Introductory paragraph
The adoptive transfer of anti-CD19 chimeric antigen receptor (CAR)-engineered T cells has shown impressive clinical responses in patients with refractory B-cell malignancies1–7. However, the therapeutic effects of CAR-T cells targeting other malignancies have not yet resulted in significant clinical benefit8–11. Although inefficient tumor trafficking and various immunosuppressive mechanisms can impede CAR-T cell effector responses, the signals delivered by the current CAR constructs may still be insufficient to fully activate antitumor T cell functions. Optimal T cell activation and proliferation requires multiple signals, including T cell receptor (TCR) engagement (signal 1), costimulation (signal 2), and cytokine engagement (signal 3)12. However, CAR gene constructs currently being tested in the clinic contain a CD3z (TCR signaling) domain and a costimulatory domain(s) but not a domain transmitting signal 313–18. Here, we have developed a novel CAR construct capable of inducing cytokine signaling upon antigen stimulation. This new generation CD19 CAR encodes a truncated cytoplasmic domain of IL-2Rβ and a STAT3-binding YXXQ motif together with CD3z and CD28 domains (28-ΔIL2RB-z (YXXQ)). The 28-ΔIL2RB-z (YXXQ) CAR-T cells showed antigen-dependent JAK-STAT3/5 pathway activation, which promoted their proliferation and prevented terminal differentiation in vitro. The 28-ΔIL2RB-z (YXXQ) CAR-T cells demonstrated superior in vivo persistence and antitumor effects in both liquid and solid tumor models compared with CAR-T cells with a CD28 or 4-1BB domain alone. Taken together, these results suggest that our new generation CAR has the potential to demonstrate superior antitumor effects with minimal toxicities in the clinic. Clinical translation of this novel CAR is warranted.
Main text
Cytokines sharing common γ chains as their receptors have a fundamental effect on T cell immunity mainly through JAK-STAT pathway activation19. Whereas IL-2, IL-7, and IL-15 predominantly induce STAT5 activation through tyrosine residues within the common γ chain, IL-2 receptor β (IL-2/IL-15), or IL-7 receptor α (IL-7), IL-21 preferentially activates STAT3 through its association motif YXXQ within the IL-21 receptor20. In addition to its critical role in memory formation and effector differentiation, IL-21 acts synergistically with other cytokines to promote T cell proliferation21–23. Indeed, the forced expression of cytokine genes in CAR-T cells improves their persistence and antitumor effects in vivo, highlighting the importance of signal 3 for CAR-T cell functions24–26. However, constitutive cytokine expression poses a risk for autonomous T cell growth, potentially causing cancers, as well as serious adverse events such as cytokine release syndrome (CRS) and neurotoxicity by CAR-T cells with excessive growth potential 27,28. In this study, we aimed to determine whether the delivery of cytokine signaling only upon antigen engagement can enhance antitumor effects of CAR-T cells.
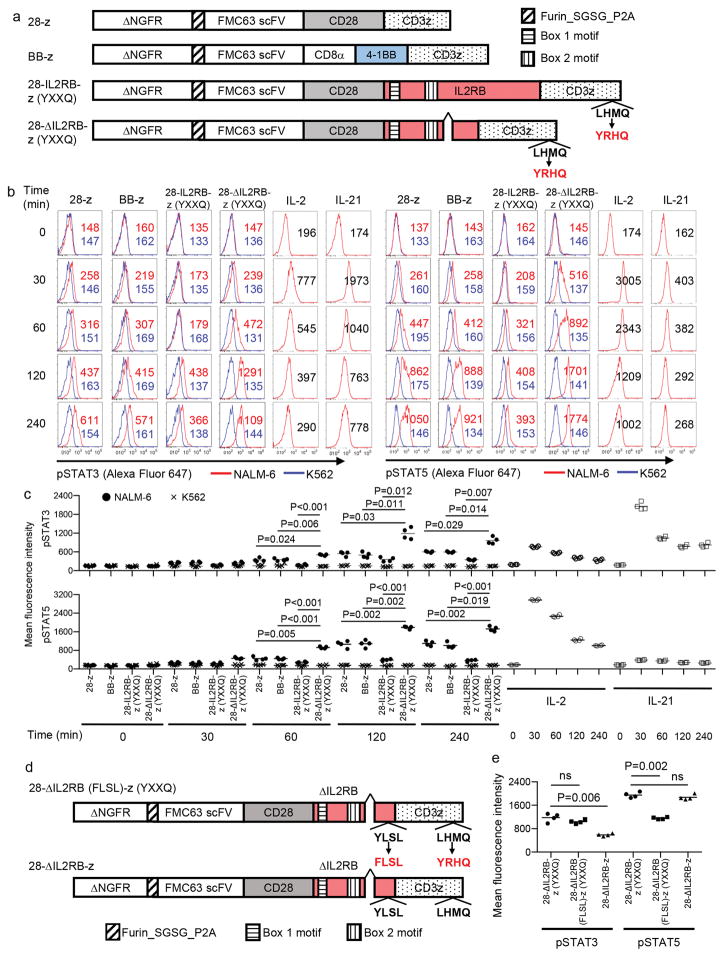
To induce JAK-STAT pathway activation in CAR-T cells in an antigen-dependent manner, we first incorporated the full-length or truncated cytoplasmic domain of IL-2 receptor β between the cytoplasmic domains of CD28 and CD3z (Fig. 1a and Supplementary Table 1). Second, we added a YXXQ motif at the C-terminus of CD3z for STAT3 recruitment [28-IL2RB-z (YXXQ) or 28-ΔIL2RB-z (YXXQ)]29. We also generated CAR constructs with CD28/CD3z (28-z) and 4-1BB/CD3z (BB-z) for comparison. An FMC63-derived single-chain variable fragment (scFv) targeting CD19 was used for antigen recognition30. All CAR constructs were N-terminally linked to truncated nerve growth factor receptor (ΔNGFR) via a P2A sequence. The 28-ΔIL2RB-z (YXXQ), but not the 28-IL2RB-z (YXXQ), CAR was efficiently expressed on the cell surface, specifically in the ΔNGFR+ T cell population, and transmitted T cell activation signals comparable to the 28-z and BB-z CARs upon encountering antigen, as demonstrated by CD69 upregulation and ERK and Akt phosphorylation (Supplementary Fig. 1, a and b and Supplementary Fig. 2, a-c). The 28-ΔIL2RB-z (YXXQ) CAR-T cells showed a significant upregulation of phospho-STAT3 (pSTAT3) and phospho-STAT5 (pSTAT5) in an antigen-dependent manner (Fig. 1, b and c and Supplementary Fig. 3, a and b). Although STAT3/5 phosphorylation was also observed in CAR-negative T cells after antigen stimulation, 28-ΔIL2RB-z (YXXQ) CAR triggered higher levels of pSTAT3/5 in CAR-positive cells, suggesting that the phosphorylation event was likely induced by cell-intrinsic signaling (Supplementary Fig. 4, a-d). Replacement of tyrosine by phenylalanine within the STAT5 association motif YLSL of the IL-2RB domain (28-ΔIL2RB (FLSL)-z (YXXQ)) significantly reduced pSTAT5 levels, and the deletion of the YXXQ motif at the C-terminus (28-ΔIL2RB-z) decreased pSTAT3 levels (Fig. 1, d and e and Supplementary Fig. 5, a-c). These results suggest the YLSL and YXXQ motifs mainly contributed to the enhanced pSTAT5 and pSTAT3 levels, respectively. Based on these data, we further analyzed the functional attributes of the 28-ΔIL2RB-z (YXXQ) CAR-T cells.
Fig. 1. Generation of the anti-CD19 chimeric antigen receptor (CAR) constructs to induce JAK-STAT pathway activation.
(a) A schematic diagram of the anti-CD19 CAR constructs used in this study. An FMC63-derived single-chain variable fragment (scFv) is linked to CD28 and CD3z (28-z); 4-1BB and CD3z (BB-z); and CD28, the cytoplasmic domain of the IL-2 receptor β chain with or without an internal deletion and CD3z with the YXXQ motif (28-IL2RB-z (YXXQ) and 28-ΔIL2RB-z (YXXQ)). All CAR-genes were linked to truncated nerve growth factor receptor (ΔNGFR) via a P2A sequence. (b, c) The indicated CAR-transduced T cells were rested in cytokine-free media overnight and stimulated with the CD19+ acute lymphoblastic leukemia cell line NALM-6 or the CD19− cell line K562 at an effector to target (E:T) ratio of 1:1. T cells mock-transduced and treated with IL-2 (300 IU/ml) or IL-21 (50 ng/ml) were used as a control. The phosphorylation of STAT3 and STAT5 within the CD8+ ΔNGFR+ T cell population was analyzed by flow cytometry at variable time points. The data shown are representative FACS plots (b) and the mean fluorescence intensity of the phosphorylated STAT3 and STAT5 (c, n=4 different donor samples; repeated measures one-way ANOVA with Tukey’s multiple comparisons test for each time point; F=63.57 (pSTAT3 at 60 min), 39.49 (pSTAT3 at 120 min), 71.41 (pSTAT3 at 240 min), 103.8 (pSTAT5 at 60 min), 201 (pSTAT5 at 120 min), 113.7 (pSTAT5 at 240 min); degree of freedom=15). (d) CAR constructs with either STAT3 or STAT5 binding domain alone. For the 28-ΔIL2RB (FLSL)-z (YXXQ) CAR, tyrosine within the YLSL motif of the IL-2 receptor β was replaced by phenylalanine to abrogate STAT5 association and the YXXQ motif was introduced by site-directed mutagenesis to the C-terminus end of CD3z (e) T cells transduced with the indicated CAR constructs were analyzed for phosphorylation of STAT3 and STAT5 120 minutes after stimulation with NALM-6 (e, n=4 different donor samples; repeated measures one-way ANOVA with Tukey’s multiple comparisons test; F=75.44 for pSTAT3, F=99.46 pSTAT5; degree of freedom=11). In c and e, horizontal lines denote mean values. ns, not significant.
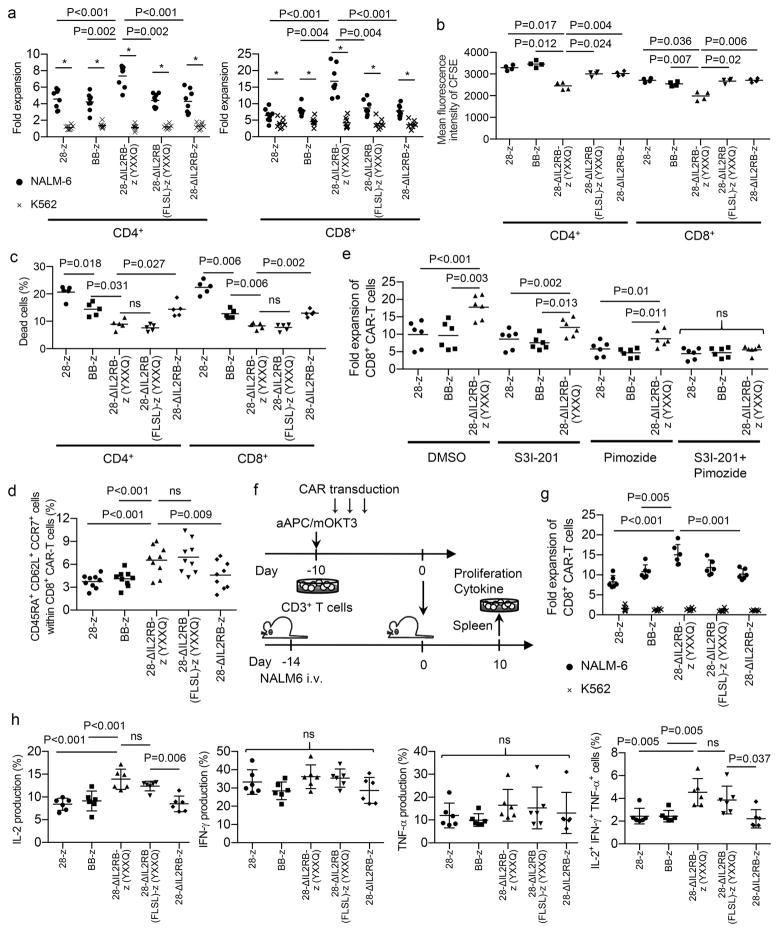
Upon in vitro antigen stimulation, the 28-ΔIL2RB-z (YXXQ) CAR-T cells achieved significantly better proliferation compared with the 28-z and BB-z CAR-T cells, regardless of cytokine supplementation, which resulted from both more rapid cellular division and less activation-induced cell death (Fig. 2, a–c and Supplementary Fig. 6, a and b). Both STAT3 and STAT5 domains were required to promote CAR-T cell proliferation. Intriguingly, the CAR-T cells with the STAT3 association motif maintained the CD8+ CD45RA+ CD62L+ CCR7+ population significantly better than the other CAR-T cells (Fig. 2d and Supplementary Fig. 7). T cells within this population mostly expressed CD27, CD28 and CD95, corresponding to a marker phenotype of stem cell-like memory T cells31. Consistent with these results, co-treatment with the specific STAT3 inhibitor S3I-201 and the STAT5 inhibitor pimozide abrogated the proliferative advantage of the 28-ΔIL2RB-z (YXXQ) CAR-T cells, and inhibition of STAT3 signaling in the 28-ΔIL2RB-z (YXXQ) CAR-T cells decreased the CD45RA+ CD62L+ CCR7+ population (Fig. 2e and Supplementary Fig. 8, a-c). Although STAT3 activation can promote PD-L1 expression in several types of tumor cells such as lymphoma and lung cancer32,33, JAK-STAT pathway activation did not have additive effects beyond antigen stimulation on the upregulation of PD-L1 or other immunoinhibitory molecules in antigen-stimulated CAR-T cells (Supplementary Fig. 9). After repeated stimulations, all CAR-T cells showed reduced proliferation and cytokine production and upregulated certain exhaustion markers (Supplementary Fig. 10 and 11). These results suggest that retrovirally transduced CAR-T cells undergo functional impairment accompanied by chronic antigen exposure, as reported previously34,35. 28-z CAR-T cells showed significantly decreased proliferation and increased expression of PD-1, LAG-3 and TIM-3 compared with the BB-z and 28-ΔIL2RB-z (YXXQ) CAR-T cells. 28-ΔIL2RB-z (YXXQ) or 28-ΔIL2RB (FLSL)-z (YXXQ) CAR-transduced CD8+ T cells maintained better proliferation, IL-2 secretion and cytokine polyfunctionality than other CAR-T cells. These attributes have been described in less differentiated memory T cells36,37. To compare CAR-T cell functions after exposure to the antigen in vivo, CAR-T cells were transplanted into NALM6-bearing NOD-scid IL2rγnull (NSG) mice (Fig. 2f). Persisting CAR-T cells were isolated from the spleen and analyzed for proliferative capacity and cytokine secretion ex vivo. Similar to the in vitro data, the 28-ΔIL2RB-z (YXXQ) CAR-T cells showed better proliferation and cytokine polyfunctionality than the 28-z and BB-z CAR-T cells (Fig. 2, g and h). These results suggest a key role of STAT3 in suppressing terminal differentiation of T cells, which is consistent with recent human and mouse studies38,39.
Fig. 2. The 28-ΔIL2RB-z (YXXQ) CAR-T cells show a superior proliferative capacity and maintain less differentiated memory T cell phenotypes following the antigen stimulation.
(a) The fold expansion of the CD4+ and CD8+ CAR-T cells was calculated 7 days after stimulation with NALM-6 or K562 (n=8; repeated measures one-way ANOVA with Tukey’s multiple comparisons test for the NALM-6 data, F=20.12 for CD4+ T cells, F=36.57 for CD8+ T cells, degree of freedom=39; paired t-test for comparison between the NALM-6 and K562 data in individual CAR-T cells, t=10.34 (CD4+-28-z), t=8.16 (CD4+-BB-z), t=13.98 (CD4+-28-ΔIL2RB-z (YXXQ)), t=10.26 (CD4+-28-ΔIL2RB (FLSL)-z (YXXQ)), t=6.33 (CD4+-28-ΔIL2RB-z), t=7.47 (CD8+-28-z), t=10.57 (CD8+-BB-z), t=9.88 (CD8+-28-ΔIL2RB-z (YXXQ)), t=12.63 (CD8+-28-ΔIL2RB (FLSL)-z (YXXQ)), t=7.44 (CD8+-28-ΔIL2RB-z), degree of freedom=7). (b) CAR-T cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) and stimulated with NALM-6. The mean fluorescence intensity of CFSE was analyzed three days following the stimulation (n=4; repeated measures one-way ANOVA with Tukey’s multiple comparisons test; F=36.84 for CD4+ T cells, F=38.44 for CD8+ T cells; degree of freedom=19). (c) The frequency of dead cells within CD4+ or CD8+ CAR-T cell population was analyzed by flow cytometry 24 hours after stimulation with NALM-6 (n=4; repeated measures one-way ANOVA with Tukey’s multiple comparisons test; F=53.83 for CD4+ T cells, F=114.1 for CD8+ T cells; degree of freedom=24). (d) The frequency of CD45RA+ CD62L+ CCR7+ cells in the CD8+ CAR-T cell population 7 days after stimulation with NALM-6 (n=9; repeated measures one-way ANOVA with Tukey’s multiple comparisons test; F=14.35; degree of freedom=44). (e) CAR-transduced T cells were stimulated with NALM-6 and cultured with or without 25 μM of S3I-201 (STAT3 inhibitor) and/or 5 μM of pimozide (STAT5 inhibitor) for 3 days. The fold expansion of the CD8+ CAR-T cells was analyzed on day 7 (n=6, repeated measures one-way ANOVA with Tukey’s multiple comparisons test for each condition; F=46.52 for DMSO, F=15.67 for S3I-201, F=19.17 for pimozide, F=1.85 for S3I-201+pimozide; degree of freedom=17). In a–e, data were collected from different donor samples. Horizontal lines indicate mean values. (f–h) NOD-scid IL2rγnull (NSG) mice were intravenously infused with NALM-6 and subsequently transplanted with CAR-T cells. The spleen cells isolated from the mice were restimulated with NALM-6 (●) or K562 (x), and the fold expansion after 7 days of culture (g) and cytokine production (h) of CD8+ CAR-T cells were analyzed. The data shown are the sum of two independent experiments (n=6 mice for each group, ordinary one-way ANOVA with Tukey’s multiple comparisons test; F=10.85 for fold expansion, F=12.42 for IL-2, F=2.17 for IFN-γ, F=0.81 for TNF-α, F=7.51 for IL-2+ IFN-γ+ TNF-α+ cells, degree of freedom=29). In g and h, horizontal lines show mean values ± s.d. ns, not significant.
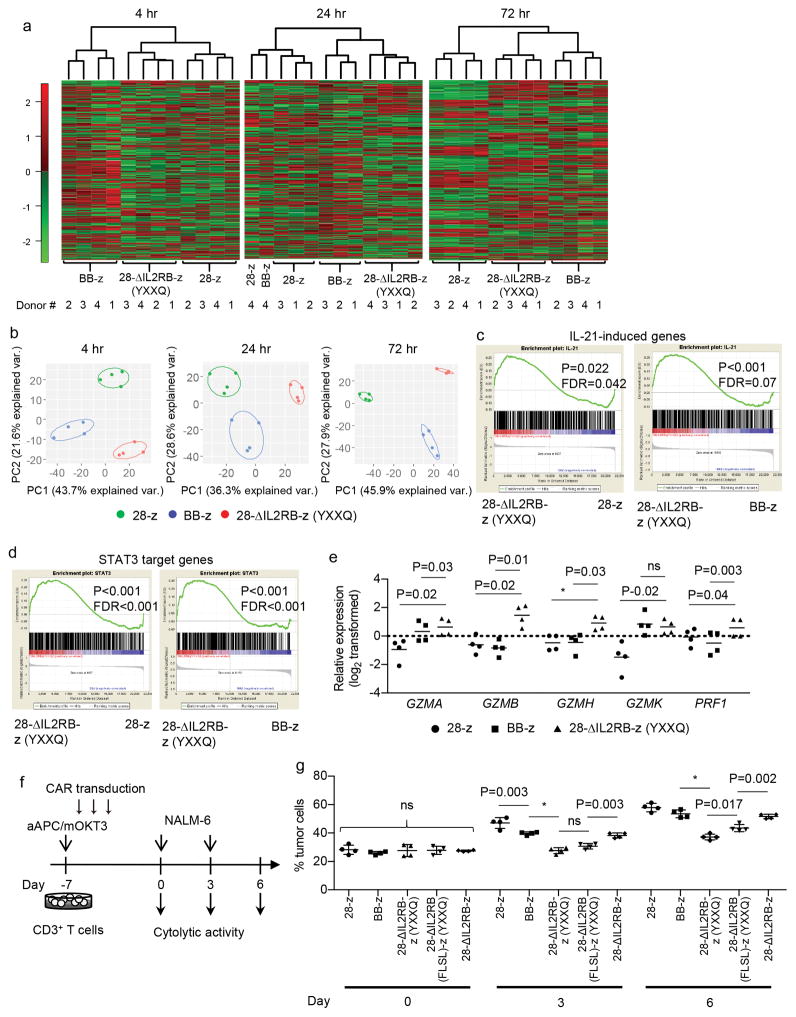
To assess the genetic mechanisms underlying the functional properties of the 28-ΔIL2RB-z (YXXQ) CAR-T cells, we compared the gene expression profiles of CD8+ CAR-T cells at 4, 24, and 72 hours following antigen stimulation in vitro. The number of differentially expressed genes among the 28-z, BB-z, and 28-ΔIL2RB-z (YXXQ) CAR-T cells progressively increased over time (Supplementary Table 2). Unsupervised hierarchical clustering and principal component analysis of those genes mostly classified different CAR-T cells into independent subclusters (Fig. 3, a and b). We further investigated whether the 28-ΔIL2RB-z (YXXQ) CAR induced gene expression profiles reflecting JAK-STAT pathway activation. Gene set enrichment analysis (GSEA) revealed significant enrichment of genes induced by IL-21, but not by IL-2, 7 and 15, in the 28-ΔIL2RB-z (YXXQ) CAR-T cells at 24 hours after stimulation (Fig. 3c, Supplementary Fig. 12a). Moreover, the 28-ΔIL2RB-z (YXXQ) CAR-T cells were enriched for expression of previously identified STAT3 target genes (Fig. 3d)40, further corroborating the preferential STAT3 pathway activation by the 28-ΔIL2RB-z (YXXQ) CAR. These profiles were not evident at 4 hours and were maintained at 72 hours after activation (Supplementary Fig. 12, b and c). Interestingly, the 28-IL2RB-z (YXXQ) CAR-T cells also showed an increased expression of multiple genes encoding effector molecules such as GZMA, GZMB, GZMH, GZMK and PRF1, which were validated by qPCR analysis (Fig. 3e). We then evaluated the cytolytic efficacy of the 28-ΔIL2RB-z (YXXQ) CAR-T cells in comparison with the other CAR-T cells. All CAR-T cells similarly demonstrated potent in vitro cytotoxicity against CD19+ cells (Supplementary Fig. 13). However, the 28-ΔIL2RB-z (YXXQ) CAR-T cells showed superior cytolytic activity following repeated antigen exposures, which progressively attenuated the effector functions of the other CAR-T cells as previously reported (Fig. 3, f and g)35.
Fig. 3. The 28-ΔIL2RB-z (YXXQ) CAR-T cells represent unique gene expression profiles and show a potent cytotoxic activity after the repetitive antigen stimulations.
(a, b) The 28-z, BB-z and 28-ΔIL2RB-z (YXXQ) CAR-transduced CD8+ T cells were stimulated with NALM-6, and their gene expression profiles were compared by microarray analysis at 4, 24 and 72 hours after stimulation (n=4 different donor samples). The data shown are unsupervised hierarchical clustering (a) and principal component analysis (b) of differentially expressed genes (raw P values<0.01 with repeated measures one-way ANOVA). (c, d) Gene set enrichment analysis (GSEA) on the expression profiles of the 28-ΔIL2RB-z (YXXQ) CAR-T cells compared with the 28-z or BB-z CAR-T cells using genes induced by the IL-21 treatment (c) or STAT3 target genes (d) as gene sets. Nominal p values and false discovery rates (FDR) to adjust for multiple hypothesis testing are presented. (e) Relative expression of the genes associated with cytolytic activity was analyzed by qPCR in the CAR-T cells stimulated for 24 hours. Expression levels were normalized to the UBC expression (n=4; log2-transformed values were compared with repeated measures one-way ANOVA with Tukey’s multiple comparisons test; F=24.52 for GZMA, F=43.68 for GZMB, F=32.5 for GZMH, F=43.43 for GZMK, F=32.54 for PRF1; degree of freedom=11). Horizontal lines denote the mean values. (f, g) The CAR-transduced T cells were repeatedly stimulated with NALM-6 at an effector to target (E:T) ratio of 1:3 (f). The cytotoxic activity against NALM-6 on days 0, 3, and 6 was evaluated by flow cytometry. The data shown are four technical replicates for individual CAR-T cells with mean ± s.d. (ordinary one-way ANOVA with Tukey’s multiple comparisons test for each round of stimulation; F=0.41 for Day 0, F=44.94 for Day 3, F=46.75 for Day 6; degree of freedom=19). Similar results were obtained in a repeat experiment. * P<0.001. ns, not significant.
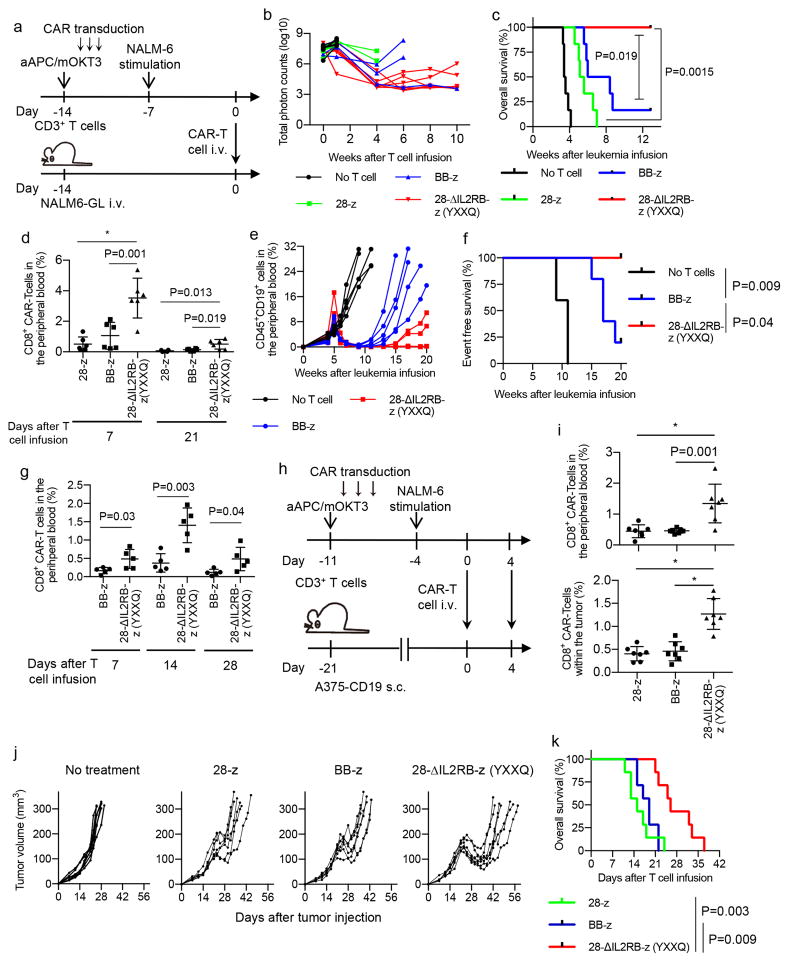
Next, we studied the antileukemic potency of the 28-ΔIL2RB-z (YXXQ) CAR-engineered T cells in vivo. NSG mice were treated with 5×106 CAR-T cells on day 14 after injection with 5×106 NALM6 cells transduced with the EGFP-firefly luciferase fusion gene (NALM6-GL) (Fig. 4a). The 28-ΔIL2RB-z (YXXQ)-transduced T cells efficiently suppressed leukemia progression compared with the 28-z and BB-z CAR-T cells, resulting in a significantly longer overall survival of the treated mice (Fig. 4, b and c and Supplementary Fig. 14). Importantly, the 28-ΔIL2RB-z (YXXQ) CAR-T cells showed significantly better persistence in the peripheral blood (Fig. 4d). Two infusions of CAR-T cells (5×106 cells per infusion, separated by two days—AU separated by how many days apart?) resulted in better control of leukemia and higher levels of secreted IL-2, IFN-γ, and TNF-α with all tested CAR constructs, although the 28-ΔIL2RB-z (YXXQ) CAR-T cells showed better persistence than the other CAR-T cells (Supplementary Fig. 15, a–g). IL-6 was below the limits of sensitivity and not detectable in this model (data not shown). Human T cells transplanted into NSG mice cause xenogeneic graft-versus-host disease (GVHD) by recognizing xenogeneic antigens through endogenous TCR engagement. The persisting CAR-positive and -negative T cells induced GVHD in some mice after leukemia eradication (Supplementary Fig. 16, a and b and Supplementary Table 3). The 28-ΔIL2RB-z (YXXQ) CAR-T cells did not increase the development of GVHD compared with the 28-z and BB-z CAR-T cells. Indeed, all CAR-T cells induced similar progressive weight loss with comparable latency when infused into tumor-free mice (Supplementary Fig. 17a). There was no significant difference in the development of lethal GVHD among 28-z, BB-z, and 28-ΔIL2RB-z (YXXQ) CAR-T cells (Supplementary Fig. 17, b and c). These results suggest that 28-ΔIL2RB-z (YXXQ) CAR does not affect endogenous TCR-mediated toxicity.
Fig. 4. T cells transduced with the 28-ΔIL2RB-z (YXXQ) CAR have superior antitumor effects in vivo.
(a–d) NOD-scid IL2rγnull (NSG) mice were intravenously infused with the NALM-6 transduced with EGFP-luciferase (NALM6-GL) (day −14) and were transplanted with 5×106 CAR-T cells (day 0) (a). (b) The quantified total photon counts analyzed by in vivo bioluminescent imaging of the luciferase activity (n=6 mice for each group). (c, d) Kaplan–Meier curve for the overall survival of the mice (c, n=6; log-rank test; p values were adjusted with Bonferroni correction) and the frequency of CD8+ CAR-T cells in the peripheral blood (d, n=4 for 28-z on day 21, n=6 for the other groups; ordinary one-way ANOVA with Tukey’s multiple comparisons test; F=17.19, degree of freedom=17 for Day 7; F=7.42, degree of freedom=15 for Day 21). Representative data of two experiments are shown. (e–g) NSG mice were intravenously infused with the CD19+ primary acute lymphoblastic leukemia (ALL) cells (day −35) and adoptively transferred with 5×105 BB-z or 28-ΔIL2RB-z (YXXQ) CAR-T cells (day 0). The serial monitoring of the CD45+ CD19+ ALL cells in the peripheral blood (e, n=5 mice for each group) and the Kaplan–Meier curve for the event-free survival of the mice are shown (f, n=5 for each group; log-rank test; p values were adjusted with Bonferroni correction). The frequency of the CD8+ CAR-T cells was analyzed in the peripheral blood on days 7, 14 and 28 (g; n=5, unpaired two-tailed t-test; t=2.58 for Day 7, t=4.28 for Day 14, t=2.46 for Day 21; degree of freedom=8). The data are representative of two experiments using different ALL samples. (h–k) NSG mice were subcutaneously injected with the melanoma cell line A375 transduced with CD19 (A375-CD19) (day −21), and treated with 5×105 CAR-T cells on day 0 and 4. (i) The frequency of the CD8+ CAR-T cells in the peripheral blood and subcutaneous tumors on day 7 (n=7, ordinary one-way ANOVA with Tukey’s multiple comparisons test; F=12.61 for peripheral blood, F=27.46 for tumor; degree of freedom=20). The serial monitoring of tumor progression (j) and overall survival (k; n=7, log-rank test; p values were adjusted with Bonferroni correction) of the mice are shown. Representative results of two experiments. In d, g, and i, horizontal lines indicate mean values ± s.d. * P<0.001.
We also compared the efficacy of the BB-z and 28-ΔIL2RB-z (YXXQ)-T cells against primary CD19+ B-acute lymphoblastic leukemia (ALL) cells in vivo. CAR-T cells significantly delayed the progression of leukemia (Fig. 4e). Similar to the results obtained with the NALM6-GL leukemic mice, the 28-ΔIL2RB-z (YXXQ) CAR-T cells showed better persistence in the peripheral blood and prevented leukemia progression for longer periods than the BB-z CAR-T cells (Fig. 4, f and g). Finally, we tested whether the JAK-STAT pathway activation in the CAR-T cells can improve their therapeutic effects against solid tumors. We adoptively transferred various CAR-engineered T cells into NSG mice subcutaneously inoculated with A375 melanoma cells transduced with CD19 (A375-CD19) (Fig. 4h). As was seen in the leukemia models, the 28-ΔILRB-z (YXXQ) CAR-T cells showed superior expansion in the peripheral blood and tumor mass, which resulted in better control of the tumor cells (Fig. 4, i–k). At the time of death, tumor cells maintained CD19 expression, and the CAR-T cells had almost disappeared within the tumor mass, suggesting that the sustained persistence of CAR-T cells is required to suppress tumor progression (Supplementary Fig. 18, a–c).
In summary, we have developed a novel CAR construct that activates the JAK-STAT pathway. The 28-ΔIL2RB-z (YXXQ) CAR triggered gene expression profiles analogous to those triggered by IL-21 treatment, which provided the T cells with distinct functional properties, including superior proliferative capacity and effector functions compared to the 28-z and BB-z CAR-T cells. Improved expansion of the 28-ΔIL2RB-z (YXXQ) CAR-T cells in vivo could potentially increase the risk for CRS, which should be cautiously addressed when this construct is clinically translated. Our novel CAR design can be utilized in any CAR-T cells, independent of antigen specificity to enhance their antitumor efficacy.
Online Methods
Construction of CAR genes
The 28-z CAR gene was generated by linking FMC63-derived scFV to the extracellular, transmembrane and cytoplasmic domain of CD28 (amino acid position 114-220) and the cytoplasmic domain of CD3z. The BB-z CAR was generated by inserting the hinge and transmembrane domain of CD8α and the 4-1BB cytoplasmic domain between the scFV and CD3z. To induce JAK-STAT signaling in the CAR, the cytoplasmic domain of full-length or truncated IL2Rβ (amino acid positions 266–337 and 530–551) was linked between CD28 and CD3z. The YXXQ motif was created by site-directed mutagenesis at the C-terminus region of CD3z. For the 28-ΔIL2RB (FLSL)-z (YXXQ) CAR, tyrosine within the YLSL motif was mutated to phenylalanine to abrogate STAT5 recruitment. All CAR genes were linked to ΔNGFR using a Furin-SGSG-P2A sequence and cloned into the pMX vector. The amino acid sequences of the CAR signaling domains are provided in Supplementary Table 1.
In vitro culture of human T cells
Peripheral blood mononuclear cells obtained from healthy donors were prepared by Ficoll-Paque PLUS density gradient centrifugation (GE Healthcare). The CD3+ and CD8+ cells were purified through negative magnetic selection using the Pan T Cell Isolation Kit and the CD8+ T Cell Isolation Kit (Miltenyi Biotec). The purified T cells were stimulated with irradiated artificial antigen-presenting cells that express a membrane-bound form of anti-CD3 mAb (clone OKT3) and the costimulatory molecules CD80 and CD83 (aAPC/mOKT3) at an effector to target (E:T) ratio of 5:1. On the following day, 100 IU/ml IL-2 and 10 ng/ml IL-15 (PeproTech) were added to the cultures. The culture media were replenished every 2–3 days. CAR-transduced T cells were restimulated with NALM-6 or K562 at an E:T ratio of 1:1 seven days after the initial stimulation. The number of CAR-T cells was determined by calculating the product of the total number of T cells and the percentage of ΔNGFR+ CD4+ or CD8+ T cells. The fold expansion was calculated by dividing the number of expanded CAR-T cells after 7 days of culture by the respective number on day 0.
Cell lines
Both aAPC/mOKT3 and K562-CD19 are derived from the human erythroleukemic cell line K562 (American Type Culture Collection; ATCC) and established as previously described41. The CD19+ acute lymphoblastic leukemia cell line NALM-6 was obtained from DSMZ (Braunschweig, Germany). The A375 melanoma cell line was obtained from the ATCC. All cell lines were routinely assessed for the presence of mycoplasma contamination using a PCR-based technology.
Retroviral transduction of T cells
PG13 packaging cells stably transduced with each retrovirus CAR plasmid were used for the infection of the T cells. The transduction was performed on days 2, 3, and 4 following the stimulation with aAPC/mOKT3 using Polybrene at a concentration of 1 μg/ml.
Flow cytometry
The following antibodies were used for the flow cytometry analysis: APC-Cy7-anti-CD4 (clone RPA-T4; BioLegend), PE-Cy7-anti-CD4 (RPA-T4; BioLegend), PE-Cy7-anti-CD8 (clone SFCI21Thy2D3; Beckman Coulter), PE-anti-CD8 (clone RPA-T8; BioLegend), Pacific-Blue-anti-CD8 (clone B9.11; Beckman Coulter), PE-anti-CD69 (clone FN50; BioLegend), FITC-anti-CD45RA (clone MEM-56; Thermo Fisher Scientific), PE-anti-CD62L (clone DREG-56; BioLegend), Pacific Blue-anti-CCR7 (clone G043H7; BioLegend), APC-Cy7-anti-CD27 (clone O323; BioLegend), APC-anti-CD28 (clone CD28.2; BioLegend), PerCP/Cy5.5-anti-CD95 (clone DX2; BD Biosciences), Alexa Fluor 488-anti-CD279 (clone EH12.2H7; BioLegend), PE-anti-CD274 (clone 29E.2A3; BioLegend), APC/Cy7-anti-CD366 (clone F38-2E2; BioLegend), PerCP/Cy5.5-anti-CD223 (clone C9B7W; BioLegend), FITC-anti-CD271 (clone ME20.4; BioLegend), PerCP/Cy5.5-anti-CD271 (clone ME20.4; BioLegend), V450-anti-CD271 (clone C40-1457; BD Biosciences), APC-anti-CD45 (clone HI30; BioLegend), FITC-anti-HLA-A2 (clone BB7.2; BioLegend), and PE-anti-CD19 (clone HIB19, BioLegend). The CAR-transduced T cells were stained with biotin-labeled protein L (GenScript), followed by streptavidin-PE (Thermo Fisher Scientific). For the carboxyfluorescein succinimidyl ester (CFSE) dilution assay, T cells were labeled with 5 μM CFSE (Thermo Fisher Scientific) before culture. Dead cells were discriminated with the LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Thermo Fisher Scientific). The stained cells were analyzed with a FACSCanto II (BD Biosciences). The data analysis was performed using the FlowJo software (Tree Star).
Analysis of phosphoproteins by intracellular flow cytometry
Phosphorylated proteins in the CAR-T cells were analyzed by intracellular flow cytometry following coculture with NALM-6 or K562 at an E:T ratio of 1:1. The cells were fixed with 1.6 % formaldehyde, followed by permeabilization by ice-cold methanol. The following antibodies were used: Alexa Fluor 647-anti-phospho-STAT3 (Tyr705) (clone 4/P-Stat3, BD Biosciences), Alexa Fluor 647-anti-phospho-STAT5 (Tyr694) (clone 47/Stat5, BD Biosciences), Alexa Fluor 647-anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (clone 20A, BD Biosciences), and anti-phospho-Akt (Thr308) (clone D25E6, Cell Signaling Technology). Alexa Fluor 647-anti-rabbit IgG (H+L) (Jackson ImmunoResearch) was used as the secondary antibody following the staining with the anti–phospho-Akt antibody. The STAT3 inhibitor S3I-201 (Selleck Chemicals) and the STAT5 inhibitor pimozide (Cayman Chemical) were used at concentrations of 25 μM and 5 μM, respectively.
Immunoblotting analysis
Immobilon-P PVDF membranes (Millipore) were probed with the primary antibodies at 4°C overnight and then washed and incubated with HRP-conjugated goat anti-mouse IgG (H+L) or anti-rabbit IgG (H+L) secondary antibody (Promega). The following primary antibodies were used: anti-STAT3 (clone D3Z2G, Cell Signaling Technology), anti-phospho-STAT3 (Tyr705) (clone D3A7, Cell Signaling Technology), anti-STAT5 (Cell Signaling Technology), anti-phospho-STAT5 (Tyr694) (clone D47E7, Cell Signaling Technology), anti-β-actin (clone C4, Santa Cruz Biotechnology, sc-47778), HRP-conjugated anti-mouse IgG (H+L) (Promega, W4021), and HRP-conjugated anti-rabbit IgG (H+L) (Promega, W4011). All images were acquired with ChemiDoc MP system (Bio-Rad) and Image Lab software (Bio-Rad). Protein levels for each blot were quantified with ImageJ software.
Cytokine production analysis
For intracellular cytokine staining, CAR-T cells were stimulated with NALM-6 at an E:T ratio of 1:1. Brefeldin A (BioLegend) was added to the culture media after 2 hours, and the cells were further cultured for 4 hours. Following the surface marker staining, the cells were fixed and permeabilized using the Cytofix/Cytoperm Kit (BD Biosciences) and stained with FITC-anti-IL-2 (clone 5344.111; BD Biosciences), PE-anti-TNF-α (clone MAb11; BioLegend), and PE-Cy7-anti-IFN-γ (clone 4S.B3; BioLegend). The frequency of cytokine-producing cells within the CD8+ ΔNGFR+ T cell population was determined by flow cytometry.
In vitro cytotoxicity assay
The cytolytic activity of the CAR-transduced T cells was analyzed as previously described42. Briefly, 1×105 CAR-T cells were cocultured with an equal number of CFSE-labeled or EGFP+ target cells for 4 hours. The cells were then harvested and stained using the LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Thermo Fisher Scientific). The absolute counts of the viable target cells were determined by flow cytometry, and the frequency of the surviving cells was calculated as a ratio to the cell counts incubated without T cells.
Microarray analysis
The CAR-transduced CD8+ T cells were cocultured with NALM-6 and purified at 4, 24, and 72 hours after stimulation with a FACSAria cell sorter (BD Biosciences). RNA was extracted from the sorted cells using the RNeasy Micro Kit (Qiagen). The gene expression profiles were analyzed using the Affymetrix Human Gene 2.0 ST Array by the Centre for Applied Genomics (TCAG) at the Hospital for Sick Children (Toronto, ON). The raw data were normalized and annotated with the Affymetrix Expression Console version 1.4.1 (Affymetrix). The differentially expressed genes among the three CAR-T cells were extracted by repeated measures one-way analysis of variance (ANOVA) using Affymetrix Transcriptome Analysis Console (Affymetrix) (P<0.01 with repeated measures one-way ANOVA). An unsupervised hierarchical clustering was performed, and a heatmap was generated using the HeatPlus software package from Bioconductor. A principal component analysis was performed using the function ‘prcomp’ in the R package ‘stats,’ and the data were shown on a 2-dimension plot using the ‘ggplot2’ package from GitHub. A gene set enrichment analysis (GSEA) was performed using the GSEA v2 software (Broad Institute). The genes induced by the cytokine treatment in the CD8+ T cells were analyzed using published gene expression data (GSE58262). Significantly upregulated genes were identified in individual cytokine-treated T cells compared with control T cells by unpaired t-test (P<0.01). STAT3 target genes were extracted according to a previous study analyzing gene expression profiles and chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq)40. The genes that were significantly downregulated in STAT3-deficient T cells compared with wild-type T cells (GSE21670, P<0.05 by unpaired t-test) and had STAT3 binding sites in the promoter regions were extracted.
Analysis of serum cytokine concentrations
The serum concentrations of human IL-2, IFN-γ, TNF-α, and IL-6 in NALM-6-bearing mice were measured using an enzyme-linked immunosorbent assay (ELISA). The following kits were used for each cytokine: Human IL-2 ELISA MAX Deluxe (BioLegend), Human IFN-gamma Quantikine ELISA Kit (R&D Systems), Human TNF-α ELISA MAX Deluxe (BioLegend), and Human IL-6 Quantikine ELISA Kit (R&D Systems). The concentration was calculated using a four-parameter logistic regression (4-PL) model.
Quantitative real-time PCR
RNA was extracted with RNeasy Micro Kit (Qiagen) and reverse-transcribed into cDNA using Superscript III (Thermo Fisher Scientific). Quantitative real-time PCR was performed using a CFX96 real-time PCR detection system (Bio-Rad) using the SYBR Select Master Mix (Thermo Fisher Scientific). The results were normalized to UBC, and the relative expression levels were calculated using the 2-ΔΔCT method. The average log2-transformed expression of all samples was normalized to 0 in each gene. The following primers were used for the real-time PCR experiments: GZMA forward, CCTGTGATTGGAATGAATATGGT, and reverse, AGGGCTTCCAGAATCTCCAT; GZMB forward, GGGCCCACAATATCAAAGAA, and reverse, GATGGGTCTTTTCACAGGGATA ; GZMH forward, CCATTCCTCCTCCTGTTGG, and reverse, TGAACAAAGGCCATGTAGGG; GZMK forward, TCTGGAACCAAATGCAAGGT, and reverse, GCAGGGTGTCAGAAGGTCTTA; PRF1 forward, CTCTCCTCGACGGAGTCCT, and reverse, TCTTGTTCCTCCTCAGAGTCG; and UBC forward, ATTTGGGTCGCGGTTCTTG, and reverse, TGCCTTGACATTCTCGATGGT.
Mouse experiments
In the murine experiments, male NSG mice bred at the Princess Margaret Cancer Centre animal facility were used. In the leukemia treatment model, animals were intravenously injected with 5 million of CD19+ NALM-6 leukemia cells transduced with pMX-EGFP-firefly luciferase (NALM6-GL) or 1 million of primary CD19+ B-ALL cells. Mice were irradiated with 1.5 Gy using X-RAD 320 (Precision X-Ray, Inc.) before the transplantation of primary ALL cells to enhance engraftment. The CD3+ T cells were retrovirally transduced with each anti-CD19 CAR gene and expanded for 2 weeks. One or two infusions of five million CAR-T cells were administered to the mice 14 days after the NALM6-GL transplantation, and a half million CAR-T cells were infused 35 days after transplantation of the primary B-ALL cells. In vivo imaging of the NALM6-GL within the mice was performed with Xenogen IVIS Spectrum and analyzed with Living Image software (Perkin Elmer). The engraftment of the primary B-ALL cells was regularly monitored by analyzing the peripheral blood. Event-free survival was defined as time until more than 25% of the human CD45+ CD19+ cells were detected in the peripheral blood. Mice were monitored at least once daily and euthanized by CO2 inhalation when they became moribund due to the leukemia progression or had more than 20% weight loss. In the solid tumor treatment model, the NSG mice were subcutaneously injected with 5×105 A375 melanoma cells transduced with CD19 (day −21) and intravenously infused with 5×105 CAR-T cells on day 0 and 4. The volume of the inoculated tumors was monitored every 2–3 days until they reached over 300 mm3 or an ulcer had formed in the tumor. In the tumor-free model, NSG mice were irradiated with 1.5 Gy and adoptively transferred with 5 or 10 million CAR-T cells the next day. The development of xenogeneic GVHD was monitored daily in each experiment. The mice were sacrificed when they exhibited one of the following symptoms: more than 20% loss of initial body weight, pronounced lethargy, hunched posture, severe diarrhea, and severe dermatitis. The mice were randomly assigned to treatment groups in each experiment. No statistical methods were used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment.
Statistics
Statistically significant differences between two groups were assessed using a two-tailed paired or unpaired t-test. Comparisons between more than two groups were carried out by an ANOVA with Tukey’s multiple comparisons test. Differences were considered statistically significant at a P-value less than 0.05. In the mice experiments, the overall survival and event-free survival of the mice treated with the T cells were depicted by a Kaplan-Meier curve, and the survival difference between the groups was compared using the log-rank test. When more than two groups were compared, the P-value was adjusted according to the number of comparisons with the Bonferroni method. All statistical analyses were performed using GraphPad Prism 6 software. No statistical method was used to predetermine the sample size.
Study approval
This study was performed in accordance with the Helsinki Declaration and approved by the Research Ethics Board of the University Health Network, Toronto, Canada. Written informed consent was obtained from all healthy donors who provided the peripheral blood samples. All animal experiments were approved by the Ontario Cancer Institute/Princess Margaret Cancer Centre Animal Care Committee at the University Health Network, and performed in accordance with Canadian Council on Animal Care guidelines.
Life Sciences Reporting Summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Data availability
The microarray data has been deposited in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE103906.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 CA148673 (NH); Ontario Institute for Cancer Research Clinical Investigator Award IA-039 (NH); BioCanRX Catalyst Program FY17CAT7 (NH); the Princess Margaret Cancer Foundation (MOB, NH); Japan Society for the Promotion of Science Postdoctoral Fellowship for Overseas Researchers (YK); the Canadian Institutes of Health Research Canada Graduate Scholarship (TG); Province of Ontario (TG, MA); and the Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship (TG).
Footnotes
Author contributions
Y.K. and N.H. designed the project. Y.K., S.T., T.G., M.A., C.-H.W., and K.S. performed the experiments. M.D.M. and M.O.B. provided critical human samples and contributed to the writing of this manuscript. Y.K. and N.H. analyzed the results and wrote the manuscript.
Competing financial interests
S.T. is an employee of Takara Bio, Inc. This study was partly sponsored by Takara Bio, Inc. The University Health Network has filed a patent application related to this study on which NH, YK, and ST are named as inventors.
References
- 1.Brentjens RJ, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5:177ra138. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grupp SA, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee DW, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davila ML, et al. Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6:224ra225. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter DL, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. 2015;7:303ra139. doi: 10.1126/scitranslmed.aac5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kochenderfer JN, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119:2709–2720. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Till BG, et al. CD20-specific adoptive immunotherapy for lymphoma using a chimeric antigen receptor with both CD28 and 4-1BB domains: pilot clinical trial results. Blood. 2012;119:3940–3950. doi: 10.1182/blood-2011-10-387969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed N, et al. Human Epidermal Growth Factor Receptor 2 (HER2) -Specific Chimeric Antigen Receptor-Modified T Cells for the Immunotherapy of HER2-Positive Sarcoma. J Clin Oncol. 2015;33:1688–1696. doi: 10.1200/JCO.2014.58.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kershaw MH, et al. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin Cancer Res. 2006;12:6106–6115. doi: 10.1158/1078-0432.CCR-06-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pule MA, et al. Virus-specific T cells engineered to coexpress tumor-specific receptors: persistence and antitumor activity in individuals with neuroblastoma. Nat Med. 2008;14:1264–1270. doi: 10.1038/nm.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kershaw MH, Westwood JA, Darcy PK. Gene-engineered T cells for cancer therapy. Nat Rev Cancer. 2013;13:525–541. doi: 10.1038/nrc3565. [DOI] [PubMed] [Google Scholar]
- 13.Kowolik CM, et al. CD28 costimulation provided through a CD19-specific chimeric antigen receptor enhances in vivo persistence and antitumor efficacy of adoptively transferred T cells. Cancer Res. 2006;66:10995–11004. doi: 10.1158/0008-5472.CAN-06-0160. [DOI] [PubMed] [Google Scholar]
- 14.Brentjens RJ, et al. Genetically targeted T cells eradicate systemic acute lymphoblastic leukemia xenografts. Clin Cancer Res. 2007;13:5426–5435. doi: 10.1158/1078-0432.CCR-07-0674. [DOI] [PubMed] [Google Scholar]
- 15.Stephan MT, et al. T cell-encoded CD80 and 4-1BBL induce auto- and transcostimulation, resulting in potent tumor rejection. Nat Med. 2007;13:1440–1449. doi: 10.1038/nm1676. [DOI] [PubMed] [Google Scholar]
- 16.Milone MC, et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol Ther. 2009;17:1453–1464. doi: 10.1038/mt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savoldo B, et al. CD28 costimulation improves expansion and persistence of chimeric antigen receptor-modified T cells in lymphoma patients. J Clin Invest. 2011;121:1822–1826. doi: 10.1172/JCI46110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim WA, June CH. The Principles of Engineering Immune Cells to Treat Cancer. Cell. 2017;168:724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng R, et al. The molecular basis of IL-21-mediated proliferation. Blood. 2007;109:4135–4142. doi: 10.1182/blood-2006-10-054973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinrichs CS, et al. IL-2 and IL-21 confer opposing differentiation programs to CD8+ T cells for adoptive immunotherapy. Blood. 2008;111:5326–5333. doi: 10.1182/blood-2007-09-113050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin G, et al. A Critical Role of IL-21-Induced BATF in Sustaining CD8-T-Cell-Mediated Chronic Viral Control. Cell Rep. 2015;13:1118–1124. doi: 10.1016/j.celrep.2015.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng R, et al. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J Exp Med. 2005;201:139–148. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markley JC, Sadelain M. IL-7 and IL-21 are superior to IL-2 and IL-15 in promoting human T cell-mediated rejection of systemic lymphoma in immunodeficient mice. Blood. 2010;115:3508–3519. doi: 10.1182/blood-2009-09-241398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintarelli C, et al. Co-expression of cytokine and suicide genes to enhance the activity and safety of tumor-specific cytotoxic T lymphocytes. Blood. 2007;110:2793–2802. doi: 10.1182/blood-2007-02-072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pegram HJ, et al. Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood. 2012;119:4133–4141. doi: 10.1182/blood-2011-12-400044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu C, et al. Cytokine-independent growth and clonal expansion of a primary human CD8+ T-cell clone following retroviral transduction with the IL-15 gene. Blood. 2007;109:5168–5177. doi: 10.1182/blood-2006-06-029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, et al. Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin Cancer Res. 2015;21:2278–2288. doi: 10.1158/1078-0432.CCR-14-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahl N, Farruggella TJ, Boulton TG, Zhong Z. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson IC, et al. Construction and characterisation of a functional CD19 specific single chain Fv fragment for immunotherapy of B lineage leukaemia and lymphoma. Mol Immunol. 1997;34:1157–1165. doi: 10.1016/s0161-5890(97)00144-2. [DOI] [PubMed] [Google Scholar]
- 31.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzec M, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1) Proc Natl Acad Sci U S A. 2008;105:20852–20857. doi: 10.1073/pnas.0810958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lastwika KJ, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 34.Eyquem J, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherkassky L, et al. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J Clin Invest. 2016;126:3130–3144. doi: 10.1172/JCI83092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabatino M, et al. Generation of clinical-grade CD19-specific CAR-modified CD8+ memory stem cells for the treatment of human B-cell malignancies. Blood. 2016;128:519–528. doi: 10.1182/blood-2015-11-683847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cieri N, et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood. 2013;121:573–584. doi: 10.1182/blood-2012-05-431718. [DOI] [PubMed] [Google Scholar]
- 38.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel AM, et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35:806–818. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durant L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler MO, et al. Ex vivo expansion of human CD8+ T cells using autologous CD4+ T cell help. PLoS One. 2012;7:e30229. doi: 10.1371/journal.pone.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jedema I, van der Werff NM, Barge RM, Willemze R, Falkenburg JH. New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood. 2004;103:2677–2682. doi: 10.1182/blood-2003-06-2070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data has been deposited in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE103906.