Abstract
Immunotherapy offers new cancer treatment options, but efficacy varies across cancer types. Colorectal cancers (CRCs) are largely refractory to immune checkpoint blockade, suggesting the presence of yet-to-be characterized immune suppressive mechanisms. Here we report that APC-loss in intestinal tumor cells or PTEN-loss in melanoma cells upregulates the expression of dickkopf-2 (DKK2), which, together with its receptor LRP5, constitutes an unconventional mechanism for tumor immune evasion. DKK2 secreted by tumor cells acts on cytotoxic lymphocytes, inhibiting STAT5 signaling by impeding STAT5 nuclear localization via LRP5 but independently of LRP6 and the Wnt-β-catenin pathway. Genetic or antibody-mediated ablation of DKK2 activates natural killer (NK) and CD8+ cells in tumors, impedes tumor progression, and cooperates with PD-1 blockade. Thus, we have identified a previously unknown tumor immune suppressive mechanism and immunotherapeutic targets particularly relevant for CRCs and a subset of melanomas.
INTRODUCTION
Significant advances, particularly in immunotherapy, have been made in treatment of cancers, a leading cause of death in humans1–6. Immune checkpoint inhibitors, including anti-PD1, anti-CTLA4, have shown clinical efficacy for some tumors, but not for many others including colorectal cancer cells (CRCs)5,7–9. While mechanisms for resistance/insensitivity to current checkpoint inhibitors have been described10, there are more mechanisms for tumor immune modulation yet to be discovered.
Natural killer (NK) cells and CD8+ T lymphocytes are the cytotoxic effector immune cells that are capable of directly killing tumor cells. The cytotoxic activity of NK and CD8+ T cells are regulated by the complex mechanisms including by cytokines. IL-15 is a key cytokine that controls all aspects of NK cell biology13. It is also important for the development and function of CD8+ intestinal intraepithelial lymphocytes (IELs)13–16. It additionally regulates effector and memory CD8+ T cell development and function and confers T cell resistance to Treg cells13,14,17,18. IL-15 signals through its receptor that consists of an IL15Rα chain, an IL2/15Rβ chain, and a common cytokine-receptor γ-chain (γc). IL-15 induces phosphorylation of STAT5 via JAK1 and JAK3. Phosphorylated STAT5 (pSTAT5) accumulates in the nucleus to regulate gene transcription. IL-15 also activates the PI3K-AKT, mTOR, and MAPK pathways. IL-15 stimulates the cytotoxic effector functions by increasing the production of perforin and granzyme B (GZMB) through these pathways13,14,19,20.
Wnt-signaling pathways control a wide range of cellular processes21–24. The Wnt-β-catenin pathway is initiated by two cell surface receptors---the low-density lipoprotein receptor related proteins 5 and 6 (LRP5/6) and frizzled25. Dysregulation of Wnt-β-catenin signaling is associated with many human diseases, including cancer21–24. Hyperactivation of the Wnt/β-catenin pathway can lead to aberrant cell growth and tumor formation. More than 80% of CRCs harbor loss of function mutations in the adenomatosis polyposis coli (APC) gene, a suppressor of the Wnt-β-catenin pathway26.
DKK223,27 inhibits Wnt-β-catenin signaling by binding to LRP5/628. DKK2 plays a less critical role in vertebrate development29–31 and adult life. Dkk2-deficiency reduces blood glucose32 and causes a moderate reduction on bone mass30. Given that DKK2 is a Wnt antagonist29,30,33–35, the conventional wisdom is that DKK2 inactivation might increase Wnt activity and lead to or accelerate cancer formation. In this study, we found, contrary to the expected, that DKK2, whose expression is upregulated in human CRCs and by APC-loss mutations, promotes tumor progression by suppressing immune effector cell activation.
RESULTS
Loss of APC drives DKK2 expression
Analysis of the Gaedcke cohort36 in the Oncomine database (www.oncomine.org) revealed that DKK2 expression was significantly upregulated in human CRC samples compared to the non-tumorous colorectal tissues (Supplementary Fig. 1a), which is consistent with a previous finding37. Analysis of the Cancer Genome Atlas Network datasets38 further revealed that DKK2 expression in the microsatellite-stable (MSS) CRCs, more than 80% of which harbor APC mutations, is significantly higher than that in the microsatellite-instable (MSI) CRCs (Supplementary Fig. 1a). In mice, the DKK2 mRNA content in the intestinal polyps of the ApcMin/+ mice, a mouse genetic intestinal tumor model39, was about four times higher than that in normal intestinal tissue (Supplementary Fig. 1b). Immunostaining of the DKK2 protein and in situ hybridization of the Dkk2 mRNA confirmed DKK2 expression upregulation in the polyps (Supplementary Fig. 1c-d). When the Apc gene in the mouse colon cancer MC38 cells was mutated by CRISPR/Cas9 , DKK2 expression was markedly upregulated in the APC-null cells (Supplementary Fig. 1e). This upregulation could be suppressed by β-catenin siRNAs (Supplementary Fig. 1f), suggesting the involvement of β-catenin in driving the DKK2 expression. APC-loss also led to DKK2 expression upregulation in human colon cancer HCT116 cells (Supplementary Fig. 1g). Therefore, we conclude that APC-loss drives DKK2 expression in both mouse and human CRC cells.
DKK2 blockade suppresses APC-loss-induced tumor formation
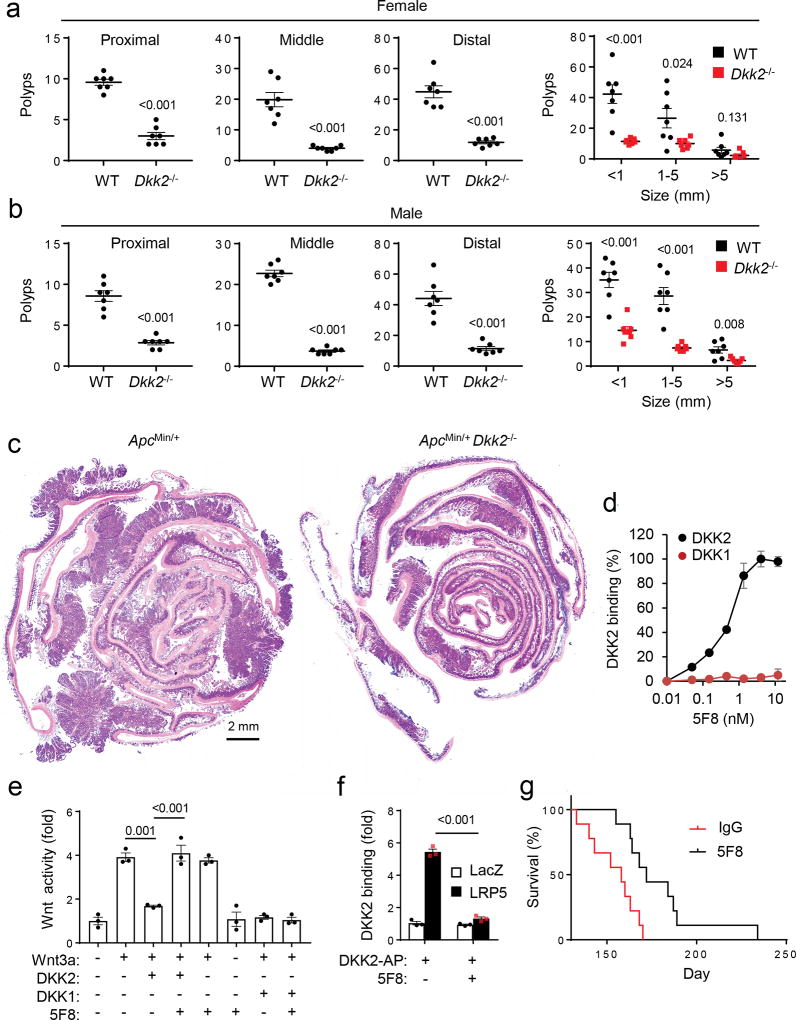
Analysis of the TCGA CRC datasets revealed correlations of high DKK2 expression with poor survival rates (Supplementary Fig. 1h). This suggests that DKK2 may play an important role in CRCs. Concordantly, DKK2-deficiency significantly reduced intestinal polyp burdens in both male and female ApcMin/+ mice (Fig. 1a-c & Supplementary Fig. 1i). A mouse monoclonal antibody (designated as 5F8) was then generated. It bound to mouse DKK2, but not to DKK1 (Fig. 1d). The antibody inhibited DKK2-, but not DKK1-, mediated antagonism of Wnt activation (Fig. 1e). Moreover, 5F8 blocked the binding of DKK2 to LRP5 (Fig. 1f). Administration of 5F8 significantly extended the survival of the ApcMin/+ mice (Fig. 1g) and reduced intestinal polyp burdens depending on the presence of DKK2 in the ApcMin/+ mice (Supplementary Fig. 2a-c). Of note, an anti-PD-1 antibody showed no significant effect on polyps burden in these mice (data not shown), which is consistent with reported lack of therapeutic efficacy of PD-1 blockade on human MSS CRCs 5,7–9. These results together support the conclusion that DKK2 blockade suppresses the formation of intestinal polyps, a form of non-malignant tumor, caused by APC-loss. Thus, DKK2 supports, rather than impeding as one might have predicted based on its known mechanism of action, tumor progression.
Figure 1. DKK2 blockade reduces tumor burdens in the ApcMin/+ mice.
(a-c). Littermates of ApcMin/+ and ApcMin/+Dkk2−/− mice were housed for 20 weeks (female) or 22 weeks (male), and their polyps numbers in intestines were counted and measured under a stereomicroscope after staining with methylene-blue (two-tailed Student t-Test for polyp number and two-way Anova for size; n=7). Representative H&E staining images of intestinal sections from an ApcMin/+ mouse and its ApcMin/+Dkk2−/− littermates are shown. (d) Binding of the anti-DKK2 antibody 5F8 to mouse DKK2 and DKK1 proteins in an ELISA assay(n=3). (e) HEK293 cells were transfected with the Wnt reporter gene TOPFlash and treated with WNT3A conditioned medium (CM), DKK2 CM or DKK1 CM, and 5F8 (18 µg/ml) as indicated (one-way Anova; n=3). (f) HEK293 cells were transfected with LacZ (a control) or LRP5 expression plasmid. The binding of DKK2-AP fusion protein to the cells in the presence or absence of 5F8 (18 µg/ml) were determined (Two-way Anova; n=3). (g) Mice (16 weeks old male) were treated with 5F8 and an isotype antibody (IgG3) (8 mg/kg, once a week, i.p.) and their survival was recorded (P=0.004; Two-sided Mantel-Cox Log-Rank test, n=9). Data are presented as means±SEM (a-b) and ±sd (d-f) with P values shown, and experiments were repeated twice (d-f).
DKK2 blockade modulates tumor immune microenvironment
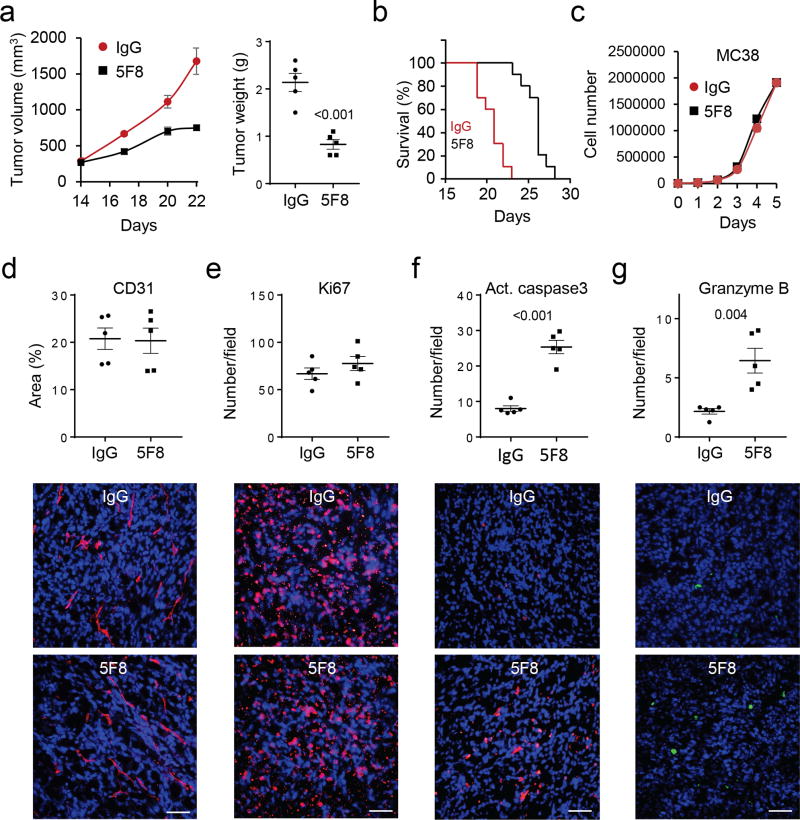
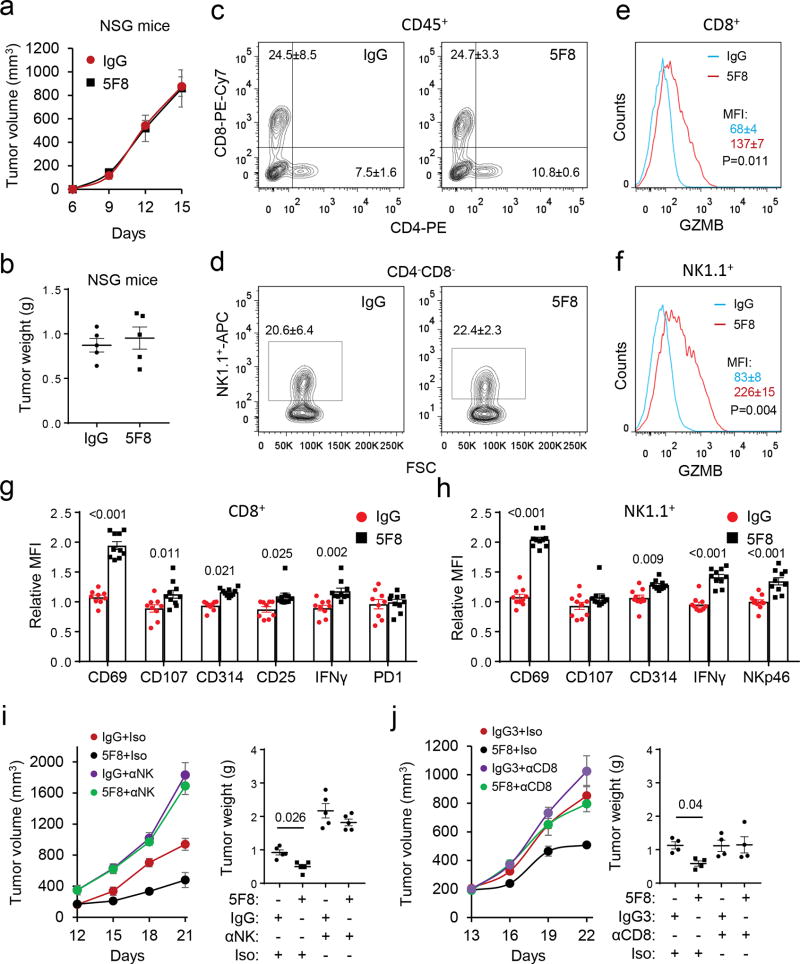
In a syngeneic mouse colon cancer model, 5F8 significantly inhibited growth of subcutaneously grafted MC38 cells in C57BL/6 mice (Fig. 2a) and extended the survival of tumor-bearing mice (Fig. 2b) compared to isotype control antibody (IgG3). Because 5F8 did not affect the growth of MC38 cells in culture (Fig. 2c), the antibody might impede tumor progression by altering tumor microenvironment. Immunohistological examination of the MC38 tumors revealed that 5F8 had no significant effects on angiogenesis (Fig. 2d) or tumor cell proliferation (Fig. 2e). However, 5F8 increased the number of apoptotic cells (Fig. 2f) and GZMB-positive cells (Fig. 2g). The increases in apoptosis and GZMB were also observed in the ApcMin/+Dkk2−/− polyps compared to the ApcMin/+ ones (Supplementary Fig. 2d-e) and in 5F8-treated polyps compared to those IgG3-treated (data not shown). DKK2 blockade did not significantly affect proliferation, infiltration of cytotoxic immune cells, or differentiation in the polyps (Supplementary Fig. 2f-i). Because GZMB is largely produced by NK and CD8+ cells and induces tumor cell apoptosis 40, the above results suggest that DKK2 blockade may modulate the immune microenvironment. In supporting this conclusion, when the MC38 cells were grafted onto the immunodeficient NSG mice, which lack mature leukocytes, 5F8 failed to show any tumor suppressive effect (Fig. 3a-b).
Figure 2. DKK2 blockade impedes tumor progression in the MC38 syngeneic tumor model.
(a-b) C57BL/6 mice inoculated with MC38 cells were treated with 5F8 (10 mg/kg, every three days, i.p.) starting at Day 14. Tumors from some of the mice were collected at Day 22 and weighed (P=0.002 and <0.001 for tumor growth at Days 20 and 22, respectively, two-way Anova; two-tailed Student t-test for tumor weight; n=5). The rest were used for evaluation of the survival (P=0.004, Two sided Log-rank Mantel-Cox test, n=10). (c) The 5F8 antibody does not affect MC38 cell growth in culture (n=4). (d-g) Effects of 5F8 treatment on tumor angiogenesis, tumor cell proliferation and apoptosis, and GZMB-positive cells were evaluated by staining the sections of tumor collected in a for CD31, Ki67, cleaved caspase 3, and GZMB, respectively. The sections were also counter-stained with DAPI. Five independent sections per tumor and 5 tumors per group were examined (two-sided Student’s t-test). Data are presented as means±SEM with P values shown (a, c-g). The scale bars are 100 µm.
Figure 3. DKK2 blockade enhances cytotoxic immune cells activation.
(a-b) MC38 cells were inoculated s.c in the NSG mice (n=5) and the treatment (10 mg/kg, every three days) commenced at Day 6. (c-h) MC38 cells were inoculated s.c in C57BL/6 mice. When tumors reach 600 mm3 in average, the mice were given one injection of the antibody (10 mg/kg, i.p.). Tumors were collected in 24 hours for flow cytometry analysis (two-tailed Student’s t-test, n=10). Panel c is pre-gated for CD45, whereas Panels d, e and g are derived from Panel c. Panels f and h are derived from Panel d. MFI, mean fluorescence intensity. The numbers of CD8+ cells are 4192±446 (IgG treated) and 4431±357 (5F8 treated) per 106 tumor cells, whereas the numbers of NK1.1+ cells are 3600±501 (IgG treated) and 3200±329 (5F8 treated). (i-j) C57BL/6 mice were inoculated s.c. with MC38 cells. Treatment of 5F8 (10 mg/kg, every three days, i.p.) commenced at Day 12 for the NK depletion experiment or at Day 13 for CD8 depletion experiment (P values for tumor growth: 0.008 for i and 0.003 for j, IgG+Iso vs 5F8+Iso; tumor weight P values are shown; n=5; two-way Anova). Iso, isotype control for cell depleting antibody. Data are presented as means±sem.
DKK2 blockade enhances NK and CD8+ cell activation
Consistent with the immunostaining results (Fig. 2g), flow cytometry analysis of tumor-infiltrated leukocytes revealed that 5F8 treatment increased GZMB in both CD8+ and NK1.1+ cells (Supplementary Fig. 3a-g). On the other hand, there were no differences between 5F8 and its isotype-treated samples in the percentage of myeloid cells (Gr1highCD11bhigh or Gr1lowCD11bhigh), CD4+, CD8+, T regulatory cells (CD4+CD25+Foxp3+), or NK1.1+ cells (Supplementary Fig. 3b-e). Analysis of leukocytes in the tumor draining lymph nodes (DLNs) showed that there were no differences in the populations of CD4+, CD8+, or NK1.1+ cells (Supplementary Fig. 3h-i), whereas there were a trend for an increase in GZMB in the CD8+ cells (Supplementary Fig. 3j) and a significant increase in GZMB in NK1.1+ cells in the 5F8-treated samples over the IgG3-treated ones (Supplementary Fig. 3k). Increases in GZMB-positive CD8+ cells were also observed in the Peyer’s patches (PPs) and DLNs for intestinal tumors in the Dkk2−/−ApcMin/+ mice over those in the ApcMin/+ mice, while there was no significant difference in the population of CD4+ or CD8+ cells (Supplementary Fig. 4a-b).
To exclude the effect of tumor size on the flow cytometry results, MC38 tumor-bearing mice were treated with antibodies for only 24 hours before analysis. At this time point, there was no obvious difference in tumor sizes. While there were still no significant differences in the populations of CD4+, CD8+, or NK1.1+ cells (Fig. 3c-d), strong increases in GZMB were observed in tumor infiltrated CD8+ and NK1.1+ cells in 5F8-treated specimens over isotype-treated ones (Fig. 3e-f). Significant increases in other activation markers of CD8+ and NK cells including CD69, CD107a, CD314, IFNγ, and CD25 on CD8+ cells and CD69, IFNγ, NKp46, and CD314 on NK cells (Fig. 3g-h) were also observed. No significant changes in Ki67, T-bet, Eomes, phospho-mTOR, phospho-S6K, Phospho-AKT, KLGR1, CD122 or CD127 in either CD8+ or NK1.1+ cells (Supplementary Fig. 4c-d) were observed. The acute 5F8 treatment also significantly increased GZMB-positive CD8+ cells in the PPs of the ApcMin/+ mice over IgG-treated, without affecting the populations of the T cells (Supplementary Fig. 4e-f).
To assess the importance of cytotoxic immune effector cells in DKK2 blockade-mediated tumor suppression, we depleted NK cells with an anti-NK1.1 antibody and CD8+ cells with an anti-CD8 antibody, respectively, in the MC38 tumor model (Supplementary Fig. 5a-c). Depletion of either NK or CD8+ cells largely diminished the tumor suppressive effect of 5F8 with NK cell depletion perhaps imparting a stronger effect (Fig. 3i-j). The cell depletion was also done in the Apcmin/+ model. While NK1.1+ cell depletion did not noticeably alter the 5F8’s effect on polyp formation, CD8+ cell depletion largely abrogated the effect of 5F8 (Supplementary Fig. 5d). These results indicate that the cytotoxic immune effector cells have significant roles in DKK2 blockade-mediated suppression of tumor formation.
DKK2 directly suppresses cytotoxic immune cells
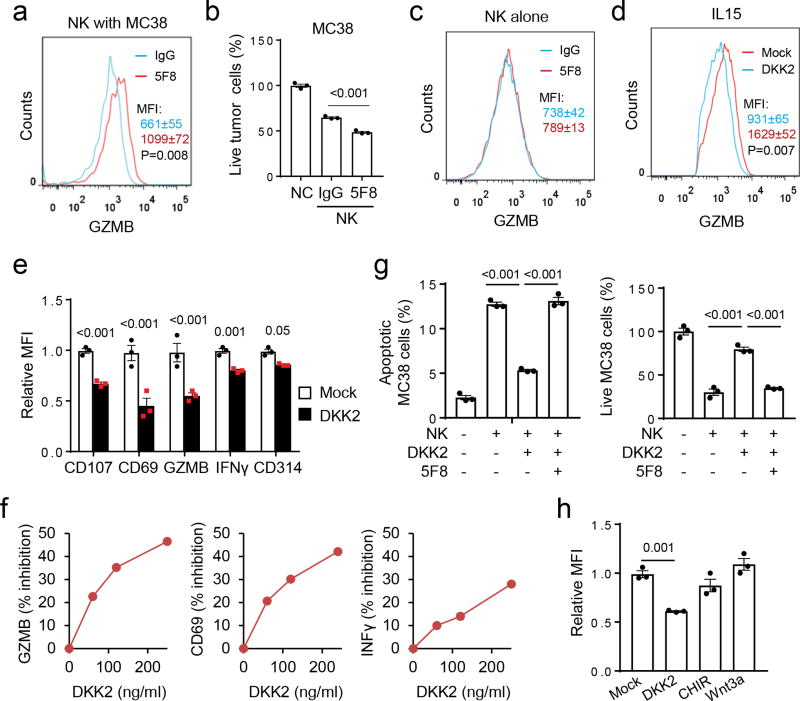
The effect of DKK2 blockade on cytotoxicity of primary NK cells was next assessed. Inclusion of 5F8 caused a marked increase in GZMB expression in the NK cells (Fig. 4a) and decreases in tumor cell viability (Fig. 4b), when IL-15-expanded primary mouse NK cells were co-cultured with the MC38 cells. By contrast, 5F8 treatment showed little effects on GZMB expression in the NK cells (Fig. 4c) or the viability of MC38 (Fig. 2c), when these cells were cultured alone.
Figure 4. DKK2 directly suppresses NK cell activation.
(a-c) Primary mouse NK cells expanded with IL-15 were added to MC38 cells that were seeded one day before in the presence of 5F8 or IgG3 (36 µg/ml) for 9 hours. GZMB expression in NK cells was examined by flow cytometry (a & c, two-tailed Student t-Test, n=4), whereas live tumor cells were determined by a Guava cell cytometer (b, one-way Anova, n=4). (d-f) Isolated primary mouse NK cells were cultured with IL-15 (50 ng/ml) for 24 hrs. DKK2 protein (400 ng/ml for d and e; varying amounts for f) were then added for another 24 hours, followed by flow cytometry analyses (P values for DKK2 vs mock, n=3, Two-sided Student’s t-test). (g) Primary NK cells were expanded in IL-15 (50 ng/ml) for 24 hours followed by treatment with or without DKK2 (400 ng/ml) or 5F8 (36 µg/ml) for 24 hrs. The NK cells were then added to MC38 cells seeded the day before at 7:1 ratio. The numbers of apoptotic MC38 cells were determined after 6 hours, and the number of live MC38 cells were determined after 9 hours of co-culture (n=3, one-way Anova). (h) Isolated primary mouse NK cells were cultured with IL-15 (50 ng/ml) for 24 hrs. DKK2 protein (400 ng/ml), Wnt3a (100 ng/ml), and GSK3 inhibitor CHIR99021 (CHIR, 1 µM) were then added for another 24 hours, followed by flow cytometry analyses (n=3, one-way Anova). Data are presented as means±sem (a-e, g,h) with P values shown (b,e,g,h), and the experiments were repeated three times (a-f) or twice(g,h).
We performed microarray gene expression analyses and did not observe significant alteration in the expression of IL-2, IL-15, MHC-I haplotypes, NKG2D ligands (RAE-1α-ε, MULT-1, and H60a-c), Fas or TRAILR1/2, all of which are important for NK cell activity, in 5F8-treated MC38 cells or tumors compared to IgG3-treated ones (data not shown). In addition, DKK2 mRNA was hardly detectable by RT-PCR in NK cells, whereas DKK2 mRNA was readily detectable in MC38 cells (Supplementary Fig. 1e). Together with the aforementioned co-culture results, we postulated that tumor cell-produced DKK2 might act directly on the NK cells. Indeed addition of recombinant DKK2 protein to primary NK cells cultured with IL-15 caused 5F8-reversible reductions in GZMB, CD69, IFNγ, CD107a, CD314 (Fig. 4d-e), KLRG1, and CD122 (Supplementary Fig. 6a). These inhibitory effects of DKK2 were dose-dependent (Fig. 4f) and also observed for NK cells stimulated with IL12 and IL18 (Supplementary Fig. 6b). Importantly, DKK2 also exerted significant impacts on the tumor killing ability, as NK cells pre-treated with DKK2 showed reduced ability to cause tumor cell apoptosis and death (Fig. 4g). Additionally, DKK2 inhibited IL-15-mediated activation of human NK and CD8+ cells isolated from peripheral bloods (Supplementary Fig. 6c-d). Furthermore, DKK2 suppressed mouse primary CD8+ cells isolated from spleens and CD8+ intraepithelial cells from intestines in response to IL-15 (Supplementary Fig. 6d-e). Thus, DKK2 could directly suppress activation of both human and mouse immune effector cells.
DKK2 inhibits NK cell activation independently of Wnt-β-catenin signaling
Next, we tested if Wnt-β-catenin signaling is responsible for NK cell regulation by DKK2, as DKK2 can inhibit this pathway. WNT3A protein induced a strong increase in the Wnt reporter gene activity, which could be inhibited by DKK2 (Supplementary Fig. 7a). WNT3A also induced β-catenin accumulation in the primary NK cells (Supplementary Fig. 7b). However, WNT3A did not alter GZMB expression in the NK cells (Fig. 4h). Concordantly, the GSK3 inhibitor CHIR99021, which increases β-catenin stability bypassing WNT and its receptors and strongly stimulated the Wnt reporter gene activity (Supplementary Fig. 7a), did not affect primary NK cells (Fig. 4h). Therefore, we conclude that DKK2-mediated inhibition of NK cell activation is likely independent of Wnt-β-catenin signaling. These results also distinguish the mechanism of action of DKK2 from recent reports indicating an involvement of Wnt-β-catenin signaling in modulation of tumor immune microenvironments 41,42.
DKK2 impedes phosphorylated STAT5 nuclear localization
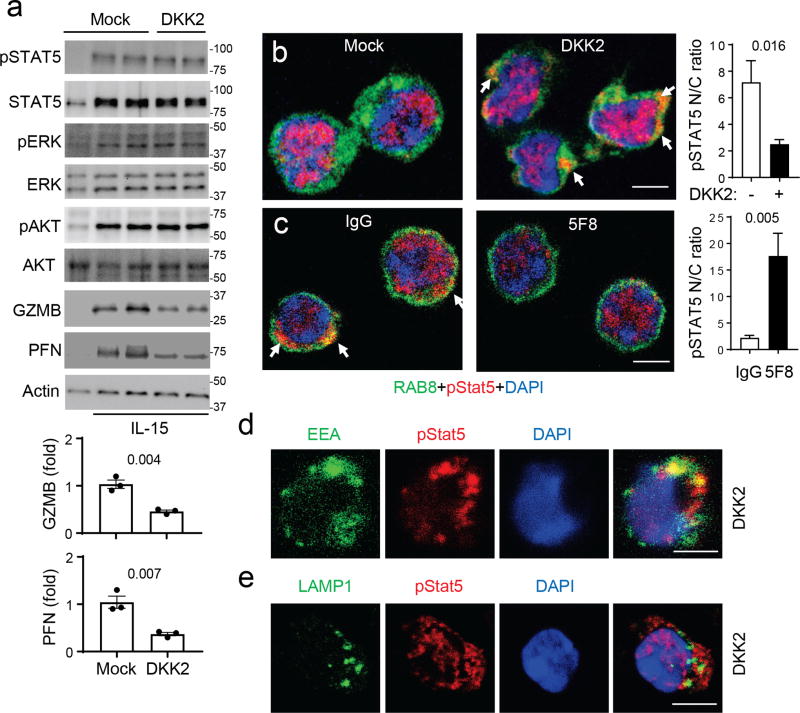
Given that DKK2 suppressed cytotoxic immune cell activation by IL-15, we examined the effects of DKK2 treatment on various signaling events stimulated by IL-15. While reductions in GZMB and perforin proteins were observed in DKK2-treated samples, no notable changes in phosphorylation of STAT5, ERK, AKT (Fig. 5a), MTOR, S6K, STAT1 or STAT4 (Supplementary Fig. 8a-b) were detected. Together with no changes in cell size by DKK2 treatment (Supplementary Fig. 8c), DKK2 does not alter the MTORC1 pathway, which can affect GZMB expression in NK cells 19,20. However, RNA-seq analysis of DKK2- and mock-treated primary NK cells suggests an alteration in STAT5 signaling by DKK2 treatment (Supplementary Fig. 8d-e & Supplementary Table I). We thus examined the localization of phospho-STAT5, a step downstream of STAT5 phosphorylation. While IL15 induces nuclear localization of phospho-STAT5 as expected, cytosolic localization of phospho-STAT5 was readily detected in DKK2-treated cells (Fig. 5b & Supplementary Fig. 8f). This result was further confirmed by subcellular fractionation (Supplementary Fig. 8g). Furthermore, NK cells isolated from 5F8-treated tumors showed reduced cytosolic localization of phospho-STAT5 over those isolated from control IgG-treated tumors (Fig. 5c & Supplementary Fig. 8h). Phospho-STAT5 was partially colocalized with early endosome marker EEA1 (Fig. 5d), but not with late endosome marker LAMP1 (Fig. 5e) in DKK2-treated NK cells, suggesting that phospho-STAT5 may be sequestered on early/recycling endosomes. Thus, we concluded that DKK2 treatment did not disrupt IL-15 signaling to STAT5 phosphorylation, rather impairing the nuclear localization of phospho-STAT5.
Figure 5. DKK2 impedes phospho-STAT5 nuclear localization.
(a-b) Primary mouse NK cells were prepared and treated as in Fig. 4d, followed by Western analysis (a) or immunostaining using anti-phospho-STAT5, anti-RAB8 (as a cytosol marker) and DAPI, followed by Alexa Fluor® 647 and FITC-labeled secondary antibodies (b). Individual color channels are shown in Supplementary Fig. 5f. Scale bars are 5 µm. Western blots for GZMB and perforin and the nuclear to cytosol (N/C) ratio of pSTAT5 staining were quantified (Two-sided Student’s t-test, n=3 for Western and n=15 for staining). Uncropped Western blots are provided in Supplementary Fig. 14. (c) Tumor-infiltrated NK cells were isolated by FACS from MC38 tumors treated with IgG3 or 5F8 for 6 days (10 mg/kg injections). The cells were fixed, permeabilized and stained with anti-RAB8, anti-p-STAT5, and DAPI, followed by Alexa Fluor-647 and FITC-labeled secondary antibodies. Individual color channels are shown in Supplementary Fig. 5h. The nuclear to cytosol (N/C) ratios of pSTAT5 staining were quantified (Student’s t-test, n=15). (d,e) Primary mouse NK cells were prepared and treated as in A, followed by immunostaining using anti-phospho-STAT5, DAPI, and anti-EEA-1 (d) or anti-LAMP1 (e), followed by Alexa Fluor® 647 and FITC-labeled secondary antibodies. Data are shown as means±sem with P values shown, and experiments were repeated three times. The scale bars are 5 µm.
DKK2 acts through LRP5, but not LRP6
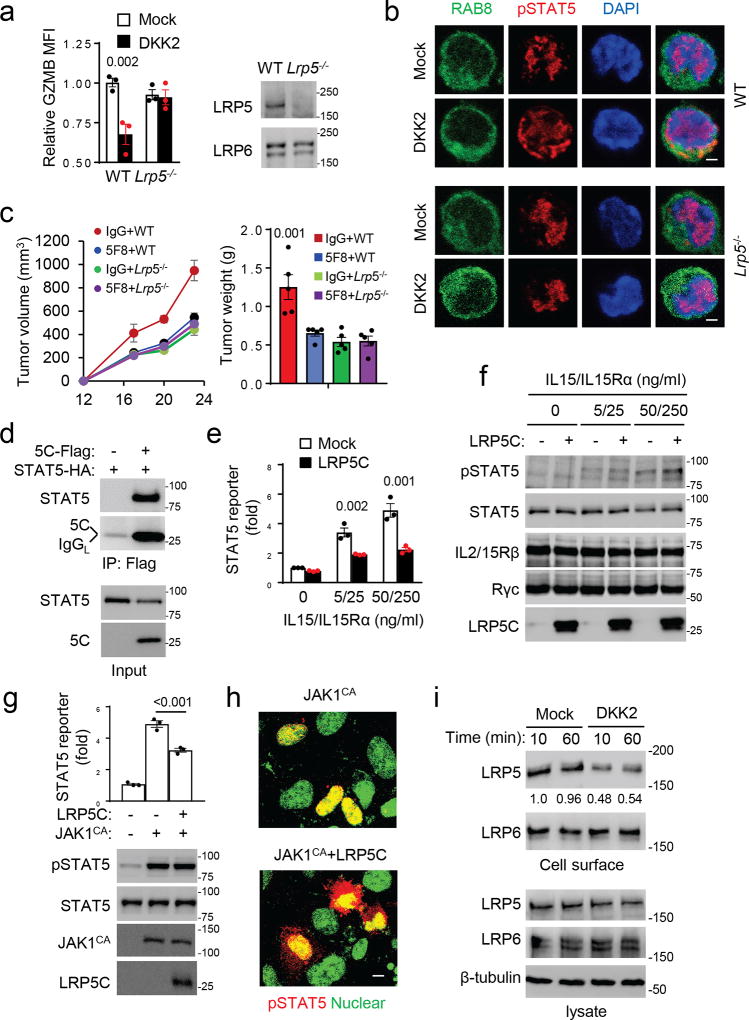
DKK2 binds to LRP5 and LRP6. While DKK2 could still inhibit the activation of primary NK cells lacking LRP6 (Supplementary Fig. 9a), it failed to inhibit LRP5-deficient NK cells (Fig. 6a). Additionally, DKK2 failed to cause impairment of phospho-STAT5 nuclear localization in NK cells lacking LRP5 (Fig. 6b). Thus, LRP5, but not LRP6, is required for DKK2’s action on NK cells. The fact that LRP5-deficiency did not affect β-catenin accumulation stimulated by WNT3A in the NK cells (Supplementary Fig. 7b) further confirms that the effect of the DKK2-LRP5 axis on NK activation is independent of Wnt-β-catenin signaling. LRP6 instead is required for Wnt-β-catenin signaling in NK cells, as WNT3A did not induce β-catenin accumulation in LRP6-deficient NK cells (Supplementary Fig. 9b).
Figure 6. LRP5 is required for DKK2-mediated inhibition of NK activation.
(a,b) Primary mouse NK cells were prepared from WT and Lrp5−/− mice and treated as Fig. 4d, followed by flow cytometry (two-way Anova) and Western analysis (a) and immunostaining (b). Immunostaining was done as Fig. 5b. (c) C57BL/6 mice receiving Lrp5f/fMX1Cre (Lrp5−/−) or Lrp5f/f (WT) bone marrows were treated with poly-I:C, followed by s.c. inoculation of MC38 cells. Treatment of 5F8 (10 mg/kg, i.p.) was given at Day 12, 16, and 20 (P<0.01 for tumor growth at Days 20 and 24, IgG+WT vs 5F8+WT; P value for tumor weight is vs 5F8+WT; n=5; two-way Anova). (d) LRP5C and STAT5 coimmunoprecipitate in transfected HEK293 cells. (e-f) HEK293 cells were infected with lentiviruses expressing JAK3, IL2/15Rβ, and the common γ subunit (Rγc). The cells were then transfected with the plasmid for LRP5 intracellular domain (LRP5C), the STAT5-luc reporter gene, and RFP (internal control) for 24 hrs. The cells were stimulated with IL-15 and IL15Rα-Fc for 6 hours before the reporter gene assay (e, two-way Anova) and Western analysis (f). (g-h) HEK293 cells were cotransfected with the STAT5 reporter gene plasmid and plasmids for activated JAK1 (JAK1CA, V658F) and LRP5C as indicated. After 24 hrs, the cells were analyzed for the reporter gene activity (two-sided Student’s t-test) or by Western (g) or immunostaining with a phospho-STAT5 antibody and DAPI (h). Immunostained cells were examined by a confocal microscope, and representative images are presented with pseudocolor. Scale bars are 8 µm. (i) HEK293 cells were treated with DKK2 for times indicated. Biotinylated cell surface proteins and cell lysate proteins were analyzed by Western blotting. The LRP5 blots were quantified and normalized against total LRP5. Data are presented as means±sem with P values (a,c,g,e), and experiments were repeated three times (a,b,d-i) or twice (c).
The importance of LRP5 in tumor progression and the anti-tumor effect of DKK2 blockade were then tested using hematopoietic-specific knockout of LRP5, which was generated by adoptive transfer of bone marrows (BMs) from the Lrp5fl/flMx1Cre mice into lethally irradiated WT C57BL/6 mice. The lack of LRP5 in hematopoietic cells impaired the MC38 tumor progression (Fig. 6c). Importantly, 5F8 did not affect the tumor progression in these LRP5 mutant mice, while it retained its effect in the control mice (Fig. 6c). Flow cytometry analysis of tumor-infiltrated leukocytes supported the conclusion that 5F8 exerts its effects on cytotoxic immune cell activation via LRP5 (Supplementary Fig. 9c-d).
We also tested the importance of NK LRP5 in tumor progression and the anti-tumor effect of DKK2 blockade using NK-specific LRP5 knockout mice (Lrp5f/fNcr1-Cre). MC38 tumor progressed slower in the Lrp5f/fNcr1-Cre mice than the control mice, and 5F8 showed marginal anti-tumor effects in these NK-specific LRP5 mutant mice (Supplementary Fig. 9e). Concordantly, NK-specific LRP5 knockout led to increases in NK cell activation markers, but had no effects on GZMB or CD69 in CD8+ cells (Supplementary Fig. 9f-g).
LRP5C interacts with and inhibits STAT5
To understand how LRP5 interferes with phosphorylated STAT5 nuclear localization, we performed co-immunoprecipitation and found that LRP5 intracellular domain (LRP5C) interacted with STAT5 (Fig. 6d). We next found that LRP5C inhibited IL-15-mediated activation of STAT5 reporter gene activity in HEK293 cells expressing JAK3, IL2/15 β and common γ receptor subunits (Fig. 6e) without affecting STAT5 phosphorylation (Fig. 6f). LRP5C also inhibited the STAT5 reporter gene activity stimulated by a constitutively active JAK1 mutant (V658F) 43 expressed in HEK293 cells without altering STAT5 phosphorylation, but impairing the nuclear localization of phospho-STAT5 (Fig. 6g-h). These data are consistent with observations made in the primary NK cells and support the conclusion that DKK2 inhibits IL-15 signaling by impeding the nuclear localization of phospho-STAT5 probably via LRP5C interaction with STAT5 as modeled in Supplementary Fig. 9h. The observation that DKK2 induces rapid internalization of LRP5 rather than LRP6 (Fig. 6i) provides a distinction between LRP5 and LRP6.
DKK2 suppresses tumor immune responses to anti-PD-1
The combination of DKK2 blockade with PD-1 blockade was tested in the MC38 tumor model. While both PD-1 and DKK2 blockade showed tumor suppressive effects, the combination yielded further anti-tumor effects (Supplementary Fig. 10a,b). Notably, a small fraction of tumors treated with the combination showed long-term regression (Supplementary Fig. 10a). Flow analysis showed that while individual blockades led to increases in GZMB in tumor-infiltrated CD8+ and NK cells, the combination blockade resulted in further increases in GZMB in these cells (Supplementary Fig. 10c-d). Intratumoral administration of recombinant DKK2 protein was performed to directly assess the effect of DKK2 on tumor immune responses elicited by PD-1 blockade. DKK2 inhibited PD-1 blockade-induced increases in the numbers of tumor-infiltrated CD45+ and CD8+ cells and activation of CD8+ and NK cells (Supplementary Fig. 10e). These results suggest that DKK2 can suppress effects of anti-PD-1 on immune effector cell activation. This conclusion is further supported by the observation that the effects of anti-PD-1 on the tumor suppression and activation of tumor-infiltrated immune effector cells become insignificant in APC-loss MC38 cell-derived tumors (Supplementary Fig. 10f,g), where DKK2 expression is upregulated (Supplementary Fig. 1e).
Analysis of the skin cutaneous melanoma cohort (TCGA, Provisional) revealed correlations of PTEN-loss and PI3K-gain of function mutations with elevated DKK2 expression (Supplementary Fig. 11a). These mutations lead to increases in phosphatidylinositol (3,4,5)-trisphosphate (PIP3) contents in cells. In addition, a trend of correlation of PD-1 resistance with increased DKK2 expression (Supplementary Fig. 11b)44 and significant correlation with PTEN-loss mutations 45 were observed in human melanomas. We thus tested the anti-tumor effect of DKK2 blockade using a YUMM1.7 mouse melanoma cells. The YUMM1.7 cells were derived from BrafV600EPten−/−Cdkn2a−/− melanoma developed in the C57BL/6 mice 46. The DKK2 mRNA content in YUMM1.7 cells, which is more than 10 folds higher than that in MC38 cells, was reduced by the PI3K inhibitor Wortmannin (Supplementary Fig. 11c), suggesting that DKK2 expression is regulated by PIP3 elevation. Importantly, 5F8 significantly impeded tumor progression and extended the survival of tumor-bearing mice in the YUMM1.7 tumor model (Supplementary Fig. 11d,e). Additionally, the DKK2 and PD-1 combination blockade significantly outperformed the individual blockade with a fraction of combination-treated mice showing stable disease (Supplementary Fig. 11d,e). Flow analysis of tumor infiltrated leukocytes showed significantly stronger activation of CD8+ and NK cells by the combination blockade than that of individual blockade (Supplementary Fig. 11f). These data support the conclusion alluded earlier that DKK2 could suppress anti-tumor immune responses elicited by PD-1 blockade.
Applicability of DKK2 blockade for treatment of other cancers
Activated KRAS mutations frequently occur in advanced human cancers. We tested DKK2 blockade in a mouse lung cancer model carrying an activating KRAS mutation. The model was generated by intra-nasal instillation of Cre-expressing adenovirus to the Apcfl/flKrasact/+ mice. Treatment of these mice with 5F8 significantly reduced tumor burden (Supplementary Fig. 12a), accompanied by increases in the numbers of CD8+ and NK cells as well as activation of these immune effector cells (Supplementary Fig. 12b-e). These results, together with the correlations of high DKK2 expression with poor survival rates and low GZMB expression for renal papillary carcinoma and bladder urothelial carcinoma based on the TCGA provisional database analysis (Supplementary Fig. 13a), suggest that DKK2 blockade may also be applicable to the treatment of cancers beyond CRCs. Given that DKK2 is generally expressed at low levels in various normal human and mouse tissues, particularly immune tissues (http://www.ebi.ac.uk/gxa/home), DKK2 blockade is unlikely a strong risk factor for autoimmunity. Indeed, neither DKK2-deficiency nor LRP5-deficiency altered various hematopoietic cell populations or NK cell development in mice housed under a specific pathogen-free condition (Supplementary Fig. 13b-e). On the other hand, DKK2 may exert more potent effects on NK cells in tumors than those observed in the in vitro assays, because LRP5 expression was upregulated in tumor infiltrated NK cells compared to that of primary NK cells from spleens (Supplementary Fig. 13f).
DISCUSSION
In this study, we uncovered a function of DKK2 in promoting tumor progression by suppressing NK and CD8+ cell activation in a LRP5-dependent manner. It remains unclear why LRP6 is not required for this DKK2 action. The ability for DKK2 to induce internalization of only LRP5 (Fig. 6i) may provide an explanation. DKK1 or Wnt3A also fails to induce internalization of endogenous LRP647,48. Three putative adaptor protein-2-binding motifs in LRP5 in contrast to just one in LRP6 may underlie the difference in their internalization48.
NK cell-expressed STAT5 has direct roles in tumor immune surveillance49. Together with NK-specific LRP5 knockout results, we believe that DKK2-mediated impediment of STAT5 nuclear localization observed in this study is the primary mechanism for DKK-mediated suppression of tumor immunity. DKK2 exerted a clear effect on STAT5 nuclear localization, but the effect is only partial (Fig. 5b). This partial effect may explain why DKK2 only affects NK cell activation rather than NK cell development, which in turn explains a lack of effect of DKK2 inhibition on NK cell number in mice, even though deficiency of STAT5 or IL15 receptor α subunit has a profound effect on NK cell development50–52. Thus, NK cell development and full activation may have different thresholds for STAT5 signaling.
DKK2 can also inhibit IL-15-mediated activation of CD8+ cells (Supplementary Fig. 6d) presumably through a similar mechanism. Of note, LRP5 expression is upregulated in human mature CD8+ cells53, suggesting that DKK2 may also exert a stronger effect on CD8+ cells in vivo. However, DKK2 does not inhibit T cell receptor-mediated activation of the primary T cells (data not shown). This explains the lack of an effect of DKK2 blockade on T cell populations. The activation of CD8+ cells by DKK2 blockade could also be a result of a combination of DKK2’s direct and NK cell-mediated indirect regulation of CD8+ cells6,11,12. Consistent with the prominent role of IL-15-STAT5 signaling in CD8+ intraepithelial cells13–15, DKK2 was able to inhibit CD8+ IELs isolated from mouse intestines (Supplementary Fig. 6e). The direct inhibition of IELs by DKK2 may play a larger role in DKK2 blockade-associated increases in GZMB-positive CD8+ cells and tumor suppression in the Apcmin/+ intestinal tumor model, which is consistent with the CD8+ cell depletion results (Supplementary Fig. 5d). Therefore, the potent anti-tumor effects of DKK2 blockade in vivo may be the results of its direct effects on NK and/or CD8+ cells, but the relative contributions of these mechanisms may be context-dependent.
This study shows that APC-loss drives DKK2 expression in CRCs, whereas PIP3 elevation mutations drive it in melanomas. Evidence also presented that DKK2 provides a resistance to PD-1 blockade in mouse models. These results suggest that DKK2 blockade may be used as single or combination therapies to treat CRCs, the MSS CRCs in particular, and melanomas harboring PIP3 elevation mutations. DKK2 blockade may also be applied for the treatment of other human cancers where DKK2 is highly expressed. These possibilities and the therapeutic potential of blockade of LRP5 warrant future investigations.
ONLINE METHODS
Mice
Dkk2−/− mice were described previously 30,32 and have been backcrossed to C57Bl background for more than 7 generation. Ncr1-Cre mice were also described previously 52. NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ), ApcMin/+ (C57BL/6J-ApcMin/J), MX1Cre [B6.Cg-Tg(Mx1-cre)1Cgn/J], KrasLSL-G12D (B6.129S4-Krastm4Tyj/J), and Apcf/f (C57BL/6-Apctm1Tyj/J) mice were acquired from the Jackson Laboratory. Wildtype C57BL/6 mice were purchased from Envigo (Harlan). The LoxP-floxed Lrp5 (Lrp5f/f) and Lrp6 (Lrp6f/f) mice were obtain from Bart Williams 54. The Lrp5f/f and Lrp6f/f mice were backcrossed to C57/BL6 for more than 7 generations before being intercrossed with MX1Cre. LRP5 and LRP6 gene disruption was induced by intraperitoneal injection of the Lrp5fl/flMX1Cre mice with 40 µl poly-I:C (10 mg/mL) every other day for 4 treatments. The mice were used for NK cell isolation three weeks after the poly-I:C treatment. For adoptive bone marrow transfer, bone marrows from the Lrp5fl/flMX1Cre mice were transferred to lethally irradiated C57/BL6 mice (8 weeks old female) via retro-orbital injection. After recovery (8 weeks), the mice were treated with poly-I:C and used in experiments three weeks after poly-I:C treatment. Mice were housed in specific pathogen free condition and cared for in accordance with US National Institutes of Health guidelines, and all procedures were approved by the Yale University Animal Care and Use Committee.
Antibodies and cells
Antibodies to phospho-Stat5 (Tyr694) (CST, 4322s), LAMP1 (sc-19992, Santa Cruz), EEA1 (BD Bioscience, 612006), phospho-AKT(serine 473) (CST, 4060), AKT1 (CST, 9272), phospho-ERK1/2 (Thr202/Tyr204) (CST, 4377), ERK1/2 (CST 9102), perforin (CST, 3693), granzyme B (CST, 4275), β-actin (CST, 3700), FLAG (Sigma Aldrich, F3165), β-catenin (BD Bioscience, 610153), LRP5 (CST, 5731), LRP6 (CST, 3395), mouse CD4-PE (eBioscience, 12-0042-82), mouse NK1.1-APC (BioLegend, 108710), mouse CD8a-PE-Cyanine7 (eBioscience, 25-0081-82), mouse CD69-PE (Biolegend ,104508), human/mouse granzyme B-FITC (BioLegend, 515403), mouse CD314 (NKG2D)-PE-Cyanine7 (eBioscience, 25-5882-81), mouse CD3e-PE (eBioscience, 12-0031-82), mouse IFNγ-PE (eBioscience, 12-7311-81), CTLA-4 / CD152 (1B8)-FITC (Thermo Fisher, HMCD15201), human CD45-eFluor® 450 (eBioscience, 48-0459-41), mouse CD107a-V450 (BD, 560648), mouse CD8a-APC (eBioscience, 17-0081-81), mouse CD25-Alexa Fluor® 488 (eBioscience, 53-0251-82), mouse CD279 (PD-1)-PE (BioLegend, 135205), mouse CD19-PE-Cyanine7 (eBioscience, 25-0193-81), mouse CD3e eFluor® 450 (eBioscience, 48-0031-82), mouse CD11b-PE (eBioscience, 12-0112-82), mouse CD27-FITC (eBioscience, 11-0271-82), Ki67 (Abcam ab,15580), Cleaved Caspase-3 (Asp175) (CST, 9661S), CD31 (Abcam ab, 28364), CD44 antibody (Abcam ab,157107), Fluorescein (FITC)-labeled AffiniPure F(ab')2 Fragment Donkey Anti-Mouse IgG (H+L) (Jackson lab, 715-096-151), Mouse Integrin alpha 4 beta 7 (LPAM-1) APC (eBioscience, 17-5887-80), Human CD56 (NCAM) APC (eBioscience, 17-0566-41), Human CD16 PE (eBioscience, 12-0167-42), Human CD3 eFluor® 450 (eBioscience, 48-0037-42), and Alexa Fluor® 647-labeled AffiniPure F(ab')2 Fragment Goat Anti-Rabbit IgG (H+L) (Jackson lab, 111-606-045). Mouse monoclonal antibody to DKK2 (5F8) was generated using standard hybridoma technology through immunization of mice with a synthetic peptide (KLNSIKSSLGGETPGC) of human DKK2 at AbMax (Beijing, China). Therapeutic anti-PD-1 antibodies are hamster mAb clone G4 55 and Clone J43 (BioXcell, BP0033-2) with polyclonal Armenian Hamster IgG (BioXcell, BE0091) as the control IgG.
HEK293 and HCT-116 cells were purchased from ATCC. YUMM1.7 was provided by Marcus Bosenberg and reported previously 46. MC38 was purchased from Kerafast (Boston, MA). Cells were all tested for mycoplasma and they were negative.
Quantitative RT-PCR
Total RNAs were isolated from cells using the RNeasy Plus Mini Kit (QIAGEN). Complementary DNAs were synthesized from the RNAs using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR was done using the iTaq Universal SYBR Green Supermix (Bio-Rad). The primer sequences are listed in Supplementary Table II. The intestinal epithelial stem cell and differentiation markers are selected based on the following references 56–59
ELISA
Recombinant mouse DKK2 or DKK1 protein (20 ng/ml, R&D) in a blocking buffer (1% BSA in PBS) was incubated in a 384-well microtiter plate for overnight at 4°C. The plate was washed with PBS twice and incubated with the blocking buffer for one hour at room temperature. The plate was then incubated with the anti-DKK2 5F8 antibody in the blocking buffer for 1 hour at room temperature. After repeatedly washing, the plate was incubated with an HRP-conjugated secondary antibody for 1 hour at room temperature. A chemiluminance substrate (Thermo Fisher 37070) was added to the plate, and the plate was read by an EnVision plate reader.
DKK2-AP binding assay
The binding assay was performed as previously described 60. In brief, HEK293 cells were transfected with LacZ or LRP5 using Lipofectamine Plus for 24 hours. Cells were washed once with a cold washing buffer (Hanks’ buffered salt solution containing 1% bovine serum albumin, 20 mM HEPES, and 0.5% NaN3) and incubated with the washing buffer containing 20% of DKK2-AP conditioned medium on ice for 2 h. The cells were then washed three times with the washing buffer and lysed with 1% Triton X-100 and 20mM Tris-HCl, pH 7.5. The lysates were heated at 65 °C for 10 min to inactivate endogenous AP and then added with a chemiluminescence AP substrate (Thermo Fisher T1015). The activity was measured by an EnVision plate reader.
Tumor Graft
MC38 or YUMM1.7 melanoma tumor cells (0.5–1×106) were mixed with BD Matrigel (Matrix Growth Factor Reduced) (BD 354230) in 100 µl and inoculated subcutaneously at the right flanks of the backs of female C57/BL mice (8–10 weeks old). Tumor growth was measured by calipers, and size was expressed as one-half of the product of perpendicular length by square width in cubic millimeters. For antibody treatment, control IgG3 antibody and anti-DKK2 antibody were diluted in PBS, and 100 µl was injected i.p. at intervals indicated in the Figures. For survival tests, mice are euthanized when the tumor size exceeding 1500 mm3 for MC38 and 1200 mm3 for YUMM1.7.
Preparation of tumor infiltrating leukocytes
Tumors were minced using scissors and scalpel blades and incubated with a digestion buffer [RPMI1640, 5% FBS, 1% PS, 25mM HEPES and 300U collagenase (Sigma C0130)] in a shaker for 2h at 37°C. Disperse cells were filtered through a 70 µm cell strainer to eliminate clumps and debris. After centrifugation for 5 minutes (500xg) at 4°C, cell pellets were resuspended in the Red Blood Cell Lysis Buffer (Sigma R7757) and incubated at RT for 5 min to remove erythrocytes. Cells were pelleted again, resuspended and incubated in 0.05% Trypsin/EDTA at 37°C for 5min, followed by DNA digestion with the addition of Type I DNase (1 µg/ml final, Sigma D4263) for 5 min. Trypsin digestion was stopped by the addition of FBS to 5%, and cells were filtered again by a 40 µm cell strainer. Finally, the cells were pelleted again and resuspended in PBS at a concentration of 2 × 107.
Flow cytometry
Cells in single cell suspension were fixed with 2% PFA (Santa-Cruz sc-281692). After washing with a Flow Cytometry Staining Buffer (eBioscience 00-4222-26), cells were stained with antibodies for cell surface markers for 1 hour on ice in the dark. For staining of intracellular proteins, the cells were washed and resuspended in the Permeabilization Buffer (BD 554723) and stained by antibodies in the Permeabilization Buffer for 1 hour on ice in the dark. The cells were then pelleted and resuspended in the Flow Cytometry Staining Buffer for flow cytometry analysis.
Tumor Sectioning and immunostaining
Tissues were fixed with 4% PFA (Santa-Cruz sc-281692) for 4–6 hours on a shaker at 4°C. They were then washed with PBS three times and perfused in 20% sucrose solution in PBS overnight at 4°C. They were subsequently mounted in the OCT embedding compound and frozen first at −20 and then at −80 °C. Tissue sections were prepared at 8 µm thickness using a cryostat and mounted onto gelatin-coated histological slides, which were stored at −80°C.
For immunostaining, slides were thawed to room temperature and fixed in pre-cold acetone for 10 minutes, followed by rehydration in PBS for 10 minutes. The slides were incubated in a blocking buffer (1% horse serum and 0.02% Tween 20 in PBS) for 1–2 hours at room temperature, followed by incubation with primary antibodies, which were diluted in an incubation buffer (1% horse serum, 0.02% Tween 20 in PBS), overnight at 4 °C. The slides were then washed three times with PBS and incubated with a secondary antibody [donkey anti-rabbit IgG H&L (DyLight® 550) preadsorbed (abcam ab96920)] in the incubation buffer for 1 hour at room temperature. After repeated washes the slides were mounted with an anti-fade mounting media containing DAPI (Thermo Fisher P36931) and visualized using a confocal microscope.
Cytotoxic effector immune cell depletion
For depletion of NK cells with the MC38 model, the anti-NK1.1 (PK136, BioXcell BE0036) or isotype control (BioXcell BE0085) was injected i.p. at 300 ug/mouse at Day -1, 5, 11 and 17 of tumor cell inoculation. For CD8 depletion, the anti-CD8α (YTS169.4, BioXcell BE0117) or isotype control (Clone LTF-2, BioXcell BE0090) was injected i.p. at 300 ug/mouse at Day 12, 15 and 19 of tumor cell inoculation.
For the Apcmin/+ model, NK1.1+ and CD8+ cells were depleted by i.p. injection of 10 mg/Kg anti-NK1.1 (PK136, BioXcell BE0036) and anti-CD8α (YTS169.4, BioXcell BE0117), respectively. Male ApcMin/+ mice were treated weekly with the control IgG3 antibody or 5F8 (8 mg/Kg) starting at the age of 15 weeks. The depletion antibodies were given six days before the therapeutic treatment at an interval of 5 days. The mice were examined four weeks after the treatment.
Preparation and treatment of mouse primary NK, CD8+ and IEL cells
Mouse primary NK and CD8+ T cells were isolated from the spleens of female mice (8 weeks old) by using the NK cell and CD8+ T cell isolation kits according to the manufacturer's instructions (Miltenyi Biotec #130-090-864 and #130-104-075), respectively. Primary NK cells were cultured in RPMI-1640 (Gibco, 11875-093) supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 µg/ml), 2-mercaptoethanol (500µM) and HEPES (10mM) at 37 °C supplemented with 5% CO2 in the presence of recombinant murine IL-15 (50 ng/ml) for 24 hours before treatment with DKK2, CHIR99021, or WNT3A. CD8+ T cells were cultured in the same culture medium and condition as NK cells, but supplemented with IL-15 (200 ng/ml) and IL-15 Rα (1 µg/ml recombinant Mouse IL-15 receptor alpha Fc chimera Protein from R&D) for 96 hours before DKK2 treatment.
Mouse primary intraepithelial lymphocytes (IELs) were prepared as described in 61,62. In brief, the small intestine was everted, divided into four pieces, and washed twice in phosphate-buffered saline (PBS) containing 100 U/ml penicillin/streptomycin. The specimens were then incubated with stirring at 37°C in prewarmed Ca2+ and Mg2+-free Hanks’ solution containing 100 U/ml penicillin-streptomycin, 5% fetal calf serum (FCS), 2 mM dithiothreitol (DTT), and 5mM EDTA for 30 min, followed by vigorous shaking for 30s. The supernatants were passed over two nylon wool columns to remove undigested tissue debris. The lymphocytes obtained were pooled and enriched on a discontinuous (40% and 70%) Percoll density gradient. Cells at the interface between the 40% and 70% fractions (IELs) were collected for treatment with IL-15 (200 ng/ml) and DKK2 (400 ng/ml), followed by flow analysis.
Preparation of human NK cells
Peripheral blood mononuclear cells from normal humans were purchased from ZenBio (SER-PBMC-200). Human NK cells were isolated from the PBMCs by using the human NK cell isolation kit according to the manufacturer's instruction (Miltenyi Biotec # 130-092-657). Human NK cells were cultured in RPMI-1640 (Gibco, 11875-093) supplemented with 10% heat-inactivated FBS, penicillin (100 U/ml), streptomycin (100 µg/ml), 2-mercaptoethanol (500µM) and HEPES (10mM) at 37 °C supplemented with 5% CO2 in the presence of recombinant human IL-15 (50 ng/ml) before treatment with recombinant human DKK2 protein.
NK and tumor cell Co-Culture
Primary NK cells were isolated from the spleens as described above and cultured in the presence of 50 ng/ml recombinant murine IL-15 for 24 hours. Meanwhile, tumor cells were plated in the 96 well plate for overnight. NK cells were added into the tumor cells at 7:1 radio in the presence of the IgG3 antibody or anti-mDKK2 5F8 for 9 hours at 37 °C. For testing the effect of DKK2 in co-culture, isolated NK cells were cultured in the presence of 50 ng/ml recombinant murine IL-15 for 24 hours and then cultured in the presence or absence of DKK2 for another 24 hours before the NK cells were added to pre-seeded MC38 cells at 7:1 (NK:MC38) radio. The numbers of live tumor cells were determined by a Guava flow cytometer (EMDmillipore), whereas the cell apoptosis was assessed by flow cytometry using an annexin v apoptosis detection kit (eBioscience, 88-8007).
Immunocytostaining
Primary NK cells were prepared as described above and treated as indicated in the Figures. They were then placed on poly-lysine coated coverslips and incubated for 30 min at room temperature. HEK293 cells grown on coverslips were transfected and stimulated as indicated in the Figures. Cells were fixed with 4%PFA for 10min at room temperature and permeabilized with ice-cold methanol for 10min at −20°C. After rinsed with PBS for 3 times, cells were blocked with a blocking buffer (5% normal donkey serum and 0.5% triton in PBS) for 1hr at room temperature. Primary antibodies were then diluted in PBS with 0.5%BSA and applied to cells with overnight incubation at 4°C. Cells were rinsed with PBS for 3 times and incubated with diluted fluorochrome-conjugated secondary antibodies (in PBS with 1% BSA) for 1hr at room temperature. Finally, cells were rinsed with PBS for 3 times and mounted with Prolong Gold Antifade solution (Thermo Fisher) for confocal microscopy.
Quantification of subcellular localization was done by determining the nuclear/cytoplasmic ratio of pSTAT5 staining intensities. The staining intensities of pSTAT5 in the nuclei, which was delineated by the DAPI staining, and in the cytosol, which was delineated by RAB8 staining, were quantified using Image J.
Immunoprecipitation
293T cells were transfected with plasmids encoding STAT5 and/or LRP5C-Flag with Lipofectamine Plus. The cells were lysed 24 hours after transfection in the lysis buffer [50 mM HEPES (pH 7.4), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 2mM MgCl2, 2mM EGTA] with the protease inhibitors cocktail (Roche) and phosphatase inhibitors (Phospo-stop from Roche) on ice. Cell lysates were centrifuged to remove insoluble materials. Immunoprecipitation was performed with an anti-Flag antibody for overnight, followed by 2-hour incubation of Protein-A/G Plus beads (Santa Cruz), at 4 degree. The beads were washed repeatedly, and bound proteins were analyzed by Western blotting.
Reporter gene assays
The Stat5 reporter assays were done in HEK293 cells for activated JAK1-induced activity or in those stably expressing JAK3, IL2/15Rβ and the common receptor γ subunit for IL15 induced activity. Cells were seeded at 8×104 cells per well in 48 well plate. The next day, cells were transfected by Lipofectamine 2000 (Invitrogen) with the pGL4.52-STAT5-Luciferase (Promega) and tagRFP (internal control) plasmids together with other plasmids expressing genes of interest. The total amount of plasmid was kept at 125ng per well. The cells were added 24 hours after transfection with IL15/IL15Rα-Fc complex or mock. Six hours later, the cells were lysed and subjected to RFP fluorescence and luciferase luminescence measurement using an Envision plate reader. The reporter gene activity is shown after being normalized against RFP readings.
The Wnt reporter gene assay was carried out in HEK293 cells that were transfected with the TOPFlash and GFP plasmids. The rest is the same as above. The reporter gene activity is shown after being normalized against GFP readings.
Generation of APC mutant cells
Gene editing of the APC genes in MC38 and HCT116 cells was done using the CRISPR-Cas9 system as previously described 63. The cells were transfected with two Cas9 plasmids expressing two guiding RNAs targeted to the APC gene. This causes homozygous C-terminal deletion of the APC protein starting at Gly-855 in MC38 cells. As these two guiding RNAs were coexpressed with GFP or RFP, respectively, the GFP+RFP+ cells were sorted directly into 96 well plates at the density of 1.2 cells/well. Homozygous mutations in the APC or Apc gene were detected by PCR and confirmed by DNA sequencing. Positive clones were pooled to avoid clonal effects. The guiding and PCR sequences are listed in Supplementary Table II.
LRP5/6 internalization assay
HEK293 cells were treated with mock or recombinant mouse DKK2 protein (250 ng/ml) in culture medium for duration indicated. The cells were washed with pre-cold PBS and cell surface proteins were biotinylated with 0.5mg/ml of EZ-Link Sulfo-NHS-SS-Biotin (Thermo Fisher, 21445) in a PBS buffer on ice for 30 min. The reaction was stopped by addition of PBS containing ice-cold 50mM NH4Cl, followed by repeated washes with ice-cold PBS. The cells were then lysed with in a buffer containing 1.25% Triton X-100, 0.25% SDS, 50mM Tris HCl PH8.0, 150mM NaCl, 5mM EDTA, 5mg/ml iodoacetamide, 10ug/ml PMSF, and the Roche proteinase inhibitor cocktail. After centrifugation, aliquots were taken as lysate controls, and the rest of supernatants were used in pull-down with NeutrAvidin beads (Thermo Fisher, 29200), followed by analysis by Western blotting.
Cell fractionation
Cell fractionation was carried out the Cell Fractionation Kit from Cell Signaling Technology (# 9038) according to its protocol. In brief, primary NK cells (2 million) were washed with cold PBS and resuspended in the cytoplasm isolation buffer (CIB). After centrifugation, the supernatant was collected as the cytosol fraction and the pellet was resuspended in the membrane isolation buffer (MIB). After centrifugation, the supernatant was collected with the membrane fraction, whereas the pellet was collected as nuclear fraction. The nuclear fraction and the pool of the membrane and cytosolic fractions were subjected for Western analysis.
RNA sequencing and data analysis
Primary NK cells were isolated from the spleens as described above and cultured in the presence of 50 ng/ml recombinant murine IL-15 for 24 hours and then cultured in the presence or absence of 500 ng/ml DKK2 for another 24 hours before mRNA was isolated and purified by using RNeasy Plus Mini Kit (Qiagen). RNA-seq libraries were prepared using the TrueSeq Stranded Total RNA Library Prep Kit (Illumina) and sequenced on Illumina HiSeq 2500 with 50 base single end read.
Raw sequence reads were mapped using TopHat in Galaxy (Version 2.1.0) with ENCODE annotation M1. The raw counts were then normalized using the trimmed mean of M values (TMM) method and compared using the Bioconductor package “edgeR”. Reads per kilobase per million (RPKM) mapped reads were also calculated from the raw counts and are deposited to GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100402). Differentially expressed genes were identified if RPKM ≥1 in at least one sample, fold change was ≥1.25. Pathway analysis of RNA sequencing results were performed at http://amp.pharm.mssm.edu/Enrichr/enrich. Gene enrichment analysis was performed with Motif Gene Set (http://software.broadinstitute.org/gsea/msigdb/index.jsp) 64.
In situ hybridization
In situ hybridization detection of Dkk2 mRNA was carried out using the following reagents acquired from Advanced Cell Diagnostics, INC based on provided protocol: RNAscope® Target Retrieval Reagents (Cat# 322000), RNAscope® Pretreat Reagents- H202 and ProteasePlus (Cat# 322330), RNAscope® 2.5 HD Detection Reagent – RED (Cat# 322360), RNAscope® Wash Buffer Reagents (Cat# 310091), BioCare EcoMount (Cat# 320409), ImmECatdge™ Hydrophobic Barrier Pen (Cat# 310018) and the mouse Dkk2 probe (#404841).
Correlation of DKK2 expression and patient survival
The DKK2 expression, overall survival, and relapse-free survival data were obtained from the TCGA provisional datasets as of July 20, 2016. The high and low DKK2 expressers were grouped using an arbitrary cutoff percentile of 15%. The Mantel-Cox Log-Rank tests were done using the GraphPad Prism 7 software.
Statistical analysis and study design
Minimal group sizes for tumor progression studies were determined by using power calculations with the DSS Researcher's Tookit with an α of 0.05 and power of 0.8. Animals were grouped unblinded, but randomized, and investigators were blinded for the qualification experiments. No samples or animals were excluded from analysis. Assumptions concerning the data including normal distribution and similar variation between experimental groups were examined for appropriateness before statistical tests were conducted. Comparisons between two groups were performed by unpaired, two tailed t-test. Comparisons between more than two groups were performed by one-way ANOVA, whereas comparisons with two or more independent variable factors by two-way ANOVA, followed by Bonferroni’s post-hoc correction using Prism 6.0 software (GraphPad). Statistical tests were using biological replicates. P<0.05 is considered as being statistically significant.
Life Sciences Reporting Summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Data Availability and Accession Code Availability Statements
The raw RNA-seq data are available in the NCBI Gene Expression Omnibus (GSE100402).
Supplementary Material
Acknowledgments
We thank Michelle Orsulak for technical assistance and Bart Willaims for providing the LRP5/6 floxed mice. The work is supported by an NIH grant (GM112182 and CA214703 to D.W.), Connecticut Bioscience Innovation Fund (to D.W.), a NSFC grant (31530094 to L.L.), the strategic priority research program of CAS (XDB19000000 to L.L), and the CAS/SAFEA International Partnership Program for Creative Research Teams (to L.L. and D.W).
Footnotes
Author contributions: D.W., L.L., Q. X., J.W., W.-J.W., W.T., M.S., and J. C. designed the experiments; Q. X., J.W., W.-J.W., S.C., M.S., W.T., and K.M. performed the experiments; D.W., L.L., Q. X., W.-J.W., M.S., J.W., A.L.M.B., L.C., and W.T. analyzed the data; X.Y. performed statistic and bioinformatic analyses; M.B., V.S., and L.S. created and provided important reagents. D.W., L.L., Q.X., J.W., W.-J.W., and W.T. wrote the manuscript; and all authors reviewed and approved the manuscript.
Competing Financial Interests: D.W. received research support from Just Biotherapeutic Asia, which licensed the intellectual property from Yale University based on the findings reported in this article.
References
- 1.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature reviews. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palucka AK, Coussens LM. The Basis of Oncoimmunology. Cell. 2016;164:1233–1247. doi: 10.1016/j.cell.2016.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung KY, et al. Phase II study of the anti-cytotoxic T-lymphocyte-associated antigen 4 monoclonal antibody, tremelimumab, in patients with refractory metastatic colorectal cancer. J Clin Oncol. 2010;28:3485–3490. doi: 10.1200/JCO.2010.28.3994. [DOI] [PubMed] [Google Scholar]
- 10.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcus A, et al. Recognition of tumors by the innate immune system and natural killer cells. Adv Immunol. 2014;122:91–128. doi: 10.1016/B978-0-12-800267-4.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crouse J, Xu HC, Lang PA, Oxenius A. NK cells regulating T cell responses: mechanisms and outcome. Trends Immunol. 2015;36:49–58. doi: 10.1016/j.it.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 14.Mishra A, Sullivan L, Caligiuri MA. Molecular pathways: interleukin-15 signaling in health and in cancer. Clin Cancer Res. 2014;20:2044–2050. doi: 10.1158/1078-0432.CCR-12-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klose CS, et al. The transcription factor T-bet is induced by IL-15 and thymic agonist selection and controls CD8alphaalpha(+) intraepithelial lymphocyte development. Immunity. 2014;41:230–243. doi: 10.1016/j.immuni.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 16.Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakobisiak M, Golab J, Lasek W. Interleukin 15 as a promising candidate for tumor immunotherapy. Cytokine Growth Factor Rev. 2011;22:99–108. doi: 10.1016/j.cytogfr.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Zarogoulidis P, et al. Interleukin-7 and interleukin-15 for cancer. J Cancer. 2014;5:765–773. doi: 10.7150/jca.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hukelmann JL, et al. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat Immunol. 2016;17:104–112. doi: 10.1038/ni.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nandagopal N, Ali AK, Komal AK, Lee SH. The Critical Role of IL-15-PI3K-mTOR Pathway in Natural Killer Cell Effector Functions. Frontiers in immunology. 2014;5:187. doi: 10.3389/fimmu.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 22.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 23.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clevers H, Nusse R. Wnt/β-Catenin Signaling and Disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 26.Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 28.Bao J, Zheng JJ, Wu D. The structural basis of DKK-mediated inhibition of Wnt/LRP signaling. Sci Signal. 2012;5:pe22. doi: 10.1126/scisignal.2003028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gage PJ, Qian M, Wu D, Rosenberg KI. The canonical Wnt signaling antagonist DKK2 is an essential effector of PITX2 function during normal eye development. Dev Biol. 2008;317:310–324. doi: 10.1016/j.ydbio.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, et al. Dkk2 has a role in terminal osteoblast differentiation and mineralized matrix formation. Nat Genet. 2005;37:945–952. doi: 10.1038/ng1614. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay M, et al. Dkk2 plays an essential role in the corneal fate of the ocular surface epithelium. Development. 2006;133:2149–2154. doi: 10.1242/dev.02381. [DOI] [PubMed] [Google Scholar]
- 32.Li X, et al. Chemical and genetic evidence for the involvement of Wnt antagonist Dickkopf2 in regulation of glucose metabolism. Proc Natl Acad Sci U S A. 2012;109:11402–11407. doi: 10.1073/pnas.1205015109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W, Glinka A, Delius H, Niehrs C. Mutual antagonism between dickkopf1 and dickkopf2 regulates Wnt/beta-catenin signalling. Curr Biol. 2000;10:1611–1614. doi: 10.1016/s0960-9822(00)00868-x. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Mao J, Sun L, Liu W, Wu D. Second cysteine-rich domain of Dickkopf-2 activates canonical Wnt signaling pathway via LRP-6 independently of dishevelled. J Biol Chem. 2002;277:5977–5981. doi: 10.1074/jbc.M111131200. [DOI] [PubMed] [Google Scholar]
- 35.Caneparo L, et al. Dickkopf-1 regulates gastrulation movements by coordinated modulation of Wnt/beta catenin and Wnt/PCP activities, through interaction with the Dally-like homolog Knypek. Genes Dev. 2007;21:465–480. doi: 10.1101/gad.406007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaedcke J, et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 2010;49:1024–1034. doi: 10.1002/gcc.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsui A, et al. DICKKOPF-4 and-2 genes are upregulated in human colorectal cancer. Cancer Sci. 2009;100:1923–1930. doi: 10.1111/j.1349-7006.2009.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 40.Afonina IS, Cullen SP, Martin SJ. Cytotoxic and non-cytotoxic roles of the CTL/NK protease granzyme B. Immunol Rev. 2010;235:105–116. doi: 10.1111/j.0105-2896.2010.00908.x. [DOI] [PubMed] [Google Scholar]
- 41.D'Amico L, et al. Dickkopf-related protein 1 (Dkk1) regulates the accumulation and function of myeloid derived suppressor cells in cancer. J Exp Med. 2016 doi: 10.1084/jem.20150950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malladi S, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell. 2016;165:45–60. doi: 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haan C, et al. Jak1 has a dominant role over Jak3 in signal transduction through gammac-containing cytokine receptors. Chem Biol. 2011;18:314–323. doi: 10.1016/j.chembiol.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Hugo W, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng W, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaur A, et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016 doi: 10.1038/nature17392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semenov MV, Zhang X, He X. DKK1 antagonizes Wnt signaling without promotion of LRP6 internalization and degradation. J Biol Chem. 2008 doi: 10.1074/jbc.M800014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim I, et al. Clathrin and AP2 are required for PtdIns(4,5)P2-mediated formation of LRP6 signalosomes. J Cell Biol. 2013;200:419–428. doi: 10.1083/jcb.201206096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gotthardt D, et al. STAT5 Is a Key Regulator in NK Cells and Acts as a Molecular Switch from Tumor Surveillance to Tumor Promotion. Cancer Discov. 2016;6:414–429. doi: 10.1158/2159-8290.CD-15-0732. [DOI] [PubMed] [Google Scholar]
- 50.Imada K, et al. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teglund S, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 52.Eckelhart E, et al. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood. 2011;117:1565–1573. doi: 10.1182/blood-2010-06-291633. [DOI] [PubMed] [Google Scholar]
- 53.Wu B, Crampton SP, Hughes CC. Wnt signaling induces matrix metalloproteinase expression and regulates T cell transmigration. Immunity. 2007;26:227–239. doi: 10.1016/j.immuni.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui Y, et al. Lrp5 functions in bone to regulate bone mass. Nat Med. 2011;17:684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirano F, et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005;65:1089–1096. [PubMed] [Google Scholar]
- 56.Belov L, Zhou J, Christopherson RI. Cell surface markers in colorectal cancer prognosis. Int J Mol Sci. 2010;12:78–113. doi: 10.3390/ijms12010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sansom OJ, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarraf P, et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat Med. 1998;4:1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 59.Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nature reviews. Cancer. 2014;14:468–480. doi: 10.1038/nrc3744. [DOI] [PubMed] [Google Scholar]
- 60.Wang K, et al. Characterization of the Kremen-binding site on Dkk1 and elucidation of the role of Kremen in Dkk-mediated Wnt antagonism. J Biol Chem. 2008;283:23371–23375. doi: 10.1074/jbc.M802376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Little MC, Bell LV, Cliffe LJ, Else KJ. The Characterization of Intraepithelial Lymphocytes, Lamina Propria Leukocytes, and Isolated Lymphoid Follicles in the Large Intestine of Mice Infected with the Intestinal Nematode Parasite Trichuris muris. The Journal of Immunology. 2005;175:6713–6722. doi: 10.4049/jimmunol.175.10.6713. [DOI] [PubMed] [Google Scholar]
- 62.Li Z, et al. Small intestinal intraepithelial lymphocytes expressing CD8 and T cell receptor gammadelta are involved in bacterial clearance during Salmonella enterica serovar Typhimurium infection. Infect Immun. 2012;80:565–574. doi: 10.1128/IAI.05078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq data are available in the NCBI Gene Expression Omnibus (GSE100402).