Abstract
Il10 forms a cytokine cluster with Il19, Il20, and Il24 in a conserved region of chromosome 1. The later genes are in the IL-20 subfamily of IL-10-related cytokines and though not as well-studied their biologic actions and expression patterns suggest they have little in common with IL-10. IL-24, like IL-10, however is uniquely expressed in T cells and is a signature gene of the Th2 lineage suggesting they could be co-regulated in certain cell types. Little is known about other cellular sources of IL-24. We investigated IL-24 and IL-10 expression in murine macrophages and NK cells and found that while co-expressed under most stimulation conditions, IL-24 and IL-10 are controlled by distinct, cell type-specific pathways. In BMM, optimal IL-24 expression required LPS + IL-4 co-stimulation and STAT6 but was independent of type I IFN receptor signaling and STAT4. Conversely, LPS-induced IL-10 was independent of IL-4/STAT6 and STAT4 but, consistent with other reports, required type I IFN receptor signaling for optimal expression. Remarkably, NK-specific IL-24 (but not IL-10) expression was dependent on both type I IFN receptor signaling and STAT4. Induction of IL-24 expression was accompanied by cell-specific recruitment of STAT6 and STAT4 to multiple sites we identified within Il24 which mediated STAT-dependent histone modifications across the gene. Collectively, our results indicate that despite being co-expressed, IL-10 and IL-24 are independently regulated by different type I IFN receptor signaling pathways in innate immune cells and provide insight into the mechanisms which fine-tune cell type-specific gene expression within the Il10 cluster.
Keywords: Interleukin 24 (IL-24), signal transducers and activators of transcription (STAT), Interleukin 10 (IL-10), epigenetics, interferon
INTRODUCTION
Cell type-specific gene expression programs are fundamental to development and disease. In the genome, related genes may form regulatory clusters which can help to synchronize shared gene expression programs as well as biological functions. Cytokine genes can be organized into gene clusters in which shared regulatory information across the locus coordinates gene expression. The Il4 locus, consisting of Il4, Il13 and Il5 is a prime example of a homologous gene cluster that, despite being disrupted by the unrelated gene Rad50, is controlled by the cooperative actions of distal cell type-specific regulatory elements across the locus (1–3) in T helper subsets. On the other hand, some cytokine gene clusters lack homology and/or have distinct gene regulatory programs despite being in relatively close proximity. For instance, Ifng and Il22 (and in humans, IL26) form a cytokine cluster despite having unique cell-specific gene expression patterns governed by discrete, gene-specific regulatory elements (4).
The IL-10 family is a relatively large group of related cytokines that is divided into subfamilies based several factors including; degree of similarity, use of shared receptor subunits, similarities in cellular targets/biological functions and location in the genome. The IL-20 subfamily, consisting of IL-19, IL-20, IL-22, IL-24 and IL-26 meet most of these criteria (5). As mentioned, while mouse Il22 co-localizes with the Ifng gene on chromosome 10, the remaining subfamily members, Il19, Il20 and Il24 are located in a highly conserved region of chromosome 1, flanked by the Il10 gene. There is little overlap in the cellular subsets which co-express IL-10 and any of its neighboring homologs. IL-24 is a notable exception because both IL-10 and IL-24 are highly inducible in Th2 cells, (6,7). Previous studies have established that IL-4/STAT6 signaling is critical for IL-24 and IL-10 expression in Th2 cells (6,8,9). However, a ChIP seq-based study identified Il24 as one of the top STAT6-target genes in Th2 cells and classified Il24, along with Il4, Gata3, and Il4ra, as a major Th2 lineage-specifying gene (10). It should be noted that the biological functions of IL-24 are still not well-defined and the biologic significance of IL-24 expression in Th2 cells has yet to be studied.
Compared to IL-10, our understanding of IL-24’s functions as well as its regulation in other cells types is limited. In polyclonally-stimulated human PBMCs or PBMC subsets (monocytes, NK cells, B cells and T cells), IL-24 was expressed predominately in T cells and monocytes (11,12). Another study found that cross-linking of the B cell receptor by CD40L plus anti-IgM stimulation triggers IL-24 expression in B cells (13). IL-24 can also be expressed in non-hematopoietic cells. IL-24 was originally cloned from melanoma cells (14). IL-24 can be expressed by both melanocytes and keratinocytes and IL-24 has been shown to have tumor suppressor activity (14–17). In addition, a recent report has linked keratinocyte-specific IL-24 expression with skin inflammation in human and mouse models (18).
We have been mapping cell-type-specific IL-10 regulatory elements as a means to better understand the molecular basis of how IL-10 expression patterns impact host inflammatory responses and disease outcomes. In this study, we sought to explore the possibility that IL-10 and IL-24 may share regulatory features by comparing and contrasting IL-10 and IL-24 regulation in cell types in which IL-24 expression is not well characterized. Specifically, we focused on two important innate sources of IL-10, macrophages and NK cells (19). We chose these cells because both macrophage-derived IL-10 as well as NK-derived IL-10 have been implicated in several disease models. For example, macrophage-derived IL-10 is required for protection against LPS-induced shock (20), while NK-derived IL-10 plays a key role in dampening inflammatory responses during infection with several pathogens including MCMV, Toxoplasma gondii, Yersinia pestis and Listeria monocytogenes (21,22). Additionally, NK-derived IL-10 has been shown to contribute to parasite burden during visceral leishmaniasis (13). Our data indicate that although IL-10 and IL-24 are largely co-expressed in NK cells and macrophages, different cell type-specific mechanisms have evolved to regulate their expression. This provides new insight into the exquisite fine-tuning of gene expression programs within the immune system which could prove useful for designing therapeutic strategies to target inflammation based on cell type and/or environmental context.
MATERIAL AND METHODS
Mice
Wild-type (WT), Stat6−/−, and Ifnra−/− mice on the C57BL/6 background were maintained at the Johns Hopkins University animal facility. Stat4−/− mice are known to lose viability when backcrossing to the C57BL/6 background. Thus, Stat4−/− mice on the BALB/c background were used. In all experiments using Stat4−/− mice, WT BALB/c mice were used as controls. Both were purchased from Jackson Laboratory and bred at our facility. All mice were maintained under specific pathogen-free conditions and were used between 8 and 12 weeks of age. All experimental procedures were approved by the Johns Hopkins University Institutional Animal Care and Use Committee.
Cytokines and antibodies
Recombinant human IL-2 was obtained from the NCI repository. Recombinant murine IL-4, IL-12 (p70), and IL-13 were purchased from Peprotech (Rocky Hill, NJ, USA). Purified Lipopolysaccharide (LPS) in lyophilized powder from Escherichia coli 0111:B4 was purchased from Sigma (St. Louis, MO, USA). Anti-STAT6 (Sc-981) and anti-STAT4 (Sc-486) monoclonal antibodies were obtained from Santa-Cruz Biotechnology (Santa Cruz, CA, USA) and anti-STAT5 was from R&D Systems (Minneapolis, MN, USA). Anti-acetylated histone H3 (AcH3; 06-599B) and anti-histone H3 lysine 27 trimethylation (H3K27me3; 07-449) were obtained from Upstate Biotechnology (Millipore, Billerica, MA, USA).
Media for cell culture
NK cell tissue culture media was prepared with RPMI 1640 medium (Lonza, Walkersville, MD, USA) supplemented with 5% or 10% heat-inactivated Fetal Bovine Serum (FBS) from Atlanta biologicals, Inc. (Flowery Branch, GA, USA), 2 mM of L-glutamine (Cellgro®, Mediatech, Inc., Manassas, VA, USA), 1X of Non-essential amino acids (Gibco®, life technologies, Grand Island, NY, USA), 1mM of Sodium pyruvate (Gibco®, life technologies, Grand Island, NY, USA), 10mM of 2-mercaptoethanol (Gibco®, Life Technologies, Grand Island, NY, USA), 100 U/ml of penicillin (Cellgro®, Mediatech, Inc., Manassas, VA, USA), and 100 mg/ml of streptomycin (CellgroR, Mediatech, Inc., Manassas, VA, USA). Culture media for Bone-marrow derived macrophages (BMM media) were made as follows: DMEM medium (Lonza, Walkersville, MD, USA), 10% of heat-inactivated FBS, 2 mM of L-glutamine, 10mM of 2-mercaptoethanol, 100 U/ml of penicillin, 100 mg/ml of streptomycin and 50 μg/ml of gentamycin (Quality Biological, Gaithersburg, MD, USA).
Isolation and culture of NK cells
Freshly isolated spleens were mashed in 5% fetal bovine serum (FBS) RPMI 1640 and passed through a cell strainer (BD) to obtain single-cell suspensions as previously described (23). The cell suspensions were pelleted by centrifugation. The red blood cells (RBC) were lysed from the pellet with ACK lysis buffer (Quality Biological, Gaithersburg, MD, USA). RBC-free cells were washed and resuspended in 5% FBS RPMI 1640. The suspensions were passed through a sterile, pre-wetted nylon wool column and incubated for 50 min at 37°C. Cells that were not bound to the nylon wool were eluted out with the 5% FBS RPMI 1640, washed, counted and routinely were between 70–95% NK1.1+CD3−. Cells were resuspended in 10% FBS RPMI media supplemented with high dose IL-2 (10,000 U/ml) and cultured for 6 days at a density of 1×106 per ml to obtain activated NK cells. Using this method for isolating/preparing NK cells we previously characterized the cultured NK cells (referred to through the manuscript as NK cells) to be phenotypically similar to freshly-isolated NK cells (23). At day 6 of culture, adherent cells were harvested from flasks with 1mM EDTA (Quality Biological, Gaithersburg, MD, USA), washed two times and starved for 3 hours to remove IL-2. The cells were seeded at a concentration of 1×106 cells/ml well in 6-well plates and stimulated with the following cytokine concentrations: IL-2 (100 UI/μl), IL-12 (10ng/ml), IL-4 (10ng/ml).
Isolation and culture of bone-marrow derived macrophages (BMM)
Bone-marrow cells were flushed from femurs and tibias of mice with the BMM culture media. The cells were then passed through a cell strainer, pelleted down and resuspended in the BMM media supplemented with 30% of L929-conditioned media from the American Type Culture collection (ATCC, Manassas, VA). Three ml of the cells at 1×106 per ml were seeded in a 6-well plate and maintained for 5 days with media change at day2 and day4. At day5, fully differentiated cells were washed three times with 1X PBS then maintained in BMM media overnight. Fresh media was added at Day6 and the cells were stimulated with LPS (100ng/ml), IL-4 (10ng/ml), or IL-13 (10ng/ml) alone or in combination.
mRNA and real time PCR
Total RNA was isolated with TRIzol® reagent (Ambion, life technologies, Carlsbad, CA, USA). One microgram of mRNA was used as a template to generate complementary DNA (cDNA) using SuperScript® First-strand Synthesis System (InvitrogrenTM, life technologies, Carlsbad, CA, USA). Quantitative real time PCR (RT-qPCR) was performed by SYBRGreen assay (Applied biosystems, life technologies, Grand Island, NY, USA). mRNA levels between samples were normalized to the murine β-2-microglobulin. The primers used for gene expression analysis were the following 5′-ACTTCAGCAGGCTGTGGG-3′ and 5′-GATGACATCACAAGCATCCG-3′ for mouse Il24, 5′-TCGGCCAGAGCCACATG-3′ and 5′-TTAAGGAGTCGGTTAGCAAGTATGTTG-3′ for mouse Il10, and 5′-AAATGCTGAAGAACGGGAAAA-3′ and 5′-ATAGAAAGACCAGTCCTTGCTGAAG-3′ for mouse β-2-microglobulin. Unless otherwise noted, data are shown as fold induction over non-stimulated cells (NS).
Chromatin Immunoprecipitation assays (ChIP)
ChIP assays were conducted as previously reported (24) using the EZ-Magna ChIP kit from Upstate Biotechnology (Millipore, Billerica, MA, USA) with minor modifications. NK cells and BMM were stimulated for 2h with the appropriate stimuli and fixed with 1% paraformaldehyde (Fisher Scientific, Fair Lawn, NJ, USA) for 10 min. Nuclei were isolated from fixed cells and sonicated in short bursts in the cold to shear the genomic DNA into manageable fragments. A sample of lysate was run on a 2% agarose gel to validate that the sonication steps yielded DNA fragments of the appropriate size. Sonicated DNA was diluted in assay diluant in the presence of protein A beads and protease inhibitors and immunoprecipated (IP) with specific antibodies for 4h at 4 ºC. DNA/ protein crosslinks were reversed by protease K digestion at 62ºC for 2h followed by incubation at 95ºC for 10min. DNA was then purified and used as template for qPCR. The primers used for qPCR are the followings: 5′-GGTCATGCTTCCCTGGAGAA-3′ and 5′-ACCCCCCTGTCTAAGAGCAAA-3′ for Site 0 published by Wei et. al. 2010 (10), 5′-CAGTTAACCCTGCTACCTTG-3′ and 5′-CAGGCCAACTTAAGCAG-3′ for Site 1, 5′-CTGCTTAAGTTGGCCTG-3′ and 5′-CATCAAGAGGTTCTAGACTC-3′ for Site 2, 5′-CCCCTGTGTGGTGTAGCTTCA-3′ and 5′-AAAGCCCTGCCTCTCATCCT-3′ for Site 3, 5′-CAGAGGCCATTCCACACA-3′ and 5′-GGGGTCAGGTATGTTAATG-3′ for Site 4. An IgG-IP control was included in each experiment and use to control for non-specific DNA binding. The results are shown as the percent input (% of Input).
Statistical analysis
Statistical differences between experimental groups were analyzed with the Mann-Whitney U test using GraphPad Prism software (version 5.0, GraphPad Software, Inc., La Jolla, CA). P values of ≥0.05 was considered statistically significant.
RESULTS
IL-24 and IL-10 are co-expressed in NK cells and macrophages but may be regulated by cell-specific mechanisms
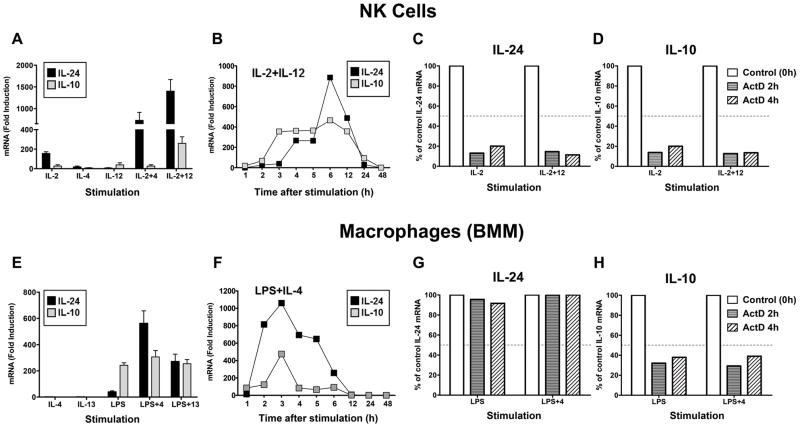
As mentioned, depending on the inflammatory trigger different IL-10-secreting cell types, including NK cells and macrophages, emerge to control host inflammatory responses. Regulation of IL-24 expression in these cells types however, is not well understood. Previously, we reported that IL-10 is regulated by IL-2 and IL-12 in NK cells (25). We found that IL-24 expression is also potently induced by these cytokines (particularly in combination) but unlike IL-10, IL-24 is synergistically upregulated by IL-2+IL-4 stimulation (Fig. 1A). We compared IL-24 and IL-10 mRNA expression patterns over time, under IL-2+IL-12 stimulation conditions, to determine if these genes are induced with similar kinetics. IL-10 expression peaked at 3h and was sustained until 12h before returning to near baseline by 24h post-stimulation (Fig. 1B). Interestingly, IL-24 mRNA appeared to be induced in two distinct phases in NK cells. The first phase occurred slightly later at 4h followed by a second burst of mRNA at 6h before returning to baseline by 24h (Fig. 1B).
Figure 1. IL-24 and IL-10 are co-expressed in cultured NK cells and BMM.
NK cells (A) were stimulated with the indicated cytokines for 6h and IL-24 (black) and IL-10 (gray) mRNA expression was determined by RT-qPCR analysis. Data represent the mean ± SEM of at least 4 independent experiments with 3–5 mice per group. (B) Kinetics of IL-24 and IL-10 mRNA expression in IL-2+IL-12-stimulated NK cells (one of two representative experiments with 3–5 mice per group). Data are presented as the relative fold increase (mRNA (Fold induction)) compared to non-stimulated cells which were assigned an arbitrary value of 1. (C, D) mRNA stability analysis of IL-24 (C) and IL-10 (D) in NK cells stimulated for 3h with IL-2 or IL-2+IL-12 (IL-2+12). Cells were then harvested (Control (0h)) or treated with actinomycin D (ActD) and incubated for and additional 2h (black bar) or 4h (gray bar). mRNA levels were measured by RT-qPCR and normalized to stimulated control cells that did not receive ActD (Control (0h). The data are presented as a percent of control of IL-24 or IL-10 mRNA expression which was assigned a value of 100%. Data are from one of two representative experiments with 3–5 mice per group. Dashed line indicates mRNA decay at 50% of the control. (E) Bone-marrow derived macrophages (BMM) were treated with the indicated stimuli for 3h and IL-24 and IL-10 mRNA expression was determined by RT-qPCR analysis. Data represent the mean ± SEM of at least 4 independent experiments with 3 mice per group. (F) Kinetics of IL-24 and IL-10 mRNA expression in LPS+IL-4-stimulated BMM (one of two representative experiments with 3 mice per group). Data are presented as the relative fold increase (mRNA (Fold induction)) compared to non-stimulated cells which were assigned an arbitrary value of 1. (G, H) mRNA stability analysis of IL-24 (G) and IL-10 (H) in BMMs stimulated for 2h with LPS or LPS+IL-4 (LPS+4). Cells were then harvested (Control (0h)) or treated with actinomycin D (ActD) and incubated for and additional 2h (black bar) or 4h (gray bar). mRNA levels were measured by RT-qPCR and normalized to stimulated control cells that did not receive ActD (Control (0h). The data are presented as a percent of control of IL-24 or IL-10 mRNA expression which was assigned a value of 100%. Data are from one of two representative experiments with 3 mice per group. Dashed line indicates mRNA decay at 50% of the control.
The accumulation of IL-24 transcripts after 6h may result from increased stability of the IL-24 mRNA which has been reported as a mechanism regulating IL-24 expression in keratinocytes (17). NK cells were stimulated as indicated for 3h (control) before treatment with the transcriptional inhibitor actinomycin D (ActD) and mRNA abundance was determined 2 and 4h later. Cytokine mRNA half-life was defined as the point at which mRNA levels fall to 50% or below of control levels (set to 100%) following ActD treatment. For both IL-24 (Fig. 1C) and IL-10 (Fig. 1D), mRNA stability in NK cells was relatively short (less than 2h) regardless of the stimulation conditions. Of note, although it is possible that cytokine receptor upregulation is a mechanism for amplifying cytokine-induced gene expression, we previously determined that these stimulation conditions do not significantly affect cytokine receptor expression (23). These data indicate that while induction of IL-24 mRNA appears to be bi-phasic, it is likely not due to stabilization of the mRNA suggesting that additional pathways may contribute to the regulation of IL-24 expression in NK cells.
In macrophages, LPS induced both IL-24 and IL-10 expression (Fig. 1E) however, LPS co-stimulation with IL-4 or, to a lesser extent, IL-13 was highly synergistic for IL-24 but not IL-10. Examination of gene expression kinetics under LPS+IL-4 stimulation revealed that both IL-24 and IL-10 mRNA levels peaked by 3h (Fig. 1F). IL-10 mRNA levels rapidly declined after 3h however, the level of IL-24 transcripts declined more slowly. Both returned to baseline by 12h after stimulation. Again, we considered if the greater abundance of IL-24 transcripts over time could be explained at least partly by stability of IL-24 mRNA. Interestingly, in contrast to NK cells, IL-24 mRNA levels remained highly stable for up to 4h in macrophages (Fig. 1G) while levels IL-10 mRNA were significantly decayed by 2h (Fig. 1H). Of note, although IL-4 was required for optimal IL-24 expression, stabilization of IL-24 transcripts occurred independently of IL-4. These findings suggest that IL-24 mRNA may be inherently more stable in macrophages and that effects of IL-4 on IL-24 expression are mediated by mechanisms other than mRNA stability. Together, these data indicate that IL-24 and IL-10 are induced by overlapping signaling pathways in NK cells and macrophages but may have developed separate mechanisms to fine-tune cell-specific gene expression.
STAT6 is required for IL-4-induced IL-24 expression in NK cells and BMM
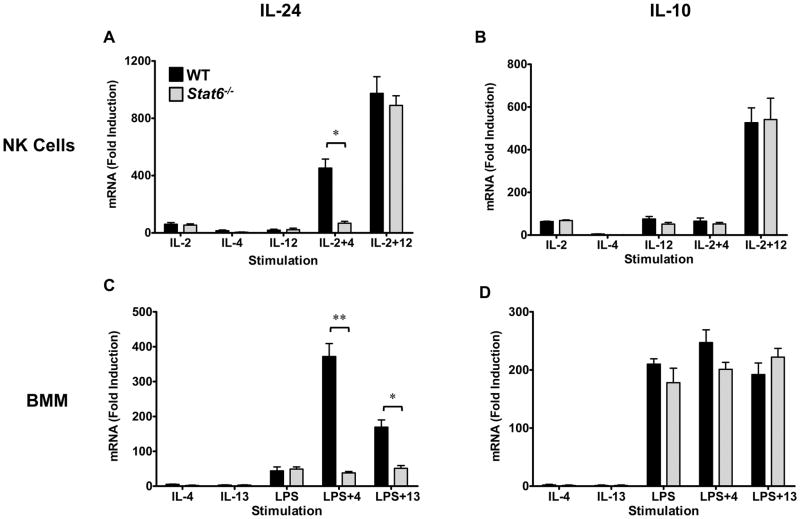
It is established that the IL-4/STAT6 pathway is a potent regulator of IL-24 expression in Th2 cells (6,8,10). Our data indicate that IL-4 also plays a role in IL-24, but not IL-10 expression in both NK cells and BMM (Fig. 1). We investigated the molecular basis of IL-4-induced IL-24 and IL-10 expression in NK cells and macrophages using Stat6−/− mice. In Stat6−/− NK cells, cytokine-induced IL-24 expression was largely intact while IL-10 expression was unaffected by STAT6 deficiency (Fig. 2A, B). However, the synergistic induction of IL-24 by IL-2+IL-4 stimulation was significantly reduced in the absence of STAT6 (Fig. 2A). In Stat6−/− macrophages, LPS-induced IL-24 remained intact but the synergistic activity of IL-4 and IL-13 was completely ablated (Fig. 2C). As in NK cells, STAT6 was largely dispensable for IL-10 expression in macrophages (Fig. 2D). These data indicate that the STAT6 pathway plays an important role in regulating IL-4/IL-13-induced IL-24 but is dispensable for IL-10 expression in both NK cells and macrophages.
Figure 2. STAT6 is required for IL-4-induced IL-24 in NK cells and BMM.
NK cells (A, B) and BMM (C, D) were generated from WT (black) and Stat6−/− mice (gray) in vitro and treated with the indicated stimuli for 6h (NK cells) or 3h (BMM). IL-24 (A, C) and IL-10 (B, D) mRNA expression was determined by RT-qPCR analysis. Data represent the mean ± SEM for 3 independent experiments with 3–5 mice per group, *p < 0.05, **p < 0.01 (Mann-Whitney U test). Data are presented as the relative fold increase (induction) compared to non-stimulated cells (for the respective strains), which were assigned a value of 1.
STAT4 is required for maximal IL-24 and IL-10 co-expression in NK cells but not BMM
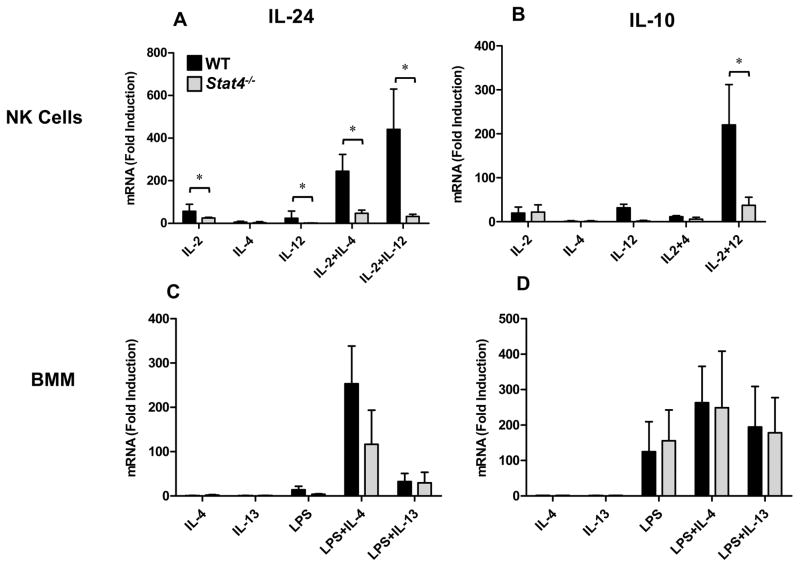
We previously demonstrated that IL-12-induced STAT4 is required for optimal induction of IL-10 expression in IL-2+IL-12-treated NK cells (25). Because IL-2+IL-12 co-stimulation also strongly induces IL-24 in NK cells, we hypothesized that STAT4 may regulate IL-24 expression under IL-12 stimulation conditions. Indeed, like IL-10, IL-24 expression was significantly diminished in Stat4−/− NK cells stimulated in the presence of IL-12 (Fig. 3A, B). Unexpectedly, the capacity for other cytokines (IL-2, IL-4) to induce IL-24 mRNA expression was also significantly reduced in the absence of STAT4 (Fig. 3A). Conversely, the effect of STAT4-deficiency on IL-10 expression was only evident under IL-12 stimulation conditions (Fig. 3B). In macrophages, STAT4-deficiency also resulted in an overall trend towards reduced IL-24 mRNA expression but was statistically different compared to WT controls (Fig. 3C). Notably, LPS-induced IL-10 expression was unaffected by STAT4 deficiency in BMM (Fig. 3D). These data suggest that IL-12-independent STAT4 signaling is required for NK-specific IL-24 expression.
Figure 3. STAT4 is essential for optimal induction of IL-24 and IL-10 in NK cells.
NK cells (A, B) and BMM (C, D) were generated from WT (black) and Stat4−/− mice (gray) in vitro and treated with the indicated stimuli for 6h (NK cells) or 3h (BMM). IL-24 (A, C) and IL-10 (B, D) mRNA expression was determined by RT-qPCR analysis. Data represent the mean ± SEM for 3 independent experiments with 3–5 mice per group, *p < 0.05, (Mann-Whitney U test). Data are presented as the relative fold increase (induction) compared to non-stimulated cells (for the respective strains), which were assigned a value of 1.
Cytokine-induced STAT recruitment to the Il24 gene in NK cells and macrophages
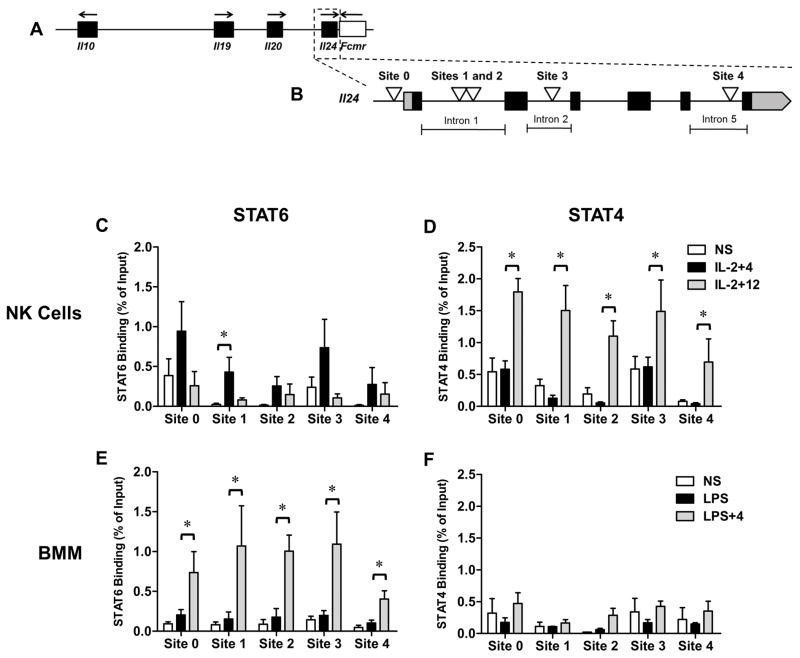
The Il24 gene, comprised of 6 exons and spanning ~5kb, flanks one end of the Il10 cluster on human/mouse chromosome 1 (Fig. 4A). Though a few STAT-motifs in Il24 have been described in T cells, relatively little is known about transcription factor recruitment to Il24 particularly in innate immune cells. Because STAT-regulated genes frequently contain multiple binding sites and can attract multiple STAT family members in a cell type-specific manner (26,27), we searched for additional STAT-binding elements around the Il24 gene utilizing bioinformatics approaches. In addition to the previously described STAT-binding motif in the proximal promoter (Site 0) (10) we identified 3 additional putative STAT-binging regions based on sequence homology. Sites 1 and 2 are separated by 23bp in intron 1 and positioned approximately 113bp 3′ to a previously described pair of tandem STAT motifs (28). Sites 3 and 4 are located in introns 2 and 5 of the Il24 gene respectively (Fig. 4B).
Figure 4. STAT6 and STAT4 recruitment to newly-identified sites the Il24 gene in NK cells and BMM.
(A) Genomic organization of the Il10 cluster (black boxes) including the 3′ flanking gene Fcmr spanning ~180kb of mouse chromosome 1. (B) Exon (black boxes)/intron structure of Il24 gene (~5.5kb) including UTRs (gray). STAT-binding sites are indicated by white triangles. Sites 1 and 2 are separated by 23bp. Genomic features not drawn to scale. (C) STAT6 and (D) STAT4 recruitment to Sites 0–4 of the Il24 gene in NK cells by ChIP. (E) STAT6 and (F) STAT4 recruitment to Sites 0–4 of the Il24 gene in BMM by ChIP. NS = non-stimulated. NK cells (C, D) or BMM (E, F) were stimulated for 2h with the indicated stimuli and ChIP analysis was performed. Anti-STAT6- or anti-STAT4-immunoprecipitated genomic DNA was subjected to RT-qPCR analysis to determine the enrichment of STAT proteins at the indicated sites. Data are presented as the relative percent of input DNA (% of Input) based on the mean ± SEM for 3–4 independent experiments with 3–5 mice per group, *p < 0.05, (Mann-Whitney U test). In each experiment an IgG IP control was used to normalize samples for non-specific DNA binding.
We examined STAT4 and STAT6 recruitment to these STAT sites in NK cells and macrophages by ChIP. In NK cells, co-stimulation with IL-4 (plus IL-2) induced STAT6 binding to all five STAT elements across the Il24 gene (Fig. 4C). The strongest enrichment of STAT6 occurred at Site 0 in the promoter followed by Site 3 in intron 2 however, only Site 1 in the 1st intron reached statistical significance. Sites 0 and 3 also displayed evidence of STAT6 binding in non-stimulated (NS) cells. Interestingly, STAT4 binding was identified at all five STAT sites in NK cells, even in the absence of IL-12 (Fig. 4D). In the presence of IL-12 however, STAT4 binding was highly enriched, particularly at Sites 0, 1 and 3. In macrophages, low levels of STAT6 binding was detected at each site under NS and LPS treatment conditions (Fig. 4E). Co-stimulation with IL-4 however, resulted in a strong induction of STAT6 binding across the locus with the weakest binding occurring at Site 4. Meanwhile, lower levels of STAT4 binding were observed in BMM that were largely unaffected by the stimulation condition (Fig 4F).
In addition to the sites described here, STAT5 has been shown to bind to a tandem repeat upstream of our Sites1/2 in the 1st intron of Il24 in T cells activated under non-polarizing conditions (28) and to a more distal upstream region in Th2 cells (29). We also found that IL-2 induced IL-24 (and to a lesser extent IL-10) in NK cells (Fig 1A) but did not observe appreciable STAT5 recruitment to the sites in Il24 tested here (data not shown). Overall, these results demonstrate that STAT6 and STAT4 are recruited to multiple sites within the endogenous Il24 gene in NK cells and macrophages.
STAT6 and STAT4 mediate cytokine-induced epigenetic modifications of Il24
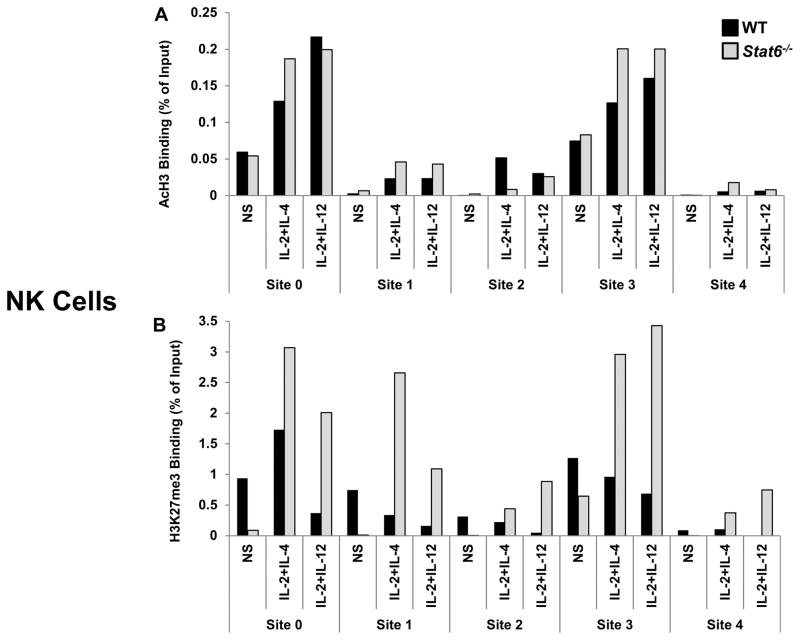
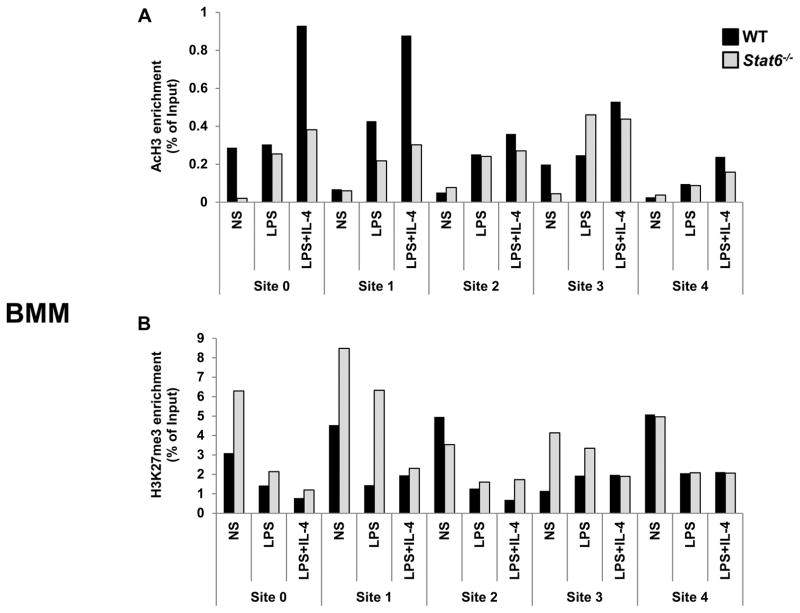
During Th2 differentiation, STAT6-dependent epigenetic remodeling of the Il24 gene occurs (8,10) but the role that STAT6 and/or STAT4 may play in other cell types have not be defined. Having established that STAT6 and STAT4 bind multiple regions of the Il24 gene in innate immune cells, we sought to examine the cell-specific impact of these STAT family members on epigenetic modifications in NK cells and macrophages. We evaluated post-translational modifications to histone proteins which are associated with permissive (acetyl-H3 [AcH3]) or repressive chromatin (H3K27me3) in WT, Stat6−/− and Stat4−/− cells by ChIP assay.
We first assessed STAT6. In NK cells, AcH3 was enriched at the promoter (Site 0) and Site 3 upon cytokine stimulation though deficiency in STAT6 had little impact on AcH3 levels (Fig. 5A). However, we observed an increased accumulation of the silent chromatin mark H3K27me3 in cytokine-stimulated Stat6−/− NK cells, most predominantly at Sites 0, 1, and 3 (Fig. 5B). In BMM, IL-4+LPS stimulation resulted in a sizeable increase of AcH3 marks specifically at Sites 0 and 1 which was dependent on STAT6 (Fig. 6A). The distribution of AcH3 across the other STAT sites in Il24 were largely similar with limited effects of stimulation in both WT and Stat6−/− cells. Site 0 and Site 1 also preferentially accumulated STAT6-dependent H3K27me3 marks in non-stimulated (NS) macrophages (Fig. 6B). Interestingly, the levels of repressive H3K27me3 marks in at Sites 0 and 1 were effectively reduced in LPS+IL-4-stimulated BMM from both WT and Stat6−/− mice.
Figure 5. STAT6-dependent epigenetic modifications of the Il24 gene in NK cells.
Cytokine-induced post-translational modifications of histone proteins in the Il24 gene in NK cells from WT (black) and Stat6−/− (gray) mice. Cells were either not stimulated (NS) or treated with the indicated stimuli. Levels of (A) AcH3 and (B) H3K27me3 were determined 2h after stimulation by ChIP analysis. Anti-acytlH3- or anti-H3K27me3-immunoprecipitated genomic DNA was subjected to RT-qPCR analysis to determine enrichment at the indicated sites. Data are presented as the relative percent of input DNA (% of Input) of one of two representative experiments with 3–5 mice per group. In each experiment an IgG IP control was used to normalize samples for non-specific DNA binding.
Figure 6. STAT6-dependent epigenetic modifications of the Il24 gene in BMM.
Activation-induced post-translational modifications of histone proteins in the Il24 gene in BMM from WT (black) and Stat6−/− (gray) mice. Cells were either not stimulated (NS) or treated with the indicated stimuli. Levels of (A) AcH3 and (B) H3K27me3 were determined after 2h stimulation by ChIP analysis. Anti-acytlH3- or anti-H3K27me3-immunoprecipitated genomic DNA was subjected to RT-qPCR analysis to determine enrichment at the indicated sites. Data are presented as the relative percent of input DNA (% of Input) of one of two representative experiments with 3 mice per group. In each experiment an IgG IP control was used to normalize samples for non-specific DNA binding.
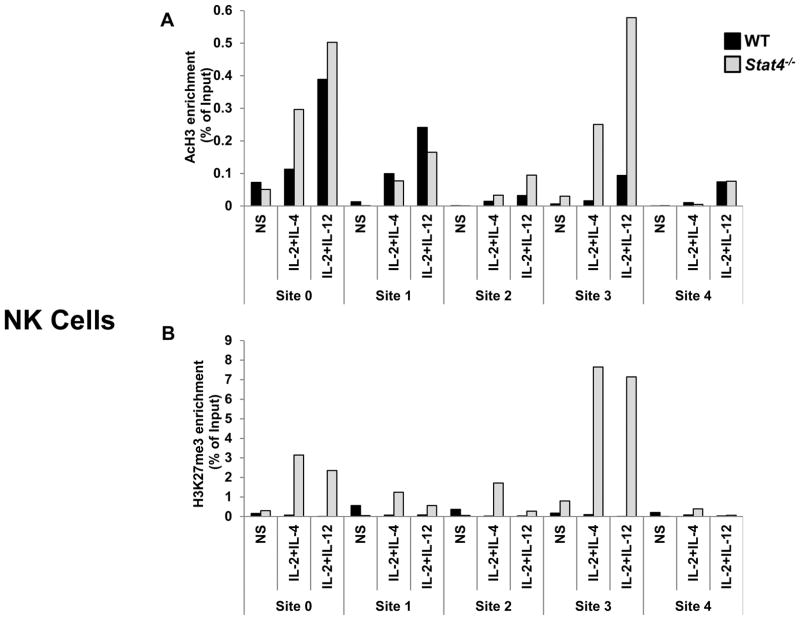
Next, we assessed the role of STAT4 in regulating epigenetic modifications to Il24. Because there was little evidence of stimulation-induced STAT4 recruitment in macrophages (Fig. 4F) and no significant differences in IL-24 expression were observed in STAT4-deficient BMM (Fig. 3C), we restricted our analyses of STAT4 to NK cells. As before, cytokine stimulation resulted in the enrichment of AcH3 marks predominantly at Sites 0 and 3 (Fig. 7A). Although STAT6-deficiency had little impact on AcH3 in NK cells (Fig. 5A), unexpectedly AcH3 levels were enhanced mostly at Site 3 in Stat4−/− NK cells. Similar to STAT6-deficient NK cells, cytokine stimulation induced a large enrichment of repressive H3K27me3 marks in Stat4−/− NK cells but were particularly concentrated at Sites 0 and 3 (Fig. 7B). Of note, this effect was observed even in the absence of IL-12 stimulation. Thus, STAT4 and STAT6 play distinct roles in IL-24 regulation in innate immune cells through binding to multiple sites in the gene and mediating epigenetic tuning of the surrounding chromatin. However, in NK cells, STAT4 and STAT6 acted cooperatively to block the accumulation of repressive H3K27me3 marks. Taken together, these data suggest that, as in Th2 cells, STAT6 is required for IL-4-induced IL-24 expression in macrophages and NK cells. Meanwhile, STAT4 is broadly required for IL-24 expression in NK cells, however, largely dispensable in BMM.
Figure 7. STAT4-dependent epigenetic modifications of the Il24 gene in NK cells.
Cytokine-induced post-translational modifications of histone proteins in the Il24 gene in NK cells from WT (black) and Stat4−/− (gray) mice. Cells were either not stimulated (NS) or treated with the indicated stimuli. Levels of (A) AcH3 and (B) H3K27me3 were determined 2h after stimulation by ChIP analysis. Anti-acytlH3- or anti-H3K27me3-immunoprecipitated genomic DNA was subjected to RT-qPCR analysis to determine enrichment at the indicated sites. Data are presented as the relative percent of input DNA (% of Input) of one of two representative experiments with 3–5 mice per group. In each experiment an IgG IP control was used to normalize samples for non-specific DNA binding.
Cell type-specific type I interferon signaling differentially regulate IL-24 and IL-10 in innate immune cells
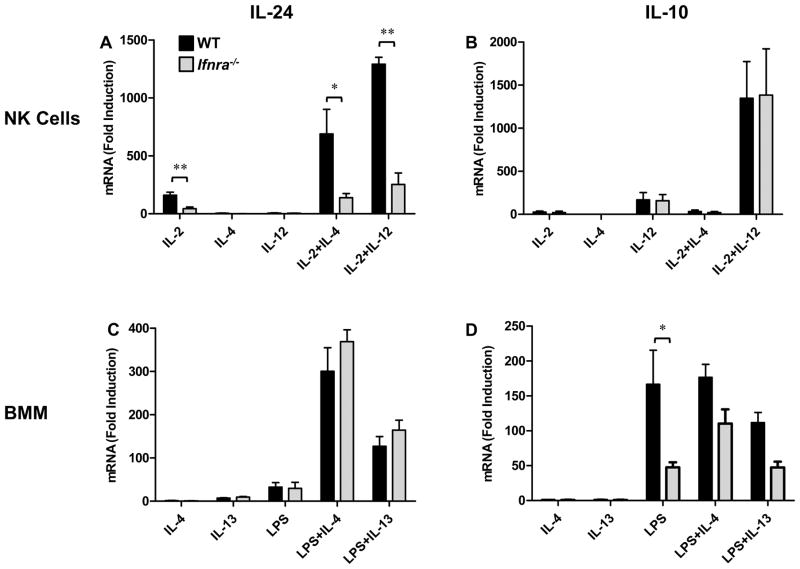
We were intrigued that IL-24 expression in NK cells was dependent on STAT4 even in the absence of IL-12 stimulation (Fig. 3A). We considered the possibility that an intermediary cytokine-induced autocrine pathway was involved in IL-24 regulation in NK cells. We examined the role of type I interferons in regulating IL-24 based on the following: 1) IL-10 expression in macrophages is partially dependent on type I interferons (30), 2) type I IFN receptor signaling can activate the STAT4 pathway (31); 3) NK cells are not an endogenous source of IL-12 (32), and 4) IFN-β has been shown to induce IL-24 expression in melanoma cells (14).
IL-24 expression from type I IFN receptor-α chain deficient (Ifnra−/−) NK cells was dramatically impaired under all treatment conditions, including co-stimulation with IL-12 (Fig. 8A). Interestingly, induction of IL-10 in NK cells was independent of type I IFN signaling (Fig. 8B). Remarkably, the role(s) of type I IFN signaling in macrophage-specific IL-24 and IL-10 expression were reversed. As reported previously, induction of IL-24 was intact in Ifnra−/− BMM, while LPS-induced IL-10 was substantially reduced (Fig. 8C, D respectively) (30). We also examined the capacity of type I IFN to directly induce cell type-specific IL-24 and IL-10 expression. In NK cells, IFN-β alone did not induce IL-24 but synergized with IL-2 to enhance IL-24 mRNA expression (Supplemental Figure 1A). This suggests that type I IFN receptor signaling alone is insufficient to regulate IL-24 in NK cells. Interestingly, although IFN-γ is also regulated by the type I IFN/STAT4 pathway in NK cells, direct stimulation with type I IFNs (IFN-α or IFN-β) was insufficient for IFN-γ induction (33,34). IFN-β had little effect on IL-10 expression in NK cells. In contrast, IFN-β-induced IL-10 but not IL-24 expression in macrophages (Supplemental Figure 1B). Taken together, these data suggest that, though Il10 and Il24 are in close genomic proximity and can be co-expressed, they are controlled by distinct cell-specific IFN signaling pathways operating in macrophages and NK cells (Fig. 9A, B respectively).
Figure 8. Type I IFN receptor signaling differentially controls IL-24 and IL-10 expression in innate immune cells.
NK cells (A, B) and BMM (C, D) prepared from WT (black) and Ifnra−/− (gray) mice were stimulated with the indicated cytokines for 6h. IL-24 (A, C) and IL-10 (B, D) mRNA expression was determined by RT-qPCR analysis. Data represent the mean ± SEM for 3 independent experiments with 3–5 mice per group, *p < 0.05, **p < 0.01 (Mann-Whitney U test). Data are presented as the relative fold increase (induction) compared to non-stimulated cells (for each respective mouse strain) which were assigned a value of 1.
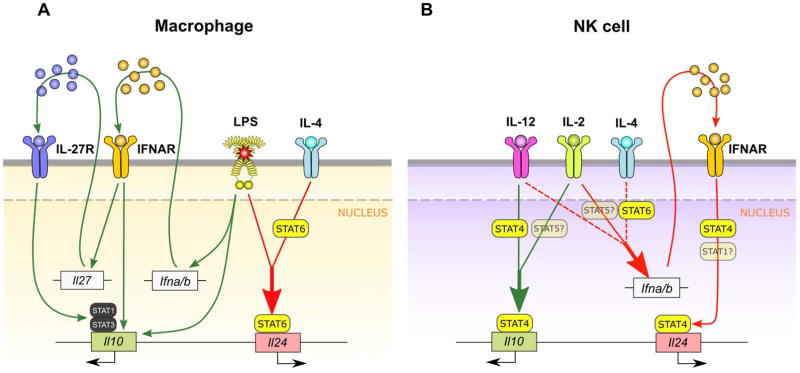
Figure 9. Model of cell type-specific type regulation of IL-24 and IL-10 in NK cells and macrophages.
Pathways leading to IL-24 induction shown with red arrows. Pathways leading to IL-10 induction shown with green arrows. (A) In macrophages, work from other groups has shown that optimal LPS-induced IL-10 expression requires signaling through both the type I IFN and IL-27 receptors (30, 49). We show that optimal LPS induction of IL-24 in macrophages is entirely independent of type I IFN receptor signaling but requires co-stimulation through the IL-4/STAT6 pathway. LPS+IL-4 stimulation induces STAT6 recruitment to multiple regions within Il24 and STAT6-dependent chromatin remodeling of the Il24 locus. Enhanced stability of IL-24 mRNA in BMM occurs independently of the IL-4/STAT6 pathway (not shown). Of note, we also determined that type I IFN receptor-mediated regulation of IL-10 in BMM is independent of STAT4. (B) In NK cells however, we identified a reversal of type I IFN’s broad role in regulating IL-10 and IL-24. Though IL-10 induction is completely independent of the type I IFN receptor, cytokine-induced IL-24 expression broadly requires both the type I IFN receptor and STAT4. The dependence on type I IFNs and STAT4 is evident even in the absence of IL-12 co-stimulation suggesting that type I IFN receptor signals through STAT4 to regulate IL-24 expression in NK cells. The alternate type I IFN/STAT4 pathway has been shown previously to regulate key functions in NK cell biology (31, 33, 47). Of note, we cannot rule out a role for IL-2-induced STAT5 or type I IFN-induced STAT1 (semitransparent) in NK-specific IL-24 or IL-10 regulation. Also, STAT4 but not type I IFN receptor, is required for the synergistic induction of IL-10 by IL-2+IL-12 (25).
DISCUSSION
Cytokines play key roles in coordinating host immune responses and must be tightly regulated in order to maintain effective immune defenses. Cytokine genes are frequently organized in clusters and considerable effort has been focused on defining the molecular basis of cytokine expression patterns as a means to understand immune development and disease risk. Intense study of the Il4 and Ifng loci has revealed important clues about cytokine gene structure/function/regulatory relationship(s) and has also offered a means to understand the molecular basis of disease risk.
Comparatively little is known regarding the genomic control of cell specific expression of the genes within the Il10 cluster (35,36). In addition, the biological properties of IL-19, IL-20 and IL-24, though still being defined, appear to have limited overlap with the anti-inflammatory activities of IL-10 (35). Of these Il10 homologs, IL-24 has been studied more intently with distinct, sometimes opposing functions compared to IL-10. For example, IL-24’s role as a tumor suppressor is well-documented (37,38) and IL-10 has been shown to antagonize this activity (39). In addition, accumulating evidence suggests that IL-24 contributes to pathological conditions in the skin (18,40,41). These functional differences coupled with the fact that IL-20 subfamily cytokine receptors are largely restricted to epithelial cells (35,40) suggests that IL-10 expression is regulated independently from IL-20 subfamily cytokines including its neighbors IL-19, IL-20 and IL-24.
IL-24 and IL-10 can be uniquely co-expressed however, most notably in Th2 cells (6) which prompted us to explore the possibility that Il24 and Il10 may share cell type-specific regulatory mechanisms. To address this, we first examined whether IL-24 and IL-10 are co-expressed in other immune cells. Since macrophage- and NK-derived IL-10 plays a distinct role in regulating host responses to inflammation and infection, we focused on NK cells and macrophages. In both NK cells and BMM, a primary characteristic distinguishing between regulation of IL-24 and IL-10 was responsiveness to IL-4. Although IL-4 alone was insufficient to induce IL-24 or IL-10 mRNA expression in either NK cells or macrophages (Fig 1A, E), IL-4 acted synergistically with IL-2 or LPS-, inducing IL-24 expression in NK cells and BMM respectively, while having little or no effect on IL-10. While IL-24 regulation has not been extensively studied in innate immune cells (42), LPS-induced IL-24 expression has been observed in human monocytes (11,12,43,44) and rat macrophages (43). In the latter, IL-4 stimulation alone modestly induced IL-24 expression in rat alveolar macrophages (43). A recent report indicated that IL-4 induces IL-24 expression in normal human airway epithelial cells. The authors also reported significantly elevated levels of IL-24 in nasal scrapings from asthmatic patients when compared to healthy controls (45). In line with our findings, others have reported that IL-4 stimulation failed to induce IL-24 expression in human monocytes or NK cells (11). Notably, the ability of IL-4 to synergize with other stimuli, such as LPS, was not examined in these studies. Overall, these data indicate that the IL-4/STAT6 pathway cooperatively regulates IL-24 expression in BMM and NK cells by amplifying gene induction signals delivered through other receptors and is largely dispensable for IL-10 expression.
STAT6 regulates both Il10 and Il24 in Th2 cells, however, Il24 is considered a Th2 lineage-specifying gene and as such is actively repressed by STAT4 in Th1 cells. Il10 on the other hand is one of only a few genes that can be positively regulated by both STAT4 and STAT6 in Th1 and Th2 cells respectively (6,8,10). Therefore, our observation that STAT4 is required for NK cell-specific IL-24 expression was particularly unexpected. To our knowledge, the STAT4 pathway has not been linked to positive IL-24 regulation. However, a recent study reported that dogs undergoing IL-12 gene therapy for cancer had elevated levels of IL-24 (as well as IFN-γ and IL-10) in the serum (46). Previously we reported a role for STAT4 in regulating IL-10 expression in NK cells exclusively under IL-12 stimulation conditions (25). Surprisingly, STAT4-deficiency resulted in broadly impaired IL-24 expression in NK cells. With that said, it is important to note that although we did not detect appreciable binding of STAT5 to the Il24 gene at the time point tested, we cannot exclude a role for STAT5 in regulating IL-24 and/or IL-10 in NK cells (Figure 9B). Additional studies will be required to determine if/how STAT5 contributes to IL-24 and IL-10 expression.
Our data suggested that an alternate IL-12-independent, STAT4-dependent pathway operates in NK cells to broadly control IL-24 expression. We suspected the type I IFN pathway for several reasons: 1) type I IFN receptor signaling in NK cells induces STAT4 phosphorylation (31,34,47), 2) type I IFNs regulate numerous NK cell functions some of which are dependent on STAT4 (48), and 3) IL-24 was originally identified through a subtractive hybridization screen of IFN-β-stimulated melanoma cells (14). In addition, type I IFNs have an established role in regulating IL-10 expression in macrophages (30,49).
Although type I IFNs can regulate both IL-24 and IL-10 under certain conditions, we were intrigued by the distinctive cell type-specific requirements for type I IFN signaling by IL-24 and IL-10 in NK cells and macrophages respectively. Consistent with other reports, LPS-induced IL-10 expression in macrophages was dependent, at least in part, on signaling through the type I IFN receptor (Fig. 8D) (30), while the induction of IL-24 remained intact in Ifnra−/− BMMs (Fig. 8C). Conversely, induction of IL-10 in NK cells was completely independent of type I IFN signaling while IL-24 expression was broadly impaired in NK cells from Ifnra−/− mice (Fig. 8B, A respectively). Importantly, type I IFN receptor signaling selectively co-opted the STAT4 pathway in NK cells to regulate IL-24 as evidenced by similar IL-24 expression patterns observed in Ifnra−/− and Stat4−/− NK cells (Fig. 8A and 3A respectively). Of note, elegant work by others has demonstrated that type I IFNs selectively engage different STAT family members in NK cells to fine-tune the response to viral infection. Specifically, STAT4 is more highly expressed than STAT1 in NK cells and is in fact constitutively associated with the type I IFN receptor (47). Upon NK cell activation, type I IFNs induce IFN-γ expression through a STAT4-dependent, STAT1-/STAT2-independent mechanism (33). However, as STAT1 levels rise, NK cell type I IFN receptors switch from STAT4 to STAT1 utilization which results in the downregulation of IFN-γ (31,47). Thus, IL-24 and IFN-γ, but not IL-10, are regulated by the IFN-STAT4 pathway in NK cells and though it is likely that similar to IFN-γ, induction of IL-24 is STAT1-independent, we cannot rule out this possibility at this time (Figure 9B). The remarkable cell type specificity of IFN signaling networks is supported by the fact that type I IFN receptor-mediated regulation of IL-10 in macrophages is independent of STAT4 (Fig. 3D). Thus, our findings indicate that although IL-24 and IL-10 are similarly expressed in innate immune cells, they are under the control of distinct, cell type-specific signaling pathways operating through the type I IFN receptor.
Based on their close proximity, structural similarity (50) and their co-regulation by STAT6 in Th2 cells (10), we hypothesized that the Il24 and Il10 genes are co-regulated in other cell types by common pathways. Our findings indicate that despite sharing overlapping expression patterns in NK cells and macrophages, Il24 and Il10 have evolved distinct regulatory requirements (Fig. 9). As such, Il24 and Il10 are “co-expressed” but not “co-regulated” in these innate immune cells. Taken together, these data provide the molecular basis for IL-24 and IL-10 co-expression in NK cells and macrophages and may offer evolutionary insight into regulation of the Il10 gene cluster. In this regard, it is important to note that while the significance of cell type-specific IL-10 expression programs has been well-documented (20,51,52), the biological roles of IL-24 are still being explored (5). In addition to it’s known antitumor activities, recent studies suggest that IL-24 contributes to skin immunopathology (18,40) however, the cellular sources of IL-24 in these contexts are not well-defined. Conversely, a biological role for IL-24 in Th2 immunity has yet to be defined despite the fact that IL-24 is a Th2 lineage-defining gene (6,10). Thus, together with our findings, it is tempting to speculate that there are additional, yet to be defined roles for IL-24 in immune regulation that are mediated by different IL-24-expressing cell subsets. Characterizing the molecular mechanisms that regulate the expression of IL-24 and other members of the IL-10 family will provide further insight into the biological roles these cytokines play in the etiology/pathology of inflammatory diseases and may point to new therapeutic strategies.
Supplementary Material
Acknowledgments
We are grateful to Stephen K. Anderson and Howard A. Young for helpful discussions. We thank Dr. Young for providing Ifnr−/− mice and for review of this manuscript, Jinxia Ma, Palak Shah and Jason Huska for lab assistance, and the Becton Dickinson Immune Function Laboratory at the Johns Hopkins Bloomberg School of Public Health for technical assistance.
This work was supported by National Institutes of Health Grants R01 AI070594 and RO1 AI113910 (to J.H.B.) and DFG Research Fellowship (He5507/1-1) to C.M.H.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
Author contributions: D.D., C.M.H. and J.H.B. designed the research; D.D., J.H., F.W., V.V, C.M.H. and J.H.B. performed the experiments, analyzed the data and prepared figures. D.D. and J.H.B. prepared and wrote the manuscript. All authors have given approval to the final version of the manuscript.
Reference List
- 1.Palstra RJ, de LW, Grosveld F. Beta-globin regulation and long-range interactions. Adv Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 2.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 3.Falvo JV, Jasenosky LD, Kruidenier L, Goldfeld AE. Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation. Adv Immunol. 2013;118:37–128. doi: 10.1016/B978-0-12-407708-9.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins PL, Henderson MA, Aune TM. Lineage-specific adjacent IFNG and IL26 genes share a common distal enhancer element. Genes Immun. 2012;13:481–488. doi: 10.1038/gene.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutz S, Wang X, Ouyang W. The IL-20 subfamily of cytokines--from host defence to tissue homeostasis. Nat Rev Immunol. 2014;14:783–795. doi: 10.1038/nri3766. [DOI] [PubMed] [Google Scholar]
- 6.Schaefer G, Venkataraman C, Schindler U. Cutting edge: FISP (IL-4-induced secreted protein), a novel cytokine-like molecule secreted by Th2 cells. J Immunol. 2001;166:5859–5863. doi: 10.4049/jimmunol.166.10.5859. [DOI] [PubMed] [Google Scholar]
- 7.Moore KW, De Waal MR, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 8.Sahoo A, Lee CG, Jash A, Son JS, Kim G, Kwon HK, So JS, Im SH. Stat6 and c-Jun mediate Th2 cell-specific IL-24 gene expression. J Immunol. 2011;186:4098–4109. doi: 10.4049/jimmunol.1002620. [DOI] [PubMed] [Google Scholar]
- 9.Mendel I, Shevach EM. The IL-10-producing competence of Th2 cells generated in vitro is IL-4 dependent. Eur J Immunol. 2002;32:3216–3224. doi: 10.1002/1521-4141(200211)32:11<3216::AID-IMMU3216>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 10.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, Peng W, O’Shea JJ, Kanno Y. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poindexter NJ, Walch ET, Chada S, Grimm EA. Cytokine induction of interleukin-24 in human peripheral blood mononuclear cells. J Leukoc Biol. 2005;78:745–752. doi: 10.1189/jlb.0205116. [DOI] [PubMed] [Google Scholar]
- 12.Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol. 2002;168:5397–5402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- 13.Maarof G, Bouchet-Delbos L, Gary-Gouy H, Durand-Gasselin I, Krzysiek R, Dalloul A. Interleukin-24 inhibits the plasma cell differentiation program in human germinal center B cells. Blood. 2010;115:1718–1726. doi: 10.1182/blood-2009-05-220251. [DOI] [PubMed] [Google Scholar]
- 14.Jiang H, Lin JJ, Su ZZ, Goldstein NI, Fisher PB. Subtraction hybridization identifies a novel melanoma differentiation associated gene, mda-7, modulated during human melanoma differentiation, growth and progression. Oncogene. 1995;11:2477–2486. [PubMed] [Google Scholar]
- 15.Jen EY, Poindexter NJ, Farnsworth ES, Grimm EA. IL-2 regulates the expression of the tumor suppressor IL-24 in melanoma cells. Melanoma Res. 2012;22:19–29. doi: 10.1097/CMR.0b013e32834d2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunz S, Wolk K, Witte E, Witte K, Doecke WD, Volk HD, Sterry W, Asadullah K, Sabat R. Interleukin (IL)-19, IL-20 and IL-24 are produced by and act on keratinocytes and are distinct from classical ILs. Exp Dermatol. 2006;15:991–1004. doi: 10.1111/j.1600-0625.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- 17.Otkjaer K, Holtmann H, Kragstrup TW, Paludan SR, Johansen C, Gaestel M, Kragballe K, Iversen L. The p38 MAPK regulates IL-24 expression by stabilization of the 3′ UTR of IL-24 mRNA. PLoS One. 2010;5:e8671. doi: 10.1371/journal.pone.0008671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumari S, Bonnet MC, Ulvmar MH, Wolk K, Karagianni N, Witte E, Uthoff-Hachenberg C, Renauld JC, Kollias G, Toftgard R, Sabat R, Pasparakis M, Haase I. Tumor necrosis factor receptor signaling in keratinocytes triggers interleukin-24-dependent psoriasis-like skin inflammation in mice. Immunity. 2013;39:899–911. doi: 10.1016/j.immuni.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Hedrich CM, Bream JH. Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol Res. 2010 doi: 10.1007/s12026-009-8150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siewe L, Bollati-Fogolin M, Wickenhauser C, Krieg T, Muller W, Roers A. Interleukin-10 derived from macrophages and/or neutrophils regulates the inflammatory response to LPS but not the response to CpG DNA. Eur J Immunol. 2006;36:3248–3255. doi: 10.1002/eji.200636012. [DOI] [PubMed] [Google Scholar]
- 21.Lee SH, Kim KS, Fodil-Cornu N, Vidal SM, Biron CA. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J Exp Med. 2009;206:2235–2251. doi: 10.1084/jem.20082387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perona-Wright G, Mohrs K, Szaba FM, Kummer LW, Madan R, Karp CL, Johnson LL, Smiley ST, Mohrs M. Systemic but not local infections elicit immunosuppressive IL-10 production by natural killer cells. Cell Host Microbe. 2009;6:503–512. doi: 10.1016/j.chom.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bream JH, Curiel RE, Yu CR, Egwuagu CE, Grusby MJ, Aune TM, Young HA. IL-4 synergistically enhances both IL-2- and IL-12-induced IFN-gamma expression in murine NK cells. Blood. 2003;102:207–214. doi: 10.1182/blood-2002-08-2602. [DOI] [PubMed] [Google Scholar]
- 24.Ranatunga DC, Ramakrishnan A, Uprety P, Wang F, Zhang H, Margolick JB, Brayton C, Bream JH. A protective role for human IL-10-expressing CD4+ T cells in colitis. J Immunol. 2012;189:1243–1252. doi: 10.4049/jimmunol.1103421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant LR, Yao ZJ, Hedrich CM, Wang F, Moorthy A, Wilson K, Ranatunga D, Bream JH. Stat4-dependent, T-bet-independent regulation of IL-10 in NK cells. Genes Immun. 2008 doi: 10.1038/gene.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang K, Robinson GW, Hennighausen L. Comprehensive meta-analysis of Signal Transducers and Activators of Transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genomics. 2013;14:4. doi: 10.1186/1471-2164-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villarino AV, Kanno Y, O’Shea JJ. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol. 2017;18:374–384. doi: 10.1038/ni.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin JX, Li P, Liu D, Jin HT, He J, Ata Ur RM, Rochman Y, Wang L, Cui K, Liu C, Kelsall BL, Ahmed R, Leonard WJ. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity. 2012;36:586–599. doi: 10.1016/j.immuni.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wurster AL, Precht P, Becker KG, Wood WH, III, Zhang Y, Wang Z, Pazin MJ. IL-10 transcription is negatively regulated by BAF180, a component of the SWI/SNF chromatin remodeling enzyme. BMC Immunol. 2012;13:9. doi: 10.1186/1471-2172-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer SS, Ghaffari AA, Cheng G. Lipopolysaccharide-mediated IL-10 transcriptional regulation requires sequential induction of type I IFNs and IL-27 in macrophages. J Immunol. 2010;185:6599–6607. doi: 10.4049/jimmunol.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack EA, Kallal LE, Demers DA, Biron CA. Type 1 interferon induction of natural killer cell gamma interferon production for defense during lymphocytic choriomeningitis virus infection. MBio. 2011:2. doi: 10.1128/mBio.00169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trinchieri G, Gerosa F. Immunoregulation by interleukin-12. J Leukoc Biol. 1996;59:505–511. doi: 10.1002/jlb.59.4.505. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 34.Freudenberg MA, Merlin T, Kalis C, Chvatchko Y, Stubig H, Galanos C. Cutting edge: a murine, IL-12-independent pathway of IFN-gamma induction by gram-negative bacteria based on STAT4 activation by Type I IFN and IL-18 signaling. J Immunol. 2002;169:1665–1668. doi: 10.4049/jimmunol.169.4.1665. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann SR, Rosen-Wolff A, Tsokos GC, Hedrich CM. Biological properties and regulation of IL-10 related cytokines and their contribution to autoimmune disease and tissue injury. Clin Immunol. 2012;143:116–127. doi: 10.1016/j.clim.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Ekmekcioglu S, Mumm JB, Udtha M, Chada S, Grimm EA. Killing of human melanoma cells induced by activation of class I interferon-regulated signaling pathways via MDA-7/IL-24. Cytokine. 2008;43:34–44. doi: 10.1016/j.cyto.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menezes ME, Bhatia S, Bhoopathi P, Das SK, Emdad L, Dasgupta S, Dent P, Wang XY, Sarkar D, Fisher PB. MDA-7/IL-24: multifunctional cancer killing cytokine. Adv Exp Med Biol. 2014;818:127–153. doi: 10.1007/978-1-4471-6458-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng M, Bocangel D, Doneske B, Mhashilkar A, Ramesh R, Hunt KK, Ekmekcioglu S, Sutton RB, Poindexter N, Grimm EA, Chada S. Human interleukin 24 (MDA-7/IL-24) protein kills breast cancer cells via the IL-20 receptor and is antagonized by IL-10. Cancer Immunol Immunother. 2007;56:205–215. doi: 10.1007/s00262-006-0175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myles IA, Fontecilla NM, Valdez PA, Vithayathil PJ, Naik S, Belkaid Y, Ouyang W, Datta SK. Signaling via the IL-20 receptor inhibits cutaneous production of IL-1beta and IL-17A to promote infection with methicillin-resistant Staphylococcus aureus. Nat Immunol. 2013;14:804–811. doi: 10.1038/ni.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shefler I, Pasmanik-Chor M, Kidron D, Mekori YA, Hershko AY. T cell-derived microvesicles induce mast cell production of IL-24: relevance to inflammatory skin diseases. J Allergy Clin Immunol. 2014;133:217–224. doi: 10.1016/j.jaci.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 42.Sahoo A, Im SH. Molecular Mechanisms Governing IL-24 Gene Expression. Immune Netw. 2012;12:1–7. doi: 10.4110/in.2012.12.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garn H, Schmidt A, Grau V, Stumpf S, Kaufmann A, Becker M, Gemsa D, Siese A. IL-24 is expressed by rat and human macrophages. Immunobiology. 2002;205:321–334. doi: 10.1078/0171-2985-00135. [DOI] [PubMed] [Google Scholar]
- 44.Nagalakshmi ML, Murphy E, McClanahan T, De Waal MR. Expression patterns of IL-10 ligand and receptor gene families provide leads for biological characterization. Int Immunopharmacol. 2004;4:577–592. doi: 10.1016/j.intimp.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Zissler UM, Chaker AM, Effner R, Ulrich M, Guerth F, Piontek G, Dietz K, Regn M, Knapp B, Theis FJ, Heine H, Suttner K, Schmidt-Weber CB. Interleukin-4 and interferon-gamma orchestrate an epithelial polarization in the airways. Mucosal Immunol. 2016;9:917–926. doi: 10.1038/mi.2015.110. [DOI] [PubMed] [Google Scholar]
- 46.Cicchelero L, Denies S, Haers H, Vanderperren K, Stock E, Van BL, de RH, Sanders NN. Intratumoural interleukin 12 gene therapy stimulates the immune system and decreases angiogenesis in dogs with spontaneous cancer. Vet Comp Oncol. 2016 doi: 10.1111/vco.12255. [DOI] [PubMed] [Google Scholar]
- 47.Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J Exp Med. 2007;204:2383–2396. doi: 10.1084/jem.20070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang EY, Guo B, Doyle SE, Cheng G. Cutting edge: involvement of the type I IFN production and signaling pathway in lipopolysaccharide-induced IL-10 production. J Immunol. 2007;178:6705–6709. doi: 10.4049/jimmunol.178.11.6705. [DOI] [PubMed] [Google Scholar]
- 50.Zdanov A. Structural analysis of cytokines comprising the IL-10 family. Cytokine Growth Factor Rev. 2010;21:325–330. doi: 10.1016/j.cytogfr.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roers A, Siewe L, Strittmatter E, Deckert M, Schluter D, Stenzel W, Gruber AD, Krieg T, Rajewsky K, Muller W. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med. 2004;200:1289–1297. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabrysova L, Howes A, Saraiva M, O’Garra A. The regulation of IL-10 expression. Curr Top Microbiol Immunol. 2014;380:157–190. doi: 10.1007/978-3-662-43492-5_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.