Abstract
Camelid single-domain antibody fragments (“nanobodies”) provide the remarkable specificity of antibodies within a single 15 kDa immunoglobulin VHH domain. This unique feature has enabled applications ranging from use as biochemical tools to therapeutic agents. Nanobodies have emerged as especially useful tools in protein structural biology, facilitating studies of conformationally dynamic proteins such as G protein-coupled receptors (GPCRs). Nearly all nanobodies available to date have been obtained by animal immunization, a bottleneck restricting many applications of this technology. To solve this problem, we report a fully in vitro platform for nanobody discovery based on yeast surface display. We provide a blueprint for identifying nanobodies, demonstrate the utility of the library by crystallizing a nanobody with its antigen, and most importantly, we utilize the platform to discover conformationally-selective nanobodies to two distinct human GPCRs. To facilitate broad deployment of this platform, the library and associated protocols are freely available for non-profit research.
Introduction
Antibodies have had a transformative impact on science and medicine due to their exceptional specificity and biochemical versatility, enabling applications in almost every aspect of biomedical inquiry. Conventional antibodies are composed of two heavy chains and two light chains. Each of these contributes to antigen binding specificity through a variable domain, termed VH and VL for the heavy and light chain, respectively. A key exception to this general architecture is found in camelids (llamas, camels, alpacas, and their relatives), which possess a parallel antibody repertoire composed solely of heavy chains1,2. Such antibodies bind to their target antigens through a single variable domain, termed VHH, which contains the entire antigen-binding surface. Unlike the antigen binding fragments of conventional antibodies (Fabs), isolated VHH domains (also called “nanobodies”) can be readily expressed in bacteria as the product of a single gene, and in many cases these fragments can even fold and retain antigen specificity in the reducing environment of the eukaryotic cytosol. Owing to their versatility, nanobodies have found applications in protein structural biology and cell biology, and as potential diagnostic and therapeutic agents2–6. Nanobodies have been particularly critical tools in studies of G protein-coupled receptors (GPCRs), where conformationally-selective nanobodies have been used extensively to stabilize these receptors in defined states for crystallographic studies that would otherwise be impossible6.
Despite the growing importance of nanobodies throughout biomedical research, the current methods for creating these powerful tools remain slow, costly, and often unreliable. The majority of nanobodies described to date have been derived from immunization of camelids, a lengthy and expensive process posing a significant barrier to entry for most laboratories. More importantly, animal-derived antibodies are frequently restricted from binding conserved epitopes due to immunological tolerance of self-antigens. As conserved epitopes often drive key protein functions like protein-protein recognition and allosteric communication, a rapid method to identify nanobodies that target such sites would provide a robust means to interrogate protein function.
Previous efforts have sought to address challenges in nanobody identification by combining phage display with a synthetic library7. However, these libraries remain available only through contract work with an expensive commercial provider, limiting their broad utilization. Moreover, with synthetic libraries it remains particularly challenging to identify nanobodies that not only bind to their target, but also specifically recognize a defined conformation – this represents one of the most important applications of animal derived nanobodies. Phage display techniques enable isolation of high affinity binders, but identification of functional clones (e.g., conformationally selective nanobodies) remains challenging, despite select successes8,9. Here, we address these challenges through the development and application of a fully synthetic yeast display nanobody library, devised using an alignment of structurally characterized nanobodies from the Protein Data Bank (PDB). The use of a cell-based selection scheme enabled by fluorescence activated cell sorting (FACS) allows straightforward and rapid identification of conformationally-selective nanobodies for two human GPCRs. To enable broad utility of this new resource, we have made the library and all associated protocols available to the scientific community.
Library construction and validation
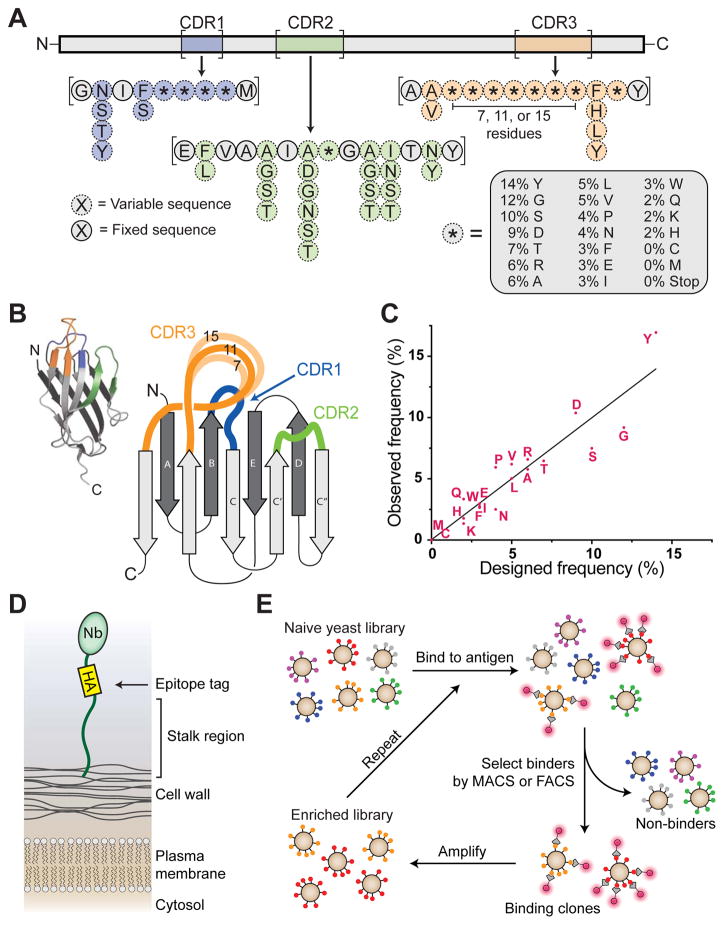
To develop a platform for rapid in vitro nanobody discovery, we first designed a synthetic nanobody library starting from a consensus framework derived from llama genes IGHV1S1–S5. This constant framework was combined with designed variation of the complementarity determining loops (CDRs) that comprise the highly variable, antigen-binding interface of the VHH. Although there are many previously described strategies for introducing variation in antibody CDR loops10, we postulated that nanobodies of known structure in the Protein Data Bank (PDB) represented an especially curated set of highly stable, biochemically well behaved variants as evidenced by their tractability in crystallographic studies. The entire set of unique nanobodies in the PDB (93 sequences at the time of analysis) was analyzed for position specific variation in the CDRs, and our design aimed to recapitulate this diversity. For the residues immediately adjacent to the CDRs, we introduced partial randomization, allowing only a few possible amino acids guided by the observed frequencies of nanobodies in the PDB. For the highly variable positions in each CDR, we introduced much more thorough randomization reflective of the high diversity at these positions. First, the frequency of each amino acid within CDR3 was determined for all nanobodies in the PDB. The resulting mixture of amino acids was modified to eliminate cysteine and methionine to avoid chemical reactivity, and was introduced into various positions in each CDR in the synthetic library. Based on analysis of nanobody sequences, we elected to introduce four such highly variable positions in CDR1 and one in CDR2. The longer CDR3 represents a critically important loop for antigen recognition – here we introduced either seven, eleven, or fifteen consecutive positions of high diversity mixture to emulate CDR3 length variation seen in the natural repertoire (Figure 1A – C).
Figure 1. Design and construction of synthetic nanobody library.
(A) Schematic of synthetic nanobody. Framework regions were fixed in sequence (gray), while portions of the CDR loops were varied (blue, green, and orange, for CDRs 1, 2, and 3, respectively). Partial randomization was achieved with mixed nucleotides to allow up to six possible residues (dotted lines), while highly variable regions were synthesized using a trimer phosphoramidite mixture (asterisks with dotted outlines). (B) Overview of a nanobody showing three-dimensional structure (inset) as well as a cartoon schematic highlighting CDR loops. Synthetic CDR3 sequences used variable lengths of 7, 11, or 15 residues of trimer phosphoramidite mixture. (C) After the library was constructed, amino acid frequencies of diversified positions in CDRs were analyzed by next-generation sequencing (NGS), showing that library frequencies closely matched target values. (D) Schematic of nanobody display on yeast. Nanobody is shown with an HA tag at the carboxy terminus followed by a long flexible stalk which covalently tethers the nanobody to the yeast cell wall. (E) Schematic of the nanobody selection process. Antigen is shown with a fluorescent tag as a glowing red circle. Yeast displaying nanobodies with affinity to antigen are shown being isolated via MACS or FACS, amplified, and undergoing iterative rounds of selection. See also Supplementary Figures 1 and 2, as well as Supplementary Table 1.
Primers encompassing the desired sequence diversity were synthesized, with high diversity regions constructed using a mixed pool of trimer phosphoramidites to match target codon frequency11,12. We then built a pooled DNA library encoding the full nanobody sequence by assembly PCR of these primers (see Methods for full details). A pervasive but often unappreciated drawback of synthetic antibody fragments is poor biochemical behavior. Prior to generating a large yeast display library, we sought to assess whether our library design would reliably produce biochemically tractable clones suitable for structural and cell biological studies. A total of 11 nanobody sequences were chosen at random from the library DNA and tested for biochemical behavior by expression and purification from Escherichia coli. Of these clones, 9 were well expressed (>20 mg/L), could be purified by routine procedures, and showed reasonably monodisperse size exclusion profiles (Supplementary Figure 1; Supplementary Table 1).
Next, we sought to develop a straightforward way to tether nanobodies and other small proteins to the surface of Saccharomyces cerevisiae and other yeast species. A variety of methods for this purpose have been reported previously, with the most common approach being fusion of a protein of interest to Aga2p, a yeast cell wall protein13. While this strategy has been widely used, it is limited by the requirement to use engineered yeast strains expressing galactose-inducible Aga1p, which tethers Aga2p to the yeast cell wall. Instead, we built a simplified system in which the protein of interest is directly connected to the cell wall through a single tether designed to replace the Aga2p-Aga1p linker protein. A synthetic tether mimicking the low complexity sequence of yeast cell wall proteins was designed and was evaluated for its ability to anchor a test protein to the yeast cell wall (see Supplementary Note for full sequence). Accessibility of this protein correlated strongly with the molecular weight of the staining reagent, suggesting steric occlusion by cell wall glycans. Consistent with this explanation, longer tethers alleviated this problem, and above 600 amino acids in length the molecular weight dependence of staining levels was negligible (Supplementary Figure 2). For library creation, we chose to use a 649 amino acid tether. In addition, we included an N-terminal engineered mating factor α pre-protein which has been reported previously to enhance antibody expression in yeast14. On the C-terminus, we included a glycosylphosphatidylinositol anchor sequence, which results in covalent tethering of the protein to the yeast cell wall15 (Figure 1D). On the basis of these results, the engineered surface display plasmid (pYDS649) was linearized and then transformed into Saccharomyces cerevisiae protease-deficient strain BJ5465 together with the DNA library, which had been amplified to include flanking sequences homologous to pYDS649 for recombination. This resulted in a yield of 5 × 108 transformants. While this is modest in comparison to some libraries, we show below that our approach yields comparable clones to those obtained by animal immunization, attesting to the value of our structure-based library design.
To characterize the resulting yeast surface displayed library, we isolated plasmid DNA from ~600,000 yeast cells and performed high throughput sequencing. Analysis of 480,000 unique sequences confirmed that the library clones possessed the desired amino acid frequencies in the hypervariable portions of the CDR loops (Figure 1C), and also confirmed that CDR-flanking region diversity resembled target frequencies. Overall, 26% of library yeast showed high expression of nanobodies as measured by flow cytometry. Taken together, these data imply that the library contains at least 108 unique full-length nanobody clones that are able to be expressed and displayed on the yeast surface.
Identification of nanobodies using magnetic cell sorting
We next sought to determine if our approach could yield nanobody clones targeting diverse antigen classes, including both soluble and membrane proteins. As an initial test, we aimed to identify nanobodies targeting human serum albumin (HSA), the most abundant protein in human plasma. Binding of therapeutic proteins and small molecules to HSA is a key determinant of drug pharmacokinetics; nanobodies targeting HSA may therefore provide a modular tool for enhancing the half-life of biologics16–18. In this first test, we exclusively employed magnetic cell sorting (MACS), a low-cost cell separation method that can be performed in even the most basic laboratory environments without specialized equipment19.
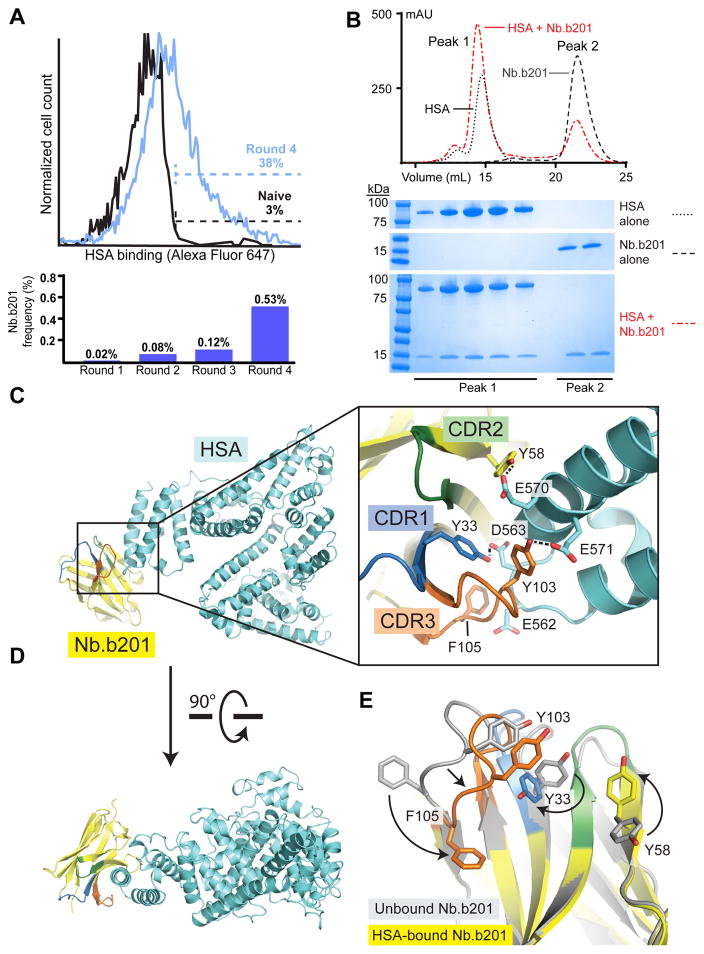
To identify novel HSA-binding nanobodies, we first prepared fluorescently-labeled HSA protein and then used this reagent to perform iterative staining and magnetic-bead based enrichments. In brief, this approach entails incubation of the yeast library with fluorescent HSA antigen, washing off excess antigen, and further staining desired clones with anti-fluorophore magnetic microbeads. Labeled cells are then isolated by magnet-based separation and amplified in standard yeast culture medium (Figure 1E). As with any library-based selection, it is of paramount importance to ensure binders recognize the antigen itself, rather than the reagents or fluorescent tags used for separation. To achieve such specificity, yeast with reactivity to beads alone were depleted from the library prior to each selection step. To decrease the probability of enriching fluorophore binders, the fluorophore tag used was alternated between Alexa Fluor 647 and fluorescein isothiocyanate (FITC) for each consecutive round of magnetic selection. After four rounds of selection (Figure 2A), single yeast colonies were isolated and stained with fluorescently labeled HSA for validation by analytical flow cytometry. For further study, we selected one clone with strong antigen binding (named Nb.b201) for in-depth characterization.
Figure 2. Validation of nanobody platform using human serum albumin (HSA) as the target antigen.
(A) Histogram of yeast library HSA binding, showing pre-selection (black) and after four rounds (blue). The percentage of HSA-binding cells is indicated. The fraction of the total library composed by Nb.b201 was assessed by deep sequencing, showing progressive enrichment of this clone. (B) Size exclusion chromatography analysis confirmed binding of purified recombinant Nb.b201 to HSA. (C) Crystal structure of nanobody:HSA complex (PDB ID 5VNW), including close-up view of CDR loops interacting with antigen. Hydrogen bonds are shown as dotted lines. (D) Rotated view of the complex (E) Comparison of nanobody structures in the antigen-bound (yellow) and antigen-free state (gray; PDB ID 5VNV), showing conformational change upon antigen binding. Changes are highlighted with arrows, and CDR loops are colored blue, green, and orange for CDRs 1, 2, and 3, respectively. See also Table 1 and Supplementary Figure 3.
As expected, Nb.b201 could be expressed and purified from E. coli using standard methods, with a yield of 26 mg of purified protein per liter of culture. We assessed HSA binding in vitro by analytical size exclusion chromatography (Figure 2B) and more quantitatively by surface plasmon resonance (SPR, Supplementary Figure 3). Both techniques confirmed specific binding of Nb.b201 to HSA. While the binding affinity was modest at 430 nM, Nb.b201 was highly specific, demonstrating no binding to the closely related mouse serum albumin (MSA, Supplementary Figure 3), and forming a stable complex with HSA as assessed by size exclusion chromatography (Figure 2B).
A major goal of our approach presented here is to facilitate structural biology efforts, so we assessed the structural basis for Nb.b201 binding to HSA. Nb.b201 crystallized readily, both in isolation and in complex with HSA. X-ray data collection yielded datasets to a resolution of 1.4 Å and 2.6 Å for the free nanobody and HSA complex, respectively (Table 1). These crystal structures revealed that Nb.b201 adopts the typical V-set immunoglobulin fold previously observed for animal-derived nanobodies (Figure 2C – E). Comparison of the two structures provides an opportunity to compare bound and free states of the same antibody fragment. Remarkably, Nb.b201 undergoes large-scale conformational changes upon antigen binding, resulting in the reorientation of the CDR3 backbone and amino acid side chains (Figure 2E). The mode of antigen recognition involves Nb.b201 binding its target largely through its CDR3 loop, with relatively few direct contacts between HSA and CDR1 or CDR2. Interestingly, the epitope recognized by Nb.b201 is a convex protrusion on the surface of HSA, in contrast to most nanobody epitopes, which are more typically concave. The interaction mode includes a mix of polar and non-polar interactions, with a recurring theme of Tyr-Glu and Tyr-Asp hydrogen bonds (Figure 2C). These results not only reveal the mechanisms of synthetic nanobody-antigen interaction, but also confirm that useful nanobodies for crystallographic applications can be obtained in a matter of 2 – 3 weeks without use of specialized equipment or large animal immunization.
Table 1.
Data collection and refinement statistics.
| Nb.b201 PDB: 5VNV |
Nb.b201 complex with HSA PDB: 5VNW |
|
|---|---|---|
| Data collection | ||
| Space group | I4122 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 106.8, 106.8, 52.2 | 66.2, 72.1, 108.5 |
| α, β, γ (°) | 90, 90, 90 | 96.2, 104.5, 104.1 |
| Resolution (Å) | 46.92-1.40 (1.48-1.40)a | 7.70 – 2.60 (2.76-2.60) |
| Rmeas(%) | 8.4 (98.4) | 8.7 (140.0) |
| <I/σ(I)> | 11.85 (1.20) | 8.99 (0.75) |
| CC1/2 | 99.8 (56.0) | 99.8 (48.7) |
| Completeness (%) | 97.3 (87.0) | 97.7 (94.4) |
| Multiplicity | 6.4 (5.2) | 3.5 (3.3) |
| Refinement | ||
| No. reflections | 29221 | 55812 |
| Rwork/Rfree (%) | 18.3/21.4 | 22.1/25.6 |
| No. atoms | ||
| Protein | 946 | 10994 |
| Ion/otherb | 39 | 77 |
| Water | 134 | 69 |
| B factors | ||
| HSA | 107.7 (Chain A), 125.3 (Chain B) | |
| Nb.b201 | 22.6 | 90.4 (Chain C), 125.3 (Chain D) |
| Solvent | 39.3 | 91.2 |
| Ramachandran statistics | ||
| Favored (%) | 97.5 | 97.4 |
| Allowed (%) | 2.5 | 2.6 |
| Disallowed (%) | 0 | 0 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.003 |
| Bond angles (°) | 0.99 | 0.500 |
Values in parentheses are for highest-resolution shell.
5VNV contains polyethylene glycol and formate ions; 5VNW contains glycerol and bound fatty acids.
Nanobodies targeting non-purified antigen
Selection of yeast or phage surface-displayed libraries is usually performed with purified and labeled protein samples. Due to the idiosyncratic nature of protein purification, it remains a critical bottleneck for protein engineering by surface display methods. This barrier proves especially steep for poorly expressed proteins, such as membrane proteins and extensively post-translationally modified protein hormones. To assess the versatility of our yeast-based nanobody discovery platform, we sought to develop methods enabling the use of non-purified antigens as selection reagents.
We focused our efforts on the metabolic hormone adiponectin, a complex protein with many post-translational modifications, which can only be expressed in eukaryotic cells, typically at low yields. Human adiponectin tagged with an amino-terminal FLAG epitope was expressed as a secreted protein in HEK293 cells, and the resulting medium containing a complex mixture of proteins was used as the selection antigen (Supplementary Figure 4A, B). Nanobody-expressing yeast were incubated with adiponectin-containing conditioned medium, washed, and then stained with fluorescently-labeled anti-FLAG M1 antibody. Adiponectin binding was confirmed for three unique clones by SPR, isolated after three rounds of MACS followed by fluorescence activated cell sorting (Figure S4C). Approaches like these may enable rapid reagent development for the large fraction of the human proteome that remains poorly characterized by biochemical methods.
Conformationally-selective GPCR-binding nanobodies
While nanobodies have contributed to a broad range of biological fields, they have had a uniquely transformative impact on our understanding of G protein-coupled receptor (GPCR) biology6. GPCRs are the largest family of transmembrane receptors in humans, playing essential roles in every aspect of physiology ranging from function of the central nervous system to regulation of metabolism and cardiovascular biology. Nanobodies have proven to be invaluable tools for detailed structural and biochemical characterization of GPCRs, enabling profound insights into receptor structure and function. As an example, a llama-derived nanobody, Nb80, that stabilizes the active conformation of the β2-adrenergic receptor (β2AR) enabled determination of the first X-ray crystal structure of the active conformation of a hormone-activated GPCR4. The conformational selectivity of Nb80 was also subsequently used to probe the localization of active β2AR in live cells, thereby revealing a new paradigm in intracellular GPCR signaling3. Another nanobody, Nb35, was instrumental in examining the structure and signaling of the heterotrimeric G protein Gs in complex with the β2AR, enabling determination of the first structure of a GPCR-G protein heterotrimer complex20. In recent years, the GPCR-binding nanobody repertoire has broadened to include a range GPCRs6. However, all GPCR-targeting nanobodies reported to date trace their origins to animal immunization – a technique which is slow, expensive, and very often unsuccessful.
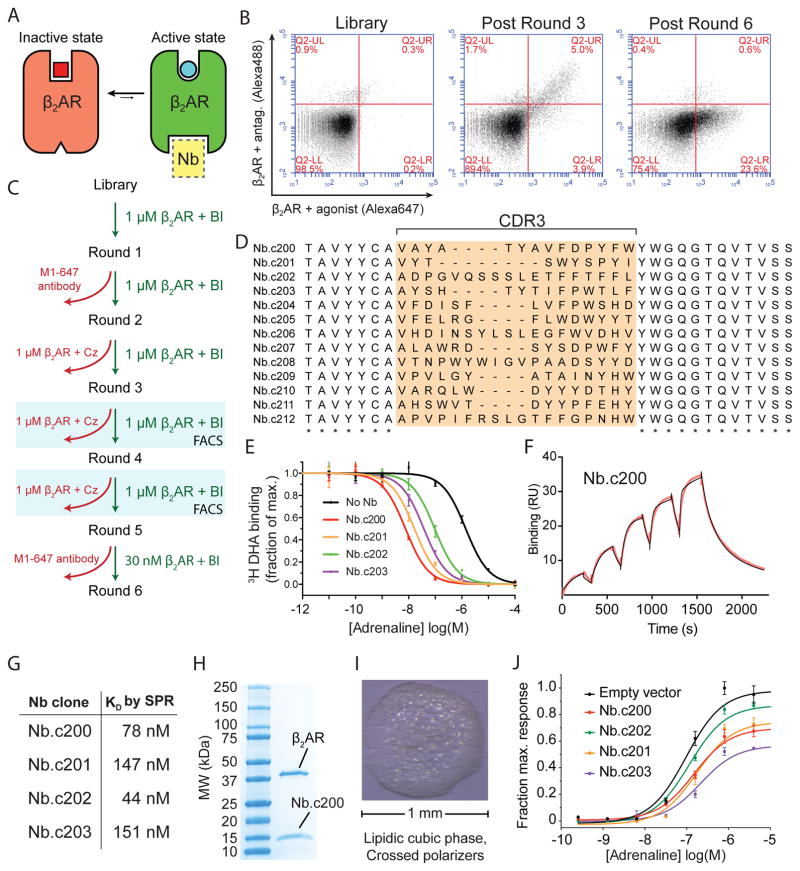
Given the broad utility of conformationally-selective antibodies in GPCR research, any in vitro platform aiming to supplant immunization-based methods must be able to generate conformationally selective GPCR binders. Hence, we sought to identify nanobodies that selectively bind the active conformation of β2AR using our synthetic nanobody library platform (Figure 3). Nanobody-displaying yeast were first selected for binding to purified, FLAG-tagged β2AR bound to the agonist BI167107 by MACS. We subsequently introduced a counterselection strategy to deplete undesired clones, which include nanobodies that bind the M1 antibody itself, the Alexa Fluor 647 fluorophore, conformationally invariant epitopes, or the inactive β2AR conformation. For example, in the third MACS round we first depleted clones that bound to β2AR occupied by the high affinity antagonist carazolol prior to enriching clones that bound agonist-occupied receptor. To enrich for the desired agonist-bound β2AR specific nanobodies, we performed two rounds of FACS. Here, the yeast library was incubated simultaneously with both agonist (BI167107) and antagonist (carazolol) occupied β2AR, with each receptor-ligand complex labeled with a specific Alexa fluorophore (Figure 3A – C). This specialized selection using FACS requires a cell-based display system. As an indication that the selected clones bind the desired epitope, we observed that a large fraction were competitive with Nb6B9, a previously affinity matured variant of Nb80 that binds with high affinity to the intracellular side of the β2AR21. Finally, we performed a round of MACS with decreased concentration of agonist-bound β2AR to enrich the highest affinity clones (Figure 3C).
Figure 3. Structural and functional modulator nanobodies targeting a GPCR.
(A) β2 adrenergic receptor interconverts between ensembles of inactive conformations (red) and active conformations (green). The active state of the receptor can be stabilized by nanobodies (yellow) that bind to the intracellular face of the receptor. (B) Results of selection summarized in flow cytometry plots. After FACS selection a significant fraction (23.6%) of clones show agonist-specific binding to the β2AR. (C) Selection schematic for isolation of active-state stabilizing nanobodies. In MACS rounds (1, 2, and 5), depletion and enrichment steps were performed sequentially. In FACS rounds, two-color sorting was used to enrich agonist-specific clones and deplete antagonist-specific and nonselective clones simultaneously. (D) Sequence analysis of β2AR conformationally selective clones, showing highly divergent CDR3 sequences. (E) Radioligand competition binding in nanodiscs confirms that synthetic nanobodies stabilize the active-state conformation β2AR. Data are shown as means +/− SEM for experiments performed in triplicate. (F) Single cycle SPR experiment showing measurement of affinity and kinetics for Nb.c200 binding to β2AR-BI167107. Experimental data are in red, and curve fit is in black. (G) Summary of nanobody affinities measured by surface plasmon resonance (see also Supplementary Figure 5). (H) Nb.c200 immobilized by metal ion affinity chromatography pulled down purified β2AR prior to crystallographic trials. (I) Nb.c200 enabled crystallization of the β2AR using the lipidic cubic phase method. (J) cAMP signaling assay to measure β2AR signaling in the presence or absence of synthetic nanobodies. Data are shown as mean +/− SEM for experiments performed in triplicate. See also Supplementary Figure 4.
Sequencing individual clones revealed that we isolated 13 unique nanobodies that bound agonist-occupied β2AR (Figure 3D; Supplementary Figure 5). We further prioritized clones with on-yeast titrations, which demonstrated that the isolated nanobodies have a range of affinities for the β2AR-BI167107 complex in the low to mid nanomolar range. We anticipate that conformationally-selective nanobodies identified from this library will find utility in biochemical, structural, and cell biological applications. Therefore, we next validated the active-state specific β2AR nanobodies in pharmacological, crystallographic, and cellular signaling experiments. Four clones were selected for further characterization and were expressed and purified from E. coli. Each of the four nanobodies increased agonist affinity of β2AR as assessed by a competition radioligand binding assay (Figure 3E). This gold-standard pharmacological assay provides definitive evidence that each nanobody stabilizes the active conformation of β2AR. Next, we determined binding affinities of each nanobody for BI167107-occupied β2AR by surface plasmon resonance (SPR) using single cycle kinetics (Figure 3F, G). Measured affinities ranged from 44 nM to 151 nM, comparing favorably to the most extensively studied llama-derived GPCR-targeting nanobody, Nb80, which binds with roughly 140 nM affinity21.
We next tested whether this conformational stabilization is sufficient to enable crystallization of an agonist-bound β2AR. Likely due to conformational dynamics in the agonist-bound state22, all previous attempts to crystallize β2AR-BI167107 complex have failed in the absence of an intracellular binding nanobody or the heterotrimeric G protein4,20,23. Due to the low throughput of crystallography experiments, we selected one nanobody, Nb.c200, for crystallization trials. The nanobody readily co-purified with β2AR (Figure 3H), and crystallized easily by the lipidic cubic phase method (Figure 3I). Of note, Nb.c200 enabled crystallization in a range of crystallization conditions rather than just a small subset.
Additionally, we assessed the potential of our synthetic nanobodies to modulate GPCR signaling in live cells, a key application of llama-derived nanobodies3,24. As with llama-derived nanobodies, we found that our synthetic nanobodies could significantly impair β2AR signaling in response to adrenaline. Among the nanobodies tested, we found that Nb.c203 was the most effective, reducing adrenaline Emax by 45% (Figure 3J). The observed variability in maximal signaling inhibition is likely reflective of both nanobody affinity and stability in the reducing environment of the cytosol. The effectiveness of Nb.c203, despite its lower affinity binding, highlights the importance of testing multiple unique clones for intrabody applications. Notably, the insurmountable inhibition observed in the presence of these nanobodies is consistent with a G protein-competitive binding mode for the nanobodies. Unlike an orthosteric inhibitor, adding excess agonist cannot overcome nanobody inhibition since the site of binding is distinct.
In order to further examine the versatility of our nanobody library, we sought to identify conformationally selective nanobodies recognizing another GPCR. As a test case, we chose the human A2A adenosine receptor (A2AR). This receptor has long been known to play important roles in the central nervous system, and has recently emerged as a target for cancer immunotherapy drug development25. Importantly, there are no active-state selective nanobodies targeting the A2AR, although an inactive-state specific Fab has been reported26. To generate active-state specific nanobody clones, we performed two rounds of MACS to generate a library of A2AR-binding nanobodies followed by one round of FACS to enrich conformationally selective binders (Figure 4A). From this pool, two clones called Nb.AD101 and Nb.AD102 were chosen for further characterization. In a flow cytometry-based binding assay, each of these clones showed a strong preference for agonist-bound A2AR (Figure 4B). To further validate these clones, we expressed and purified them and measured binding to receptor in an in vitro pull-down assay. Consistent with cell-based staining experiments, these nanobodies showed binding exclusively to agonist-occupied receptor (Figure 4C), confirming the generalizability of the approach.
Figure 4. Isolation and characterization of agonist-bound-A2AR specific nanobodies.
(A) FACS enrichment for conformationally selective nanobodies. (B) Staining and flow analysis of individual clones for specific recognition of agonist UK 432097-bound A2AR. Low frequency events in off-target quadrants likely arise from nonspecific binding and weak binding to basally active receptor molecules. (C) On-yeast binding of UK 432097- or antagonist ZM 241385-bound receptor for Nb.AD101 and Nb.AD102 expressing yeast. (D) Pulldown of purified Nb.AD101 (Lane 2) and Nb.AD102 (Lane 3) with UK 432097-bound A2AR or ZM 241385-bound A2AR (Lanes 4 and 5).
Taken together, these data show that conformationally-selective nanobodies identified by our in vitro platform recapitulate all key features of llama-derived clones. In addition, the large diversity of conformationally-selective clones identified from the synthetic library may enable a larger repertoire of applications; some may perform better as crystallographic chaperones, others may be more stable in the reducing intracellular environment, while others may have increased specificity required for identifying a specific protein in a complex milieu.
Discussion
Antibodies and their minimal binding fragments have transformed biomedical research. Nanobodies, derived from camelid heavy chain antibodies, have emerged as particularly powerful research tools in large part due to their ability to detect and stabilize specific conformations for GPCRs and other biochemically challenging proteins. However, despite their broad utility, nanobodies have only been exploited for a subset of potential applications due to challenges associated with nanobody discovery. Foremost among these is the requirement for camel, llama, or alpaca immunization – a slow and expensive process requiring large quantities of purified protein and access to large animal husbandry and veterinary facilities. Moreover, this process frequently yields only small numbers of nanobody clones, many or all of which may not possess the desired activity. Our approach reported here addresses these challenges by providing a fully in vitro platform for nanobody discovery based on an engineered library displayed on S. cerevisiae.
To assess the utility of our library, we selected nanobodies targeting a diverse array of antigens: soluble proteins, non-purified proteins, and two human GPCRs. In each case, our platform enabled identification of nanobody clones capable of binding to the target antigen with the desired activity. To date, all nanobody selection efforts conducted with this library have led to the successful isolation of clones with sub-micromolar affinity, with the exception of an effort targeting FLAG peptide, a short unstructured sequence. Strikingly, with the addition of a FACS enrichment to our selection methodology the affinities of our synthetic β2AR binding clones were comparable to or better than the most extensively studied GPCR-targeting nanobody, the llama-derived Nb80.
While there are many potential selection strategies that enable nanobody identification, we demonstrate that a combination of magnetic-bead and FACS enrichments provide a rapid approach to the identification of nanobodies selective for a particular target or to a specific conformation. Early MACS rounds enable depletion of large pools of non-binding clones, rapidly reducing library diversity by two to three orders of magnitude. However, MACS-based counter-selection strategies suffer from poor precision, and identification of clones specific for a target or conformational states often requires many iterations for sufficient enrichments. Given the throughput of modern FACS instruments, we opt to leverage the precision afforded by fluorescence-based selections as early in the selection scheme as possible, usually after two rounds of magnetic sorting to reduce library diversity to a manageable level. Though it is possible that this approach may miss the single highest affinity clone in the library, nanobodies are readily amenable to affinity maturation to improve affinity, even for difficult targets such as GPCRs21. Importantly, our library generates nanobody binders that are amenable to crystallization and subsequent structural determination and, thus, it demonstrates considerable promise as a tool to aid structural studies.
From soluble protein binders to conformationally-selective GPCR stabilizers, our platform allows straightforward, rapid, and low-cost isolation of nanobodies with even highly specialized functionalities. To facilitate broad deployment of this resource, we have made the library publicly available free of charge for non-profit use. Detailed protocols for nanobody binder selection are included in the Supplementary Note, and can be adapted or modified to enable more diverse applications. We envision that rapid and simple nanobody discovery will be an enabling resource for applications throughout biomedical research.
Methods
Cell lines
Expi293 cells (ThermoFisher Scientific) were used to create a stable cell line secreting FLAG-Adiponectin. Cells were maintained using a bioreactor (CELLine Flasks, Wheaton). The cell growth medium of the lower and upper chamber comprised Expi293 Expression Medium (Thermo Fisher) supplemented with 10% FBS (Zen-Bio), 10 μg/mL gentamicin (VWR) and FreeStyle 293 Expression Medium (Life Technologies), supplemented with 10 μg/mL gentamicin (VWR), respectively. Human β2AR and A2AR were purified from infected Sf9 cells grown in ESF 921 media (Expression Systems). HEK293T cells (ATCC) for β2AR signaling assays were grown at 37 °C and 5% CO2 in DMEM (+4.5 g/L glucose, +L-glutamine, no sodium pyruvate; VWR) supplemented with 10% fetal bovine serum (USA Scientific).
Nanobody library construction
The DNA library of nanobodies was constructed by two-step overlap extension PCR (OE-PCR). A set of ten primers; P1_for, P2_rev, …, P10_rev (Keck Biotechnology Resource Laboratory; Supplementary Table 2), were dissolved at 100 μM concentration and mixed in an equimolar ratio to prepare three mixed pools containing each primer at a concentration of 10 μM. The three mixed pools, ‘mix short’, ‘mix medium’, and ‘mix long’, differed in the P9 primers, using P9a_for, P9b_for, or P9c_for to introduce a CDR3 region of variable length; 7, 11, or 15 randomized residues respectively. 1 μL of each mixed pool at 10 μM concentration and at 5-fold sequential dilutions was subsequently used to prepare 50 μl OE-PCR reactions using Phusion polymerase. 0.4 μM of each primer, or a 25-fold diluted stock, was found to produce optimal yields for OE-PCR for each of the three mixed pools. The full length nanobody DNA product from each pool was mixed in an 1:2:1 molar ratio of short to medium to long CDR3 regions, referred to as the nanobody DNA library pool hereafter, recapitulating the length distribution frequencies observed in camelid VHH domains. Validation of the biochemical tractability of these synthetic nanobodies was carried out in Escherichia coli by amplifying the resulting mixture with primers pET26b_NbLib_GA_for and pET26b_NbLib_GA_rev and cloning into pET26b.
The nanobody DNA library pool was successively amplified for yeast transformations with pYDSFor1-pYDSRev1, pyDSFor2-pYDSRev2, and pYDSFor3-pYDSRev2 primers. 500 mL of BJ5465 yeast (MATa ura352 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL) were grown to OD600 1.5 and transformed with 245 μg nanobody insert DNA and 50 μg of pYDS649 plasmid, digested with NheI-HF and BamHI-HF (New England BioLabs), using an ECM 830 Electroporator (BTX-Harvard Apparatus). Dilutions of transformed yeast were then plated on –Trp dropout medium as single colonies to obtain an estimate of library diversity.
Three cultures of 5×105 yeast were inoculated in Trp dropout medium (US Biological) and grown overnight at 30 °C for whole library next generation sequencing reactions. From these cultures, PCRs were performed on 8×106 pelleted yeast to amplify nanobody genes using three sets of primers with different barcodes: Batch1For-Batch1Rev, Batch2For-Batch2Rev, and Batch3For- Batch3Rev. A sequencing library was prepared using PrepX (Wafergen) and sequencing was performed by a MiSeq (Illumina) with 10% PhiX. Next generation sequencing of HSA binding nanobody selection rounds was performed similarly starting with 2×105 yeast cells in the initial inoculum and using the primers: Rd1For-Rd1Rev, Rd2For-Rd2Rev, Rd3For-Rd3Rev, Rd4For-Rd4Rev.
Optimization of yeast display plasmid is summarized in Supplementary Figure 2. SIRPα mutant CV1 and CD47 proteins used for the assay were expressed and purified as described previously27. Analytical yeast staining experiments were performed as described below, using yeast strain BJ5465 as in other experiments.
Isolation of nanobody binders from library
Human serum albumin
Initially, 2.5 × 109 induced yeast for the first round and 1.4 – 5 × 107 induced yeast for subsequent rounds were washed and resuspended in buffer (20 mM HEPES pH 7.5, 150 mM sodium chloride, 0.1% (w/v) ovalbumin, 1 mM EDTA) and then incubated with anti-Alexa Fluor 647 or anti-FITC microbeads (Miltenyi) at 4 °C for 30 min. Each round of MACS selection began with a pre-clear step which involved passing the yeast, through an LD column (Miltenyi) to remove yeast expressing nanobodies that bound nonspecifically to magnetic beads. After pre-clearing, human serum albumin (HSA) binding nanobodies were enriched over four rounds of MACS selection by staining the yeast alternately with Alexa Fluor 647 or FITC labeled HSA (Sigma) and microbeads, then passing them through an LS column (Miltenyi). During these four selection rounds, yeast were stained with successively lower concentrations of HSA: 1 μM, 250 nM, 75 nM,and 15 nM, in order to enrich for binders with higher affinities. After MACS selection, yeast were plated as single colonies which were picked and grown as clonal populations in a 96 well plate. Following galactose induction of nanobodies, yeast were stained with Alexa Fluor 488 labeled HSA and analyzed by flow cytometry with an Accuri C6 (BD Biosciences) to screen for nanobody binders.
Human adiponectin
The adiponectin-containing supernatant taken from the bioreactor cell culture was used as the selection reagent, with no purification performed. An approximate concentration of secreted FLAG-adiponectin in the supernatant was estimated by dividing the average yield of pure adiponectin obtained for each harvest by volume of supernatant.
For the first round of selection, 5 × 109 induced yeast were washed and incubated in buffer (20 mM HEPES pH 7.5, 150 mM sodium chloride, 2 mM CaCl2, 0.1% BSA, 1.8% maltose) with Alexa647-labelled anti-FLAG M1 and anti-647 microbeads. Clones binding nonspecifically to the staining reagents were removed by passage through an LD column and the remaining yeast from the flow-through were incubated with unpurified FLAG-adiponectin at ~500 nM for 30 min at 4 °C. Cells were then stained with anti-FLAG-647 and anti-647 microbeads. Binding clones were enriched via magnetic selection in an LS column, and cultured overnight in –TRP medium at 30 °C. Rounds 2 and 3 of selection were performed similarly with 3 × 107 induced yeast and cells were washed, stained with 647- or FITC-labelled M1, and labelled with anti-Alexa Fluor 647 or anti-FITC microbeads before magnetic separation during rounds 2 and 3, respectively.
Following round 3, induced cells incubated with ~500 nM unpurified adiponectin and stained simultaneously with M1-647 and M1-FITC to differentiate dye-, antibody-, and adiponectin-binding clones. FACS was used to enrich for cells that exhibited adiponectin-dependent 647- and FITC staining. Nanobodies from individual clones were sequenced and purified and adiponectin binding was confirmed and characterized by SPR with purified adiponectin.
Human β2-adrenergic receptor
To isolate agonist-specific β2AR binding nanobodies, two rounds of MACS were performed with 1 μM purified, FLAG-tagged β2AR bound to the high affinity agonist BI167107 and FITC or Alexa Fluor 647 labeled anti-FLAG antibody in a selection buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl, 0.1% dodecylmaltoside (DDM, Anatrace), 0.01% CHS, 0.05% BSA, 5 mM CaCl2, and 10 mM maltose. For round 2 of selection the yeast library was precleared, as detailed above, with anti-FLAG antibody labeled with Alexa Fluor 647 to deplete antibody or fluorophore binders. Round 3 utilized MACS again, but counterselection was performed against β2AR bound to the high affinity antagonist carazolol to deplete nanobody clones that bind conformationally invariant or inactive-state epitopes. In subsequent rounds, we utilized FACS to more specifically select nanobodies that selectively bind β2AR in the active conformation. In round 4, yeast were simultaneously incubated with 1 μM β2AR-BI167107 complex labeled with Alexa Fluor 647 NHS ester and 1 μM β2AR-carazolol complex labeled with Alexa Fluor 488 NHS ester. Yeast displaying selectivity for agonist (high Alexa Fluor 647 signal and low Alexa Fluor 488 signal) were collected, expanded in growth media, induced and subjected to another round of FACS. The conformational selection procedure was repeated as in round 4, however the fluorophores coupled to agonist and antagonist were switched to deplete non-specific clones. A final MACS round was performed at 30 nM β2AR-BI167107 to isolate the highest affinity nanobodies. Nanobodies from individual clones were subsequently sequenced and purified for biochemical and biophysical studies.
Human adenosine A2A receptor
Two rounds of MACS were performed with 1.5 μM of FLAG-tagged A2AR bound to the agonist 5′-N-ethylcarboxamidoadenosine (NECA) and M1 anti-FLAG antibody labeled with FITC or 647 in a selection buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 0.1% MNG, 0.01% CHS, 2 mM CaCl2, and 0.2% maltose. Each round began with a pre-clearing step as detailed above. To isolate conformationally selective binders, the second round yeast were stained for 20 minutes simultaneously with 500 nM each of A2AR bound to the agonist UK 432097 and A2AR bound to the antagonist ZM 241385 which were labeled with secondary anti-FLAG M1 488 and anti-FLAG M1 647, respectively. Yeast that selectively bound receptor in complex with the agonist UK 432097 were isolated with FACS. Individual clones were grown and stained with both receptor populations to confirm binding selectivity and on-yeast binding titrations were performed to approximate binding affinities. Nanobodies were then purified and binding was confirmed in vitro with an anti-FLAG pulldown.
Protein expression and purification
The ADIPOQ gene was cloned into a pTARGET vector so that an HA signal peptide directs secretion of N-terminally FLAG-tagged adiponectin into the medium. Cells from the lower chamber of the bioreactor were harvested by centrifugation at 4000 g for 10 minutes. The supernatant was diluted 1:1 with buffer (20 mM HEPES pH 7.5, 150 mM sodium chloride, 2 mM CaCl2 2 mM) before loading onto FLAG resin. FLAG resin was first washed with 20 mM HEPES pH 7.5, 300 mM NaCl, 2mM CaCl2 and then with 20 mM HEPES pH 7.5, 150 mM NaCl, 2mM CaCl2. Adiponectin was eluted with buffer comprised of 150 mM NaCl, 20 mM Hepes pH 7.5, FLAG peptide 0.2 mg/mL, and 5mM EDTA and then dialyzed overnight into 20 mM HEPES pH 7.5, 150 mM sodium chloride.
Human β2AR fused to an amino-terminal hemagglutinin signal peptide and FLAG-tag as well as a carboxy-terminal 1D4 tag was purified from infected Sf9 cells. Cells were collected 64 hours after baculoviral infection and stored at −80 °C. For receptor preparations used during nanobody selections, purification was performed using ligand-affinity chromatography as described previously28. For biophysical assays, we used a simpler purification method omitting ligand-affinity chromatography. Cells were lysed in a buffer comprised of 20 mM HEPES pH 7.5, 2 mM MgCl2, 2 μl benzonase, and 1 μM ICI-118,551. The resulting lysate was centrifuged, and the pellets were resuspended in solubilization buffer containing 20 mM HEPES pH7.5, 200 mM NaCl, 10% glycerol, 1% - MNG, 0.1% CHS, and 1 μM ICI-118,551 followed by incubation at 4 °C for 2 h. The insoluble fraction was separated by centrifugation at 39,000x g for 20 min, and the supernatant was loaded over 5 mL of homemade anti-FLAG M1 resin. FLAG resin was subsequently washed with 20 mM HEPES pH 7.5, 200 mM NaCl, 10% glycerol, 0.1% MNG, and 0.01% CHS. β2AR was eluted in the same buffer with a lower concentration of detergent and CHS (0.01% and 0.001%, respectively) supplemented with 5 mM EDTA and 0.2 mg/mL FLAG peptide. Receptor was further purified by size exclusion chromatography in the presence of agonist BI167107. For crystallographic experiments, β2AR fused to an amino-terminal T4 lysozyme29 was similarly purified.
Isolated nanobody genes were cloned into the periplasmic expression vector pET26b containing a C-terminal 6×histidine tag and expressed from BL21 (DE3) E. coli. For each purification, E. coli were grown in Terrific Broth (Research Products International) and induced after reaching OD600 0.6–0.8 with 1 mM IPTG (Gold Biotechnology) at 25 °C for about 15 hours. Induced cells were pelleted and resuspended in 100 mL of buffer consisting of 0.5 M sucrose, 0.2 M Tris pH 8, 0.5 mM EDTA and osmotically shocked by adding 200 mL of water with 45 minutes of stirring, releasing periplasmic nanobody. The lysate was brought to a concentration of 150 mM NaCl, 2 mM MgCl2, and 20 mM imidazole and then centrifuged 20,000 × G for 20 min at 4 °C. The supernatant was applied to a gravity column with 3 mL of Ni Sepharose 6 Fast Flow (GE Healthcare). The resin was washed with a high salt buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 20 mM Imidazole) and then washed with NH7.5 buffer (20 mM HEPES pH7.5, 100 mM NaCl) + 20 mM imidazole and nanobodies were eluted with NH7.5 + 400 mM imidazole. Nanobodies used for crystallography were treated to carboxypeptidase A (Sigma) and B (Roche) to remove the histidine tag, dialyzed into HBS buffer (10 mM HEPES pH 7.4, 150 mM NaCl), and co-purified with their respective binding partners over an S200 size-exclusion column (GE Healthcare).
Human A2AR was fused at the N-terminus to an HA signal sequence followed by a FLAG epitope and at the C-terminus it was truncated at Ala317 with the insertion of a 9×His tag. The receptor was expressed in Sf9 insect cells following generation of BestBac Baculovirus (Expression Systems) mediated by the pVL1393 vector. Membranes from three 1 L cultures were prepared essentially as described previously30 and solubilized in 1% DDM, 0.1% CHS, 20 mM HEPES, pH 7.5, 800 mM NaCl, 10% glycerol, 4 mM theophylline, 5 μg/ml leupeptin, 5 μg/ml benzamidine, 2 mg/ml iodoacetamide, and 100 μM TCEP. Following centrifugation, the supernatant incubated overnight with Ni2+ chelating sepharose under rotation at 4 °C. The resin was transferred to a column and washed in 5 column volumes of ice-cold wash buffer (0.1% DDM, 0.01% CHS, 20 mM HEPES, pH 7.5, 800 mM NaCl, 10% glycerol, 4 mM theophylline, 5 μg/ml leupeptin, 5 μg/ml benzamidine) containing 25 mM imidazole and repeated with 50 mM imidazole. The A2AR was eluted in the same buffer with 200 mM imidazole added. A2AR was loaded onto a anti-FLAG M1 antibody column (Sigma) at room temperature in presence of 2 mM CaCl2. Theophylline was washed out in exchange for UK-432097 (30 μM) or ZM-241385 (30 μM) in buffer containing 0.1 % DDM, 0.01 % CHS, 20 mM HEPES, pH 7.5, 100 mM NaCl, 10% glycerol, and 2 mM CaCl2. The elution was performed at room temperature in the same buffer with the exclusion of CaCl2 and in presence of 5 mM EDTA and 0.2 mg/mL FLAG peptide. The purified receptor was dialyzed overnight at 4 °C into the same buffer used for anti-FLAG column washes excluding CaCl2. Following addition of 15 % glycerol 1 nmol aliquots of A2AR were flash frozen in liquid nitrogen.
Characterization of nanobody binding
Binding of Nb.b201 with HSA or MSA (Sigma) was qualitatively assessed by mixing the proteins in a 1.2:1 ratio, respectively, and separating through a Superdex 200 10/300 gel filtration column (GE Healthcare) in HBS buffer.
For SPR of Nb.b201, EZ-Link NHS-PEG4-Biotin (Thermo Fisher Scientific) labeled recombinant HSA from S. cerevisiae (HSAr; Sigma) or MSA were immobilized to a Series S Sensor Chip SA using a Biacore T200 (GE Healthcare). Dilutions of Nb.b201 in running buffer (10 mM HEPES pH 7.5, 150 mM NaCl, 0.03% Tween) were made and the sample was injected at a flow rate of 30 μl/minute with a contact time of 120 s and dissociation time of 300 s. For SPR of β2AR binding nanobodies, β2AR was labeled with NHS-PEG4-Biotin and immobilized on a Sensor Chip CAP (GE Healthcare). Dilutions of β2AR were made in running buffer (10 mM HEPES pH 7.5, 150 mM NaCl, 0.01% MNG, 0.001% CHS, 20 nM BI167107) and the sample was injected at a flow rate of 30 μl/min with a contact time of 240 s and dissociation time of 700 s.
For SPR of Nb.AQ103, EZ-Link NHS-PEG4-Biotin (Thermo Fisher Scientific) labeled FLAG-Adiponectin was immobilized to a Series S Sensor Chip SA using a Biacore T200 (GE Healthcare). Dilutions of Nb.AQ103 in running buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.05% Tween) were made and the sample was injected at a flow rate of 30 μl/minute with a contact time of 150 s and dissociation time of 500 s.
For SEC-MALS, a 1.4:1 molar mix of Nb.b201 and HSAr, as well as Nb.b201 and HSAr alone controls, were injected into an AdvanceBio 300 Å column (Agilent) using an Agilent 1260 Infinity Isocratic Liquid Chromatography System and characterized using a Wyatt DAWN HELEOS II Multi-Angle static Light Scattering detector.
For HIS-pull down assay, 5 μM Nb.AQ103 was incubated with Ni2+ resin at 4 °C for 1 h, then the excess was removed with wash with HBS and then incubated with adiponectin globular domain at 4 °C for 1 h. The reaction was washed with HBS and imidazole 20 mM and eluted with HBS in presence of 250 mM imidazole.
Validation of conformationally-selective β2AR nanobodies
For radioligand binding studies, β2AR was reconstituted into high-density lipoprotein (HDL) nanodiscs containing a 3:2 molar ratio of POPC/POPG as described previously31. Binding experiments were performed with 150 fmol β2AR. Saturation-binding was first performed using 3H-dihydroalprenolol (3H-DHA; PerkinElmer), yielding a KD of 0.7 nM. Competition binding experiments to measure adrenaline affinity in the presence and absence of nanobodies were carried out in a binding buffer comprised of 20 mM HEPES pH 7.5, 150 mM NaCl, 0.1% BSA, and 1 mM EDTA containing 2 nM 3H-DHA, 150 fmol β2AR, and a range of adrenaline concentrations 10−4 – 10−11 M. Nanobodies were used at a final concentration of 5 μM. Reactions were incubated for 1.5 hr at room temperature prior to rapid filtration with a Brandel 96-well harverster onto a filter pre-treated with 0.1% polyethyleneimine. Bound 3H-DHA was measured by liquid scintillation counting. Measurements were performed in triplicate and are expressed as mean +/− SEM.
β2AR cAMP signaling assay
β2AR signaling was measured using a transcriptional CRE-SEAP (secreted embryonic alkaline phosphatase) reporter to indirectly measure cAMP production32. Briefly, HEK293T cells were seeded at 3.3×104 cells/well in 96-well plates the day before transfection in 200 μL/well of DMEM. The following day, medium was aspirated from the cells and replaced with 50 μL of serum-free DMEM, and cells at 70% confluency were transfected in triplicate with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Each well was transfected with 10 ng of pcDNA3.1(+) encoding β2AR, 20 ng of CRE-SEAP reporter plasmid (BD Biosciences), and 20 ng of pEGFP-N1. Plasmids included conformationally-selective β2AR nanobodies (Nb.c200 - c203), a negative control nanobody (Nb.BV025), and the pEGFP-N1 empty vector. After six hours of incubation at 37 °C, the transfection mix was aspirated from the cells and 200 μL of serum-free DMEM was added with the indicated final concentrations of adrenaline. The cells were incubated at 37 °C for 48 hours, then at 70 °C for 2 hours. To determine SEAP activity, the substrate 4-methylumbelliferyl phosphate (Sigma Aldrich) was prepared at 1.2 mM in 2 M diethanolamine bicarbonate pH 10 and mixed with an equal volume of cell supernatant at room temperature for 10 minutes. Fluorescence was measured on an EnVision 2103 Multilabel Reader (Perkin Elmer) with an excitation wavelength of 360 nm and an emission wavelength of 449 nm. For each condition, the baseline fluorescence value was determined from cells not treated with adrenaline and was subtracted from every value in the data set. β2AR signaling was calculated as a fraction of the maximum observed response (pEGFP-N1 empty vector) and plotted using GraphPad Prism.
Protein crystallization
Nb.b201 for crystallographic study was purified from E. coli as described above. The Nb.b201 complex with HSA was prepared by mixing human serum albumin (Sigma) with Nb.b201 at a ratio of 1:1.15, followed by size exclusion chromatography purification. Purified complex was concentrated to 120 mg/mL and mixed in 200 nL + 200 nL drops with Morpheus HT-96 screen (Molecular Dimensions). Crystals were grown in a sitting drop format, and were obtained directly from the screen without further optimization. The precipitant solution consisted of 0.02 M each of 1,6-hexanediol, 1-butanol, 1,2-propanediol, 2-propanol, 1,4-butanediol, 1,3-propanediol; 100 mM Tris(base)/BICINE pH 8.5, 20% (v/v) PEG 500 MME, 10% w/v PEG 20,000. Crystals of Nb.b201 alone were obtained when the complex was mixed in a 1:1 drop with a solution comprised of 4.0 M Potassium formate, 0.1 M BIS-TRIS propane pH 9.0, 2% w/v Polyethylene glycol monomethyl ether 2,000. Crystals were soaked with 20% glycerol as a cryoprotectant prior to flash freezing in liquid nitrogen.
For β2AR/Nb.c200 crystallization, the two-syringe mixing method was used to reconstitute the sample in lipidic cubic phase33. Crystals were obtained from MemMeso LCP screen (Molecular Dimensions) conditions consisting of 100 mM MES pH 6, 100 mM NaCl, 100 mM MgCl2, and 40% PEG 200. Crystals were also obtained in a variety of additional conditions.
Data collection and structure refinement
Data collection was performed at Advanced Photon Source GM/CA beamlines 23ID-B and 23ID-D. Diffraction data were collected at an energy of 12 keV with 0.2 sec exposure per frame. Each frame covered a 0.2 degree oscillation, and beam intensity was attenuated 100 to 1000 fold depending on crystal size. The structure of Nb.b201 in isolation was solved by molecular replacement in Phaser using the structure of the β2 adrenergic receptor binding nanobody Nb80 as a search model (PDB ID: 3P0G)4. The structure of the HSA:Nb.b201 complex (PDB ID: 5VNW) was solved by molecular replacement using the structure of Nb.b201 (PDB ID: 5VNV) and the structure of HSA (PDB ID: 3JRY)34 as search models.
Structural refinement for Nb.b201 in isolation was performed by alternate manual building in Coot35 and reciprocal space refinement in Phenix 36. In the final stages, TLS refinement was used to model anisotropic B factors. Refinement for HSA:Nb.b201 complex was performed similarly, with the additional inclusion of non-crystallographic symmetry restraints in the first two rounds of refinement. As in the case of Nb.b201 in isolation, TLS refinement was used in final stages of refinement. Crystallographic data analysis was performed with xds and phenix.refine, using standard metrics to assess structure quality. Full details of crystallographic statistics are summarized in Table 1.
Data availability
Atomic coordinates and structure factors are deposited in the Protein Data Bank under PDB ID: 5VNV and PDB ID: 5VNW, for the structures of Nb.b201 alone and in complex with human serum albumin, respectively. All other data will be made available upon request.
Supplementary Material
Acknowledgments
Financial support for this work was provided by the Vallee Foundation (A.C.K.), the Smith Family Foundation (A.C.K.), National Institutes of Health grants 5DP5OD021345 (A.C.K.), 1DP5OD023048 (A.M.), and 1DP5OD023088 (A.M.R.), the Lundbeck Foundation (S.G.F.R.), and the Danish Independent Research Council (S.G.F.R.).
References
- 1.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 2.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 3.Irannejad R, et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staus DP, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manglik A, Kobilka BK, Steyaert J. Nanobodies to Study G Protein-Coupled Receptor Structure and Function. Annu Rev Pharmacol Toxicol. 2017;57:19–37. doi: 10.1146/annurev-pharmtox-010716-104710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moutel S, et al. NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife. 2016;5 doi: 10.7554/eLife.16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, Sidhu SS, Wells JA. Two-state selection of conformation-specific antibodies. Proc Natl Acad Sci U S A. 2009;106:3071–3076. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizk SS, et al. Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins. Nat Struct Mol Biol. 2011;18:437–442. doi: 10.1038/nsmb.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams JJ, Sidhu SS. Synthetic antibody technologies. Curr Opin Struct Biol. 2014;24:1–9. doi: 10.1016/j.sbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Kayushin A, Korosteleva M, Miroshnikov A. Large-scale solid-phase preparation of 3′-unprotected trinucleotide phosphotriesters--precursors for synthesis of trinucleotide phosphoramidites. Nucleosides Nucleotides Nucleic Acids. 2000;19:1967–1976. doi: 10.1080/15257770008045471. [DOI] [PubMed] [Google Scholar]
- 12.Kayushin AL, et al. A convenient approach to the synthesis of trinucleotide phosphoramidites--synthons for the generation of oligonucleotide/peptide libraries. Nucleic Acids Res. 1996;24:3748–3755. doi: 10.1093/nar/24.19.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 14.Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng. 2009;103:1192–1201. doi: 10.1002/bit.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orlean P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics. 2012;192:775–818. doi: 10.1534/genetics.112.144485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makrides SC, et al. Extended in vivo half-life of human soluble complement receptor type 1 fused to a serum albumin-binding receptor. J Pharmacol Exp Ther. 1996;277:534–542. [PubMed] [Google Scholar]
- 17.Van Roy M, et al. The preclinical pharmacology of the high affinity anti-IL-6R Nanobody(R) ALX-0061 supports its clinical development in rheumatoid arthritis. Arthritis Res Ther. 2015;17:135. doi: 10.1186/s13075-015-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tijink BM, et al. Improved tumor targeting of anti-epidermal growth factor receptor Nanobodies through albumin binding: taking advantage of modular Nanobody technology. Mol Cancer Ther. 2008;7:2288–2297. doi: 10.1158/1535-7163.MCT-07-2384. [DOI] [PubMed] [Google Scholar]
- 19.Kim CC, Wilson EB, DeRisi JL. Improved methods for magnetic purification of malaria parasites and haemozoin. Malar J. 2010;9:17. doi: 10.1186/1475-2875-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ring AM, et al. Adrenaline-activated structure of beta2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manglik A, Kobilka B. The role of protein dynamics in GPCR function: insights from the beta2AR and rhodopsin. Curr Opin Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbaum DM, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staus DP, et al. Regulation of beta2-adrenergic receptor function by conformationally selective single-domain intrabodies. Mol Pharmacol. 2014;85:472–481. doi: 10.1124/mol.113.089516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijayan D, Young A, Teng MWL, Smyth MJ. Targeting immunosuppressive adenosine in cancer. Nat Rev Cancer. 2017 doi: 10.1038/nrc.2017.86. [DOI] [PubMed] [Google Scholar]
- 26.Hino T, et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiskopf K, et al. Engineered SIRPalpha variants as immunotherapeutic adjuvants to anticancer antibodies. Science. 2013;341:88–91. doi: 10.1126/science.1238856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manglik A, et al. Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou Y, Weis WI, Kobilka BK. N-terminal T4 lysozyme fusion facilitates crystallization of a G protein coupled receptor. PLoS One. 2012;7:e46039. doi: 10.1371/journal.pone.0046039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- 33.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protocols. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hein KL, et al. Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin. J Struct Biol. 2010;171:353–360. doi: 10.1016/j.jsb.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/s0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors are deposited in the Protein Data Bank under PDB ID: 5VNV and PDB ID: 5VNW, for the structures of Nb.b201 alone and in complex with human serum albumin, respectively. All other data will be made available upon request.