Abstract
The PU.1 transcription factor plays a critical role in the regulation of T cell development, so a report that it is dispensable for fetal thymopoiesis is puzzling. In order to understand this paradox, we examined the requirement for PU.1, encoded by Spi1, during fetal, neonatal, and adult thymopoiesis in a PU.1 hypomorphic mouse generated by deletion of the Spi1 14Kb upstream regulatory element and by analysis of patterns of gene expression in fetal and adult T cell progenitors. Our data demonstrate that the initiation of thymopoiesis during early gestation is less dependent on PU.1 compared to T cell differentiation in adults and that fetal T cell progenitors express lower levels of Spi1 compared to their adult counterparts. We also show that expression of the core network of T lineage transcription factors regulated by PU.1 differs in fetal and adult T cell progenitors. In particular, PU.1 regulated genes that promote T cell differentiation are differentially expressed in fetal versus adult ETPs. These results indicate that the transcriptional differences between the fetal and adult T cell developmental programs are driven in part by differential levels of PU.1 expression and that this likely underlies the differences in the properties of fetal and adult T cell progenitors.
Introduction
The stages of development leading to the generation of mature T cells within the thymus are well defined (1–3). The most immature cells in that organ are referred to as Early T Lineage Progenitors (ETPs) (4), and their progeny mature through CD4− CD8− double negative (DN) stages which undergo progressive recombination of genes encoding the T Cell Receptor (TCR) and differentiate into double positive (DP) cells that co-express CD4 and CD8. These cells then become single positive (SP) CD4 or CD8 T cells that, after appropriate selection, exit the thymus and colonize secondary lymphoid tissues. The majority of newly produced T cells express the αβ TCR, but a minor subpopulation utilizes the γδ TCR (5). Because the thymus does not contain self-renewing stem cells, sustained thymopoiesis is dependent on the migration of T cell precursors from the bone marrow. While multiple precursors may have the potential to enter the thymus, most adult thymocytes are thought to be derived from thymus seeding lymphoid primed multipotential precursors (6).
T cell development initiates in the fetus and occurs in distinct waves distinguished by the source and properties of the thymus seeding cells. Cells with T lineage potential are generated at least a day prior to the emergence of hematopoietic stem cells (HSCs) at embryonic age (E)10.5 in the yolk sac (YS). These pre-HSC YS progenitors can produce both αβ and γδ T cells (7, 8) and likely contribute to the initial wave of T cell development recently described by Ramond et al. that occurs between E11 and E15 (9). The progenitors that seed the thymus in this developmental window are T lineage restricted and have limited capacity for expansion while those that emerge in a second wave after E16 have B and myeloid potential and are highly proliferative (9). Whether this latter wave sustains adult thymopoiesis or, a third, adult wave of T cell development occurs in unclear.
In addition to the differences in T cell development between the two fetal waves, numerous distinctions between fetal and adult T cell development exist. For example, γδ T cells that express the Vγ3 receptor are preferentially generated during fetal thymopoiesis (10, 11), and the requirement for cytokines such as IL-7 may be distinct (12). The transit time through the thymus also varies, with fetal progenitors generating mature T cells in four days while this process requires over ten days in the adult (13, 14). Finally, evidence from various knockout strains of mice indicate that the transcriptional regulation of fetal and adult thymopoiesis differs (15, 16). How the transcriptional regulation of T cell development in the different waves compares and explains the distinct properties of progenitors arising at different times during development remains to be determined.
PU.1 is a pioneer transcription factor (17) that coordinates the expression of the core network of T cell regulatory genes (18). In view of this, data indicating that it is dispensable for fetal (19), but not adult (20), thymopoiesis are puzzling. We thus considered that a detailed re-examination of this paradoxical observation could provide new insights into the transcriptional regulation of thymopoiesis in the fetus and adult as well as a basis for understanding differences in the properties of fetal and adult T cell progenitors. A systematic analysis of the various waves of fetal and adult T cell development in PU.1 knockout mice is not possible, because these strains die in utero or soon after birth (21, 22). However, the availability of a PU.1 hypomorphic mouse, generated by deletion of an upstream regulatory element (URE) 14 kb from the Spi1 (formerly Sfpi1) transcription start site, obviates this limitation (20). These URE deficient (UREΔ/Δ) mice survive into adulthood even though their hematopoietic cells exhibit an 80% reduction in Spi1 expression. In this study, we analyzed fetal and adult thymopoieis in UREΔ/Δ mice and examined the expression of PU.1 regulated genes in E15.5 and adult T cell progenitors.
The studies of UREΔ/Δ mice demonstrate that wild type levels of PU.1 are dispensable for emergence of the pre-HSC and early fetal waves of thymopoiesis but not for those initiating during late gestation and in the adult. We also found that these developmental differences correlated with lower expression of Spi1 in fetal compared to adult ETPs from wild type mice. Finally, we used a recently published database (23) to show that expression of genes regulated by PU.1, and in particular those that control T cell progenitor proliferation and differentiation, differed between fetal and adult ETPs. These results provide a genetic foundation for the layered immune system hypothesis (24) and a basis for understanding why the functional properties of T cell progenitors arising in the different waves of development are distinct.
Materials and Methods
Mice and genotyping
UREΔ/Δ (Sfpitm1.3Dgt/J) mice with a deletion of the URE located 14kb upstream of the Spi1 gene were obtained from The Jackson Laboratory (Bar Harbor, ME), bred in the UCLA Division of Laboratory Animal Medicine, and genotyped using the protocol recommended by the Jackson Laboratory. Preliminary analyses indicated that thymopoiesis in UREΔ/+ heterozygotes and C57BL/6 (B6) mice were comparable, so both strains were used interchangeably as positive controls. B6 mice were obtained from the Jackson Laboratory or the UCLA Division of Laboratory Animal Medicine. All experiments on male and female mice were conducted according to UCLA Institutional Animal Care and Use Committee guidelines.
Flow Cytometry
YS, fetal liver (FL), bone marrow (BM), and thymic cell suspensions were prepared as previously described (25, 26). Hematopoietic Stem Cells (HSCs) in FL and BM and ETPs, DN2, DN3, DN4, CD4+ CD8+, CD4+ CD8− (CD4+) and CD4− CD8+ (CD8+) thymocytes were resolved using specific combinations of FITC, PE, PerCP/Cy5.5, PECy7, APC efluor780, Pacific Blue, and 605NC conjugated antibodies (Fig. S1A, S1B and Supplemental Table 1). The lineage cocktail used to deplete mature cells for purification of ETPs included anti-CD3ε, CD8α, Gr-1, CD11b, IgM, NK1.1, TCRβ, TCRγδ, and TER119 antibodies, whose clone number and source are listed in Table S1. Anti-CD11b was omitted in the Lin cocktail used for HSCs purification. Progenitors were purified using Aria Cell sorters (BD Biosciences) located in the Jonsson Comprehensive Cancer Center flow cytometry core and analyses were performed on an LSRII (BD Biosciences) located in the Broad Stem Cell Research Center flow cytometry core at UCLA. FL and/or BM controls were included in all experiments to account for instrumental variations during analyses and used as positive controls when assessing CD135 and CD127 expression on ETPs.
In vitro cultures
Thymic organ cultures were established as previously described (27), placed in humidification chambers incubated at 37°C and 5% CO2 and fed once a week with fresh medium. Thymocytes were recovered by mechanical disruption of the thymic lobes and T cell production was assessed by flow cytometry.
The T cell potential of YS, FL, MPPs and HSCs was tested following seeding onto confluent layers of OP9-DL1 stromal cells (27) in RPMI-1640 supplemented with 10% FCS, 100 μg of streptomycin, 100 U/ml penicillin, 10 μg/ml gentamicin, 0.1 mM nonessential amino acids, 0.1 mM nonessential vitamins, 1 mM sodium pyruvate, 5 × 10−5 M 2-ME, 1 mM sodium pyruvate, 30nM L-ascorbic acid (1-ascorbic acid-2-phosphate sesquimagnesium salt pAsc; SIGMA), (28), 20 ng/ml FLT3, and 20 ng/ml IL-7 (Biosource) in 12 well plates. YS and FL were seeded at 2 embryo equivalents per well, while purified progenitors were seeded at 200 cells per well. Cultures were placed in a 37°C and 5% CO2 in humidified incubator and fed three times weekly with fresh medium. T cell production was assessed at 13 and 21 days following culture initiation as follows: On day thirteen, hematopoietic cells produced in culture were harvested and split into 2 samples. One sample was tested for production of γδ, TCRVγ2+ and Vγ3+ T cells by immunofluorescence, while the other was reseeded over fresh OP9-DL1 stroma. Seven days later, the cultures were terminated and CD4 and CD8 T cell production was assessed by flow cytometry.
Quantitative RT-PCR
RNA was extracted with the RNeasy Plus microkit and cDNA was synthesized with the RT2 First Strand kit as recommended by the manufacturer (both from QIAGEN). Reactions were run in 20 μL using TaqMan Universal PCR Master Mix, no AmpErase UNG and the following Taqman primers: mGapdh (Mn99999915-g1) and mSpfi1 (Mn00488142-m1) as recommended by the manufacturer (all from ThermoFisher Scientific). Data were acquired with a MyIQ (BioRad) using the Bio-Rad IQ5 2.0 software and relative levels of expression were calculated using the Pfaffl method using Gapdh as a reference gene. Amplification efficiencies were routinely found to be between 95% and 105%.
Microarray data analysis
Microarray raw data files downloaded from NCBI’s Gene Expression Omnibus (accession number GSE24142; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24142) deposited by Belyaev et al (23) were processed in R (http://www.R-project.org). The .CELL files from the Affymetrix platform were imported (ReadAffy package) and subjected to RMA normalization (rma package). Final expression estimates in logarithmic scale were obtained with the exprs package. We then compared expression levels (log2 intensities) of the genes listed in Supplemental Tables 2 in E15.5 and 6 week old B6 DN1 ETP, DN2 and DN3 populations. These were included in the 4,970 probes that passed the p<0.05 filter following ANOVA test as described by Belyaev et al. (23).
Statistical analysis
Data are expressed as a mean ± SEM as indicated in the figure legends. Differences between groups were tested by a two-tailed, unpaired Student’s t test (α = 0.05).
Results
The initial waves of fetal T cell development emerge in UREΔ/Δ mice
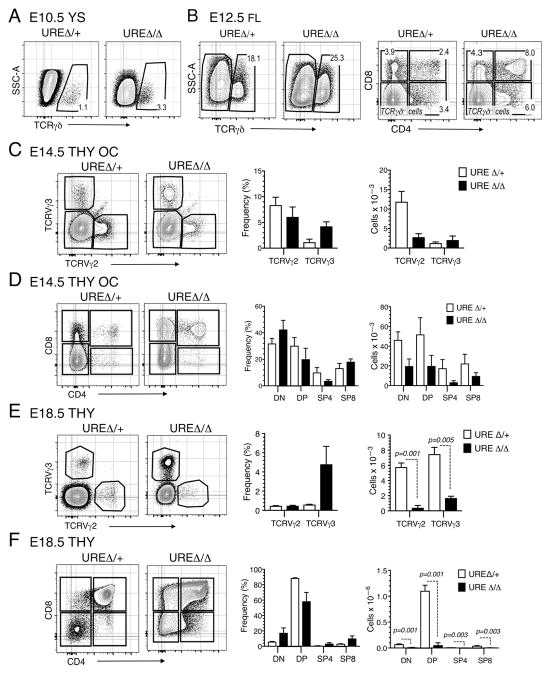
Progenitors first colonize the thymus at around E11.5 (29), and the vast majority of the cells present at that time and for several days thereafter are immature DN cells. We used several approaches, including the OP9-DL1 culture system and thymic organ culture (27), to assess their thymopoietic potential. T cell progenitors arise prior to the emergence of HSCs on E10.5 (7, 8), and this wave of development is intact in UREΔ/Δ mice. E10.5 YS cells produced TCRγδ T cells (Fig. 1A), consistent with a previous report from our laboratory showing that they could also generate CD4 and/or CD8 expressing lymphocytes (30). By E12.5 the FL contains HSCs as well as progenitors committed to multiple hematopoietic lineages, and we found that FL from UREΔ/Δ mice included progenitors that could generate CD4+ CD8+, as well as CD4+ and CD8+ thymocytes following seeding on OP9-DL1 stroma as well (Fig. 1B). Finally, when we tested the thymopoietic potential of progenitors within the E14.5 thymus to mature into T cells by placing thymic lobes in organ culture for three weeks, both TCRVγ2 and TCRVγ3, DP, as well as SP4 and SP8 thymocytes were produced (Fig. 1C, 1D).
Fig. 1. The initial waves of fetal T cell development emerge in UREΔ/Δ embryos.
(A) FACS plots showing production of TCRγδ+ cells from E10.5 YS from UREΔ/Δ and UREΔ/+ embryos following 13 days of culture over OP9-DL1 stroma. (B) FACS plots showing the frequency of TCRVγ2+, Vγ3+, CD4+ CD8+, CD4+, and CD8+ cells generated from E12.5 UREΔ/Δ and UREΔ/+ FL following seeding on OP9-DL1 stroma. Frequencies of gated populations are indicated. Each panel is representative of 2 independent experiments. (C) FACS plots and graphs showing the frequencies and numbers of TCRVγ2+, Vγ3+ cells harvested from E14.5 UREΔ/Δ and UREΔ/+ thymic lobes placed in organ cultures for 3 weeks. (D) FACS plots and graphs showing the frequencies and numbers of DN, DP, SP4 and SP8 cells generated in E14.5 UREΔ/Δ and UREΔ/+ thymic lobes placed in organ cultures for 3 weeks. Data are representative of 4 independent experiments. E14.5 UREΔ/Δ: n = 5–6 lobes; E14.5 UREΔ/+: n = 5–8 lobes. (E) FACS plots and graphs showing the frequencies and numbers of DN, DP, SP4 and SP8 cells generated in E18.5 UREΔ/Δ or UREΔ/+ thymic lobes. Mean ± SEM and t test p values are shown. E18.5 UREΔ/Δ: n = 8–10 lobes; E18.5 UREΔ/+: n = 4 lobes.
Taken together, these results indicate that the initial fetal waves of T cell development can emerge in UREΔ/Δ mice. However, a slight deficit in cell production was evident when comparing E14.5 UREΔ/Δ and UREΔ/+ thymic lobes. Specifically, fewer TCRVγ2+, CD4+ CD8+ and CD4+ T lineage cells were generated in the UREΔ/Δ thymus (Fig. 1C, 1D). These data suggest that as a result of the lower levels of PU.1, due to the deletion of the Spi1 14Kb URE, the potential of progenitors to generate normal numbers of T lineage cells is altered.
Late-gestation and adult thymopoiesis are compromised in UREΔ/Δ mice
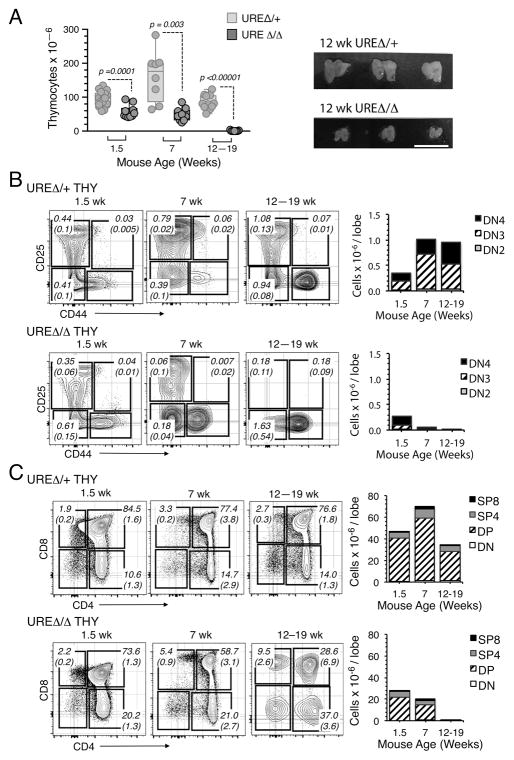
In contrast to the above results, the pattern of thymopoiesis in UREΔ/Δ mice at subsequent times differed significantly. For example, while all stages of T cell development were present in thymic lobes from E18.5 UREΔ/Δ embryos, the trend towards diminished cell production in all T cell subsets was accentuated (Fig. 1E, 1F). This was also the case during neonatal thymopoiesis. While the number of thymocytes increases exponentially in the weeks after birth (31), this occurred at a significantly lower level in UREΔ/Δ mice. Thymocyte numbers were significantly reduced in the thymus of 1.5 and 7 wk old UREΔ/Δ mice, and by 12 weeks after birth that organ was hypoplastic and contained less than one million cells (Fig. 2A). Examination of T cell production in the UREΔ/Δ thymus at these times indicated that there was a progressive exhaustion of DN (Fig. 2B) and DP (Fig. 2C) subsets and a trend towards terminal differentiation into SP cells (Fig. 2C).
Fig. 2. Thymopoesis wanes by 12 weeks of age in UREΔ/Δ mice.
(A) Left panel: Evolution of total thymic cellularity in UREΔ/Δ and UREΔ/+ mice with age. Each symbol represents a mouse. Means ± SEM and p values are shown. Right panel: Photomicrograph of thymuses from 12 wk old UREΔ/Δ and UREΔ/+ mice (Bar = 1.0 cm). (B) Left panels: FACS plots showing the frequencies of Lin− DN2, DN3 and DN4 cells in the thymus of 1.5, 7 and 12–19 wk old UREΔ/Δ and UREΔ/+ mice. Right panels: Number of Lin− DN2, DN3 and DN4 thymocytes in UREΔ/Δ and UREΔ/+ thymii with age. Numbers in plots indicate the frequency and (SEM) for each population in total thymus. t test comparisons for DN2, DN3, and DN4 subpopulations in UREΔ/Δ and UREΔ/+ thymuses yielded values of at least p<0.0001. (C) Left panels: FACS plots showing the frequencies DN, DP, SP4 and SP8 cells in the thymus of 1.5, 7 and 12–19 wk old UREΔ/Δ and UREΔ/+ mice. Right panels: Number of DN, DP, SP4 and SP8 thymocytes in UREΔ/Δ and UREΔ/+ thymii with age. Numbers in plots indicate the frequency and (SEM) for each population in total thymus. t test comparisons for DN, DP, SP4 and SP8 thymocytes in UREΔ/Δ and UREΔ/+ thymuses yielded values of at least p<0.0005. The data in Figures 2B and 2C are based on analysis of the following numbers of mice: 1.5 wk: UREΔ/+ (n=14) and UREΔ/Δ (n=10); 7 wk: UREΔ/+ (n=5) and UREΔ/Δ (n=6); 12–19 wk: UREΔ/+ (n=9) and UREΔ/Δ (n=9).
T cell developmental potential wanes in the UREΔ/Δ thymus by late gestation
The above results indicate that the potential to generate T cell progenitors in UREΔ/Δ mice is lost between late gestation and young adult life. In order to determine more precisely when this occurs, we tested the potential of HSCs harvested from UREΔ/Δ and control mice of increasing age to initiate T lineage cells in culture over OP9-DL1 stroma. It has been reported that HSCs are not maintained in PU.1 deficient mice (32). However, no significant differences in the frequency or total number of Lin− CD117+ Sca-1+ CD150+ cells were observed between the UREΔ/Δ and control mice used in this study as previously reported (30).
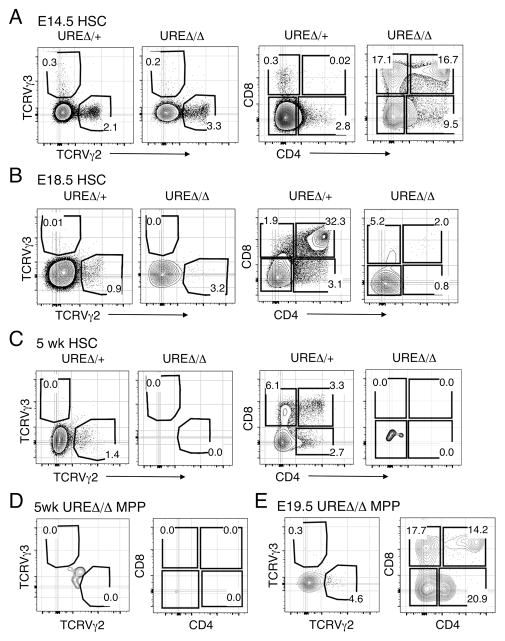
HSCs purified from E14.5 UREΔ/Δ FL produced γδTCR and CD4 and CD8 expressing cells (Fig. 3A). In contrast, HSCs from the E18.5 UREΔ/Δ FL had limited to non-existent potential to do so (Fig. 3B), and HSCs harvested from the BM of 5 wk old UREΔ/Δ mice lacked T cell potential (Fig. 3C). We also observed that Lin− CD117+ Sca-1+ CD150− mulipotential precursors (MPPs) from E19.5 UREΔ/Δ FL differentiated into CD4 and CD8 expressing cells on OP9-DL1 stroma but MPPs from the BM of 5 wk old UREΔ/Δ mice failed to do so (Fig. 3D, 3E). The inability of HSCs from UREΔ/Δ mice to generate thymocytes correlated with their lower expression of Spi1 (Supplemental Fig. 1C), which has been previously observed (20). We also observed that MPPs from adult UREΔ/Δ mice expressed lower levels of Spi1 compared to their age matched controls (Supplemental Fig. 1C).
Fig. 3. T cell potential of UREΔ/Δ HSCs wanes with age.
(A) FACS plots showing the frequencies of TCRVγ2+, Vγ3+, and CD4+ CD8+, as well as CD4+ and CD8+ cells produced by HSCs from E14.5 UREΔ/Δ and UREΔ/+ FL seeded on OP9-DL1 stroma. (B) FACS plots showing the frequency of TCRVγ2+, Vγ3+, and CD4+ CD8+, as well as CD4+ and CD8+ cells generated by HSCs isolated from E18.5 UREΔ/Δ and UREΔ/+ FL following seeding on OP9-DL1 stroma. (C) FACS plots showing the frequency of TCRVγ2+, Vγ3+, and CD4+ CD8+, as well as CD4+ and CD8+ cells generated by HSCs from BM of 5 wk old UREΔ/Δ and UREΔ/+ mice following seeding on OP9-DL1 stroma. (D) FACS plots showing the frequency of TCRVγ2+, Vγ3+, and CD4+ CD8+, as well as CD4+ and CD8+ cells generated by MPPs isolated from BM of 5 wk old UREΔ/Δ following seeding on OP9-DL1 stroma. (E) FACS plots showing the frequency of TCRVγ2+, Vγ3+, and CD4+ CD8+, as well as CD4+ and CD8+ cells generated by MPPs isolated from E19.5 UREΔ/Δ FL following seeding on OP9-DL1 stroma. TCR Vγ production was assessed following 13 days in culture and CD4+ and/or CD8+ cell production following 21 days in culture, as described in the Material and Methods. Frequencies of gated populations are indicated in each plot. Data representative of 2 independent experiments for each population tested. Number of animals used per experiment: E14.5: UREΔ/Δ (n=9) and UREΔ/Δ (n=8), E18.5: UREΔ/Δ (n=4) and UREΔ/+ (n=5), 5 wk: UREΔ/Δ (n=3) and UREΔ/+ (n=3).
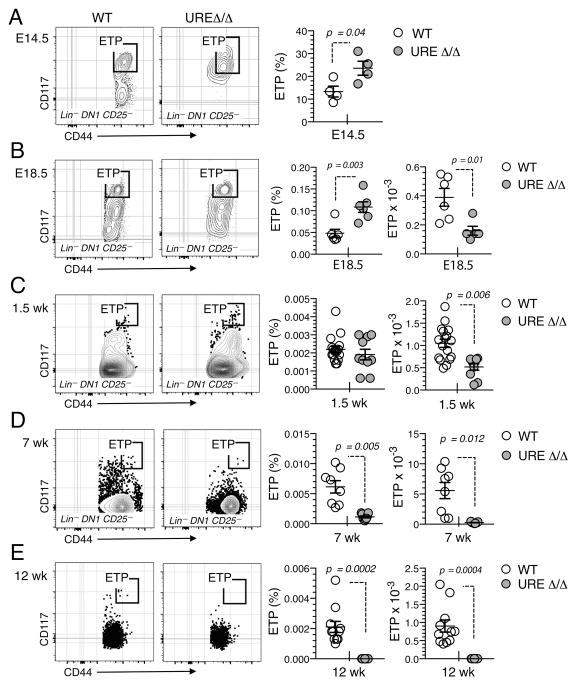
The timing of when T cell potential was lost in UREΔ/Δ mice correlated with ETP production. ETPs were present in the E14.5 UREΔ/Δ thymus (Fig. 4A). They were also observed in the thymus of E18.5 UREΔ/Δ fetuses and 1.5 wk old UREΔ/Δ neonates, but at significantly lower numbers than in UREΔ/+ mice (Fig. 4B, 4C). However, ETPs were barely detectable in the thymus of UREΔ/Δ mice after seven weeks of age (Fig. 4D, 4E). This latter observation conflicts with a previous report showing an enlarged ETP compartment in adult UREΔ/Δ mice (20). However, in contrast to other studies which have used more stringent purification strategies (33, 34), that report defined ETPs as CD3− CD4− CD8− c-kit+ CD25− cells. In view of the relative increase of DN1 thymocytes we observed in the 12–19 wk old UREΔ/Δ thymus (Fig. 2B, 2C), it is likely that the cells analyzed in the previous study (20) were non-T lineage cells able to express Spi1 independently of the 14Kb URE (35).
Fig. 4. ETP frequency and number decline with age in UREΔ/Δ mice.
(A) FACS plots and graphs showing the frequency of ETPs in the E14.5 UREΔ/Δ and UREΔ/+ (WT) thymus. Mean ± SEM and t test p values are shown. (B) FACS plots and graphs showing the frequency and number of ETPs in thymic lobes from E18.5 UREΔ/Δ and UREΔ/+ (WT) embryos. Mean ± SEM and p values are shown. Cell suspensions were prepared by pooling thymic lobes from multiple embryos in each experiment: E14.5 UREΔ/Δ and WT: n=5–8 lobes; E18.5 UREΔ/Δ: n=8–10 lobes and E18.5 WT: n=4 lobes. Each symbol represents an independent experiment. (C) FACS plots and graphs showing the frequency and number of ETPs in the thymus of 1.5 wk old UREΔ/Δ and UREΔ/+ mice. (D) FACS plots and graphs showing the frequency and number of ETPs in the thymus of 7 wk old UREΔ/Δ and UREΔ/+ mice. (E) FACS plots and graphs showing the frequency and number of ETPs in the thymus of 12 wk old UREΔ/Δ and UREΔ/+ mice. Each symbol represents a mouse. Mean ± SEM and p values are shown; data are representative of 2 to 4 independent experiments.
CD135 expressing ETPs do not emerge in UREΔ/Δ mice
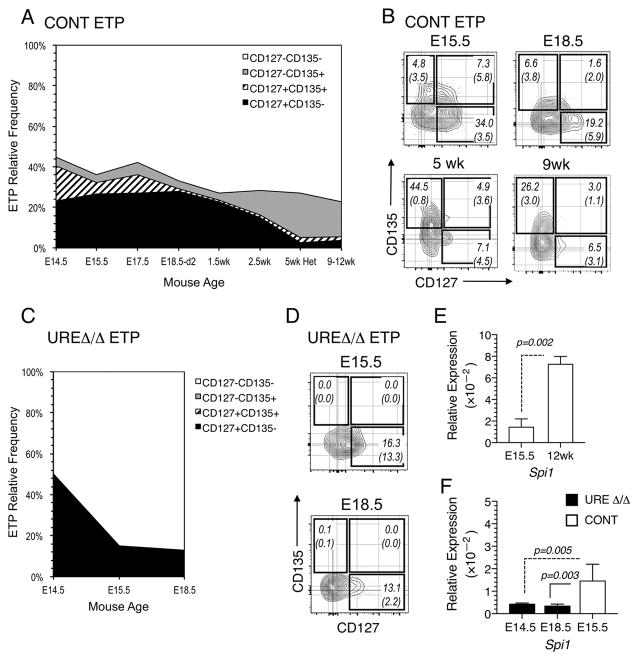
The expression of the Interleukin-7 (CD127) and the FLT3 (CD135) receptors have been used to subdivide fetal ETPs into multiple subpopulations (9). A subpopulation of ETPs in the adult thymus express CD135, which is important for the overall efficiency of post-natal thymopoiesis (36). The absence of adult thymopoiesis in UREΔ/Δ mice raised the possibility that the generation of CD135+ T cell progenitors was compromised, particularly since PU.1 has been shown to regulate expression of this receptor (37).
We first established when the different subpopulations of ETPs emerged and how their distribution evolved in control mice of increasing age. While CD127+ CD135+ as well as CD127+ CD135− ETPs were most abundant during early gestation, their frequencies declined with advancing age and they comprised a very small fraction of ETPs in neonatal and adult mice (Fig. 5A, 5B). In contrast, the frequency of CD127− CD135+ ETPs was low in the fetus, but this population ultimately predominated in the adult (Fig. 5A, 5B). A similar analysis in UREΔ/Δ mice revealed a complete absence of CD135 expressing ETPs and showed that only CD127+ CD135− ETPs emerged (Fig. 5C, 5D). These results suggest that T cells in UREΔ/Δ mice are derived from CD127+ ETPs.
Fig. 5. ETPs emerge in successive waves distinguished by CD127 and CD135 expression.
(A) Graph showing the relative frequencies of CD135+ CD127−, CD135+ CD127+, CD135− CD127+ and CD135− CD127− ETPs in the thymus of B6 control (CONT) mice at the indicated fetal, neonatal and adult ages. Mean frequencies and (SEM) are indicated. (B) FACS plots showing CD135 and CD127 expressing ETP populations in the thymus of B6 control (CONT) mice at the indicated fetal and adult ages. (C) FACS plots showing CD135 and CD127 expressing ETP populations in the thymus of UREΔ/Δ mice at the indicated fetal ages. Mean frequencies and (SEM) are indicated. (D) FACS plots showing the frequency of CD135+ CD127−, CD135+ CD127+, CD135− CD127+ and CD135− CD127− ETPs in the thymus of UREΔ/Δ mice at the indicated fetal ages. Data are representative of 3 to 4 independent experiments in which thymic lobes from 5 to 10 fetal/neonatal mice were pooled. Day 15, 2.5, and 5 wk values were obtained with 7 to 10 mice per age group. (E) Expression of Spi1 relative to Gapdh in ETPs from E15.5 and 12 wk old B6 mice. The data are based on analysis of ETPs purified from four independently obtained fetal and two adult thymus pools. Each fetal pool was prepared from 6–10 fetuses and each adult pool was prepared from 4–6 mice. (F) Expression of Spi1 relative to Gapdh in ETPs from E14.5 and E18.5 UREΔ/Δ and E15.5 B6 mice. The data are based on analysis of ETPs purified from two independently obtained E14.5 UREΔ/Δ, two E18.5 UREΔ/Δ, and four B6 fetal thymus pools. Each fetal pool was prepared from 4–10 fetuses. qPCR reactions were performed two to three times on each sample in E and F. Mean ± SEM and p values are shown.
The fetal and adult T cell developmental program is differentially regulated
Taken together, the above results raised the possibility that fetal and adult T cell progenitors might normally express different levels of Spi1. To determine if this was the case, we measured Spi1 expression in ETPs from E15.5 fetal and 12 wk old B6 mice and found that levels were significantly lower in the fetal cells (Fig. 5E). We also observed that levels of Spi1 in E14.5 and E18.5 UREΔ/Δ ETPs were significantly lower than in ETPs from E15.5 B6 control mice (Figure 5F).
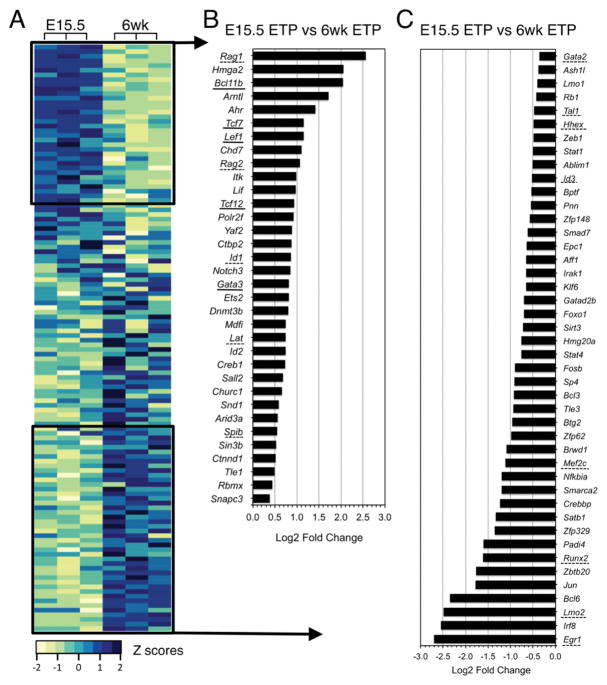
In view of these results, we determined how the expression of additional transcription factors that are up- and down-regulated during ETP differentiation (38) compares between fetal and adult T cell progenitors. We did so using expression data from a previously published microarray analysis that profiled ETPs and DN2, DN3, and DN4 thymocytes from wild type mice (Figure 6A; Supplemental Table 2). Belyaev et al. (23) showed that fetal and adult T cell progenitors have different transcriptional signatures and proposed that these might provide a genetic basis for the accelerated development that is observed during fetal development (13, 14). Our reanalysis confirmed their conclusions and further revealed that transcription factors such as Bcl11b, Lef1, Tcf12(HEB), Tcf7(TCF1) and Gata3, were expressed at higher levels in fetal compared to adult ETPs (Fig. 6B). Additional genes such as Lmo2, Runx2, Mef2c, Gata2 and Tal1, which have been implicated in T lineage specification and commitment and may also regulate ETP self-renewal and commitment (1, 18), were expressed at lower levels (Fig. 6C). Similar trends in the network of T lineage transcription factors were observed in DN2 and DN3 thymocytes from fetal and adult mice (Supplemental Fig 2A–2F, Supplemental Table 2).
Fig. 6. Expression of key T lineage transcription factors differs between fetal and adult ETPs.
(A) Heatmap showing expression of selected transcription factors in ETPs from E15.5 and 6 wk old B6 mice. Log2 intensities were obtained from microarray data deposited by Belyaev et al (GSE24142) (23). The complete gene list is included in Supplemental Table 2. All genes were included in the 4,970 probes that passed the p<0.05 filter following ANOVA test as described by Belyaev et al. (23). Z-scores are shown for 3 independent samples for each mouse age. Dark blue: high expression levels; pale green: low expression levels. (B) Log2 fold change of transcription factors with higher expression in fetal than adult ETPs (E15.5 versus 6 wk). (C) Log2 fold change of the transcription factors with lower expression in fetal than adult ETPs (E15.5 versus 6 wk). Genes whose expression is regulated by PU.1 are underlined.
We were particularly interested in genes regulated by PU.1 and found that many of the transcription factor genes whose expression differed between fetal and adult ETPs were included in this category (Fig. 6B) (18). We also found that, in addition to those transcription factors, several PU.1 target genes were differentially expressed between fetal and adult ETPs (18) (Fig. 6 and Supplemental Fig. 2G, 2H; Supplemental Table 2). For example, Flt3, a known PU.1 target (37), was expressed at lower levels in fetal ETPs while others such as Notch3, Nrarp and Dtx1, were expressed at higher levels (Supplemental Fig. 2G, 2H; Supplemental Table 2). Taken together, these data suggest, in agreement with Belyaev et al., that as a result of their intrinsic low levels of Spi1, fetal ETPs are primed to progress through the T cell developmental program faster than adult progenitors (23).
Discussion
Schemes depicting the transcriptional network that regulates T cell development have been formulated, but the possibility that expression of component transcription factors may not be uniform throughout development is generally not considered. We explored this issue, in view of the reported differences between fetal and adult thymopoiesis, by analyzing PU.1 expression and function in a PU.1 hypomorphic mouse (20) and assessing expression of key T cell transcription factors in fetal and adult progenitors.
The results demonstrate fundamental differences in the requirement for PU.1 during fetal and adult T cell development. Specifically, the threshold of Spi1 expression required for the emergence of ETPs in the initial waves of fetal T cell development is lower compared to levels required during late gestation and in the adult. In this regard, we show that the absence of thymopoiesis in late gestation fetal and adult UREΔ/Δ mice occurs because ETPs cannot be generated from their upstream precursors that include HSCs and MPPs.
We demonstrate that differential expression of Spi1 between fetal and adult ETPs is a feature of normal T cell development. In this regard, we found that fetal ETPs from wild type, B6 mice expressed lower levels of Spi1 compared to their adult counterparts. We also observed that ETPs from E14.5 and E18.5 UREΔ/Δ thymuses expressed lower levels of Spi1 than ETPs from E15.5 wild type thymuses, indicating that the 14 Kb URE is active in these fetal populations. Additional studies to define the precise functions of this regulatory element, as well as that of other potential regulators of Spi1 expression, are needed to understand how Spi1 expression is regulated in fetal versus adult hematopoietic progenitors.
Ramond et al. described an early wave of fetal T cell development that occurs between E11 and E15 (9), and we propose that it is sustained in part by progenitors that arise in the pre-HSC wave (7, 8). Emergence of this T cell developmental wave is largely PU.1 independent, since it is intact in UREΔ/Δ mice. Interestingly, B lineage cells that develop in the pre-HSC wave are completely absent in UREΔ/Δ mice (30), suggesting that the B and T cell progenitors that emerge in this stem cell independent wave have differential PU.1 requirements and may not share a common upstream lymphoid progenitor. This hypothesis is supported by the observation that the earliest thymus seeding cells identified in the fetus lack B cell potential (9). We also observed that thymocytes were generated from E14.5 HSCs in UREΔ/Δ mice, and lymphoid potential in MPPs was observed at least until E19.5 in these animals. The progenitors arising at this time may sustain the second wave of T cell development, arising at E16, described by Ramond et al (9).
In contrast, thymopoiesis in UREΔ/Δ mice was not sustained beyond seven weeks of age and neither E18.5 HSCs nor HSCs or MPPs from adult UREΔ/Δ mice had T cell developmental potential. These results suggest that in addition to the pre-HSC and mid-gestation (i.e., E14.5) waves, an additional wave responsible for adult T cell development must exist. The existence of this third wave of thymopoiesis, which was predicted based on studies of avian T cell development (39, 40), would not have been observed by Spain and colleagues (19) because the PU.1 deficient mice they analyzed did not survive beyond two weeks after birth. Therefore, we propose that the T cell production they detected initiated in the pre-HSC and/or E14.5 waves, which we demonstrate do not require wild type levels of PU.1.
Although the pre-HSC and E14.5 waves of T cell development emerged in UREΔ/Δ mice, they differed from those arising in UREΔ/+ mice in two ways. First, a reduced number of ETPs and thymocytes were generated in UREΔ/Δ compared to age matched control mice. Although ETPs are not a self-sustaining population, they nevertheless undergo limited replication to expand their pool and allow for the generation of a significant number of T lineage cells. Because fetal UREΔ/Δ ETPs expressed lower levels of Spi1 than normal fetal progenitors, our data indicate that the level of PU.1 in UREΔ/Δ cells was sufficient for ETP emergence and survival but not for their optimal self-renewal and/or proliferation. These results are in agreement with studies showing that PU.1 regulates the length of cell cycle in progenitors (41) and that inhibition of PU.1 function results in a decrease in ETP proliferation accompanied by accelerated differentiation (18). This latter result is particularly relevant to the observation that increased proportions of DP and CD8 cells were present in fetal UREΔ/Δ thymus.
Second, we observed that CD135 expressing ETPs did not emerge in UREΔ/Δ mice. Whether this reflects the absence of a distinct ETP subset or simply lack of expression of this determinant by a homogeneous population of progenitors cannot be determined from our data. However, T cell progenitors from the UREΔ/Δ fetus expressed CD127. This result is consistent with a report form Masuda et al. who reported that fetal T cell progenitors expressed this cytokine receptor along with the paired immunoglobulin-like receptor (PIR) (42). Many of these PIR+ CD127+ cells could be found in the fetal circulation at E11-14, suggesting that they could include cells generated in the pre-HSC wave. PU.1 is known to regulate CD127 expression (43), but this may not be the case in fetal T cell progenitors. In this regard, the Ets family transcription factor GABP has been shown to regulate CD127 expression through binding to the GGAA motif located 5′ of the Il7r translation start codon (44). The observation that Gabpa deficient fetal thymocytes do not express CD127 supports the conclusion that GABP rather than PU.1 regulates Il7r expression in fetal T cell progenitors.
In addition to revealing the existence of differentially regulated waves of T cell development, our data show that differences in expression of Spi1 between fetal and adult ETPs are a feature of normal T cell development. This latter finding raised the question of whether expression of the network of transcription factors regulated by PU.1 would also differ between fetal and adult T cell progenitors. This was a logical assumption in view of studies showing that experimentally antagonizing PU.1 expression and function affects the expression patterns of these genes in T cell progenitors (18). Our analyses showed that this was the case, and that fetal progenitors expressed higher levels of PU.1 regulated genes that promoted T cell differentiation compared to adult ETPs.
In summary, our results indicate that the dosage of pioneer transcription factors such as PU.1 and that of additional regulators of T cell differentiation distinguishes the fetal versus adult T cell developmental program and is likely to be a major determinant of the functional differences between pre- and post-natal T cell progenitors. Our results also provide a framework for understanding observations showing that mice lacking expression of specific transcription factors exhibit differential effects on fetal versus adult T cell development (15, 16).
Supplementary Material
Acknowledgments
This work was supported by grant AI021256 from the National Institutes of Health. The flow cytometry core in the Jonsson Comprehensive Cancer Center is supported by NIH grants CA16042 and AI28697.
Abbreviations
- B6
C57BL/6
- BM
bone marrow
- DN
CD4− CD8− double negative thymocytes
- DP
CD4+ CD8+ double positive thymocytes
- E
embryonic day
- ETP
Early T Lineage Progenitors
- FL
fetal liver
- HSC
Hematopoietic Stem Cell
- MPP
multipotent progenitor
- OP9-DL1
OP9-Delta1 stromal cells
- URE
upstream regulatory element
- YS
Yolk sac
References
- 1.Rothenberg E, Moore J, Yui M. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhandoola A, von Boehmer H, Petrie H, Zuniga-Flucker J. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Petrie H, Zuniga-Pflucker J. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 4.Allman D, Sambandam A, Kim S, Miller J, Pagan A, Well D, Meraz A, Bhandoola R. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 5.Chien Y, Meyer C, Bonneville M. gd T cells: first line of defense and beyond. Annu Rev Immunol. 2014;32:121–155. doi: 10.1146/annurev-immunol-032713-120216. [DOI] [PubMed] [Google Scholar]
- 6.Luc S, Luis T, Boukarabila H, Macaulay I, Buza-Vidas N, Bouriez-Jones T, Lutteropp M, Woll P, Loughran S, Mead A, Hultquist A, Brown J, Mizukami T, Matsuoka S, Ferry H, Anderson K, Duarte S, Atkinson D, Soneji S, Domanski A, Farley A, Sanjuan-Pla A, Carella C, Patient R, de Bruijn M, Enver T, Nerlov C, Blackburn C, Godin I, Jacobsen S. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshimoto M, Porayette P, Glosson N, Conway S, Carlesso N, Cardoso A, Kaplan M, Yoder M. Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood. 2012;119:5706–5714. doi: 10.1182/blood-2011-12-397489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böiers C, Carrelha J, Lutteropp M, Luc S, Green J, Azzoni E, Woll P, Mead A, Hultquist A, Swiers G, Perdiguero E, MI, Melchiori L, Luis TKS, Bouriez-Jones T, Deng Q, Pontén A, Atkinson D, Jensen C, Sitnicka E, Geissmann F, Godin I, Sandberg R, de Bruijn M, Jacobsen S. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell. 2013;13:535–548. doi: 10.1016/j.stem.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Ramond C, Berthault C, Burlen-Defranoux O, de Sousa A, Guy-Grand D, Vieira P, Pereira P, Cumano A. Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat Immunol. 2013;15:27–35. doi: 10.1038/ni.2782. [DOI] [PubMed] [Google Scholar]
- 10.Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, Chien Y, Weissman I. A developmental switch in thymic lymphocyte maturaion potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–874. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 11.Havran W, Allison J. Origin of Thy1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature. 1990;344:68–70. doi: 10.1038/344068a0. [DOI] [PubMed] [Google Scholar]
- 12.Crompton T, Outram S, Buckland J, Owen M. Distinct roles of the interleukin-7 receptor alpha chain in fetal and adult thymocyte development revealed by analysis of interleukin-7 receptor alpha -deficient mice. Eur J Immunol. 1998;28:1859–1866. doi: 10.1002/(SICI)1521-4141(199806)28:06<1859::AID-IMMU1859>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 13.Hosokawa H, Rothenberg E. Cytokines, transcription factors, and the initiation of T-cell development. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe Y, Aiba Y, Katsura Y. T cell progenitors in the murine fetal liver: Differences from those in the adult bone marrow. Cell Immunol. 1997;177:18–25. doi: 10.1006/cimm.1997.1094. [DOI] [PubMed] [Google Scholar]
- 15.Schilham M, Wilson A, Moere R, Benaissa-Trouw B, Cumano A, Clevers H. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161:3984–3991. [PubMed] [Google Scholar]
- 16.Wang JH, Nichogiannopouloua A, Wub L, Suna L, Sharpe A, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 17.Zaret K, Carroll J. Pioneer transcription factors: establishing competence for gene expression. Genes Develop. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Champhekar A, Damle S, Freedman G, Carotta S, Nutt S, Rothenberg E. Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Develop. 2015;29:832–848. doi: 10.1101/gad.259879.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spain L, Guerriero A, Kunjibettu S, Scott E. T cell development in PU.1-deficient mice. J Immunol. 1999;163:2681–2687. [PubMed] [Google Scholar]
- 20.Rosenbauer F, Owens B, Yu L, Tumang J, Steidl U, Kutok J, Clayton L, Wagner K, Scheller M, Iwasaki H, Liu C, Hackanson B, Akashi K, Leutz A, Rothstein T, Plass C, Tenen D. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 21.Scott E, Simon M, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 22.McKercher S, Torbett B, Anderson K, Henkel G, Vestal D, Baribault H, Klemsz M, Feeney A, Wu G, Paige C, Maki R. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 23.Belyaev N, Biro J, Athanasakis D, Fernandez-Reyes D, Potocnik A. Global transcriptional analysis of primitive thymocytes reveals accelerated dynamics of T cell specification in fetal stages. Immunogenetics. 2012;64:591–604. doi: 10.1007/s00251-012-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herzenberg LA, Herzenberg L. Toward a layered immune system. Cell. 1989;59:953–954. doi: 10.1016/0092-8674(89)90748-4. [DOI] [PubMed] [Google Scholar]
- 25.Min H, Montecino-Rodriguez E, Dorshkind K. Effects of aging on the common lymphoid progenitor to pro-B cell transition. J Immunol. 2006;176:1007–1012. doi: 10.4049/jimmunol.176.2.1007. [DOI] [PubMed] [Google Scholar]
- 26.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 27.Ramsdell F, Zúñiga-Pflücker J, Takahama Y. Current Protocols in Immunology. John Wiley & Sons, Inc; 2001. In Vitro Systems for the Study of T Cell Development: Fetal Thymus Organ Culture and OP9-DL1 Cell Coculture. [DOI] [PubMed] [Google Scholar]
- 28.Manning J, Mitchell B, Appadurai D, Shakya A, Pierce L, Wang H, Nganga V, Swanson P, May J, Tantin D, Spangrude G. Vitamin C promotes maturation of T-cells. Antioxid Redox Signal. 2013;19:2054–2067. doi: 10.1089/ars.2012.4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen J, Ritter M. Tissue interaction in the development of thymus lymphocytes. J exp Med. 1969;129:431–442. doi: 10.1084/jem.129.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montecino-Rodriquez E, Fice M, Casero D, Barber C, Berent-Maoz B, Dorshkind K. Distinct genetic programs orchestrate the emergence of distinct waves of B1 and B-2 progenitor development. Immunity. 2016;45:527–533. doi: 10.1016/j.immuni.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pernit C, Vasseur F. Cell proliferation and differentiation in the fetal and early postnatal mouse thymus. J Immunol. 1989;142:3369–3377. [PubMed] [Google Scholar]
- 32.Staber P, Zhan G, Ye M, Welne R, Nombela-Arrieta C, Bach C, Kerenyi M, Bartholdy B, Zhang H, Alberich-Jordà M, Lee S, Yang H, Ng F, Zhang J, Leddin M, Silberstein L, Hoefler G, Orkin S, Göttgens B, Rosenbauer F, Huang G, Tenen D. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell. 2013;49:934–946. doi: 10.1016/j.molcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zúñiga-Pflücker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Min H, Montecino-Rodriguez E, Dorshkind K. Reduction in the developmental potential of intrathymic T cell progenitors with age. The Journal of Immunology. 2004;173:245. doi: 10.4049/jimmunol.173.1.245. [DOI] [PubMed] [Google Scholar]
- 35.Zarnegar MA, Chen J, Rothenberg EV. Cell-Type-Specific Activation and Repression of PU.1 by a Complex of Discrete, Functionally Specialized cis-Regulatory Elements. Molecular and Cellular Biology. 2010;30:4922–4939. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambandam A, Maillard I, Zediak V, Xu L, Gerstein R, Aster J, Pear W, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 37.Carotta S, Dakic A, D’Amico A, Pang S, Greig K, Nutt S, Wu L. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Mortazavi A, Williams B, Wold B, Rothenberg E. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jotereau F, Le Douarin N. Demonstration of a cyclic renewal of the lymphocyte precursor cells in the embryonic chick thymus. J Immunol. 1982;129:1869–1877. [PubMed] [Google Scholar]
- 40.Coltey M, Jotereau F, Le Douarin N. Evidence for a cyclic renewal of lymphocye precursor cells in the embryonic chick thymuis. Cell Differ. 1987;22:71–82. doi: 10.1016/0045-6039(87)90414-3. [DOI] [PubMed] [Google Scholar]
- 41.Kueh H, Champhekar A, Nutt S, Elowitz M, Rothenberg E. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 2013;341:670–673. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuda K, Kubagawa H, Ikawa T, Chen CC, Kakugawa K, Hattori M, Kageyama R, Cooper MD, Minato N, Katsura Y, Kawamoto H. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. The EMBO Journal. 2005;24:4052. doi: 10.1038/sj.emboj.7600878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeKoter RP, Schweitzer BL, Kamath MB, Jones D, Tagoh H, Bonifer C, Hildeman DA, Huang KJ. Regulation of the Interleukin-7 Receptor α Promoter by the Ets Transcription Factors PU.1 and GA-binding Protein in Developing B Cells. Journal of Biological Chemistry. 2007;282:14194–14204. doi: 10.1074/jbc.M700377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue HH, Bollenbacher J, Rovella V, Tripuraneni R, Du YB, Liu CY, Williams A, McCoy JP, Leonard WJ. GA binding protein regulates interleukin 7 receptor α-chain gene expression in T cells. Nature Immunology. 2004;5:1036. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.