Abstract
Genome-scale network reconstructions have helped uncover the molecular basis of metabolism. Here we present Recon3D, a computational resource that includes three-dimensional (3D) metabolite and protein structure data and enables integrated analyses of metabolic functions in humans. We use Recon3D to functionally characterize mutations associated with disease, and identify metabolic response signatures that are caused by exposure to certain drugs. Recon3D represents the most comprehensive human metabolic network model to date, accounting for 3,288 open reading frames (representing 17% of functionally annotated human genes), 13,543 metabolic reactions involving 4,140 unique metabolites, and 12,890 protein structures. These data provide a unique resource for investigating molecular mechanisms of human metabolism. Recon3D is available at http://vmh.life.
It is widely recognized that progress in the biomedical sciences is hampered by the difficulty of integrating multiple disparate data types to obtain a coherent understanding of physiological and disease states1. A genome-scale network reconstruction represents a curated knowledge-base containing many different data types and sources, including high-quality genome annotation, assessment of biochemical properties of gene products, and a wide array of physiological functional information. Computational genome-scale models integrate large-scale omics data from these knowledge-bases to aid in the interpretation and prediction of biological functions2. In recent years, human metabolic network reconstructions3–6 have generated insights into inborn errors of metabolism7, cancer8 and human microbiome co-metabolism9,10.
Using metabolic reconstructions, information about chemical reactions is stored and continually updated in a standardized biochemical and genetic representation through a well-established process11. Over the past ten years, updating the human metabolic network reconstruction has focused on expansion of metabolic reaction coverage. From the first human reconstruction, Recon14, to the most recent version, Recon23, the content has been expanded from 1,496 genes (corresponding to 3,311 reactions) to 1,675 genes (7,785 reactions). Various other reconstructions have been released and community driven-efforts have been made to ensure interoperability of these resources3,5. Our knowledge about human metabolism is continuously increasing and the deluge of ‘omics’ data provides ample opportunity for updating current knowledge-bases of human metabolism3–6. In addition to updating metabolic coverage, expanding human reconstructions to include different types, such as metabolite and protein structures as well as atom- transitions, thereby enables a broader scope of biomedical questions to be addressed.
Historically, systems biology has focused on characterizing catalytic or regulatory roles of proteins in metabolism without placing emphasis on the three-dimensional structure of the proteins themselves. For example, studies on genetic variation have mainly focused on frequency of occurrence12 or sequence-based attributes13. Only recently have mutations been explored in the context of their three-dimensional location or spatial relationship14–17. Exploring mutations in 3D extends beyond nucleotide sequence identity18, as mutations that may be far away from each other in linear sequence may actually be proximal in the folded state. In recent years, increasingly accessible protein and metabolite data have enabled the progression of systems biology to a 3D perspective. In one study, protein structures were mapped to the metabolic network of Escherichia coli, to reveal the role of ribosome pausing in co-translational protein folding19. In another study, human population variation was studied by integrating protein structures with the human erythrocyte metabolic network to understand the adverse effects of drugs on genetic variants20 and identify new pathways related to drug perturbation. These studies highlight the value of integrating different types of data to address complex biological questions.
We present Recon3D, an updated and expanded human metabolic network reconstruction that integrates pharmacogenomic associations, large-scale phenotypic data, and structural information for both proteins and metabolites. Recon3D contains over 6,000 more reactions than Recon2, all of which were manually curated to remove redundant or blocked reactions. We use Recon3D to prioritize putative disease-causing genetic variants by mapping single nucleotide variants (SNVs) to protein structures. We show that deleterious mutations are more likely to cluster together into functional hotspots than non-deleterious mutations. In contrast to previous models, these mutation hotspots identify ACAT1 as a cancer-related gene. Furthermore, we demonstrate how structural information can be used to investigate the potential mechanisms by which drugs exert an effect on metabolism. The Recon3D Resource (http://vmh.life) provides new avenues for investigating the molecular basis of disease and may aid the development of treatment strategies, biomarkers, and drug repurposing.
RESULTS
Increasing the scope of the human metabolic network reconstruction
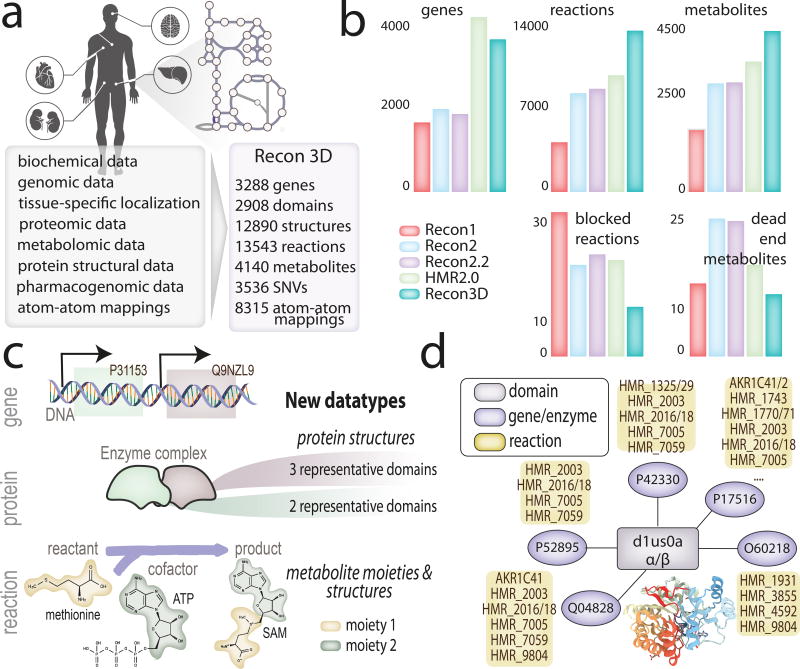
We expanded Recon 23 by using ten metabolomic data sets to identify new metabolites and transport and catalyzing reactions (1,865 reactions). We added reactions from HMR 2.021 (2,478), a drug module22 (721), a transport module23 (51), host-microbe reactions10 (24), and absorption and metabolism of dietary compounds (20). Overall, 66 metabolic subsystems, including lipoprotein (44 reactions) and bile acid (216 reactions), were expanded and 10 new subsystems were added (Supplementary Figure 1–3; Supplementary Data Files 1–2; Supplementary Note 1). We further refined numerous aspects of the reconstruction, including 2,181 gene-protein-reaction (‘GPR’) associations, reaction/metabolite duplication, reaction directionality, and thermodynamic feasibility (see Online Methods). The metabolic scope was extended by 82% (total 13,543) reactions and 58% (total 4,140) unique metabolites (Figure 1(a-b)). Recon3D is the most comprehensive metabolic resource currently available (Supplementary Table 1(a-c)). Out of the 20,266 human proteins documented in UniProt24 (queried July 2016), 19,213 are functionally annotated (i.e., not hypothetical) and 17% of this subset is metabolic, well-characterized, and included in Recon3D.
Figure 1. The properties and content of the Recon3D knowledge-base.
(a) Recon3D includes information on 3,288 open reading frames that encode metabolic enzymes catalyzing 13,543 reactions on 4,140 unique metabolites, protein structural information from Protein Data Bank (PDB)27, metabolite structures from CHEBI70 and is capable of performing flux-balance analysis to integrate and interpret a variety of emerging data types including linking mutations identified from human variation data or cancer genome atlases. (b) A comparison of the genes, reactions, metabolites, blocked reactions, and dead end metabolites among Recon predecessors3–5 and HMR2.06. (c) Relationships between genes, their encoding proteins, and the reactions they catalyze, (i.e., GPRs), are now described in the context of their specific 3D configurations, interactions, and properties. New data types include representative structural domains32 of proteins, metabolite structures along with their conserved moieties, and atom-atom mappings. Atom-level transitions were analyzed for 8,315 reactions (Supplementary Note 3). (d) Domain connectivity explored across the network to identify domains that are shared across multiple proteins, or involved in multiple catalyzing reactions. An example is the alpha/beta protein domain (d1su0a_), which is present in eight different genes (described by Uniprot accession number). The proteins encoded by these genes belong to the reductase family; they catalyze different reactions in various metabolic subsystems, ranging from glycolysis and the pentose phosphate pathway to xenobiotics metabolism and glycerophospholipid metabolism. Recon3D can be queried and downloaded from http://bigg.ucsd.edu/ or http://vmh.life. Users can visualize protein structures in networks via www.rscb.org or visualize network simulation results using the interactive ReconMap built on the Google Maps API (http://vmh.life/#mapnavigator).
Genome-scale network reconstructions can be converted into computational models that enable predictive biology2. We derived a computational model from Recon3D by removing reactions that were stoichiometrically inconsistent and that were flux inconsistent, (i.e., reactions that could not carry flux under the applied reaction bounds; Supplementary Note 2; Supplementary Data 1). After performing standard quality-control tests, the resulting generic Recon3D model contained 10,600 reactions (78% of the reconstruction reactions) and was able to reproduce literature-consistent energy (ATP) yields from different carbon sources (Supplementary Data Files 3–10), to fulfill metabolic functions describing cellular and whole body metabolism, and to replicate the predictions of infant growth from a previous study25 (Supplementary Figure 4).
Enabling a three-dimensional view of metabolism
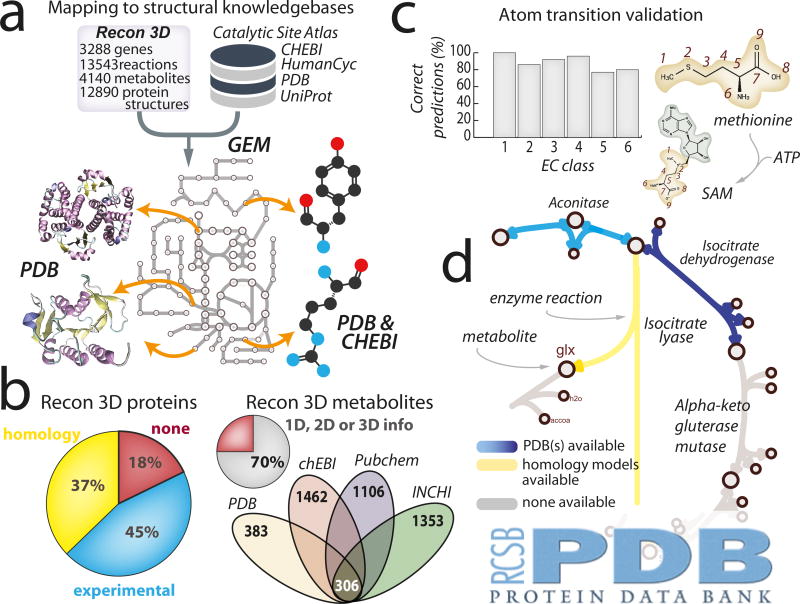
Using a recently established approach26, the metabolic network content of Recon3D was expanded to include three-dimensional protein structures from the Protein Data Bank (PDB)27 as well as homology models (Figure 2(a); Supplementary Figure 5; Supplementary Data Files 11–13; Online Methods). In addition, we mapped content from a variety of external database resources (Online Methods), to include metabolite structures (Supplementary Data File 14). We obtained high-quality structural coverage for over 80% of the human metabolic proteome (Figure 2(a); Supplementary Figure 6; Supplementary Tables 2–4) and 85% of the metabolome (Figure 2(b)). Furthermore, we used 2,369 unique metabolite structures to trace algorithmically atom transitions28 (from each substrate to product atom) for 7,804 (87%) internal, mass-balanced reactions of the Recon 3D derived model (Online Methods; Supplementary Note 3) . The prediction accuracy of the algorithms was validated by comparison with 512 manually curated atom mapped reactions (Figure 2(c); Supplementary Figure 7). The atom mappings enable identification of conserved moieties, which are the fundamental structural units of any chemical reaction network. Hence, we provide an invaluable bridge between metabolic modeling and chemoinformatics. For the first time, relationships between human metabolic genes, their encoded proteins, and the reactions they catalyze can be described in the context of specific 3D configurations, interactions, and properties (Figure 1(c-d)).
Figure 2. Linking human metabolic network to protein structural databases, cheminformatics platforms, and the Protein Data Bank.
(a) The metabolic content in Recon3D was cross-referenced with sequence and structure-based databases, such as UniProt24 and PDB27. The links in the metabolic network, which represent reactions, were mapped to three-dimensional (3D) structures through their encoding genes. The nodes in the network, which represent metabolites, were also linked to structural representations (3D, 2D, or 1D connectivity specifications). (b) Structural coverage of both proteins and metabolites in Recon3D is given by the pie charts, which indicate that over 80% of the metabolic proteome (2,793/3,297 genes) and 85% of the unique metabolome (2369/2797) has structural information. In the case of metabolite structures, the combination of structural data from multiple sources allows for the total structural coverage to exceed 70%. (c) Validation of atom-atom mapping by comparison with curated atom mappings for each major class of reaction. Recon3D is the first metabolic network reconstruction to contain atomic-level details. (d) An example of the type of visualization that can be found at the RCSB PDB website: http://www.rcsb.org/. The systems biology interface provides users with the ability to visualize metabolic network maps, that have been annotated to highlight which reactions are associated with experimental crystallographic structures (blue), homology models (yellow), or metabolite structures.
Web visualization of protein structures in metabolic networks
Using Recon3D, we have implemented the first web-based visualization of 3D macromolecular structures in the context of their neighboring chemical reactions, metabolites, and their metabolic subsystems, (e.g., glycolysis, citric acid cycle, amino acid metabolism, and carbohydrate metabolism, among others). This tool utilizes a recently developed global human network map29, together with network visualization software (see Online Methods) and conversion tools (Supplementary Note 4 and Supplementary Figures 8–11) and is available through the RCSB PDB website: http://www.rcsb.org/. The systems biology interface provides users with the ability to visualize networks that have been annotated to highlight which reactions are associated with experimental crystallographic structures, homology models, or metabolite structures (Figure 2(d)). Dataframes for Recon3D are found in the github repository: https://github.com/SBRG/Recon3D.
Gene variation in 3D
We probed mutations in the context of representative protein domains (i.e., common structural regions redundant across the proteome). Such domains (e.g., tim-barrel motif) are often linked directly to their encoding gene’s function, and thus provide a new way to directly assess the functional impact of a mutation.
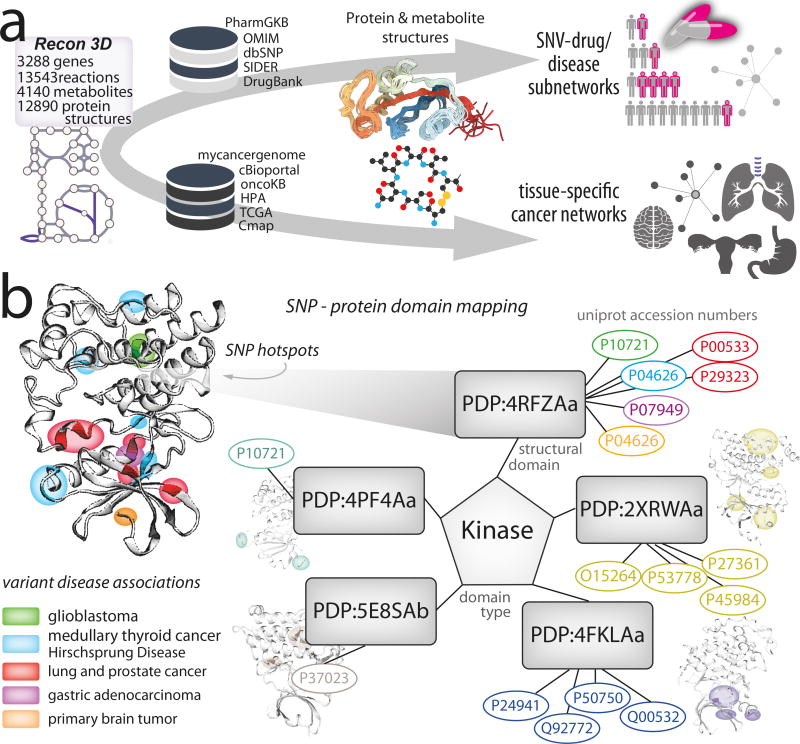
We used Recon3D to map missense mutations from Single Nucleotide Polymorphism (SNP) database (dbSNP)30, UniProt24, PharmGKB31, among others, to the metabolic network using a previously established pipeline20 (Figure 3(a)). We chose to focus on SNPs that were known to be deleterious or potentially harmful. In total, we mapped 3,536 SNPs to 655 genes within Recon3D. We identified representative protein domains for this set of genes using a structure-based clustering algorithm32. We tallied the number of SNPs (or single nucleotide variants, SNVs) occurring in each protein domain and found the gene to domain ratio to be less than one (i.e., domain redundancy; Supplementary Figure 12(a)). This analysis resulted in the identification of specific regions within protein domains that are commonly mutated (mutation hotspots), share common disease associations, and are prone to malfunction. As shown in Figure 3(b), six genes share the Bruton’s Tyrosine Kinase representative domain (PDP:4RFZa, PF007714) and, when mutated, are affiliated with diseases such as cancer. This kinase domain is known for its role in non-small cell lung cancer33 and the SNPs associated with lung cancer cluster in one specific region of the protein (see the red-colored mutation hotspot in Figure 3(b)).
Figure 3. Linking human metabolic network to gene variation and cancer knowledge-bases.
(a) Recon3D, as a Resource, provides information on three important layers of data related to disease biology: (i) amino acid location of mutations (or SNVs/SNPs) in the set of metabolic genes; (ii) the three-dimensional structure of proteins with sequence variants; and (iii) the relationships between mutations and the onset of disease. Information was cross-referenced from Recon3D to human variation and pharmacogenomics databases, such as dbSNP30, PharmGKB31, and cancer-specific databases, such as the Cancer Genome Atlas (TCGA), the Human Protein Atlas (HPA), and CMap. We mapped single nucleotide variants (SNVs) and single nucleotide polymorphisms (SNPs) to the genes in Recon3D. Within the set of genes with genetic variation, we focused on cases where (1) protein structural data was available; (2) SNPs/SNVs were considered to be deleterious or potentially harmful (655 genes). (b) Using this information, we probed characteristics of missense mutations and their three-dimensional spatial relationships. For each protein, we identified its representative protein structural domain (or a fold or set of folds unique to a given protein or multiple proteins). For example, for kinases, we identify various representative domains (five are shown here) that are associated with one or multiple genes (given by UniProt accession numbers). To this end, these five representative domains constitute “structure-based protein templates” shared among a group of genes. As illustrated, numerous mutations are found in 3D localized “hotspots” (or regions of the domain that experience high mutation burden). Interestingly, these mutation hotspots appear to be associated with specific diseases, such as primary brain cancer, glioblastoma, and other cancers in the case of Bruton’s Tyrosine Kinase (BTK) kinase domain scaffold (PDP:4RFZAa). All domains are determined by structural alignment32 and those featured here are named by the Protein Domain Parser (PDP) and the corresponding PDB structure (and chain) selected as the representative domain (see Online Methods; Supplementary Note 3). Colors map genes to the region (hotspot) of their respective variant(s) and the diseases associated with that variant.
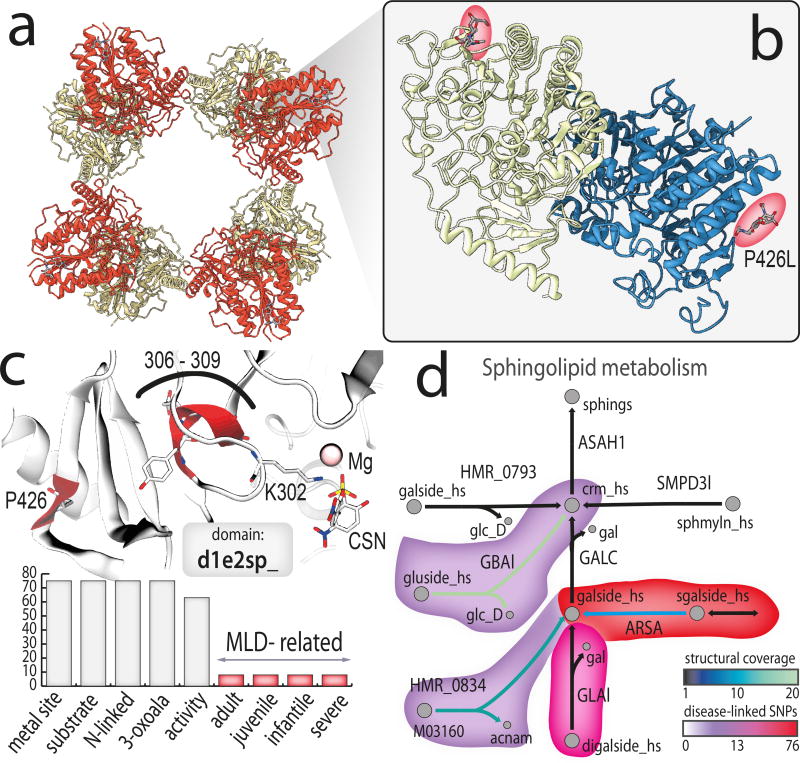
The power of exploring gene variation in the context of both protein and network structure is further illustrated by Aryl sulfatase A (ARSA). Within the subset of SNPs that map to the representative domain of ARSA (SCOP: d1e2sp_), the mutation P428L (P426L in PDB 1e2s; dbSNP rs28940893) is associated with Metachromatic Leukodystrophy Disease (MLD)34. This mutation influences the biological assembly of ARSA, in which the native homo-octamer state (Figure 4(a)) is disfavored relative to the dimeric state (Figure 4(b)). Other SNPs associated with the most severe form of MLD are located in the vicinity of the metal binding site, a mutation hotspot (Figure 4(c)). ARSA is also located within a “network hotspot,” with other deleterious SNPs dispersed throughout the neighborhood of surrounding reactions (Figure 4(d)). All mappings between SNPs, PDB, their representative domain, hotspots, and disease relevance are provided in Supplementary Data Files 15–20.
Figure 4. An example of bridging systems biology and structural biology through Recon3D.
(a) Arylsulfatase A (ARSA) highlights an example of how the intersection of systems, structural, and pharmacogenomic information provides additional understanding of human disease variants. The macromolecular assembly in the native state contains a homo-octamer (four complexes of homodimers; PDB entry 1auk). (b) Identifying the location of a variant (e.g., P426L, dbSNP rs28940893) within the protein three-dimensional structure reveals mechanistic details of disease progression. This mutation, which is associated with a mild form of Metachromatic Leukodystrophy (MLD), weakens the interaction between monomers, causing the biological assembly to favor the homo-dimer state over the homo-octamer state. (c) Clustering all SNPs that fall within a 5–10Å vicinity of other mutations, we find that the largest cluster falls within 10 Å of both the metal-binding site and the substrate-binding site (residues 306 to 309 in PDB entry e2sp). These specific cases all cause a severe form of MLD in adults, juveniles, and infants. The distribution of structural and disease properties associated with all 76 SNPs that map to the representative domain of this protein (d1e2sp_) is given by the bar chart. The majority of cases map to the calcium binding domain, substrate binding domain, and have a significant effect on enzyme activity. (d) ARSA and its neighborhood of surrounding reactions link to a number of disease-associated mutations, indicating that this is a “network hotspot” for deleterious or potentially harmful mutations. In many cases, the proteins catalyzing these reactions also have available protein structural content (shown by a heat map and reaction link color), enabling 3D visualization of other SNPs in proteins in neighboring reactions. Figures for protein structures were generated using ChimeraX, the next generation version of Chimera. Reactions are drawn with minimal number of metabolites and cofactors for clarity.
Oncogenic mutations cluster in structurally equivalent positions in the human proteome
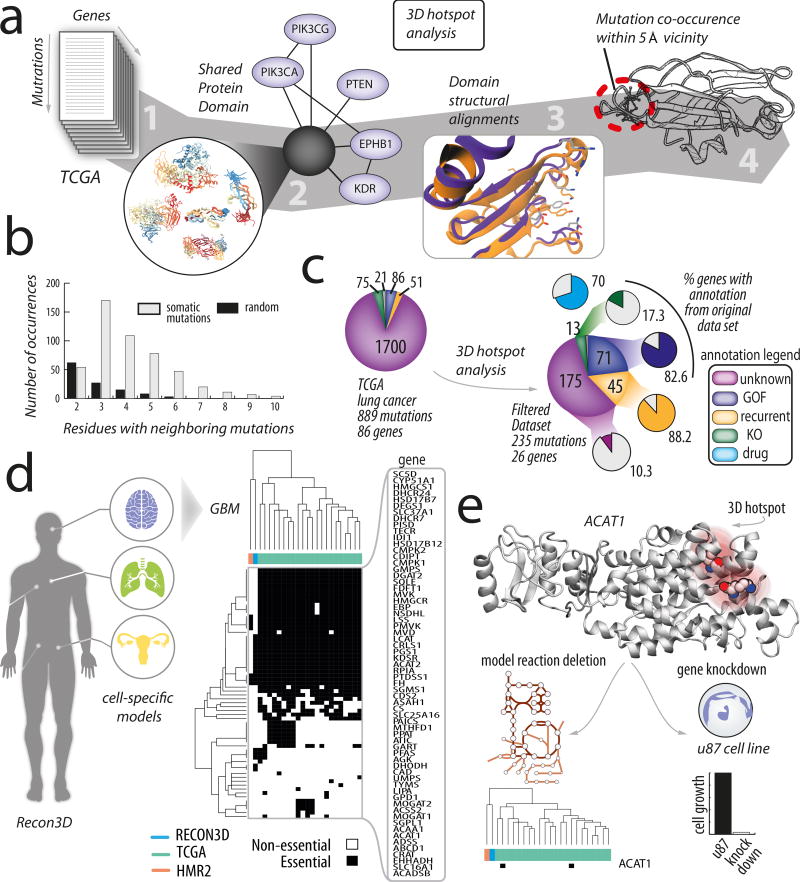
The first application of Recon3D demonstrates its capability to discriminate pathogenic mutations from passenger mutations. We studied 889 somatic cancer mutations from 88 genes (which were previously analyzed35) from whole-exome sequence data from 178 tumour–normal pairs of lung squamous cell carcinoma36. Furthermore, we obtained detailed annotations about each of the mutations from cBioportal37, including whether a gene is a known oncogene37,38 (KO) or the mutation is recurrent12, has gain-of-function (GOF), and has a drug association. Using Recon3D, we mapped each of the mutations to their corresponding protein, its representative domain(s) and network reaction(s) (Figure 5(a)).
Figure 5. Protein structure-guided discovery of mutation hotspots across structurally-related genes.
Synchronization of protein structural domains, metabolic networks, and somatic mutation landscapes allows for stratification of variants into informative and meaningful sub-clusters. (a) The 3D hotspot analysis workflow. A list of genes with mutations35 is cross-referenced with databases such as TCGA. In this example, we studied mutations taken from whole-exome sequence data from 178 tumour–normal pairs of lung squamous cell carcinoma36. We then assembled protein structural information for this subset of genes with somatic mutations and evaluated the number of representative protein domains for this set of genes. In total, 86 genes associated with 889 missense mutations had available experimental crystallographic structures and could be linked to representative structural domains. We tallied the mutations occurring within 5 and 10 Å spheres for each representative domain. The domains with multiple mutations in a specific 3D location were termed “mutation hotspots.” (b) We compared the frequency of mutation co-occurrence (in a 5 Å sphere) in randomly selected residues (grey) within the same set of proteins with those taken from the lung cancer dataset (black). This comparison strongly suggests that somatic mutations are more likely to be found neighboring other mutations than what is expected by chance (p, val < 0.02). (c) Selecting the top 25% of mutations (235/889) with the highest number of neighboring mutations (within the same 5Å region in a representative protein domain) brings about a striking commonality that many are associated with known oncogenic roles. Information about various mutations was taken from several databases providing detailed annotations (which are color-coded in the plot), including recurrent sequence hotspots (R)12, known oncogenes (KO)37,38 (www.oncokb.org), as well as drug (Olaparib/BYL-719), Memorial Sloan Kettering level of evidence (3B), and other cancer subtype (endometrial/breast) associations (www.mycancergenome.org). For example, of all the mutations in this dataset with gain-of-function (GOF) oncogenic associations, 83% are found in the subset of mutations selected for on the basis of 3D localization. Similarly high percentages are recovered for other characteristic annotations, including the frequency of occurrence (88%), association with endometrial cancer (100%), and associated with breast cancer (40%). Intriguingly, percentage of mutations with unknown effects is greatly reduced from 90% in the total dataset (bottom pie chart; 889 mutations across 86 genes) compared to 10% in the 3D filtered subset (top pie chart; 235 mutations across 26 genes). Random selection of 235 mutations (averaged across 10,000 trials) demonstrates that the probability of recovering the same percentage of mutations with known oncogenic roles is very low (shown by the white outlined bars). (d) We combined the 3D hotspot analysis with metabolic modeling and focused on the somatic landscape of glioblastoma multiforme65. Gene knockdowns were performed in various models, including Recon3D, HMR2.0, and cell-specific (GBM) and patient-specific models. (e) The majority of models predicted ACAT1 to be non-essential. Yet, when analyzing the mutations in this gene in 3D, we find a mutation hotspot. The importance of this gene is further confirmed by experiment, demonstrating its importance to GBM growth40. This example suggests that protein structure could facilitate model predictions by highlighting genes of interest using complementary information.
Analysis of all cancer mutations in the context of their representative protein domains suggests that oncogenic mutations cluster in structurally-equivalent positions within representative domains. For the 88 genes, we counted the number of mutations that occur within 5 Å of another mutation within the representative domain (referred to as the 3D hotspot analysis; Online Methods; Supplementary Note 5). In some cases, mutations from different genes co-occurred in the same region of a shared domain, suggesting that domain plays an important role in oncogenesis. Mutations co-occurring in the same location of other mutations are significantly more likely to be associated with somatic mutations, when compared to a random selection (p < 0.02 using a two-tailed t-test; Figure 5(b)). All data mapping related to the somatic cancer mutations can be found in Supplementary Data Files 21–23).
Filtering mutations based on their spatial relationships brings about several significant biomedical implications. When mutations are rank-ordered by the number of neighboring mutations, we can filter the mutations with known roles in oncogenesis (based on known annotations37; Figure 5(c)). For example, we find that selecting the top 25% of the data recovers 82 and 88 percent based on co-occurrence aids in identifying known oncogenic mutations and GOF mutations, respectively (compared to 1.6 and 2.9 percent when selected at random; Figure 5(c); for a sensitivity analysis, see Supplementary Note 5). Furthermore, striking similarities in protein structure, based on three-dimensional structure alignments, indicate that not only do mutations co-occur in shared domains, they also occur in structurally-similar proteins within the same dataset (Supplementary Figure 12(b)). These findings suggest that cancer mutations cluster in functionally-relevant parts of protein domains and that this property could guide the discovery of novel biomarkers and drug targets.
We combined our approach with metabolic modeling to understand whether structural information could improve the predictive power of the model. We focused on glioblastoma multiforme (GBM), a malignant brain tumour, and studied the mutational landscape of metabolic genes (Figure 5(a)). Genes were selected based on the rate of mutation found in exome samples of 291 glioblastomas as well as involvement in cholesterol metabolism39 (Online Methods; Supplementary Note 5). Gene knockdowns were performed and the essential genes were compared across different generic and cell-type specific human metabolic models (Recon3D, HMR2, and HMR-derived TCGA-derived models8 (Online Methods). Notably, the majority of models predicted the gene ACAT1 (GeneID 38) to be non-essential (Figure 5 (d); Supplementary Figure 13). Yet, a 3D hotspot analysis of the mutations in this gene suggested that this gene may be important in cancer (Figure 5 (e)). This finding was recently validated, confirming that inhibition of ACAT1 suppresses GBM growth by blocking SREBP-1-mediated lipogenesis40. This result highlights the potential for structure-based analysis in genome-scale models to identify important genes for cell growth.
Co-occurring mutations across shared protein domains are significantly more deleterious
We used Recon3D to identify potentially deleterious mutations in a large-scale population study. We analyzed SNP data from multiple gene variation databases (dbSNP30, UniProt24, PharmGKB31) and assessed whether the 3D location of variants in a gene could, in general, discern whether mutations were deleterious or tolerated41. We mapped over 10,000 SNPs to their 3D structural coordinates using our 3D hotspot analysis workflow and computed the number of mutations co-occurring in 5 and 10 Angstrom spheres in common protein domains. 1,385 unique genes had 3,649 SNPs that mapped to regions of a protein where structural data exist. We computed the number of mutational co-occurrences across this set of SNPs and found that deleterious mutations are much more likely to neighbor other deleterious mutations (p < 0.05 using a two-tailed t-test) than those predicted to be tolerated (p > 0.1, using a two-tailed t-test; Supplementary Figure 14, Supplementary Tables 5–6). These added features enable predictive power over any existing model, in that mutational data can be assessed in the context of protein structure and compared with network-level, genome-wide model knockdowns (e.g., Figure 5(e)). Prior reconstructions are unable to identify structural changes that affect complex assembly or other intrinsic protein properties. Such details can now be explicitly studied using Recon3D. To this end, Recon3D provides new inroads for metabolic models to explore disease-relevant mutations.
Elucidating relationships between drug indications and their metabolic responses
Drug interventions influence the behavior of metabolic networks42, but the impact of drug treatment on metabolic responses and the mechanisms underlying these responses are poorly understood.
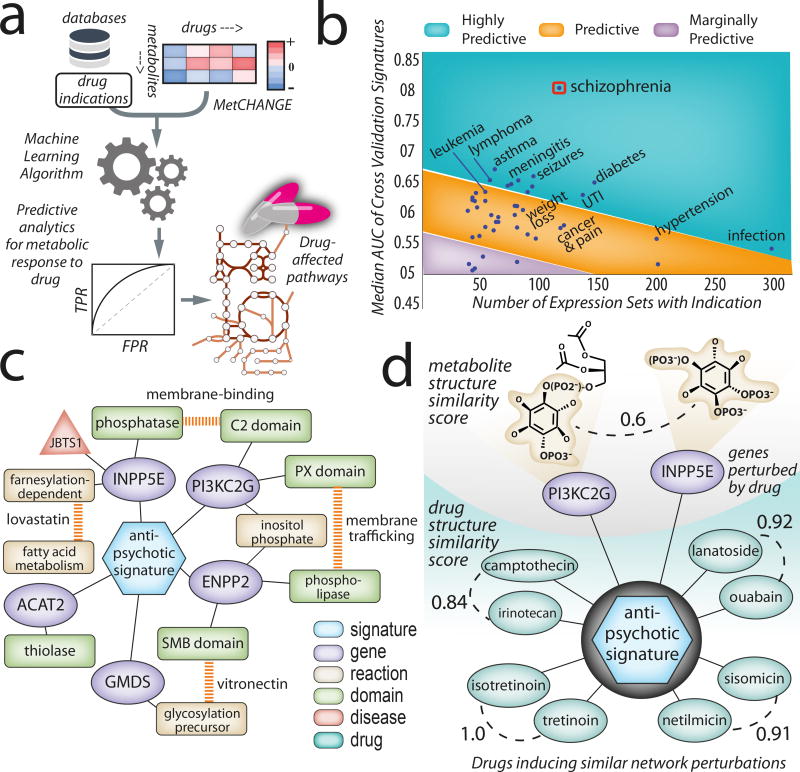
We used Recon3D to combine large-scale data on drugs, their indications, and their effects on gene expression. This data was used to guide and inform genome-scale constraint-based modeling analyses42,43 to identify the metabolic pathways most perturbed in a given condition (Supplementary Figure 15). More specifically, we used a machine-learning approach to assess similarities in metabolic responses to a given drug. Using a genetic algorithm, the area under the curve (AUC) of the receiver operating characteristic (ROC) curve was maximized to predict the indication of the drug based on the type and degree of perturbation (Figure 6(b); Supplementary Data File 24; Online Methods; Supplementary Note 6). Finally, we use the structural information in Recon3D to provide insights into the possible mechanisms by which the drugs exert their effects on metabolic pathways.
Figure 6. Identification of metabolic signatures linked to drug indications.
(a) A machine-learning based approach to predict metabolic responses to drugs. Drug indications were taken from the Side Effect Resource (SIDER) database45 for all available drugs overlapping with drug-treated gene expression profiles from the Connectivity Map (CMap) database44. A total of 47 drug indications were analyzed in the context of the metabolic network, based upon 1,459 expression sets from cell culture responses to 334 drugs (see Supplementary Data File 26). (b) Cross validation results of metabolic gene expression signatures trained against drug indications versus the number of expression sets with the indication used in training. Results were empirically grouped as highly predictive, predictive, and marginally or poorly predictive based on AUC. Results were plotted with consideration to dataset size, showing that the signature is conserved over a greater number of drugs and amount of noise. Schizophrenia appeared as a clear outlier with greater predictability for a relatively large number of expression sets and drugs (13 drugs used in training), indicating that the gene signature is highly conserved (median AUC of 0.8). (c) Analysis of the antipsychotic signature in the context of known metabolic effects in schizophrenia and antipsychotic therapy. Genes that cluster based on the antipsychotic drug indication signature are linked to structure, biochemical, and disease properties through Recon3D. Such connectivity networks provide a first glimpse at whether genes share similar biological functions or domain archetypes. (d) Perturbations in genes that cluster based on metabolite/drug similarity. Computing structural alignments of the drugs inducing the antipsychotic drug indication signature indicates that certain pairs are likely to have similar bioactivities (based on tanimoto coefficient > 0.8). Chemically similar drugs cluster into four structurally distinct groups that differ on the basis of drug class. Drugs within these four groups all induce the same drug indication signature despite being radically different in structure (tanimoto coefficient < 0.2).
We first grouped 6,040 transcriptomic profiles (exposed to over 1,200 drug compounds in breast, leukemia, and prostate cancer cell lines from the Connectivity Map, or CMap44) by drug indication, using information from SIDER45 (Supplementary Table 7). A total of 47 drug indications were analyzed in the context of the metabolic network, using a previously described machine-learning approach42 (Supplementary Figure 15). The analysis revealed that indication-specific drugs induced similar patterns of gene expression changes or “gene indication signatures.” Our findings suggest that metabolic responses are significantly conserved for a wide range of drugs (Figure 6(b)), with the most conserved pathway perturbations occurring for antipsychotic drugs (median AUC of 0.80; Supplementary Data File 25). For this specific case, the gene indication signature is composed of nine genes that have been previously associated with schizophrenia (Supplementary Table 8). We also find associations between changes in lipid and cholesterol pathways and common antipsychotic drug side effects (weight gain, cardiovascular risk, and anti-inflammatory effects). Notably, some drugs with entirely different indications shared similar pathway-level changes with antipsychotic drugs and had previously been tested as adjunctive schizophrenia treatments46 (Supplementary Table 9). A list of the drugs with the most predictable metabolic responses is provided in Supplementary Data File 26.
We then used protein and metabolite structural data in Recon3D to probe for mechanistic insights into drug response. In general, understanding mechanistic details entails identifying single or multiple targets of drug binding (or off-binding) and the respective downstream effects. Information in Recon3D can be visualized as a topological network to indicate shared features across nodes (genes) in a gene indication signature. Displayed in Figure 6(c) is one connected hub of genes (antipsychotic gene indication signature) and several features for comparison: protein structural domains, metabolites, biochemical reactions, and disease relevance. For this signature, we found several overlapping features, such as the metabolic subsystems targeted by known drugs (e.g., lovastatin and fatty acid metabolism) and the function of certain protein domains (e.g., an influence in membrane binding/trafficking). Despite these shared domain functions, minimal structural alignment of the protein domains and metabolites indicates that the majority of genes in this signature are not direct drug targets, but may play a role in compensatory signalling pathways that mediate drug effects synergistically. Finally, structural alignment of the drug compounds themselves yielded unexpected results; drugs that induced the same pattern of perturbation (both drugs with known antipsychotic action and unrelated drug indications) were found to be structurally diverse (Figure 6(d)). This finding is surprising given that drug discovery efforts tend to emphasize small changes in molecular structure to tune a desired biochemical effect. Here, we find that structurally diverse molecules exert similar effects on metabolic pathways, highlighting the potential of Recon3D for drug repurposing and the design of multi-targeted therapies that support a new polypharmacological paradigm in drug research47,48.
DISCUSSION
Recon3D is the first network reconstruction to include protein and metabolite structures as well as atom-atom mappings. Recon3D provides functional insights into genetic variation and the mechanisms underlying the effects of drugs on metabolic response in humans. It also serves as a computable knowledge-base with clear functional connectivity between genes and biochemical pathways. Pairing Recon3D with biomedical data provides a compelling avenue for studying disease at scale.
The ability of Recon3D to integrate multiple layers of biological data will provide a tool for obtaining a meaningful and coherent understanding of variation and the influence it exerts at both the level of individual proteins and within complex pathways. The inclusion of these multiple disparate data types offers new opportunities for network reconstruction: (i) it introduces atomic scale properties, such as ligand binding interactions; (ii) it provides new avenues for precision medicine by exploring human variation14,15, and (iii) it enables the probing of genetic variation via changes in the molecular properties of proteins20. In this way, individual sequence variations can be explicitly represented and the functional connections among disease, genetic perturbation, and drug action can be probed systematically.
Recon3D enables straightforward data integration, as its content has been linked to external databases (KEGG, PDB, CHEBI, PharmGKB, UniProt). This knowledge-base can be converted into a genome-scale model, which can be computationally interrogated and characterized. Constraint-based methods43 can be used to assess network properties and bioinformatics tools32 can be used to assess protein or metabolite properties. Our findings present preliminary, yet compelling, support for the potential of Recon3D to complement traditional structure-based approaches for empowering applications in drug discovery and target validation. We have shown that a systematic exploration of mutations in the context of their three-dimensional spatial relationship provides a unique means for filtering out functionally relevant mutations and determining potential genes of interest. Furthermore, analysis of in vitro drug-treated gene expression profiling in the context of the human metabolic network provides insight into the broad metabolic response to different drug therapies.
The Recon3D knowledge-base is instrumental to gene variation analyses as it provides a framework for integrating structure-function relationships, and assessing specific and proteome-wide effects of sequence variation. Integrated frameworks like Recon3D enable understanding of how mutations or binding events lead to downstream responses and could aid in the identification of novel targets when coupled to structural bioinformatics16, molecular dynamics simulations20,49, and kinetic modeling50. In contrast, current metabolic models are not able to contextualize the effect of a sequence variant (beyond gene deletions) and therefore cannot be used to study disease-relevant mutations. Recon3D will potentially aid in translating biomedical knowledge, from large-scale omic data to drug discovery, target identification, and clinical biomarker development. Future efforts are likely to extend to personalized or precision medicine healthcare applications, where drug responses can be assessed in the context of individual patient-specific genomes. Recon3D is available via two databases3,51: http://bigg.ucsd.edu/ and http://vmh.life
ONLINE METHODS
Metabolic reconstruction
Recon 3D has been assembled using multiple data sources, i.e., HMR 2.006 (2,478 reactions), metabolomics data sets (1,865 reactions), a drug module22 (721 reactions), a transport module (51 reactions), host-microbe reactions (24 reactions), absorption and metabolism of dietary compounds (20 reactions), and others (1004 reactions). The ‘others’ category included reactions that captured metabolism in specific human organs, (e.g., kidney), as well as novel metabolic pathways of lipoproteins, bile acids, and sphingolipids. The expansion of Recon 2 was performed in an iterative manner (Supplement Figure 1). With each addition, there followed extensive model debugging and manual curation for flux consistency and refinement.
Recon 2 was expanded in two stages: (i) additions of new reactions and (ii) network refinements for building high-quality flux-consistent model (Supplementary Figure 1). The total number of novel additions included 6163 reactions, 1589 metabolites, and 1654 genes completing Recon 3D. These new reactions were mostly from transport (32%), lipid metabolism (24%), exchange (19%), xenobiotic (11%), and amino acid (7%) metabolism (Supplementary Figure 2B-C). Other major additions include those required for debugging the network for flux consistency (10% of newly added reactions), reactions representing organ-specific metabolism (7%), transport module (2% of newly added reactions), and those representing lipoprotein metabolism (2% of newly added reactions), novel dietary compounds and their associated reactions (1% of newly added reactions), and reactions capturing interaction between gut microbes and host (1% of newly added reactions). For details on the precise metabolic pathways, see Supplementary Note 1.
The largest contribution for new metabolic genes were those from: (i) lipid metabolism (10%), (ii) carbohydrate metabolism (5%), (iii) transport processes (5%), (iv) amino acid (3%), and (v) nucleotide metabolism and vitamin metabolism (1%) (Supplementary Figure 2). The miscellaneous category mostly contained genes from HMR 2.0 (99%) (Supplementary Figure 2). The largest contribution for new metabolites were lipid (42%) and amino acid (19%) classes. Novel metabolites added in other subsystems include miscellaneous and xenobiotics (18%), carbohydrates (2%), vitamins (1.4%), and nucleotide (0.3%) metabolism (Supplementary Figure 2).
Once reactions and genes were added to Recon3D, the reconstruction was subjected to various quality control/quality assurance tests (Supplement Figure 1). These include: (i) checking for reaction and metabolite duplicity, (ii) modification of gene-protein-reaction associations, (iii) modification of metabolite formulae to pH 7.2 along with mass-charge balancing of reactions, (iv) a leak test, checking for stoichiometric and flux consistency and checking for thermodynamic feasibility52, (v) debugging and curation for removal of dead-end metabolites, and (vi) checking for network accomplishment of defined functions/tests (Supplement Figure S1).
To check reaction and metabolite duplicity, we took several approaches. First, Quek et al53 reported 95 duplicate metabolites, 71 of which were replaced (Supplementary Data File 9, and Supplementary Note 1). Second, the reaction and metabolite duplicity was checked for HMR reactions and metabolites (prior to inclusion in Recon 3). The metabolite formulae, particularly those received from HMR 2.0, were adjusted to an internal pH of 7.2, using mol files28 and COBRA toolbox54 and ChemAxon software (https://chemicalize.com/). This led to correct assignment of reaction stoichiometry and mass-charge-balancing of reactions. Third, gene-protein-reaction associations were curated and corrected for 2,180 reactions (Supplementary Data Files 6–7, and Supplementary Note 1). Finally, we performed additional QC/QA tests, (e.g., functional leaks, production of matter from water and oxygen, etc).
The COBRA toolbox54 was used to identify a subset of 10,600 reactions involving 5,835 metabolites, representing the stoichiometrically consistent flux balance model. The final model was tested for 431 model objectives, representing essential biochemical functions of the human body. The model debugging was mostly done by the addition of extracellular and intracellular transport reactions. Examples include the addition of novel transport proteins for bile acids and folate intermediates. Novel intracellular transport proteins, i.e., mitochondrial pyruvate carriers (MPC1, GeneID: 51660 and MPC2, GeneID: 25874) were added for phenylpyruvate that operates in a proton symport mechanism55. These transport reactions connected the intracellular and extracellular compartments of the model, enabling flux consistency. Manual curation of the relevant scientific literature was followed to obtain complete information on the respective biochemical pathway. A typical example includes the addition of 4-methyl-thio-oxo-butyrate (an intermediate of methionine metabolism) into the network. Upon literature curation, addition of the alternative route of methionine transamination and decarboxylation reactions were identified and added (Supplementary Data File 1).
In total, out of the 20,266 human proteins documented in UniProt24 (queried July 2016), 19,213 are functionally annotated (i.e., not hypothetical) and 17% of this subset is metabolic, well-characterized, and included in Recon3D. Please refer to Supplement Note 1 and Data Files 1–10 for detailed information on the network building and refinements.
GEM-PRO reconstruction
We followed the previously described procedure26 to map, assess, and refine PDB or homology models for integration into genome-scale models. For Recon 3D, additions to the gene identifier mapping workflow were made to address inconsistencies in gene isoforms across database entries and the ability to link isoforms to available homology models. In addition, QC/QA steps were taken in order to ensure the correct sequence was being retrieved (Supplementary Figure 5; Supplementary Note 3). For PDB structures with missing residues, we have filled in the gaps by querying previously generated databases of I-TASSER homology models56,57, and manually generating homology models for genes that were not part of these databases using a previously defined protocol58. In the final master GEM-PRO data frame (Supplementary Data File 11), we note where available homology models have been mapped to their respective genes. For most homology modeling procedures, the amino acid sequence of a protein is all that is required to generate a homology model of a protein. It is important to note that certain PDB structures with unresolved residues or gaps in the structure can also be homology modeled to enhance the structural coverage of the amino acid sequence. Any sequences longer than 600 amino acids long were not homology modeled. We assessed the overall quality of the information coming from homologous templates in terms of (i) which organism the protein was crystallized from, (ii) the resolution of the PDB template, and (iii) the deposition date. We used these properties to compare the templates that were used to construct homology models in the previous GEM-PRO models with those of the recently updated versions (Supplementary Tables 2–4; Supplementary Figure 6).
To identify structures for the given set of metabolites in Recon 3D, we evaluated a number of databases where metabolite structures are publicly available, such as PDB (ligand-expo: http://ligand-expo.rcsb.org/, http://ligand-expo.rcsb.org/ld-search.html), PubChem59 Url (https://pubchem.ncbi.nlm.nih.gov/), and ChEBI Url (http://www.ebi.ac.uk/chebi/). We downloaded structures in various formats: 2D structure in .mol format (ChEBI), 3D structure in .sdf format (PubChem59), and in .pdb/.xyz format (RCSB). Supplementary Data File 14 provides all the information content processed for metabolites in Recon 3D, which includes SMILEs and INCHI descriptors, Kyoto Encyclopedia of Genes and Genomes (KEGG)60 IDs, CID IDs, CID file names, ChEBI file names, ChEBI IDs, and experimental coordinate file URL locations and the ideal coordinate file name. The ChEBI mapping procedure contained the following steps: (i) identification of the particular metabolite from ChEBI using the source link (the metabolite name will be the starting point of search which is taken from the metabolite names in the Supplementary Data File 14); (ii) checking the molecular formula and charge (neutral or charged) of the metabolite in the ChEBI database; (iii) capturing the ChEBI link, ChEBI ID, SMILES, and INCHI into the respective fields in the dataset spreadsheet; (iv) 2D-structure is downloaded in .mol format. The same overall search was conducted in Pubchem and PDB (Ligand expo) with slight variations as to the initial search inputs and file type outputs.
The dataset of human single nucleotide polymorphisms (SNPs) and single nucleotide variants (SNVs) was collected from UniProt from a subset of protein altering variants from the 1000 Genomes Project. Furthermore, all SNPs/SNVs for model genes were downloaded directly from dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) via the Ensembl BioMart interface61. We then selected all variants that were characterized to be “damaging” or “possibly damaging” as a predicted functional impact using the PolyPhen2 bioinformatics tool41. Functional annotations of the missense mutations were also annotated using SIFT (http://sift.jcvi.org/). In addition, we linked the missense variants to their gene-drug associations (clinically relevant pharmacogenomics interactions) using the PharmGKB pharmacogenomics database (https://www.pharmgkb.org/). All annotated gene-drug pairs contain information such as dosing guidelines, drug label annotations and each pair is generally specified in more than 1 type of annotation (dosing guideline, drug label, clinical annotation, variant annotation, VIP, or pathway). These selected pharmacogenomic associations allow us to understand whether certain missense variants have functional effects on drug therapies. All selected missense variants and their drug associations have been provided as Supplementary Data Files 15 and 16.
More details on the process and procedure for network reconstruction, protein and metabolite structure integration, identification of representative protein domains, linking to pharmacogenomics databases, linking to cancer genome atlases, mutation hotspot analyses, and comparison of tissue-specific cancer and pharmacogenomic/gene variation networks are all provided in Supplementary Note 3–4
Atom-atom mapping
Generation of atom mapping data requires chemical structures, reaction stoichiometry and an atom mapping algorithm. Atom mappings were predicted using the Reaction Decoder Tool62, and the DREAM algorithm63 for 7,535 (86%) mass balanced reactions with implicit and explicit hydrogens, respectively, while Reaction Decoder Tool and the CLCA algorithm64 were used to predict atom mappings for a further 269 reactions with incompletely specified metabolites (e.g., R group) with implicit and explicit hydrogens, respectively. We compared these predictions for internal reactions to a set of 512 reactions with atom mappings that we and others manually curated (Supplementary Note 3). This reaction set is representative of all six top level EC numbers. Based on this comparison, we observed that the predicted atom mappings are highly accurate for most of the reaction types28 (Supplementary Figure 7).
3D mutation hotspot analysis
We filtered a set of mutations (whose genes are associated with experimental protein structures) based on whether the location of the mutated residue itself was resolved (e.g., certain protein domains are unresolved due to flexibility or unstructured regions of the protein being challenging to crystallize). Once the subset of mutations was established to (i) be linked to genes with experimental protein structures and (ii) be located within regions of the protein that were experimentally determined, we carried out 3D structure alignments between all proteins and their representative domains (mapping to representative protein domains is described previously in the section entitled “mapping and alignment of PDBs to their representative domains”). In contrast to sequence alignments, 3D structure alignments find a best fit in terms of the three-dimensional shape or geometry of two proteins. Therefore, any two proteins that have different sequences but share a common domain architecture can be successfully aligned in 3D space. Similar to sequence alignments, the 3D structural alignment provides a direct residue-to-residue mapping for residues that share structurally equivalent positions in a common/shared domain motif. Once this residue-to-residue mapping was established for all proteins in our dataset, we located 3D “hotspot” mutations by tallying all residues in the representative domains that map to mutated residues in a given protein of interest. To this end, certain residues in a representative domain may have multiple hits if more than one gene is linked to that representative domain and the same structurally equivalent residue is mutated across various genes. Supplementary Data File 17 provides the mapping between the residue number of the Uniprot missense variant > the PDB residue number > the PDB chain where the residue is located > the representative domain ID linked to a given PDB chain > the structurally equivalent residue within that representative domain.
Mapping cancer mutations in 3D
We used the TCGA level 3 variant data in the cBioPortal (http://www.cbioportal.org/). For this study, we used high level (processed) data from a subset of pre-analyzed mutations from 178 tumour–normal pairs of lung squamous cell carcinoma36. When the MutSig1.0 approach was applied on this dataset35, it identified 450 genes as significantly mutated. Starting from this set of genes, we identified a subset of 86 genes that have Uniprot accession numbers and protein structural information. Within this set of genes, we found that 889 somatic cancer mutations map to residues that have been successfully resolved in the crystallographic structures of proteins. We used the list of 86 genes to query the cBioportal web-based dataset and downloaded various information including: somatic cancer mutations, cancer study sample IDs, amino acid mutations, annotations (coming from various sources, such as http://oncokb.org/ and https://www.mycancergenome.org/), type of mutation, copy number changes, overlapping mutations in COSMIC, the predicted functional impact score (from Mutation Assessor), variant allele frequency in the tumor sample, and total number of nonsynonymous mutations in the sample. A summary of cancer data sets used in this study is given in Supplementary Data File 21 and a detailed summary of all somatic mutations for this set of genes is provided in Supplementary Data Files 22–23. The 3D hotspot analysis was carried out as detailed above and mutations were rank-ordered on the basis of how many mutations fell within a 5Å sphere (i.e., number of nearest neighbors). We performed a sensitivity analysis to understand whether the selection of data points had an effect on the significance of these results.
The above 3D hotspot analysis approach was also applied to 22 genes from which cancer mutations have already been analyzed65 (exome samples of 291 glioblastomas) and 92 genes involved in cholesterol metabolism, owing to the fact that cholesterol biosynthesis plays an important role in GBM39.
Statistical Tests
We performed a sensitivity analysis to understand whether the selection of data points had an effect on the significance of these results. We find that the 3D hotspot analysis is more likely to select somatic mutations compared to a random selection. Data points (50–700) were selected so that 0.065–0.91 of the total data set was covered. We performed the 3D hotpot analysis across the different selections and found p values to range 0.017 - 0.049 compared to 0.182–0.241, using a random residue selection.
For annotations of mutations that are known oncogenes (KO) and known hotspots (HS), selection of the data based on 3D hotspot analysis is significant, regardless the number of data (or % of data) selected (pval < 0.05). Compared to a random selection, our computed (using a two-tailed t-test) p value is > 0.1. We also performed a sensitivity analysis using the slices of the total data set as mentioned above (50–500 data points) and computed the total number of known oncogenes and known hotspots (from previously published analyses), using the 3D hotspot analysis compared to a random selection. We find that the percentage of data selected is significantly higher using the 3D hotspot analysis. For KO, 37–83% of the data is selected using 3D hotspot compared to 0.046–0.43 at random. Similarly, for HS, 72.5–88.3% of the data is selected using 3D hotspot analysis compared to 9.8–64%. See Supplementary Note 5 for more information.
Gene deletion simulations in GBM
In silico single gene deletion (SGD) simulations were performed as previously described66. Given a certain GEM, the simulation of a SGD was performed by formulating the linear program problem (1) for each gene g in the GEM:
max vobj subject to:
0 < vobj < γ
S · v = 0
−1000 ≤ vj ≤ +1000 ∀j ∈ {Exchange reaction indexes for medium metabolites}
vr = 0 where r ∈ {Reaction indexes univocally encoded by gene g}
where vobj is the flux through the biomass equation, γ is an arbitrary number set to 1, S is the stoichiometric matrix of the GEM (that is a m × n matrix where m is the number of metabolites and n is the number of reactions and each (i,j) entry is the stoichiometric coefficient of the metabolite corresponding to row i in the reaction corresponding to column j), v is the vector containing the values of the fluxes through each reaction in the GEM, and j indexes each exchange reaction known to be present in a rich mammalian medium (Ham’s medium, HAM; see Supplementary Note 5 for more details). The simulation was carried out for the following GEMs: Recon3D, HMR2.00, and 22 personalised GEMs for glioblastoma multiforme (GBM) previously reconstructed using HMR2.00 as a template from as many GBM expression profiles retrieved at The Cancer Genome Atlas67.
Drug perturbation analysis
To compute metabolic pathways with gene expression perturbed by drugs, the human metabolic network model was first converted into an irreversible network. Then, the MetChange algorithm42 was run using gene expression presence/absence p-values from the Connectivity Map (Cmap) database44 build 02. Drug indications were taken from Side Effect Resource (SIDER) database68 for all available drugs overlapping with the Cmap database. Synonyms were aggregated when present as with side effects. A minimum of 10 drugs for each indication were required for the inclusion in the analysis, corresponding to a much greater number of expression sets for each indication. A total of 48 drug indications were analyzed for 1459 expression sets corresponding to 334 drugs. A genetic algorithm (Supplementary Figure 15)) was then implemented as described in Supplementary Note 6. Details of the gene indication signatures can be found in Supplementary Note 6.
All other details on reproducibility and statistics can be found in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
The results here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. This work was funded by the Novo Nordisk Foundation Center for Biosustainability and the Technical University of Denmark (grant number NNF10CC1016517), the National Institutes of Health (grant GM057089 to B.O.P.) and by the Luxembourg National Research Fund (FNR) through the National Centre of Excellence in Research (NCER) on Parkinson’s disease and the ATTRACT programme (FNR/A12/01), by the European Union’s Horizon 2020 research and innovation programme under grant agreement No 668738, by the Institutional Strategy of the University of Tübingen (German Research Foundation DFG, ZUK 63), and by Google Inc. (Summer of Code 2016). RCSB PDB is funded by the National Science Foundation (NSF DBI-1338415), the Department of Energy, and the National Institutes of Health (NIGMS and NCI). This research used resources of the National Energy Research Scientific Computing Center. The authors gratefully acknowledge Professor Paul Mischel and Wenjing Zheng for experimental help and discussions on GBM, Professor Nathan Lewis, Professor Andy McCammon, Professor Jill Mesirov, Professor Janet M. Thornton, Dr. Jon Monk and Dr. Josh Lerman for scientific discussions and Zak King for help with Escher integration in RCSB PDB, Marc Abrams for manuscript editing, Veronika Kohler and Anja E. Kärcher-Dräger for drawing the platelet and RBC map in Escher, and Fatima Monteiro and Miguel A.P. Oliveira for help in reconstructing the dopamine subsystem.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, E.B., I.T., D.Z.; Methodology, (Reconstruction of metabolic network: S.S., IT, RMTF, ADD, AH, MKA ; Reconstruction of GEM-PRO: EB, NM AS; 3D-hotspot analysis: EB, AP, AS, PWR; Machine learning: D.Z.; PDB visualization: AA, AP, AD, RMTF, SKB; atom-atom mapping: GAPG, RMTF; Model testing and validation: IT, RMTF, SS, MKA, DZ, AN, FG; Cell-specific and infant model simulations: MKA, AN, FG); Investigation, EB., DZ, GAPG; Writing – Original Draft, EB, BOP; Writing – Review & Editing: all authors; Funding Acquisition: IT, RMTF, SKB, JN, and BOP; Resources, IT, RMTF, SKB, JN, and BOP; Supervision: IT, RMTF, SKB, and BOP.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
DATA AND CODE AVAILABILITY
Recon3D is available available as a metabolic reconstruction at http://vmh.life.
To facilitate the future use of the Recon 3D GEM-PRO model, the procedure to collect sequence and structure information as described above has been consolidated into a shareable JSON file, which we call the “minimal” GEM-PRO needed to start structural analyses. This model assigns a single representative structure per gene in the reconstructed metabolic model, and is available at https://github.com/SBRG/Recon3D. The accompanying software package required for reading and working with the GEM-PRO JSON is available at https://github.com/SBRG/ssbio. This entire repository can be cloned to a user’s computer and contains Jupyter notebooks in the root directory to guide a user through the content available in the Recon 3D GEM-PRO model (Recon3D_GP - Loading and Exploring the GEM-PRO.ipynb) as well as to update the model with revised sequence information or newly deposited structures in the PDB (Recon3D_GP - Updating the GEM-PRO.ipynb). This repository also includes all sequence and structure files mapped per gene, metadata downloaded through UniProt and the PDB, as well as the ability to rerun the QC/QA pipeline with different parameters such as sequence identity and resolution cutoffs. These notebooks also include basic visualization features enabled with the NGL viewer package69.
All other scripts related to gene deletion simulations and infant growth simulations can be found at https://github.com/SBRG/Recon3D.
References
- 1.Bui AAT, Van Horn JD. NIH BD2K Centers Consortium. Envisioning the future of ‘big data’ biomedicine. J. Biomed. Inform. 2017;69:115–117. doi: 10.1016/j.jbi.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Brien EJ, Monk JM, Palsson BO. Using Genome-scale Models to Predict Biological Capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiele I, et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proceedings of the National Academy of Sciences. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swainston N, et al. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pornputtapong N, Nookaew I, Nielsen J. Human metabolic atlas: an online resource for human metabolism. Database. 2015;2015:bav068. doi: 10.1093/database/bav068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argmann CA, Houten SM, Zhu J, Schadt EE. A Next Generation Multiscale View of Inborn Errors of Metabolism. Cell Metab. 2016;23:13–26. doi: 10.1016/j.cmet.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gatto F, Nielsen J. Pan-cancer analysis of the metabolic reaction network. bioRxiv. 2016;050187 doi: 10.1101/050187. [DOI] [PubMed] [Google Scholar]
- 9.Ji B, Nielsen J. New insight into the gut microbiome through metagenomics. Adv. Genomics Genet. 2015;5:77–91. [Google Scholar]
- 10.Heinken A, Thiele I. Systems biology of host–microbe metabolomics. WIREs Syst Biol Med. 2015;7:195–219. doi: 10.1002/wsbm.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller ML, et al. Pan-Cancer Analysis of Mutation Hotspots in Protein Domains. Cell Syst. 2015;1:197–209. doi: 10.1016/j.cels.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laskowski RA, et al. Integrating population variation and protein structural analysis to improve clinical interpretation of missense variation: application to the WD40 domain. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddv625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu B, et al. Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat. Genet. 2016;48:827–837. doi: 10.1038/ng.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Z, Xie L, Xie L, Bourne PE. Delineation of Polypharmacology across the Human Structural Kinome Using a Functional Site Interaction Fingerprint Approach. J. Med. Chem. 2016;59:4326–4341. doi: 10.1021/acs.jmedchem.5b02041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porta-Pardo E, Godzik A. Mutation Drivers of Immunological Responses to Cancer. Cancer Immunol Res. 2016;4:789–798. doi: 10.1158/2326-6066.CIR-15-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 19.Ebrahim A, et al. Multi-omic data integration enables discovery of hidden biological regularities. Nat. Commun. 2016 doi: 10.1038/ncomms13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mih N, Brunk E, Bordbar A, Palsson BO. A Multi-scale Computational Platform to Mechanistically Assess the Effect of Genetic Variation on Drug Responses in Human Erythrocyte Metabolism. PLoS Comput. Biol. 2016;12:e1005039. doi: 10.1371/journal.pcbi.1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mardinoglu A, et al. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014;5:3083. doi: 10.1038/ncomms4083. [DOI] [PubMed] [Google Scholar]
- 22.Sahoo S, Haraldsdóttir HS, Fleming RMT, Thiele I. Modeling the effects of commonly used drugs on human metabolism. FEBS J. 2015;282:297–317. doi: 10.1111/febs.13128. [DOI] [PubMed] [Google Scholar]
- 23.Sahoo S, Aurich MK, Jonsson JJ, Thiele I. Membrane transporters in a human genome-scale metabolic knowledgebase and their implications for disease. Front. Physiol. 2014;5:91. doi: 10.3389/fphys.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Famiglietti ML, et al. Genetic variations and diseases in UniProtKB/Swiss-Prot: the ins and outs of expert manual curation. Hum. Mutat. 2014;35:927–935. doi: 10.1002/humu.22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson A, Mardinoglu A, Nielsen J. Predicting growth of the healthy infant using a genome scale metabolic model. npj Systems Biology and Applications. 2017;3:3. doi: 10.1038/s41540-017-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunk E, et al. Systems biology of the structural proteome. BMC Syst. Biol. 2016;10:26. doi: 10.1186/s12918-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berman J, Westbrook HM, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;106:16972–16977. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preciat Gonzalez GA, et al. Comparative evaluation of atom mapping algorithms for balanced metabolic reactions: application to Recon 3D. J. Cheminform. 2017;9:39. doi: 10.1186/s13321-017-0223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noronha A, et al. ReconMap: an interactive visualization of human metabolism. Bioinformatics. 2017;33:605–607. doi: 10.1093/bioinformatics/btw667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whirl-Carrillo M, et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–55. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 33.Kris MG, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 34.von Bülow R, et al. Defective oligomerization of arylsulfatase a as a cause of its instability in lysosomes and metachromatic leukodystrophy. J. Biol. Chem. 2002;277:9455–9461. doi: 10.1074/jbc.M111993200. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villa GR, et al. An LXR-Cholesterol Axis Creates a Metabolic Co-Dependency for Brain Cancers. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geng F, et al. Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP-1-Mediated Lipogenesis. Clin. Cancer Res. 2016;22:5337–5348. doi: 10.1158/1078-0432.CCR-15-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7, Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zielinski DC, et al. Pharmacogenomic and clinical data link non-pharmacokinetic metabolic dysregulation to drug side effect pathogenesis. Nat. Commun. 2015;6:7101. doi: 10.1038/ncomms8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 45.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer A, Sananbenesi F, Mungenast A, Tsai L-H. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol. Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Xie L, Xie L, Kinnings SL, Bourne PE. Novel computational approaches to polypharmacology as a means to define responses to individual drugs. Annu. Rev. Pharmacol. Toxicol. 2012;52:361–379. doi: 10.1146/annurev-pharmtox-010611-134630. [DOI] [PubMed] [Google Scholar]
- 48.Hopkins AL. Network pharmacology. Nat. Biotechnol. 2007;25:1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 49.Brunk E, Rothlisberger U. Mixed Quantum Mechanical/Molecular Mechanical Molecular Dynamics Simulations of Biological Systems in Ground and Electronically Excited States. Chem. Rev. 2015;115:6217–6263. doi: 10.1021/cr500628b. [DOI] [PubMed] [Google Scholar]
- 50.Bordbar A, et al. Personalized Whole-Cell Kinetic Models of Metabolism for Discovery in Genomics and Pharmacodynamics. Cell Systems. 2015;1:283–292. doi: 10.1016/j.cels.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 51.King ZA, et al. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44:D515–22. doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noor E, Haraldsdóttir HS, Milo R, Fleming RMT. Consistent estimation of Gibbs energy using component contributions. PLoS Comput. Biol. 2013;9:e1003098. doi: 10.1371/journal.pcbi.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quek L-E, et al. Reducing Recon 2 for steady-state flux analysis of HEK cell culture. J. Biotechnol. 2014;184:172–178. doi: 10.1016/j.jbiotec.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 54.Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, Haraldsdottir HS, Keating SM, Vlasov V, Wachowiak J, et al. Creation and analysis of biochemical constraint-based models: the COBRA Toolbox v3.0. doi: 10.1038/s41596-018-0098-2. arXiv:1710.04038 [q-bio.QM] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dawson PA, Lan T, Rao A. Bile acid transporters. J. Lipid Res. 2009;50:2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu D, Zhang Y. Ab Initio structure prediction for Escherichia coli: towards genome-wide protein structure modeling and fold assignment. Sci. Rep. 2013;3 doi: 10.1038/srep01895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou H, Gao M, Kumar N, Skolnick J. SUNPRO: Structure and function predictions of proteins from representative organisms. 2012 [Google Scholar]
- 58.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim S, et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44:D1202–13. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kinsella RJ, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahman SA, et al. Reaction Decoder Tool (RDT): extracting features from chemical reactions. Bioinformatics. 2016;32:2065–2066. doi: 10.1093/bioinformatics/btw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.First EL, Gounaris CE, Floudas CA. Stereochemically consistent reaction mapping and identification of multiple reaction mechanisms through integer linear optimization. J. Chem. Inf. Model. 2012;52:84–92. doi: 10.1021/ci200351b. [DOI] [PubMed] [Google Scholar]
- 64.Kumar A, Maranas CD. CLCA: maximum common molecular substructure queries within the MetRxn database. J. Chem. Inf. Model. 2014;54:3417–3438. doi: 10.1021/ci5003922. [DOI] [PubMed] [Google Scholar]
- 65.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gatto F, Miess H, Schulze A, Nielsen J. Flux balance analysis predicts essential genes in clear cell renal cell carcinoma metabolism. Sci. Rep. 2015;5:10738. doi: 10.1038/srep10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gatto F, Nielsen J. Pan-cancer analysis of the metabolic reaction network. bioRxiv. 2016:050187. doi: 10.1101/050187. [DOI] [PubMed] [Google Scholar]
- 68.Kuhn M, Letunic I, Jensen LJ, Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rose AS, Hildebrand PW. NGL Viewer: a web application for molecular visualization. Nucleic Acids Res. 2015;43:W576–9. doi: 10.1093/nar/gkv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hastings J, et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 2013;41:D456–63. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.