Abstract
Peptides folded through interwoven disulfides display extreme biochemical properties and unique medicinal potential. Their exploitation was hampered by the limited amounts isolatable from natural sources and the expense of chemical synthesis. We developed reliable biological methods for high-throughput expression screening and large-scale production of these peptides. 46 were successfully produced in multimilligram quantities, and over 600 more were deemed expressible by stringent screening criteria. Many showed extreme resistance to temperature, proteolysis, and/or reduction, and all displayed inhibitory activity against at least one of 20 ion channels tested, confirming biological functionality. Crystal structures of 12 were determined, confirming proper cystine topology, and the utility of crystallography for studying these molecules, but highlighted the need for rational classification. Previous attempts at categorization have focused on limited subsets siloed around distinct motifs. Stepping back, we present a global definition, classification, and analysis of over 700 structures of cystine-dense peptides, unifying these molecules.
INTRODUCTION
Proteins are differentiated from peptides by size, with peptides generally fewer than ~50 residues long. Peptides, lacking the ability to form sufficient cooperative interactions, usually do not adopt stable, defined structures, achieved in proteins through well-packed hydrophobic cores. Exceptions include peptides that alternately organize around cores of tightly-packed disulfides (Fig. 1a), often conferring extreme thermal, chemical, and proteolytic stability1–4. Archetypes of such peptides, with cores of at least three cystines, include “inhibitor cystine knot peptides”, or knottins, and the closely-related “cyclic cystine knot peptides”, or cyclotides5,6. Examples include venom toxins from cone snails, spiders, and scorpions; plant protease inhibitors; and antimicrobial defensins. Knottins and cyclotides are topologically pseudoknotted, with one cystine (the “knotting” cystine) crossing through the macrocycle formed by the other two cystines (the “bracketing” cystines) and the interconnecting backbone, often with additional, accessory cystines (Fig. 1b). Proteins can also be pseudoknotted through intrachain cystines, eg., “growth factor cystine knots” (GFCKs; Fig. 1c)7,8. However, GFCK intrachain cystines do not dominate the fold of the protein, which includes a conventional hydrophobic core, distinct from knottins and cyclotides.
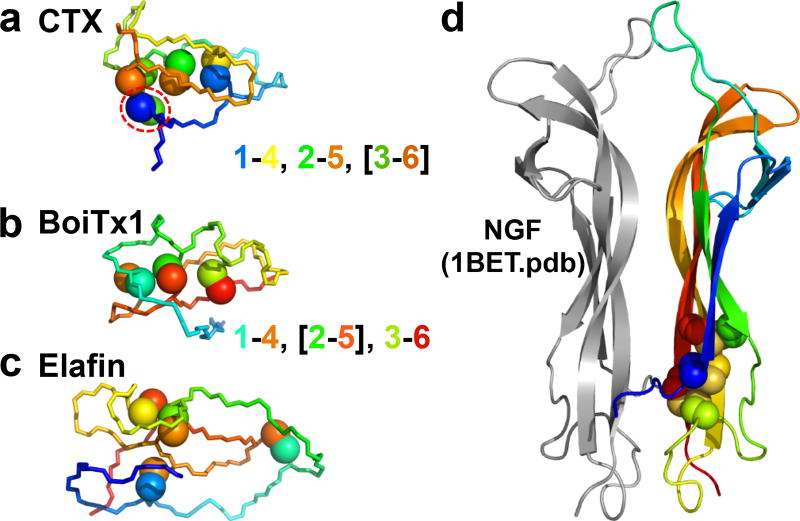
Figure 1. CDPs versus GFCKs.
a, The crystal structures of the archetypical scorpion venom knottin, Chlorotoxin (CTX), b, a distinct knotted peptide, alpha-KTx 3.10 (BoiTx1), and c, a non-knotted, cystine-containing peptide, Elafin, all determined as part of this work, are shown in a backbone representation, with cysteine side-chain sulfur atoms shown as spheres, demonstrating the degree that cystines dominate the core of these structured peptides. The backbone and cysteine side-chain atoms are colored from blue to red, N- to C-terminus, highlighting the cystine connectivity and pseudoknot topology characteristic of knottins and related peptides. The topology is represented as u-v, [w-x], y-z, where w-x is the knotting cystine, and u-v and y-z are the bracketing cystines. CTX, like many knotted CDPs, has an accessory cystine, circled in red, in addition to the three core cystines defining the pseudoknot element. d, A typical dimeric GFCK, human Nerve Growth Factor (NGF), shown in a cartoon representation, with one monomer colored gray, and the other monomer colored as in a.
Natural knottins and cyclotides have demonstrated many properties beyond stability useful for clinical applications, including the potential for oral delivery, cell penetration, and tumor homing3,4,9. Inherent pharmacological properties can also include analgesic, antihelminthic, antimicrobial, antitumor, insecticidal, or ion channel modulatory activities5. These peptides are intermediate in size between protein biologics and conventional small-molecule drugs, potentially small enough to penetrate a variety of tissues and solid tumors, but large enough to enable protein-like ligand specificity and affinity. Approved drugs exploiting these properties include ziconotide (Prialt), based on a cone snail venom knottin, linaclotide (Linzess), based on E. coli heat-stable enterotoxin (STa), and plecanatide (Trulance), based on human uroguanylin10.
Overall, the minimal common elements defining this class of molecules are short sequences, constituting independent folding domains, with a high density of at least three cystines. We refer to this categorization as “cystine-dense peptides” (CDPs), drawing a distinction with larger proteins with cystine-knotted elements, like GFCKs. Inspired by their unique folds, the potential of these peptides for clinical application, and the success of ziconotide, linaclotide, plecanatide, and tozuleristide (Tumor Paint)11, a tumor-homing, fluorophore conjugate of the scorpion knottin Chlorotoxin (CTX), we sought to more fully explore and exploit these molecules. However, while these peptides are generally amenable to chemical synthesis, and some have been expressed recombinantly, a major impediment to exploiting this unique pharmaco-molecular space has been the lack of a reliable, high-throughput expression platform. Here we report biologic production of over 700 CDPs, with exhaustive biochemical characterization of 46, demonstrating unique properties. We have optimized crystallographic methods for elucidating CDP structures. Structural analyses highlighted another impediment to understanding this fold space: the absence of a unified, global scheme for classifying CDPs. As there are many examples of structured, cystine-rich peptides that are not knottins or cyclotides, or even knotted, we propose a structure-based classification scheme unifying and framing an analysis of a complete catalogue of available CDP structures.
RESULTS
CDP sequence-based definition
Inspection of the Protein Data Bank (PDB)12 and homologous sequences from natural sources suggested a CDP-defining motif: (i) six or more cysteine residues in a span from 13 to 81 residues long not recognizable as a cytoplasmic protein or domain, a zinc finger protein, or a GFCK; and (ii) a constrained distribution of cysteines, Cys-X[0-15]-Cys-X[0-15]-Cys-X[0-15]-Cys-X[0-15]-Cys-X[0-15]-Cys (“X”: any amino acid). To confirm formation of cystines, the candidate CDP should be embedded in a sequence with a recognizable leader peptide, eg., using SignalP13, or annotated as a secreted or integral membrane protein, or experimentally shown to contain specific cystines. CDPs may be embedded in larger proteins, or in tandem arrays (some examples have over 4014,15), but should comprise an independent folding unit. Applying these rules to the PDB yielded a continuum of structures, though adding the criterion of a minimal “cysteine density”, with at least 12% cysteine content in the span including the bounding cysteines, satisfactorily separated CDPs from small proteins with emergent hydrophobic cores. This threshold density is approximately 10-fold higher than the average for all proteins16,17. There were 775 experimentally-determined structures in the PDB as of April, 2017 with domains conforming to this motif, excluding wholly synthetic, designed sequences, including 422 knotted CDPs, 203 non-knotted CDPs with three cystines, and 150 non-knotted CDPs with more than three cystines (Supplementary Table 1).
CDP biologic production
To develop a biologic expression platform for CDPs, a target set of 100 was selected (Supplementary Table 2). We concentrated on CDPs similar or identical to previously studied CDPs, largely spider and scorpion venom components, in order to validate success when achieved, but also included some simpler, two-cystine, non-CDP sequences (eg., Hefutoxin), to phase in the magnitude of the challenge. Initial attempts to produce CDPs in bacterial expression systems were abandoned in favor of mammalian secretion pathway-based systems, which incorporate folding chaperones and extensive quality control machinery to dramatically improve success rates. CDPs produced in mammalian cells were also free of contaminating endotoxins, streamlining in vivo applications. CDPs were ultimately most successfully produced using a variation of the Daedalus lentivirus transduction system18, in HEK293 cells, with Siderocalin (Scn) fusion protein partners19 to foster folding and increase yield (Supplementary Figure 1). An improved version of Tobacco Etch Virus (TEV) protease20–23, SuperTEV, was also developed to complement Daedalus expression (Supplementary Figure 2). SuperTEV showed identical activity to TEV, but was more stable, did not require reducing agents for stability, and could be functionally expressed in mammalian cells.
Since isolated CDPs displayed anomalous behavior by reduced/non-reduced comparative polyacrylamide gel electrophoresis (PAGE) and size exclusion chromatography (SEC), reverse phase chromatography (RPC) was used for preparative purification after cleavage, final purity assessment, and biochemical characterization. RPC is extremely sensitive to any chemical heterogeneity in a preparation, and can also detect the presence of chemically identical, but conformationally distinct, isoforms. Expression of a CDP was considered successful only if the final RPC traces showed only a single, dominant peak under both oxidizing and reducing conditions, indicating a single folding state, and the absence of proteolysis or heterogeneity of any kind, including conformational (Fig. 2a, Supplementary Dataset 1). Prior studies of CDP synthesis (eg. 24) yielded preparations that were biologically functional but displayed more complex RPC traces, argued to indicate that multiple conformers were present that do not interchange on the RPC timescale. Distinct, metastable conformers of a CDP may not, for instance, all interact equivalently with a target ion channel, complicating analyses of less-stringently produced peptides. We chose to apply the most rigorous threshold for success, requiring absolute homogeneity. 46 of the 100 targets were successfully produced under these stringent criteria (Supplementary Dataset 1). For example, CDP #11 showed near complete resistance to reduction, but CDP #17 is a doublet under non-reducing conditions, failing our quality-control criteria (Fig. 2a). This system is amenable to both high-throughput, pilot-scale expression screening on automated robotic platforms and affordable preparative-scale production, with final yields of up to 10 mg/L at the 2 L culture scale. The high-throughput platform processed hundreds of CDPs per week, yielding up to 20 µg at the 1 mL culture scale, enough to evaluate proper folding by analytical RPC, applying the same stringent criteria as above. We determined that another 678 CDPs (55%) were expressible out of an additional list of 1,232 targets using the high-throughput approach (Supplementary Table 3). These additional targets more broadly sampled CDPs from plants, arthropods, and other taxa; 872 had less than 75% identity to a CDP in the PDB.
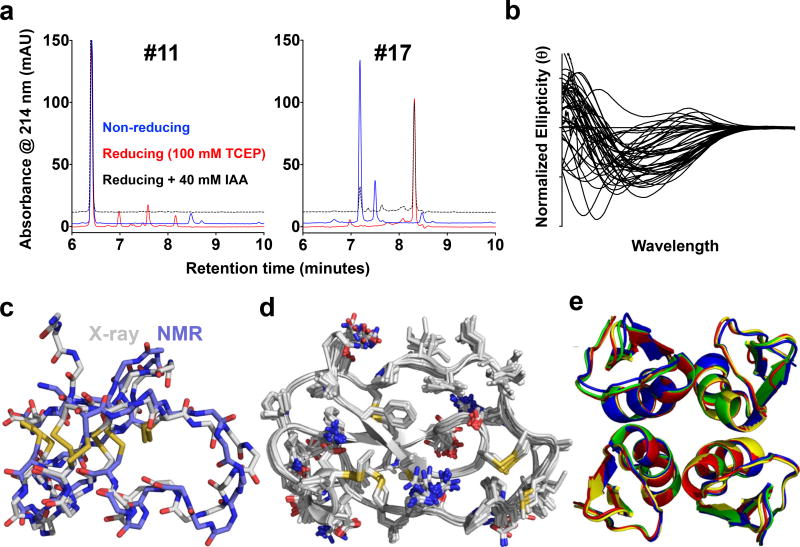
Figure 2. CDP biochemical and crystallographic analyses.
a, Example comparative RPC analyses of two scorpion potassium channel toxins, CDP #11 (left) and #17 (right), produced using the Daedalus-based biologic production platform, under non-reducing conditions (blue), reducing conditions (red; by addition of 100 mM tris(2-carboxyethyl)phosphine (TCEP)), and thiol-blocking treatment after reduction (dashed; by addition of 40 mM iodoacetamide (IAA) after addition of TCEP). b, Normalized CD spectra from all 46 successfully-expressed CDPs showed a wide range of secondary structure compositions. c, A superposition of two structures of CTX (target #28), shown as licorice-stick representations of the peptide backbone plus cysteine side-chains, colored by atom type, determined by NMR (1CHL.pdb33, periwinkle carbons) or by crystallography (reported here, gray carbons). d, A superposition of all 18 independent views of the structure of Elafin (target #4, a human class 1–3, 2–5, 4–6 non-knotted CDP) in the crystal structure reported here, shown as a cartoon ribbon representation of the peptide backbone plus licorice-stick representations of all side-chains, colored by atom type. The structure of Elafin had been previously determined by crystallography (1FLE.pdb34), though only in complex with elastase, and alone by NMR (2REL.pdb35). e, A superposition of all five independent views of the tetramer of the potassium channel toxin gamma-KTx 2.2 hitchin from the venom of the Manchurian scorpion (target #48), from two different crystal forms, shown in a cartoon representation, colored by tetramer.
CDP biochemical characterization
To further explore the previously reported extreme biochemical stability of CDPs4,6,7, we tested the 46 successfully produced CDPs under a battery of conditions that would be expected to denature or degrade conventional globular proteins, including extended incubation under reducing conditions or at high temperatures, and proteolytic digestion (Supplementary Dataset 1). Most of the 46 showed resistance to some combination of these conditions, but a handful showed truly exceptional stability, including two knotted CDPs that were completely resistant to all conditions tested (Supplementary Dataset 1). Circular dichroism (CD) spectra were collected from the 46 successfully produced CDPs to evaluate secondary structure content (Fig. 2b, Supplementary Dataset 1). The range observed included spectra showing very limited secondary structure content, consistent with known homologous structures.
CDP crystallography
The ultimate confirmation of proper cystine formation in a CDP is experimental determination of its three-dimensional structure. Most isolated CDP structures available in the PDB were determined by NMR, partly due to the perception that CDPs are inherently difficult to crystallize. Crystallography has been used previously mostly, but very effectively, to determine structures of complexes between CDPs and binding-partner proteins. However, access to an efficient expression system allowed large-scale production of highly-purified CDPs that can be highly concentrated (≥80 mg/mL), thereby enhancing crystallizability. 12 of the 46 successfully-expressed CDPs were crystallized (26% success rate, one in two crystal forms), and were used to determine structures (Tables 1 and 2, Supplementary Table 4, Supplementary Fig. 3). Initial phases were determined either by molecular replacement (MR) or sulfur single-wavelength anomalous diffraction (sSAD)25, using Cu Kα radiation to maximize the anomalous signal. With optimized procedures, sSAD proved to be an extremely effective approach for CDP structural analysis, providing completely unbiased initial phases, and more direct, detailed information about sulfur substructure.
Table 1.
Data collection and refinement statistics for structures phased by molecular replacement
| Target #4 (6ATU) |
Target #28 (6ATW) |
Target #34 (6AVA) |
Target #48, form #1 (6AUP) |
Target #48, form #2 (6AU7) |
Target #49 (6AVC) |
Target #83 (6AV8) |
|
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P 41 | P 21 21 21 | P 43 21 2 | P 21 21 21 | C 1 2 1 | P 32 2 1 | P 65 |
| Cell dimensions | |||||||
| a, b, c (Å) | 71.3, 71.3, 214.4 | 22.5, 26.3, 48.0 | 41.4, 41.4, 88.1 | 58.9, 80.4, 94.2 | 50.2, 48.1, 50.3 | 43.5, 43.5, 32.1 | 50.64, 50.64, 20.10 |
| α, β, γ, (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 107.0, 90.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.00 – 2.40 (2.49 – 2.40) | 50.00 – 1.53 (1.56 – 1.53) | 50.00 – 2.20 (2.28 – 2.20) | 50.00 – 1.95 (1.98 – 1.95) | 50.00 – 1.90 (1.93 – 1.90) | 50.00 – 1.88 (1.95 – 1.88) | 50.00 – 1.89 (1.92 – 1.89) |
| Rmerge | 0.72 (0.49) | 0.11 (0.13) | 0.16 (0.37) | 0.16 (0.64) | 0.07 (0.23) | 0.10 (0.41) | 0.06 (0.29) |
| Rmeas | 0.08 (0.53) | 0.10 (0.13) | 0.16 (0.37) | 0.16 (0.65) | 0.08 (0.21) | 0.10 (0.42) | 0.06 (0.31) |
| I/σ(I) | 29.91 (5.87) | 40.15 (11.78) | 44.9 (26.7) | 46.8 (8.5) | 13.0 (1.9) | 59.3 (16.2) | 58.11 (4.63) |
| CC1/2 | (0.94) | (0.93) | (0.99) | (0.96) | (0.20) | (0.98) | (0.94) |
| Completeness (%) | 99 (97) | 87 (11) | 100 (100) | 98 (80) | 83 (17) | 85 (38) | 97.5 (80.3) |
| Redundancy | 7.2 (6.8) | 13.1 (2.3) | 58.4 (59.5) | 48.3 (23.7) | 3.2 (1.5) | 57.7 (40.4) | 25.9 (6.2) |
| Refinement | |||||||
| Resolution (Å) | 50.00 – 2.40 (2.39 – 2.45) | 24.02 – 1.53 (1.57 – 1.53) | 37.50 – 2.20 (2.26 – 2.20) | 31.32 – 1.95 (2.02 – 1.95) | 48.07 – 1.90 (1.95 – 1.90) | 24.41 – 1.88 (1.95 – 1.88) | 43.85 – 1.89 (1.94 – 1.89) |
| No. reflections | 39448 (2852) | 3857 (52) | 4048 (293) | 32452 (2732) | 7568 (195) | 2574 (110) | 2207 (138) |
| Rwork / Rfree | 0.20 / 0.25 | 0.13 / 0.18 | 0.17 / 0.24 | 0.19 / 0.24 | 0.20 / 0.22 | 0.20 / 0.26 | 0.23 / 0.25 |
| No. atoms | |||||||
| Protein | 6304 | 289 | 556 | 4561 | 1147 | 272 | 228 |
| Ligand/ion | |||||||
| Sulfate | - | - | - | 100 | 29 | - | - |
| Glycerol | - | - | - | 24 | 3 | - | - |
| Water | 312 | 55 | 77 | 120 | 43 | 37 | 18 |
| B factors | |||||||
| Protein | 44.53 | 13.84 | 15.07 | 15.84 | 23.07 | 15.95 | 16.28 |
| Ligand/ion | - | - | - | 26.05 | 44.74 | - | - |
| Water | 36.8 | 28.1 | 24.28 | 13.36 | 24.27 | 35.17 | 35.18 |
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.013 | 0.016 | 0.009 | 0.018 | 0.014 | 0.011 | 0.012 |
| Bond angles (°) | 1.12 | 1.54 | 1.33 | 1.97 | 1.62 | 1.52 | 1.32 |
Values in parentheses are for highest-resolution shell.
Table 2.
Data collection and refinement statistics for structures phased by sSAD
| Target #19 (6ATS) |
Target #56 (6AVD) |
Target #60 (6ATN) |
Target #63 (6ATL) |
|
|---|---|---|---|---|
| Data collection | ||||
| Space group | C 1 2 1 | P 1 21 1 | P 41 21 2 | P 1 21 1 |
| Cell dimensions | ||||
| a, b, c (Å) | 46.1, 21.3, 20.4 | 21.5, 26.2, 31.1 | 46.6, 46.6, 44.4 | 27.7, 23.2, 46.3 |
| α, β, γ (°) | 90.0, 92.6, 90.0 | 90.0, 99.2, 90.0 | 90.0, 90.0, 90.0 | 90.0, 94.4, 90.0 |
| Peak | Peak | Peak | Peak | |
|
|
||||
| Wavelength | ||||
| Resolution (Å)a | 50.00 – 1.95 (1.98 – 1.95) | 50.00 – 1.80 (1.83 – 1.80) | 50.00 – 1.76 (1.79 – 1.76) | 50.00 – 1.80 (1.83 – 1.80) |
| Rmerge | 0.04 (0.05) | 0.06 (0.13) | 0.05 (0.19) | 0.11 (0.31) |
| Rmerge | 0.04 (0.04) | 0.07 (0.13) | 0.07 (0.26) | 0.11 (0.32) |
| I/σ(I) | 41.8 (25.8) | 14.2 (6.2) | 25.0 (2.7) | 35.3 (12.7) |
| CC1/2 | (0.99) | (0.98) | (0.94) | (0.98) |
| Completeness (%) | 90 (45) | 83.9 (67.9) | 44.7 (2.2) | 99.9 (100) |
| Redundancy | 2.7 (1.6) | 1.8 (1.2) | 2.1 (1.0) | 11.6 (11.0) |
| Refinement | ||||
| Resolution (Å) | 23.03 – 1.90 (1.95 – 1.90) | 30.70 – 1.80 (1.84 – 1.80) | 32.93 – 1.76 (1.81 – 1.76) | 24.58 – 1.77 (1.83 – 1.77) |
| No. reflections | 1370 (51) | 2623 (125) | 4611 (158) | 4274 (73) |
| Rwork / Rfree | 0.14 / 0.16 | 0.17 / 0.23 | 0.17 / 0.20 | 0.14 / 0.18 |
| No. atoms | ||||
| Protein | 197 | 289 | 283 | 538 |
| Ligand/ion | ||||
| Sulfate | - | 6 | - | 15 |
| Citric Acid | - | - | - | 11 |
| Water | 33 | 40 | 17 | 67 |
| B factors | ||||
| Protein | 14.8 | 10 | 25.1 | 15.8 |
| Ligand/ion | - | 29.4 | - | 15.8 |
| Water | 30.1 | 26.2 | 36.13 | 29.4 |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.014 | 0.009 | 0.013 | 0.016 |
| Bond angles (°) | 1.26 | 1.52 | 1.44 | 1.97 |
Values in parentheses are for highest-resolution shell.
These new structures all showed the expected overall cystine connectivity and topology, based on comparisons with previously-determined, homologous structures, validating the expression platform. However, many detailed structural differences were found when comparing these crystal structures with previous NMR structures (eg., CTX; Fig. 2c), particularly in the arrangement of the cystine core, the key defining element of CDP structure. The disparity in structural details likely also explains the inability to use the CTX NMR structure as deposited as an MR search model to phase the crystallographic data, which required computational remodeling with Rosetta26 to generate a successful search model. A number of CDP crystals yielded multiple independent views of their structures (Supplementary Table 4), providing additional information about structural rigidity/flexibility (Fig. 2d), and quaternary structure (Fig. 2e). In the most extreme cases, the whey acidic protein (WAP)-type, four-disulfide core (WFDC) peptide Elafin (target #4, a human, non-knotted CDP) crystallized with 18 copies in the asymmetric unit (AU), demonstrating the extraordinary structural rigidity possible in CDPs. The gamma-KTx 2.2 potassium channel toxin (target #48, a knotted CDP from the venom of the Manchurian scorpion) crystallized in the same tetrameric state, with 20 copies total in the AUs of two different crystal forms. While prior NMR structural analyses had suggested inherent flexibility or the presence of multiple conformers in solution for some CDPs (eg., Supplementary Fig. 3), the high degree of structural conservation observed among multiple views in CDP crystal structures conversely argues for the adoption of a single, rigid structure for these examples.
CDP ion channel modulation
Many natural venom and toxin CDPs have been reported to modulate ion channel activity4, and our initial target set of CDPs included many related molecules. In order to assess channel activity, both to verify production of functional CDPs and to identify additional specificities/activities, the selectivity profiles of 37 successfully-expressed CDPs were assessed using a commercial electrophysiological assay on a panel of 20 human ion channels (Supplementary Figure 4). All showed some activity towards at least a subset of ion channels, predominantly potassium channels. While many of the 37 had not been previously characterized or had been tested on a more focused set of channels, agreement between our results and limited prior results was very good, reinforcing the utility of our CDP production/characterization platform.
CDP structure-based classification
Coupling optimized CDP crystallography with robust expression platforms enables fully determining the boundaries of CDP fold space, if CDPs can be identified on the basis of sequence, and classified on the basis of structure. Several practical problems, however, were encountered in applying our CDP-defining motif to larger databases to fully catalogue candidate CDPs on the basis of sequence. To ensure cystine formation, the cluster of cysteines defining a candidate CDP should be localized in a protein ectodomain. While the presence of recognizable leader peptides worked well for Type I transmembrane proteins, problems were encountered with reliably localizing CDPs to ectodomains in Type II and Type III transmembrane proteins, and annotations were found to lack sufficient standardization to confidently substitute. Identifying the bounding cysteines in a cluster of cysteines in a sequence was also problematic, as the number of cysteines in CDPs can be quite variable, and can even be an odd number, as some CDPs are covalently linked to another peptide through an interchain cystine (eg., 1BUN.pdb27). Likewise, no sequence-based rules were discerned that could rigorously identify the full sequence boundaries of CDPs outside of the bounding cysteines. Therefore, it was not possible to prospectively generate a complete CDP catalogue based solely on sequence. Previous proposed structural classification schemes28–30, sometimes also relying on inconsistent functional annotations, were too narrow to be applied globally, also precluding a unified classification based solely on structure.
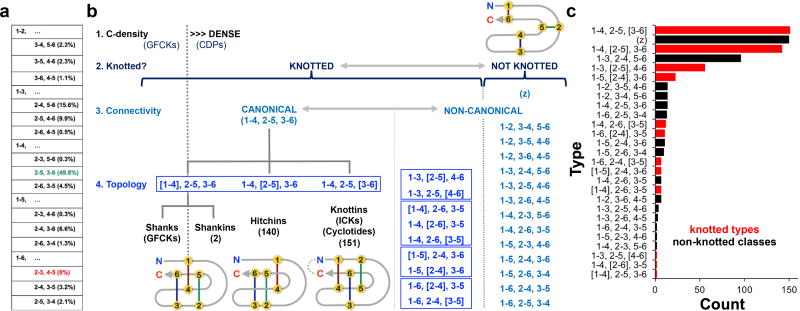
Our CDP definition does not require that the fold contain a cystine pseudoknot, but many CDPs do, defining a subset: the knotted CDPs. CDPs are defined as having at least three cystines, the minimum required to generate a pseudoknotted topology. Stepping back, inspection of available CDP structures did establish a global, unified, structure-based CDP classification scheme by focusing solely on the arrangement of the three core cystines defining a CDP. In contrast, previous approaches drew distinctions on the total number of cystines, inter-cysteine loop lengths, or secondary structure content. The first level of our proposed classification was determined by cystine connectivity (Fig. 3a). Numbering the cysteines in the three-cystine core or knotting element sequentially from 1 to 6 yields 15 theoretically possible connectivity classes, with archetypical knottins and most GFCKs falling into the 1–4, 2–5, 3–6 class. This class was, by far, the most frequently observed among knotted CDP structures in the PDB (298 examples), and thus is referred to as “canonical”. Four other connectivity classes were observed in deposited knotted CDP structures, with variable representation, and nine additional connectivity classes were observed exclusively in deposited non-knotted CDPs with three cystines (Fig. 3b). One connectivity class (1–6, 2–3, 4–5) was not observed in any natural CDPs, though was found in wholly synthetic, designed CDPs (eg., 5JI4.pdb31). Non-knotted CDPs with more than three cystines cannot be assigned to comparable connectivity classes, as the focus subset of three cystines cannot be defined and numbered in the same way in the absence of a knotting element, and were lumped together in a separate class, “z”.
Figure 3. Structure-based CDP classification scheme.
a, The 15 potential connectivities linking six cysteines pairwise are shown, arranged by the five possible pairings in the first cystine (ie., 1–2, 1–3, etc., numbering the focus set of six cysteines from 1 to 6, N- to C-terminal). Subsequent pairings in the remaining cystines are shown in descending rows. Connectivities with corresponding experimentally-determined structures in the PDB are indicated in black or green, highlighting the lack of any natural CDPs with a 1–6, 2–3, 4–5 connectivity in the PDB (indicated in red). The 1–4, 2–5, 3–6 connectivity pattern (highlighted in green) was by far the most commonly observed: 312 examples, 298 knotted and 14 non-knotted. Percentage class distributions of the 621 knotted CDPs, plus non-knotted, three-cystine CDPs, are shown in parentheses. Cystine connectivity cannot be determined for the subgroup of 150 non-knotted CDPs with more than three cystines. b, The overall CDP classification scheme is outlined, showing the hierarchical relationship of cysteine density (GFCKs vs. CDPs), pseudoknotting (the knotted CDP subset of CDPs), and type classifications, based on cystine connectivity plus knotting topology. Common connectivities are grouped within boxes, highlighting that only five of the 15 possible connectivities were observed among known knotted CDP structures. Example cartoon schematics of cystine connectivity/topology (numbered yellow circles indicate cysteines; number observed in the PDB is indicated in parentheses) are shown for the canonical shankins, hitchins, and knottins, and the simplest non-knotted CDP type, (1–6, 2–5, 3–4; at upper right). Only types observed in the PDB as of April, 2017 are indicated, showing the non-random distribution of knotted CDP types: only twelve of the possible 45 (15 connectivities times three pseudoknotted topologies) were observed. c, The class and type distribution of the 775 CDP structures, knotted (red) and non-knotted (black), was tabulated, highlighting the dominance of knottins and hitchins among knotted CDPs.
The second level of classification, applicable to knotted CDPs, was based on cystine topology: which cystine, of the three core cystines comprising the knotting element, pseudoknots the fold, ignoring variable accessory cystines (Fig. 3b). In any connectivity class, denoted as u-v, w-x, y-z to indicate the core cystine connectivity, there are three theoretical topologies, each with a different knotting cystine. CDP connectivity class plus knotting topology generates structure-based knotted CDP type, represented as u-v, w-x, [y-z], where the knotting cystine is indicated by brackets. Non-knotted CDPs with three cystines are denoted solely by connectivity class (u-v, w-x, y-z). Using this nomenclature, archetypical knottins were classified as type 1–4, 2–5, [3–6] knotted CDPs (151 examples), which is distinct from the [1–4], 2–5, 3–6 topology observed in GFCKs, despite a common connectivity. The second most commonly observed knotting topology in this connectivity class was 1–4, [2–5], 3–6 (145 examples), but there were only two CDP structures displaying the third possible, GFCK-like, topology in this class: [1–4], 2–5, 3–6. Following the knottin nomenclature, we refer to the many known type 1–4, [2–5], 3–6 knotted CDPs as “hitchins” (a hitch is a kind of knot), type [1–4], 2–5, 3–6 GFCKs as “shanks” (a shank is another kind of knot used to shorten a rope), and rare type [1–4], 2–5, 3–6 knotted CDPs as “shankins”. Though far fewer knotted CDP structures have non-canonical connectivities, examples of nine additional knotted types were found. The distribution between different connectivity classes (non-knotted CDPs) and types (knotted CDPs) was very uneven, dominated by knottins, z-class, and hitchins (Fig. 3c). This proposed scheme provides an unambiguous method for CDP structural classification and comparison independent of source organism, sequence similarity, or functional annotation. Advantages include avoiding broad annotations, like “defensin”, which denotes cysteine-rich, cationic, antimicrobial peptides, but which also encompasses a wide range of structurally-dissimilar knotted and non-knotted CDPs, including many hitchins and knottins. While robust and global, our proposed classification scheme, however, cannot be applied in the absence of experimentally determining, or reliably modeling, CDP structure. However, structure type can correlate with function: hitchins predominated among multiply-resistant CDPs.
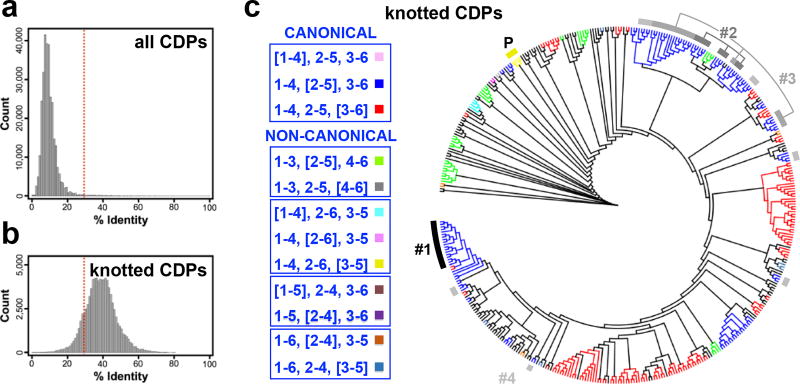
Global, pair-wise sequence comparisons among all 775 CDPs in the PDB showed very limited similarity (Fig. 4a), precluding grouping or clustering across all CDPs to discern sequence-structure relationships. However, limiting the analysis to knotted CDPs revealed sequence similarity scores meaningful for potentially identifying structural homology (Fig. 4b). However, sequence-based phylograms of knotted CDPs failed to reveal global sequence/structure clustering, with, for example, knottin and hitchin branches inseparably interwoven (Fig. 4c). Exceptions included limited subsets of obviously structurally-related knotted CDPs, such as the pacifastins, which are type 1–4, 2–6, [3–5] knotted CDP serine protease inhibitors from arthropods (global superposition root mean square deviation (RMSD) = 0.35 Å; Fig. 5a). In addition to the pacifastin cluster, available CDP structures were aligned to group CDPs into four more recognizably similar clusters: a cluster (#1) of αββ three-cystine hitchins from scorpions (RMSD = 0.55 Å; Fig. 5b); a cluster (#2) of βαββ four-disulfide containing hitchins from scorpions (RMSD = 0.60 Å; Fig. 5c); a cluster (#3) of βαββ four-disulfide containing hitchins from plants (RMSD = 0.46 Å; Fig. 5d); and a cluster (#4) of three-disulfide containing hitchins from various taxa (RMSD = 0.65 Å; Figs. 5e). These results echo previous studies identifying knotted CDP sub-type structural clusters, eg., the Möbius and Bracelet clusters of the type 1–4, 2–5, [3–6] cyclotides6, and, more broadly, the “cysteine-stabilized αβ defensins”29. These results also validated our proposed classification focused on core cystines, by revealing hitchin structural homologies despite variable accessory cystines.
Figure 4. CDP sequence/structure relationships.
a, Distribution of pairwise, non-self, non-redundant, sequence identities from an N-by-N alignment of the 775 CDP structures in the PDB or b, the subset of 422 knotted CDPs. The dashed red line indicates the threshold of the 30% identity “twilight zone”, where structural homology cannot be assumed, or reliably modeled36,37. The sequence diversity in CDP space was broad enough to preclude useful clustering, or correlating sequence with structure. However, limiting the analysis to knotted CDPs showed much better sequence clustering, at identity levels supporting potential linkage of sequence with structure. c, A sequence-based phylogram of the 422 knotted CDPs in the PDB, colored by knotted CDP structure-based type, tabulated at left. In the legend, CDP type is divided by canonical vs. non-canonical cystine connectivity, with common connectivities boxed. The locations of pacifastin (yellow arc, labeled “P”) and hitchin (black to gray arcs, labeled by cluster number) clusters defined by structural homology (Fig. 5) are indicated, showing strong sequence similarity between members of the pacifastin and hitchin cluster #1 subsets, but weaker similarities between other hitchin clusters, and knotted CDP types in general. Sequence identity between the knottins in the sub-branch associated with the cluster #1 hitchins and the cluster #1 hitchins is ~26%, where the minimal pairwise identity within the cluster #1 hitchins is ~50%. Note the interspersion of knotted CDP types throughout the phylogram, with types typically clustering only very locally.
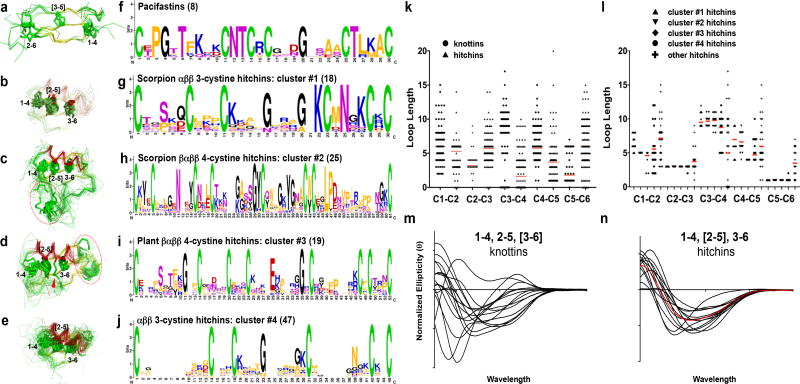
Figure 5. Structure/sequence clustering of select subgroups of knotted CDPs.
a, b, c, d, and e, Structure-based superpositions of selected CDP subsets, with structures represented as α-carbon backbones, colored by secondary structure (α: red; β: yellow; coil: green), and with cystines shown in licorice-stick representation, are paired with derived MEME38-identified sequence motifs, shown as sequence logo plots39 (f, g, h, i, and j). Five recognizably similar CDP clusters are shown: a, b, pacifastins; c, d, αββ three-cystine hitchins from scorpions (#1); βαββ four-cystine hitchins from scorpions (#2); e, f, βαββ four-cystine hitchins from plants (#3); and i, j, three-cystine hitchins from various taxa (#4). Cysteine numbering of the type-defining cystines is indicated in a, b, c, d, and e, and additional, accessory cystines are circled in red. One “four-cystine” hitchin in panel e (1N4N.pdb40) has a fifth cystine (red arrow), but clearly belonged within this cluster, based on the structure superpositions shown. Number of knotted CDPs in each cluster is indicated in parentheses in f, g, h, i, and j. The distributions of the lengths of loop sequences connecting the focus set of six cysteines in the knotting elements of k, knottins versus hitchins, and l, hitchin subtypes, is plotted. Average loop lengths are indicated with red bars. m, Normalized CD spectra from all 14 successfully-expressed knottins (left), and, n, from all 16 successfully-expressed hitchins (right), are shown. The red line is an averaged CD spectrum calculated from the set of 14 hitchin spectra.
Structure-based sequence alignments yielded motifs potentially useful for prospectively identifying candidate structural homologs of the pacifastins (Fig. 5f), extending prior studies32, and one hitchin subset (Fig. 5g). Sequence conservation for the other three hitchin subsets was more limited, or even unrecognizable, despite considerable structural similarity (Figs. 5h through 5j). Even more simplistic sequence-based metrics, such as inter-cysteine loop lengths, failed to robustly differentiate between knottins and hitchins (Fig. 5k), or between hitchin subsets (Fig. 5l). Contrasting the knottins, these hitchin subset clusters all displayed conserved secondary structure content, with very similar αββ or βαββ folds, echoed in their respective CD spectra (Figs. 5m, 5n). Normalized CD spectra from all 14 successfully-expressed knottins showed a wide range of secondary structure compositions, where spectra from all 16 successfully-expressed hitchins showed a fairly narrow range of secondary structure compositions, consistent with the greater degree of structural homology observed among hitchins in general.
Taxonomy-based phylogenies (Supplementary Fig. 5), though likely affected by experimenter selection bias, showed very uneven distributions, with knotted CDPs dominated by examples from arachnids, magnoliopsids, mammals, and gastropods, respectively, at the class level. Non-knotted CDPs were predominately from mammalian (class) and primate (order) sources, while knotted CDPs were predominately from arachnids, mostly scorpions.
DISCUSSION
We have developed an efficient, reliable platform for large-scale production and high-throughput expression screening of endotoxin-free CDPs. The pipeline incorporates multiple steps to stringently validate proper folding, including structure determinations by x-ray crystallography, and assay biological function, including ion channel inhibition. Starting from a purely sequence-based CDP definition, we have also proposed a robust, purely structure-based classification system for CDPs, framing an exhaustive analysis of these molecules. Classified CDPs encompassed a wide range of diverse molecules, including epidermal growth factor (EGF)-like domains, low density lipoprotein (LDL)-like domains, tumor necrosis factor receptor (TNFR)-like domains, transforming growth factor receptor (TGFR)-like domains, trefoil/plexin domains, notch repeat-like domains, resistin-like domains, osmotin-like domains, thaumatin-like proteins, disintegrins, anaphylatoxins, insect antifreeze proteins, and chitin-binding penaeidins. The most surprising result of analyses of CDP sequence/structure relationships based on this classification was the very limited correlations between CDP sequence and structure type, which severely restricts prospective mapping of structures, based only on sequence. The next challenges are to develop more sophisticated sequence-based tools to completely catalogue CDP sequence space, useful for guiding a broader sampling of CDP structure space, more evenly across taxa, to confidently identify its boundaries. Focused studies to parse the roles of the few conserved residues in determining CDP folds are now possible with high-throughput platforms for expressing panels of sequence variants to deeply sample effects on folding. These tools combine to enable future exploration of CDP space for advancing basic science, through the study of an exceptional protein fold family, and clinical application, through production and manipulation of molecules possessing uniquely useful properties.
ONLINE METHODS
CDP production and purification
For expression of CDPs in mammalian cell lines, the Daedalus18 expression cassette was modified to include, from N- to C-terminus: a murine Igκ leader peptide sequence, an optimized FLAG epitope sequence41, a hexahistidine purification tag sequence, the Scn fusion partner19 sequence, the TEV scission sequence (ENLYFQ|…), a short glycine/serine spacer sequence, and the CDP sequence. A minimum of three amino acids before the first CDP cysteine residue was found to be essential for efficient protease cleavage, necessitating a short glycine/serine spacer. Since CDPs are found in non-mammalian hosts, but the Daedalus system is based on production in mammalian cells, the targeted CDP sequences were checked and corrected for cryptic N-glycosylation sequences, eg. in STa, by mutation to generate fully-functional peptides. Difficulties in defining CDP boundaries outside of the bounding cysteines likely also limited overall success rate. CDP-encoding Daedalus lentiviruses were produced by transient transfection of suspension-adapted HEK293T cells (ATCC CRL-3216) with psPAX2 (Addgene 12260), pMD2.G (Addgene 12259), and Scn/CDP fusion-encoding vectors using linear 25-kDa polyethyleneimine (PEI; Polysciences). Cells were checked for mycoplasma contamination (MycoProbe; R&D Systems). For preparative-scale production of CDPs, transfected cells were cultured in 5 ml of FreeStyle 293 Expression Medium (Thermo Fisher), and the culture was fed with 5 ml of FreeStyle media supplemented with 6 mM valproic acid (Sigma-Aldrich) after 24 h. Lentivirus was harvested 44 h after feeding through a 0.45 µM Steriflip filter (Millipore). HEK293F cells (Thermo Fisher) were transduced with 1 ml of lentivirus stock added dropwise in 125 ml shake flasks with 1×107 cells in 9 ml of FreeStyle medium. After 6 h, the culture was fed with 20 ml of Freestyle medium. Transduced cells were expanded until a total culture size of 4 L at ~5×106 cells/ml was reached or viability began to drop. CDP fusion proteins were purified by immobilized metal affinity chromatography (IMAC)42 with HisTrap FF Crude columns (GE) on an ÄKTA Pure FPLC system (GE). The CDP was cleaved from the Scn fusion partner by protease digestion, separated from unwanted digestion products by RPC (0.1% w/w TFA in water vs. 0.1% w/w TFA in acetonitrile) on a Tricorn 10/150 column packed with Source 15RPC resin (GE) using an ÄKTA Pure FPLC system, and lyophilized for storage.
The incorporated TEV cleavage sequence in the fusion construct leaves an exogenous GS dipeptide stump from the linker at the N-terminus of the recombinant CDP. Expressed CDPs were confirmed by direct-infusion electrospray mass spectrometry (ES-MS) on an LTQ-OrbiTrap mass spectrometer (Thermo Electron). For ES-MS, CDPs were dissolved in water at 1 mg/mL, and desalted and purified by C18 ZipTip chromatography (EMD Millipore). Cystine formation was confirmed by analyzing the m/z monoisotopic distribution and determination of net charge, and by crystallography. Calculated and observed m/z values are listed in Supplementary Table 2.
High-throughput, pilot-scale expression screening was carried out in an analogous but scaled fashion, with viral production performed in 2 mL deep well blocks (Axygen), producing 1 ml of lentivirus stock per well. 50–100 µL of the lentivirus stock was used to transduce ~2×106 HEK293F cells in 1 mL of Freestyle medium in deep well blocks. Transduction was confirmed by flow cytometry after 36 h, using a NovoCyte cytometer (ACEA), and cultures were fed with 6 mM valproic acid after 120 h. Culture supernatants were harvested after 7 days and transferred to a Protino purification plate (Machery-Nagel) containing 100 µL of Ni-NTA IMAC resin (GE) per well, washed, and eluted. Eluted CDPs were cleaved and analyzed by RPC, as above. For high-throughput expression screening of the broader set of 1,232 CDPs, termini were chosen three residues beyond the bounding cysteines in the embedding sequence, or the native termini of the peptide if the sequence did not extend that far.
Biochemical characterization
In order to determine resistance to high temperatures, CDPs were incubated at 0.5 mM in PBS at 75° C or 100° C for 1 h, pelleted, and the supernatant analyzed by RPC. To determine resistance to proteolytic digestion, CDPs were mixed with 50 U of porcine pepsin (Sigma-Aldrich P7012) in Simulated Gastric Fluid43 at pH 1.05, or 50 U of porcine trypsin (Sigma-Aldrich 6567) in PBS, incubated for 30 m at 37° C, and analyzed by RPC. Oxidized and reduced forms (adding 10 mM DTT) were compared. CD spectra were measured with a Jasco J-720W spectropolarimeter using a 1.0 mm path length cell, with CDPs diluted into 20 mM phosphate buffer, pH 7.4, at a concentration of 15–25 µM.
Crystallization and crystallography
CDPs were resuspended at a target concentration of 80 mg/mL. Crystallization screening was performed at room temperature by vapor diffusion, with 1:1 protein solution:reservoir solution sitting drops, set up using the Nextal JCSG+, PEGs, and AmSO4 factorial suites (Qiagen) and sub-microliter robotics (TTP Labtech mosquito). Diffraction data were collected from single crystals using a Rigaku MicroMax-007 HF home source or beamline 5.0.1 at the Advanced Light Source (Lawrence Berkley National Laboratory, Berkeley, CA). For sSAD phasing, Bijvoet pair measurement was optimized by collecting data through 5° wedges with alternating phi rotations of 180°, in 1° oscillations. Data were reduced and scaled with HKL200044. Initial phases were determined by MR using PHASER45 in the CCP4 program suite46 using homologous structures from the PDB as search models (as is, or after computational refinement using the Rosetta suite26), or by sSAD25, determining sulfur substructures with SHELX47. Iterative cycles of model building and refinement were performed with COOT48 and REFMAC49; structure validation was performed with MolProbity50. Crystallization and cryopreservation conditions, and additional phasing and structure validation information, for 10 of the 12 CDP structures are detailed in Supplementary Table 4. The crystal structure determinations of CDPs #11 and #29 are described elsewhere (Cook Sangar, M., Girard, E., Hopping, G., Yin, C., Pakiam, F., Brusniak, M.-Y., Gewe, M. G., Mehlin, C., Strand, A., Correnti, C., Strong, R. K., Simon, J., and Olson, J. M., manuscript under consideration).
Ion channel activity assays
The selectivity profile of 37 CDPs selected from the 46 successfully expressed peptides on a panel of 20 ion channels was assessed using a commercial electrophysiological assay platform (IonWorks Barracuda, Charles River)51,52. Peptides were screened in duplicate at final concentrations of 20 µM and 200 nM, determined by amino acid analysis, diluted into HEPES buffered physiological saline (HBPS). HEK293 or CHO cells from the American Type Culture Collection (ATCC) were transfected with the appropriate ion channel before use. HEK293 cells were grown in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12, and CHO cells were cultured in Ham’s F-12 media. All cultures were supplemented with 10% fetal bovine serum, 100 U/ml penicillin G, 100ug/ml streptomycin sulfate, and the appropriate selection antibiotics. Prior to use, cells were washed twice with Hank’s Balanced Salt Solution, and treated with Accutase cell detachment solution (Sigma-Aldrich). Cells were then washed with HBPS twice, and resuspended in HBPS. Peptides and controls were incubated with cells for at least five minutes prior to assay. The IonWorks software package was used for data acquisition and analysis, and the data was corrected for leak current. The decrease in current amplitude in the presence of the peptides was used to calculate the percentage of blocking as follows:
where [CDP] is the concentration of tested CDP, IC50 is the concentration of the test CDP producing half-maximal inhibition, N is the Hill coefficient, %VC is the percentage of the current run-down (the mean current inhibition at the vehicle control). and %Block is the percentage of ion channel current inhibited at each concentration of a test article. Controls, matched to particular channels, include ondansetron, mecamylamine, mibefradil dihydrochloride, picrotoxin, E-4031, BaCl2, 4-aminopyridine, verapamil, lidocaine, memantine, and capsazepine (sourced from Sigma-Aldrich or Tocris). Nonlinear least squares fits were solved with the XLfit add-in for Microsoft Excel.
Structure and sequence analyses
Sequence alignments and identity scores were determined with CLUSTALW53 using dynamic BLOSUM matrix selection. The phylogenetic analysis was performed with the MUSCLE alignment algorithm in the Geneious version 10.2.3 software package, using default settings, the Jukes-Cantor genetic distance model, and the UPGMA tree-building method54,55. Three-dimensional structure alignments were performed with Theseus56,57, and structure-based sequence alignment was performed with PROMALS3D58,59. RMSDs are quoted as the Theseus global superposition maximum likelihood values.
Data Availability
Crystal structure atomic coordinates and diffraction data have been deposited into the PDB with accession codes 6ATL, 6ATN, 6ATS, 6ATU, 6ATW, 6AU7, 6AUP, 6AV8, 6AVA, 6AVC, and 6AVD. Source data for Figs. 3a, 3b, 3c, 5f, 5g, 5h, 5i, 5j, 5k, and 5l are available with the paper online. Other data supporting this study are available upon request.
Supplementary Material
Acknowledgments
We thank J. Carter for substantial logistical support, and J. Simon, A. Mhyre, and N. Nairn for helpful discussions. The project was supported by NIH grants R01CA135491-07 (J.M.O.) and T32-H600035 (C.D.B.), Project Violet, the Wissner-Slivka Foundation, the Kismet Foundation, the Sarah M. Hughes Foundation, Strong4Sam, Yahn Bernier and Beth McCaw, Len and Norma Klorfine, Anne Croco, and Pocket Full of Hope.
Footnotes
AUTHOR CONTRIBUTIONS
C.E.C., M.M.G., C.M., M.-Y.B., J.M.O., and R.K.S. designed experiments, analyzed data, administered the project, and wrote the manuscript. C.E.C., C.M., A.D.B., W.A.J., W.d.v.d.S., and S.M.T. developed the CDP production and analysis pipeline. M.M.G., P.B.R., and R.K.S. performed crystallographic structure determinations and analyses. C.E.C., M.M.G., M.-Y.B., and S.E.B. performed phylogenetic and taxonomic analyses. C.E.C., A.D.B., W.A.J., M.C., S.E.B., W.d.v.d.S., S.M.T., A.W., and M.K.C. produced and analyzed CDPs. C.E.C., M.M.G., K.P., C.D.B., and R.K.S. designed and developed SuperTEV.
COMPETING FINANCIAL INTERESTS STATEMENT
J.M.O. is a founder and shareholder of Blaze Bioscience, Inc.
References
- 1.Molesini B, Treggiari D, Dalbeni A, Minuz P, Pandolfini T. Plant cystine-knot peptides: pharmacological perspectives. British Journal of Clinical Pharmacology. 2017;83:63–70. doi: 10.1111/bcp.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herzig V, King GF. The Cystine Knot Is Responsible for the Exceptional Stability of the Insecticidal Spider Toxin omega-Hexatoxin-Hv1a. Toxins. 2015;7:4366–4380. doi: 10.3390/toxins7104366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinwarth M, Nasu D, Kolmar H, Avrutina O. Chemical synthesis, backbone cyclization and oxidative folding of cystine-knot peptides: promising scaffolds for applications in drug design. Molecules. 2012;17:12533–12552. doi: 10.3390/molecules171112533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolmar H. Natural and engineered cystine knot miniproteins for diagnostic and therapeutic applications. Current Pharmaceutical Design. 2011;17:4329–4336. doi: 10.2174/138161211798999465. [DOI] [PubMed] [Google Scholar]
- 5.Postic G, Gracy J, Perin C, Chiche L, Gelly JC. KNOTTIN: the database of inhibitor cystine knot scaffold after 10 years, toward a systematic structure modeling. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould A, Ji Y, Aboye TL, Camarero JA. Cyclotides, a novel ultrastable polypeptide scaffold for drug discovery. Current Pharmaceutical Design. 2011;17:4294–4307. doi: 10.2174/138161211798999438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz E. Cystine knot growth factors and their functionally versatile proregions. Biol. Chem. 2017;398:1295–1308. doi: 10.1515/hsz-2017-0163. [DOI] [PubMed] [Google Scholar]
- 8.Iyer S, Acharya KR. Tying the knot: the cystine signature and molecular-recognition processes of the vascular endothelial growth factor family of angiogenic cytokines. The FEBS Journal. 2011;278:4304–4322. doi: 10.1111/j.1742-4658.2011.08350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kintzing JR, Cochran JR. Engineered knottin peptides as diagnostics, therapeutics, and drug delivery vehicles. Curr. Opin. Chem. Biol. 2016;34:143–150. doi: 10.1016/j.cbpa.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 10.Al-Salama ZT, Syed YY. Plecanatide: First Global Approval. Drugs. 2017;77:593–598. doi: 10.1007/s40265-017-0718-0. [DOI] [PubMed] [Google Scholar]
- 11.Veiseh M, et al. Tumor paint: a chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Cancer Res. 2007;67:6882–6888. doi: 10.1158/0008-5472.CAN-06-3948. [DOI] [PubMed] [Google Scholar]
- 12.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 14.Liou YC, Tocilj A, Davies PL, Jia Z. Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein. Nature. 2000;406:322–324. doi: 10.1038/35018604. [DOI] [PubMed] [Google Scholar]
- 15.Liang Z, Sottrup-Jensen L, Aspan A, Hall M, Soderhall K. Pacifastin, a novel 155-kDa heterodimeric proteinase inhibitor containing a unique transferrin chain. Proc. Natl. Acad. Sci. USA. 1997;94:6682–6687. doi: 10.1073/pnas.94.13.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moura A, Savageau MA, Alves R. Relative amino acid composition signatures of organisms and environments. PLoS One. 2013;8:e77319. doi: 10.1371/journal.pone.0077319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandaranayake AD, et al. Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Res. 2011;39:e143. doi: 10.1093/nar/gkr706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finton KA, et al. Autoreactivity and exceptional CDR plasticity (but not unusual polyspecificity) hinder elicitation of the anti-HIV antibody 4E10. PLoS Pathog. 2013;9:e1003639. doi: 10.1371/journal.ppat.1003639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabrita LD, et al. Enhancing the stability and solubility of TEV protease using in silico design. Protein Sci. 2007;16:2360–2367. doi: 10.1110/ps.072822507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapust RB, et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 23.Cesaratto F, Lopez-Requena A, Burrone OR, Petris G. Engineered tobacco etch virus (TEV) protease active in the secretory pathway of mammalian cells. J. Biotechnol. 2015;212:159–166. doi: 10.1016/j.jbiotec.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 24.Wingerd JS, et al. The tarantula toxin beta/delta-TRTX-Pre1a highlights the importance of the S1-S2 voltage-sensor region for sodium channel subtype selectivity. Scientific Reports. 2017;7:974. doi: 10.1038/s41598-017-01129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Liu Q, Hendrickson WA. Robust structural analysis of native biological macromolecules from multi-crystal anomalous diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1314–1332. doi: 10.1107/S0907444913001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leaver-Fay A, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong PD, McDonald NQ, Sigler PB, Hendrickson WA. Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action. Structure. 1995;3:1109–1119. doi: 10.1016/s0969-2126(01)00246-5. [DOI] [PubMed] [Google Scholar]
- 28.Zhu LM, Gao B, Zhu SY. Origin of neurotoxins from defensins. Sheng Li Xue Bao. 2015;67:239–247. [PubMed] [Google Scholar]
- 29.Tarr DE. Establishing a reference array for the CS-alphabeta superfamily of defensive peptides. BMC Res. Notes. 2016;9:490. doi: 10.1186/s13104-016-2291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, Gao B, Zhu S. New fungal defensin-like peptides provide evidence for fold change of proteins in evolution. Biosci. Rep. 2017;37:BSR20160438. doi: 10.1042/BSR20160438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhardwaj G, et al. Accurate de novo design of hyperstable constrained peptides. Nature. 2016;538:329–335. doi: 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simonet G, Claeys I, Broeck JV. Structural and functional properties of a novel serine protease inhibiting peptide family in arthropods. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002;132:247–255. doi: 10.1016/s1096-4959(01)00530-9. [DOI] [PubMed] [Google Scholar]
- 33.Lippens G, Najib J, Wodak SJ, Tartar A. NMR sequential assignments and solution structure of chlorotoxin, a small scorpion toxin that blocks chloride channels. Biochemistry. 1995;34:13–21. doi: 10.1021/bi00001a003. [DOI] [PubMed] [Google Scholar]
- 34.Tsunemi M, Matsuura Y, Sakakibara S, Katsube Y. Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution. Biochemistry. 1996;35:11570–11576. doi: 10.1021/bi960900l. [DOI] [PubMed] [Google Scholar]
- 35.Francart C, Dauchez M, Alix AJ, Lippens G. Solution structure of R-elafin, a specific inhibitor of elastase. J. Mol. Biol. 1997;268:666–677. doi: 10.1006/jmbi.1997.0983. [DOI] [PubMed] [Google Scholar]
- 36.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 37.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 38.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology; ISMB. 1994;2:28–36. [PubMed] [Google Scholar]
- 39.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janssen BJ, Schirra HJ, Lay FT, Anderson MA, Craik DJ. Structure of Petunia hybrida defensin 1, a novel plant defensin with five disulfide bonds. Biochemistry. 2003;42:8214–8222. doi: 10.1021/bi034379o. [DOI] [PubMed] [Google Scholar]
- 41.Knappik A, Pluckthun A. An improved affinity tag based on the FLAG peptide for the detection and purification of recombinant antibody fragments. Biotechniques. 1994;17:754–761. [PubMed] [Google Scholar]
- 42.Kim Y, et al. Chapter 3. High-throughput protein purification for x-ray crystallography and NMR. Advances in Protein Chemistry and Structural Biology. 2008;75:85–105. doi: 10.1016/S0065-3233(07)75003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Yadav V, Smart AL, Tajiri S, Basit AW. Toward oral delivery of biopharmaceuticals: an assessment of the gastrointestinal stability of 17 peptide drugs. Mol. Pharm. 2015;12:966–973. doi: 10.1021/mp500809f. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. In: Meth. Enzymol. Carter CW Jr, Sweet RM, editors. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Graef JD, et al. Validation of a high-throughput, automated electrophysiology platform for the screening of nicotinic agonists and antagonists. Journal of Biomolecular Screening. 2013;18:116–127. doi: 10.1177/1087057112457414. [DOI] [PubMed] [Google Scholar]
- 52.Gillie DJ, Novick SJ, Donovan BT, Payne LA, Townsend C. Development of a high-throughput electrophysiological assay for the human ether-a-go-go related potassium channel hERG. J Pharmacol. Toxicol. Methods. 2013;67:33–44. doi: 10.1016/j.vascn.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 54.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Re.s. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Theobald DL, Steindel PA. Optimal simultaneous superpositioning of multiple structures with missing data. Bioinformatics. 2012;28:1972–1979. doi: 10.1093/bioinformatics/bts243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pei J, Grishin NV. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 2014;1079:263–271. doi: 10.1007/978-1-62703-646-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structure atomic coordinates and diffraction data have been deposited into the PDB with accession codes 6ATL, 6ATN, 6ATS, 6ATU, 6ATW, 6AU7, 6AUP, 6AV8, 6AVA, 6AVC, and 6AVD. Source data for Figs. 3a, 3b, 3c, 5f, 5g, 5h, 5i, 5j, 5k, and 5l are available with the paper online. Other data supporting this study are available upon request.