Abstract
Hyperspectral imaging (HSI) holds the potential for the noninvasive detection of cancers. Oral cancers are often diagnosed at a late stage when treatment is less effective and the mortality and morbidity rates are high. Early detection of oral cancer is, therefore, crucial in order to improve the clinical outcomes. To investigate the potential of HSI as a non-invasive diagnostic tool, an animal study was designed to acquire hyperspectral images of in vivo and ex vivo mouse tongues from a chemically induced tongue carcinogenesis model. A variety of machine-learning algorithms, including discriminant analysis, ensemble learning, and support vector machines, were evaluated for tongue neoplasia detection using HSI and were validated by the reconstructed pathological gold-standard maps. The diagnostic performance of HSI, autofluorescence imaging, and fluorescence imaging were compared in this study. Color-coded prediction maps were generated to display the predicted location and distribution of premalignant and malignant lesions. This study suggests that hyperspectral imaging combined with machine-learning techniques can provide a non-invasive tool for the quantitative detection and delineation of squamous neoplasia.
Detection and delineation of tongue neoplasia with hyperspectral imaging validated by the pathological gold standard.
Keywords: hyperspectral imaging (HSI), squamous neoplasia, 4NQO-induced tongue carcinogenesis model, supervised learning, linear support vector machine (SVM), linear discriminant analysis (LDA), ensemble LDA, random forest
Graphical abstract
Early detection of oral cancer is crucial to improve the survival and life of quality of patients. However, oral cancers are often diagnosed at a late stage when treatment is less effective and the mortality and morbidity rates are high. This article demonstrated the feasibility of using hyperspectral imaging as a noninvasive diagnostic tool to detect premalignant and malignant tongue cancers noninvasively in a preclinical animal model.
1. Introduction
Oral cancers are among the most common cancers globally, with an estimated 300,400 new cases and 145,400 deaths in 2012, and thus posing a significant health problem 1. Although the oral cavity is easily accessible, lesions in the oral cavity often go unnoticed. In the United States, approximately 65% of patients diagnosed with oral cavity and pharynx cancer have regional spread or distant metastases 2. If these cancers are diagnosed at a localized stage, most of the patients can be cured with a five-year survival rate of 83% 2. Late-stage cancer identification can lead to poor survival with speech, swallowing, and cosmetic problems. Moreover, patients who survive the initial occurrence of oral cancer have an increased risk of developing a second primary tumor 3. Therefore, early detection of pre-malignant and malignant lesions remains the most promising approach for improving the clinical outcomes 4.
The conventional screening method for oral cancer begins with visual inspection and palpation of the tissue surface using incandescent light. Next, highly suspicious tissue regions are biopsied for pathological examination to make a definitive diagnosis 5. However, visual examination is highly dependent on a clinician's training and experience. It is often a difficult task, even for an experienced physician, to identify the most malignant tissue region for biopsy due to the heterogeneity of the lesions. In addition, biopsy is invasive, costly and label-intensive. Pathology diagnosis is inconsistent due to intra- and inter-observer variations6, 7. Noninvasive alternatives are, therefore, desirable in order to improve the early diagnosis of oral cancer, and thus decreasing the morbidity and mortality of the disease.
Most of the lesions in the oral cavity are squamous-cell carcinoma, which originates from the oral mucosal lining of the oral cavity8. The easy access of the oral lesions makes it possible to detect superficial tumors with optical imaging which offers a variety of noninvasive tools utilizing intrinsic and extrinsic tissue contrast for the early detection of oral cancer 5. The intrinsic approaches involve the development of optical devices to specifically probe the alterations of tissue absorption and scattering properties by reflectance imaging and to reveal the levels of endogenous chromophores, such as reduced nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD), and with autofluorescence accompanying malignant progression. The extrinsic methods rely on the exogenous molecular imaging agents which are topically applied on the tissue surface to enhance the detection of fluorescence changes. One study reported the use of a point spectroscopy device combining intrinsic fluorescence, diffuse reflectance and scattering signals to collect signals from multiple sites within the oral cavity. A 96% sensitivity and 96% specificity for the distinction of cancerous/dysplastic from normal tissue, and 65% sensitivity and 90% specificity for the discrimination of dysplastic from cancerous tissue were reported9. Although the results were promising, only a very limited number of measurements were made from selected sites of the oral cavity. In addition, it would be difficult to screen the entire oral cavity with small optic fibers, and this limits the clinical use of spectroscopy for oral cancer screening. In another study, multispectral imaging with reflectance wavelengths corresponding to hemoglobin spectral features was shown to maximize microvasculature visualization and the contrast of oral tissue 10. However, only two wavelengths at 530 nm and 600 nm were explored, and without quantitative evaluations of their diagnostic performance.
Two, fluorescence vital-dyes have been previously tested for the detection of oral neoplasia 11. The first dye is a fluorescent deoxyglucose molecule called 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose (2-NBDG, Cayman Chemical, Ann Arbor, MI, USA), which can be used to assess the metabolic activity of cells. The second fluorescence dye, proflavine, can non-specifically stain cellular structures and enable observation of the nuclear morphology. The standard deviation of the proflavine fluorescence intensity and the mean fluorescence intensity of 2-NBDG have been reported to discriminate non-neoplastic regions of interest from neoplasia, including moderate dysplasia, severe dysplasia, and carcinoma, with a sensitivity and specificity of 91% 11. However, this method was shown to be only 42% sensitive when used to classify mild dysplasia.
Hyperspectral imaging (HSI) is an emerging optical technique that integrates wide-field imaging with spectroscopy to simultaneously acquire both spectral and spatial information, and this makes it possible to interrogate large tissue surfaces in a non-contact and noninvasive way. A wavelength-scanning hyperspectral imaging system generates a three-dimensional (3-D) dataset called hypercube by spectrally splitting the light reflected from a sample surface with a dispersive device and then collecting the light using a two-dimensional (2-D) detector array12. The alterations of the reflectance spectrum are associated with the tissue structural and biochemical changes. Each pixel in a hypercube has a spectral fingerprint that is associated with the structural and biochemical properties of tissue. To fully exploit the diagnostic potential of hyperspectral imaging, machine-learning techniques offer quantitative tools to mine the rich, spectral-spatial information in a hypercube. In our previous studies, we have demonstrated the feasibility of using HSI for the noninvasive detection of prostate cancer 13 and head and neck cancers (HNC) 14-18 in subcutaneous, xenograft tumor models.
To further evaluate the diagnostic potential of HSI, we designed a longitudinal study for both intrinsic and extrinsic HSI of a 4-Nitroquinoline-1-oxide (4NQO)-induced, oral carcinogenesis model. 4NQO, a synthetic water-soluble carcinogen, has been widely used in murine models to investigate all stages of oral carcinogenesis 19. One advantage of this model is that 4NQO-induced lesions exhibit similar histological and molecular changes as in human oral carcinogenesis 19. This animal model mimics human, oral neoplastic transformation with reproducible isolation of all stages, including dysplasia, carcinoma in situ (CIS), and squamous-cell carcinoma (SCC), and therefore providing an excellent opportunity to investigate the application of HSI for the noninvasive detection of squamous neoplasia.
The goal of our study is to develop supervised learning methods for the distinction of neoplasia (dysplasia, CIS, and SCC) and non-neoplastic tongue tissue with hyperspectral imaging and to validate the diagnostic performance of HSI using the histopathology gold standard. Since HSI can provide images relying on intrinsic tissue contrast alone (reflectance, autofluorescence) or using exogenous contrast agents mapping the expression of biomarkers (fluorescence), we compared the diagnostic performance of label-free HSI with autofluorescence and vital-dye fluorescence imaging of 2-NBDG and proflavine for the detection and delineation of squamous neoplasia.
2. Materials and Methods
2.1 Instrumentation
We used a hyperspectral imaging system called CRI Maestro (PerkinElmer Inc., Waltham, MA, USA). This instrument is comprised of a flexible fiber-optical light system with a Xenon light source, a solid-state liquid crystal tunable filter (LCTF) as the wavelenth dispersion device, a spectrally optimized lens, and a 12-bit charge-coupled device (CCD) as the area detector20, 21. This system can be used to acquire both reflectance images and fluorescence.
2.2 Mouse Tongue Carcinogenesis Model
Thirty, female CBA/J mice were purchsed from Jackson Laboratoray (Bar Harbor, ME, USA) and were then separated into an experimental group (N = 24) and a control group (N = 6). Mice in the experimental group were treated with drinking water mixed with 4NQO powder (Sigma Aldrich, Saint Louis, MO, USA) for 16 consecutive weeks (concentration: 100 μg/mL) in order to induce tongue carcinogenesis. Mice in the control group received normal drinking water without 4NQO. The mice in each group were monitored weekly for body weight and water consumption. The experiment was terminated at 24 weeks. All of the animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee (IACUC) of Emory University.
2.3 Data Acquisition
During the experiment, we randomly selected several mice from both the experimental group and the control group at weeks 8, 12, 20, and 24 for hyperspectral imaging using the following procedures. First, we acquired white and dark reference hypercubes before imaging the animals 16, 22-24. Second, we anesthetized the selected mouse with ketamine after the reference hypercubes were acquired. To image the tongue in vivo, we placed the mouse in a supine position, gently pulled out the mouse tongue and taped it on a sterilized imaging stage. Next, we acquired reflectance hyperspectral images of the in vivo tongue from 450-950 with 5 nm intervals and then acquired autofluorescence images of the tongue with 455nm excitation and a 490-nm long-pass emission filter. After in vivo imaging was complete, we euthanized the mouse by cervical dislocation and procured the tongue specimen for a series of ex vivo imaging, including HSI, autofluorescence imaging and fluorescence imaging of 2-NBDG (Cayman Chemical, Ann Arbor, MI, USA) and proflavine (Sigma Aldrich, St. Louis, MO, USA). Both 2-NBDG and proflavine were topically applied on the resected tongues using the procedures described in11. Briefly, tongue specimens were first incubated in a 160 μM solution of 2-NBDG in 1× PBS for 20 min at 37°C, and were then washed once with PBS. Fluorescence images of 2-NBDG were obtained using the blue excitation and 490 long-pass emission. Next, tongues were incubated in a 0.01% w/v solution of proflavine in 1× PBS for 2 min at room temperature, and were then washed once with PBS. Fluorescence images of proflavine were obtained using blue excitation and 490 nm emission. The fluorescence signal from proflavine staining is much brighter than that of 2-NBDG, and thus allowing for imaging proflavine-stained tissue after 2-NBDG staining.
2.4 Histology Correlation
Immediately after ex vivo imaging of the dissected tongue, the ventral surface of the tongue was inked to allow the identification and correct orientation of the tongue during subsequent processing. The inked tongue specimens were placed in 10% buffered formalin overnight for fixation. Each fixed tongue was further processed through dehydration, clearing and wax infiltration. Next, the specimen was embedded in a cassette to form a paraffin block which was then clamped into a microtome for tissue sectioning. Tongue tissues procured at weeks 8 and 12 were sectioned sagitally into a series of 5-μm slices. Specimens procured at weeks 20 and 24 were first bisected longitudinally along the midline groove and then embedded in paraffin blocks which were sagitally sectioned into a series of 5-μm slices. The interval between the two tissue sections was 200 μm for week 12, and 100 μm for week 20 and 24. Immediately after sectioning, tissue slices were laid onto each glass for histological staining with hematoxylin and eosin (H&E), and the H&E stained slides were digitized for pathology diagnosis.
2.4.1 Pathology Diagnosis
An clinically experienced pathologist (SM) reviewed the H&E slides and graded each tongue slice according to the most severe pathology it contained. The dorsal surface of each tongue slice was further segmented into regions of normal (including healthy and hyperplasitic tissue), dysplasia, CIS and SCC, as demonstrated in Figure 1. The grading and diagnosis of tongue cancer is based on the presence of architectural and cytological changes of the epithelial layer based on microscopic examination of H&E stained sections 25.
Figure 1.
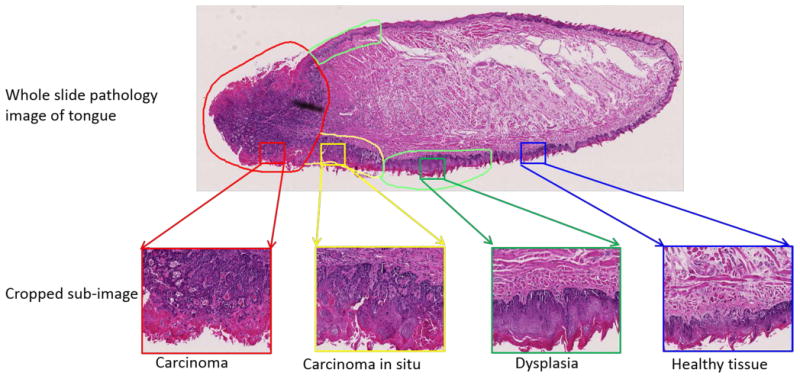
An example of pathology grading and annotation in a whole slice image of tongue specimen.
2.4.2 Generation of Pathology Gold Standard Maps
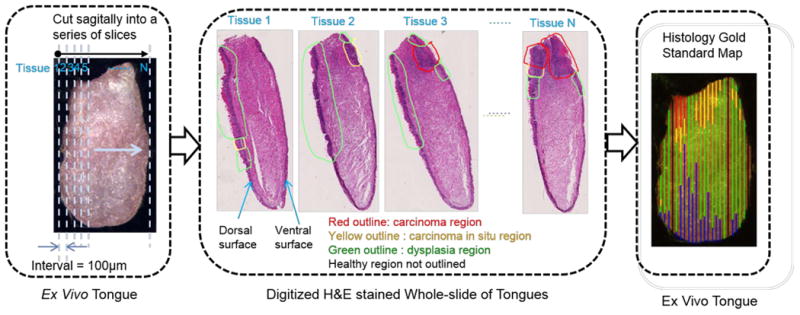
It was a very challenging task to map the pathology diagnosis onto the dorsal surface of the tongue as the gold standard in order to validate the cancer prediction results as hyperspectral images and fluorescence images were the projections of the dorsal tongue surface, while the histology slides were the cross-sections of the tongue with both the dorsal and ventral surfaces. To reconstruct the pathology map, we tracked each step of the histological processing so as to ensure correct tongue orientation during embedding, numbering of each tissue slice, and the specific layout of tissue slices on each glass slide during sectioning. As shown in Figure 2, we sectioned each tongue specimen into sagittal slices of 5 μm with 100 μm or 200 μm intervals between sections. Since each H&E slice corresponded to one straight line along the midline of the dorsal tongue surface, we measured the width of each pathological region along the dorsal surface and then mapped these pathology readings back proportionally onto the corresponding regions of the tongue images. The last image in Figure 2 shows an example of the two-dimensional, color-coded pathology map overlaid with an ex vivo tongue image. Some regions were labelled as black lines on the pathology map and thus indicating the missing gold standard.
Figure 2. Schematic diagram of the pathological gold standard map generation.
2.5 Classification of Hyperspectral Image
2.5.1 Pre-processing of Hyperspectral Image
The pre-processing of hyperspectral images involves two steps. First, we converted the raw data into a percent reflectance value in a pixel-wise manner, as described in 16,22-24. The purpose of this step was to eliminate the illumination non-uniformity and the influence of the dark current. Next we identified and removed glare pixels from the normalized hyperspectral images. Glare regions were formed due to specular reflection from the moist tongue surfaces which did not contain useful diagnostic information. We observed that as glare pixels were characterized with very bright reflectance intensity, they fell into the long tail region in the intensity histogram of the sum image for all the spectral bands. To identify a threshold to detect these pixels, we developed an adaptive threshholding method which fitted the histogram of the sum image of all the spectral bands with a loglogistic distribution and then used the method described in 24 to compute the intensity threshold for glare pixel detection and removal.
2.5.2 Feature Extraction
Reflectance Hyperspectral Imaging
For reflectance hyperspectral images, we evaluated and compared the pixel-wise and block-based feature extraction (FE) methods: (1) the pixel-wise spectral method extracted the normalized reflectance intensity of each pixel on the gold standard lines of the pathology map, and each pixel was labelled with a pathology type; and (2) A block-based spectral method first gridded each tongue hypercube (M × N × K M: image height, N: image width, K(x0003D) 101: number of wavelengths) into many blocks with a size m × n × k (m = 5, n = 5, K = 101) and then computed the mean spectra of all the pixels from each block as the spectral feature. The physical size of each pixel is 26 micron. Each block is labelled with the most frequent pathology type in this block.
Autofluorescence Imaging
For autofluorescence imaging, we extracted the average fluorescence intensity from 500 to 720 nm with 5 nm increments within each block for classification. We also extracted the red-to-green (red: 650 nm, green: 510 nm) fluorescence for comparison, which was reported to be effective in distinguishing neoplastic and normal areas of the oral cavity 26.
Vital-dye Fluorescence Imaging
For 2-NBDG and proflavine imaging, we extracted the average fluorescence intensity of 2-NBDG or proflavine from 500 to 720 nm with a 5-nm increment within each block for classification. Moreover, the mean fluorescence intensity at the maximum emission band (540 nm) of 2-NBDG and the standard deviation of fluorescence intensity at the peak emission (515 nm) of proflavine were extracted as features for comparison. These two features were shown to discriminate between neoplastic and non-neoplastic regions of interest with 91% sensitivity and specificity 11.
2.5.3 Supervised Classification
Supervised classification can be used to ascertain the underlying patterns of training data and build predictive models to differentiate between neoplastic and non-neoplastic tissue from hyperspectral images of mouse tongues. The neoplastic class included lesions histopathologically diagnosed as dysplasia, CIS and SCC. To build a robust prediction model with low bias and low variances, we evaluated seven classifiers, i.e. linear and quadratic discriminant analysis classifiers (LDA and QDA), ensemble learning methods including ensemble LDA, random forests, RUSBoost and support vector machines (SVMs) with linear and radial basis kernel (RBF). These classifiers were implemented in Matlab R2015b. Their characteristics were briefly summarized as follows:
Discriminant Analysis (LDA and QDA)
Discriminant analysis intends to find the projections of the original high dimensional space to a lower dimensional space in order to maximize the class separation 27. Both LDA and QDA classification models assume that the data has a multivariate normal distribution 27,28. The difference between them is that the LDA model assumes the same covariance matrix for each class with varying means, while QDA assumes an individual mean and covariance matrix for each class 28,29. LDA is advantageous in several aspects. First, it does not require tuning of free parameters. Secondly, it has been shown to perform well in classification tasks even when the assumption of the common covariance matrix among groups and normality is violated 30. Finally, the decision boundary obtained by LDA is equivalent to the binary SVM on the set of support vectors 31.
Ensemble LDA
Ensemble learning methods build a high-quality ensemble predictor by combining results from many weak learners, such as decision trees and discriminant learners. Ensemble LDA improves the accuracy of the LDA with random subspace ensembles 32 which involve training multiple LDA learners with randomly selected subsets of features without replacing and classifying the testing data by taking the average of the scores predicted by the weak learners. Two parameters in this model need to be tuned for optimal classification performance, i.e. the number of feature dimensions to sample in each learner and the number of learners in the ensemble.
RUS Boost
RUS Boost is an ensemble learning method that is especially effective at classifying an imbalanced dataset 33. Random Under Sampling (RUS) is a commonly used data sampling method that randomly removes examples from the majority class in order to adjust the class distribution of the training data set. AdaBoost is a popular boosting technique that has been shown to improve the classification performance of weak learners. RUSBoost combines RUS with AdaBoost techniques in order to improve the classification performance for a skewed dataset. This method first generates a balanced training dataset for each weak learner in the ensemble, by taking N, the number of instances in the minority class as the basic unit for sampling and selecting a subset of the majority classes with N instances, and then following the boosting procedure to iteratively create an ensemble of weak learners and make predictions according to the weighted vote of individual weak learners. Here, a decision tree is chosen as the weak learner. We control the depth of a decision tree by adjusting the number of observations per leaf node and the maximal number of branch node splits per tree. We also select the optimal number of tree learners in the ensemble by cross validation.
Random Forest
A random forest is a classification algorithm that uses an ensemble of decision trees 34. Random forest combines bagging and random feature selection to yield an ensemble that can achieve both low bias and low variance. Each tree is grown on a training set randomly drawn with replacement from the original dataset. In each bootstrap training set, approximately one-third of the instances are left out for out-of-bag estimates. At each node of the tree, a random subset of features was selected from the input features to split on. The final classification decision is made by computing the mean class probabilities from all the trees. To generate the forest trees, two parameters need to be optimized, including the number of trees to be produced and the number of features to be selected. Random forest classifier has been widely used in the remote sensing field due to its classification accuracy 35. Our previous work also demonstrated the successful use of random forests for tongue cancer detection36.
SVM
SVM is a binary classifier that intends to find the optimal separating hyperplane which maximizes the distance between the closest training sample and the hyperplane. The simplest form of SVM utilizes a linear decision function to separate linearly separable data. Nonlinear SVM generalizes the linear SVM by non-linearly mapping the input data through kernel transformation into a higher dimensional feature space where they are linearly separable. SVM has been widely used in the remote sensing field for hyperspectral image classification due to its high classification accuracy, very good generalization capability, and effectiveness within the presence of heterogenous classes even with few training samples available 37. In this study, we chose the LIBSVM software package 38 for both linear and kernel SVMs.
2.5.4 Performance Evaluation
We assessed the performance of the classifiers with a variety of metrics, including receiver operating characteristic (ROC) curves, the areas under the ROC curve (AUC), accuracy, sensitivity and specificity. The ROC curve offers a graphic interpretation of the trade-off between sensitivity and specificity for a range of possible cut-off points. AUC of a classifier is equivalent to the probability that the classifier will rank a randomly chosen positive instance higher than a randomly chosen negative instance. At the optimal operating point of an ROC curve, we can obtain the accuracy, sensitivity and specificity from the confusion matrix as defined in 15, 16.
3. Results
3.1 Mouse Tongue Carcinogenesis Model
Over the course of the neoplastic progression, a number of mice from 4NQO-treated group and the control group were selected for hyperspectral imaging and were euthanized after imaging at various periods of observation. Mice in the control group were healthy throughout the experiment, while mice exposed to 4NQO with observations up to 24 weeks developed premalignant and malignant tongue lesions, as shown in Table 1. At eight weeks after the 4NQO treatment, six of the six mice developed epithelial dysplasia of the tongue. CIS was induced in one of six mice at 12 weeks, in two of five mice at 20 weeks, and in one of three mice at 24 weeks. SCC was detected in three of five at 20 weeks and in two of three mice at 24 weeks. Four of 24 mice from the 4NQO-treated group died during the experiment and were not included in Table 1. 4NQO treatment for up to 16 weeks induced heterogeneous lesions in the mouse tongue as well as the oesophagus.
Table 1. Histological diagnosis of all the mice in the experimental group.
| Week | Number of Mice | |||
|---|---|---|---|---|
|
| ||||
| Dysplasia | CIS | SCC | Total | |
| 8 | 6 | 6 | ||
| 12 | 5 | 1 | 6 | |
| 20 | 2 | 3 | 5 | |
| 24 | 1 | 2 | 3 | |
3.2 Characteristics of Hypercube
Each hypercube consists of a series of two-dimensional, grayscale spectral images at wavelengths ranging from 450 nm to 950 nm. Figure 3 (a) shows some representative spectral bands from a reflectance hypercube of an ex vivo tongue. It can be very difficult to visually identify lesions on tongue surfaces from individual grayscale images. Different spectral bands obtained optical information at varying tissue depths. The image intensities varied across the wavelength region, and thus reflecting the variations of tissue absorption and scattering properties at different wavelengths. Figure 3 (b) shows a representative average spectral curve of neoplastic and non-neoplastic tissue. The dominant chromophores in the VIS region are hemoglobin which has been used to visualize increased vasculature in the oral cavity during malignant progression 39,40. It seems that the most prominent difference between different pathologies are in the reflectance intensity. However, there are many intra- and inter-mouse variances in the spectral curves and these factors further complicate the detection of neoplasia.
Figure 3.
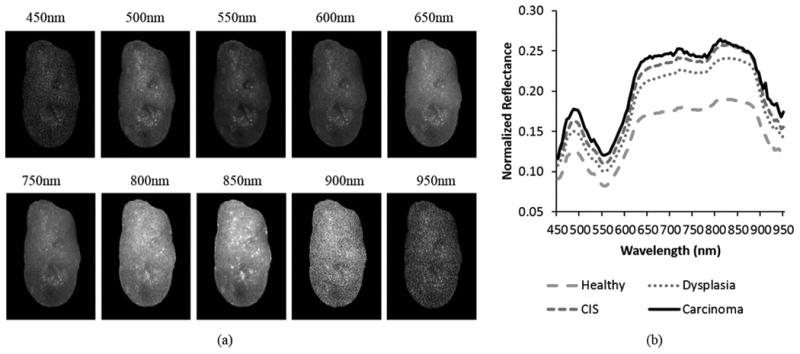
Characteristics of the reflectance hypercube for an ex vivo tongue. (a) Representative spectral bands at multiple wavelengths. (b) Average spectra of healthy, dysplasia, CIS, and carcinoma tissue.
3.3 Predictive Analysis
Dataset
Imaging data of ten mice fulfilling the following criteria were selected for quantitative analysis: (1) the mouse has detailed gold standard of the epithelium for the dorsal tongue outlined by the pathologist; (2) the mouse has all six types of images available, including in vivo reflectance hyperspectral images, autofluorescence images as well as ex vivo reflectance hyperspectral images, autofluorescence, 2-NBDG and proflavine fluorescence images; (3) Satisfactory resection without severe damage to the tongue regions; and (4) Good imaging quality without significant motion artifacts. Images acquired at eight weeks were excluded from analysis due to severe motion artifacts and tissue folds in pathology slices. All of the selected mice developed heterogeneous lesions on different sites of their tongues following exposure to 4NQO for up 16 weeks.
Comparison of Feature Extraction Methods
To begin with, we compared the predictive performance of pixel-wise feature extraction and block-based feature extraction with all of the spectral bands of ex vivo tongues. As shown in Figure 4, the block-based method was found to be more accurate than the pixel-wise method in terms of all the performance metrics and classifiers (p<0.05). In addition, as the block size was chosen as 5×5, the number of samples for training and testing using the block-based method was approximately four times less than the number of samples using the pixel-based method and thus significantly reducing the computation time. Therefore, the results of block-based method are reported in the following sections.
Figure 4.
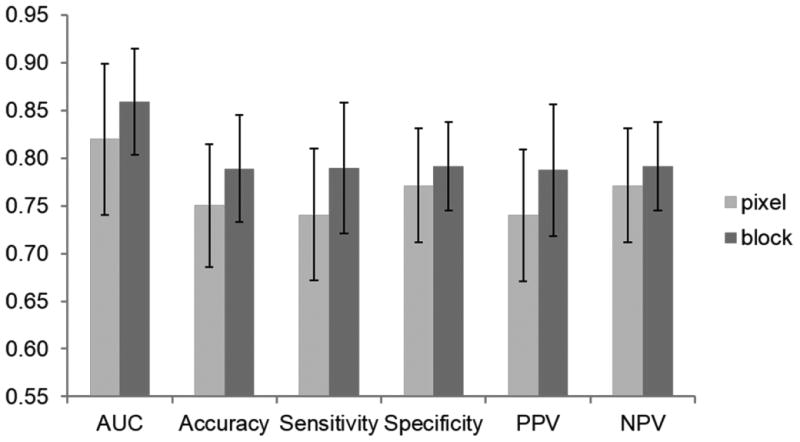
Classification performance of pixel-wise and block-wise feature extraction methods.
Optimal Classifier Selection
For cancer detection in ex vivo tongues, we selected a classifier with minimal generalization errors by comparing the predictive performance of seven, different classifiers. Since nested cross validation (CV) has been proven to give an almost unbiased estimate of the true error on independent (unseen) test dataset, we conducted nested CV with k-fold internal CV on the training dataset in order to tune model parameters and we used leave-one-out external CV to estimate the generalization error 41, 42, 43. For each fold of the external CV, we built a training model on the data from nine out of ten mice and tested the model on the remaining mouse data. On each of the training datasets, we performed an additional k-fold CV to search for the optimal model parameters. We tuned the optimal parameters of the individual classifiers as follows:
(1) Ensemble LDA
We determined two parameters in a sequential search on the training dataset. First, the optimal number of features to sample in each weak learner was searched with five-fold CV in the range of 1:10:101 (the spectral dimension of each hypercube is 101). Next, the optimal number of weak learners used in the ensemble was searched from 1 to 500 with five-fold CV;
(2) RUSBoost
We first searched for the minimum number of observations per leaf node Nleaf (x02208) 10[0.50,0.67,0.83,1.00,1.17,1.33,1.50,1.67,1.83,2.00] with five-fold CV. Next we did a grid search to identify the maximum number of branch node splits per tree (2[0:log(Ntr–1)], Ntris the number of training samples) and the optimal number of tree learners (1:1200) in the ensemble by five-fold CV;
(3) Random forest
We tuned two parameters to grow the optimal classification trees. The optimal number of decision trees were selected from 1 to 2000 that produced the lowest out-of-bag error. The number of features to be selected was fixed to be the square root of the total number of input features 35;
(4) Linear SVMs
We conducted a three-fold CV to search for the optimal cost parameter C (x02208) 2 [-10, 9, …0, …9,10];
(5) SVMs with Gaussian RBF kernel
We performed a grid search with 3-fold CV to determine the optimal combination of the cost parameter C and kernel parameter γ over the range of C (x02208) 2 [-5, 4, …0, …4,5] and C γ 2 [-5, 4, …0, …4,5]
As shown in Table 2, among all the classifiers, linear SVM, ensemble LDA, and LDA performed the best with an AUC value of 0.86, while SVM with RBF kernel only yielded an AUC value of 0.80. Ensemble LDA slightly enhanced the performance of LDA, which was better than QDA and random forest. RUSBoost had the highest sensitivity, although the variance was very high. In the remaining results, we only reported classification with linear SVM.
Table 2. Predictive performance of different classification models for ex vivo tongue cancer detection.
| Classifier | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Linear SVM | 0.86±0.06 | 79%±6% | 79%±7% | 79%±5% |
| Ensemble LDA | 0.86±0.06 | 79%±6% | 78%±7% | 79%±5% |
| LDA | 0.86±0.06 | 78%±6% | 78%±8% | 80%±4% |
| Random Forest | 0.84±0.08 | 77%±7% | 77%±7% | 77%±8% |
| QDA | 0.82±0.05 | 76%±5% | 76%±6% | 75%±4% |
| RBF SVM | 0.80±0.07 | 75%±7% | 77%±9% | 75%±8% |
| RUSBoost | 0.79±0.12 | 71%±15% | 80%±6% | 69%±20% |
3.4 Diagnostic Performance of Reflectance, Autofluorescence and Fluorescence Imaging
In this study, we compared the diagnostic performance of reflectance HSI and autofluorescence imaging of both in vivo and ex vivo tongues, as well as 2-NBDG fluorescence and proflavine fluorescence imaging of the ex vivo tongues. As shown in Table 3, among all the imaging methods, 2-NBDG fluorescence imaging yielded the best performance, with an average AUC, sensitivity and specificity of 0.91, 85% and 84%, respectively, for ex vivo tongue neoplasia detection. Label-free HSI without any contrast agent achieved an average AUC, sensitivity and specificity of 0.86, 79% and 79%. Although the AUC values of autofluorescence and proflavine fluorescence imaging were slightly better than that of the reflectance method, the differences were not statistically significant (p<0.05). As for neoplasia detection for in vivo tongues, label-free HSI based on reflectance produced similar performance compared with that of autofluorescence imaging. Features extracted from one or two spectral band, including the red-to-green ratio of autofluorescence intensity, the mean intensity of 2-NBDG fluorescence at 540 nm, and the standard deviation of proflavine fluorescence at 515 nm, did not provide better prediction performance than multiband autofluorescence or fluorecence features.
Table 3.
Diagnostic performance of hyperspectral reflectance imaging, autofluorescence imaging, 2-NBDG fluorescence imaging and proflavine fluorescence imaging for the distinction of non-neoplastic and neoplastic tissue of ex vivo tongue and the diagnostic performance of hyperspectral reflectance imaging and autofluorescence imaging of in vivo tongue
| Imaging Method | AUC | Accuracy | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| In Vivo | HSI | 0.84±0.05 | 78%±5% | 78%±5% | 78%±5% |
| Autofluorescence | 0.84±0.06 | 78%±5% | 78%±4% | 77%±9% | |
| Autofluorescence(R/G) | 0.43±0.10 | 48%±6% | 48%±6% | 47%±12% | |
|
| |||||
| Ex Vivo | 2-NBDG | 0.91±0.04 | 84%±3% | 85%±3% | 84%±6% |
| Proflavine | 0.89±0.03 | 82%±3% | 83%±3% | 81%±5% | |
| HSI | 0.86±0.06 | 79%±6% | 79%±7% | 79%±5% | |
| Autofluorescence | 0.87±0.03 | 80%±4% | 80%±6% | 81%±3% | |
| Autofluorescence(R/G) | 0.49±0.19 | 50%±14% | 50%±17% | 54%±12% | |
| 2-NBDG (Mean) | 0.75±0.23 | 70%±17% | 70%±18% | 71%±19% | |
| Proflavine (Std) | 0.49±0.15 | 54%±9% | 56%±12% | 49%±20% | |
Furthermore, we examined how many samples of dysplasia, CIS, and SCC were correctly classified in the neoplastic group, as shown in Table 4. With 2-NBDG fluorescence imaging, 84% of normal tissue, 84% of dysplasia, 91% CIS, and 100.0% SCC were correctly classified. Ex vivo, label-free HSI was able to classify 79% of normal tissue, 79% of dysplasia, 85% of CIS and 71% of SCC, while in vivo reflectance HSI was able to classify 78% of normal tissue, 75% of dysplasia, 86% of CIS and 81% of SCC.
Table 4. Classification accuracy of different imaging methods for the distinction of different pathologies.
| Imaging Method | Normal | Dysplasia | CIS | SCC | |
|---|---|---|---|---|---|
| In Vivo | HSI | 78% | 75% | 89% | 81% |
| Autofluorescence | 75% | 76% | 83% | 91% | |
|
| |||||
| Ex Vivo | 2-NBDG | 84% | 84% | 91% | 100% |
| HSI | 79% | 79% | 85% | 71% | |
| Proflavine | 81% | 82% | 87% | 98% | |
| Autofluorescence | 81% | 78% | 88% | 83% | |
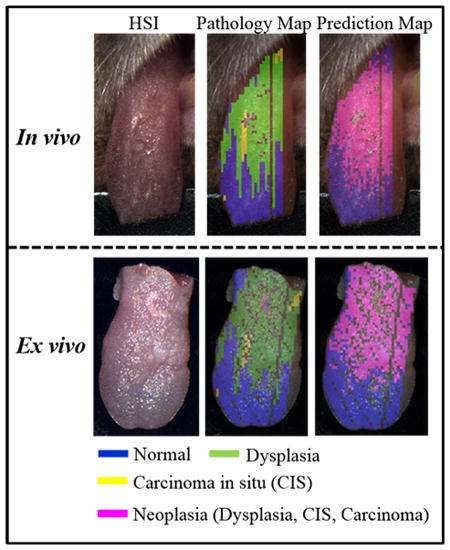
Figure 5 shows that the diagnostic performance of HSI varied among different mouse tongues with varying ROC curves. Figure 6 shows a representative example of tongue cancer detection with in vivo and ex vivo imaging. As shown in the first and third column, label-free HSI can accurately detect and delineate the premalignant and malignant lesions with an AUC of 0.88 and 0.92 across the heterogenous in vivo and ex vivo tongues respectively. The prediction color map matched well with the gold standard color map. The diagnostic performance of label-free HSI was found to be better than that of autofluorescence imaging and comparable to that of fluorescence imaging of proflavine and 2-NBDG stains in terms of the AUC value for the example tongue.
Figure 5.
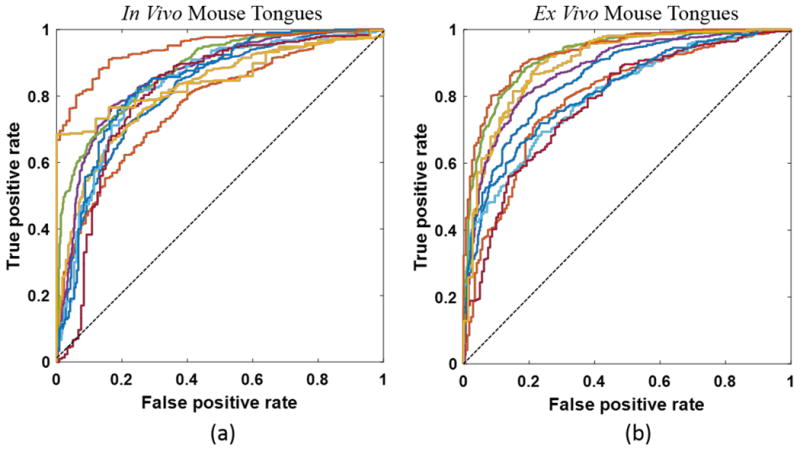
ROC curve of the diagnostic performance for HSI in both in vivo (a) and ex vivo (b) tongues.
Figure 6.
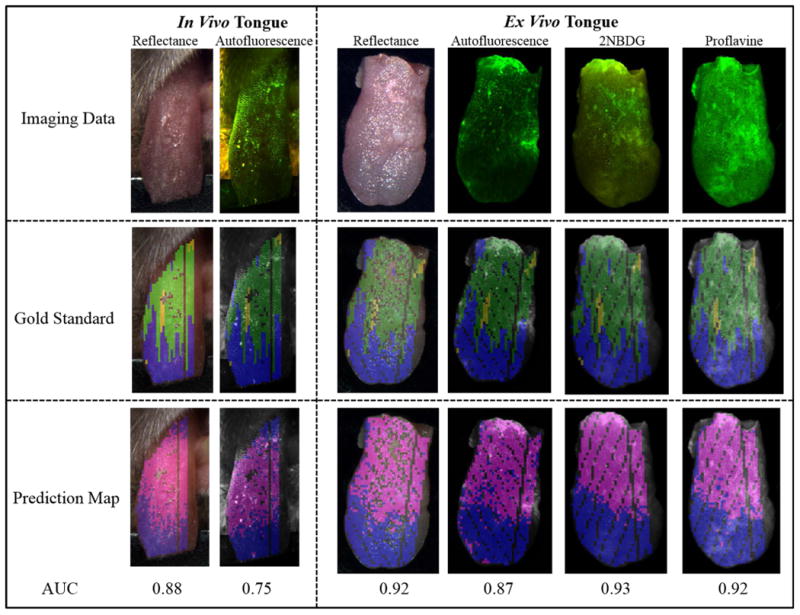
Example Cancer Prediction Results for in vivo and ex vivo tongues with different imaging methods. The first rows are the RGB composite images of all imaging data. The second and third rows are gold standard color maps and predicting color maps corresponding to the block-based feature extraction method, respectively. In the gold standard color map, blue, green, and yellow color indicated normal, dysplasia, and CIS. In the prediction color map, magenta and cyan represent neoplastic and non-neoplastic tissue, respectively. The numbers under the prediction map are the AUC values of the corresponding prediction models. In the gold standard and prediction map, glare pixels excluded from the feature extraction and classification, as well as tissue regions missing the gold standard, were left unlabeled.
4. Discussion
Early detection of oral lesions could improve the clinical outcomes because treatment is most effective at an early stage. Furthermore, identification of local recurrences and second primary tumors is important for surveillance of patients who have survived their initial oral cancer. HSI is a noninvasive optical imaging modality that holds the potential to improve oral cancer diagnosis and reduce cancer-related mortality and morbidity. In this proof-of-principle study, we demonstrated the diagnostic capability of HSI for the detection and delineation of premalignant and malignant lesions in a chemically-induced carcinogenesis murine model which was validated by the reconstructed pathological gold standard maps for hyperspectral, autofluorescence and fluorescence images of ex vivo and in vivo tongues. We found that the prediction results of ex vivo imaging were slightly better than the results of in vivo imaging. This might be due to the fact that it was much more difficult to accurately align the ex vivo histology gold standard with in vivo tongue images due to the stretching and motion of the tongue during in vivo imaging, in conjunction with the tissue deformations during the histological preparation phase.
In terms of feature extraction, we utilized the full reflectance spectrum from 450 to 950 nm in order to capture all of the available spectral-spatial information provided by HSI. We found that spatial averaging of spectral bands over a small block was more accurate than using the spectrum from an individual pixel. This may be attributed to two reasons: (1) spatial averaging of the spectra made the spectral features more robust to noises and registration errors between the pathology and hyperspectral images; and (2) spatial averaging reduced the spectral redundance in neighboring pixels due to the cross-talks inherent in the wide-field imaging approach. Feature selection can be conducted in our future work to rank the diagnostic value of individual wavelengths. In addition, development of spectral-spatial feature extraction may further improve the diagnostic performance of label-free HSI.
We evaluated and compared a variety of machine learning classifiers in this animal model in order to separate neoplasia from non-neoplastic tongue tissue. Linear SVM performed the best among all of the seven classifiers included in the study. The classification performance of LDA, ensemble LDA and random forest were comparable to that of linear SVM. According to 38, linear SVM is a special case of RBF SVM, as linear SVM with parameter C has the same performance as the RBF SVM with some parameter (C, γ). In addition, the choice of linear or kernel SVM depends on the sample size and feature dimension. Linear SVM is faster than RBF SVM as we only need to search for one parameter, i.e. the penalty parameter C. Here we showed that linear SVM gave better classification results than RBF SVM for the detection of tongue neoplasia. Our previous work 16,17 demonstrated that the SVMs with the RBF kernel also produced accurate diagnosis for hyperspectral imaging of xenograft head and neck cancer models in mice. LDA is a fast, efficient, yet accurate classifier that has been widely used in many fields, including the classification of multispectral and hyperspectral images. Our results show that ensemble LDA and LDA produced better classification performance than random forest and QDA. As reported in 44, LDA performed better than the K-nearest neighbor (KNN) classifier, decision tree (DT), QDA, ensemble LDA, ensemble KNN, ensemble-DT, etc.) in classifying multispectral images of burn wound tissue in a swine model. LDA was also shown to be more accurate than decision trees for classifying multispectral reflectance and autofluorescene images for discriminating cancers and precancers from normal tissue 40.
We grouped dysplasia, CIS and SCC as the neoplastic group and used binary classifiers to distinguish neoplasia and non-neoplastic groups. We showed that dysplastic lesions (including mild dysplasia and moderate dysplasia) were more difficult to differentiate from normal tissue than CIS, most likely due to the histological similarity of normal tissue and dysplasia, as well as the difficulty in making an accurate histological diagnosis of mild dysplasia. Only two out of 10 tongues developed small regions of SCC and these regions constituted only a very small part of the dataset. Therefore, it was more difficult to make a robust and accurate prediction of SCC.
As shown in Figure 6, it is very difficult to directly visualize the abnormal tissue transformations from the RGB images of the tongue. However, label-free HSI was able to detect and delineate lesions on the dorsal surface of the tongue compared to the pathology gold standard, which suggested that wide-field HSI can better capture the neoplastic changes over the heterogenous tongue surface than point-based spectroscopy methods. It was also found that most of the misclassification errors of HSI occurred in the interface of different pathologies, such as the normal and dysplasia interface and the dysplasia and CIS interface. As seen in the gold standard map, there were finger-like, protruding dysplasia regions in the interface of dysplasia and normal tongue regions, and where wide-field imaging may not be as sensitive as spectroscopy. In these local regions, spectroscopy-based technlogy may complement HSI to assist in the identification of the small tumor foci around the tumor-normal interface.
Furthermore, this study compared the diagnostic performance of label-free HSI with autofluorescence and fluorescence of two topical dyes in the 4NQO-induced tongue carcinogenesis model. Although autofluorescence and proflavine fluorescence performed slightly better than HSI, the differences were not statistically significant (p>0.05). In addition, HSI is more favorable for future clinical translation in humans because no contrast agent is needed. Reflectance signal obtained by HSI is a complex interplay of tissue scattering, absorption which is associated with the underlying tissue structure and biochemical properties. Autofluorescence imaging reveals the reduced signal from neoplastic tissue, likely due to decreased collagen crosslinks in the stroma 26. One FDA-approved, commercial device, called VELscope, detects neoplastic tissue based on their fluorescence visualization loss (FVL) under blue-violet light (400-460nm). Unfortunately, the sensitivity of the VELscope for detecting malignancy and dysplasia has been reported to be from 30% to 100%, and the specificity ranges from 15.3% to 100% 5. The mean intensity of 2-NBDG imaging which reflected increased metabolic activity of neoplastic cells, and the standard deviation of proflavine fluorescence intensity which revealed neolastic transformation by disorganized fluorescence intensity patterns from nuclei accompanying carcinogenesis, were found to be the best discriminative features for neoplasia detection 11. However, our results suggested that classification with multiband fluorescence intensity was more accurate than the single band in both autofluorescence and fluorescence imaging. One limitation of the topically applied dye on ex vivo tissue lies in the fact that the cut edge of the tissue or any damaged areas on the tissue surface could lead to high fluorescence intensity due to nonspecific uptake, which may confound the detection of neoplastic regions. In the example case, tissue regions close to the cut edge exhibited very high fluorescence intensity which did not cause confusion because the cut edge region was also neoplastic.
Our ultimate goal is to provide an affordable, noninvasive and accurate tool for oral cancer screening in human patients. There is still a long way to go in order to achieve this goal. Moving forward, we are working on the evaluation of the diagnostic utility HSI on fresh human surgical specimens of tongue SCC and have achieved promising preliminary results. In the future, development of a portable HSI instrument with higher spectral resolution and validation of the HSI device in a large population of human patients could enable clinical translation from bench to bedside and potentially improve oral cancer detection and diagnosis.
5. Conclusion
This proof-of-concept study suggested that hyperspectral imaging is a promising optical modality for quantitative and noninvasive detection and delineation of squamous neoplasia in an animal model. In this study, we designed an animal study to acquire multimodal, hyperspectral images of in vivo and ex vivo mouse tongues from a chemically induced, oral carcinogenesis model. We reconstructed the pathological gold standard map for the dorsal surface of the tongue in order to validate tumor prediction results. We implemented and validated a variety of machine learning classifiers, including discriminant analysis, ensemble learning methods, and support vector machines, compared the diagnostic performance in hyperspectral reflectance, autofluorescence, and fluorescence images, and generated prediction maps that displayed the location and distribution of neoplasia. In the future, we would like to apply the HSI and machine learning techniques to the diagnosis of human tongue tumors. Further development of hyperspectral imaging and image quantification methods could provide a noninvasive tool to improve the early detection of oral cancers.
Acknowledgments
This research was supported in part by NIH grants (CA176684, CA156775, and CA204254). Research reported in this publication was supported in part by Developmental Funds from the Winship Cancer Institute of Emory University under award number P30CA138292. We thank Ms. Jennifer Shelton from the Pathology Core Lab at Winship Cancer Institute of Emory University for her help with histological processing of the tongue specimen.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. International journal of cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.E. Surveillance, and End Results (SEER) Program. All Races, Both Sexes by SEER Summary Stage 2000. National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch; 2016. SEER*Stat Database in SEER 18 2005-2011. ( www.seer.cancer.gov) http://seer.cancer.gov/statfacts/html/oralcav.html. [Google Scholar]
- 3.Morris LGT, Sikora AG, Patel SG, Hayes RB, Ganly I. Journal of Clinical Oncology. 2011;29:739–746. doi: 10.1200/JCO.2010.31.8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messadi DV. In J Oral Sci. 2013;5:59–65. doi: 10.1038/ijos.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rashid A, Warnakulasuriya S. Journal of Oral Pathology & Medicine. 2015;44:307–328. doi: 10.1111/jop.12218. [DOI] [PubMed] [Google Scholar]
- 6.van den Brekel MWM, Lodder WL, Stel HV, Bloemena E, Leemans CR, van der Waal I. Head & Neck. 2012;34:840–845. doi: 10.1002/hed.21823. [DOI] [PubMed] [Google Scholar]
- 7.Ismail SM, Colclough AB, Dinnen JS, Eakins D, Evans DM, Gradwell E, O'Sullivan JP, Summerell JM, Newcombe RG. British Medical Journal. 1989;298:707–710. doi: 10.1136/bmj.298.6675.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel RL, Miller KD, Jemal A. CA: a cancer journal for clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 9.Müller MG, Valdez TA, Georgakoudi I, Backman V, Fuentes C, Kabani S, Laver N, Wang Z, Boone CW, Dasari RR, Shapshay SM, Feld MS. Cancer. 2003;97:1681–1692. doi: 10.1002/cncr.11255. [DOI] [PubMed] [Google Scholar]
- 10.Roblyer D, Richards-Kortum R, Sokolov K, El-Naggar AK, Williams MD, Kurachi C, Gillenwater AM. J Biomed Opt. 2008;13:024019. doi: 10.1117/1.2904658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellebust A, Rosbach K, Wu JK, Nguyen J, Gillenwater A, Vigneswaran N, Richards-Kortum R. J Biomed Opt. 2013;18:126017. doi: 10.1117/1.JBO.18.12.126017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu G, Fei B. Journal of biomedical optics. 2014;19:10901. doi: 10.1117/1.JBO.19.1.010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akbari H, Halig LV, Schuster DM, Osunkoya A, Master V, Nieh PT, Chen GZ, Fei B. Journal of biomedical optics. 2012;17:076005. doi: 10.1117/1.JBO.17.7.076005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pike R, Patton SK, Lu G, Halig LV, Wang D, Chen ZG, Fei B. Proc SPIE 9034. 2014;9034:90341w. doi: 10.1117/12.2043848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu G, Halig L, Wang D, Chen ZG, Fei B. Proc SPIE 9034. 2014:903413. doi: 10.1117/12.2043796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu G, Halig L, Wang D, Qin X, Chen ZG, Fei B. Journal of biomedical optics. 2014;19:106004. doi: 10.1117/1.JBO.19.10.106004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pike R, Lu G, Wang D, Chen ZG, Fei B. Biomedical Engineering, IEEE Transactions on PP. 2015:1–1. [Google Scholar]
- 18.Chung H, Lu G, Tian Z, Wang D, Chen ZG, Fei B. Proc SPIE 9788. 2016;9788:978813. doi: 10.1117/12.2216559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanojia D, Vaidya MM. Oral Oncology. 2006;42:655–667. doi: 10.1016/j.oraloncology.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Lu G, Halig L, Wang D, Chen ZG, Fei B. Proc SPIE 9036. 2014:90360s. doi: 10.1117/12.2043805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu G, Qin X, Wang D, Chen ZG, Fei B. Proc SPIE 9417. 2015:94170Q. doi: 10.1117/12.2082299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun DW. Hyperspectral imaging for food quality analysis and control. Elsevier; 2010. [Google Scholar]
- 23.Park B, Lu R. Hyperspectral imaging technology in food and agriculture. Springer; New York: 2015. [Google Scholar]
- 24.Lu G, Wang D, Qin X, Halig L, Muller S, Zhang H, Chen A, Pogue BW, Chen ZG, Fei B. Journal of biomedical optics. 2015;20:126012. doi: 10.1117/1.JBO.20.12.126012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speight PM. Head and neck pathology. 2007;1:61–66. doi: 10.1007/s12105-007-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roblyer D, Kurachi C, Stepanek V, Williams MD, El-Naggar AK, Lee JJ, Gillenwater AM, Richards-Kortum R. Cancer Prevention Research. 2009;2:423–431. doi: 10.1158/1940-6207.CAPR-08-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher RA. Annals of Eugenics. 1936;7:179–188. [Google Scholar]
- 28.Guo Y, Hastie T, Tibshirani R. Biostatistics (Oxford, England) 2006 doi: 10.1093/biostatistics/kxj035. [DOI] [PubMed] [Google Scholar]
- 29.Landgrebe D. IEEE Signal Processing Magazine. 2002;19:17–28. [Google Scholar]
- 30.Li T, Zhu S, Ogihara M. Knowledge and Information Systems. 2006;10:453–472. [Google Scholar]
- 31.Shashua A. Neural Processing Letters. 1999;9:129–139. [Google Scholar]
- 32.Tin Kam H. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1998;20:832–844. [Google Scholar]
- 33.Seiffert C, Khoshgoftaar TM, Hulse JV, Napolitano A. IEEE Transactions on Systems, Man, and Cybernetics - Part A: Systems and Humans. 2010;40:185–197. [Google Scholar]
- 34.Breiman L. Machine Learning. 2001;45:5–32. [Google Scholar]
- 35.Belgiu M, Drăguţ L. ISPRS Journal of Photogrammetry and Remote Sensing. 2016;114:24–31. [Google Scholar]
- 36.Lu G, Qin X, Wang D, S M, H Z, Chen A, Chen ZG, Fei B. Proceedings of SPIE. 2016;9788:978812. doi: 10.1117/12.2216553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melgani F, Bruzzone L. IEEE Transactions on Geoscience and Remote Sensing. 2004;42:1778–1790. [Google Scholar]
- 38.Chang CC, Lin CJ. ACM Trans Intell Syst Technol. 2011;2:1–27. [Google Scholar]
- 39.Subhash N, Mallia JR, Thomas SS, Mathews A, Sebastian P, Madhavan J. J Biomed Opt. 2006;11:014018. doi: 10.1117/1.2165184. [DOI] [PubMed] [Google Scholar]
- 40.Roblyer D, Kurachi C, Stepanek V, Schwarz RA, Williams MD, El-Naggar AK, Lee JJ, Gillenwater AM, Richards-Kortum R. J Biomed Opt. 2010;15:066017. doi: 10.1117/1.3516593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varma S, Simon R. BMC Bioinformatics. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kothari S, Phan JH, Young AN, Wang MD. BMC medical imaging. 2013;13:9. doi: 10.1186/1471-2342-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kothari S. Georgia Institute of Technology. 2013 [Google Scholar]
- 44.Squiers JJ, Li W, King DR, Mo W, Zhang X, Lu Y, Sellke EW, Fan W, DiMaio JM, Thatcher JE. Proc SPIE 9785. 2016;9785:97853L-97853L–97810. [Google Scholar]


