Abstract
Rare variants are considered underlying causes of complex diseases. The complex and severe group of disorders called neural tube defects (NTDs) results from failure of the neural tube to close during early embryogenesis. Neural tube closure requires the coordination of numerous signaling pathways, including the precise regulation of retinoic acid (RA) concentration which is controlled by enzymes involved in RA synthesis and degradation. Here we used a case-control mutation screen study to reveal rare variants in retinoid related genes in a Han Chinese NTD population by sequencing six genes in 355 NTD cases and 225 controls. NTD-specific rare variants were found in exonic regions and upstream regions. The RA-responsive genes CYP26A1, CRABP1 and ALDH1A2 harbored NTD-specific rare variants in their upstream regions. Unexpectedly, the majority of missense variants in NTD cases were found in CYP26B1 which encodes a RA degradation enzyme, whereas no missense variants in this gene were found in controls. Functional analysis indicated that the CYP26B1 NTD variants were inefficient in the degradation of RA using assays of RA-induced transcription and RA-initiated neuronal differentiation. Our study supports the contribution of rare variants in RA related genes to the etiology of human NTDs.
1. INTRODUCTION
Rare variants are causal genetic factors for complex diseases (Bruel et al., 2017; Manolio et al., 2009). Neural tube defects (NTDs; MIM# 601634) are complex congenital malformations of the central nervous system, caused by failure of neural tube closure during embryogenesis. NTDs include cranial defects such as anencephaly and encephalocele and caudal defects including spina bifida. Genetic studies of mice have identified more than 240 genes involved in neural tube closure (Harris & Juriloff, 2010; Wilde, Petersen, & Niswander, 2014; Wilson & Maden, 2005) and this information has provided a framework to explore the genetic causes of NTDs in humans over the past decade (X. Chen et al., 2017; De Marco et al., 2014; Kibar et al., 2007; Qiao et al., 2016). Genes that function in particular pathways involved in planar cell polarity (PCP), ciliogenesis, the glycine cleavage system and one-carbon metabolism have been explored in a relatively large number of cases (Allache et al., 2015; C. Cai & Shi, 2014; De Marco et al., 2014; Dowdle et al., 2011; Hopp et al., 2011; Jones, Fiozzo, Waters, McKnight, & Brown, 2014; Kibar et al., 2007; Y. Lei et al., 2013; Y. Lei et al., 2014; Y. P. Lei et al., 2010; Marini et al., 2011; Merello et al., 2015; Miao et al., 2016; Narisawa et al., 2012; Qiao et al., 2016; Roberson et al., 2015; Robinson et al., 2012; Shaheen et al., 2015; Yang et al., 2013; Zhang et al., 2015). PCP gene variants have been associated with the most severe NTD phenotype, craniorachischisis, as well as more limited NTDs such as myelomeningocele. Rare variants in cilia genes have been associated with human cranial NTD phenotypes (X. Chen et al., 2017; Miao et al., 2016) and multiple potentially damaging mutations in cilia genes and the TMEM superfamily were found in Meckel-Gruber syndrome (MKS) patients, a prenatal lethal disorder characterized by the traits of occipital encephalocele, polydactyly and polycystic kidney (Dowdle et al., 2011; Hopp et al., 2011; Jones et al., 2014; Roberson et al., 2015; Shaheen et al., 2015; Zhang et al., 2015). Several NTD-specific missense mutations and splicing variants were identified in genes which function in the glycine cleavage system and one-carbon metabolism (Marini et al., 2011; Narisawa et al., 2012; Saxena, Gupta, Pandey, Gangopadhaya, & Pandey, 2011; Tilley, Northrup, & Au, 2012). These pathways have been relatively well-explored for their association with NTDs in human, but many NTD cases have no known genetic etiology.
The vitamin A derivative retinoic acid (RA) is essential for the patterning and differentiation of neural progenitor cells in the hindbrain and spinal cord (Duester, 2008; Guan, Chang, Rolletschek, & Wobus, 2001; Maden, 2007). It is critical to maintain an appropriate balance of the essential vitamin A and RA, as too much or too little is associated with a range of birth defects (Alles & Sulik, 1990; W. H. Chen, Morriss-Kay, & Copp, 1995; Iulianella, Beckett, Petkovich, & Lohnes, 1999; Lee et al., 2012; Maden, Gale, Kostetskii, & Zile, 1996). During embryogenesis, levels of RA are established by the synthesizing enzyme ALDH1A2 and the degradative enzymes CYP26A1/CYP26B1/CYP26C1 (Figure 1A) (Niederreither et al., 2002; Pennimpede et al., 2010). Moreover, RA signaling is exquisitely controlled locally and this is accomplished by the tissue-specific expression of retinoid metabolism genes to allow localized production and destruction of RA. RA acts as a ligand and associates with two families of nuclear receptors that bind DNA through RA response elements (RARE) and directly regulate transcription. RA receptors (RARA, RARB and RARG) bind the abundant form of RA known as all-trans-RA and the retinoid X receptors (RXRA, RXRB and RXRG) bind an isomer known as 9-cis-RA (Chawla, Repa, Evans, & Mangelsdorf, 2001).
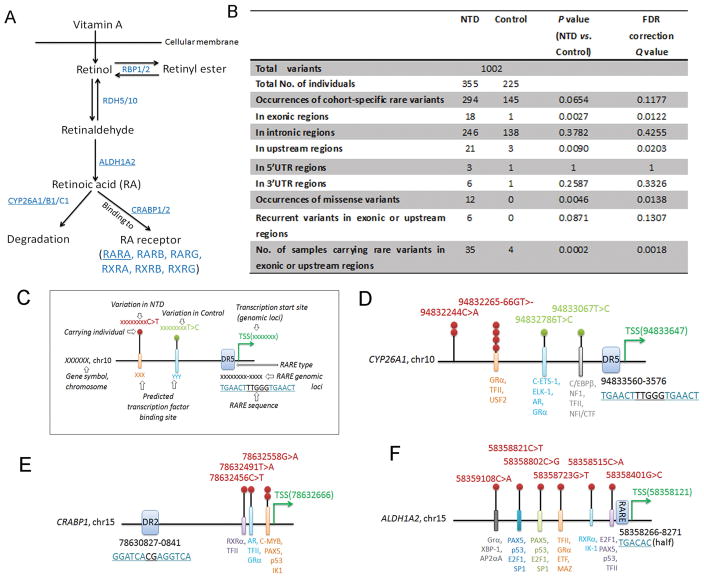
FIGURE 1. Retinoid related genes harbor rare variants in humans with neural tube defects.
A) A schematic of the synthesis and degradation of retinoic acid from vitamin A with the enzymes involved in retinoid metabolism and binding indicated in blue font. The genes sequenced in the present study are denoted by underlining of the encoded proteins. B) Rare variants were found in all sequenced genes in the present cohort of NTD cases relative to controls. P value was obtained via Fisher’s exact test and false discovery rate (FDR) correction. C) Illustration for each symbol in D-F. D-F) Schematics show the genomic positions of rare variants found in upstream regions of CYP26A1, CRABP1, and ALDH1A2 genes relative to the retinoic acid response-element (RARE) and predicted transcription factor binding sites.
The relationship between genetic polymorphisms in retinoid metabolism genes and increased risk of NTDs in humans is well-documented. A polymorphism discovery screen was carried out for ALDH1A2 (MIM# 603687), CYP26A1 (MIM# 602239), CYP26B1 (MIM# 605207), CRABP1 (MIM# 180230) and CRABP2 (MIM# 180231) genes in 230 individuals with lumbosacral myelomeningocele (including spina bifida aperta or spina bifida cystica), and a significant association between three polymorphisms in ALDH1A2 and this spinal NTD was identified (Deak et al., 2005). In another study, a deletion (g.3116delT) in the CYP26A1 gene which creates a premature stop codon was found in a sample of 40 spina bifida patients of Caucasian origin from southern Italy (Rat et al., 2006). Moreover, investigation of the three RA receptors encoded by RARA (MIM# 180240), RARB (MIM# 180220) and RARG (MIM# 180190) in 329 affected family trios and 281 affected family duos of patients with meningomyelocele from Texas, identified SNPs in each of these genes and these were associated with a protective effect against meningomyelocele (Tran et al., 2011). It is important to test the functional significance of rare variants to begin to develop a deeper mechanistic understanding of NTDs in humans. This has been done for some of the studies above, such as testing how PCP variants affect cell behavior or how truncated CYP26A1 affects RA metabolism (Rat et al., 2006)
To increase our knowledge as to the importance of RA related genes in a broader range of NTDs in human, we analyzed a Han Chinese population that included 355 NTD cases ranging from cranial to caudal defects and 225 ethnicity-matched controls. We focused on genes in six RA metabolism and signaling related genes (underlined in Figure 1A). This case-control mutation screen showed a strong correlation between rare variants located in or upstream of RA related genes in cranial and caudal NTDs. Strikingly, CYP26B1 harbored numerous missense variants within the gene body. We also provide experimental evidence that these CYP26B1 variants disrupt function as assessed by the transcription of RA target genes and RA-mediated differentiation of neuroepithelial cells.
2. SUBJECTS AND METHODS
2.1 Subjects
As described in our previous paper (S. Chen et al., 2016), we performed high-throughput sequencing of genomic DNA samples collected from subjects with NTD in the Han Chinese population ranging in age from gestational week (GW) 12 to 10-years old and from multiple local hospitals in six provinces in China. Ethnicity-matched controls were collected from non-medically related terminations and were free of any NTDs. We enrolled individuals with NTDs that had been assessed by clinical geneticists and placed them into at least one of the following diagnostic groups: anencephaly, spina bifida (aperta or cystica), craniorachischisis, and encephalocele (Tables 1 and 2). In total there were 19 individuals with craniorachischisis; 17 anencephaly; 55 anencephaly with spina bifida aperta; 2 anencephaly with spina bifida cystica; 27 encephalocele and spina bifida aperta; 57 encephalocele; 91 spina bifida aperta; 26 spina bifida occulta; 42 spina bifida cystica, and 19 spina bifida without further clinical details.
Table 1.
Rare variants in exonic regions in the present NTD cohort
| Gene | Variant type | Genome position | cDNA change | Protein change | SIFT Score | POLYPhen Score | Age | Sex | Clinical Phenotype |
|---|---|---|---|---|---|---|---|---|---|
| CYP26A1 | Silent | chr10:94836414 | 1113A>G | Pro371= | 13GW | F | Frontal encephalocele; cervical spina bifida aperta | ||
| CYP26A1 | Silent | chr10:94836369 | 1068C>T | Ile356= | 20GW | F | Lumbosacral spinal bifida occulta; hydrocephaly | ||
| CYP26A1 | Silent | chr10:94836369 | 1068C>T | Ile356= | 28GW | M | Lumbosacral spina bifida aperta; hydrocephaly | ||
| CYP26B1 | Silent | chr2:72360332 | 966G>C | Leu322= | 33GW | M | Thoracic Lumbar sacral spina bifida; left equinovarus | ||
| CYP26B1 | Missense | chr2:72362036 | 715G>A | Ala239Thr | 0.01 | 0.98 | 22GW | F | Lumbar spina bifida cystica; hydrocephaly; double equinovarus |
| CYP26B1 | Missense | chr2:72362036 | 715G>A | Ala239Thr | 0.01 | 0.98 | 20GW | M | Occipital encephalocele; lack of pulmonary lobe; ASD |
| CYP26B1 | Missense | chr2:72362036 | 715G>A | Ala239Thr | 0.01 | 0.98 | 28GW | M | Lumbosacral spina bifida aperta; lack of left kidney; right polycystic kidney |
| CYP26B1 | Missense | chr2:72362039 | 712C>G | Gln238Glu | 0.01 | 0.08 | 24GW | M | Frontal encephalocele; ASD; hydrocephaly |
| CYP26B1 | Splicing/Mis sense | chr2:72362274 | 704G>A | Arg235Gln | 0.01 | 0.76 | 37GW | F | Occipital encephalocele; cervical thoracic spina bifida aperta; short neck |
| CYP26B1 | Missense | chr2:72362389 | 589C>A | Leu197Met | 0 | 0.99 | 16GW | M | Thoracic lumbar spina bifida aperta; hydrocephaly; lack of right pulmonary lobe; overlong lower limbs; right equinovarus |
| CYP26B1 | Missense | chr2:72362389 | 589C>A | Leu197Met | 0 | 0.99 | 31GW | M | Craniorachischisis; short neck; adrenal gland fusion; dextrocardia |
| CYP26B1 | Missense | chr2:72362389 | 589C>A | Leu197Met | 0 | 0.99 | U | U | Encephalocele |
| CYP26B1 | Missense | chr2:72362395 | 583C>T | Arg195Trp | 0 | 1 | 24GW | F | Craniorachischisis; short neck; lack of left upper limb; thoracocyllosis; lack of pulmonary lobe; renal shift |
| CYP26B1 | Silent | chr2:72362471 | 507G>C | Leu169= | U | U | Spina bifida | ||
| CYP26B1 | Silent | chr2:72371316 | 231G>A | Arg77= | U | U | Spina bifida | ||
| CRABP1 | Missense | chr15:78635856 | 265G>A | Glu89Lys | 0.01 | 0.41 | U | U | Anencephaly |
| ALDH1A2 | Missense | chr15:58256102 | 1067C>G | Thr356Ser | 0.32 | 0 | 20GW | F | Lumbosacral spinal bifida occulta; hydrocephaly |
| ALDH1A2 | Splicing/Mis sense | chr15:58254374 | 1087A>G | Ile363Val | 0.89 | 0 | 22GW | F | Occipital encephalocele |
ASD: Atrial septal defect; F: Female; M: Male; GW: Gestational week. U: Unknown; CYP26B1(NC_000002.11; NM_019885.3; NP_063938.1); CYP26A1(NC_000010.10; NM_000783.3; NP_000774.2); CRABP1(NC_000015.9; NM_004378.2; NP_004369.1); ALDH1A2 (NC_000015.9; NM_003888.3; NP_003879.2). In cDNA change, nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1
Table 2.
Rare variants in upstream regions in the present NTD cohort
| Gene | Genome position (build hg19) | Ref allele | Alt allele | Age | Sex | Clinical Phenotype |
|---|---|---|---|---|---|---|
| CYP26A1 | chr10:94832244 | C | A | 20GW | M | Lumbosacral spina bifida aperta; hydrocephaly |
| CYP26A1 | chr10:94832244 | C | A | 18GW | M | Anencephaly; occipital cervical spina bifida aperta; lack of pulmonary lobe; no neck |
| CYP26A1 | chr10:94832265-94832266 | GT | - | 24GW | M | Anencephaly; occipital cervical spina bifida aperta; no neck; lack right pulmonary lobe |
| CYP26A1 | chr10:94832265-94832266 | GT | - | 21GW | F | Occipital encephalocele; cervical thoracic spina bifida aperta; lack of pulmonary lobe; mediastinal cyst |
| CYP26A1 | chr10:94832265-94832266 | GT | - | 15GW | M | Occipital encephalocele |
| CYP26A1 | chr10:94832265-94832266 | GT | - | 31GW | M | Spinal bifida occulta; hydrocephaly; double equinovarus; thoracocyllosis; lack of pulmonary lobe |
| CYP26B1 | Chr2:72375908 | G | A | 34GW | F | Thoracic lumbar spinal bifida occulta |
| RARA | chr17:38464726 | T | C | 26GW | M | Lumbosacral spina bifida aperta; hydrocephaly; double equinovarus |
| RARA | chr17:38464601 | C | T | 37GW | F | Occipital encephalocele; short neck, |
| ALDH1A2 | chr15:58358723 | G | T | 20GW | M | Anencephaly; double adrenal glands malformation |
| ALDH1A2 | chr15:58359108 | C | A | Unknown | Unknown | Spina bifida cystica |
| ALDH1A2 | chr15:58358515 | C | A | 28GW | M | Lumbosacral spinal bifida aperta; hydrocephaly |
| ALDH1A2 | chr15:58358821 | C | T | 28GW | M | Lumbosacral spinal bifida aperta; hydrocephaly |
| ALDH1A2 | chr15:58358802 | C | G | 2 years | F | Thoracic spina bifida aperta |
| ALDH1A2 | chr15:58358401 | G | C | 2 years | F | Lumbosacral spinal bifida occulta |
| CRABP1 | chr15:78632491 | T | A | 20GW | M | Lumbosacral spina bifida aperta; hydrocephaly1 |
| CRABP1 | chr15:78632456 | C | T | 19GW | F | Anencephaly; occipital cervical spina bifida aperta; short neck |
| CRABP1 | chr15:78632558 | G | A | 24GW | F | Lumbosacral spina bifida aperta; hydrocephaly |
| CRABP1 | chr15:78632558 | G | A | Unknown | Unknown | Anencephaly; occipital cervical spina bifida aperta |
| CRABP2 | chr1:156676423 | C | A | 39GW | M | Lumbosacral spina bifida cystica; hydrocephaly |
| CRABP2 | chr1:156676091 | C | T | 16GW | M | Anencephaly; occipital cervical thoracic spina bifida aperta; no neck; thoracocyllosis; double adrenal gland dysplasia |
F: Female; M: Male; GW: Gestational week
CYP26B1(NC_000002.11; NM_019885.3; NP_063938.1); CYP26A1(NC_000010.10; NM_000783.3; NP_000774.2); CRABP1(NC_000015.9; NM_004378.2; NP_004369.1); ALDH1A2 (NC_000015.9; NM_003888.3; NP_003879.2); CRABP2(NC_000001.10; NM_001878.3; NP_001869.1)RARA(NC_000017.10; NM_000964.3; NP_000955.1)
The study was approved by the Committee of Medical Ethics of the Capital Institute of Pediatrics (Beijing, China) (SHERLLM 2009002). We carried out the study in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans and in accordance with the approved guidelines. Written informed consent was obtained from the parents. The enrolled pregnant women were diagnosed by local clinicians using ultrasonography.
2.2 Genomic DNA sequencing
Similar to our previous paper (S. Chen et al., 2016), target sequencing was performed on CYP26B1 (NC_000002.11; NM_019885.3; NP_063938.1); CYP26A1 (NC_000010.10; NM_000783.3; NP_000774.2); CRABP1 (NC_000015.9; NM_004378.2; NP_004369.1); ALDH1A2 (NC_000015.9; NM_003888.3; NP_003879.2); CRABP2 (NC_000001.10; NM_001878.3; NP_001869.1); RARA (NC_000017.10; NM_000964.3; NP_000955.1). Genomic DNA was prepared using a Truseq DNA Sample preparation kit (Illumina Inc, San Diego, CA), libraries constructed with Agilent Custom SureSelect Enrichment Kit, and run on an Agilent Custom enrichment array (Probe Code: BI426526171). Sequence reads were aligned to the UCSC human genome GRCh37/hg19 using BWA (v0.5.9) (Li & Durbin, 2009). The average depth of coverage in the present sequencing was 22.5X. Based on cDNA sequence, nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1. Single-nucleotide variants (SNVs) were called using GATK (Samtools pileup (MAPQ30))(version 0.1.17) (Li et al., 2009) and Varscan (Koboldt et al., 2009) (minimum coverage=1, minimum alterative allele reads=1, minimum variation frequency > 0.03) and short indels (insertions and deletion) were called using Varscan with a loose standard (minimum coverage=2, minimum alternative allele reads=2, minimum variation frequency > 0.1). Genotypes were called using Bayes. Variants were annotated with ANNOVAR (Wang, Li, & Hakonarson, 2010). Rare variants were filtered out using the dbSNP in NCBI, the 1000 Genomes Project, the NHLBI Exome Sequencing Project, and shared variants in cases and controls.
2.3 Neuroepithelial cell line and plasmid transfection
NE-4C cells (ATCC CRL-2935), a neuroepithelial cell line isolated from E9 mouse embryonic brain, were grown at 37°C and 5% CO2 in MEM (41090, Invitrogen) supplemented with 6% FBS, 1X MEM non-essential amino acids (11140, Invitrogen), and 1X GlutaMax (35050, Invitrogen). All-trans RA (Sigma Aldrich) was prepared as a 1 mM stock (1000X) in dimethyl sulfoxide (DMSO) and used at a final concentration of 1μM. DMSO was used as a vehicle control for all RA experiments. Human CYP26B1 cDNA clone in pCMV6-AC-GFP vector was purchased (RG221075, Origene) and Site-Directed mutagenesis (Agilent Technologies) was used to introduce the NTD-specific variants. Plasmids containing wildtype (WT) or mutated CYP26B1 tagged with GFP were sequence verified. WT or mutant CYP26B1 constructs were co-transfected with pCMV-AN-RFP (PS100033, Origene) into NE-4C cells using the Xfect Transfection Reagent (Clontech) according to the manufacturer’s protocol (2μg CYP26B1 plasmid and 2μg pCMV-AN-RFP for co-transfection per well of 12-well plate). For functional assays, WT or mutant CYP26B1 constructs were transfected as 7.5μg plasmid per well of a 6-well plate or 1μg plasmid per well of 24-well plate. For analysis of downstream gene expression (Figure 3), cells were transfected with CYP26B1 constructs, 1μM RA added 24 hours later, and cells harvested 24 hours after RA addition. For analysis of neural differentiation, cells were transfected, 1μM RA added 24 hours later, and cells were stained for neural markers 7–8 days after RA addition.
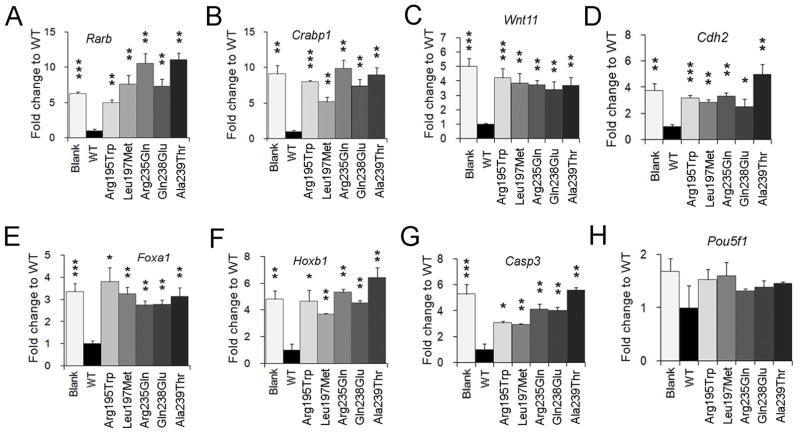
FIGURE 3. CYP26B1 NTD-variants attenuate function as assessed by modulation of RA-mediated gene transcription.
A–H) Real-time quantitative PCR assays were performed to test the mRNA levels of genes that are downstream of RA signaling and involved in neural development. NE-4C cells were transfected with CYP26B1 WT or mutant plasmids and treated for 24 hours with RA. Gapdh gene was used as a loading control. The level of expression is indicated as fold change relative to WT vector, which was set to 1. All experiments were from three biological replicates. ***: P < 0.0001; **: P < 0.01; *: P < 0.05; Student’s t-test.
2.4 Western blotting
Proteins extracts were subjected to polyacrylamide gel electrophoresis and electrotransfer to PVDF membranes. The membranes were blocked with 5% non-fat milk and then probed with the indicated antibodies in TBS-Tween, 5% (w/v) non-fat dry milk overnight at 4°C. Antibodies used were against 2H8 anti-turboGFP (Origene, TA150041, 1:2000), anti-turboRFP clone OTI2D9 (Origene, TA150061, 1:1000), anti-Gapdh (Sigma, G9545, 1:2000), anti-RARA(c-20) (Santa Cruz, sc-551, 1:50), anti-RARB(C-19) (Santa Cruz, sc-552, 1:50), anti-RARG(C-19) (Santa Cruz, sc-550; 1:50), and β-tubulin (Sigma, T3952, 1:2000).
2.5 Immunofluorescence staining
NE-4C cells were fixed with 4% formaldehyde in PBS for 15 min at room temperature, rinsed with PBS, and then treated for 10 min with 0.1% Triton X-100 at 4°C. The cells were blocked with 4% fetal bovine serum in PBS for 1 hour and incubated with primary antibodies at 4°C overnight. Antibodies against Tuj1 (Covance, PRB-435p, 1:1000), anti-Map2 (Sigma, M2320, 1:200), and anti-Tbr2 (Abcam, ab23345, 1:500) were used. Images were obtained on a Zeiss LSM510 Meta laser scanning confocal microscope using Zen software. Imaris 8.0 software was used for cell counting quantitative analysis. For each group, three to four biological replicates were performed. The number of Hoechst-labeled cells in a field was counted and the percentage of Tuj1, Map2, or Tbr2 positive cells were quantified and finally the values in all mutants were compared to the WT.
2.6 Real-time quantitative PCR
RNA was isolated according to manufacturer’s instructions using the High Pure RNA Isolation Kit (Roche) and cDNA was generated using random hexamers and the Transcriptor First Strand 18 cDNA Synthesis Kit (Roche). qPCR was performed on a LightCycler 480 (Roche) using the LightCycler 480 Probes Master reagent (Roche) and Universal Probe Library (Roche) according to manufacturer’s protocol. All primers and probes are listed in Supp. Table S1. Data were collected and analyzed with LightCycler 480 Software (Roche, Version 1.5.1). All experiments were from three biological replicates.
2.7 Statistical Analysis
For genetic variation analyses, Fisher’s exact test was used to compare the difference between cases and controls and false discovery rate (FDR) correction was performed (Benjamini, Drai, Elmer, Kafkafi, & Golani, 2001). The student’s t-test was used for Western blot and immunofluorescent staining experiments (*= P<0.05, **= P<0.01, ***= P<0.0001).
3. RESULTS
3.1 Retinoid related genes harbor disease-specific rare variants in exonic and upstream regions in NTD cases
Vitamin A is processed by the retinoid metabolism pathway to RA (Figure 1A) and signaling through this pathway is critical for neural patterning, neural differentiation, and neural tube closure (Copp & Greene, 2010; Duester, 2008; Maden, 2007). We performed a case-control mutation screen by sequencing the RA pathway genes ALDH1A2, CRABP1, CRABP2, RARA, CYP26A1 and CYP26B1 in 355 NTD cases affecting cranial and caudal central nervous system and 225 controls from the Han Chinese population.
For these six genes in control and NTD cases we found 1002 variants in total. NTD-specific rare variants were slightly more than control-specific rare variants (Figure 1B; P = 0.0654; Q = 0.1177). Although there was no significant difference between NTD cases and controls for rare variants present in intronic, 5′UTR or 3′UTR regions, the number of NTD-specific rare variants increased significantly within exonic regions and upstream regions (P = 0.0027; Q = 0.0122 for exonic and P = 0.0090; Q = 0.0203 for upstream). Moreover, the occurrence of missense variants in the 6603bp of coding regions sequenced was significantly higher in NTD cases than in controls (Figure 1B; 5.12 missense variants per Mb DNA sequenced in NTDs [12/355/6603bp] vs. 0 per Mb in controls [0/225/6603bp], P = 0.0046; Q = 0.0138). All missense variants discovered were heterozygous. When viewed as recurrent rare mutations in exonic and upstream regions, the number of NTD-specific variants were not significantly different from control (6/355 vs. 0/225, P = 0.0871; Q = 0.1307; Figure 1B). However, the number of individuals carrying rare variants in exonic regions or upstream regions were dramatically more in NTD cases than in controls (Figure 1B, 35/355 NTD individuals vs. 4/225 controls, P = 0.0002; Q = 0.0018). The rare variants in exonic regions and upstream regions are listed in Table 1 and Table 2.
For the rare variants identified in upstream regions, defined as 2000bp upstream of transcription start site, the majority were found in CYP26A1, CRABP1, and ALDH1A2. In the upstream regions, we found six occurrences of two NTD-specific rare variants in CYP26A1, four occurrences of three rare variants in CRABP1 and six occurrences of six rare variants in ALDH1A2 (Table 2 and Figure 1D–1F). It is interesting that these are all experimental-based RA-responsive genes with RARE binding sites located in their upstream regions (Li et al., 2015; Loudig et al., 2000; Ohoka, Yokota-Nakatsuma, Maeda, Takeuchi, & Iwata, 2014) or RXR binding sites predicted by PROMO (Farre et al., 2003; Messeguer et al., 2002) (Figure 1D–1F). Although the genomic distances between clustered NTD-specific rare variants and RAREs are about 130 bp (ALDH1A2), 1294 bp (CYP26A1), and 1664 bp (CRABP1) (Figure 1D–1F), it is possible that the NTD-specific rare variants could impact their transcriptional regulation by RA, an idea that remains to be tested. Additionally, using PROMO (Farre et al., 2003; Messeguer et al., 2002) to predict transcription factor binding sites in the human genome, these variants could disrupt binding (Figure 1D–1F), highlighting the possible functional importance of these variants in the regulation of genes expression. Furthermore, we found a deletion in CYP26A1 (CYP26A1:NC_000010.10:g.94832265_94832266del) occurred four times (Figure 1D) and a total of three recurrent rare mutations were found in the upstream regions of CYP26A1 and CRABP1 (Figure 1D–1E), thus emphasizing the possible association of these variants with NTD prevalence. All variants reported were submitted to the ClinVar (Submission ID: SUB3259094; https://submit.ncbi.nlm.nih.gov/subs/clinvar_file/SUB3259094/overview)
We next correlated exonic or upstream variants with the clinical phenotype. The 18 occurrences of NTD-specific rare variants in exonic regions were present in the following cases: nine spina bifida, four encephalocele, two craniorachischisis, two spina bifida with encephalocele, and one anencephaly (Table 1). The 21 occurrences in upstream regions were present in twelve spina bifida, five anencephaly with spina bifida, two encephalocele, one encephalocele with spina bifida, and one anencephaly (Table 2). Thus, rare variants in retinoid metabolism genes can be associated with multiple types of NTDs (Tables 1 and 2). Interestingly, some cases who carry missense variants in CYP26B1 and/or variants in upstream regions of CYP26A1, RARA, ALDH1A2, and CRABP2 exhibit pulmonary hypoplasia or urinary malformation or limb malformation (Table 1 and 2), consistent with the phenotypes seen in mice with genetic-deficiencies of these genes (F. Chen et al., 2010; Lee et al., 2012; Maclean, Dolle, & Petkovich, 2009; Malpel, Mendelsohn, & Cardoso, 2000), emphasizing the importance of RA for neural tube, lung, urinary system and limb development.
In total, 158 common variants were found in the present study. Following Hardy-Weinberg equilibrium modeling in control and filtering, four single nucleotide polymorphisms (SNPs) were associated with risk of human NTDs (Supp. Table S2). Amongst them, CRABP2 rs12039622:T>C and CYP26B1 rs72374435:C>T appear as risk factors for the disease, whereas ALDH1A2 rs4646579:G>C and rs4238328:G>A may play a protective role (Supp. Table S2).
3.2 RA degradation enzyme CYP26B1 gene harbors the most NTD-specific missense rare variants
We found the CYP26B1 gene harbored the most NTD-specific rare variants within the exonic regions of the six genes analyzed (Table 3, 12 exonic variants in CYP26B1 vs. 6 for the remaining five genes) and more than the single CYP26B1 exonic variant identified in controls (Table 3, P = 0.0215, Q = 0.129 after FDR correction). Moreover, 9 of the 12 exonic variants in the CYP26B1 gene in NTD cases were missense or predicted splicing mutations, significantly more than in control (Table 1; NTD vs. Control: 9/355 vs. 0/225; P = 0.0152). The two ALDH1A2 and one CRABP1 exonic variants were also missense or predicted splicing mutations whereas the three CYP26A1 exonic variants were silent mutations (Table1).
Table 3.
Summary of rare variants harbored in exonic regions and upstream regions of sequenced genes involved in retinoid related metabolism and signaling
| Gene symbol | In exonic | In upstream | ||||||
|---|---|---|---|---|---|---|---|---|
| NTD | Control | P value | FDR correction Q value | NTD | Control | P value | FDR correction Q value | |
| CYP26A1 | 3 | 0 | 0.2878 | 0.8634 | 6 | 2 | 0.7170 | 1 |
| CYP26B1 | 12 | 1 | 0.0215 | 0.129 | 1 | 0 | 1 | 1 |
| CRABP1 | 1 | 0 | 1 | 1 | 4 | 0 | 0.3037 | 0.9111 |
| CRABP2 | 0 | 0 | 1 | 1 | 2 | 0 | 0.5249 | 1 |
| RARA | 0 | 0 | 1 | 1 | 2 | 1 | 1 | 1 |
| ALDH1A2 | 2 | 0 | 0.5249 | 1 | 6 | 0 | 0.0871 | 0.5226 |
FDR: False discovery rate
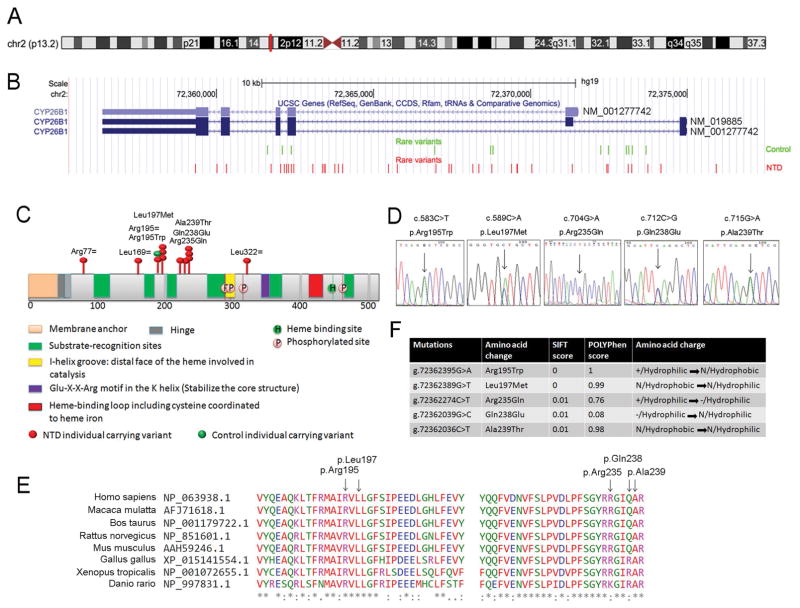
The CYP26B1 gene is located on chromosome 2 in the p13.2 genomic region (Figure 2A). According to the database from Exome Aggregation Consortium (ExAC), this gene is intolerant for missense mutations (z = 2.06) and extremely intolerant for loss of function mutations (pLI = 0.95) (Supp. Table S3) (Lek et al., 2016). In our case-control study, the coding and non-coding regions of the gene were examined. In total there were 45 occurrences of NTD-specific and 16 occurrences of control-specific rare variants in the CYP26B1 gene (Figure 2B, P = 0.0701). Visualization of the rare variants in the context of the human coding sequence shows accumulation of NTD-specific rare variants in exonic regions (Figure 2B), but there does not appear to be a specific genomic region that is more causally associated with NTDs. When viewed within the context of the CYP26B1 protein, the exonic NTD-specific missense mutations clustered in a 120 amino acid region (chr2:72362036-chr2:72362395) around two substrate-recognition sites (Figure 2C). Sanger sequencing was performed to confirm the rare variants (Figure 2D and not shown). Three NTD-specific variants would be expected to yield silent mutations (NP_063938.1:p.Arg77=, p.Leu169= and p.Leu322=), four to yield missense mutations (observed as eight occurrences: NP_063938.1:p.Arg195Trp, p.Leu197Met, p.Gln238Glu and p.Ala239Thr), and one splicing/exonic mutation (NC_000002.11:g.72362274C>T NP_063938.1:p.Arg235Gln). In contrast, only one silent mutation was found in control (NP_063938.1:p.Arg195=, shown in green in Figure 2C). Amongst them, the variants NC_000002.11g.72362395G>A NP_063938.1:p.Arg195Trp, NC_000002.11g.72362389G>T NP_063938.1:p.Leu197Met and NC_000002.11g.72362274C>T NP_063938.1:p.Arg235Gln have been reported in ExAC and the allele frequencies are 0.0001, 0.0001 and 0.0002 respectively (Lek et al., 2016). The variant NC_000002.11g.72362039G>C p.Gln238Glu and the recurrent variant g.72362036C>T p.Ala239Thr (3 occurrences) were found only in the NTD cases and were not present in the ExAC database. Clustal omega was used to analyze the conservation of the sequence across species at the position of these missense variants (Sievers et al., 2011). The results indicated that the amino acid residues p.Arg195, p.Leu197 and p.Ala239 are invariant from zebrafish to human, while p.Arg235 and p.Gln238 are conserved in mammals (Figure 2E). Notably, even when individuals carried the same missense mutation in CYP26B1, the clinical phenotypes varied and ranged from spinal to cranial defects (Table 1), suggesting that these heterozygous missense mutations in CYP26B1 might contribute to the NTD but are not the unique determinant of the phenotype.
FIGURE 2. Rare variants found in the CYP26B1 gene in the NTD case:control cohort.
A) CYP26B1 is located on chromosome 2 in the p13.2 genomic region. B) CYP26B1 contains 5–6 exons depending on alternative splicing. All cohort-specific rare variants found in the present study were visualized in the UCSC genome browser (GRCh37/hg19). Green font indicates rare variants identified in controls, red font indicates NTD-specific rare variants. C) Schematic of the CYP26B1 protein showing functional domains and the position of rare variants found in exonic regions indicated by red oval for NTD-specific variants or green oval for the single control individual. D) Sanger sequencing traces to confirm the potential damaging rare variants found in the CYP26B1 gene in NTD individuals. E) Conservation analysis by Clustal omega and location of NTD-specific variants. F) Predicted effect of the rare variants relative to protein function and amino acid charge.
3.3 Predictions of the impact of rare variants on protein function
Rare missense or predicted splicing mutations have the potential to impact protein function and the studies above showed a significant burden of these potentially damaging NTD-specific variants in the CYP26B1 gene. Therefore we focused our attention on testing the impact of NTD-specific missense or splicing mutations in CYP26B1 gene. We first evaluated the predicted splicing/missense mutation NM_019885:c.704G>A, Arg235Gln, which occurred at the end of exon3 (Supp. Figure S1). Using Human Splice Finder 3.0 (Desmet et al., 2009), this variant is located in a splice donor site and could potentially influence splicing; moreover, it could create a new exonic splicing silencer (ESS) site. Furthermore, this variant in exon3 would be expected to create a missense mutation (p.(Arg235Gln)) (Supp. Figure S1B and S1C). This potential to alter both splicing and protein sequence lead us to evaluate the only other splicing mutation seen, which was in the ALDH1A2 gene. This occurred in exon10 (NM_003888) and the variant is predicted to create an exonic ESS site or an alteration of an exonic splicing enhancer (ESE), as well as to create a p.(Ile363Val) missense mutation. Therefore, we classified both of these as splicing/missense variants in Table 1. We then evaluated the predicted effect of the individual missense variants on amino acid charge and hydrophobicity and used SIFT and Polyphen algorithims to predict the effect on protein function (Table 1 and Figure 2F). These tools predict that all five missense variants tested below would impact protein function.
3.4 Functional analysis of CYP26B1 variants demonstrates loss of function in modulation of RA-mediated gene transcription
To experimentally test the impact of these predicted damaging mutations, we transiently expressed GFP-tagged wildtype (WT) CYP26B1 cDNA or mutant constructs containing each NTD specific-variant in a neuroepithelial cell line NE-4C and assessed protein expression, effect on RA-mediated transcription, and neural differentiation. The NE-4C neuroepithelial cell line is derived from embryonic day 9 mouse cranial neural tissue during the period of neural tube closure. Plasmids carrying WT or mutant CYP26B1 (all constructs fused to GFP) and an RFP-expression plasmid to control for transfection efficiency were transiently transfected into the NE-4C cells. The relative expression levels of GFP indicated that there were no significant difference between WT and mutant constructs (Supp. Figure S2A). Western blot assays indicated that the protein level of RFP was similar for the WT and all mutant constructs and that the GFP antibody detected a prominent band at 84kD, the predicted size of the CYP26B1-GFP fusion protein for all constructs (CYP26B1 57kD plus GFP 27kD; Supp. Figure S2B). These constructs were used in NE-4C cells for the following functional studies.
RA can promote differentiation of neural stem cells and NE-4C cells respond to RA treatment by activating the expression of both direct and indirect RA-responsive genes within 24 hours. The CYP26B1 gene encodes a key modulator of intracellular RA levels via RA degradation. Therefore, we postulated that introduction of the WT CYP26B1 would result in RA degradation and minimal induction of RA-responsive genes. In contrast, if the NTD variant creates a loss of CYP26B1 function, then it would fail to degrade RA and would give similar results as blank controls upon RA induction. If the CYP26B1 variant retains partial function, then there may be a level of downstream RA-responsive gene transcription intermediate between WT and blank control. To test CYP26B1 function we transiently transfected the CYP26B1 expression plasmids into NE-4C cells and then treated the cells with RA (1μM) for 24 hours. Using real-time quantitative PCR, we assessed the mRNA levels of genes that are activated by RA and/or are implicated in neural tube closure or neural differentiation (Balmer & Blomhoff, 2002; Wilde et al., 2014). When WT CYP26B1 was introduced, the expression of all of the genes shown in Figure 3A–G was greatly reduced relative to blank control. This indicates that WT CYP26B1 reduces the level of RA in the cell leading to a lower RA-mediated transcriptional response. In contrast, all the CYP26B1 variants showed expression profiles similar to the blank control (Figure 3A–G), indicating that the CYP26B1 variants fail to modulate RA signaling, essentially acting as loss of function alleles. Genes that were suppressed by WT CYP26B1 but not by the CYP26B1 variants were the direct RA targets Rarb and Crabp1 (Figure 3A and 3B), which contain RARE sites and act in a feedback loop as components of the RA signaling pathway (Bastien & Rochette-Egly, 2004; Wilde, Siegenthaler, Dent, & Niswander, 2017); Wnt11 (Figure 3C), which encodes a ligand for non-canonical Wnt signaling pathway (Hardy et al., 2008); Cdh2 encoding a cell adhesion protein (Figure 3D); the transcription factor gene Foxa1 (Figure 3E) which plays a role in dorsal-ventral patterning of the neural tube (Maier, Lo, & Harfe, 2013); the homeobox gene Hoxb1 (Figure 3F), which patterns progenitor cells in the hindbrain (Gaufo, Flodby, & Capecchi, 2000; Zigman, Laumann-Lipp, Titus, Postlethwait, & Moens, 2014) and consistent with the increase in Hoxb1 mRNA levels in RA-treated human neural stem cells (Colleoni et al., 2011); and Casp3 (Figure 3G) encoding the pro-apoptotic protein. In contrast, the mRNA level of Pou5f1 (Figure 3H), which encodes the embryonic stem cell pluripotency protein Oct4 is not affected by WT or NTD-specific CYP26B1 variants. These changes in RA-mediated transcriptional activity occur without apparent influence on RA receptor protein expression as indicated by western blot analysis of Rara, Rarb and Rarg relative to WT (Supp. Figure S3). Taken together, these results suggest that the CYP26B1 variants introduce loss of function mutations that attenuate CYP26B1 function in the degradation of RA and the modulation RA activity in the context of transcriptional regulation of downstream genes.
3.5 CYP26B1 variants impact modulation of neural differentiation
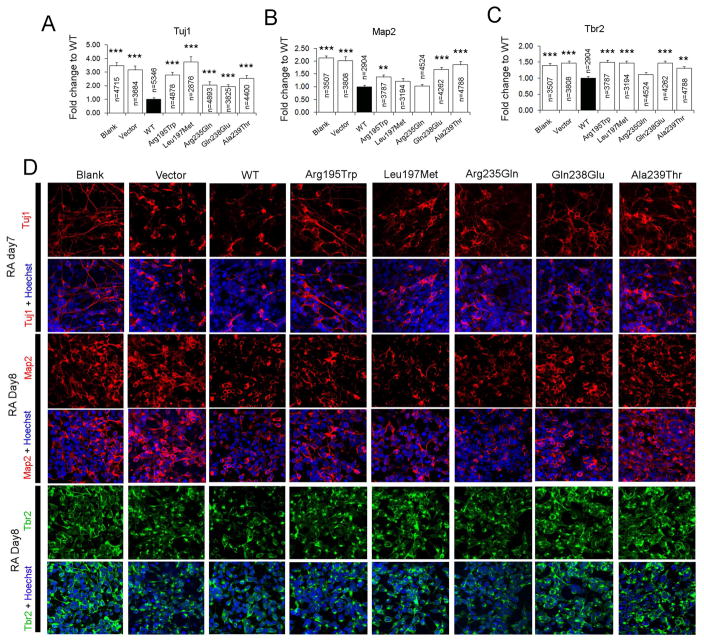
A well-known function of RA is to promote the differentiation of neural progenitor cells to neurons. To test the ability of CYP26B1 constructs to affect neuronal differentiation, we treated CYP26B1 transiently transfected NE-4C cells with 1μM RA to induce differentiation. We then assessed neuronal differentiation 7–8 days after RA treatment by quantifying the number of cells that stained positively for the neuronal markers Tuj1, Map2, and Tbr2. WT CYP26B1 suppressed neuronal differentiation as assessed with all three neuronal markers (Figure 4). In contrast, all CYP26B1 variants showed attenuated ability to suppress differentiation and were more similar to blank or empty vector controls. For all mutant constructs, the number of Tuj1-positive cells was significantly higher than for WT CYP26B1 (Figure 4A and 4D). For Map2 and Tbr2 we noted some intermediate or non-significant responses suggesting that some variants may retain partial function, in particular p.(Arg235Gln), whereas other variants such as p.(Arg195Trp) and p.(Ala293Thr) showed significant loss of function phenotypes in all assays (Figure 4B–4D).
FIGURE 4. CYP26B1 variants disrupt modulation of neural differentiation.
Quantification of the immunofluorescent staining results of neuronal markers Tuj1 (A), Map2 (B) or Tbr2 (C) positive cells 7–8 days after RA treatment of NE-4C cells (Blank), or NE-4C cells transiently transfected with vector (Vector), wildtype CYP26B1 (WT), or different CYP26B1 variants. The percentage of immunopositive cells relative to all cells (Hoechst labeled) in a field of view were scored and then compared to cells transfected with WT CYP26B1. ***: P < 0.0001; **: P < 0.01; Student’s t-test. D) Typical images of the immunofluorescent staining used for calculations in A–C. For each genotype and neuronal marker, three to four replicates were performed.
4. DISCUSSION
Our NTD case:control study has identified a significant number of rare mutations in retinoid metabolism genes associated with NTDs in humans. Furthermore, our functional experiments demonstrate that the NTD-specific missense mutations in CYP26B1 create loss of function mutations. CYP26B1 acts to degrade RA and modulate RA signaling levels. Consistent with this, WT CYP26B1 decreases the expression of direct and indirect targets of RA signaling and inhibits RA-mediated neural differentiation. In contrast, the NTD-specific CYP26B1 variants show significantly attenuated function in these assays. These RA-mediated activities are relevant for the process of neural tube closure and hence we suggest that impairment in the balance of RA signaling due to CYP26B1 variants can contribute to NTD risk.
In our cohort, all variants found were genetically heterozygous, with the variant present on only one allele. From our sequencing data of six RA related genes, CYP26B1 was most burdened of NTD-specific damaging mutations. All three CYP26 enzymes are dynamically expressed in mice from gastrulation through the time of neural tube closure. These genes are critical in modulating RA signaling during neural tube formation as loss of all three genes results in duplication of the neural tube in Cyp26a1b1c1−/− embryos and this defect is more pronounced than in Cyp26a1c1−/− embryos (Uehara, Yashiro, Takaoka, Yamamoto, & Hamada, 2009), implying that Cyp26b1 separately contributes to early neural tube formation. Moreover, Cyp26b1 is the most highly expressed of the CYP26 isoforms within the hindbrain, although Cyp26b1−/− mutants do not show a significant defect in hindbrain patterning in mice (Maclean et al., 2009).
Our functional experiments of the rare variants in the CYP26B1 gene found in our NTD cohort indicate they are partial or complete loss of function alleles. We postulate this leads to insufficient regulation of RA-mediated transcription and neural stem cell differentiation due to failure of RA degradation, which leads to supraphysiological RA accumulation. Excess RA can elicit NTDs in rat and mouse, ranging from caudal spina bifida aperta, to encephalocele and a combination of these NTDs (Alles & Sulik, 1990; W. Cai et al., 2007). In our present cohort, 3 of 9 individuals carrying CYP26B1 missense rare variants displayed lower spina bifida, 3 individuals were affected with encephalocele, and 1 was affected with the combination of spina bifida and encephalocele: consistent with the rodent phenotypes. Notably, two individuals exhibited craniorachischisis, highlighting the possibility that supraphysiological RA might be associated with a craniorachischisis phenotype, although there is no evidence that excess RA induces this phenotype in animal models. By contrast, lack of RA synthesis elicits craniorachischisis in mice (Niederreither, Subbarayan, Dolle, & Chambon, 1999), emphasizing the importance of proper levels of RA in convergence and extension of the neural plate during early neural tube closure. It has been proposed that supraphysiological RA leads to spina bifida due to excessive cell death in the hindgut and mesenchyme ventral to the neuroepithelium of the posterior neuropore, and a disparity in growth between the ventral and dorsal regions of the tail bud causes the relatively faster growing dorsal region to evert, preventing closure of the caudal neural tube (Alles & Sulik, 1990). Our data show that the NTD-specific missense constructs cannot appropriately decrease the expression of Casp3 (Figure 3G), consistent with the idea that apoptosis may be increased when levels of RA are in excess. Additionally, Hoxb1 is a crucial RA-responsive (Colleoni et al., 2011) regulator of hindbrain lumen morphogenesis (Zigman et al., 2014) and the NTD-specific mutant constructs are not capable of modulating Hoxb1 expression (Figure 3F) and this could contribute to possible CYP26B1 phenotypes.
Our human NTD sequencing data also identified a significant number of NTD-specific rare variants in upstream regions of CYP26A1, CRABP1 and ALDH1A2 (Figure 1C–1E and Table 2). Cyp26a1−/− mouse embryos exhibit exencephaly and caudal spina bifida (Abu-Abed et al., 2001). The upstream region of Cyp26a1 is highly conserved and has DR5 RARE elements which are bound by RAR receptors to regulate transcription (Loudig et al., 2000). Similarly, in the upstream region of CRABP1 there is a DR2 RARE element (Figure 1E) and NTDs in humans have been associated with ectopic occupation of CRABP1 promoters by RXR receptors and histone alterations (Li et al., 2015). Aldh1a2 null mouse embryos exhibit craniorachischisis (Niederreither et al., 1999). In view of our findings that NTD-specific rare variants are located 130–1664 bp upstream of the RARE for these three genes, we postulate that these allelic variants impact the binding of RA receptors and/or the transcriptional regulation of these key genes. Furthermore, transcription factor binding predictions suggest the possible impact of NTD-specific variants on the expression of these genes (Figure 1D–1F).
In summary, our data provide a dataset of novel rare variants in retinoid related genes that are genetically correlated with NTD. Our experimental results emphasize the loss of function characteristics of these rare mutations encoding for the RA degradation enzyme CYP26B1. Our findings together with genetic association studies by others (Deak et al., 2005; Rat et al., 2006; Tran et al., 2011) highlight this functional pathway in the genetic contribution of rare mutations to human NTDs.
Supplementary Material
Acknowledgments
Grant sponsor: This work was supported by NIH HD81117; the National Natural Science Foundation of China, Beijing, China (No. 81471163, 81701441).
We are grateful to Sofia Pezoa for her comments on the manuscript.
Footnotes
DISCLOSURE STATEMENT
The authors declare no conflict of interest.
References
- Abu-Abed S, Dolle P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15(2):226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allache R, Wang M, De Marco P, Merello E, Capra V, Kibar Z. Genetic studies of ANKRD6 as a molecular switch between Wnt signaling pathways in human neural tube defects. Birth Defects Res A Clin Mol Teratol. 2015;103(1):20–26. doi: 10.1002/bdra.23273. [DOI] [PubMed] [Google Scholar]
- Alles AJ, Sulik KK. Retinoic acid-induced spina bifida: evidence for a pathogenetic mechanism. Development. 1990;108(1):73–81. doi: 10.1242/dev.108.Supplement.73. [DOI] [PubMed] [Google Scholar]
- Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43(11):1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Bruel AL, Franco B, Duffourd Y, Thevenon J, Jego L, Lopez E, … Thauvin-Robinet C. Fifteen years of research on oral-facial-digital syndromes: from 1 to 16 causal genes. J Med Genet. 2017;54(6):371–380. doi: 10.1136/jmedgenet-2016-104436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Shi O. Genetic evidence in planar cell polarity signaling pathway in human neural tube defects. Front Med. 2014;8(1):68–78. doi: 10.1007/s11684-014-0308-4. [DOI] [PubMed] [Google Scholar]
- Cai W, Zhao H, Guo J, Li Y, Yuan Z, Wang W. Retinoic acid-induced lumbosacral neural tube defects: myeloschisis and hamartoma. Childs Nerv Syst. 2007;23(5):549–554. doi: 10.1007/s00381-006-0289-y. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chen F, Cao Y, Qian J, Shao F, Niederreither K, Cardoso WV. A retinoic acid-dependent network in the foregut controls formation of the mouse lung primordium. J Clin Invest. 2010;120(6):2040–2048. doi: 10.1172/JCI40253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang Q, Bai B, Ouyang S, Bao Y, Li H, Zhang T. MARK2/Par1b Insufficiency Attenuates DVL Gene Transcription via Histone Deacetylation in Lumbosacral Spina Bifida. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-0164-0. [DOI] [PubMed] [Google Scholar]
- Chen WH, Morriss-Kay GM, Copp AJ. Genesis and prevention of spinal neural tube defects in the curly tail mutant mouse: involvement of retinoic acid and its nuclear receptors RAR-beta and RAR-gamma. Development. 1995;121(3):681–691. doi: 10.1242/dev.121.3.681. [DOI] [PubMed] [Google Scholar]
- Chen X, An Y, Gao Y, Guo L, Rui L, Xie H, … Zhang T. Rare Deleterious PARD3 Variants in the aPKC-Binding Region are Implicated in the Pathogenesis of Human Cranial Neural Tube Defects Via Disrupting Apical Tight Junction Formation. Hum Mutat. 2017;38(4):378–389. doi: 10.1002/humu.23153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni S, Galli C, Gaspar JA, Meganathan K, Jagtap S, Hescheler J, … Lazzari G. Development of a neural teratogenicity test based on human embryonic stem cells: response to retinoic acid exposure. Toxicol Sci. 2011;124(2):370–377. doi: 10.1093/toxsci/kfr245. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Greene ND. Genetics and development of neural tube defects. J Pathol. 2010;220(2):217–230. doi: 10.1002/path.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco P, Merello E, Piatelli G, Cama A, Kibar Z, Capra V. Planar cell polarity gene mutations contribute to the etiology of human neural tube defects in our population. Birth Defects Res A Clin Mol Teratol. 2014;100(8):633–641. doi: 10.1002/bdra.23255. [DOI] [PubMed] [Google Scholar]
- Deak KL, Dickerson ME, Linney E, Enterline DS, George TM, Melvin EC … Group NTDC. Analysis of ALDH1A2, CYP26A1, CYP26B1, CRABP1, and CRABP2 in human neural tube defects suggests a possible association with alleles in ALDH1A2. Birth Defects Res A Clin Mol Teratol. 2005;73(11):868–875. doi: 10.1002/bdra.20183. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle WE, Robinson JF, Kneist A, Sirerol-Piquer MS, Frints SG, Corbit KC, … Reiter JF. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet. 2011;89(1):94–110. doi: 10.1016/j.ajhg.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134(6):921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31(13):3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaufo GO, Flodby P, Capecchi MR. Hoxb1 controls effectors of sonic hedgehog and Mash1 signaling pathways. Development. 2000;127(24):5343–5354. doi: 10.1242/dev.127.24.5343. [DOI] [PubMed] [Google Scholar]
- Guan K, Chang H, Rolletschek A, Wobus AM. Embryonic stem cell-derived neurogenesis. Retinoic acid induction and lineage selection of neuronal cells. Cell Tissue Res. 2001;305(2):171–176. doi: 10.1007/s004410100416. [DOI] [PubMed] [Google Scholar]
- Hardy KM, Garriock RJ, Yatskievych TA, D’Agostino SL, Antin PB, Krieg PA. Non-canonical Wnt signaling through Wnt5a/b and a novel Wnt11 gene, Wnt11b, regulates cell migration during avian gastrulation. Dev Biol. 2008;320(2):391–401. doi: 10.1016/j.ydbio.2008.05.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res A Clin Mol Teratol. 2010;88(8):653–669. doi: 10.1002/bdra.20676. [DOI] [PubMed] [Google Scholar]
- Hopp K, Heyer CM, Hommerding CJ, Henke SA, Sundsbak JL, Patel S, … Harris PC. B9D1 is revealed as a novel Meckel syndrome (MKS) gene by targeted exon-enriched next-generation sequencing and deletion analysis. Hum Mol Genet. 2011;20(13):2524–2534. doi: 10.1093/hmg/ddr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iulianella A, Beckett B, Petkovich M, Lohnes D. A molecular basis for retinoic acid-induced axial truncation. Dev Biol. 1999;205(1):33–48. doi: 10.1006/dbio.1998.9110. [DOI] [PubMed] [Google Scholar]
- Jones D, Fiozzo F, Waters B, McKnight D, Brown S. First-trimester diagnosis of Meckel-Gruber syndrome by fetal ultrasound with molecular identification of CC2D2A mutations by next-generation sequencing. Ultrasound Obstet Gynecol. 2014;44(6):719–721. doi: 10.1002/uog.13381. [DOI] [PubMed] [Google Scholar]
- Kibar Z, Torban E, McDearmid JR, Reynolds A, Berghout J, Mathieu M, … Gros P. Mutations in VANGL1 associated with neural-tube defects. N Engl J Med. 2007;356(14):1432–1437. doi: 10.1056/NEJMoa060651. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, … Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25(17):2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LM, Leung CY, Tang WW, Choi HL, Leung YC, McCaffery PJ, … Shum AS. A paradoxical teratogenic mechanism for retinoic acid. Proc Natl Acad Sci U S A. 2012;109(34):13668–13673. doi: 10.1073/pnas.1200872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Zhu H, Duhon C, Yang W, Ross ME, Shaw GM, Finnell RH. Mutations in planar cell polarity gene SCRIB are associated with spina bifida. PLoS One. 2013;8(7):e69262. doi: 10.1371/journal.pone.0069262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Zhu H, Yang W, Ross ME, Shaw GM, Finnell RH. Identification of novel CELSR1 mutations in spina bifida. PLoS One. 2014;9(3):e92207. doi: 10.1371/journal.pone.0092207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei YP, Zhang T, Li H, Wu BL, Jin L, Wang HY. VANGL2 mutations in human cranial neural-tube defects. N Engl J Med. 2010;362(23):2232–2235. doi: 10.1056/NEJMc0910820. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T … Exome Aggregation, C. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bai B, Zhang Q, Bao Y, Guo J, Chen S, … Zhang T. Ectopic cross-talk between thyroid and retinoic acid signaling: A possible etiology for spinal neural tube defects. Gene. 2015;573(2):254–260. doi: 10.1016/j.gene.2015.07.048. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N … Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14(9):1483–1497. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Maclean G, Dolle P, Petkovich M. Genetic disruption of CYP26B1 severely affects development of neural crest derived head structures, but does not compromise hindbrain patterning. Dev Dyn. 2009;238(3):732–745. doi: 10.1002/dvdy.21878. [DOI] [PubMed] [Google Scholar]
- Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8(10):755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- Maden M, Gale E, Kostetskii I, Zile M. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6(4):417–426. doi: 10.1016/s0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- Maier JA, Lo Y, Harfe BD. Foxa1 and Foxa2 are required for formation of the intervertebral discs. PLoS One. 2013;8(1):e55528. doi: 10.1371/journal.pone.0055528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpel S, Mendelsohn C, Cardoso WV. Regulation of retinoic acid signaling during lung morphogenesis. Development. 2000;127(14):3057–3067. doi: 10.1242/dev.127.14.3057. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, … Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini NJ, Hoffmann TJ, Lammer EJ, Hardin J, Lazaruk K, Stein JB, … Rine J. A genetic signature of spina bifida risk from pathway-informed comprehensive gene-variant analysis. PLoS One. 2011;6(11):e28408. doi: 10.1371/journal.pone.0028408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merello E, Mascelli S, Raso A, Piatelli G, Consales A, Cama A, … Marco PD. Expanding the mutational spectrum associated to neural tube defects: literature revision and description of novel VANGL1 mutations. Birth Defects Res A Clin Mol Teratol. 2015;103(1):51–61. doi: 10.1002/bdra.23305. [DOI] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18(2):333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- Miao C, Jiang Q, Li H, Zhang Q, Bai B, Bao Y, Zhang T. Mutations in the Motile Cilia Gene DNAAF1 Are Associated with Neural Tube Defects in Humans. G3 (Bethesda) 2016;6(10):3307–3316. doi: 10.1534/g3.116.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa A, Komatsuzaki S, Kikuchi A, Niihori T, Aoki Y, Fujiwara K, … Kure S. Mutations in genes encoding the glycine cleavage system predispose to neural tube defects in mice and humans. Hum Mol Genet. 2012;21(7):1496–1503. doi: 10.1093/hmg/ddr585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dolle P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet. 2002;31(1):84–88. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21(4):444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Ohoka Y, Yokota-Nakatsuma A, Maeda N, Takeuchi H, Iwata M. Retinoic acid and GM-CSF coordinately induce retinal dehydrogenase 2 (RALDH2) expression through cooperation between the RAR/RXR complex and Sp1 in dendritic cells. PLoS One. 2014;9(5):e96512. doi: 10.1371/journal.pone.0096512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, Petkovich M. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res A Clin Mol Teratol. 2010;88(10):883–894. doi: 10.1002/bdra.20709. [DOI] [PubMed] [Google Scholar]
- Qiao X, Liu Y, Li P, Chen Z, Li H, Yang X, … Wang H. Genetic analysis of rare coding mutations in CELSR1-3 in Chinese Congenital Heart and Neural Tube Defects. Clin Sci (Lond) 2016 doi: 10.1042/CS20160686. [DOI] [PubMed] [Google Scholar]
- Rat E, Billaut-Laden I, Allorge D, Lo-Guidice JM, Tellier M, Cauffiez C, … Broly F. Evidence for a functional genetic polymorphism of the human retinoic acid-metabolizing enzyme CYP26A1, an enzyme that may be involved in spina bifida. Birth Defects Res A Clin Mol Teratol. 2006;76(6):491–498. doi: 10.1002/bdra.20275. [DOI] [PubMed] [Google Scholar]
- Roberson EC, Dowdle WE, Ozanturk A, Garcia-Gonzalo FR, Li C, Halbritter J, … Reiter JF. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J Cell Biol. 2015;209(1):129–142. doi: 10.1083/jcb.201411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, Escuin S, Doudney K, Vekemans M, Stevenson RE, Greene ND, … Stanier P. Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum Mutat. 2012;33(2):440–447. doi: 10.1002/humu.21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena AK, Gupta J, Pandey S, Gangopadhaya AN, Pandey LK. Prevalence of cystathionine beta synthase gene mutation 852Ins68 as a possible risk for neural tube defects in eastern India. Genet Mol Res. 2011;10(4):2424–2429. doi: 10.4238/2011.October.7.4. [DOI] [PubMed] [Google Scholar]
- Shaheen R, Almoisheer A, Faqeih E, Babay Z, Monies D, Tassan N, … Alkuraya FS. Identification of a novel MKS locus defined by TMEM107 mutation. Hum Mol Genet. 2015;24(18):5211–5218. doi: 10.1093/hmg/ddv242. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, … Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley MM, Northrup H, Au KS. Genetic studies of the cystathionine beta-synthase gene and myelomeningocele. Birth Defects Res A Clin Mol Teratol. 2012;94(1):52–56. doi: 10.1002/bdra.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PX, Au KS, Morrison AC, Fletcher JM, Ostermaier KK, Tyerman GH, Northrup H. Association of retinoic acid receptor genes with meningomyelocele. Birth Defects Res A Clin Mol Teratol. 2011;91(1):39–43. doi: 10.1002/bdra.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara M, Yashiro K, Takaoka K, Yamamoto M, Hamada H. Removal of maternal retinoic acid by embryonic CYP26 is required for correct Nodal expression during early embryonic patterning. Genes Dev. 2009;23(14):1689–1698. doi: 10.1101/gad.1776209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde JJ, Petersen JR, Niswander L. Genetic, epigenetic, and environmental contributions to neural tube closure. Annu Rev Genet. 2014;48:583–611. doi: 10.1146/annurev-genet-120213-092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde JJ, Siegenthaler JA, Dent SY, Niswander LA. Diencephalic Size Is Restricted by a Novel Interplay Between GCN5 Acetyltransferase Activity and Retinoic Acid Signaling. J Neurosci. 2017;37(10):2565–2579. doi: 10.1523/JNEUROSCI.2121-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L, Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev Biol. 2005;282(1):1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Yang XY, Zhou XY, Wang QQ, Li H, Chen Y, Lei YP, … Wang HY. Mutations in the COPII vesicle component gene SEC24B are associated with human neural tube defects. Hum Mutat. 2013;34(8):1094–1101. doi: 10.1002/humu.22338. [DOI] [PubMed] [Google Scholar]
- Zhang M, Cheng J, Liu A, Wang L, Xiong L, Chen M, … Lu Y. A missense mutation in TMEM67 causes Meckel-Gruber syndrome type 3 (MKS3): a family from China. Int J Clin Exp Pathol. 2015;8(5):5379–5386. [PMC free article] [PubMed] [Google Scholar]
- Zigman M, Laumann-Lipp N, Titus T, Postlethwait J, Moens CB. Hoxb1b controls oriented cell division, cell shape and microtubule dynamics in neural tube morphogenesis. Development. 2014;141(3):639–649. doi: 10.1242/dev.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.