Abstract
Malaria remains a major burden on global health, with roughly 200 million cases worldwide and more than 400,000 deaths per year. Besides biomedical research and political efforts, modern information technology is playing a key role in many attempts at fighting the disease. One of the barriers toward a successful mortality reduction has been inadequate malaria diagnosis in particular. To improve diagnosis, image analysis software and machine learning methods have been used to quantify parasitemia in microscopic blood slides. This article gives an overview of these techniques and discusses the current developments in image analysis and machine learning for microscopic malaria diagnosis. We organize the different approaches published in the literature according to the techniques used for imaging, image preprocessing, parasite detection and cell segmentation, feature computation, and automatic cell classification. Readers will find the different techniques listed in tables, with the relevant articles cited next to them, for both thin and thick blood smear images. We also discussed the latest developments in sections devoted to deep learning and smartphone technology for future malaria diagnosis.
INTRODUCTION
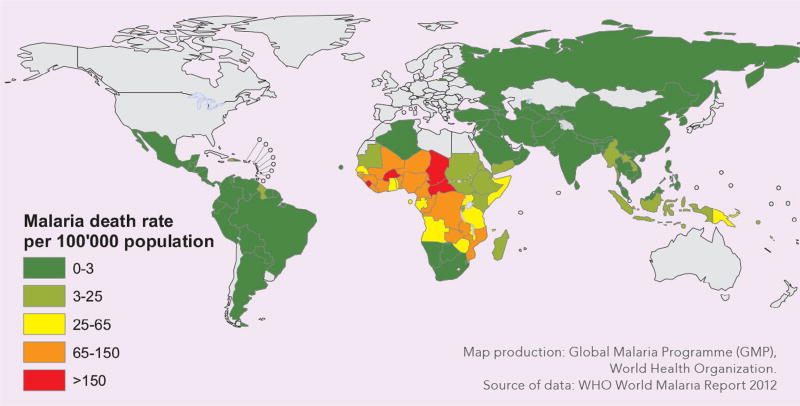
Malaria is caused by protozoan parasites of the genus Plasmodium that are transmitted through the bites of infected female Anopheles mosquitoes and that infect the red blood cells. Most deaths occur among children in Africa, where a child dies almost every minute from malaria, and where malaria is a leading cause of childhood neuro-disability. According to the World Malaria Report 2016,1 an estimated 3.2 billion people in 95 countries and territories are at risk of being infected with malaria and developing disease, and 1.2 billion are at high risk (>1 in 1000 chance of getting malaria in a year). There were about 214 million cases of malaria globally in 2016 and about 438,000 malaria deaths. The burden was heaviest in the African region, where an estimated 92%2 of all malaria deaths occurred, and in children aged under 5 years, who accounted for more than two thirds of all deaths (see also the malaria death rates from an earlier WHO report in Fig 1). Typical symptoms of malaria include fever, fatigue, headaches, and, in severe cases, seizures and coma, leading to death.
Fig 1.
Worldwide malaria death rates (Source: WHO World Malaria Report 2012).
Hundreds of millions of blood films are examined every year for malaria, which involves manual counting of parasites and infected red blood cells by a trained microscopist. Accurate parasite counts are essential not only for malaria diagnosis. They are also important for testing for drug-resistance, measuring drug-effectiveness, and classifying disease severity. However, microscopic diagnostics is not standardized and depends heavily on the experience and skill of the microscopist.1 It is common for microscopists in low-resource settings to work in isolation, with no rigorous system in place that can ensure the maintenance of their skills and thus diagnostic quality.1 This leads to incorrect diagnostic decisions in the field.1 For false-negative cases, this leads to unnecessary use of antibiotics, a second consultation, lost days of work, and in some cases progression into severe malaria. For false-positive cases, a misdiagnosis entails unnecessary use of anti-malaria drugs and suffering from their potential side effects, such as nausea, abdominal pain, diarrhea, and sometimes severe complications.
This sober analysis of malaria diagnosis has prompted efforts to perform malaria diagnosis automatically. Automatic parasite counting has several advantages compared with manual counting: (1) it provides a more reliable and standardized interpretation of blood films, (2) it allows more patients to be served by reducing the workload of the malaria field workers, and (3) it can reduce diagnostic costs. Several key processing steps are typically required to quantify parasitemia automatically. First, digital blood slide images need to be acquired, which often requires preprocessing to normalize for lighting or staining variations. In a second step, blood cells or parasites need to be detected. For blood cells, this typically implies cell segmentation to identify individual cells in cell clumps to obtain accurate cell counts. In a third step, after cell detection and segmentation, features are computed to describe the typical visual appearance of infected and uninfected blood cells. In a final classification step, a classifier, who has been trained on an independent and typically manually annotated training set, then discriminates between infected and uninfected cells. Once the number of infected and uninfected cells is known, computation of parasitemia is a straightforward mathematical equation, which includes clinical parameters such as hematocrit value, for example.
The prospects of automating malaria diagnosis with its obvious advantages has attracted many researchers, especially in the last decade. The publications reflect all the major developments we have seen in the areas of automatic pattern recognition and machine learning in the last years. Our article will give an overview of the articles that have been published, using the processing steps mentioned above as a framework and guide. This is not the first survey article on the subject. In fact, several survey articles have already been published before, which bear testimony to both the importance of automated malaria diagnosis and the research dynamics and rapid system development. We refer readers in particular to the following surveys for additional information about the background of automatic malaria diagnosis and the image processing and machine learning methods used for automated microscopy diagnosis of malaria.3–5 In addition, more specific surveys have been published on cell features for malaria parasite detection,6 on malaria diagnosis,7 on malaria diagnostic tools,8 and on alternatives to conventional microscopy.9 The purpose of our article is not to replace these surveys, but rather to complement them and to provide the latest update of the state of the art in image analysis and machine learning for malaria diagnosis as it presents itself at the end of the year 2017. With about 170 literature citations, we have collected more references compared with the other surveys. We had the goal to include also maybe lesser known publications to provide a historical documentation of the work done. In addition, we included a section on deep learning, which is the latest development in malaria diagnosis and which arguably has the potential to render many of the old approaches obsolete, similar to the development in other imaging application areas. There have also been many developments in hardware for automatic malaria diagnosis, which are however out of the scope of this article and deserve a separate article14,17,66,67,138. Nevertheless, we devote a section to rapid diagnostic tests (RDTs) for malaria diagnosis because they are also widely used in the field. The bulk of our articles have been collected from the Journal of Microscopy, Malaria Journal, and PLOS ONE, including a few articles from Nature and others. We have also collected publications from Institute of Electrical and Electronics Engineers (IEEE) conferences and other proceedings published by Springer and Elsevier. Furthermore, we have organized the articles into sections for preprocessing, cell detection and segmentation, feature computation, and classification. We have also added a separate section about deep learning and an extensive section about mobile smartphone applications for malaria diagnosis. A discussion of the latest developments and our conclusion mark the end of this article.
MALARIA
There are 5 Plasmodium species that cause malaria in human: Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale, and Plasmodium knowlesi. The 2 most common species are P. falciparum and P. vivax. P. falciparum is the most severe form and is responsible for most malaria-related deaths globally.1
P. falciparum is the most prevalent malaria parasite in sub-Saharan Africa, accounting for 99% of estimated malaria cases in 2016. Outside of Africa, P. vivax is the predominant parasite in the WHO Region of the Americas, representing 64% of malaria cases, and is above 30% in the WHO Southeast Asia and 40% in the Eastern Mediterranean regions.10
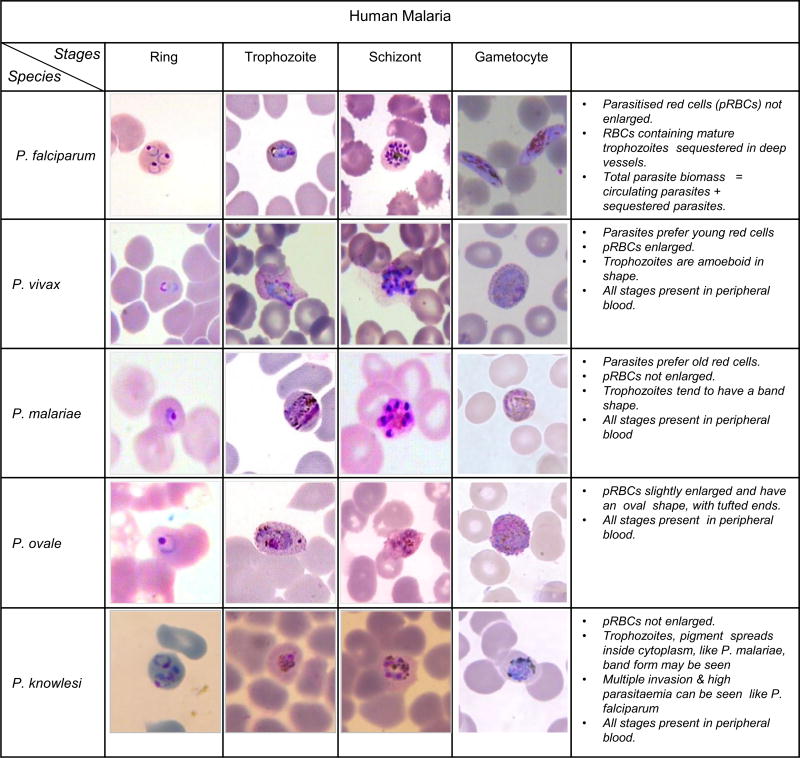
Each of these parasite species goes through stages during their development cycle (48 hours), which gives the parasites a different visual appearance that can be observed under the microscope. In chronologic order, these stages are the ring stage, trophozoite stage, schizont stage, and gametocyte stage. Fig 2 shows typical examples of all stages for each species.
Fig 2.
Five different human malaria Plasmodium species and their life stages in thin blood film (Source: K. Silamut and CDC).
In nonsevere malaria, mostly the young stages (<24 hours old) of P. falciparum are present in the peripheral blood, whereas for severe malaria all stages can be present in the peripheral blood. For P. falciparum, the trophozoite-infected red blood cells disappear from the peripheral blood circulation by attachment to the walls of capillaries inside vital organs, which is a process called sequestration. If the capillaries are blocked for newly infected cells by already attached cells, more mature parasite stages (trophozoites and schizonts) will be visible in the peripheral blood, which indicates a severe infection and a bad prognosis.
For P. falciparum, ring stages have a visible cytoplasm and 1 or 2 small chromatin dots. The infected blood cells are not enlarged but can feature multiple infections. P. falciparum trophozoites are rarely seen in peripheral blood smears. The cytoplasm of mature trophozoites tends to be more dense than younger rings, trophozoites can appear round in shape with brown malarial pigment inside, (Centers for Disease Control and Prevention (CDC)). P. falciparum schizonts are also seldomly seen in peripheral blood. They are displaying more than 2 and up to 32 nuclei (merozoites) with dark brown pigment clumped in the middle. Gametocytes of P. falciparum have a crescent or sausage shape, and can be seen in the blood smear 1 week after a parasite infection. The chromatin is visible as a single mass or is diffuse. For more information about P. falciparum morphology, see for example References11,12. Similar observations can be made for the stages of the other parasite species. For example, for P. vivax, host cells are often enlarged and have irregular shape. Trophozoites are amoeboid in shape with malaria pigment seen, and for severe infections multiple infections of single blood cells are not uncommon. For P. malariae, host cells are not enlarged. Trophozoites have a strong tendency to form a band with malarial pigment scattered along across the diameter of infected red blood cells. Multiple infections are extremely rare for P. malariae. On the other hand, for P. ovale, host cells are slightly enlarged and have an oval shape with tufted ends, often fimbriated. Parasites are slightly enlarged and trophozoites are amoeboid in shape with malarial pigment. Multiple infections of a single cell are more common than for P. vivax. For P. knowlesi, infected red blood cells do not appear enlarged. The parasite erythocytic cycle is only 24 hours, which is shorter than P. falciparum’s cycle (48 hours) and much shorter than P. malariae’s cycle (72 hours), which will lead to the same stage seen in peripheral blood every day at a given time. The morphology of P. knowlesi parasites is similar to P. malariae. Trophozoites can feature malarial pigment spread inside, band form may be seen like P. malariae, but their cytoplasm is more irregular, and multiple parasites infecting 1 single red blood cell can be seen like in P. falciparum.
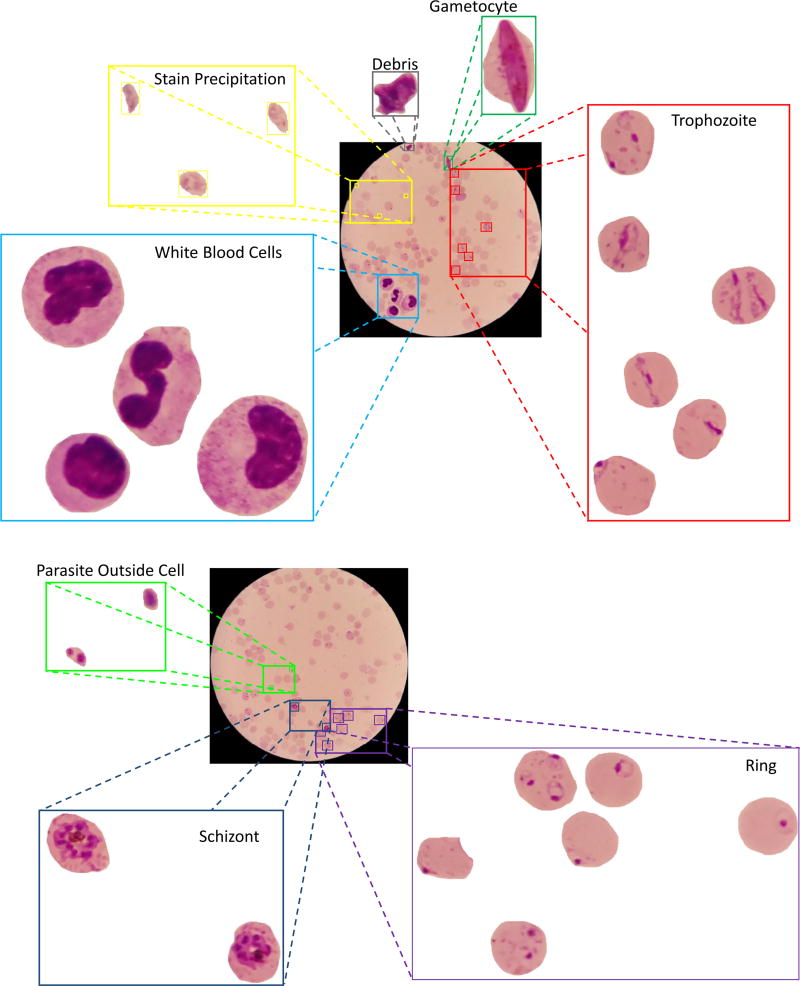
Fig 3 shows 2 examples of different parasite stages in the same thin blood slide image. In the first slide image, P. falciparum trophozoites and gametocytes can be seen together with white blood cells. The latter are larger and have a pronounced nucleus compared with the many red blood cells in the image. In the second image, P. falciparum ring stages are together with schizonts. In addition, other objects such as parasite outside cells and staining noise are visible in both images. Staining noise in particular can be confused with parasites by an unexperienced microscopist.
Fig 3.
Parasite stages in a single thin blood smear.
MALARIA DIAGNOSIS
Malaria is a curable disease, with drugs available for treatment, including drugs that can help prevent malaria infections in travelers to malaria-prone regions. However, there exists no effective vaccine against malaria yet, although this is an area of active research and field studies. Once infected, malaria is a rapidly progressing disease, with a serious risk of developing into severe and cerebral malaria with neurologic symptoms for P. falciparum infections. Therefore, a timely diagnosis of malaria is very important. Although malaria can be diagnosed in many different ways, there is room for improvement for current malaria diagnostic tests including reducing cost, increasing specificity, and improving ease of use. Because automated malaria diagnosis for resource-poor settings is the main topic of this survey, we have devoted 2 subsections to light microscopy and RDTs, which are by far the 2 most heavily used diagnostic means in these areas. We also briefly discuss the other options for malaria diagnosis, although they are arguably less suited for the conditions in remote malaria regions. For more information about malaria diagnosis, we refer readers to the surveys in Ref 7 and 9 and the following references:8,13,14.
Detecting the presence of parasites is the key to malaria diagnosis. In addition, identifying the parasite species and presence of potentially mixed infections is important, as well as the observation of the stage development of P. falciparum parasites in relation to the severity of the disease. Counting parasites for determining the level of parasitemia is not only important for identifying an infection and measuring its severity, it also allows monitoring patients by measuring drug efficacy and potential drug resistance.
Light microscopy
The current gold-standard method for malaria diagnosis in the field is light microscopy of blood films, which is the main focus of this article. Although other forms of diagnosis exist and have become popular in recent years, in particular RDTs, microscopy remains the most popular diagnostic tool, especially in resource-poor settings. With microscopy, all parasite species can be detected. It allows computing the level of parasitemia, clearing a patient after a successful treatment, and monitoring drug resistance. Furthermore, it is less expensive than other methods and widely available. However, its biggest disadvantages are the extensive training required for a microscopist to become a proficient malaria slide reader, the high cost of training and employing, maintaining skills, and the large component of manual work involved.
To diagnose malaria under a microscope, a drop of the patient’s blood is applied to a glass slide, which is then immersed in a staining solution to make parasites more easily visible under a conventional light microscope, usually with a 100× oil objective. Two different types of blood smears are typically prepared for malaria diagnosis: thick and thin smears.15 A thick smear is used to detect the presence of parasites in a drop of blood. Thick smears allow a more efficient detection of parasites than thin smears, with an 11 times higher sensitivity.5 On the other hand, thin smears, which are the result of spreading the drop of blood across the glass slide, have other advantages. They allow the examiner to identify malaria species and recognize parasite stages more easily.
The actual microscopic examination of a single blood slide, including quantitative parasite detection and species identification, takes a trained microscopist 15–30 minutes. Considering that hundreds of thousands of blood slides are manually inspected for malaria every year, this amounts to a huge economic effort required for malaria diagnosis.
Rapid diagnostic tests
The main advantage of microscopic malaria diagnosis lies in its low direct cost, which gives it a distinct advantage in resource-poor settings.1 Other existing diagnostic methods, and any new method, have to prove that they can provide the same ease of use and price point as microscopy given the limited financial resources typically available in malaria-prone regions. Arguably the only and main competitor in this sense are RDTs. They detect evidence of malaria parasites (antigens) and take about 10–15 minutes to process. Their detection sensitivity is lower but comparable with manual microscopy, and they do not require any special equipment and require only minimal training.
Although RDTs are currently more expensive than microscopy in high-burden areas,16 a valid question is whether these tests can replace microscopy in the near future. At the time of this writing, according to WHO,1 more countries use microscopy more than they use RDTs.2 RDTs are used more in rural areas where microscopy is not available. About 47% of malaria tests in malaria endemic countries worldwide were made by RDT.2
The use of RDTs, however, does not eliminate the need for malaria microscopy. A major disadvantage is that RDTs do not provide quantification of the results. Therefore, at this point in time, microscopy and RDTs are more complementing each other than one replacing the other.
Other tests
Several methods for diagnosing malaria are available. Important criteria are cost per test, sensitivity and specificity of the method, time per test, and the required skill level of the user. Furthermore, quantification of the number of infected red blood cells is important as a prognostic indicator.17
Polymerase chain reaction (PCR). A molecular method called PCR has shown higher sensitivity and specificity than conventional microscopic examination of stained peripheral blood smears.7 In fact, it is considered the most accurate among all tests. It can detect very low parasite concentrations in the blood and can differentiate species. However, PCR is a complex high-cost technology that takes many hours to process by trained staff. According to Tangpukdee et al.,7 PCR is not routinely implemented in developing countries because of the complexity of the testing and the lack of resources to perform these tests adequately and routinely. Quality control and equipment maintenance are also essential for the PCR technique, so that it may not be suitable for malaria diagnosis in remote rural areas or even in routine clinical diagnostic settings.
Fluorescent microscopy. Quantitative buffy coat is a laboratory test to detect infection with malaria or other blood parasites, using fluorescent microscopy. A fluorescent dye makes parasites visible under ultraviolet light. According to Adeoye and Nga,18 this test is more sensitive than the conventional thick smear. Nowadays, portable fluorescent microscopes with fluorescent reagent to label parasites, are available commercially. Although the quantitative buffy coat technique is simple, reliable, and user friendly, it requires specialized instrumentation, is more costly than conventional light microscopy, and is poor at determining species and numbers of parasites.7
Flow cytometry. This is a laser-based cell counting and detection method that allows to profile thousands of cells per second. Although flow cytometry offers automated parasitemia counts, this is offset by a rather low sensitivity. Flow cytometry is less suitable as a diagnostic technique in the field, when a direct answer is required for treatment decisions. However, in developed countries, it can be applied in the clinical setting for accurate counting of parasite numbers, for instance in the follow-up of drug treatment.19
STAINING METHODS
More than 100 years ago, Giemsas stain (1902) was applied for the first time for the diagnosis of malaria. Since then, it received increased attention. Because of its low cost, its high sensitivity, and specificity, it is currently widely used in microscopical malaria examinations.20 However, Giemsa staining requires multiple reagents, experienced personal, and is labor-intensive and time-consuming (it typically requires at least 45 minutes to stain a slide20).
Other stains have been used, too, like Field stain that significantly reduces the staining time, although it requires drying of samples before and during staining.21 However there are also disadvantages with Field’s stain, especially in under-resourced health centers in which the stain might be used. Poor blood preparations often result in the generation of artifacts commonly mistaken for malaria parasites, such as bacteria, fungi, stain precipitation, dirt, and cell debris. These can frequently cause false-positive readings.
Another stain is Leishman’s stain (1901), which has a high sensitivity, is cheap, and relatively easy to perform. Among the other stains being used is, for example, the Wright-Giemsa stain, which is a combination of Wright and Giemsa stain, and where the former facilitates the differentiation of blood cell types.
In 1970s, Sodeman et al.22 investigated the effect of fluorochrome staining in identifying the malaria parasites at low-level infection. It has been shown that fluorochrome staining is more sensitive and less time-consuming than Romanowsky and Giemsa staining methods23–25 but requires considerable practice and training, and suffers from artifacts including photobleaching and phototoxicity.26,27 Moreover, fluorescence microscopes are more expensive than standard light microscopes, which is a factor in tropical resource-poor regions where malaria is endemic.22,24,28
Table I shows the blood smear types and staining techniques used for the approaches published in the literature. Clearly, the vast majority of publications has been for thin smears. Certainly, 1 reason for this lies in the fact that thin smears allow to determine the parasite species and stages more easily, in addition to the parasitemia. So, in some sense, thin smears are more versatile and contain more information. Another important reason is probably that the presence of red blood cells gives the problem of parasite detection more structure, and makes the problem easier to a certain degree, as parasites need to be detected only inside cells. For thick films, parasite detection may be harder because of noise and staining artifacts that can lead to false positives. Nevertheless, because of the importance of thick smears for practical malaria diagnosis, it is very likely that more approaches for thick films will be implemented in the future. However, if convincing optical hardware solutions are found to scan multiple fields in thin smears and achieve a sensitivity comparable with thick smears, then this may be a moot point.44,142
Table I.
Blood smear types and staining methods for malaria diagnosis
| Blood smear |
Staining |
|---|---|
| Thin | Giemsa8,19,29–106 |
| Leishman98,107–120 | |
| Leishman-Methylene blue121 | |
| Combination of DNA and RNA fluorescent122 | |
| Wright123–125 | |
| Fluorochrome13,22,24,25,28,126 | |
| Romanowsky23 | |
| Acridine orange (AO)17 | |
| DAPI/Mitotracker127 | |
| Toluidine blue14 | |
| Unstained128–131 | |
| Thick | Giemsa8,55,132–143 |
| Leishman98 |
Table I also shows that the majority of approaches, for both thin and thick smears, have adopted the most popular stain in practice, Giemsa. Although stains like Leishman provide very good results for malaria parasites, Giemsa stain has proved to be the best all-round stain for the routine diagnosis of malaria. It has the disadvantage of being relatively expensive, but this is outweighed by its stability over time and its consistent staining quality over a wide range of temperatures.
AUTOMATED DIAGNOSIS OF MALARIA
This section provides the core information of our survey, namely a compilation of references that should cover the vast majority of articles ever published on automated microscopy for malaria diagnosis, with the bulk of the articles published in the last 10 years. The work that has been done in this area is quite diverse. Nevertheless, a system for automated cell microscopy usually implements a sequence of key processing steps that can serve as a guideline. Therefore, each of the following subsections will focus on 1 specific aspect of the processing pipeline.
The first step is usually the acquisition of digital images of blood smears, which largely depends on the equipment and materials being use. The Image acquisition section breaks down the different approaches for the different types of microscopy, blood slides (thin or thick), and staining.
Following image acquisition, most systems perform one or several preprocessing methods to remove noise and to normalize lighting and color variations inherent in the image acquisition and staining process. The Preprocessing section sorts the publications according to the preprocessing methods implemented.
The next step usually involves the detection and segmentation (outlining) of individual blood cells and maybe other objects that can be visible in a blood slide image, such as parasites or platelets. The section titled Red blood cell detection and segmentation gives an overview of all the segmentation methods that have been used for microscopic malaria diagnosis.
For most articles, cell segmentation is followed by the computation of a set of features, which describe the visual appearance of the segmented objects in a mathematical succinct way. The section titled Feature extraction and selection presents the different features and potential feature selection strategies that can be found in the literature.
In the last step, a mathematical discrimination method that classifies the segmented objects into different classes based on the computed features is implemented. For example, labeling each red blood cell as either infected or uninfected is a key classification task performed in this step, which then allows to compute the parasitemia. The section titled Parasite identification and labeling lists all the classification methods used in the literature for malaria diagnosis.
Later in the article, in the section titled Deep Learning, we will present references for the latest classification trend, deep learning, which skips the feature computation step and sometimes even the segmentation step. Furthermore, in the section titled Mobile Smatphones for Malaria Diagnosis, we will discuss how smartphones can be used for microscopic malaria diagnosis and list the systems that have already been implemented and published.
Image acquisition
Table II lists all published systems according to the type of microscopy used. Because light microscopy is the most common form of malaria diagnosis in resource-poor settings, where automation will also have the largest impact on health care and economy, it is not surprising that most authors implemented systems for standard microscopy. We have also added all other imaging techniques that we found in the literature and for which automated systems have been developed. For more detailed information about these approaches, we refer to the references listed in the table and the reference list at the end of this article.7–9,13,14,159
Table II.
Malaria image acquisition
| Imaging techniques |
|---|
| Light microscopy30–32,35–60,63–65,68–70,72–77,79–87,89,90,92,94,95,97–99,103–106,108–112,114,116–121,124,125,129,133,135–137,139–155 |
| Binocolor microscopy71,91,100,101 |
| Fluorescent microscopy13,22,24,25,28,126,127 |
| Polarized microscopy156 |
| Multi-spectral and multi-modal microscopy131,157 |
| Image-based cytometer29 |
| Sub-pixel resolving optofluidic microscopy (SROFM)14 |
| Quantitative phase imaging (QPI)128 |
| Quantitative cartridge-scanner system17 |
| Scanning electron microscopy (SEM)130 |
| Fiber array-based Raman imaging61,158 |
| Serial block-face scanning electron microscopy (SBFSEM)62 |
| SightDx digital imaging scanning66 |
Preprocessing
Table III lists all preprocessing approaches that have been applied to automatic analysis of digital blood slide images.
Table III.
Image preprocessing techniques applied to enhance malaria blood smear images
| Blood smear |
Challenges | Preprocessing methods | Remarks |
|---|---|---|---|
| Thin | Noise reduction | Mean filtering88,160 | |
| Median filtering29,31,34,36,39–42,45–48,63,65,71,73,87,103,108,114,116,117,119,147 | Remove impulse noise and preserve edges | ||
| Geometric mean filtering112,161 | |||
| Wiener filtering57 | |||
| Gamma equalization147 | |||
| SUSAN nonlinear filtering91,100 | |||
| Gaussian low-pass filtering69,95,135 | |||
| Nonlinear diffusion filtering58 | |||
| Gamma transformation123 | |||
| Interscale orthogonal wavelet-based thresholding162 | |||
| Perona-Malik denoising model50 | |||
| Morphological operations36,40,41,45,54,60,81,84,90,104,115,119,124,153 | Remove unwanted small objects, hole filling, closing and opening | ||
| Low image contrast | Laplacian filtering46,65,76 | Edge detection | |
| Adaptive/local histogram equalization46,47,50,64,68,82,87,133,135,163 | Enhance image resolution | ||
| Forward discrete curvelet transform87 | |||
| Contrast stretching techniques39,49,119,133,134 | Contrast enhancement | ||
| Uneven illumination | Low-pass filtering59,60,77,135 | Remove high frequency components | |
| Morphological top-hat operation60,90,104 | Remove nonuniform illumination effects | ||
| Cell staining variation | Linear model35 | ||
| Color normalization85 | Illumination correction | ||
| Gray world color normalization33,79,86,93,112,114,116,161 | Normalization of image color profile | ||
| Thick | Noise reduction | Median filtering136,139 | |
| Contrast enhancement133,134 | |||
| Gaussian low-pass filter61 | |||
| Histogram Equalization61 | |||
| Laplacian spatial filter142 |
Preprocessing is mainly applied to improve the quality of the image and to reduce variations in the images that would unnecessarily complicate the subsequent processing steps. Three key objectives can be identified: noise removal, contrast improvement, illumination and staining correction.
For noise removal, the most popular approaches have been well-established filters, such as mean and median filters, or Gaussian low-pass filtering. In addition, applying morphologic operations is very popular. For contrast improvement, contrast stretching techniques and histogram equalization in particular, have been the most popular approaches. For illumination and staining variations, color normalization techniques have been applied, including the popular use of grayscale colors.
Red blood cell detection and segmentation
Table IV shows the different segmentation techniques applied to thin smears. The vast majority of these techniques are thresholding techniques, such as Otsu thresholding in combination with morphologic operations. However, these techniques may not be dominating because of their superior performance compared with other methods, but rather because of their relative simplicity. Other methods include Hough transform, which makes assumptions about the blood cell shape, and unsupervised k-means pixel clustering. Cell segmentation needs to be accurate to compute the correct parasitemia. However, touching cells in particular complicate the identification and segmentation of individual cells. For this problem, methods like watershed and active contours have been applied.
Table IV.
Segmentation techniques for thin blood smears
| Blood smear |
Segmentation techniques | Remarks |
|---|---|---|
| Thin | Otsu thresholding36,40,46–48,57,65,81,103,104,108,118–120,127,144,164 | Calculates optimum threshold assuming that image contains bimodal histogram |
| (Adaptive) histogram thresholding29,35,42,44,50,53,71,75,89,96,107,124–126,128–130,139,150 | Difficult to determine the thresholding value | |
| Zack thresholding115 | Triangle-based method particularly effective with a weak peak in the image histogram | |
| Poisson distribution thresholding102 | Finding a threshold that separates foreground and background using minimum error | |
| Morphological operation32,34,37,38,41,43,45,60,63,74,84,85,87,90,101,135,160,165 | Mathematical morphology operations including granulometry, opening, closing, etc. | |
| Edge detection algorithm64,82,149 | Works well for high-contrast images with sharp edges, false edge detections should be filtered out | |
| Hough transform44,69,124,125,129,163 | Requires red blood cells circular measures including radius, shape | |
| K-means clustering39,49,83,166 | Unsupervised learning technique that iteratively assigns pixels to K clusters using their feature descriptors | |
| Watershed algorithm72,81,105,145,165 | Extract continuous boundary regions but oversegmentation is the typical issue | |
| Marker-controlled watershed108,111,112,114,116,130,161,164 | Mostly applied to separate touching cells | |
| Active contour models52,68,111,113,167 | Level-set based approaches that ensures topological flexibility, computationally expensive | |
| Rule-based segmentation64 | Requires knowledge about cells shape, size, color, etc. | |
| Fuzzy rule-based segmentation95 | Building rules is not easy when uncertainty is high | |
| Fuzzy divergence segmentation109,117 | ||
| Neural network106 | Requires discriminative and strong features to distinguish foreground and background pixels | |
| Template matching35 | ||
| Adaptive Gaussian mixture model distance transform73 | ||
| Distance transform168 | ||
| Ada-boost17 | ||
| Look-up table77 | ||
| Normalized-cut algorithms162 | Computationally expensive |
Table V shows the different segmentation techniques in the literature for thick smears. The segmentation situation for thick smears is different in that white blood cells and parasites need to be segmented. However, white blood cells are bigger than red blood cells and have more texture, which makes their segmentation much easier. Furthermore, white blood cells just need to be identified and not to be processed or classified further. In addition, parasites are very small and their reliable identification is most important. Therefore, the detection of these objects is practically more important than their segmentation, which may explain again the dominance of thresholding techniques and morphologic operations.
Table V.
Segmentation techniques for thick blood smears
| Blood smear | Segmentation techniques | Remarks |
|---|---|---|
| Thick | Otsu thresholding104,136 | Calculates optimum threshold assuming that image contains 2 classes following bimodal histogram |
| Histogram threshold132,135,137,141–143 | Difficult to determine the thresholding value, usually fused with other methods to improve performance | |
| Morphological operations104 | Mathematical morphology operations including granulometry, opening, closing, etc. are useful to characterize and represent blood cells circular shape, size, boundaries, skeletons, texture, gradient, etc. | |
| Fuzzy C-means147 |
Feature extraction and selection
Table VI lists the different features used in the literature to describe the appearance of red blood cells, infected and uninfected, in thin smears. Obviously, because parasites have been stained, color features are most natural and indeed used by many articles. In addition, several texture and morphologic features have been used to describe the inside of red blood cells. The idea is that in case of infected cells, these features can pick up the typical appearance of ring structures with visible cytoplasm and other unique parasite characteristics. Generally speaking, most of the features used are tried and trusted features that have already been applied in other, often nonmedical, application domains. For example, Haralick’s texture features, local binary patterns, co-occurence matrices, histogram of gradients, and many others have been successfully used across a wide range of applications. This also includes morphologic shape features and moments.
Table VI.
Feature computation for malaria parasite classification in thin blood smears6
| Blood smear |
Features type |
Feature | Remarks |
|---|---|---|---|
| Thin | Color | RGB14,23,29,30,40,44,45,59,60,63,72–75,77–79,81–84,93,95,97,103,106,114,120,123,126,131,141,150,154,162,167 | Provide color information |
| HSV38,39,42,47,52,53,59,95,107,115,162 | |||
| YCbCr160,162,166 | |||
| LAB49,57,95 | |||
| Intensity36,41,43,46,48,59,68,69,85,90,99,111,131,153 | |||
| Color correlogram, color co-occurrence matrix35,79,93,119 | |||
| Texture | Haralick52,108,125 | Characterize the overall shape and size of the erythrocyte without taking the density into account | |
| Gray-level run length matrices (GLRLM)112,116,119,125 | |||
| GLCM17,90,112,116,130 | |||
| Local binary pattern (LBP)31,52,112,116,119 | |||
| Fractal95,116,117,119 | |||
| Wavelet transform141 | |||
| Gradient texture30,40,76,105,141,164 | |||
| Gray-level co-occurrence matrix52,90,101,117 | |||
| Entropy88,94,112,116,124,169 | |||
| SIFT31 | |||
| Multiscale Laplacian of Gaussian and Gabor102 | |||
| Morphologic | Shape (area, perimeter, compactness ratio, eccentricity, bending energy, etc.)17,23,38,42,43,46,58,60,63,71,72,74,78,79,81,84,86,87,90,93,94,96,97,101,106,112,118,122,125,127,128,130,139, 143,144,153,161,168,169 | Encodes the spatial distribution of the intensity in a particular region | |
| Moments (zero, central, Hu)46,79,88,92,93,112,116,124,125,141,161 | |||
| Area granulometry37,40,60,65,100,143,150 |
Most notably, here is the use of different color spaces, which leads to sets of more malaria-specific features, depending on the color space used. Although most articles remain in the standard RGB color space, we think that there is a perfectly good reason to use a different color space better suited to extract the typical staining colors, which often range from a blue or purple to brownish shade. The HSV color space is favored by many articles, and several other articles use the green channel of RGB to extract staining-related color information in gray scale.
Table VII shows the features used for thick smears. Because of the smaller number of publications for thick smears, a smaller number of features has been experimented with in the literature. Nevertheless, authors have used similar, if not identical, features compared with the ones used for thin smears, experimenting with established features as well as different color spaces.
Table VII.
Feature computation for malaria parasite classification in thick blood smears6
| Blood smear |
Features type |
Feature | Remarks |
|---|---|---|---|
| Thick | Color | RGB136 | Provide color information |
| HSV142 | |||
| LAB98 | |||
| Intensity132,135 | |||
| Texture | Haralick55 | Characterize the overall shape and size of the erythrocyte without taking the density into account | |
| Morphologic | Shape (area, perimeter, compactness ratio, eccentricity, bending energy, etc.)55,136,137 | Encodes the spatial distribution of the intensity in a particular region | |
| Moment (zero, central, Hu)55,137 |
Some articles compute a large set of many different features, and then for practicality reasons cut down on these features by selecting the most discriminative feature subset using feature selection strategies. Specifically, the feature selection techniques used to reduce feature dimensionality include principal component analysis, F-statistic, 1-way analysis of variance, information gain, and support vector machine-based recursive feature elimination.98,111,112,114,116,119,125
However, such classical approaches to feature computation and selection run the serious danger of being superseded soon by techniques not relying on handcrafted features, such as deep learning in particular, which we will discuss in the section titled Deep Learning.
Parasite identification and labeling
Table VIII lists all classification methods that have been used for either discriminating between infected and uninfected red blood cells in thin smears or identifying parasites in thick smears.
Table VIII.
Classification methods
| Blood smear |
Classification methodology | |
|---|---|---|
| Thin | Unsupervised | K-mean clustering68 |
| Quaternion Fourier transform (QFT)56 | ||
| Supervised | Thresholding35,42,47,57,69,71,75,80,82,85,96,105,118 | |
| Bayesian classifier45,79,93,112,117,130 | ||
| Annular ring ratio method43,54 | ||
| Naive Bayes tree36,111,119,128 | ||
| Logistic regression tree108,111,128,161 | ||
| Linear programming155 | ||
| Euclidean distance classifier102 | ||
| K-nearest neighbors classifier40,49,60,77,79,128,144 | ||
| Decision tree58,64,76,89,101,127 | ||
| Template matching23,74 | ||
| Ada-boost17,129 | ||
| Nearest mean classifier (NM)128 | ||
| Fuzzy interface system109 | ||
| Normalized cross-correlation32 | ||
| Support vector machine (SVM)29,31,46,49,59,65,81,112,117,122,125,136,149,165,167 | ||
| Linear discriminant (LD)40,128 | ||
| Crowd source games30 | ||
| Neural network53,84,86,87,90,95,97,99,100,106,111,114,116,124,150,161,169 | ||
| Deep learning51,52,124,164,170 | ||
| Thick | Unsupervised | K-mean clustering98 |
| Supervised | Naive Bayes tree111 | |
| Randomized tree classifier137 | ||
| Nearest mean classifier (NM)98 | ||
| Thresholding132,139,142,143 | ||
| Support vector machine (SVM)55,136 | ||
| Neural network141 | ||
| Genetic algorithm55 |
Virtually all classification methods popular in the last decade have been applied to malaria diagnosis, ranging from decision trees and basic artificial neural networks over support vector machines to random tree classifiers. Very few articles have developed classification technologies specifically for cell discrimination or parasite detection. Most of the malaria-specific domain knowledge lies in the interplay of segmentation, features, and classification.
Comparing the performance of the published systems is very hard. The systems have been evaluated on blood slides from entirely different origins with largely varying parameters for image acquisition and slide preparation. Very often the evaluation set is too small or too limited to allow making a statement about the general system performance. Currently, there exists no publicly available image benchmark set, small or large, which could be used for fair comparisons of systems. Therefore, although many articles are reporting quite high performance numbers in terms of accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve, we prefer not to compare these numbers in this survey article.
We can observe a trade-off between the processing pipeline’s run-time performance and its accuracy. Typically, as the accuracy of a technique increases, its computational complexity increases all the same. For example, sophisticated level-set methods for cell segmentation perform better than Otsu thresholding but also require a longer runtime. Furthermore, feature computation can affect system efficiency. Some articles therefore apply feature selection methods to reduce feature dimensionality and remove nondiscriminative features, which can improve both accuracy and efficiency. Finally, the runtime of cell classification depends on the classification architecture used. For example, a support vector machine’s classification is much faster than the classification by a deep neural network. Although many articles do not report runtimes for their systems, we think that most of the cited systems will perform their task many times faster than a microscopist, or at least will perform faster than a human after a little optimization of their implementation. We have also found 2 articles in which the authors developed dedicated hardware devices with motorized stage units to increase throughput.142,164
In combination with software, this will fully automate the slide screening process so that a microscopist does not need to move the microscope dish to take an image of the next field. This will also result in a higher throughput that can increase the sensitivity of the system by allowing to inspect more fields in the same time.
To improve system accuracy, there seems to be a trend to follow the mainstream classification method at the time of publication to take advantage of the latest classification architecture and performance improvements it brings. Consequently, we are now seeing the first deep learning articles entering the scene, as listed in the next section.
DEEP LEARNING
Deep learning is the latest trend in machine learning, which has already boosted the performance in many nonmedical areas. Deep learning can be seen as an extension of the well-known multilayer neural network classifiers trained with back-propagation, except that many more layers are used. There are also different kind of layers that are used in typical successions. Deep learning typically requires large training sets. This is the reason why medical applications have been among the last applications to adopt deep learning, as annotated training images are significantly harder to obtain because of expert knowledge requirements and privacy concerns. The first article to apply deep learning to malaria diagnosis is by Liang et al.,51 who use a convolutional neural network to discriminate between infected and uninfected cells in thin blood smears, after applying a conventional level-set cell segmentation approach. This is an ideal application for deep learning because images of segmented red blood cells are a natural input for a convolutional neural network. Deep learning does not require the design of handcrafted features, which is one of its biggest advantages. Other authors who have applied deep learning to cell segmentation are Dong et al.124,170 and Gopakumar et al.,164 who used convolutional neural networks, Bibin et al.,52 who used deep belief networks, and recently Hung et al.173 who presented and end-to-end framework using faster Region-based Convolutional Neural Network.
Because deep learning is the overarching machine learning technique nowadays, we can expect many more publications to appear soon for cell classification, cell staging, cell segmentation, and other sub-problems in automated malaria diagnosis.
MOBILE SMARTPHONES FOR MALARIA DIAGNOSIS
The ideal hardware solution for microscopic malaria diagnosis in resource-poor settings would be a small portable slide reader into which a blood slide could be inserted and which would then output the parasitemia. Although modern technology is heading this way, we are still far from having a field-usable device. In particular, the relatively high optical magnification needed (up to 1000×) for malaria diagnosis in combination with oil immersion is a major miniaturization obstacle, unless alternatives are found. The next best solution are small camera-equipped computing devices, such as smartphones, which can be attached to a magnifying device and can then compute the parasitemia automatically, using image analysis and machine learning. Modern smartphones have become powerful computing devices and their cameras provide sufficient resolution for malaria diagnosis. Moreover, Android phones have become relatively cheap and are often already in the possession of health-care workers, even in resource-poor settings. Although cellular network connectivity can help with the information exchange between field workers and hospital, it is not immediately needed for malaria diagnosis and the actual cell counting. Small magnifying devices that can be attached to a smartphone’s camera, allowing true optical magnification compared with mere digital zooming, are commercially available. However, from the authors’ experience, these devices are still lacking in the image quality provided. Therefore, a more practical approach is to simply attach the smartphone to the eyepiece of a regular microscope with an adapter so that blood slide pictures can be taken with the smartphone’s camera.
A few experimental set-ups along these lines have been reported in the literature. In Ref 126 Breslauer et al. built a mobile phone-mounted light microscope and demonstrated its potential for clinical use by imaging P. falciparum-infected and sickle red blood cells in brightfield and Mycobacterium tuberculosis-infected sputum samples in fluorescence with LED excitation. In all cases, resolution exceeded that necessary to detect blood cell and microorganism morphology. For tuberculosis samples, they took advantage of the digitized images to demonstrate automated bacillus counting via image analysis software.
In Ref 156 Pirnstill and Cote present a cost-effective, optical cellphone-based transmission polarized light microscope system for imaging the malaria pigment known as hemozoin, which is a disposal product of the parasite’s blood digestion. It can be difficult to determine the presence of the pigment from background and other artifacts, even for skilled microscopy technicians. The pigment is much easier to observe using polarized light microscopy. However, implementation of polarized light microscopy lacks widespread adoption because the existing commercial devices have complicated designs, require sophisticated maintenance, tend to be bulky, can be expensive, and would require re-training for existing microscopy technicians. The cellphone-based polarimetric microscopy design presented by Pirnstill and Cote shows the potential to have both the resolution and the specificity to detect malaria in a low-cost, easy-touse, modular platform.
Rosado et al. presented an image processing and analysis methodology using supervised classification to assess the presence of P. falciparum trophozoites and white blood cells in Giemsa-stained thick blood smears.136 Using a support vector machine and a mix of geometric, color, and texture features, their automatic detection of trophozoites achieved a sensitivity of 80.5% and a specificity of 93.8%, whereas their white blood cell detection achieved 98.2% sensitivity and 72.1% specificity.
In Ref 137 Quinn et al. presented their 3-dimensional printable design of an adapter to attach a smartphone to a microscope, although all images for their experiments were taken with a dedicated microscope camera, which offered a higher pixel resolution than their smartphone camera. They presented a workflow for automated analysis of thick blood smears, which involved the computation of morphologic and moment features and an ensemble tree classifier trained on these features to discriminate between abnormal patches containing parasites and normal patches. The performance they reported was 97% area under the receiver operating characteristic curve.
Skandarajah et al. built a custom mobile phone microscope that is compatible with phones from multiple manufacturers.123 They demonstrated that quantitative microscopy with micron-scale spatial resolution can be carried out with multiple phones and that image linearity, distortion, and color can be corrected as needed. Specifically, they showed that phones with greater than 5 megapixel cameras are capable of nearly diffraction-limited resolution over a broad range of magnifications, including those relevant for single cell imaging. Furthermore, they found that automatic focus, exposure, and color gain standard on mobile phones can degrade image resolution and reduce accuracy of color capture if uncorrected, and they devise procedures to avoid these barriers to quantitative imaging.
Dallet et al. describe a mobile application platform for Android phones that can diagnose malaria from Giemsastained thin blood film images.54 The main imaging component consists of elaborate morphologic operations that can detect red and white blood cells, and identify parasites in the infected cells. The application also recognizes the different life stages of parasites and calculates the level of parasitemia. The application takes less than 60 seconds to give a diagnosis, and has been tested and verified on several version and types of Android mobile phones and tablets.
The authors of this survey article have developed a smartphone application to compute parasitemia in Giemsastained thin blood film images.51,167,171 To segment individual red blood cells, we applied marker-controlled watershed to thin blood smears to efficiently detect and segment individual cells, separate touching cells, and meet the demand of real-time processing. In the cell detection step, we apply a multiscale Laplacian of Gaussian filter on the green channel of an RGB color slide image. The local extrema of the Laplacian of Gaussian response indicate the approximate centroids of the individual cells that will serve as the approximate centroids for the marker-controlled watershed segmentation step. The cell foreground mask is estimated using Otsu thresholding, and cell edges are extracted by computing the gradient magnitude over the minimum values of the green and blue channels. Then, in the segmentation step, we apply watershed transform on cell markers, foreground masks, and edge information to segment and separate touching cells. For cell classification, we follow a deep learning approach and use a convolutional neural network for discriminating infected from uninfected cells.51,171
Cesario et al. discuss mobile support for vector-borne diseases in areas where specialist health care is scarce.93 They focus on the image analysis and classification component of a system that aims to reduce the chance of misdiagnosing less common diseases as malaria and to assist health professionals. Their article largely describes work in progress toward the image analysis and classification component, but feedback from healthcare professionals has been generally positive.
Herrera et al. tested the diagnostic performance of a device for automated interpretation of RDTs, which uses smartphone technology and image analysis software.138 The diagnostic performance of the device was comparable with visual interpretation of RDTs, without significant differences for P. falciparum and P. vivax. Providing standardized automated interpretation of RDTs in remote areas, in addition to almost real-time reporting of cases and enabling quality control, would greatly benefit large-scale implementation of RDT-based malaria diagnostic programs.
In similar work, Mudanyali et al. demonstrated a cellphone-based RDT reader platform that can work with various lateral flow immuno-chromatographic assays and similar tests.172 Their compact and cost-effective digital RDT reader attaches to the existing camera unit of a cellphone, where RDTs can be inserted to be imaged. Captured raw images of these RDTs are then digitally processed through a software application running on the cellphone for validation of the RDT and for automated reading of its diagnostic result. In addition, this smart RDT reader platform running on cellphones provides real-time spatio-temporal statistics for the prevalence of various infectious diseases, which allows tracking epidemics.
DISCUSSION
From the very different methods published during the last 10 years, we can see that there has been a lot of experimenting done to reach the current state of the art. However, despite the large number of publications, the performance numbers that have been published are very unsatisfying from a clinician’s point of view. It is actually very hard to quantify the current state of the art. Many of the articles just present performance numbers in terms of sensitivity and specificity for classification, representing only 1 operating point among many on a receiver operating characteristic, which would present a more complete evaluation of any method for different sensitivity requirements. Furthermore, the data used for evaluation have very often been simply too small to allow a convincing statement about a system’s performance. Many different training and test sets have been used to evaluate the proposed methods, but the lack of uniformity and standardization across all articles makes a fair comparison almost impossible. Extensive field studies on patient level or for tracking disease severity over time are needed to establish a baseline for standardized comparisons in the future.
A well-performing system will require the interplay of several factors, such as the characteristics of the microscope, the type of staining, the slide preparation, and the image analysis and machine learning software. However, no clear winners for each of these factors have emerged yet.
Nevertheless, progress has been made as can be seen by the natural development of methods used for image analysis and machine learning. In fact, this development has largely followed the development in other fields and has adopted major techniques and successfully applied them to malaria diagnosis. Many of these methods are general-purpose methods that are independent from the application domain. This being said, there has been a lot of fine-tuning of these methods to make them perform better for blood smear images, and more so for the image analysis methods than for machine learning. There is certainly the potential that some of these methods gain importance outside malaria diagnosis, in particular for preprocessing and for detecting and segmenting red blood cells in other applications.
For example, the filters used for preprocessing, as listed in Table III, are a good example of known methods applied to malaria diagnosis. The same holds for the detection and segmentation methods in Tables IV and V, with established methods like k-means clustering, Hough transform and active contour models, among others. Watershed in particular was a preferred technique to split touching cells. For feature computation, we can find the whole gamut of features used in other computer vision areas, ranging from the first Haralick features and chain codes to established and widely used local binary patterns and other texture measures. The same holds for the classification methods in Table VIII, which nicely reflect the historical development of classification methods over the last 10 years. We can see the older decision tree methods, followed by the then-popular Ada-boost classification strategy and support vector machines, culminating in the modern deep learning networks.
CONCLUSION
We wrote this survey article on image analysis and machine learning methods to give an update on the latest development in automated malaria diagnosis with image analysis and machine learning. This is a very dynamic area of research that has seen an extensive number of publications in the last decade. However, with the advent of new deep learning approaches, which have already left a deep impression, the research is seeing a new exciting development that is nothing short of a revolution. So far, only a few articles have been published, but it is already evident that this will be the dominating technique in the foreseeable future. This will render many of the former classification approaches dispensable. Moreover, because deep learning takes the difficult task of designing features for classification from the user, many of the handcrafted features used so far may become useless. In addition, because deep learning can be used not only for cell classification but also for cell segmentation, many of the cell segmentation approaches presented so far could become outdated very soon. Even the preprocessing techniques, which play an important role, are not safe from this development. One way of thinking is that neural networks can learn how to process different staining and lighting variations if only enough training data are being presented to the network. Given the recent developments and future possibilities, there is in fact a good chance that most of the articles referenced in our and other surveys will become a mere historical side note very soon, describing the state of the art before the advent of deep learning. All of the deep learning articles published so far have concentrated on thin blood smears, but it is very likely that we will see articles for thick films very soon. Given the wide acceptance of deep learning, the importance of large annotated data image repositories for training is now widely understood, leading to a great support of data acquisition efforts. This will likely lead to larger test suites on patient level, allowing for more standardized evaluations and extensive field testing. Given these developments, automated microscopy is very much in the race toward a cheap, simple, and reliable method for diagnosing malaria.
Acknowledgments
This research is supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and Lister Hill National Center for Biomedical Communications (LHNCBC). Mahidol-Oxford Tropical Medicine Research Unit is funded by the Wellcome Trust of Great Britain.
Abbreviations
- GLRLM
Gray Level Run Length Matrix
- HoG
Histogram of Gradient
- HSV
Hue Saturation Value
- IEEE
Institute of Electrical and Electronics Engineers
- LBP
Linear Binary Pattern
- LED
Light Emitting Diode
- NIH
National Institute of Health
- NLM
National Library of Medicine
- NM
Nearest Mean
- P
Plasmodium
- PCR
Polymerase Chain Reaction
- PLOS
Public Library of Science
- QFT
Quaternion Fourier Transform
- QPI
Quantitative Phase Imaging
- RDT
Rapid Diagnostic Test
- RGB
Red Green Blue
- RNA
RiboNucleic Acid
- SBFSEM
Serial Block-Face Scanning Electron Microscopy
- SEM
Scanning Electron Microscope
- SightDx
Sight Diagnostics
- SROFM
Sub-pixel Resolving Optofluidic Microscope
- SUSAN
Smallest Univalue Segment Assimilating Nucleus
- SVM
Support Vector Machine
- WHO
World Health Organization
Footnotes
Conflicts of Interest: All authors have read the journals policy on disclosure of potential conflicts of interest and have none to declare. All authors have read the journals authorship agreement and the manuscript has been reviewed and approved by all authors.
References
- 1.WHO. Malaria microscopy quality assurance manual-version 2. World Health Organization; 2016. [Google Scholar]
- 2.WHO. World malaria report 2016. World Health Organization; 2016. [Google Scholar]
- 3.Tek FB, Dempster AG, Kale I. Computer vision for microscopy diagnosis of malaria. Malar J. 2009;8:153. doi: 10.1186/1475-2875-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das D, Mukherjee R, Chakraborty C. Computational microscopic imaging for malaria parasite detection: a systematic review. J Microsc. 2015;260:1–19. doi: 10.1111/jmi.12270. [DOI] [PubMed] [Google Scholar]
- 5.Jan Z, Khan A, Sajjad M, Muhammad K, Rho S, Mehmood I. A review on automated diagnosis of malaria parasite in microscopic blood smears images. Multimedia Tools Appl. 2017:1–26. [Google Scholar]
- 6.Devi SS, Sheikh SA, Laskar RH. Erythrocyte features for malaria parasite detection in microscopic images of thin blood smear: a review. Int J Interact Multimedia Artificial Intell. 2016;4:34–9. [Google Scholar]
- 7.Tangpukdee N, Duangdee C, Wilairatana P, Krudsood S. Malaria diagnosis: a brief review. Korean J Parasitol. 2009;47:93. doi: 10.3347/kjp.2009.47.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT) Am J Trop Med Hyg. 2007;77(6_Suppl):119–27. [PubMed] [Google Scholar]
- 9.Hänscheid T. Diagnosis of malaria: a review of alternatives to conventional microscopy. Int J Lab Hematol. 1999;21:235–45. doi: 10.1046/j.1365-2257.1999.00220.x. [DOI] [PubMed] [Google Scholar]
- 10.WHO. Malaria microscopy quality assurance manual-version 2. World Health Organization; 2017. [Google Scholar]
- 11.Silamut K, White N. Relation of the stage of parasite development in the peripheral blood to prognosis in severe falciparum malaria. Trans Royal Soc Trop Med Hyg. 1993;87:436–43. doi: 10.1016/0035-9203(93)90028-o. [DOI] [PubMed] [Google Scholar]
- 12.Silamut K, Phu NH, Whitty C, et al. A quantitative analysis of the microvascular sequestration of malaria parasites in the human brain. Am J Pathol. 1999;155:395–410. doi: 10.1016/S0002-9440(10)65136-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsel SM, Gustafson SA, Friedlander E, et al. Malaria overdiagnosis in Cameroon: diagnostic accuracy of Fluorescence and Staining Technologies (FAST) Malaria Stain and LED microscopy versus Giemsa and bright field microscopy validated by polymerase chain reaction. Infect Dis Poverty. 2017;6:32. doi: 10.1186/s40249-017-0251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SA, Leitao R, Zheng G, Yang S, Rodriguez A, Yang C. Color capable sub-pixel resolving optofluidic microscope and its application to blood cell imaging for malaria diagnosis. PLoS ONE. 2011;6:e26127. doi: 10.1371/journal.pone.0026127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowling M, Shute G. Acomparative study of thick and thin blood films in the diagnosis of scanty malaria parasitaemia. BullWorld Health Organ. 1966;34:249. [PMC free article] [PubMed] [Google Scholar]
- 16.WHO. [Accessed January 16, 2018];Determining cost effectiveness of malaria rapid diagnostic tests in rural areas with high prevalence. Available at http://www2.wpro.who.int/sites/rdt.
- 17.Vink J, Laubscher M, Vlutters R, et al. An automatic vision-based malaria diagnosis system. J Microsc. 2013;250:166–78. doi: 10.1111/jmi.12032. [DOI] [PubMed] [Google Scholar]
- 18.Adeoye G, Nga I. Comparison of Quantitative Buffy Coat technique (QBC) with Giemsa-stained Thick Film (GTF) for diagnosis of malaria. Parasitol Int. 2007;56:308–12. doi: 10.1016/j.parint.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Janse CJ, Van Vianen PH. Flow cytometry in malaria detection. Methods Cell Biol. 1994;42:295–318. doi: 10.1016/s0091-679x(08)61081-x. [DOI] [PubMed] [Google Scholar]
- 20.Keiser J, Utzinger J, Premji Z, Yamagata Y, Singer BH. Ann Trop Med Parasitol. Vol. 96. Taylor & Francis; 2002. Acridine orange for malaria diagnosis: its diagnostic performance, its promotion and implementation in Tanzania, and the implications for malaria control; pp. 643–54. [DOI] [PubMed] [Google Scholar]
- 21.Houwen B. Clin Lab Med. Vol. 22. Elsevier; 2002. Blood film preparation and staining procedures; pp. 1–14. [DOI] [PubMed] [Google Scholar]
- 22.Shute G, Sodeman T. Identification of malaria parasites by fluorescence microscopy and acridine orange staining. BullWorld Health Organ. 1973;48–50:591–6. [PMC free article] [PubMed] [Google Scholar]
- 23.Suwalka I, Sanadhya A, Mathur A, Chouhan MS. International conference on computing, communication and applications. IEEE; 2012. Identify malaria parasite using pattern recognition technique; pp. 1–4. [Google Scholar]
- 24.Kawamoto F. Rapid diagnosis of malaria by fluorescence microscopy with light microscope and interference filter. Lancet. 1991;337:200–2. doi: 10.1016/0140-6736(91)92159-y. [DOI] [PubMed] [Google Scholar]
- 25.Wongsrichanalai C, Kawamotob F. Fluorescent microscopy and fluorescent labelling for malaria diagnosis. 2014 [Google Scholar]
- 26.Diaspro A, Chirico G, Usai C, Ramoino P, Dobrucki J. Handbook of biological confocal microscopy. Springer; 2006. Photobleaching; pp. 690–702. [Google Scholar]
- 27.Waters JC. Accuracy and precision in quantitative fluorescence microscopy. Rockefeller University Press; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guy R, Liu P, Pennefather P, Crandall I. The use of fluorescence enhancement to improve the microscopic diagnosis of falciparum malaria. Malar J. 2007;6:89–96. doi: 10.1186/1475-2875-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang D, Subramanian G, Duan J, et al. A portable image-based cytometer for rapid malaria detection and quantification. PLoS ONE. 2017;12:e0179161. doi: 10.1371/journal.pone.0179161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mavandadi S, Dimitrov S, Feng S, et al. Distributed medical image analysis and diagnosis through crowd-sourced games: a malaria case study. PLoS ONE. 2012;7:e37245. doi: 10.1371/journal.pone.0037245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linder N, Turkki R, Walliander M, et al. A malaria diagnostic tool based on computer vision screening and visualization of Plasmodium falciparum candidate areas in digitized blood smears. PLoS ONE. 2014;9:e104855. doi: 10.1371/journal.pone.0104855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammed HA, Abdelrahman IAM. International conference on communication, control, computing and electronics engineering (ICCCCEE) IEEE; 2017. Detection and classification of malaria in thin blood slide images; pp. 1–5. [Google Scholar]
- 33.Tek FB, Dempster AG, Kale I. 14th signal processing and communications applications. IEEE; 2006. A colour normalization method for giemsa-stained blood cell images; pp. 1–4. [Google Scholar]
- 34.Di Rubeto C, Dempster A, Khan S, Jarra B. 15th international conference on pattern recognition. Vol. 3. IEEE; 2000. Segmentation of blood images using morphological operators; pp. 397–400. [Google Scholar]
- 35.Halim S, Bretschneider TR, Li Y, Preiser PR, Kuss C. 9th international conference on control, automation, robotics and vision. IEEE; 2006. Estimating malaria parasitaemia from blood smear images; pp. 1–6. [Google Scholar]
- 36.Anggraini D, Nugroho AS, Pratama C, Rozi IE, Pragesjvara V, Gunawan M. International conference on advanced computer science and information system. IEEE; 2011. Automated status identification of microscopic images obtained from malaria thin blood smears using Bayes decision: a study case in Plasmodium falciparum; pp. 347–52. [Google Scholar]
- 37.Kareem S, Morling RC, Kale I. International symposium on circuits and systems (ISCAS) IEEE; 2011. A novel method to count the red blood cells in thin blood films; pp. 1021–4. [Google Scholar]
- 38.Kareem S, Kale I, Morling RC. Asia Pacific conference on circuits and systems (APCCAS) IEEE; 2012. Automated malaria parasite detection in thin blood films: a hybrid illumination and color constancy insensitive, morphological approach; pp. 240–3. [Google Scholar]
- 39.Nasir AA, Mashor M, Mohamed Z. EMBS conference on biomedical engineering and sciences (IECBES) IEEE; 2012. Segmentation based approach for detection of malaria parasites using moving k-means clustering; pp. 653–8. [Google Scholar]
- 40.Malihi L, Ansari-Asl K, Behbahani A. 8th Iranian conference on machine vision and image processing (MVIP) IEEE; 2013. Malaria parasite detection in giemsa-stained blood cell images; pp. 360–5. [Google Scholar]
- 41.Berge H, Taylor D, Krishnan S, Douglas TS. International symposium on biomedical imaging: from nano to macro. IEEE; 2011. Improved red blood cell counting in thin blood smears; pp. 204–7. [Google Scholar]
- 42.Di Ruberto C, Dempster A, Khan S, Jarra B. International conference on pattern recognition. Vol. 3. IEEE; 2000. Automatic thresholding of infected blood images using granulometry and regional extrema; pp. 441–4. [Google Scholar]
- 43.Kareem S, Kale I, Morling RC. International conference on computer modeling and simulation. IEEE; 2012. Automated P. falciparum detection system for post-treatment malaria diagnosis using modified annular ring ratio method; pp. 432–6. [Google Scholar]
- 44.Zou L, Chen J, Zhang J, Garcia N. International conference on digital image computing: techniques and applications. IEEE; 2010. Malaria cell counting diagnosis within large field of view; pp. 172–7. [Google Scholar]
- 45.Mushabe MC, Dendere R, Douglas TS. International conference engineering in medicine and biology society (EMBC) IEEE; 2013. Automated detection of malaria in Giemsa-stained thin blood smears; pp. 3698–701. [DOI] [PubMed] [Google Scholar]
- 46.Savkare S, Narote S. International conference on communication, information & computing technology. IEEE; 2015. Automated system for malaria parasite identification; pp. 1–4. [Google Scholar]
- 47.Mehrjou A, Abbasian T, Izadi M. International conference on robotics and mechatronics (ICRoM) IEEE; 2013. Automatic malaria diagnosis system; pp. 205–11. [Google Scholar]
- 48.Gatc J, Maspiyanti F, Sarwinda D, Arymurthy AM. International conference on advanced computer science and information systems. IEEE; 2013. Plasmodium parasite detection on red blood cell image for the diagnosis of malaria using double thresholding; pp. 381–5. [Google Scholar]
- 49.Nanoti A, Jain S, Gupta C, Vyas G. International conference on inventive computation technologies. Vol. 1. IEEE; 2016. Detection of malaria parasite species and life cycle stages using microscopic images of thin blood smear; pp. 1–6. [Google Scholar]
- 50.Maiseli B, Mei J, Gao H, Yin S. International conference on mechatronics and control (ICMC) IEEE; 2014. An automatic and cost-effective parasitemia identification framework for low-end microscopy imaging devices; pp. 2048–53. [Google Scholar]
- 51.Liang Z, Powell A, Ersoy I, et al. International conference on bioinformatics and biomedicine (BIBM) IEEE; 2016. CNN-based image analysis for malaria diagnosis; pp. 493–6. [Google Scholar]
- 52.Bibin D, Nair MS, Punitha P. Malaria parasite detection from peripheral blood smear images using deep belief networks. Int J Appl Eng Res. 2017;5:9099–108. [Google Scholar]
- 53.Adi K, Pujiyanto S, Gernowo R, Pamungkas A, Putranto AB. Identifying the developmental phase of Plasmodium falciparum in malaria-infected red blood cells using adaptive color segmentation and back propagation neural network. Int J Appl Eng Res. 2016;11:8754–9. [Google Scholar]
- 54.Dallet C, Kareem S, Kale I. International conference on circuits and systems. IEEE; 2014. Real time blood image processing application for malaria diagnosis using mobile phones; pp. 2405–8. [Google Scholar]
- 55.Elter M, Haßlmeyer E, Zerfaß T. International conference on engineering in medicine and biology society. IEEE; 2011. Detection of malaria parasites in thick blood films; pp. 5140–4. [DOI] [PubMed] [Google Scholar]
- 56.Fang Y, Xiong W, Lin W, Chen Z. International conference on engineering in medicine and biology society. IEEE; 2011. Unsupervised malaria parasite detection based on phase spectrum; pp. 7997–8000. [DOI] [PubMed] [Google Scholar]
- 57.May Z, Aziz SSAM, Salamat R. International conference on signal and image processing applications. IEEE; 2013. Automated quantification and classification of malaria parasites in thin blood smears; pp. 369–73. [Google Scholar]
- 58.Sheikhhosseini M, Rabbani H, Zekri M, Talebi A. Automatic diagnosis of malaria based on complete circle–ellipse fitting search algorithm. J Microsc. 2013;252:189–203. doi: 10.1111/jmi.12081. [DOI] [PubMed] [Google Scholar]
- 59.Díaz G, González FA, Romero E. A semi-automatic method for quantification and classification of erythrocytes infected with malaria parasites in microscopic images. J Biomed Inform. 2009;42:296–307. doi: 10.1016/j.jbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 60.Tek FB, Dempster AG, Kale I. Parasite detection and identification for automated thin blood film malaria diagnosis. Comput Vis Image Underst. 2010;114:21–32. [Google Scholar]
- 61.Brückner M, Becker K, Popp J, Frosch T. Fiber array based hyperspectral Raman imaging for chemical selective analysis of malaria-infected red blood cells. Anal Chim Acta. 2015;894:76–84. doi: 10.1016/j.aca.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 62.Sakaguchi M, Miyazaki N, Fujioka H, Kaneko O, Murata K. Three-dimensional analysis of morphological changes in the malaria parasite infected red blood cell by serial block-face scanning electron microscopy. J Struct Biol. 2016;193:162–71. doi: 10.1016/j.jsb.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 63.Di Ruberto C, Dempster A, Khan S, Jarra B. Analysis of infected blood cell images using morphological operators. Image Vision Comput. 2002;20:133–46. [Google Scholar]
- 64.Sio SW, Sun W, Kumar S, et al. MalariaCount: an image analysis-based program for the accurate determination of parasitemia. J Microbiol Methods. 2007;68:11–8. doi: 10.1016/j.mimet.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 65.Savkare S, Narote S. Automatic system for classification of erythrocytes infected with malaria and identification of parasite’s life stage. Proc Technol. 2012;6:405–10. [Google Scholar]
- 66.Srivastava B, Anvikar AR, Ghosh SK, et al. Computer-visionbased technology for fast, accurate and cost effective diagnosis of malaria. Malar J. 2015;14:526. doi: 10.1186/s12936-015-1060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prescott WR, Jordan RG, Grobusch MP, et al. Performance of a malaria microscopy image analysis slide reading device. Malar J. 2012;11:155. doi: 10.1186/1475-2875-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Purwar Y, Shah SL, Clarke G, Almugairi A, Muehlenbachs A. Automated and unsupervised detection of malarial parasites in microscopic images. Malar J. 2011;10:364. doi: 10.1186/1475-2875-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma C, Harrison P, Wang L, Coppel RL. Automated estimation of parasitaemia of Plasmodium yoelii-infected mice by digital image analysis of Giemsa-stained thin blood smears. Malar J. 2010;9:348. doi: 10.1186/1475-2875-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Proudfoot O, Drew N, Scholzen A, Xiang S, Plebanski M. Investigation of a novel approach to scoring Giemsa-stained malaria-infected thin blood films. Malar J. 2008;7:62. doi: 10.1186/1475-2875-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daniel T, Pierre E, Emmanuel T, Philippe B. Automated diagnosis of malaria in tropical areas using 40× microscopic images of blood smears. Int J Biometrics Bioinformatics. 2016;10:12. [Google Scholar]
- 72.Kim JD, Nam KM, Park CY, Kim YS, Song HJ. Automatic detection of malaria parasite in blood images using two parameters. Technol Health Care. 2016;24(s1):S33–9. doi: 10.3233/THC-151049. [DOI] [PubMed] [Google Scholar]
- 73.Abbas N, Saba T, Mohamad D, Rehman A, Almazyad AS, Al-Ghamdi JS. Machine aided malaria parasitemia detection in Giemsa-stained thin blood smears. Neural Comput Appl. 2016:1–16. [Google Scholar]
- 74.Di Ruberto C, Dempster A, Khan S, Jarra B. Visual form 2001. Springer; 2001. Morphological image processing for evaluating malaria disease; pp. 739–48. [Google Scholar]
- 75.Prasad K, Winter J, Bhat UM, Acharya RV, Prabhu GK. Image analysis approach for development of a decision support system for detection of malaria parasites in thin blood smear images. J Digit Imaging. 2012;25:542–9. doi: 10.1007/s10278-011-9442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh P, Bhattacharjee D, Nasipuri M, Basu DK. Computer information systems–analysis and technologies. Springer; 2011. Medical aid for automatic detection of malaria; pp. 170–8. [Google Scholar]
- 77.Daz G, Gonzalez F, Romero E. Infected cell identification in thin blood images based on color pixel classification: comparison and analysis. Progr Pattern Recogn Image Anal Appl. 2007:812–21. [Google Scholar]
- 78.Raviraja S, Bajpai G, Sharma SK. 3rd Kuala Lumpur international conference on biomedical engineering. Springer; 2007. Analysis of detecting the malarial parasite infected blood images using statistical based approach; pp. 502–5. [Google Scholar]
- 79.Tek FB, Dempster AG, Kale I. Malaria parasite detection in peripheral blood images. BMVC. 2006:347–56. [Google Scholar]
- 80.Le MT, Bretschneider TR, Kuss C, Preiser PR. A novel semiautomatic image processing approach to determine Plasmodium falciparum parasitemia in Giemsa-stained thin blood smears. BMC Cell Biol. 2008;9:15–27. doi: 10.1186/1471-2121-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Savkare S, Narote S. Automatic detection of malaria parasites for estimating parasitemia. Int J Comput Sci Sec. 2011;5:310–5. [Google Scholar]
- 82.Suradkar PT. Detection of malarial parasite in blood using image processing. Int J Eng Innov Technol. 2013;2:124–6. [Google Scholar]
- 83.Aimi Salihah AN, Yusoff M, Zeehaida M. Colour image segmentation approach for detection of malaria parasites using various colour models and k-means clustering. WSEAS Trans Biol Biomed. 2013;10:41–55. [Google Scholar]
- 84.Khan MI, Acharya B, Singh BK, Soni J. Content based image retrieval approaches for detection of malarial parasite in blood images. Int J Biometrics Bioinformatics. 2011;5:97. [Google Scholar]
- 85.Khatri K, Ratnaparkhe V, Agrawal S, Bhalchandra A. Image processing approach for malaria parasite identification. National conference on growth of technologies in electronics, telecom and computers Indias perception. 2013:5–7. [Google Scholar]
- 86.Hirimutugoda Y, Wijayarathna G. Image analysis system for detection of red cell disorders using artificial neural networks. Sri Lanka J Bio Med Info. 2010;1:35–42. [Google Scholar]
- 87.Razzak MI. Malarial parasite classification using recurrent neural network. Int J Image Process. 2015;9:69. [Google Scholar]
- 88.Ajala F, Fenwa O, Aku M. Comparative analysis of different types of malaria diseases using first order features. Int J Appl Info Syst. 2015;8:20–6. [Google Scholar]
- 89.Pamungkas A, Adi K, Gernowo R. Identification of Plasmodium falciparum development phase in malaria infected red blood cells using adaptive color segmentation and decision tree based classification. Int J Appl Eng Res. 2015;10:4043–55. [Google Scholar]
- 90.Razzak MI. Automatic detection and classification of malarial parasite. Int J Biometrics Bioinformatics. 2015;9:1–12. [Google Scholar]
- 91.Ahirwar N, Pattnaik S, Acharya B. Advanced image analysis based system for automatic detection and classification of malarial parasite in blood images. Int J Info Technol Knowledge Manage. 2012;5:59–64. [Google Scholar]
- 92.Anand PR, Bajpai G, Bhaskar V, Job SM. 4th Kuala Lumpur international conference on biomedical engineering. Springer; 2008. Detection of the malarial parasite infected blood images by 3D-analysis of the cell curved surface; pp. 166–9. [Google Scholar]
- 93.Cesario M, Lundon M, Luz S, Masoodian M, Rogers B. Proceedings of the 13th international conference of the NZ chapter of the ACM’s special interest group on human-computer interaction. ACM; 2012. Mobile support for diagnosis of communicable diseases in remote locations; pp. 25–8. [Google Scholar]
- 94.Chavan SN, Sutkar AM. Malaria disease identification and analysis using image processing. Int J Latest Trends Eng Technol. 2014;3:218–23. [Google Scholar]
- 95.Chayadevi M, Raju G. Usage of art for automatic malaria parasite identification based on fractal features. Int J Video Image Process Network Sec. 2014;4:7–15. [Google Scholar]
- 96.Ghate DA, Jadhav C. Automatic detection of malaria parasite from blood images. Int J Comput Sci Appl. 2012;1 [Google Scholar]
- 97.Gitonga L, Memeu DM, Kaduki KA, Kale MAC, Muriuki NS. Determination of plasmodium parasite life stages and species in images of thin blood smears using artificial neural network. Open J Clin Diagnostics. 2014;4:78. [Google Scholar]
- 98.Khan NA, Pervaz H, Latif AK, et al. International joint conference on computer science and software engineering. IEEE; 2014. Unsupervised identification of malaria parasites using computer vision; pp. 263–7. [Google Scholar]
- 99.Premaratne SP, Karunaweera ND, Fernando S, Perera WSR, Rajapaksha R. A neural network architecture for automated recognition of intracellular malaria parasites in stained blood films. Proceedings of APAMI & CJKMI-KOSMI conference. 2003:1–4. [Google Scholar]
- 100.Soni J. Advanced image analysis based system for automatic detection of malarial parasite in blood images using SUSAN approach. Int J Eng Sci Technol. 2011;3:5260–74. [Google Scholar]
- 101.Soni J, Mishra N, Kamargaonkar N. Automatic difference between RBC and malaria parasites based on morphology with first order features using image processing. Int J Adv Eng Technol. 2011;1:290–7. [Google Scholar]
- 102.Suryawanshi MS, Dixit V. Improved technique for detection of malaria parasites within the blood cell images. Int J Sci Eng Res. 2013;4:373–6. [Google Scholar]
- 103.Walliander M, Turkki R, Linder N, et al. Automated segmentation of blood cells in Giemsa stained digitized thin blood films. Diagn Pathol. 2013;8:S37. [Google Scholar]
- 104.Von Mühlen A. Computer image analysis of malarial Plasmodium vivax in human red blood cells. California Polytechnic State University; San Luis Obispo: 2004. [Google Scholar]
- 105.Špringl V. Automatic malaria diagnosis through microscopy imaging. 2009 [Google Scholar]
- 106.Memeu DM. A rapid malaria diagnostic method based on automatic detection and classification of plasmodium parasites in stained thin blood smear images. 2014 [Google Scholar]
- 107.Makkapati VV, Rao RM. International conference on acoustics, speech and signal processing. IEEE; 2009. Segmentation of malaria parasites in peripheral blood smear images; pp. 1361–4. [Google Scholar]
- 108.Das D, Ghosh M, Chakraborty C, Maiti AK, Pal M. International conference on image information processing (ICIIP) IEEE; 2011. Probabilistic prediction of malaria using morphological and textural information; pp. 1–6. [Google Scholar]
- 109.Ghosh M, Das D, Chakraborty C, Ray AK. International conference on image information processing (ICIIP) IEEE; 2011. Plasmodium vivax segmentation using modified fuzzy divergence; pp. 1–5. [Google Scholar]
- 110.Makkapati VV, Rao RM. International conference on engineering in medicine and biology society. IEEE; 2011. Ontology-based malaria parasite stage and species identification from peripheral blood smear images; pp. 6138–41. [DOI] [PubMed] [Google Scholar]
- 111.Das D, Maiti A, Chakraborty C. Automated system for characterization and classification of malaria-infected stages using light microscopic images of thin blood smears. J Microsc. 2015;257:238–52. doi: 10.1111/jmi.12206. [DOI] [PubMed] [Google Scholar]
- 112.Das DK, Ghosh M, Pal M, Maiti AK, Chakraborty C. Machine learning approach for automated screening of malaria parasite using light microscopic images. Micron. 2013;45:97–106. doi: 10.1016/j.micron.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 113.Bibin D, Punitha P. Multimedia processing, communication and computing applications. Springer; 2013. Stained blood cell detection and clumped cell segmentation useful for malaria parasite diagnosis; pp. 195–207. [Google Scholar]
- 114.Devi SS, Sheikh SA, Talukdar A, Laskar RH. Malaria infected erythrocyte classification based on the histogram features using microscopic images of thin blood smear. Ind J Sci Technol. 2016;9 [Google Scholar]
- 115.Damahe LB, Krishna R, Janwe N. Segmentation based approach to detect parasites and RBCs in blood cell images. Int J Comput Sci Appl. 2011;4:71–81. [Google Scholar]
- 116.Das DK, Maiti AK, Chakraborty C. Textural pattern classification of microscopic images for malaria screening. Adv Ther Eng. 2012:419–46. [Google Scholar]
- 117.Ghosh M, Das D, Chakraborty C, Ray AK. Quantitative characterisation of Plasmodium vivax in infected erythrocytes: a textural approach. Int J Artificial Intell Soft Comput. 2013;3:203–21. [Google Scholar]
- 118.Kumar A, Choudhary A, Tembhare P, Pote C. Enhanced identification of malarial infected objects using Otsu algorithm from thin smear digital images. Int J Latest Res Sci Technol ISSN. 2012:2278–5299. [Google Scholar]
- 119.Maity M, Maity AK, Dutta PK, Chakraborty C. Aweb-accessible framework for automated storage with compression and textural classification of malaria parasite images. Int J Comput Appl. 2012;52:31–9. [Google Scholar]
- 120.Panchbhai VV, Damahe LB, Nagpure AV, Chopkar PN. RBCs and parasites segmentation from thin smear blood cell images. Int J Image Graph Signal Process. 2012;4:54. [Google Scholar]
- 121.Mulay HD, Murthy TD, Nerune SM, Amrutha M. New methylene blue stain for malaria detection on thin smears. J Krishna Inst Med Sci. 2017;6:76–81. [Google Scholar]
- 122.Eshel Y, Houri-Yafin A, Benkuzari H, et al. Evaluation of the Parasight platform for malaria diagnosis. J Clin Microbiol. 2017;55:768–75. doi: 10.1128/JCM.02155-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Skandarajah A, Reber CD, Switz NA, Fletcher DA. Quantitative imaging with a mobile phone microscope. PLoS ONE. 2014;9:e96906. doi: 10.1371/journal.pone.0096906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dong Y, Jiang Z, Shen H, et al. EMBS international conference on biomedical & health informatics (BHI) IEEE; 2017. Evaluations of deep convolutional neural networks for automatic identification of malaria infected cells; pp. 101–4. [Google Scholar]
- 125.Muralidharan V, Dong Y, Pan WD. International conference on biomedical and health informatics (BHI) IEEE; 2016. A comparison of feature selection methods for machine learning based automatic malarial cell recognition in wholeslide images; pp. 216–9. [Google Scholar]
- 126.Breslauer DN, Maamari RN, Switz NA, Lam WA, Fletcher DA. Mobile phone based clinical microscopy for global health applications. PLoS ONE. 2009;4:e6320. doi: 10.1371/journal.pone.0006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moon S, Lee S, Kim H, et al. An image analysis algorithm for malaria parasite stage classification and viability quantification. PLoS ONE. 2013;8:e61812. doi: 10.1371/journal.pone.0061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Park HS, Rinehart MT, Walzer KA, Chi JTA, Wax A. Automated detection of P. falciparum using machine learning algorithms with quantitative phase images of unstained cells. PLoS ONE. 2016;11:e0163045. doi: 10.1371/journal.pone.0163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang Z, Ong LS, Fang K, et al. 38th annual international conference of the engineering in medicine and biology society (EMBC) IEEE; 2016. Image classification of unlabeled malaria parasites in red blood cells; pp. 3981–4. [DOI] [PubMed] [Google Scholar]
- 130.Bhowmick S, Das DK, Maiti AK, Chakraborty C. Structural and textural classification of erythrocytes in anaemic cases: a scanning electron microscopic study. Micron. 2013;44:384–94. doi: 10.1016/j.micron.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 131.Omucheni DL, Kaduki KA, Bulimo WD, Angeyo HK. Application of principal component analysis to multispectral-multimodal optical image analysis for malaria diagnostics. Malar J. 2014;13:485. doi: 10.1186/1475-2875-13-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Toha S, Ngah U. International conference on signal processing, communications and networking. IEEE; 2007. Computer aided medical diagnosis for the identification of malaria parasites; pp. 521–2. [Google Scholar]
- 133.Salamah U, Sarno R, Arifin A, et al. International conference on knowledge creation and intelligent computing. IEEE; 2016. Enhancement of low quality thick blood smear microscopic images of malaria patients using contrast and edge corrections; pp. 219–25. [Google Scholar]
- 134.Hanif N, Mashor M, Mohamed Z. International colloquium on signal processing and its applications (CSPA) IEEE; 2011. Image enhancement and segmentation using dark stretching technique for Plasmodium falciparum for thick blood smear; pp. 257–60. [Google Scholar]
- 135.Arco JE, Górriz JM, Ramírez J, Álvarez I, Puntonet CG. Digital image analysis for automatic enumeration of malaria parasites using morphological operations. Expert Syst Appl. 2015;42:3041–7. [Google Scholar]
- 136.Rosado L, da Costa JMC, Elias D, Cardoso JS. Automated detection of malaria parasites on thick blood smears via mobile devices. Proc Comput Sci. 2016;90:138–44. [Google Scholar]
- 137.Quinn JA, Andama A, Munabi I, Kiwanuka FN. Automated blood smear analysis for mobile malaria diagnosis. Mobile Point Care Monitors Diagno Device Design. 2014;31:115. [Google Scholar]
- 138.Herrera S, Vallejo AF, Quintero JP, Arévalo-Herrera M, Cancino M, Ferro S. Field evaluation of an automated RDT reader and data management device for Plasmodium falciparum/Plasmodium vivax malaria in endemic areas of Colombia. Malar J. 2014;13:87. doi: 10.1186/1475-2875-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frean JA. Reliable enumeration of malaria parasites in thick blood films using digital image analysis. Malar J. 2009;8:218. doi: 10.1186/1475-2875-8-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Luengo-Oroz MA, Arranz A, Frean J. Crowd sourcing malaria parasite quantification: an online game for analyzing images of infected thick blood smears. J Med Internet Res. 2012;14:1–19. doi: 10.2196/jmir.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yunda L, Ramirez AA, Millán J. Automated image analysis method for p-vivax malaria parasite detection in thick film blood images. Sistemas Telemática. 2012;10:9–25. [Google Scholar]
- 142.Kaewkamnerd S, Uthaipibull C, Intarapanich A, Pannarut M, Chaotheing S, Tongsima S. An automatic device for detection and classification of malaria parasite species in thick blood film. BMC Bioinformatics. 2012;13:S18. doi: 10.1186/1471-2105-13-S17-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Frean J. Microscopic determination of malaria parasite load: role of image analysis. Micrsocopy Sci Technol Appl Educ. 2010:862–6. [Google Scholar]
- 144.Devi SS, Roy A, Sharma M, Laskar R. 2nd international conference on computational intelligence and networks (CINE) IEEE; 2016. kNN classification based erythrocyte separation in microscopic images of thin blood smear; pp. 69–72. [Google Scholar]
- 145.Sharif JM, Miswan M, Ngadi M, Salam MSH, bin Abdul Jamil MM. International conference on biomedical engineering. IEEE; 2012. Red blood cell segmentation using masking and watershed algorithm: a preliminary study; pp. 258–62. [Google Scholar]
- 146.Thung F, Suwardi IS. International conference on electrical engineering and informatics. IEEE; 2011. Blood parasite identification using feature based recognition; pp. 1–4. [Google Scholar]
- 147.Somasekar J, Reddy BE. Segmentation of erythrocytes infected with malaria parasites for the diagnosis using microscopy imaging. Comput Elect Eng. 2015;45:336–51. [Google Scholar]
- 148.Khot S, Prasad R. Proceedings of the 3rd international conference on frontiers of intelligent computing: theory and applications. Springer; 2015. Optimal computer based analysis for detecting malarial parasites; pp. 69–80. [Google Scholar]
- 149.Kumarasamy SK, Ong S, Tan KS. Robust contour reconstruction of red blood cells and parasites in the automated identification of the stages of malarial infection. Machine Vision and Applications. 2011;22:461–9. [Google Scholar]
- 150.Ross NE, Pritchard CJ, Rubin DM, Duse AG. Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Med Biol Eng Comput. 2006;44:427–36. doi: 10.1007/s11517-006-0044-2. [DOI] [PubMed] [Google Scholar]
- 151.Somasekar J, Reddy ARM, Reddy LS. Global trends in information systems and software applications. Springer; 2012. An efficient algorithm for automatic malaria detection in microscopic blood images; pp. 431–40. [Google Scholar]
- 152.Payne D. Use and limitations of light microscopy for diagnosing malaria at the primary health care level. Bull World Health Organ. 1988;66:621. [PMC free article] [PubMed] [Google Scholar]
- 153.Somasekar J, Reddy BE, Reddy EK, Lai CH. An image processing approach for accurate determination of parasitemia in peripheral blood smear images. Int J Comput Appl. 2011:23–8. [Google Scholar]
- 154.Memeu DM, Kaduki KA, Mjomba A, Muriuki NS, Gitonga L. Detection of plasmodium parasites from images of thin blood smears. Open J Clin Diagnostics. 2013;3:183. [Google Scholar]
- 155.Parkhi V, Pawar P, Surve A. Computer automation for malaria parasite detection using linear programming. Int J Adv Res Elec Elec Instrum Eng. 2013;2:1984–8. [Google Scholar]
- 156.Pirnstill CW, Coté GL. Malaria diagnosis using a mobile phone polarized microscope. Sci Rep. 2015;5:13368. doi: 10.1038/srep13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.DABO-NIANG S, Zoueu J. Combining kriging, multispectral and multimodal microscopy to resolve malaria-infected erythrocyte contents. J Microsc. 2012;247:240–51. doi: 10.1111/j.1365-2818.2012.03637.x. [DOI] [PubMed] [Google Scholar]
- 158.Chen F, Flaherty BR, Cohen CE, Peterson DS, Zhao Y. Direct detection of malaria infected red blood cells by surface enhanced Raman spectroscopy. Nanomedicine. 2016;12:1445–51. doi: 10.1016/j.nano.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Frean J. Improving quantification of malaria parasite burden with digital image analysis. Trans Royal Soc Trop Med Hyg. 2008;102:1062–3. doi: 10.1016/j.trstmh.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 160.Tulsani H, Saxena S, Yadav N. Segmentation using morphological watershed transformation for counting blood cells. Int J Comput Info Technol. 2013;2:28–36. [Google Scholar]
- 161.Das D, Chakraborty C, Mitra B, Maiti A, Ray A. Quantitative microscopy approach for shape-based erythrocytes characterization in anaemia. J Microsc. 2013;249:136–49. doi: 10.1111/jmi.12002. [DOI] [PubMed] [Google Scholar]
- 162.Mandal S, Kumar A, Chatterjee J, Manjunatha M, Ray AK. Annual India conference. IEEE; 2010. Segmentation of blood smear images using normalized cuts for detection of malarial parasites; pp. 1–4. [Google Scholar]
- 163.Maitra M, Gupta RK, Mukherjee M. Detection and counting of red blood cells in blood cell images using Hough transform. Int J Comput Appl. 2012;53:18–22. [Google Scholar]
- 164.Gopakumar GP, Swetha M, Sai Siva G, Subrahmanyam S. Convolutional neural network-based malaria diagnosis from focus stack of blood smear images acquired using custom-built slide scanner. J Biophotonics. 2017 doi: 10.1002/jbio.201700003. [DOI] [PubMed] [Google Scholar]
- 165.Wahyuningrum RT, Indrawan AK. A hybrid automatic method for parasite detection and identification ofPlasmodium falciparumin thin blood images. Int J Acad Res. 2012;4:44–50. [Google Scholar]
- 166.Abbas N, Mohamad D. Microscopic RGB color images enhancement for blood cells segmentation in YCbCr color space for k-means clustering. J Theor Appl Inf Technol. 2013;55:117–25. [Google Scholar]
- 167.Poostchi M, Ersoy I, Bansal A, et al. Image analysis of blood slides for automatic malaria diagnosis. In NIH-IEEE Strategic Conference on Healthcare Innovations and Point-of-Care Technologies for Precision Medicine (HI-POCT) 2015 MoPoster04.22. [Google Scholar]
- 168.Nguyen NT, Duong AD, Vu HQ. International conference on knowledge and systems engineering. IEEE; 2010. A new method for splitting clumped cells in red blood images; pp. 3–8. [Google Scholar]
- 169.Annaldas S, Shirgan S, Marathe V. Automatic identification of malaria parasites using image processing. Int J Emerg Eng Res Technol. 2014;2:107–12. [Google Scholar]
- 170.Dong Y, Jiang Z, Shen H, Pan WD. SoutheastCon, 2017. IEEE; 2017. Classification accuracies of malaria infected cells using deep convolutional neural networks based on decompressed images; pp. 1–6. [Google Scholar]
- 171.Sivaramakrishnan R, Antani S, Jaeger S. Visualizing deep learning activations for improved malaria cell classification. Medical informatics and healthcare. 2017:40–7. [Google Scholar]
- 172.Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A. Integrated rapid-diagnostic-test reader platform on a cellphone. Lab Chip. 2012;12:2678–86. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hung J, Carpenter A. 2017 IEEE Conference on Computer Vision and Pattern RecognitionWorkshops (CVPRW) IEEE; 2017. Applying Faster R-CNN for Object Detection on Malaria Images; pp. 808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]