Abstract
We report the high-yield heterologous expression of bioactive θ-defensin RTD-1 inside Escherichia coli cells by making use of intracellular protein trans-splicing in combination with a high efficient split-intein. RTD-1 is a small backbone-cyclized polypeptide with three disulfide bridges and a natural inhibitor of anthrax lethal factor protease. Recombinant RTD-1 was natively folded and able to inhibit anthrax lethal factor protease. In-cell expression of RTD-1 was very efficient and yielded ≈ 0.7 mg of folded RTD-1 per gram of wet E. coli cells. This approach was used to generate of a genetically-encoded RTD-1-based peptide library in live E. coli cells. These results clearly demonstrate the possibility of using genetically-encoded RTD-1-based peptide libraries in live E. coli cells, which is a critical first step for developing in-cell screening and directed evolution technologies using the cyclic peptide RTD-1 as a molecular scaffold.
Keywords: RTD-1, θ-defensins, anthrax lethal factor, genetically-encoded libraries, backbone-cyclized peptides, protein splicing, split-intein
TOC
1. Introduction
Defensins are cysteine-rich antimicrobial peptides that are important in the innate immune defense of mammals.1–3 Although defensins have been of interest for their antimicrobial activities, they are also involved in other defense mechanisms, which include toxin neutralization, modulation of the immune system and anti-cancer properties.3,4 The defensins found in mammals are positively charged peptides containing mostly β-sheet structures and six conserved cysteines that form three intramolecular disulfides. According to their overall structure, mammal defensins have been classified in α-, β- and θ-defensins.5
The overall fold of α- and β-defensins is quite similar except for having different disulfide connectivities, and the presence of an N-terminal α-helix segment in β-defensins that is not present in α-defensins.5 In contrast, θ-defensins are backbone cyclized peptides formed by the head-to-tail covalent assembly of two nonapeptides derived from α-defensin related precursors.1 Interestingly, θ-defensins still remain to date the only known cyclic polypeptides expressed in animals.6
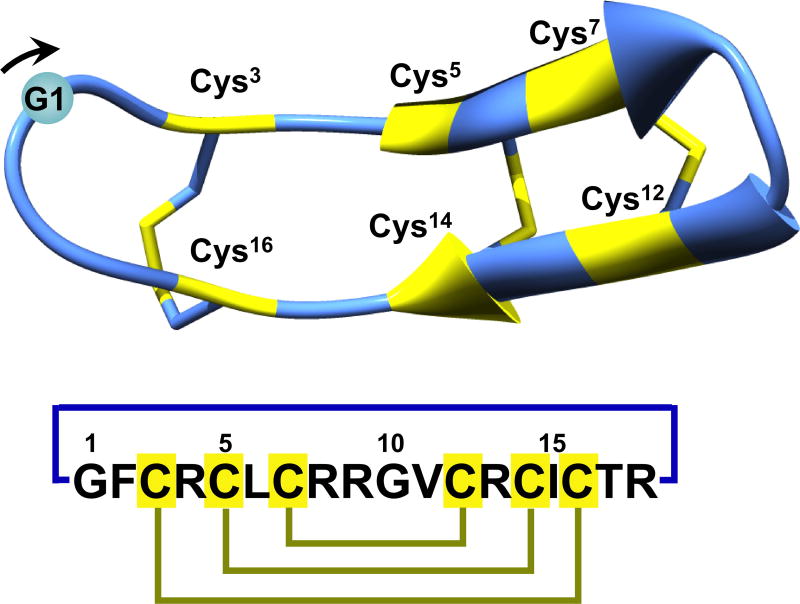
Rhesus θ-defensin-1 (RTD-1) was the first θ-defensin to be isolated from an extract of Rhesus macaques leukocytes.1 The structure of RTD-1 is different from that of α- and β-defensins, exhibiting a backbone-cyclized double β-hairpin-like fold with two antiparallel β-strands stabilized by three disulfide bonds in a ladder configuration (Fig. 1).7 Other less abundant θ-defensins have been also been found in Rhesus macaques and have been named RTD-2 to RTD-6.6,8,9 θ-Defensins have been also found in other primate species.10–12 Interestingly, although humans possess genes encoding θ-defensins, they lost the ability to produce these circular peptides as a result of the introduction of a stop codon mutation within the signal sequence that prevents subsequent translation.13
Figure 1.
Primary and tertiary structures of rhesus θ-defensin RTD-1 (PDB ID code: 1HVZ).7 The backbone cyclized peptide (connecting bond shown in blue) is stabilized by the three disulfide bonds in a ladder formation (disulfide bonds shown in yellow). Molecular graphics were created using Yasara (www.yasara.org).
θ-Defensins have been shown to possess antibacterial activity against both Gram-positive and Gram-negative,1 however, this activity strongly depends on the experimental conditions, mostly buffer composition and presence of serum proteins, used in the antibacterial assays.14 For example, the antimicrobial activity of RTD-1 is negatively affected by the presence of 10% human serum or high ionic strength buffers.14 θ-Defensins also possess anti-fungal1 and anti-HIV13,15 activities. For example, chemically-produced human θ-defensins (called retrocyclins), which are derived from the human pseudogene sequences, have been shown to protect human cells from infection by HIV-113 and have been evaluated as potential topical anti-HIV agent for the prevention of HIV transmission,16–18 showing some promise when compared to other topical anti-HIV drugs in pre-clinical development.15
It has been speculated that the anti-HIV activity of θ-defensins may be linked to their ability to bind gp120 and CD4 glycoproteins, which protects cells from HIV-1 infection.19 θ-Defensins have also been shown to inactivate germinating anthrax spores and act as a non-competitive inhibitor of anthrax lethal factor (LF) protease and other metalloproteases like TNF-α converting enzyme (TACE).20,21 A recent study demonstrated that the biological activity of RTD-1 against LF and TACE can be significantly improved using chemical molecular evolution techniques.21 Naturally occurring θ-defensins have been also shown to possess anti-inflammatory properties in animal models,22 likely due to the ability to inhibit key metalloproteases involved in the inflammatory response like TNF-α.
The circular backbone topology of θ-defensins is required for their biological activity as their linear counterparts show a decreased biological activity.21 The presence of three disulfides and a circular backbone topology makes θ-defensins a particularly stable peptide framework that shows high resistance to proteolytic degradation in serum and plasma.22,23 Altogether, these data suggest that the θ-defensin is an ideal molecular framework for the development of novel peptide-based therapeutics.22,24
In nature, RTD-1 is produced by binary ligation of two protein precursors containing truncated α-defensin sequences,1 however the complete biochemical mechanism including the enzymes required for their biosynthesis have not been fully elucidated yet. RTD-1 can also be chemically synthesized in a ‘one-pot’ cyclization-folding reaction using an intramolecular version of native chemical ligation (NCL).25 This approach has been recently employed for the chemical production of RTD-1 based libraries to optimize its biological activity against LF and TACE proteases.21 RTD-1 can be also biosynthesized employing standard bacterial expression systems using a modified protein splicing unit in combination with an in-cell intramolecular NCL reaction to perform the backbone cyclization of the linear peptide precursor.26
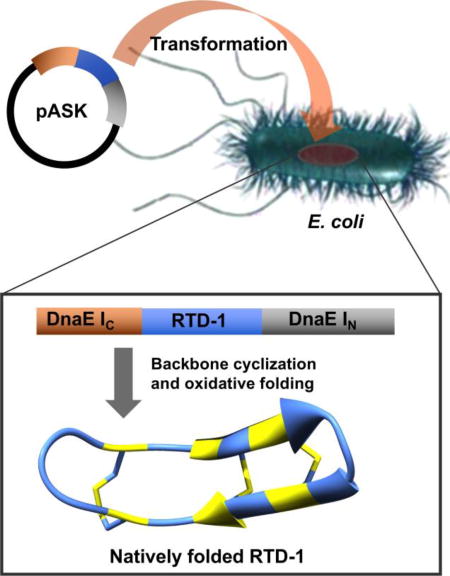
In this work, we report an alternative and efficient approach for the recombinant expression of RTD-1 using protein trans-splicing (PTS) to facilitate the in-cell cyclization process and improve the expression yield (Fig. 2). RTD-1 obtained by in-cell PTS adopted a native structure and was fully active inhibiting LF protease. In-cell expression of RTD-1 by PTS was also highly efficient, reaching intracellular concentrations ≈ 360 µM (≈70 mg of RTD-1 per 100 g of wet cells). Intriguingly and despite the antimicrobial properties described for RTD-1, no cellular toxicity was observed when overexpressed in E. coli cells. The natively folded RTD-1 peptide was isolated from the insoluble fraction of the cell lysate. This approach was also used successfully to prepare a genetically-encoded library based on RTD-1, which should allow the screening of new peptides based on the θ-defensin molecular scaffold for improving or finding novel biological activities.
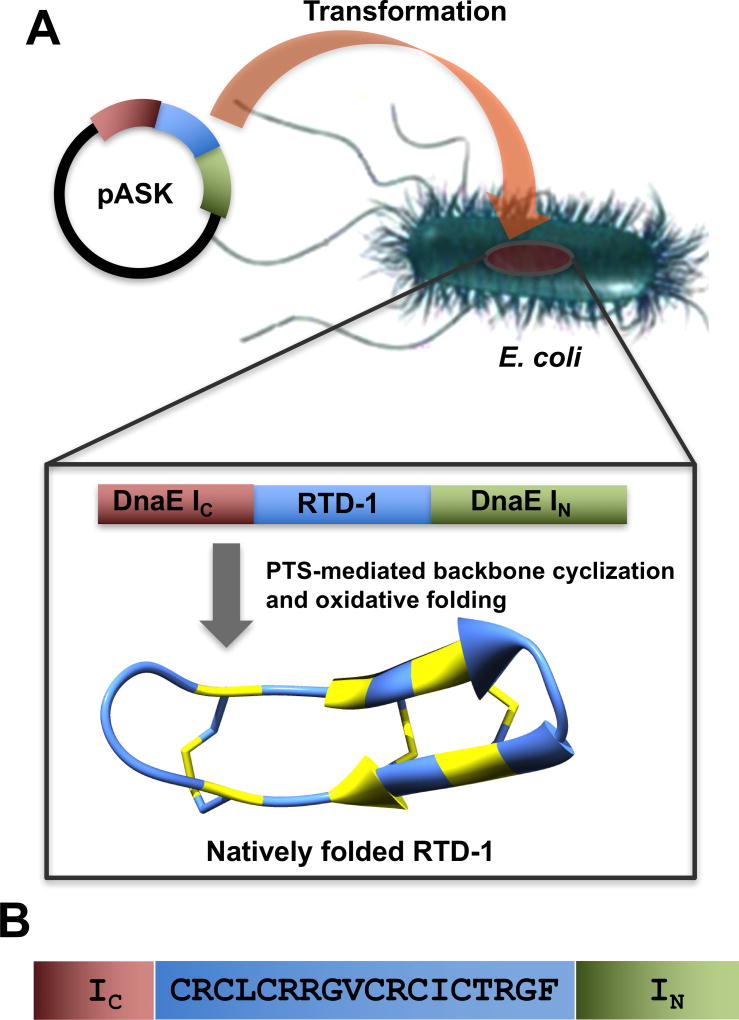
Figure 2.
Biosynthesis of the θ-defensin RTD-1 in E. coli cells. A. In-cell production of natively folded RTD-1 in bacterial cells using PTS-mediated backbone cyclization and oxidative folding. B. Sequences of the linear precursor used for in-cell production of RTD-1. The precursor contains an N-terminal containing linear precursor of RTD-1 (cyclization site is located between residues 2 and 3) fused to the N- and C-termini of the Npu DnaE IN and IC polypeptides, respectively. The His tag located at the N-terminus of the constructs is not shown for clarity.
2. Results and Discussion
Expression of peptide RTD-1 in living bacterial cells was explored by using of PTS in combination with the ultra-fast and highly efficient Nostoc punctiforme PCC73102 (Npu) DnaE split-intein to facilitate the in-cell cyclization process and to increase the expression yield. The DnaE split intein has one the highest reported rates of protein trans-splicing (t1/2 ≈ 60 s) and has a high splicing yield27,28. Also, in contrast with other split-inteins, the Npu DnaE split intein has been also shown to have minimal sequence requirements at the intein-extein junctions,27–31 which should allow the production of native RTD-1 without the need to add extra extein residues at the intein-exteins junctions to facilitate the trans-splicing reaction.
PTS is a post-translational modification similar to protein splicing with the difference that the intein self-processing domain is split into two fragments, the N- (IN) and C-intein (IC) polypeptides. These intein fragments are not active individually, but they can form an active protein splicing or intein domain in trans when they bind to each other under appropriate conditions.32,33 Accordingly, PTS-mediated backbone cyclization can be accomplished by fusing the IN and IC fragments to the C- and N-termini, respectively, of the linear polypeptide precursor to be cyclized. Once the two split-intein fragments bind to each other in an intramolecular fashion, the concomitant trans-splicing reaction yields a backbone-cyclized polypeptide (Fig. 2).33 This approach has been recently used successfully for the efficient heterologous expression of other disulfide rich peptides such as the trypsin inhibitor SFTI-1 and several cyclotides using bacterial and yeast expression systems.29–31
RTD-1 contains six cysteine residues that could be potentially used for cyclization. However, the Cys residues located in positions 3 and 7 are the only ones in the sequence of RTD-1 that do not have a charged or β-branched residue N-terminally adjacent, which should facilitate the kinetics of the cyclization reaction without affecting the splicing activity of the intein (Fig. 1). Both cyclization sites have shown to provide very good cyclization yields in the chemical synthesis and recombinant expression of RTD-1.21,26,33 In this work, we used the native residue Cys3 to facilitate the PTS-mediated backbone cyclization (Fig. 2). Accordingly, the corresponding RTD-1 linear precursor was fused in-frame at the C- or N-termini directly of the Npu DnaE IN and IC polypeptides, respectively (Fig. 2). A His-tag was also added at the N-terminus of the construct to facilitate identification and purification.
In-cell expression of the RTD-1 using PTS-mediated backbone cyclization was achieved by transforming the plasmid encoding the RTD-1 precursor construct into Origami 2(DE3) cells to facilitate oxidative folding. The split-intein construct was over expressed for 18 h at room temperature. Under these conditions the RTD-1 protein precursor (quantified based on the IN and IC fragments) was expressed at very high level (≈0.7 mg of RTD-1 per gram of wet E. coli cells) and was almost completely cleaved (Fig. 3A). In order to purify the intein-precursor protein, cells were also induced for a shorter time (2 h at 37 °C), however, no precursor could be detected under these new conditions either (Fig. 3A). It is worth noting that induction at 37 °C for 2 h produced around three and six times less precursor protein and RTD-1 peptide, respectively. The extremely high reactivity of the RTD-1-intein precursor prevented us from performing a full characterization of the precursor protein including kinetic studies of the trans-splicing induced reaction in vitro. The high reactivity of this Npu DnaE-base intein construct, highlights the low sequence requirements of this split-intein, which has been shown to provide high splicing activity and expression yields for the production of other Cys-rich backbone cyclized polypeptides, including cyclotides and the backbone cyclized Bow-Birkman trypsin inhibitor SFTI-1 (Table 1).29–31,34
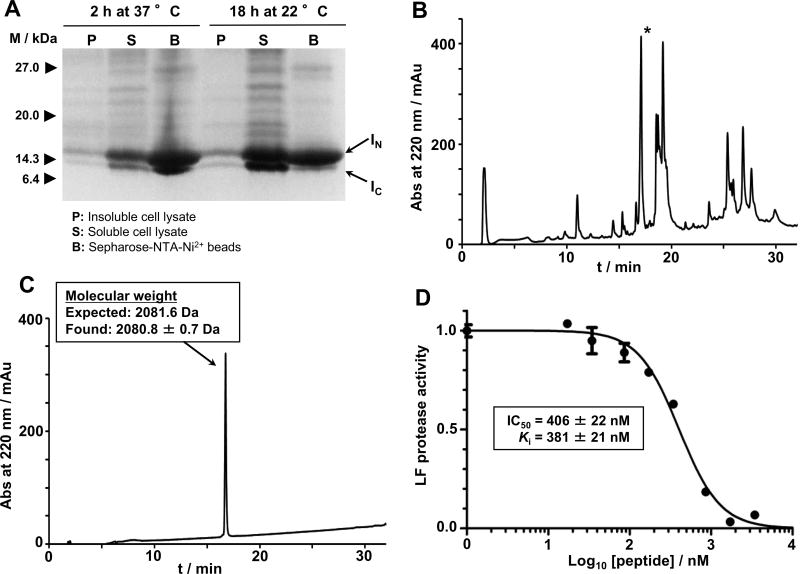
Figure 3.
In-cell expression of RTD-1 in E. coli cells using Npu DnaE intein-mediated PTS. A. SDS-PAGE analysis of the recombinant expression of RTD-1-intein precursor in Origami2(DE3) cells for in-cell production of RTD-1 at different induction conditions. B. Analytical HPLC trace of the solubilized cell lysate pellet obtained by inducing bacterial expression of the RTD-1 precursor for 18 h at 22 °C. The peak corresponding to the folded RTD-1 is indicated with an asterisk. C. Analytical HPLC and observed molecular weight by ES-MS of purified RTD-1. The calculated mass corresponds to the average isotopic mass. D. Biological activity of purified RTD-1. Inhibition assay of RTD-1 against anthrax LF protease. Different concentrations of RTD-1 were tested against LF protease. At each concentration, residual LF activity was measured and divided by the activity of LF in the absence of inhibitor. Activity was measured as the rate LF protease cleaves a fluorescence LF substrate43 and determined by the rate of fluorescence signal decaying (see the experimental section).
Table 1.
The Npu DnaE split-intein shows high sequence tolerance at the N- and C-extein/intein junctions while keeping high protein trans-splicing activity. The table shows the N- and C-extein sequences found in the Npu Dna split-intein and in several protein constructs used for the production of different backbone cyclized polypeptides. Only the last and first five residues of the N- and C-extein are shown, respectively. The first and last residues of the IN and IC polypeptides, respectively, are highlighted in bold.
| Spliced protein | IC/C-extein junction |
N-extein/IN junction |
Splicing efficiency |
References |
|---|---|---|---|---|
| Npu DnaE pola | N-CFNKS | EVFEY-C | ≈80%a | 27,28 |
| MCoTI-Ib | N-CGSGS | RGNGY-C | >98% | 29,30 |
| SFTI-Ib | N-CTKSI | FPDGR-C | >98% | 31 |
| N-CFPDG | SIPPI-C | >98% | ||
| RTD-1b | N-CRCLC | CTRGF-C | ≥95% | This work |
Intermolecular protein trans-splicing results in the production of full-length DnaE Pol
Intramolecular protein trans-splicing results in the production of backbone cyclized polypeptides
Next, we tried to isolate the natively folded RTD-1 generated in-cell. Interestingly, almost all of the folded RTD-1 produced inside living E. coli cells was found in the insoluble fraction of the cell lysate as quantified by HPLC (Fig. 3B) and LC-MS/MS (Fig. S3). θ-Defensins have been described to interact with model phospholipid membranes showing low nM affinity for glycoproteins (gp120 and cd4) and glycolipids (galactosylceramide).19 This could explain the presence of recombinant RTD-1 in the insoluble fraction of the cellular lysate, which is likely rich in insoluble membrane fragments containing glycolipids and/or membrane-bound peptidoglycan precursors. In fact, α- and β-defensins have been shown to bind to peptidoglycan precursors.35
Recombinant RTD-1 was purified by HPLC (Fig. 3C) by extracting the folded RTD-1 peptide from the insoluble fraction of the cell lysate with an 8 M guanidine hydrochloride solution. Interestingly, the RTD-1 peptide could not be extracted using detergents. The biological activity of pure RTD-1 was tested using an anthrax LF protease inhibitory assay (Fig. 3D).21 In vitro produced RTD-1 was fully active being able to inhibit anthrax LF protease with an IC50 of 406 ± 22 nM, which corresponds to a Ki ≈ 380 nM under the conditions used in the inhibitory assay. This Ki value is very similar to previously reported values,20,21,36 thus confirming the biological activity of the recombinantly produced θ-defensin.
NMR spectroscopy also confirmed that in-cell produced RTD-1 adopted a native θ-defensin fold.7,26 Both NOESY and ROESY spectra clearly showed NH-Hα connectivities below the water signal (≈ 4.7 ppm), which is consistent with the peptide adopting a β-sheet secondary structure as in the native θ-defensin fold (Fig. S1).37
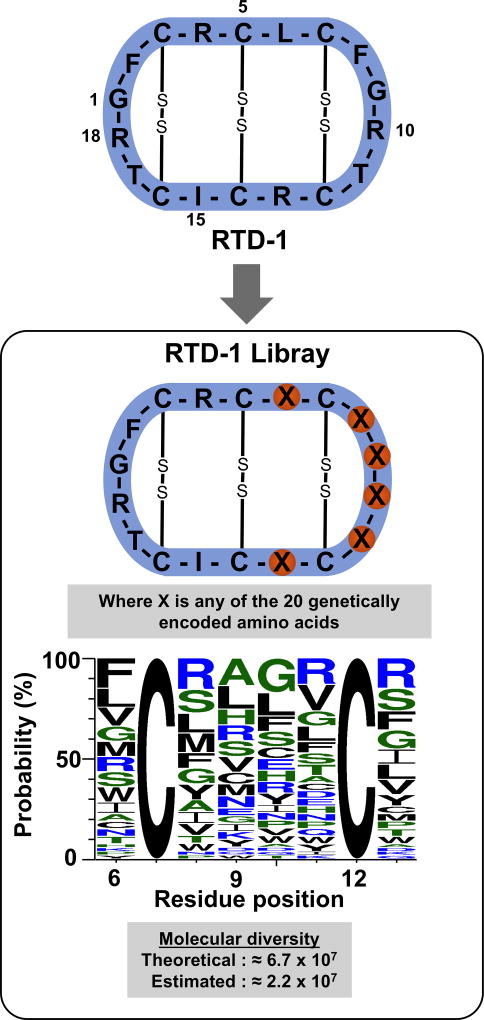
Encouraged by these results, we decided to construct a genetically-encoded library using the θ-defensin RTD-1 as molecular framework and PTS-mediated backbone cyclization. We randomized positions 6, 8, 9, 10, 11 and 13 in the RTD-1 peptide using all the 20 genetically-encoded amino acids (Fig. 4). This was accomplished at the DNA level by synthesizing a degenerate synthetic oligonucleotide template encoding the precursor RTD-1 polypeptide and using an NNK (where N = A, C, G or T and K = G or T) codon scheme for the randomized positions. This scheme uses 32 codons to encode all 20 amino acids and encodes only 1 stop codon (amber codon) giving a theoretical diversity for the library of ≈6.7 × 107 sequences. The real diversity of the library was estimated using the sequences of 60 clones (Table S1) and the WebLOGO algorithm38 providing a diversity of ≈ 2.2 × 107 sequences, ≈33% of the theoretical value (Fig. 4).
Figure 4.
Design and diversity characterization of the library. The genetically-encoded library based on the RTD-1 scaffold was produced at DNA level by synthesizing a degenerate synthetic oligonucleotide template encoding the precursor RTD-1 polypeptide and using a NNK (where N = A, C, G or T and K = G or T) codon scheme for the randomized positions located in residues 6, 8, 9, 10, 11 and 13. The library was characterized by sequencing 60 randomly picked clones and using the WebLOGO algorithm.38 The picture shows the WebLOGO picture of the θ-defensin peptide segment where the mutations were introduced. The Cys residues were conserved to allow the corresponding peptides adopting a native θ-defensin fold.
Next, 12 clones not containing Cys residues or stop codons at the randomized positions were selected and tested for bacterial expression in Origami 2(DE3) cell using the best expression conditions found for RTD-1. After expression at room temperature for 18 h, the corresponding cells were lysed and the amount of intein precursor and intein fragments was quantified by SDS-PAGE (Fig. S2 and Table S2). Only one clone of the twelve analyzed did not express any detectable amount of intein (clone #41, Table S2). The rest of the clones expressed the intein precursor with slightly better (clones #10, #14, #18 and #29, average expression yield of 134 ± 34% when compared to wt-RTD-1) or slightly worse (clones #2, #5, #9, #33, #37, #52 and #60, average expression yield of 50 ± 17% when compared to wt-RTD-1) yields than the clone encoding the wild-type RTD-1 precursor (Table S2). All the clones expressing the intein precursor spliced almost quantitatively (splicing yield >95%) with only clones #18 and #29 providing splicing yields slightly less efficient (≈90%). While these differences in splicing reactivity are very small, they may be attributed to the proximity of some of the randomized positions to the IC-N-extein junction. For example, residues at positions 6 and 8 are four and six residues away from the IC-N-extein junction.
Next, the amount of the corresponding θ-defensins present in the insoluble and soluble fractions of the corresponding clone cell lysates was quantified by LC-MS/MS (Table S2). All the clones except clone #37, which did not yield any detectable peptide with the correct molecular weight, produced detectable amounts of peptides with the correct mass for a backbone cyclized structure containing three disulfides. Clones #9, #18, #29, #33 and #52 provided better production yield of the corresponding oxidized backbone cyclized peptide than the wild-type θ-defensin RTD-1 (≈0.68 mg of peptide per gram of wet cells), with expression yields ranging from ≈1.09 mg (clone #33) to ≈2.45 mg (clone #18) of the corresponding peptide per gram of wet cells. Clones #14 and #60 provided similar yields for the corresponding oxidized cyclic peptides to that of the wild-type RTD-1. On the other hand, clone #2 provided an expression yield around ≈5 times smaller than that of the wild-type RTD-1, while clones #5 and #10 barely expressed any detectable peptide with expression yields around 100 times smaller than that of the wild-type RTD-1. The expression level of the intein precursors for these clones ranged from 40% to 130% of that of the RTD-1 precursor, respectively, and no precursor intein was found in the insoluble fraction of the corresponding cell lysates (data not shown), which strongly suggests that the low expression yield for these peptides could be attributed to the poor cyclization and folding propensity of these sequences.
Intriguingly, most of the cyclic peptides that had good expression yields were found in the insoluble fraction of the cell lysate (Table S2). Only clones #10, #14, #29, #52 and #60 provided detectable levels of the corresponding cyclic oxidized peptides in the soluble fraction of the cell lysate, with percentages ranging from ≈30% (#29) to ≈3% (clones #14 and #52) of the total peptide amount. As observed for RTD-1, the fact that the peptides found in the insoluble fraction of the cell lysate had molecular weights consistent with the presence of three disulfide bonds and a backbone-cyclized topology and no intein-precursors were found in the insoluble fraction, strongly suggests that they may also bind different components present in the insoluble fraction of the cell lysate. This result should not be totally unexpected since all the members of this library share ≥66% of the sequence of RTD-1 as only 6 residues of the of the original 18 residues in RTD-1 were randomized. As for RTD-1, the fact that the peptides found in the insoluble fraction of the cell lysate had molecular weights consistent with the presence of three disulfide bonds and a backbone-cyclized topology strongly suggests that they may bind with different components present in the insoluble fraction of the cell lysate.
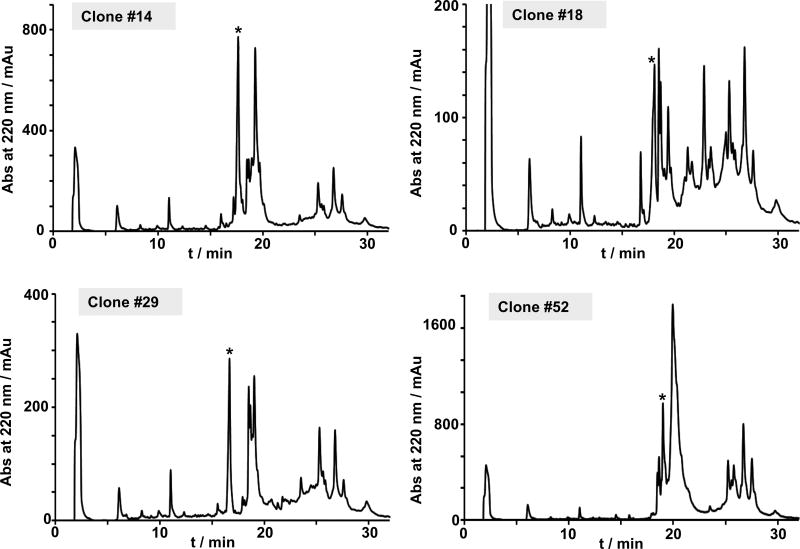
Four of the clones expressing the corresponding cyclic peptides in high yield (clones #14, #18, #29, and #52) were expressed in larger scale and the insoluble fraction of the cell lysate analyzed by analytical HPLC, ES-MS and LC-MS/MS (Figs S3 and S4). In all the cases a peak corresponding to the fully oxidized backbone-cyclized polypeptide was detected in the corresponding cell lysate crudes (Fig. 5). In addition, analysis by LC-MS/MS of the crude cell lysates showed very clean traces indicating the presence of a major backbone-cyclized folded specie for the different clones analyzed (Fig. S3), which is indicative of a stable tertiary structure.
Figure 5.
In-cell expression of selected individual clones from the RTD-1 library in E. coli cells. Analytical HPLC traces of the solubilized cell lysate pellet obtained by inducing bacterial expression of selected RTD-1 analogs. Expression of the precursor was carried out for 18 h at 22 °C. An asterisk indicates the desired folded cyclic peptide.
Clones #14, #18, #29 and #52 were also tested for its ability to inhibit anthrax LF protease (Table 2 and Fig. S5). Interestingly, clones #14, #18 and #52 inhibited LF protease with IC50 values ≈ 1 µM, about two times less potent than RTD-1. On the other hand, clone #29 had a very weak anti-LF activity with an IC50 value above 10 µM. These results are consistent with these clones adopting a native θ-defensin fold, as the reduced version of RTD-1 has been shown to be unable to inhibit LF protease. Furthermore, NMR characterization of these peptides also showed clear evidence of a well-folded structure with high β-sheet structure content (data not shown) confirming again their native θ-defensin structure.
Table 2.
Anti-LF activity of selected RTD-1 analogs obtained from the genetically-encoded library based on the θ-defensin RTD-1. Mutated residues are shown in bold in the RTD-1 analogs obtained from the library. The wild-type θ-defensin RTD-1 is also shown for comparison purposes.
| Peptide | Sequence | IC50 / nM | Ki / nM |
|---|---|---|---|
| RTD-1 | GFCRCLCRRGVCRCICTR | 406 ± 22 | 381 ± 21 |
| Clone #14 | GFCRCACLHGVCRCICTR | 971 ± 48 | 946 ± 47 |
| Clone #18 | GFCRCFCGRSSCDCICTR | 1,198 ± 84 | 1,173 ± 92 |
| Clone #29 | GFCRCNCASEHCTCICTR | > 10,000 | ND |
| Clone #52 | GFCRCPCSRYGCWCICTR | 971 ± 60 | 946 ± 58 |
3. Conclusions
In summary, we have shown that PTS-mediated backbone cyclization using the highly efficient Npu DnaE split-intein can be employed for the extremely efficient production of native folded θ-defensin RTD-1 inside live E. coli cells. We estimate that the in-cell production of RTD-1 was around 60 times more efficient than the previously reported method using intein-mediated intramolecular-NCL (also known as expressed protein ligation (EPL)39), thereby providing an attractive alternative for the production of these type of backbone-cyclized defensins in bacterial expression systems. The use of PTS-mediated backbone cyclization using the Npu DnaE split-intein has been also shown to be more efficient in the recombinant production of other backbone-cyclized polypeptides, such as cyclotides29,30,34 and the backbone-cyclized Bow-Birkman trypsin inhibitor SFTI-1.31 This highlights the high sequence tolerance at the extein-intein junctions of this split-intein (Table 1). For example, the second residue of the C-extein can be an aromatic (Phe), positive charged (Arg) or β-branched (Thr) residue without affecting the splicing activity of this intein. Similarly, this intein also tolerates the presence of aromatic (Tyr, Phe), positive charged (Arg) and β-branched (Ile) residues at the last position of the N-extein. This intein can also tolerate well sequences rich in Pro and Cys residues at both intein-extein junctions, which is remarkable. The use of highly tolerant split-inteins is key for the efficient production of genetically-encoded of libraries of backboned-cyclized polypeptides, which should prevent any sequence-based bias during the selection process of the corresponding library.
The in-cell production of bioactive backbone-cyclized peptides like RTD-1 containing a network of disulfide bonds may have tremendous potential for drug discovery due to the high stability of these scaffolds to chemical and biological degradation.23 Previous studies have shown that the θ-defensin scaffold may provide an ideal scaffold for improving21 and/or introducing novel biological activities.23,40 In fact, we have also shown in this work that this θ-defensin can be used in combination with PTS-based backbone-cyclization for the production of large genetically-encoded combinatorial libraries inside of living E. coli cells. As proof of principle, we created in this work an RTD-1-based library with a complexity of ≈ 2.2 × 107. Around 50% of 12 randomly selected clones from the library expressed in E. coli the corresponding folded backbone-cyclized peptide in high yield. These libraries, when coupled to an appropriate in-cell reporter system, should provide a high throughput approach for the selection of novel peptide sequences with new biological activities.
4. Experimental Section
General methods and reagents
Analytical reverse phase (RP)-HPLC was performed on a HP1100 series instrument with 214 nm, 220 nm and 280 nm detection using a Vydac C18 column (5 mm, 4.6 × 150 mm) at a flow rate of 1 mL/min. Semipreparative RP-HPLC was performed on a Waters Delta Prep system fitted with a Waters 2487 Ultraviolet-Visible (UV-vis) detector using a Vydac C18 column (5 µm, 10 × 250 mm) at a flow rate of 5 mL/min. All runs used linear gradients of 0.1% aqueous trifluoroacetic acid (TFA, solvent A) vs. 0.1% TFA, 90% MeCN in H2O (solvent B). UV-vis spectroscopy was carried out on an Agilent 8453 diode array spectrophotometer. Electrospray mass spectrometry (ES-MS) was performed on an Applied Biosystems API 3000 triple quadrupole mass spectrometer using Analyst 1.4.2. Calculated masses were obtained using Analyst 1.4.2. Protein samples were analyzed by SDS-PAGE 4–20% tris(hydroxymethyl)-aminomethane (Tris)-glycine gels (Lonza, Rockland, ME). The gels were then stained with Pierce (Rockford, IL) Gelcode Blue, photographed/digitized using a Kodak (Rochester, NY) EDAS 290, and quantified using NIH Image-J software (http://rsb.info.nih.gov/ij/). The integrity of all plasmids was confirmed by DNA sequencing. DNA sequencing was performed by Retrogen Inc., and the sequence data was analyzed with DNAStar (Madison, WI) Lasergene v8.0.2. Amino acid analysis was performed at the Amino Acid Laboratory in the Department of Molecular Biosciences, School of Veterinary Medicine, University of California at Davis. All chemicals were obtained from Sigma-Aldrich (Milwaukee, WI) unless otherwise indicated.
Cloning of pASK-TS-RTD
Synthetic DNA oligos (Integrated DNA Technologies, Coralville, IA) encoding RTD-1 (5’ - CGA ACT GCC GTT GCC TGT GCC GTC GTG GTG TTT GCC GTT GCA TCT GCA CCC GTG GTT TCT GTT TAT CAT ATG AAA CGG A - 3’ and 5’ - GAT CTC CGT TTC ATA TGA TAA ACA GAA ACC ACG GGT GCA GAT GCA ACG GCA AAC ACC ACG ACG GCA CAG GCA ACG GCA GTT - 3’) were annealed and ligated into BstBI and BglII treated pASK-TS-MCoTI31 to give plasmid pASK-TS-RTD.
Cloning of pASK-TS-RTD-L1
The dsDNA encoding the RTD-1 linear precursor with the residues at positions 6, 8, 9, 10, 11 and 13 randomized was amplified by polymerase chain reaction (PCR) using 5’ - GGC TTC ATA GCT TCG AAC TGC CGT TGC NNK TGC NNK NNK NNK NNK TGC NNK TGC ATC TGC ACC CGT GGT TTC TGT TTA TCA TAT GAA ACG GAG ATC TTG ACA GTA GA - 3’ (where N = A, C, G or T and K=G or T) as template. The 5’-primer (5’ - GGG TTC ATA GCT TCG AAC TGC C - 3’) encoded a BstB I restriction site. The 3’-primer (5’ - TCT ACT GCT AAG ATC TCC GTT TCA TAT - 3’) encoded a Bgl II. The PCR products were digested with BstB I and Bgl II, and ligated into a BstB I, Bgl II-treated pASK-TS-MCoTI to give the library plasmid pASK-TS-RTD-L1. The library plasmid was transformed into electrocompetent DH5-a cells to quantify the cell library size. To characterize the library at the DNA level, 60 individual clones were randomly picked and the RTD-1 encoding region was sequenced. Library complexity was calculated using the WebLOGO algorithm.38
Expression and purification of the RTD-1 peptide
Origami 2(DE3) cells (Novagen) were transformed with plasmids pASK-TS-RTD or individual library plasmids. Expression was carried out in 1 L of 2XYT medium with carbenicillin (100 mg/L) at room temperature. Briefly, 5 mL of an overnight starter culture were used to inoculate 1 L of 2XYT. Cells were grown to an OD600≈ 0.6 at 37° C. Protein expression was induced by addition of anhydrotetracycline hydrochloride (AHT) to a final concentration of 200 µg/L at room temperature for overnight. The cells were harvested by centrifugation, resuspended in 30 mL of lysis buffer (1 mM PMSF, 10 mM imidazole, 50 mM sodium phosphate, 300 mM NaCl buffer at pH 8.0 containing 5% glycerol) and then lysed by sonication. The lysate was clarified by centrifugation at 15,000 rpm in a Sorval SS-34 rotor for 30 min. The clarified supernatant was incubated with 1 mL Ni-NTA agarose beads (Qiagen), previously equilibrated with column buffer (20 mM imidazole, 50 mM sodium phosphate, 300 mM NaCl buffer at pH 8.0) at 4° C for 1 h with gentle rocking. The beads were separated from the cell lysate by centrifugation and then extensively washed with column buffer containing 0.1% Triton X-100 followed by washes with column buffer without detergent. Quantification of the level of expression was performed by quantifying the IC and IN polypeptides by SDS-PAGE analysis (Fig. S2). The insoluble fraction of the cell lysate was first washed three times with column buffer containing 0.1% Triton X-100 and then twice with just column buffer. The resulting pellet was dissolved in minimal amount of 8 M GdmCl in water. Both the soluble cell lysate and solubilized cell lysate pellet were extracted using C18 SepPak cartridges (Waters, Milford, MA) with elution in MeCN:H2O (3:2 vol.) containing 0.1% TFA and lyophilized. RTD-1 peptides were purified by semi-preparative HPLC using a linear gradient of 18%–32% MeCN in H2O (+0.1% TFA) over 30 min except for RTD-1 analog #52 where a linear gradient of 22%–35% MeCN in H2O (+0.1% TFA) was used instead. Purified RTD-1 and library peptides were characterized by analytical HPLC and ES-MS (Fig. S4).
LC-MS/MS peptide quantification
The soluble cell lysate and solubilized cell lysate pellet were extracted using C18 SepPak cartridges as described above. The lyophilized samples were dissolved in H2O containing 0.1% formic acid and analyzed by HPLC-tandem mass spectrometry using a C18-HPLC column (5 mm, 2.1 × 100 mm), and H2O-MeCN buffers containing 0.1% formic acid as mobile phase. The typical analysis used a linear gradient of 0%–90% MeCN in H2O over 10 min. Detection was performed on an API 3000 ES-MS using a multiple reaction-monitoring (MRM) mode. MRM analysis was performed using peaks at the 3rd and 4th charge states (e.g. for RTD-1, m/z = 521.5 and 694.6 were used). Data were collected and processed using Analyst software (Applied Biosystems). Quantification was performed using the same molecular ion in Q3. The calibration curve using pure RTD-1 was found to be linear in the range of 5–200 ng. The RTD-1 analogs were quantified using the RTD-1 calibration curve. Loss of RTD-1 during sample preparation was quantified by spiking a control sample (bacterial lysate) with different known amounts of purified RTD-1 and then analyzed by HPLC-MS/MS.
HPLC peptide quantification
RTD-1 peptide analogs were quantified by HPLC at 214 nm using a relative extinction coefficient calculated as previously described.21,41 A solution of pure peptide RTD-1 of known concentration estimated by amino acid analysis was used as internal control for HPLC quantification.
Anthrax LF protease inhibition assay
Lethal factor (LF) protease and the FRET-based substrate containing fluorescent proteins CyPet and YPet linked by a consensus sequence (RRKKVYPYPMEGTIA) with the linker (GGS)6 at both sites of the consensus sequence were expressed and purified as previously described.42,43 LF inhibition assay was performed in LF reaction buffer (10 µM CaCl2, 10 µM MgCl2, 20 µM ZnCl2, 20 mM sodium phosphate, and 100 mM NaCl at pH 7.2). Samples of 50 nM LF in LF reaction buffer (100 µL) were pre-incubated with different concentrations of RTD-1 ranging from 1 nM to 10 µM. After incubation at room temperature for 30 min, residual LF activity was measured by adding the FRET-based substrate to a final concentration of 10 nM and the decrease in FRET signal was measured every 2 min for 3 h.43 FRET was measured using an Envision 2103 plate reader (PerkinElmer) using an excitation wavelength of 405 nm. The relative FRET change was calculated using: FRC = It535/I0535, where I0 and It are the fluorescence intensities at time zero and at a particular time (t), at 535 nm. The initial velocities for the hydrolysis of substrate Lethal Factor in the presence of different concentrations of RTD-1 were fitted to a log(inhibitor) versus response with a variable Hill-slope using the software package Prism (GraphPad Software). Ki was calculated using the equation for non-competitive tightly bound inhibitors; where Ki = IC50 − E0/2 (E0 = enzyme concentration).44,45
NMR spectroscopy
NMR samples were prepared by dissolving the RTD-1 peptide into 80 mM sodium phosphate buffer at pH 6.0 in 90% H2O/10% 2H2O (v/v) or 100% D2O to a concentration of approximately 0.2 mM as previously described.26 All 1H NMR data were recorded on a Bruker Avance II 500 MHz spectrometer equipped with a TCI cryoprobe. Data were acquired at 27 °C, and 2,2-dimethyl-2-silapentane-5-sulfonate, DSS, was used as an internal reference. 2D 1H{1H}-TOCSY, 1H{1H}-NOESY and 1H{1H}-ROESY experiments were performed according to standard procedures46 with spectral widths of 14 ppm in proton dimension. The carrier frequency was centered on the water signal, and the solvent was suppressed by using WATERGATE pulse sequence. 2D-TOCSY (spin lock time 80 ms) and 2D-NOESY/2D-ROESY (mixing time 250 ms) spectra were collected using 4096 t2 points and 512 t1 blocks of 64 transients. Spectra were processed using Topspin 3.2 (Bruker).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Research Grant R01-GM113363 (J.A.C.), Department of Defense Congressionally Directed Medical Research Programs in Lung Cancer Grant LC150051 (J.A.C), BROAD Medical Research Program-Crohn's & Colitis Foundation of America Grant #483566 (J.A.C.), Lupus Research Institute (J.A.C) and Whittier Foundation (J.A.C).
References
- 1.Tang YQ, Yuan J, Osapay G, et al. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 1999;286(5439):498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 2.Tanabe H, Yuan J, Zaragoza MM, et al. Paneth cell alpha-defensins from rhesus macaque small intestine. Infect Immun. 2004;72(3):1470–1478. doi: 10.1128/IAI.72.3.1470-1478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6(6):551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 4.Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2(9):727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 5.Rosengren KJ, Daly NL, Fornander LM, et al. Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4. J Biol Chem. 2006;281(38):28068–28078. doi: 10.1074/jbc.M604992200. [DOI] [PubMed] [Google Scholar]
- 6.Leonova L, Kokryakov VN, Aleshina G, et al. Circular minidefensins and posttranslational generation of molecular diversity. J Leukoc Biol. 2001;70(3):461–464. [PubMed] [Google Scholar]
- 7.Trabi M, Schirra HJ, Craik DJ. Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from Rhesus macaque leukocytes. Biochemistry. 2001;40(14):4211–4221. doi: 10.1021/bi002028t. [DOI] [PubMed] [Google Scholar]
- 8.Tran D, Tran PA, Tang YQ, Yuan J, Cole T, Selsted ME. Homodimeric theta-defensins from rhesus macaque leukocytes: isolation, synthesis, antimicrobial activities, and bacterial binding properties of the cyclic peptides. J Biol Chem. 2002;277(5):3079–3084. doi: 10.1074/jbc.M109117200. [DOI] [PubMed] [Google Scholar]
- 9.Tongaonkar P, Tran P, Roberts K, et al. Rhesus macaque theta-defensin isoforms: expression, antimicrobial activities, and demonstration of a prominent role in neutrophil granule microbicidal activities. J Leukoc Biol. 2011;89(2):283–290. doi: 10.1189/jlb.0910535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen TX, Cole AM, Lehrer RI. Evolution of primate theta-defensins: a serpentine path to a sweet tooth. Peptides. 2003;24(11):1647–1654. doi: 10.1016/j.peptides.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 11.Garcia AE, Osapay G, Tran PA, Yuan J, Selsted ME. Isolation, synthesis, and antimicrobial activities of naturally occurring theta-defensin isoforms from baboon leukocytes. Infect Immun. 2008;76(12):5883–5891. doi: 10.1128/IAI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stegemann C, Tsvetkova EV, Aleshina GM, Lehrer RI, Kokryakov VN, Hoffmann R. De novo sequencing of two new cyclic theta-defensins from baboon (Papio hamadryas) leukocytes by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2010;24(5):599–604. doi: 10.1002/rcm.4424. [DOI] [PubMed] [Google Scholar]
- 13.Cole AM, Hong T, Boo LM, et al. Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc Natl Acad Sci U S A. 2002;99(4):1813–1818. doi: 10.1073/pnas.052706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran D, Tran P, Roberts K, et al. Microbicidal properties and cytocidal selectivity of rhesus macaque theta defensins. Antimicrob Agents Chemother. 2008;52(3):944–953. doi: 10.1128/AAC.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penberthy WT, Chari S, Cole AL, Cole AM. Retrocyclins and their activity against HIV-1. Cell Mol Life Sci. 2011;68(13):2231–2242. doi: 10.1007/s00018-011-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munk C, Wei G, Yang OO, et al. The theta-defensin, retrocyclin, inhibits HIV-1 entry. AIDS Res Hum Retroviruses. 2003;19(10):875–881. doi: 10.1089/088922203322493049. [DOI] [PubMed] [Google Scholar]
- 17.Owen SM, Rudolph D, Wang W, et al. A theta-defensin composed exclusively of D-amino acids is active against HIV-1. J Pept Res. 2004;63(6):469–476. doi: 10.1111/j.1399-3011.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 18.Gallo SA, Wang W, Rawat SS, et al. Theta-defensins prevent HIV-1 Env-mediated fusion by binding gp41 and blocking 6-helix bundle formation. J Biol Chem. 2006;281(27):18787–18792. doi: 10.1074/jbc.M602422200. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Cole AM, Hong T, Waring AJ, Lehrer RI. Retrocyclin, an antiretroviral theta-defensin, is a lectin. J Immunol. 2003;170(9):4708–4716. doi: 10.4049/jimmunol.170.9.4708. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Mulakala C, Ward SC, et al. Retrocyclins kill bacilli and germinating spores of Bacillus anthracis and inactivate anthrax lethal toxin. J Biol Chem. 2006;281(43):32755–32764. doi: 10.1074/jbc.M603614200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Gould A, Aboye T, et al. Full Sequence Amino Acid Scanning of theta-Defensin RTD-1 Yields a Potent Anthrax Lethal Factor Protease Inhibitor. J Med Chem. 2017;60(5):1916–1927. doi: 10.1021/acs.jmedchem.6b01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaal JB, Tran D, Tran P, et al. Rhesus macaque theta defensins suppress inflammatory cytokines and enhance survival in mouse models of bacteremic sepsis. PLoS One. 2012;7(12):e51337. doi: 10.1371/journal.pone.0051337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conibear AC, Bochen A, Rosengren KJ, et al. The cyclic cystine ladder of theta-defensins as a stable, bifunctional scaffold: A proof-of-concept study using the integrin-binding RGD motif. Chembiochem. 2014;15(3):451–459. doi: 10.1002/cbic.201300568. [DOI] [PubMed] [Google Scholar]
- 24.Tongaonkar P, Trinh KK, Schaal JB, et al. Rhesus macaque theta-defensin RTD-1 inhibits proinflammatory cytokine secretion and gene expression by inhibiting the activation of NF-kappaB and MAPK pathways. J Leukoc Biol. 2015;98(6):1061–1070. doi: 10.1189/jlb.3A0315-102R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aboye TL, Li Y, Majumder S, Hao J, Shekhtman A, Camarero JA. Efficient one-pot cyclization/folding of Rhesus theta-defensin-1 (RTD-1) Bioorg Med Chem Lett. 2012;22(8):2823–2826. doi: 10.1016/j.bmcl.2012.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gould A, Li Y, Majumder S, et al. Recombinant production of rhesus theta-defensin-1 (RTD-1) using a bacterial expression system. Mol Biosyst. 2012 doi: 10.1039/c2mb05451e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwai H, Zuger S, Jin J, Tam PH. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580(7):1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 28.Zettler J, Schutz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583(5):909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Jagadish K, Borra R, Lacey V, et al. Expression of fluorescent cyclotides using protein trans-splicing for easy monitoring of cyclotide-protein interactions. Angew Chem Int Ed Engl. 2013;52(11):3126–3131. doi: 10.1002/anie.201209219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jagadish K, Gould A, Borra R, et al. Recombinant Expression and Phenotypic Screening of a Bioactive Cyclotide Against alpha-Synuclein-Induced Cytotoxicity in Baker's Yeast. Angew Chem Int Ed Engl. 2015;54(29):8390–8394. doi: 10.1002/anie.201501186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Aboye T, Breindel L, Shekhtman A, Camarero JA. Efficient recombinant expression of SFTI-1 in bacterial cells using intein-mediated protein trans-splicing. Biopolymers. 2016;106(6):818–824. doi: 10.1002/bip.22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sancheti H, Camarero JA. "Splicing up" drug discovery. Cell-based expression and screening of genetically-encoded libraries of backbone-cyclized polypeptides. Adv Drug Deliv Rev. 2009;61(11):908–917. doi: 10.1016/j.addr.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboye TL, Camarero JA. Biological synthesis of circular polypeptides. J Biol Chem. 2012;287(32):27026–27032. doi: 10.1074/jbc.R111.305508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jagadish K, Camarero JA. Recombinant Expression of Cyclotides Using Split Inteins. Methods Mol Biol. 2017;1495:41–55. doi: 10.1007/978-1-4939-6451-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilmes M, Cammue BP, Sahl HG, Thevissen K. Antibiotic activities of host defense peptides: more to it than lipid bilayer perturbation. Nat Prod Rep. 2011;28(8):1350–1358. doi: 10.1039/c1np00022e. [DOI] [PubMed] [Google Scholar]
- 36.Kawakami T, Ohta A, Ohuchi M, Ashigai H, Murakami H, Suga H. Diverse backbone-cyclized peptides via codon reprogramming. Nat Chem Biol. 2009;5(12):888–890. doi: 10.1038/nchembio.259. [DOI] [PubMed] [Google Scholar]
- 37.Wishart DS, Sykes BD, Richards FM. Simple techniques for the quantification of protein secondary structure by 1H NMR spectroscopy. FEBS Lett. 1991;293(1–2):72–80. doi: 10.1016/0014-5793(91)81155-2. [DOI] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berrade L, Camarero JA. Expressed protein ligation: a resourceful tool to study protein structure and function. Cell Mol Life Sci. 2009;66(24):3909–3922. doi: 10.1007/s00018-009-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rapireddy S, Nhon L, Meehan RE, et al. RTD-1mimic containing gammaPNA scaffold exhibits broad-spectrum antibacterial activities. J Am Chem Soc. 2012;134(9):4041–4044. doi: 10.1021/ja211867j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuipers BJ, Gruppen H. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J Agric Food Chem. 2007;55(14):5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- 42.Kwon Y, Welsh K, Mitchell AR, Camarero JA. Preparation of peptide p-nitroanilides using an aryl hydrazine resin. Org Lett. 2004;6(21):3801–3804. doi: 10.1021/ol048417n. [DOI] [PubMed] [Google Scholar]
- 43.Kimura RH, Steenblock ER, Camarero JA. Development of a cell-based fluorescence resonance energy transfer reporter for Bacillus anthracis lethal factor protease. Anal Biochem. 2007;369(1):60–70. doi: 10.1016/j.ab.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Cer RZ, Mudunuri U, Stephens R, Lebeda FJ. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009;37(Web Server issue):W441–445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha S. Tight-binding inhibitors-I. Kinetic behavior. Biochem Pharmacol. 1975;24(23):2177–2185. doi: 10.1016/0006-2952(75)90050-7. [DOI] [PubMed] [Google Scholar]
- 46.Cavanagh J, Rance M. Suppression of cross relaxation effects in TOCSY spectra via a modified DISI-2 mixing sequence. J Magn Res. 1992;96:670–678. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.