodium chloride) containing 100 mg/L ampicillin, from which plasmid pCW′_NcoI_CYP725A4_hNPR was isolated. The isolated plasmid was digested with NcoI and HindIII (FastDigest™, Thermo Scientific) at 37°C for 15 min, which confirmed the presence of the NcoI site and removed hNPR (~2 kb). The resulting linearized plasmid served as vector for an In-Fusion® cloning reaction to clone NcoI-coTCPR-HindIII DNA fragment (~2 kb), which was prepared by PCR as described above using pCWb_TCPR_NcoI_F1gs, and pCWb_TCPR_HindIII_R1 stp as forward and reverse primers (Table 2), and synthetic coCYP725A4/coTCPR as template (Fig. S4 in Supplementary Material).
To replace hNPR with CYP725A4/TCPR, the mutated plasmid pCW′_NcoI_coCYP725A4_hNPR was digested with NcoI and HindIII as described above to remove the hNPR gene, which was replaced with purified NcoI-coCYP725A4/coTCPR-HindIII DNA fragment using an In-Fusion® cloning kit. NcoI-coCYP725A4/coTCPR-HindIII fragment was prepared by PCR as above using primers pCWb_coT5H_NcoI_F1 and pCWb_TCPR_HindIII_R1 stp (Table 2) with coCYP725A4/coTCPR as the DNA template (Fig. S5 in Supplementary Material).
The plasmid pCW′_coCYP725A4_coCYP725A4/coTCPR was digested with NdeI and XbaI to remove coCYP725A4 (~1.5 kb) and generate linearized pCW′_coCYP725A4/coTCPR plasmid. A Taxus x media cytochrome b5 that was codon optimized (coTxCb5) and synthesized by BioBasic Inc. (Markham, Ontario, Canada) was amplified by PCR using primers pCWb_TxCb5.6_NdeI_F1 and pCWb_TxCb5.6_XbaI_R1 stp (Table 2) to generate NdeI-coTxCb5.6-XbaI DNA fragment (~500 bp). The latter was then ligated into linearized pCW′_coCYP725A4/coTCPR using an In-Fusion® cloning kit (Fig. S6 in Supplementary Material).
2.3. Heterologous expression of TS and CYP725A4 in E. coli
For production of the taxadiene isomers, pGG_coTS79 was cotransformed with another plasmid pIRS into OverExpress™ C41 (DE3) E. coli cells (Lucigen, USA) to obtain the IRSGT strain (Table 3). A single isolated IRSGT colony was grown at 37°C for 16 h in 5 mL LB media supplemented with 34 mg/L chloramphenicol and 50 mg/L spectinomycin. For production of T5α-ol, IRSGT cells were transformed with different CYP725A4 expression plasmids (prepared as described in Section 2.2) and each transformant was selected on LB agar plates containing 34 mg/L chloramphenicol, 50 mg/L spectinomycin, and 100 mg/L ampicillin to obtain IRSGTC-1, IRSGTC-2, IRSGTC-3, IRSGTC-4, and IRSGTC-5 E. coli strains (Table 3). A single isolated colony of each IRSGTC strain was grown at 37°C for 16 h in 5 mL LB medium supplemented with 34 mg/L chloramphenicol, 50 mg/L spectinomycin, and 100 mg/L ampicillin. Overnight cultures (500 μL) of IRSGT and IRSGTC strains were transferred to 50 mL LB in a 250-mL conical flask and grown at 37°C until the absorbance measured at 600 nm reached 0.6–0.8. The flask was brought to 25°C prior to the addition of 1 mM IPTG, 0.5 mL of sodium phosphate buffer (pH 7), and 1 mL of 80% glycerol. The flask was then placed at 30°C for three days, with shaking at 250 rpm.
Table 3.
Recombinant lines of OverExpress™ C41 (DE3) E. coli cells (genotype: F−ompT hsdSB (rB− mB−) gal dcm (DE3)) used for the expression of different gene constructs
| Bacterial cell lines | Gene inserts and constructs |
|---|---|
| IRSGT | dxs, dxr, idi, ggpps, coTS79 |
| IRSGTC-1 | dxs, dxr, idi, ggpps, coTS79, coCYP725A4/coTCPR |
| IRSGTC-2 | dxs, dxr, idi, ggpps, coTS79, coCYP725A4_hNPR |
| IRSGTC-3 | dxs, dxr, idi, ggpps, coTS79, coCYP725A4_coTCPR |
| IRSGTC-4 | dxs, dxr, idi, ggpps, coTS79, coCYP725A4_coCYP725A4/coTCPR |
| IRSGTC-5 | dxs, dxr, idi, ggpps, coTS79, coTxCb5_coCYP725A4/coTCPR |
2.4. GC-MS analysis
Hexane (5 mL) was added to 50 mL bacterial culture and stirred for 1 h. The mixture was transferred to a 50-mL conical tube and centrifuged at 3,220 x g for 5 min to separate the organic from the aqueous layer. The upper hexane layer was collected in a glass vial, and 1 μl was injected (in splitless mode) into a VF-5ms capillary column (30 m x 0.25 mm I.D. with 0.25 μm film thickness) installed on a Varian 3900 GC coupled to a Saturn 2100T ion trap MS. Helium (ultra-pure) was used as the carrier gas at a flow rate of 1.0 mL/min. The oven temperature was programmed to start at 50°C for 1 min, then increased to 220°C at a rate of 10°C/min with a 10 min hold at 220°C, followed by a 20°C/min increase to 300°C with a 2.5 min hold at 300°C. The injector temperature was set at 200°C.
3. Results
3.1. Expression of TS in E. coli
An N-terminal truncation of TS from Taxus brevifolia (GenBank accession U48796) was codon optimized for efficient expression in E. coli (coTS79, see Fig. S7 in Supplementary Material for the nucleotide sequence). This gene was coexpressed with GGPPS from Abies grandis and three isoprenoid genes from E. coli, namely 1-deoxy-D-xylulose-5-phosphate synthase (dxs), 1-deoxy-D-xylulose-5-reductase (dxr) and isopentenyl diphosphate isomerase (idi).
3.2. Formation of multiple deprotonation products during heterologous expression of TS
TS catalyzes a multistep cyclization reaction that occurs through a series of carbocation transformations, with deprotonation of a taxen-4-yl cation to give either endotaxadiene or exotaxadiene, depending on which proton is abstracted (Fig. S8 in Supplementary Material). Deprotonation of other carbocation intermediates can also occur, forming monocyclic (cembrenes) [25–27] and dicyclic (verticillenes) diterpenes [26–28] (Fig. S8). TS can also deprotonate the bridgehead methine C3 of taxen-4-yl cation resulting in taxa-4(3),11(12)-diene isomer [7, 29]. To investigate this multispecificity, TS-expressing transgenic bacterial cultures were extracted with hexane and analyzed by gas chromatography-mass spectrometry (GC-MS).
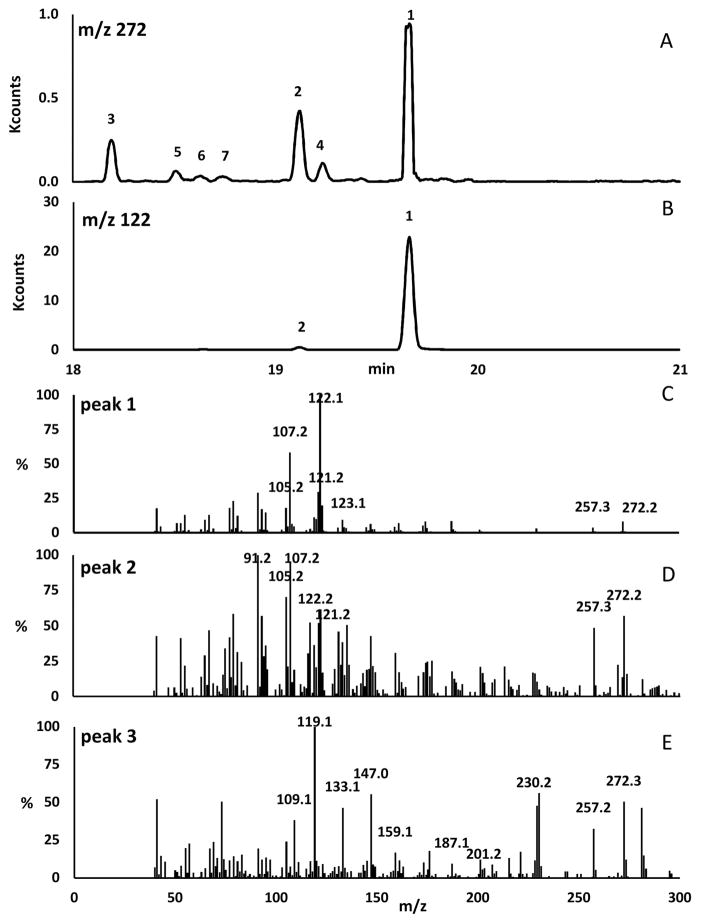
Transgenic bacteria transformed with five genes (dxs, dxr, idi, ggpps, and ts) showed the presence of both a major and a minor product, which were identified by selective ion monitoring at m/z 272 and 122 as endotaxadiene and exotaxadiene correspondingly (Fig. 2, A and B). While m/z 272 corresponds to the molecular mass of diterpenes (C20H32), m/z 122 is a characteristic ion of endotaxadiene due to a C-ring cleavage fragment with the formula C9H14 [30]. The mass spectrum of putative endotaxadiene detected at 19.65 min (Fig. 2, C) showed ion peaks at m/z 122 (100%), 107 (58%), 121 (30%), 123 (20%), 105 (18%), 272 (8%) and 257 (4%) which is consistent with previous publications [7, 31]. Similarly, the mass spectrum of putative exotaxadiene detected at 19.12 min (Fig. 2, D) showed ion peaks at m/z 91 (100%), 107 (95%), 105 (70%), 122 (62%), 272 (57%), 121 (52%), 257 (49%), 123 (17%) as reported previously [7, 31]. Alongside, putative verticilla-3,7,12(13)-triene (verticillene) and other unknown taxane products (TX1, TX2, TX3, and TX4) were also observed by selective ion monitoring at m/z 272 (Fig. 2, A). The possible presence of verticillene was consistent with the mass spectrum of peak 3 at 18.19 min (Fig. 2, E) having a base peak at m/z 119 [7]. Each of the other four taxane products could be one of the remaining proposed deprotonated intermediates (Fig. S8, cembrene A, verticilla-4(20),7,11-triene, verticilla-3,7,11(12)-triene), and taxa-3(4),11(12)-diene) but it is difficult to identify each of them based solely on their mass spectra which are very similar to each other (Fig. S9 in the Supplementary Material).
Fig. 2.
GC-MS analysis of diterpenes produced by E. coli expressing TS, GGPPS and MEP pathway enzymes. (A) Selective ion monitoring chromatogram at m/z 272; (B) Selective ion monitoring chromatogram at m/z 122; (C) Mass spectrum of peak 1 putatively identified as endotaxadiene; (D) Mass spectrum of peak 2 putatively identified as exotaxadiene; (E) Mass spectrum of peak 3 putatively identified as verticillene.
3.3. Expression of CYP725A4, CPRs and Cb5 in E. coli
The protein sequences of CYP725A4 and CPR from T. cuspidata (TCPR) (GenBank accession AY289209 and AY571340, respectively) were truncated to remove transmembrane regions at the N-terminal, corresponding to 42 and 74 amino acids, respectively, as was done previously by others [22]. The amino acid sequence MALLLAVF derived from bovine P450 was added to the N-terminal of CYP725A4 to enhance its expression and direct the protein to the bacterial plasma membrane [32]. A fusion protein was constructed consisting of CYP725A4 and TCPR connected by a GSTGS linker peptide (see Fig. S10 in Supplementary Material for the nucleotide sequence) to establish a self-sufficient enzyme, similar to soluble bacterial P450BM3 (Bacillus megaterium). pCWori vector was used to express codon optimized CYP725A4 and TCPR, either as a fusion protein or as two separate proteins encoded by a single bicistronic transcript. This was accomplished by using the bicistronic plasmid pCW′_2A6_hNPR as a starting point to make our constructs. To express only the fusion protein, the linked CYP725A4/TCPR construct replaced the bicistronic CYP2A6 and hNPR in the original pCW′_2A6_hNPR plasmid. All the newly constructed pCWori-based plasmids were cotransformed with pGG_coTS79 and pIRS to effect the production of T5α-ol in E. coli.
3.4. Monooxygenated products detected from heterologous expression of CYP725A4
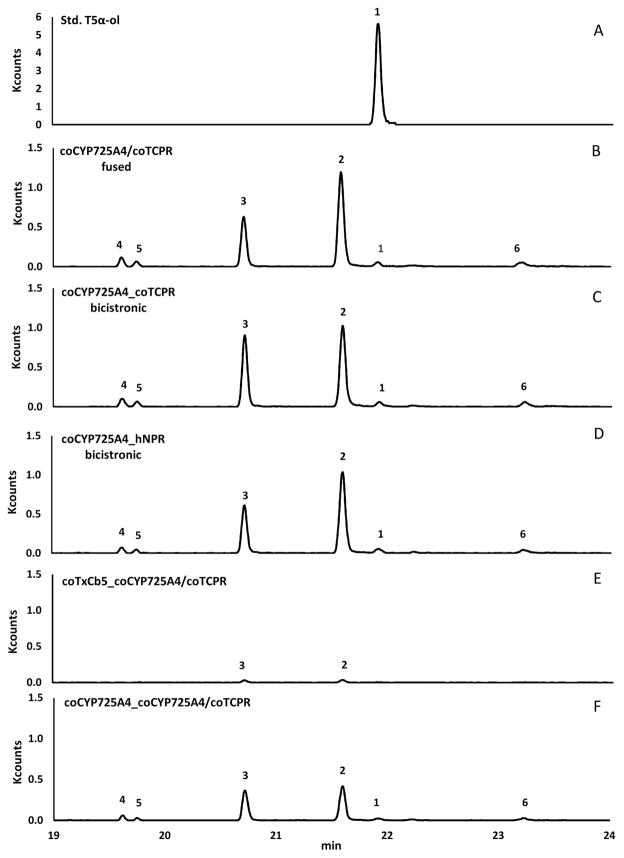
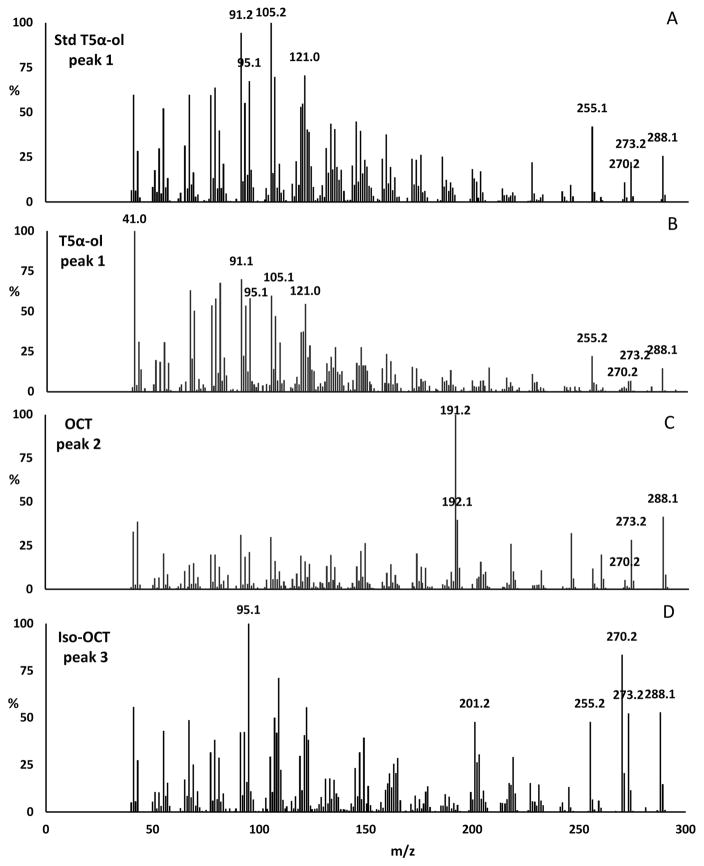
Transgenic E. coli expressing seven heterologous enzymes (DXS, DXR, IDI, GGPPS, TS, CYP725A4, and TCPR or hNPR) showed two major and four minor compounds as detected by GC-MS selective ion monitoring at m/z 288 (molecular mass of a monooxygenated diterpene) (Fig. 3, B–D). A chemically synthesized T5α-ol [33] was used as a reference standard (Fig. 3, A) to identify the putative T5α-ol generated by CYP725A4, which was detected as a minor peak (Fig. 3, B–D). This minor peak (1) at 21.91 min shared the same retention time and mass spectrum (characteristic ions at m/z 288 (P+), 273 (P+-CH3), 270 (P+-H2O) and 255 (P+-H2O-CH3) with ratio ~ 5:2:1:7) as that of the synthetic standard (Fig. 4, A and B). This result showed that our metabolically engineered E. coli produced T5α-ol, but only in small quantities. The major peak (2) at 21.59 min was tentatively identified as 5(12)-oxa-3(11)-cyclotaxane (OCT), with a mass spectrum having a base peak at m/z 191 (Fig. 4, C), which is consistent with prior publications [15–17, 22, 34]. The second major peak (3) at 20.71 min showed similar characteristic ions at m/z 288, 273, 270, 255, but in the ratio of ~ 1:1:1.5:1 (Fig. 4, D), and was tentatively identified as 5(11)-oxa-3(11)-cyclotaxane (iso-OCT), based on characterizations from recent publications [15–17]. We also observed three unknown compounds, designated here as TXO1, TXO2, and TXO3, that were presumed to be monooxygenated diterpenes based on their mass spectra (Fig. S11 in Supplementary Material). The mass spectral profiles of the latter unknown products are different from those previously observed in CYP725A4-expressing bacteria [15, 16].
Fig. 3.
GC-MS analysis of monooxygenated diterpenes. Selective ion monitoring at m/z 288 detects monooxygenated taxanes, including chemically synthesized taxadiene 5α-ol (A) as well as those formed by recombinant bacteria expressing GGPPS, coTS79 and coCYP725A4/coTCPR fusion protein (B), recombinant bacteria expressing GGPPS, coTS79, coCYP725A4, and coTCPR (C), recombinant bacteria expressing GGPPS, coTS79, coCYP725A and hNPR (D), recombinant bacteria expressing GGPPS, coTS79, coTxCb5 and coCYP725A4/coTCPR fusion protein (E), and recombinant bacteria expressing GGPPS, coTS79, coCYP725A4 and coCYP725A4/coTCPR fusion protein (F). Peak 1 = T5α-ol; Peak 2 = OCT; Peak 3 = iso-OCT; and Peaks 4, 5 and 6 are presumed to be monooxygenated taxanes named TXO1, TXO2, and TXO3, respectively.
Fig. 4.
Mass spectra of tentatively identified monooxygenated diterpenes detected by GC-MS analysis at m/z 288. Peak 1 = T5α-ol; Peak 2 = OCT; Peak 3 = iso-OCT.
3.5. Effect of different CPRs and Cb5 on CYP725A4 activity
Unlike animals, some plants may have more than one CPR isoform. CPRs in plants can be differentially regulated as in the case of the two CPR genes in Arabidopsis thaliana, where one CPR is found to be constitutively expressed, while the other CPR is enhanced during environmental stress [35], which suggests different isoforms of CPR being used for different P450-mediated reactions. So far, only one CPR has been isolated from Taxus and shown to work with P450 enzymes in the paclitaxel pathway including CYP725A4 [12], but it is also possible that other Taxus CPRs exist which may be preferred by these P450s. By contrast, a single P450 reductase (hNPR) in humans provides the electrons to presumably all 57 P450 enzymes encoded in the human genome. Here, we compared the human P450 reductase and the native Taxus P450 reductase (TCPR) in their effects on the products formed by CYP725A4, and looked at the effect of unlinking the P450 reductase from CYP725A4 to compare our results with those from previous work [15–17, 22].
The same products were observed regardless of the P450 reductase used (Fig. 3, C and D), suggesting that CYP725A4 can accept electrons from other non-native P450 reductases. Based on the peak areas in GC-MS chromatograms, the ratio of OCT:iso-OCT is the same (~1.5:1) between the linked CYP725A4/TCPR and bicistronic CYP725A4_hNPR protein constructs (Fig. 3, B and D). The ratio of OCT:iso-OCT shifted to approximately 1:1 in the case of bicistronic CYP725A4_TCPR protein (Fig. 3, C). The difference in the product profiles generated by linked and bicistronically expressed reductases had been reported previously, although changes in the levels of T5α-ol had apparently been observed [16]. In our case, the levels of T5α-ol was not affected by changing the source of CPR, nor by the unlinking of CPR from CYP725A4 (Fig. 3, B–D).
The involvement of Cb5 was also explored since studies have shown that Cb5 can inhibit [36, 37], stimulate [38, 39], or otherwise have no effect [40] on the catalytic activities of P450s. We searched the transcriptome of Taxus x media (http://medplants.ncgr.org/) and found a protein sequence similar to Cb5. This sequence was codon optimized (see Fig. S12 in Supplementary Material for the nucleotide sequence) and synthesized for coexpression with CYP725A4 and TCPR. The bicistronic coexpression of Cb5 with the CYP725A4/TCPR fusion protein resulted in apparently reduced activity of CYP725A4, with relatively lower amounts of OCT and iso-OCT formed (Fig. 3, E).
In our last attempt to increase the production of T5α-ol, we coexpressed CYP725A4 with the CYP725A4/TCPR fusion protein in the same bicistronic plasmid. The rationale behind this experiments is that a single CPR is shared among multiple P450s in plant membranes [13]. By coexpressing CYP725A4 with CYP725A4/TCPR, the TCPR can potentially provide electrons to both the linked and unlinked CYP725A4 and perhaps lead to higher levels of T5α-ol. Instead, we found slightly reduced OCT and iso-OCT levels, while T5α-ol and all three unknown monooxygenated products were still about the same (Fig. 3, F).
4. Discussion
Prior to the in vitro enzymatic characterization of TS in 1995 [9], it was widely accepted that the biosynthesis of paclitaxel goes through taxa-4(20),11(12)-diene (designated by us as “exotaxadiene” in reference to the ‘exo’cyclic double bond), based on the common occurrence of 4(20)- and 11(12)- double bonds in taxanes of yew [18]. It was thus surprising (at that time) when Croteau’s group reported that the partially purified TS from yew formed another taxadiene isomer taxa-4(5),11(12)-diene (“endotaxadiene” containing an ‘endo’cyclic double bond) from geranylgeranyl diphosphate (GGPP), instead of exotaxadiene [9]. This result was later confirmed repeatedly with the heterologous expression of TS in bacteria [7, 8, 41, 42], yeast [43, 44], and plants [45–50], all of which reported the formation of endotaxadiene as the main product.
The first heterologous expression of TS in bacteria in 2000 [7], used truncated forms of TS (e.g. 60 or 79 amino acids were removed from the N-terminus) cloned into a pSBET vector to obtain an active soluble enzyme, which was partially purified for further characterization. In vitro assays of the recombinant TS showed (besides endotaxadiene) other minor products, including exotaxadiene, verticillene, and taxa-3(4),11(12)-diene (referred to as iso-endotaxadiene), which we have also seen in our TS-expressing E. coli. We have used a similarly truncated version of TS (without the 79 N-terminal amino acids) but ours was codon optimized for bacterial expression to avoid codons rarely used by E. coli. Without codon optimization, the rare arginine codon tRNA will need to be coexpressed with the original Taxus TS sequence to obtain high levels of expression. This was previously accomplished using the pSBET expression vector, which carries the argU gene encoding the rare arginine codon tRNA [7]. With codon optimization, we did not have to use pSBET, and instead opted for the dual expression vector pACYCDuet (in the form of pGG_DEST), which enabled the coexpression of both GGPPS and TS in the same plasmid, as was done previously by others [22].
The first attempt to produce taxadiene in E. coli (in 2001) expressed three other enzymes besides TS namely, DXP synthase (DXS), IPP isomerase (IDI), and GGPPS, which yielded 1.3 mg endotaxadiene per liter of culture [8]. Following the same approach, we also expressed other enzymes upstream of TS, including GGPPS, IDI, DXR, and DXS. Three enzymes (IDI, DXR, and DXS) were found to be bottleneck steps in the MEP pathway and had been shown to increase diterpene production in bacteria when coexpressed with a diterpene synthase [23]. The plasmid pIRS, which contained idi, dxr, and dxs genes, allowed us to express three MEP pathway enzymes (instead of just two) together with GGPPS and TS in E. coli, and detect the formation of endotaxadiene as the major product, as previously observed by others [8, 22]. This taxadiene producing E. coli had enabled us to begin to independently investigate CYP725A4, which had been variously reported to produce either T5α-ol only [10], OCT only [34], both OCT and T5α-ol in equal amounts [22], or more recently T5α-ol, OCT and iso-OCT, together with unidentified products [15–17].
Heterologous coexpression of TS and CYP725A4 was first accomplished in yeast (Saccharomyces cerevisiae), which resulted in only low levels of T5α-ol (25 μg/L) being detected [44]. When the same genes were expressed by Rontein et al. in tobacco trichomes and S. cerevisiae, only OCT was observed instead of T5α-ol [34]. Later work in recombinant E. coli reported the formation of both T5α-ol and OCT under optimized conditions in relatively equal amounts [22]. The latter publication was considered a breakthrough in paclitaxel metabolic engineering because of the high levels of taxadiene produced (up to 1 g per liter) [51]. T5α-ol did not reach the same production level as the taxadiene, but was reportedly equal to the amount of OCT (~58 mg per liter). In our case, the T5α-ol produced by a similarly transformed E. coli was ~10-fold less than OCT. Upon further investigation, we noticed that the GC-MS peak thought to be T5α-ol in the previous paper [22] did not have the same mass fragmentation pattern nor chromatographic characteristics as our authentic standard T5α-ol, and is instead more similar to iso-OCT. Thus, we propose that the compound thought to be T5α-ol in the breakthrough paper is likely iso-OCT, which is consistent with our current data and those of others [15–17]. Hence, efforts to increase the production of T5α-ol in E. coli is less optimistic than previously projected [15, 16].
The lower amounts of T5α-ol relative to OCT and iso-OCT (Fig. 3) are consistent with data from a recent publication investigating the role of an epoxide intermediate in T5α-ol formation [15]. Taxadiene-4(5)-epoxide has been shown to rearrange under acidic conditions to T5α-ol, OCT and iso-OCT [52], while OCT spontaneously forms iso-OCT upon storage [16]. It is therefore possible that taxadiene-4(5)-epoxide is the initial product of CYP725A4 and the products observed are the result of proton-induced nonenzymatic rearrangements, which can easily be interpreted as enzyme promiscuity [15–17]. Alternatively, if allylic rearrangements did occur in the active site of CYP725A4, thus generating multiple products, then CYP725A4 may justifiably be considered a promiscuous enzyme. However, it is not yet clear if such promiscuity is an artifact of heterologous expression or an inherent property of the enzyme [15, 16]. At any rate, this catalytic promiscuity may help to explain the different results obtained from studies that evaluate the product pools derived from constructed pathways that include heterologous expression of CYP725A4.
We expressed CYP725A4 with TCPR either as a fusion protein, similar to what was done by Ajikumar et al. [22], or as two separate proteins using a bicistronic plasmid. Ajikumar et al. used a pTrc plasmid [22], while we used pCWori to express CYP725A4 and TCPR either as a fusion protein or as two separate proteins. We also used pCWori to express CYP725A4 with a human P450 reductase (hNPR) as two separate proteins. The use of these two different expression plasmids is not expected to affect the catalytic properties of CYP725A4. Our results showed that human reductase (hNPR) is just as effective as TCPR in supporting CYP725A4 activity in vivo, and T5α-ol is just a minor product in all cases (Fig. 3, B–D). Ajikumar’s group recently coexpressed rat and stevia P450 reductases with CYP725A4 in E. coli (expressing TS and GGPPS) which also showed T5α-ol as a minor product relative to OCT [16]. When we expressed CYP725A4 and TCPR as separate proteins instead of a fusion protein, iso-OCT levels increased but the levels of T5α-ol and OCT remained the same (Fig. 3, B and C). Hence, changing the source and the way P450 reductases interact with CYP725A5 did not increase T5α-ol production in any of our experiments.
Jennewein et al. [12] investigated the effects of different P450 reductases and Cb5 on a Taxus P450 enzyme, specifically taxoid 10β-hydroxylase (T10H). By conducting in vitro enzyme assays on microsomes from T10H-expressing yeast cells, with or without coexpressing TCPR, they found that T10H was more active with coexpressed TCPR than with the endogenous yeast P450 reductase [12]. They also found that P450 reductases from spearmint and mungbean could effectively replace TCPR to support T10H activity in microsomes prepared from T10H-expressing insect cells, while rabbit P450 reductase was only about 60% as effective in vitro as P450 reductases of plant origin [12].
Cb5 has been shown to enhance the activities of P450s [38, 53–55] so we coexpressed a Cb5 gene from Taxus x media, with CYP725A4 and TCPR (along with TS, GGPPS, DXS, DXR, and IDI). This resulted in reduced levels of OCT and iso-OCT, and the apparent absence of T5α-ol, which is in contrast to the reported increase in the in vitro activity of T10H in the presence of rat Cb5 [12]. Assuming the expression of CYP725A4 and other genes had not been affected by Cb5, it can be hypothesized that this particular Cb5 interacts with CYP725A4 and negatively affects its activity, although further experimentation is required to support this premise. However, if such interaction did occur, it is worth exploring how other Cb5 genes would affect CYP725A4 activity, with an eye for those that can enhance the production of T5α-ol while reducing the formation of OCT and iso-OCT.
The ability of P450 enzymes to catalyze hydroxylation reactions appears to be influenced by their interaction with its redox partner CPR. Upon heterologous expression in yeasts, for example, plant P450s had been shown to prefer plant CPRs over endogenous host CPR as reflected by increased P450 activity [12, 56]. The presence of another interacting partner, Cb5, has also been shown to influence product formation. For example, Cb5 from Artemisia annua combined with CPR expression could produce a higher concentration of artemisinic acid when enzymes from the artemisinin pathway were reconstructed in yeast [55]. In the case of CYP725A4, our results showed that Taxus Cb5 negatively affected T5α-ol formation while TCPR alone promoted T5α-ol formation. Hence, different redox partners affected CYP725A4 activity and product formation in recombinant bacteria. This opens the possibility that other P450-interacting proteins in Taxus influence the product outcome of CYP725A4 in planta, which may explain the apparent absence of OCT and iso-OCT in Taxus.
We also tried coexpressing CYP725A4 with the CYP725A/TCPR fusion protein to see if this will enhance T5α-ol production. The idea behind this experiment is that the linked TCPR may be able to support both the CYP725A4 linked to it and a separate CYP725A4 protein. However, we only observed decreased levels of all products (but still in the same proportion), which suggested that the free CYP725A4 interacted with TCPR linked to another CYP725A4, but this interaction hindered the transfer of electrons to both the free and the linked CYP725A4. Thus, in future experiments that will include other P450 genes in the paclitaxel pathway, it would be better to express TCPR separately from CYP725A4 rather than as a fusion protein, as suggested previously [21].
Given the low levels of T5α-ol produced by CYP725A4 when expressed in E. coli, others have tried mutating TS so it mainly produces exotaxadiene instead of endotaxadiene [41]. This approach should work in theory since exotaxadiene is a viable substrate for CYP725A4 and does not generate OCT and iso-OCT as side products [10, 15]. However, the TS mutants generated so far have not yet led to the same breakthrough as the production of 1 gram taxadiene per liter of bacterial culture. An alternative approach would be to find naturally occurring TS isoforms that produce mainly exotaxadiene, or a new taxadiene hydroxylase that produces only T5α-ol, which may perhaps be found in the newly sequenced genomes of taxane producing organisms [57]. It is hard to predict which approach will eventually break through the current bottleneck in this field, but it is now clear that the next challenge for us is to produce T5α-ol (or an alternative paclitaxel precursor) without the side products. Only then can we realize the potential of metabolic engineering to provide an alternative source of paclitaxel.
Supplementary Material
Acknowledgments
We thank Dr. Reuben J. Peters from Iowa State University for pIRS and pGG_DEST plasmids, Dr. F. Peter Guengerich from Vanderbilt University for the pCWori/2A6/hNPR plasmid (pCW′_2A6_hNPR), and Dr. Phil S. Baran from the Scripps Research Institute for the synthetic taxadiene-5α-ol. This work was supported by the National Institutes of Health (NIH-1R15CA139416-01).
Abbreviations
- Cb5
cytochrome b5
- CPR
cytochrome P450 reductase
- CYP
cytochrome P450
- DMAPP
dimethylallyl diphosphate
- DXS
1-deoxy-D-xylulose-5-phosphate synthase
- DXR
1-deoxy-D-xylulose-5-phosphate reductase
- GC-MS
gas chromatography-mass spectrometry
- GGPP
geranylgeranyl diphosphate
- GGPPS
geranylgeranyl diphosphate synthase
- hNPR, human NADPH
P450 reductase
- IDI
isopentenyl diphosphate isomerase
- IPP
isopentenyl diphosphate
- IPTG
isopropyl-β-D-1-thiogalactopyranoside
- LB
Lysogeny Broth
- MEP
methylerythritol phosphate
- NCBI
National Center for Biotechnology Information
- T5α-ol
taxadiene-5α-ol
- OCT
5(12)-oxa-3(11)-cyclotaxane
- iso-OCT
5(11)-oxa-3(11)-cyclotaxane
- TS
taxadiene synthase
References
- 1.Suffness M, Wall ME. In: Taxol: Science and Applications. Suffness M, editor. CRC press; Florida: 1995. pp. 3–25. [Google Scholar]
- 2.Arbuck SG, Blaylock BA. In: Taxol: Science and Applications. Suffness M, editor. CRC press; Florida: 1995. pp. 379–415. [Google Scholar]
- 3.Holton RA, Biediger RJ, Boatman PD. In: Taxol: Science and Applications. Suffness M, editor. CRC press; Florida: 1995. pp. 97–121. [Google Scholar]
- 4.Tabata H. Adv Biochem Eng Biotechnol. 2004;7:1–23. doi: 10.1007/b13538. [DOI] [PubMed] [Google Scholar]
- 5.Hefner J, Ketchum RE, Croteau R. Arch Biochem Biophys. 1998;360:62–74. doi: 10.1006/abbi.1998.0926. [DOI] [PubMed] [Google Scholar]
- 6.Hezari M, Lewis NG, Croteau R. Arch Biochem Biophys. 1995;322:437–444. doi: 10.1006/abbi.1995.1486. [DOI] [PubMed] [Google Scholar]
- 7.Williams DC, Wildung MR, Jin AQ, Dalal D, Oliver JS, Coates RM, Croteau R. Arch Biochem Biophys. 2000;379:137–146. doi: 10.1006/abbi.2000.1865. [DOI] [PubMed] [Google Scholar]
- 8.Huang Q, Roessner CA, Croteau R, Scott AI. Bioorg Med Chem. 2001;9:2237–2242. doi: 10.1016/s0968-0896(01)00072-4. [DOI] [PubMed] [Google Scholar]
- 9.Koepp AE, Hezari M, Zajicek J, Vogel BS, LaFever RE, Lewis NG, Croteau R. J Biol Chem. 1995;270:8686–8690. doi: 10.1074/jbc.270.15.8686. [DOI] [PubMed] [Google Scholar]
- 10.Jennewein S, Long RM, Williams RM, Croteau R. Chem Biol. 2004;11:379–387. doi: 10.1016/j.chembiol.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Hefner J, Rubenstein SM, Ketchum RE, Gibson DM, Williams RM, Croteau R. Chem Biol. 1996;3:479–489. doi: 10.1016/s1074-5521(96)90096-4. [DOI] [PubMed] [Google Scholar]
- 12.Jennewein S, Park H, DeJong JM, Long RM, Bollon AP, Croteau RB. Biotechnol Bioeng. 2005;89:588–598. doi: 10.1002/bit.20390. [DOI] [PubMed] [Google Scholar]
- 13.Jensen K, Moller BL. Phytochemistry. 2010;71:132–141. doi: 10.1016/j.phytochem.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Waskell L, Kim J-JP. In: Cytochrome P450: Structure, Mechanism and Biochemistry. Ortiz de Montellano PR, editor. Springer; Switzerland: 2015. pp. 33–68. [Google Scholar]
- 15.Edgar S, Zhou K, Qiao K, King JR, Simpson JH, Stephanopoulos G. ACS Chem Biol. 2015;11:460–469. doi: 10.1021/acschembio.5b00767. [DOI] [PubMed] [Google Scholar]
- 16.Biggs BW, Rouck JE, Kambalyal A, Arnold W, Lim CG, De Mey M, O’Neil-Johnson M, Starks CM, Das A, Ajikumar PK. ACS Chem Biol. 2016;11:1445–1451. doi: 10.1021/acschembio.5b00968. [DOI] [PubMed] [Google Scholar]
- 17.Yadav V. J Mol Catalysis B: Enzymatic. 2014;110:154–164. [Google Scholar]
- 18.Floss HG, Mocek U. In: Taxol: Science and Applications. Suffness M, editor. CRC press; Florida: 1995. pp. 191–208. [Google Scholar]
- 19.Kaspera R, Croteau R. Phytochem Rev. 2006;5:433–444. doi: 10.1007/s11101-006-9006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Croteau R, Ketchum RE, Long RM, Kaspera R, Wildung MR. Phytochem Rev. 2006;5:75–97. doi: 10.1007/s11101-005-3748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, Ajikumar PK. PNAS. 2016;113:3209–3214. doi: 10.1073/pnas.1515826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, Peters RJ. Appl Microbiol Biotechnol. 2010;85:1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillam EM, Aguinaldo AM, Notley LM, Kim D, Mundkowski RG, Volkov AA, Arnold FH, Soucek P, DeVoss JJ, Guengerich FP. Biochem Biophys Res Commun. 1999;265:469–472. doi: 10.1006/bbrc.1999.1702. [DOI] [PubMed] [Google Scholar]
- 25.Chow SY, Williams HJ, Huang Q, Nanda S, Scott AI. J Org Chem. 2005;70:9997–10003. doi: 10.1021/jo0517489. [DOI] [PubMed] [Google Scholar]
- 26.Hong YJ, Tantillo DJ. J Am Chem Soc. 2011;133:18249–18256. doi: 10.1021/ja2055929. [DOI] [PubMed] [Google Scholar]
- 27.Schrepfer P, Buettner A, Goerner C, Hertel M, Rijn JV, Wallrapp F, Eisenreich W, Sieber V, Kourist R, Bruck T. PNAS. 2016;113:E958–E967. doi: 10.1073/pnas.1519680113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin Y, Williams DC, Croteau R, Coates RM. J Am Chem Soc. 2005;127:7834–7842. doi: 10.1021/ja050592r. [DOI] [PubMed] [Google Scholar]
- 29.Williams DC, Carroll BJ, Jin Q, Rithner CD, Lenger SR, Floss HG, Coates RM, Williams RM, Croteau R. Chem Biol. 2000;7:969–977. doi: 10.1016/s1074-5521(00)00046-6. [DOI] [PubMed] [Google Scholar]
- 30.Lin X, Hezari M, Koepp AE, Floss HG, Croteau R. Biochemistry. 1996;35:2968–2977. doi: 10.1021/bi9526239. [DOI] [PubMed] [Google Scholar]
- 31.Schmeer H, Jennewein S. Methods Mol Biol. 2010;643:165–184. doi: 10.1007/978-1-60761-723-5_12. [DOI] [PubMed] [Google Scholar]
- 32.Barnes HJ, Arlotto MP, Waterman MR. PNAS. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilde NC, Isomura M, Mendoza A, Baran PS. J Am Chem Soc. 2014;136:4909–4912. doi: 10.1021/ja501782r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rontein D, Onillon S, Herbette G, Lesot A, Werck-Reichhart D, Sallaud C, Tissier A. J Biol Chem. 2008;283:6067–6075. doi: 10.1074/jbc.M708950200. [DOI] [PubMed] [Google Scholar]
- 35.Mizutani M, Ohta D. Plant Physiol. 1998;116:357–367. doi: 10.1104/pp.116.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan ET, Coon MJ. Drug Metab Dispos. 1984;12:358–364. [PubMed] [Google Scholar]
- 37.Zhang H, Hamdane D, Im SC, Waskell L. J Biol Chem. 2008;283:5217–5225. doi: 10.1074/jbc.M709094200. [DOI] [PubMed] [Google Scholar]
- 38.Yamazaki H, Shimada T, Martin MV, Guengerich FP. J Biol Chem. 2001;276:30885–30891. doi: 10.1074/jbc.M105011200. [DOI] [PubMed] [Google Scholar]
- 39.McLaughlin LA, Ronseaux S, Finn RD, Henderson CJ, Roland Wolf C. Mol Pharmacol. 2010;78:269–278. doi: 10.1124/mol.110.064246. [DOI] [PubMed] [Google Scholar]
- 40.Yamazaki H, Gillam EM, Dong MS, Johnson WW, Guengerich FP, Shimada T. Arch Biochem Biophys. 1997;342:329–337. doi: 10.1006/abbi.1997.0125. [DOI] [PubMed] [Google Scholar]
- 41.Edgar S, Li FS, Qiao K, Weng JK, Stephanopoulos G. ACS Synth Biol. 2016 doi: 10.1021/acssynbio.6b00206. [DOI] [PubMed] [Google Scholar]
- 42.Huang KX, Huang Q, Wildung MR, Croteau R, Scott AI. Protein Expr Purif. 1998;13:90–96. doi: 10.1006/prep.1998.0870. [DOI] [PubMed] [Google Scholar]
- 43.Engels B, Dahm P, Jennewein S. Metab Eng. 2008;10:201–206. doi: 10.1016/j.ymben.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Dejong JM, Liu Y, Bollon AP, Long RM, Jennewein S, Williams D, Croteau RB. Biotechnol Bioeng. 2006;93:212–224. doi: 10.1002/bit.20694. [DOI] [PubMed] [Google Scholar]
- 45.Besumbes O, Sauret-Gueto S, Phillips MA, Imperial S, Rodriguez-Concepcion M, Boronat A. Biotechnol Bioeng. 2004;88:168–175. doi: 10.1002/bit.20237. [DOI] [PubMed] [Google Scholar]
- 46.Cha MJ, Shim SH, Kim SH, Kim OT, Lee SW, Kwon SY, Baek KH. BMB reports. 2012;45:589–594. doi: 10.5483/bmbrep.2012.45.10.085. [DOI] [PubMed] [Google Scholar]
- 47.Kovacs K, Zhang L, Linforth RS, Whittaker B, Hayes CJ, Fray RG. Transgenic Res. 2007;16:121–126. doi: 10.1007/s11248-006-9039-x. [DOI] [PubMed] [Google Scholar]
- 48.Anterola A, Shanle E, Perroud PF, Quatrano R. Transgenic Res. 2009;18:655–660. doi: 10.1007/s11248-009-9252-5. [DOI] [PubMed] [Google Scholar]
- 49.Li M, Jiang F, Yu X, Miao Z. Biomed Res Int. 2015;2015:1–8. doi: 10.1155/2015/504932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasan MM, Kim HS, Jeon JH, Kim SH, Moon B, Song JY, Shim SH, Baek KH. Plant Cell Rep. 2014;33:895–904. doi: 10.1007/s00299-014-1568-9. [DOI] [PubMed] [Google Scholar]
- 51.Liu T, Khosla C. Science. 2010;330:44–45. doi: 10.1126/science.1195014. [DOI] [PubMed] [Google Scholar]
- 52.Barton NA, Marsh BJ, Lewis W, Narraidoo N, Seymour GB, Fray R, Hayes CJ. Chem Sci. 2016;7:3102–3107. doi: 10.1039/c5sc03463a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vetten ND, Horst JT, Schaik HPV, Boer AD, Mol J, Koes R. PNAS. 1999;96:778–783. [Google Scholar]
- 54.Vik D, Crocoll C, Andersen TG, Burow M, Halkier BA. Plant Signaling & Behavior. 2016;11:1–6. doi: 10.1080/15592324.2016.1160189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 56.Eberle D, Ullmann P, Werck-Reichhart D, Petersen M. Plant Mol Biol. 2009;69:239–253. doi: 10.1007/s11103-008-9420-7. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Zhao H, Barrero RA, Zhang B, Sun G, Wilson IW, Xie F, Walker KD, Parks JW, Bruce R, Guo G, Chen L, Zhang Y, Huang X, Tang Q, Liu H, Bellgard MI, Qiu D, Lai J, Hoffman A. BMC Genomics. 2014;15:69–82. doi: 10.1186/1471-2164-15-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.