Abstract
The T-box transcription factors T-bet and Eomesodermin (Eomes) instruct discrete stages in natural killer (NK) cell development. However, their role in the immune response of mature NK cells against pathogens remains unexplored. We used an inducible deletion system to elucidate the cell-intrinsic role of T-bet and Eomes in mature NK cells during the course of mouse cytomegalovirus (MCMV) infection. We show both T-bet and Eomes to be necessary for the expansion of virus-specific NK cells, with T-bet upregulation induced by IL-12 signaling and STAT4 binding to a conserved enhancer region upstream of the Tbx21 loci. Interestingly, our data suggests maintenance of virus-specific memory NK cell numbers and phenotype was dependent on T-bet, but not Eomes. These findings uncover a non-redundant and stage-specific influence of T-box transcription factors in the antiviral NK cell response.
Introduction
Although natural killer (NK) cells are innate lymphocytes, they are now appreciated to possess developmental and functional traits in common with T and B cells of adaptive immunity (1). In particular, a subset of NK cells expressing the activating Ly49H receptor, which can bind the MCMV-encoded glycoprotein m157, will undergo a clonal-like proliferation and generate long-lived memory cells (2) similar to antigen-specific CD8+ T cells encountering pathogen-derived peptides presented on MHC class I. Specific pro-inflammatory cytokines and transcription factors have been shown to promote adaptive NK cell responses in settings of infection and cancer (3). Specific defects in the clonal expansion of Ly49H+ NK cells have been shown to cripple host control of MCMV (3). Despite recent advances in defining the rapid clonal expansion of effector NK cells, the mechanisms governing the formation and maintenance of memory NK cells during viral infection remain to be elucidated.
T-box family transcription factors T-bet and Eomes have wide-ranging effects that direct lymphocyte immunity. Although recent studies have documented the importance of T-bet and Eomes in NK cell development and function (4–8), their influence on pathogen-specific NK cell responses is unknown. Using an inducible deletion system where T-bet and Eomes can be individually deleted in mature NK cells, we have discovered a stage-specific and non-redundant role for T-box transcription factors during NK cell homeostasis, antiviral response, and generation of long-lived memory.
Materials and Methods
Mice, tamoxifen treatment, and MCMV infection
All of the mice used in this study were bred and maintained at MSKCC in accordance with IACUC guidelines. Mixed bone marrow chimeric mice were generated and adoptive transfer studies were performed as previously described (2). Mice were infected by intraperitoneal (IP) injections of MCMV (Smith strain) with 7.5 × 103 plaque-forming units. Mice were administered 8 mg tamoxifen dissolved in 200 μL olive oil by oral gavage days 0, 1, and 3. Control mice received 200 μL olive oil.
Flow cytometry and cell sorting
Single cell suspensions were prepared from indicated organs and Fc receptors were blocked with 2.4G2 mAb before staining with the indicated surface or intracellular antibodies (BD, BioLegend, or eBioscience). Flow cytometry was performed on an LSR II (BD). Cell sorting was performed on an Aria II cytometer (BD). All data were analyzed with FlowJo software (TreeStar). NK cell enrichment and adoptive transfers were performed as previously described (9).
ChIP and qRT-PCR
Chromatin immunoprecipitation (ChIP) and quantitative reverse-transcription (qRT)-PCR were performed as described previously (10). The following qRT-PCR primers were used for ChIP studies: CNS1 pTbx21, For: 5′-CTAAGCAGGCACTCCATCAGTTG-3′, Rev: 5′-GTCCTTCCTCCGCTGTTCTATTC-3′; CNS2 pTbx21, For: 5′-TAGCGGAAAGCGAGATGGTG-3′, Rev: 5′-AGTGAAGGAGTTCTGTGGTTCTGG -3′; CNS3 pTbx21, For: 5′-GAGCCGACATACTGACATTCTGC-3′, Rev: 5′-CATTCTCCTCTCCCACCATCTTG-3′; Ifng promoter, For: 5′-GCTCTGTGGATGAGAAAT-3′, Rev: 5′-GCTCTGTGGATGAGAAAT-3′; Gene desert 50 kB upstream of Foxp3, For: 5′-TAGCCAGAAGCTGGAAAGAAGCCA-3′, Rev: 5′-TGATACCCTCCAGGTCCAACCATT-3′; Utf1, For: 5′-AGTCGTTGAATACCGCGTTGCTG-3′, Rev: 5′-CTGTTGAGATGTCGCCCAAGTGC-3′; Zpf42, For: 5′-AGAGGGCGGTGTGTACTGTGGTG-3′, Rev: 5′-CTTCTTCTTGCACCCGGCTTGAG-3′.
ChIP-Seq and RNA-seq and sequencing analysis
ChIP-seq, RNA-seq, and analysis of sequencing data were all performed, as previously described (11). The GEO accession numbers for the ChIP-seq and RNA-seq data reported in this paper are GSE106137 and GSE106138, can be found in the GEO database (http://www.ncbi.nlm.nih.gov/geo/).
Statistical methods
All plots depict mean ± s.e.m, except RNA-seq counts, which show mean ± s.d. Two-tailed unpaired Student’s t-test was used to derive statistical differences, unless otherwise indicated. A p value > 0.05 was considered significant. Plots and statistical analyses were produced in GraphPad Prism or R (v 3.3.3).
Results and discussion
T-box transcription factors are dispensable for NK cell homeostasis
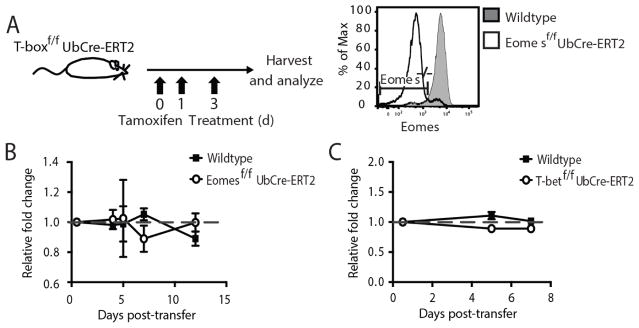
To investigate the role of individual T-box transcription factors on the homeostasis and antiviral response of mature NK cells without impacting their development, we generated mice containing a floxed Eomes or T-bet (Tbx21) gene and a Cre recombinase-human estrogen receptor ligand-binding domain (ERT2) fusion protein expressed downstream of the ubiquitin promoter (Ub) (Eomesf/f x UbCre-ERT2 or Tbx21f/f x UbCre-ERT2 mice, respectively). Treatment of Eomesf/f x UbCre-ERT2 or Tbx21f/f x UbCre-ERT2 mice or cells with tamoxifen causes the specific excision of the floxed T-box transcription factor gene and loss of protein in cells of interest (Fig. 1A and data not shown). Cells where the genes encoding Eomes or Tbx21 are ablated following tamoxifen treatment will be referred to as Eomes−/− or T-bet−/−, respectively.
Figure 1. Eomes and T-bet are dispensable for NK cell homeostasis in both normal and lymphopenic mice.
(A) Schematic of tamoxifen treatment. Experimental mice were given a regimen of tamoxifen on days 0, 1, and 3, and control mice received oil alone. Panel shows expression of Eomes in NK cells three weeks after Eomesf/f x UbCre-ERT2 mice were given tamoxifen. Gates show Eomes−/− population in mice receiving tamoxifen. WT (CD45.1) and (B) Eomesf/f x UbCre-ERT2 (CD45.2) or (C) Tbx21f/f x UbCre-ERT2 (CD45.2) NK cells were co-transferred into WT mice (CD45.1×2) and treated with tamoxifen or oil at day 0 PT. The relative fold change of the floxed and WT NK cell ratio relative to their starting ratio is shown in panels B and C. Data are mean ± SEM representative of two independent experiments with at least n=3 biological replicates per condition. * p < 0.05 and ns, not significant, paired Student t-test.
We first tested whether Eomes expression was required for the homeostasis of mature NK cells. We isolated purified NK cells (Supp. Fig 1A–B) from the spleen of Eomesf/f x UbCre-ERT2 mice (CD45.2) and co-transferred them with an equal number of WT NK cells (CD45.1) into WT recipients (CD45.1 0× CD45.2), which were immediately treated with a tamoxifen regimen to efficiently induce Eomes deletion. Greater than one week following tamoxifen treatment, we found equal numbers of Eomes−/− and WT NK cells (Fig. 1B), suggesting Eomes is not necessary for the maintenance of total NK cell numbers under steady state conditions. However, Eomes−/− NK cells exhibited a slight increase in the percentage TRAIL+ cells and a decrease in the percentage Ly49H+ cells in both the spleen and liver (Supp. Fig. 1C). Using CD27 and CD11b as markers of NK cell maturation (12), no significant changes were observed between Eomes−/− and WT splenic NK cells (Supp. Fig. 1D).
When Tbx21f/f x UbCre-ERT2 NK cells were transferred with WT NK cells and recipients treated with tamoxifen, equal numbers of co-transferred populations were recovered one week later as well (Fig. 1C), suggesting T-bet, like Eomes, is also not essential for the maintenance of mature NK cell numbers at steady state, and that perhaps T-box transcription factors compensate for the loss of one another during basal homeostasis. In contrast to Eomes deletion, NK cells that were deleted of T-bet exhibited a significant decrease in the percentage of TRAIL+ cells, and increase in the percentage of Ly49H+ NK cells in the spleen and liver (Supp. Fig. 1E). A significant increase in the percentage of CD27+ CD11b+ cells was also observed (Supp. Fig. 1F). In all experiments, a control set of mice receiving oil alone showed no numerical or phenotypic differences between floxed and WT cells (data not shown). Similar observations were made when resting splenic NK cells from floxed and WT mice were co-transferred into lymphopenic Rag2−/− x Il2rg−/− mice, followed by tamoxifen treatment (data not shown). Altogether, these data are consistent with the previously ascribed role of Eomes and T-bet in repressing and promoting TRAIL expression, respectively, and in the maintenance of a mature phenotype (5).
Eomes and T-bet are required for the clonal expansion of virus-specific NK cells
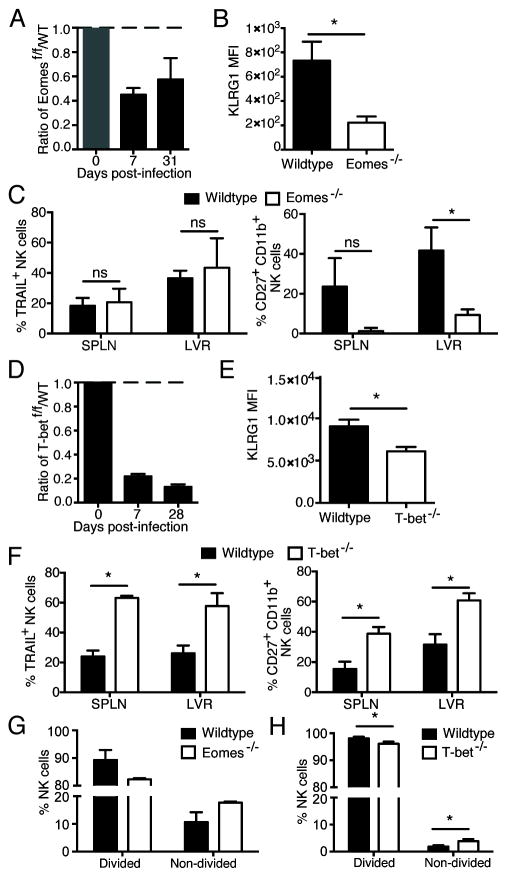
To investigate the effects of T-box transcription factors on the various stages of the NK cell response against viral infection, we first induced deletion of Eomes or T-bet immediately before infection in order to test for their role in clonal expansion of virus-specific NK cells. We isolated splenic Ly49H+ NK cells from Eomesf/f x UbCre-ERT2 or Tbx21f/f x UbCre-ERT2 mice (CD45.2) and co-transferred them with an equal number of WT Ly49H+ NK cells (CD45.1) into Ly49h−/− hosts that were treated with tamoxifen (or oil as a control). Host mice were then infected with MCMV four days after adoptive transfer. We found Eomes to be critical for the expansion of Ly49H+ NK cells during MCMV infection, as Eomes−/− NK cells exhibited a marked impairment compared to WT NK cells (Fig. 2A and Supp. Fig 2). No significant difference in expansion between WT and Eomes−/− NK cells was found in the control group receiving oil alone (Supp. Fig 2). Activated NK cells deficient in Eomes showed a significant decrease in KLRG1 expression (Fig. 2B) at day 7 post-infection (PI), but similar levels of FLICA staining (data not shown), which detects cells undergoing apoptosis. Although deletion of Eomes did not affect TRAIL expression in splenic or liver NK cells at day 7 PI, a decrease in the CD27+ CD11b+ NK cell population was observed (Fig. 2C).
Figure 2. T-box transcription factors are necessary for the expansion of antigen-specific NK cells.
WT (CD45.1) and Eomesf/f x UbCre-ERT2 (CD45.2) or Tbx21f/f x UbCre-ERT2 (CD45.2) NK cells were co-transferred into Ly49h−/− mice and treated with tamoxifen or oil at day −4 PI. Mice were infected with MCMV at day 0 PI. (A and D) The populations of tamoxifen-treated Eomesf/f x UbCre-ERT2 or Tbx21f/f x UbCre-ERT2 NK cells and their WT counterparts relative to oil-treated populations are shown for each time point. (B and E) MFI of KLRG1 staining at day 7 PI in the spleen is shown. (C and F) Assessment of phenotypic markers at day 7 PI for the spleen and liver are shown. (G and H) Ly49H+ NK cells from WT and Eomesf/f x UbCre-ERT2 or Tbx21f/f x UbCre-ERT2 mice were labeled with CTV, transferred into a Ly49h−/− host which was treated with tamoxifen (or oil) at day −4 PI and infected with MCMV at day 0. Bar graph shows percentages of divided and non-divided NK cells at day 4 PI for each group. Data are mean ± SEM representative of two to three independent experiments with at least n=4 biological replicates per condition. * p < 0.05 and ns, not significant, paired Student t-test.
Similar to Eomes deletion, when T-bet was inducibly-ablated following transfer, effector and memory Ly49H+ NK cell numbers were severely diminished compared to WT controls (Fig. 2D and Supp Fig. 2). Unlike T-bet deletion in mature NK cells during basal homeostasis (Figure 1), virus-specific NK cells lacking T-bet exhibited an increase in the percentage of TRAIL+ NK cells and a less mature phenotype (CD27+ CD11b+ KLRG1lo) at day 7 PI (Fig. 2E–F), highlight a role for T-bet in the differentiation and terminal maturation of anti-viral effector NK cells. An increase in CD27 expression may be due a loss in gene repression by T-bet, and not a reversion to a phenotypically distinct immature population (13). A similar phenotype was observed at day 28 PI (data not shown). When deletion of Eomes or T-bet was initiated at day 3 PI instead of at day −4 PI, similar results were observed (data not shown). Altogether, these findings uncover a novel and stage-specific role for T-bet in differentiating NK cells, where T-bet promotes TRAIL expression in resting NK cells but represses its expression in activated NK cells.
To determine whether the decreased expansion of T-bet−/− or Eomes−/− NK cells during viral infection was due to diminished proliferation, we adoptively co-transferred equal numbers of cell trace violet (CTV)-labeled splenic NK cells from WT and Eomesf/f x UbCre-ERT2 or Tbx21f/f x UbCre-ERT2 mice into Ly49h−/− recipients that were immediately treated with tamoxifen (or oil as a control). Compared to their WT counterparts, a greater percentage of NK cells lacking individual T-box transcription factors had not divided in response to MCMV infection (Fig. 2G–H), suggesting that T-bet and Eomes activity in mature NK cells is necessary for the execution of a robust proliferative program. Altogether, these data highlight a non-redundant role for T-box transcription factors during mature NK cell priming for the vigorous proliferation and proper differentiation of virus-specific NK cells.
IL-12 and STAT4 upregulate T-bet in NK cells during MCMV infection
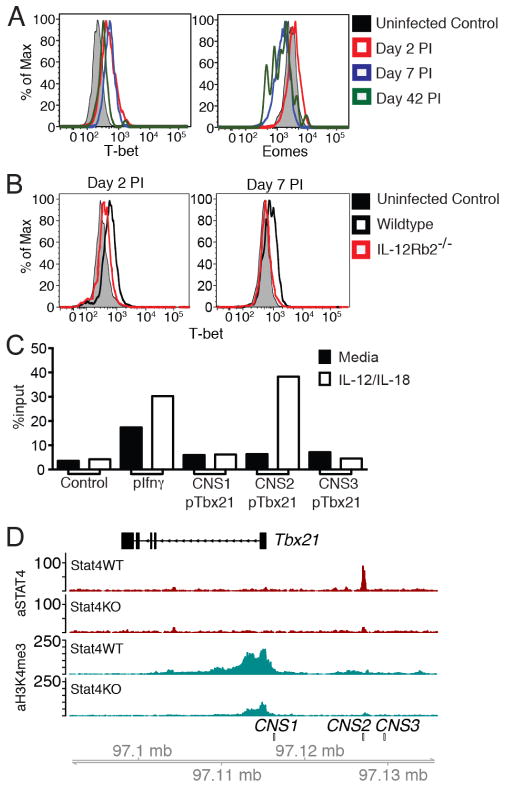
To gain insight into when T-box transcription factors may be exerting influences on the proliferation of virus-specific NK cells, the amount of T-bet and Eomes in Ly49H+ NK cells were measured at different time points during MCMV infection. We found that T-bet expression is upregulated in NK cells at day 2 PI and remains high at effector (day 7 PI) and memory (day 42 PI) time points, whereas Eomes expression is largely unchanged at day 2 PI and downregulated at day 7 and 42 PI (Fig. 3A). The rapid modulation of T-bet levels immediately following MCMV infection suggests a role for pro-inflammatory cytokine signals in its upregulation. Because IL-12 has been described to upregulate T-bet expression in CD8+ T cells (14, 15), we evaluated the expression of T-bet in NK cells from WT:Il12rb2−/− bone marrow chimeric mice infected with MCMV. In contrast to WT NK cells, we found that T-bet levels were not upregulated at day 2 and 7 PI in Il12rb2−/− NK cells (Fig. 3B). Eomes expression in Il12rb2−/− NK cells did not differ from WT NK cells at various time points during MCMV infection (Supp. Fig. 2C).
Figure 3. IL-12 and STAT4 signals upregulate T-bet expression in NK cells during MCMV infection.
(A) WT NK cells were transferred into Ly49h−/− mice and infected with MCMV. Expression of T-bet and Eomes at the indicated time points is shown. (B) Expression of T-bet in WT:Il12rb2−/− bone marrow chimeras is shown for the indicated time points. (C) STAT4 binding at Tbx21, Ifng, and control promoters as assessed through ChIP followed by qPCR in purified WT NK cells stimulated for 18h with IL-12 and IL-18. STAT4 occupancy as percent of input is shown for target (Tbx21) and control DNA (negative control: average of gene desert 50 kb upstream of Foxp3, Utf1, and Zfp42 promoters; positive control: Ifng promoter). Data in A–C are representative of three independent experiments with at least n=3 biological replicates per condition. (D) Zoomed-in histograms of STAT4 ChIP-seq and H3K4me3 ChIP-seq reads mapped to the Tbx21 loci in activated WT or Stat4−/− NK cells. Data are representative of two independent experiments with n=15–20 pooled mice per group per experiment.
Given that IL-12 induces T-bet, we predicted that the signal transducer and activator of transcription 4 (STAT4), which acts downstream of the IL-12 receptor, may directly bind to the gene loci encoding T-bet (Tbx21) in activated NK cells. Analysis of the Tbx21 promoter and enhancer regions revealed several putative STAT4 binding sites (Supp. Fig. 2D), one of which (site 2) is embedded within a conserved noncoding site (CNS). We performed STAT4 chromatin immunoprecipitation (ChIP) on resting and IL-12 + IL-18-activated NK cells, and found substantial enrichment of STAT4 at site 2 (Fig. 3C–D), an enhancer ~11 Kb upstream of the Tbx21 transcriptional start site. In addition, we demonstrate an increase in the ‘permissive’ histone modification H3K4me3 at the promoter region in activated WT but not Stat4−/− NK cells, which correlates with an increase in Tbx21 transcript levels in the WT but not Stat4−/− NK cells at d2 PI (Fig. 3D and Supp. Fig. 2E). Thus, together these data support the direct regulation of T-bet expression in NK cells by IL-12 signaling and STAT4 activity during MCMV infection.
T-bet, but not Eomes, is required for the maintenance of virus-specific memory NK cells
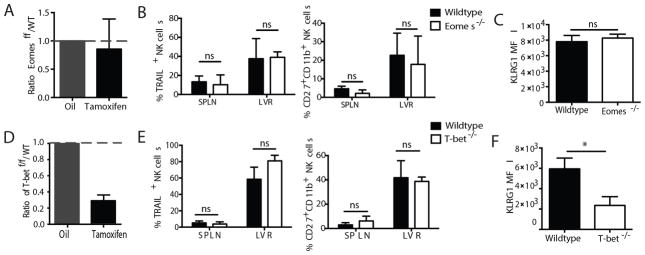
Lastly, we investigated the role of Eomes and T-bet on the maintenance of memory NK cells following MCMV infection. We isolated NK cells from the spleen of Eomesf/f x UbCre-ERT2 or Tbx21f/f x UbCre-ERT2 mice (CD45.2) and co-transferred them with equal numbers of WT NK cells (CD45.1) into Ly49h−/− recipients that were immediately infected with MCMV. At day 19 PI, when memory NK cell numbers were comparable between the two groups, we administered tamoxifen to induce gene deletion in the Eomesf/f x UbCre-ERT2 NK cells. We found that the ratio of WT to Eomes-deleted NK cells did not change for several weeks after tamoxifen administration (Fig. 4A). Furthermore, no changes were observed in the frequency of TRAIL-expressing or CD27+ CD11b+ NK cells (Fig. 4B), and unlike effector NK cells, memory NK cells did not suffer a decrease in KLRG1 expression (Fig. 4C). This lack of a memory maintenance defect in Eomes−/− NK cells is consistent with the observation that Eomes expression was found to be downregulated in virus-specific memory NK cells (Fig. 3A),
Figure 4. T-bet, but not Eomes, is required for the maintenance of memory NK cells.
WT (CD45.1) and Eomesf/f x UbCre-ERT2 (CD45.2, A–C) or Tbx21f/f x UbCre-ERT2 (CD45.2, D–F) NK cells were co-transferred into Ly49h−/− mice, which were infected with MCMV and treated with tamoxifen (or oil alone) at 3 weeks PI. (A and D) The relative fold change of floxed and WT populations two weeks following tamoxifen treatment relative to their starting ratio is shown. (B and E) Assessment of phenotypic markers for the spleen and liver are shown. (C and F) MFI of KLRG1 staining in the spleen is shown. Data are mean ± SEM representative of three independent experiments with at least n=3 biological replicates per condition. * p < 0.05 and ns, not significant, paired Student t-test.
In contrast to Eomes ablation, induced deletion of T-bet in memory NK cells led to a significant 3 to 4-fold decrease in the ratio of T-bet−/− to WT cells in mice receiving tamoxifen compared with controls (Fig. 4D), suggesting a role for T-bet in the persistence of virus-specific memory NK cells. Although only slight changes were observed in the frequency of TRAIL-expressing or CD27+ CD11b+ NK cells (Fig. 4E and Supp. Fig. 2F–G), a significant decrease in the expression of KLRG1 was observed (Fig. 4F and Supp. Fig. 2G). Previous reports have suggested T-bet to be an important factor in the terminal maturation of NK cells (4), and our data suggests that T-bet is continuously required for the maintenance of a mature NK cell phenotype. Altogether, these data uncover a unique role for T-bet, but not Eomes, in the maintenance of number and phenotype of virus-specific memory NK cells, further highlighting the divergent function of these T-box family transcription factors at different stages of the antiviral response.
Although Eomes and T-bet may play a complementary role in the induction of a proliferative program in NK cells responding to viral infection, our data suggests that only T-bet acts to maintain antigen-experienced NK cells in a mature state. We propose that in addition to TRAIL marking immature NK cells (5, 16) and possibly a distinct NK cell lineage in the liver (6), it may also serve to track NK cell differentiation, as loss of TRAIL was accompanied with increased KLRG1 expression. Interestingly T-bet, but not Eomes, was found to be critical in the maintenance of memory NK cell numbers and phenotype. In contrast to the continuous importance of individual T-box transcription factors throughout the lifespan of NK cells, Nfil3 is a transcription factor required for NK cell development, but dispensable in mature NK cell homeostasis or response to infection (17). How T-bet and Eomes are driving the clonal proliferation of NK cells via a non-redundant mechanism remains to be elucidated. The underlying mechanisms by which T-bet promotes survival of memory NK cells, possibly by regulating IL-15 responsiveness (7), also require further investigation. Nonetheless, these findings highlighting a stage-specific influence of individual T-box transcription factors in NK cells responding to MCMV infection.
Our current findings reveal distinct molecular events that influence NK cell responses against viral infection, including the maintenance of a memory NK cell pool, which may inform clinical approaches that couple NK cell-mediated immunity to the treatment of human diseases.
Supplementary Material
Acknowledgments
S.M. was supported by MSTP (T32GM07739), NIH (T32AI007621), and the Cancer Research Institute (CRI) grants. C.M.L. was supported by a T32 award (CA009149) from the NIH. O.P. was supported by NIH grants T32GM007367 and F30AI113963. S.L.R. was supported by NIH grants R01AI061699 and R01AI113365. J.C.S. was supported by the Searle Scholars Program, the CRI, the Burroughs Wellcome Fund, the American Cancer Society, and NIH grants R01AI100874, R01AI130043, and P30CA008748.
Footnotes
The authors declare no financial conflicts of interest.
References
- 1.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol. 2011 doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol. 2016;16:112–123. doi: 10.1038/nri.2015.9. [DOI] [PubMed] [Google Scholar]
- 4.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 5.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, Marvel J, Yoh K, Takahashi S, Prinz I, de Bernard S, Buffat L, Walzer T. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 8.Walzer T, Chiossone L, Chaix J, Calver A, Carozzo C, Garrigue-Antar L, Jacques Y, Baratin M, Tomasello E, Vivier E. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol. 2007;8:1337–1344. doi: 10.1038/ni1523. [DOI] [PubMed] [Google Scholar]
- 9.Sun JC, Madera S, Bezman NA, Beilke JN, Kaplan MH, Lanier LL. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med. 2012;209:947–954. doi: 10.1084/jem.20111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaulieu AM, Zawislak CL, Nakayama T, Sun JC. The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat Immunol. 2014;15:546–553. doi: 10.1038/ni.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rapp M, Lau CM, Adams NM, Weizman OE, O’Sullivan TE, Geary CD, Sun JC. Core-binding factor beta and Runx transcription factors promote adaptive natural killer cell responses. Science immunology. 2017:2. doi: 10.1126/sciimmunol.aan3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176:1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 13.van Helden MJ, Goossens S, Daussy C, Mathieu AL, Faure F, Marcais A, Vandamme N, Farla N, Mayol K, Viel S, Degouve S, Debien E, Seuntjens E, Conidi A, Chaix J, Mangeot P, de Bernard S, Buffat L, Haigh JJ, Huylebroeck D, Lambrecht BN, Berx G, Walzer T. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med. 2015;212:2015–2025. doi: 10.1084/jem.20150809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood. 2007;110:2494–2500. doi: 10.1182/blood-2006-11-058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. Journal of immunology. 2006;177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- 16.Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, Yagita H, Kinoshita K, Okumura K, Smyth MJ. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood. 2005;105:2082–2089. doi: 10.1182/blood-2004-08-3262. [DOI] [PubMed] [Google Scholar]
- 17.Firth MA, Madera S, Beaulieu AM, Gasteiger G, Castillo EF, Schluns KS, Kubo M, Rothman PB, Vivier E, Sun JC. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J Exp Med. 2013;210:2981–2990. doi: 10.1084/jem.20130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.