Abstract
Eukaryotic translation initiation factor 2B (eIF2B) is the guanine nucleotide exchange factor of the GTPase eIF2, which brings the initiator Met-tRNAi to the ribosome in the form of the eIF2-GTP·Met-tRNAi ternary complex (TC). The activity of eIF2B is inhibited by phosphorylation of its substrate eIF2 by several stress-induced kinases, which triggers the integrated stress response (ISR). The ISR plays a central role in maintaining homeostasis in the cell under various stress conditions, and its dysregulation is a causative factor in the pathology of a number of neurodegenerative disorders. Over the past three decades, virtually every aspect of eIF2B function has been the subject of uncertainty or controversy: from the catalytic mechanism of nucleotide exchange, to whether eIF2B only catalyzes nucleotide exchange on eIF2 or also promotes binding of Met-tRNAi to eIF2-GTP to form the TC. Here, we provide the first complete thermodynamic analysis of the process of recycling of eIF2-GDP to the TC. The available evidence leads to the conclusion that eIF2 is channeled from the ribosome (as an eIF5·eIF2-GDP complex) to eIF2B, converted by eIF2B to the TC, which is then channeled back to eIF5 and the ribosome. The system has evolved to be regulated by multiple factors, including post-translational modifications of eIF2, eIF2B, and eIF5, as well as directly by the energy balance in the cell, through the GTP:GDP ratio.
Graphical Abstract
Eukaryotic translation initiation factor 2B (eIF2B) is one of the main targets in the regulation of protein synthesis in the cell. It is the guanine nucleotide exchange factor (GEF) of the GTPase eIF2, which when bound to GTP, brings the initiator Met-tRNAi to the ribosome, in the form of the eIF2-GTP·Met-tRNAi ternary complex (TC). eIF2 consists of α, β, and γ subunits, with eIF2γ being the actual GTPase, and eIF2α and -β serving accessory functions. Upon start codon recognition, the GTPase-activating protein (GAP) eIF5 promotes GTP hydrolysis. eIF2-GDP has a lower affinity for Met-tRNAi and is released from the ribosome. eIF2B catalyzes the conversion of eIF2-GDP back to eIF2-GTP and the binding of Met-tRNAi to produce a new TC.1–3
The activity of eIF2B is regulated by phosphorylation of its substrate eIF2, by binding of nucleotides and cofactors to eIF2B, and by phosphorylation of eIF2B itself. In humans, several kinases phosphorylate eIF2α at serine 51 (S51) in response to various types of stress, including viral infection (PKR), unfolded proteins in the ER (PERK), amino acid starvation (GCN2), and heme deficiency (HRI), in what is collectively known as the integrated stress response (ISR). Phosphorylated eIF2-GDP [eIF2(α-P)-GDP] is a competitive inhibitor of eIF2B. Inhibition of eIF2B activity causes downregulation of global protein synthesis and triggers the ISR by inducing the production of a set of transcription factors. The result is the activation of both pro-apoptotic pathways and pro-survival pathways aimed at restoring homeostasis (Figure 1). Because the stressors themselves can cause cell death, either an insufficient or overly aggressive stress response can lead to apoptosis. Dysregulated ISR is a causative factor in the pathology of a number of neurodegenerative disorders, including Alzheimer’s disease and prion disease.3–8
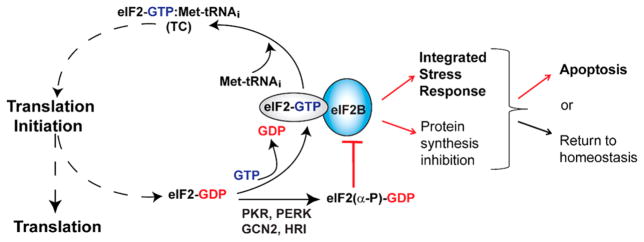
Figure 1.
Functions and regulation of eIF2B. eIF2 brings the Met-tRNAi to the ribosomal translation initiation complex, in the form of the eIF2-GTP·Met-tRNAi ternary complex (TC). Upon start codon recognition, eIF2 hydrolyzes GTP, and eIF2-GDP is released. eIF2B catalyzes nucleotide exchange and Met-tRNAi binding to form a new TC. Phosphorylation of the α subunit of eIF2 by several stress-activated kinases turns eIF2-GDP from a substrate into an inhibitor of eIF2B. Inhibition of eIF2B activity causes a decrease in the level of global protein synthesis and at the same time triggers the integrated stress response (ISR), which involves both pro-apoptotic and pro-survival pathways. The ultimate fate of the cell is either restoration of homeostasis or apoptosis, depending on the interplay between pro-survival and pro-apoptotic processes in the cell.
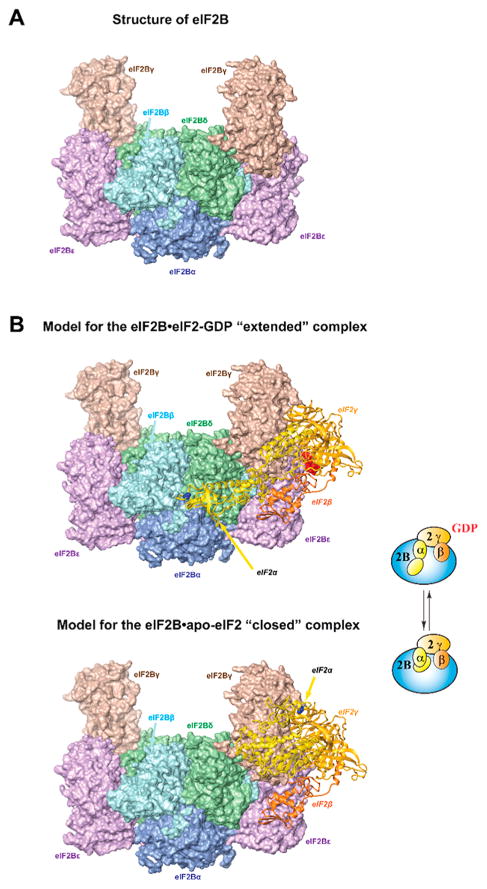
eIF2B has five subunits, α–ε, recently shown by us and others to assemble into an ~600 kDa decamer.9–13 eIF2Bγ and -ε are homologous to each other and form the catalytic subcomplex, eIF2Bγε (eIF2Bcat). The eIF2Bε C-terminal domain (eIF2Bε-CTD) is the catalytic domain. eIF2Bα, -β, and -δ (homologous to each other, but not to eIF2Bγε) form the hexameric regulatory subcomplex, eIF2Bα2(βδ)2 (eIF2Breg), to which two eIF2Bcat complexes bind (Figure 2A).11
Figure 2.
Structural basis for the mechanisms of eIF2B action and regulation. (A) Structure of Schizosaccharomyces pombe eIF2B,11 viewed from the eIF2α-binding pocket. The individual subunits are labeled and color-coded. Only one each of the eIF2Bα, -β, and -δ subunits are visible. (B) Model for the eIF2B·eIF2-GDP complex in an extended conformation from ref 45 (top). eIF2 subunits are shown as ribbons. The side chain of S51 in eIF2α is colored blue. GDP is colored red. Model of the eIF2B·apo-eIF2 complex in a closed conformation from ref 45 (bottom). Only the position of eIF2α-NTD is changed: it now contacts eIF2α-CTD and is near eIF2γ. The equilibrium between the GDP-bound and apo states of the eIF2B·eIF2 complex is shown schematically on the right.
While it has been reported that eIF2Bε has some catalytic activity in vitro, and the eIF2Bcat subcomplex has activity comparable to that of intact eIF2B,14 four of the five eIF2B subunits (β, γ, δ, and ε) are essential in Saccharomyces cerevisiae. Their essential function must be related to eIF2 nucleotide exchange, because the lethality of an eIF2B deletion is suppressed by co-overexpression of eIF2 and tRNAi. The resulting cells have a slow-growth phenotype, which is partly ameliorated by a mutation in eIF2γ that increases the rate of spontaneous GDP dissociation.15 The lethal phenotype of eIF2Bγ and eIF2Bε depletion can be suppressed by overexpression of only eIF2, without overexpressing tRNAi, while overexpressing only tRNAi is sufficient to suppress the lethality of eIF2Bδ depletion.16 Therefore, the essential functions of eIF2Bγ and eIF2Bε appear to be related to nucleotide exchange, while that of eIF2Bδ appears to be related to binding of Met-tRNAi to eIF2-GTP. eIF2Bβ depletion causes co-depletion of eIF2Bδ, and suppressing the lethal phenotype of eIF2Bβ depletion requires overexpression of both eIF2 and tRNAi. Therefore, it is not clear whether the essential function of eIF2Bβ is in only nucleotide exchange or also in Met-tRNAi binding.16
eIF2Bα deletion is not lethal in S. cerevisiae. Instead, it reduces the affinity of eIF2B for eIF2(α-P)-GDP and renders it resistant to inhibition by eIF2α phosphorylation: General control nonderepressible (Gcn−) phenotype, characterized by the inability to induce ISR under conditions of amino acid starvation (reviewed in refs 5 and 6). eIF2α and its phosphorylated form (eIF2α-P) bind in a pocket on eIF2Breg formed by the N-terminal domains (NTDs) of eIF2Bα, -β, and -δ (Figure 2).11 In S. cerevisiae, Gcn− mutations have been found in the eIF2Bα, -β, and -δ subunits: on the surfaces now known to contact eIF2α and at the interfaces between eIF2Bα2 and eIF2Bβδ. The former group of mutations directly lowers the eIF2B affinity for eIF2α, while the latter likely destabilizes the binding of eIF2Bα to the rest of eIF2B or changes the architecture of eIF2Breg and its eIF2α-binding pocket, thus indirectly affecting the affinity of eIF2B for eIF2α.11
Mutations that decrease eIF2B activity induce the ISR in the absence of eIF2α phosphorylation (General control derepressed, Gcd−). Gcd− mutations have been isolated not only in the four essential eIF2B subunits but also in eIF2Bα. Furthermore, some Gcn− mutations, when combined with eIF2Bα deletion, cause a Gcd− phenotype.3,5,16,17 Therefore, eIF2α binding in the eIF2Breg pocket appears to be important not only for inhibition by eIF2(α-P)-GDP but also for catalysis. However, catalysis and inhibition differ in their sensitivity to the strength of the interaction, with inhibition being affected first, while catalysis may become impaired only if eIF2α binding in the eIF2Breg pocket is severely disrupted. A role for the eIF2α–eIF2Breg interaction in catalysis is also supported by the observation that overexpression of eIF2β γ and tRNAi in S. cerevisiae suppresses the lethality of both eIF2B deletion and eIF2α deletion.15
EIF2B FUNCTIONS AND MECHANISM OF ACTION
Slow Rate of Dissociation of GDP from eIF2
The rate of dissociation of GDP from eIF2 is ~1 × 10−1 min−1 for S. cerevisiae eIF218,19 and even slower, ~5 × 10−3 min−1, for mammalian eIF2.20,21 Because translation is initiated on a time scale of seconds, eIF2, like many other GTPases, needs a GEF to accelerate GDP release. GEFs promote GDP dissociation by destabilizing the GDP-bound state of the GTPase.22,23 Accordingly, the eIF2B·eIF2 complex has an affinity for GDP (~1 μM) that is lower than that of eIF2 (~10 nM),20,24 and eIF2B has greater affinity for apo-eIF2 than for eIF2-GDP.20,24–27 Binding of eIF2 to eIF2B and binding of eIF2 to GDP are thermodynamically coupled. Thermodynamically coupled interactions exert reciprocal effects on each other, which stems from the Conservation of Energy Principle.28 For instance, if eIF2B lowers the affinity of GDP for eIF2 ~100-fold, GDP must lower the affinity of eIF2B for eIF2 ~100-fold, and the experimentally determined ratio of the KDs of eIF2B for eIF2-GDP and eIF2B for apo-eIF2 is indeed ~100:1.20,24–27
While the destabilizing effect of eIF2B on binding of GDP to eIF2 is not a subject of controversy, a recent article by Jennings and co-authors reported, based on an affinity pull-down assay, that eIF2B has the same affinity for eIF2-GDP, apo-eIF2, and eIF2-GTP.29 This result is at odds with previous reports20,24–27 and appears to indicate that an enzyme (eIF2B) has the same affinity for the substrate (eIF2-GDP) and the reaction intermediate (apo-eIF2), making it difficult to rationalize how eIF2B would be able to catalyze GDP dissociation. Furthermore, the affinity pull-down assay is not a standard method for KD determination.
eIF2-GDP:eIF2-GTP Equilibrium Ratio of 10:1
The affinity of eIF2 for GDP is ~100-fold greater than that for GTP, while the GDP concentration in the cell is only ~10-fold lower. Therefore, the equilibrium eIF2-GDP:eIF2-GTP ratio is ~10:1.20,24 An enzyme cannot change the equilibrium between a substrate and a product; it only accelerates the process of reaching the equilibrium. Therefore, if eIF2-GTP is the product of the exchange reaction, the most eIF2B can achieve is 10% eIF2-GTP.20,24 Such a challenge is overcome by coupling the energetically unfavorable step with a more favorable one, Met-tRNAi binding in the case of eIF2B. Because the equilibrium between eIF2-GDP and TC is ~1:1, coupling nucleotide exchange with Met-tRNAi binding yields a 1:1 substrate:-product ratio.20,24 Coupling could be achieved in two different ways. (i) Binding of Met-tRNAi to eIF2-GTP could continually remove eIF2-GTP and yield sufficient rates of TC formation by mass action, even though only 10% of eIF2 is bound to GTP. (ii) eIF2B catalyzes both nucleotide exchange and binding of Met-tRNAi to eIF2-GTP. Under this scenario, the TC is the product of eIF2B catalysis while eIF2-GTP is a reaction intermediate. Both of these scenarios have precedents. For instance, bacterial translation elongation factor Ts (EF-Ts, also known as EF-1B) appears to work as a “stand-alone” GEF and releases EF-Tu-GTP, followed by aa-tRNA binding. However, the equilibrium ratio of GTP- and GDP-bound EF-Tu (EF-1A) is more favorable than that of eIF2.30 In contrast, eukaryotic translation elongation factor 1B (eEF1B) has a high affinity for eEF1A-GTP, remains bound to it, and is released only upon aa-tRNA binding.31–33 Analysis of the available evidence shows that eIF2B works like eEF1A: it has high affinity for eIF2-GTP, the rate of eIF2-GTP release is too slow to be physiologically relevant (hallmark of a reaction intermediate), and the rate of GDP to GTP exchange is stimulated by the presence of Met-tRNAi, 34,35 consistent with scenario (ii) described above. Assays for eIF2B activity typically use exchange of unlabeled GDP for labeled GDP, because the rate of release of eIF2-GDP from eIF2B is approximately an order of magnitude faster than the rate of eIF2-GTP release.36,37 Exchange rates in the presence of both GTP and Met-tRNAi are comparable to those in the presence of GDP, because the rate of release of the TC from eIF2B is comparable to the rate of release of eIF2-GDP.34 Therefore, eIF2-GTP is a reaction intermediate, whereas the TC is the actual product of eIF2B catalysis.34,35 Unfortunately, this observation has largely been ignored in the field in recent years. The yeast genetic data on eIF2Bβ and -δ depletion,16 described above, also indicate that eIF2B may play a role not only in nucleotide exchange but also in binding of Met-tRNAi to eIF2-GTP. In fact, eIF2B appears to channel eIF2 even further, because it forms a relatively stable complex with the TC34,35 (see below).
Catalytic Mechanism of eIF2B
Like other GEFs,22,23 eIF2B acts as a GDP dissociating factor: it destabilizes binding of GDP to eIF2, which allows GTP to bind.38 However, the eIF2B catalytic mechanism had been the subject of a decades-long controversy, which has not completely died down even now. The confusion stems from the observation that eIF2B itself binds GTP, raising the idea that catalysis could involve direct transfer of GTP from eIF2B to eIF2.36,39 However, such a mechanism has not been observed in other GEFs,22,23 and Williams and co-authors showed that eIF2B itself does not need GTP to promote dissociation of GDP from eIF2.38 Furthermore, eIF2Bε-CTD (which has not been reported to bind GTP) has some GEF activity on its own.40 Therefore, it is clear that eIF2B can promote nucleotide exchange without direct transfer of GTP from eIF2B to eIF2. Nevertheless, the idea that binding of GTP to eIF2B can stimulate exchange through direct transfer or at least by somehow increasing the GTP concentration around eIF2 seems to persist and is difficult to rule out experimentally because GTP, like a number of other small molecules, can have an allosteric effect on eIF2B activity.36,41–43 However, this idea can be ruled out on the basis of thermodynamic analysis. Unlike other types of catalysis, nucleotide exchange does not involve formation or breaking of covalent bonds. Therefore, it does not appear to be possible to find even a theoretical scenario under which transfer of a GTP molecule from eIF2B to eIF2 would be energetically or kinetically beneficial for catalysis, nor is there a scenario under which binding of GTP to eIF2B would be strong enough to increase the effective concentration of GTP near eIF2 but weak enough for GTP to be transferred to eIF2 faster than the rate of binding of free GTP. Furthermore, the GTP concentration in the cell is ~0.5 mM, and GTP binding is hardly a rate-limiting step. Therefore, unless and until at least a theoretical mechanism is found, via which eIF2B-bound GTP could directly contribute to the rate of nucleotide exchange, it is safe to assume that the role of binding of GTP to eIF2B is allosteric.
Regulation of eIF2B Activity by eIF2α Phosphorylation
eIF2(α-P)-GDP is a competitive inhibitor of eIF2B, and it is generally assumed that phosphorylated eIF2α has a higher affinity for eIF2Breg and locks the eIF2B·eIF2(α-P)-GDP complex into an inhibited state that presumably has an architecture that is different from that of the eIF2B·eIF2-GDP complex.5,6,8,16,43,44 Recently, we showed that phosphorylation does not directly affect the affinity of eIF2α for eIF2Breg but instead acts indirectly by destabilizing an autoregulatory intramolecular interaction between eIF2α-NTD and -CTD, which enables eIF2α-NTD to bind to eIF2Breg.45 The available evidence indicates that the binding of phosphorylated and unphosphorylated eIF2α-NTD to eIF2B is similar, and we proposed a model in which the eIF2B·eIF2 complex exists in equilibrium between a nucleotide-bound state and an apo state. In the nucleotide-bound state, eIF2α is in an “extended” conformation, where eIF2α-NTD is bound in the eIF2Breg pocket and does not contact eIF2α-CTD. In the apo state, eIF2α is in a “closed” conformation, where eIF2α-NTD and -CTD contact each other and eIF2α-NTD is not in the eIF2Breg pocket (Figure 2B). eIF2B favors the apo state when eIF2α is not phosphorylated, thus promoting GDP dissociation. According to our model, eIF2α phosphorylation destabilizes the “closed” conformation of eIF2α and thus promotes the extended GDP-bound state at the expense of the “closed” apo state of the eIF2B·eIF2(α-P) complex. As a result, eIF2B has the same affinity for eIF2(α-P)-GDP as for the reaction intermediate, apo-eIF2(α-P), and thus has no catalytic activity for eIF2(α-P)-GDP.45 Therefore, to inhibit eIF2B activity, eIF2α phosphorylation does not need to change the overall architecture of the eIF2B·eIF2 complex into a putative new inhibited state; it only needs to change the relative stability of the complexes of the enzyme with the substrate (eIF2-GDP) and reaction intermediate (apo-eIF2).
CHANNELING OF EIF2
It was suggested as early as the 1980s that eIF2 may be channeled from eIF2B to the ribosomal translation initiation complex.35 While this idea has mostly been forgotten, evidence emerged in recent years that channeling of eIF2 could in fact be even more extensive. eIF2-GDP was reported to be released from the ribosome in complex with its GAP, eIF5, which competes with eIF2B for binding to eIF2-GDP.19,26,46 It was also recently reported that eIF2B and eIF5 compete for the TC,29 although the authors did not suggest that eIF2 is channeled from eIF2B to eIF5.
Stable eIF5·eIF2-GDP Complex
eIF2-GDP is released from the ribosome in a stable complex with eIF5.46–48 eIF5-CTD, which binds to eIF2β-NTT, is homologous to the catalytic eIF2Bε-CTD, and the two compete for binding to eIF2β. The concentrations of eIF2 and eIF5 are comparable (see, e.g., the PaxDb protein abundance database49); therefore, if there is no pool of free eIF2-GDP, eIF2B must displace eIF5 from eIF2-GDP, before it can promote nucleotide exchange. Thus, eIF5 acts as a GDP dissociation inhibitor (GDI), and eIF2B acts as a GDI dissociation factor (GDF).19,26,46 In S. cerevisiae, eIF5 inhibits by 2-fold the already slow rate of dissociation of GDP from eIF2,19 whereas human eIF5 has no effect on GDP dissociation.21 It should be noted that the role of GDIs is primarily to prevent binding of the GEF to the GTPase and thus GEF-catalyzed nucleotide exchange, rather than further decelerating an already slow spontaneous GDP dissociation.50 While the two-fold effect of yeast eIF5 on GDP dissociation from eIF2 could have some accessory role, it is clear that such an effect would be modest, compared to the inhibitory effect of eIF5 on eIF2B binding. Binding of eIF2B and eIF5 to eIF2 is not strictly competitive, because eIF2B· eIF2·eIF5 complexes were observed in S. cerevisiae when eIF5 was overexpressed. It appears that in these complexes, eIF5-CTD is bound to eIF2β-NTT, because a mutation at the eIF2β-binding surface of eIF5-CTD abrogated complex formation.46
Stable eIF2B·eIF2-GTP Complex and eIF2B·TC Complex
The slow rate of dissociation of eIF2-GTP from eIF2B was already discussed above. Because eIF2-GTP is an intermediate and not a product, it does not need to be released from eIF2B until it binds Met-tRNAi, forming the TC, which has lower affinity for eIF2B and can be released. Remarkably, the eIF2B·TC complex is also relatively stable, with rates of TC release on the order of ~5 min−1,34 and is able to withstand analytical centrifugation.35 Adding ribosomal initiation complexes stimulated eIF2B activity, and it was suggested that the TC may be channeled from eIF2B directly to the ribosome.35 If the rate of dissociation of the TC from eIF2B is too slow, then eIF2B needs to be displaced from the TC by another molecule. Because binding of eIF5 to eIF2 appears to be independent of the presence of a nucleotide or Met-tRNAi, or even has a slightly higher affinity for the TC,21,51 eIF5 would be able to compete more efficiently with eIF2B for TC binding than for binding to apo-eIF2 or eIF2-GTP. Unlike eIF5, the multifactor complex (MFC), composed of at least eIF3, eIF1, and eIF5, has a higher affinity for TC than for eIF2,21 and therefore, the MFC [or the 43S ribosomal preinitiation complex (PIC)] would be even more effective in displacing eIF2B.
Jennings and co-authors recently suggested that Met-tRNAi and eIF2B compete for eIF2-GTP.29 While Met-tRNAi clearly lowers the affinity of eIF2-GTP for eIF2B and vice versa,29,34 the distinction between weaker binding and competition effectively comes down to the relative KDs. Because the concentrations of both eIF2B and Met-tRNAi are in the range of 1 μM, if the KD for binding of eIF2B to the TC is in the nanomolar range (lower than the eIF2B concentration), then a significant amount of the eIF2B·TC complex exists. Conversely, if the KD for binding of eIF2B to the TC is in the micromolar range or greater, then the equilibrium will be in favor of free eIF2B and the TC. Previous reports indicate stable binding, consistent with a KD in the nanomolar range.34,35 Because binding of Met-tRNAi to eIF2-GTP in the presence of eIF2B, and vice versa, was observed at nanomolar concentrations,29 these results are also fully consistent with the existence of the eIF2B·TC complex in the cell. The authors proposed that eIF5 and eIF2B compete for the TC and that the eIF5·TC complex is the final product,29 which is the same conclusion as the one we reach here (see also the thermodynamic analysis below). The difference lies in how the eIF5·TC complex is formed. Jennings and co-authors propose that eIF2-GTP is first released from eIF2B, then binds Met-tRNAi to form the TC, and finally eIF5 binds the TC and “protects” it from eIF2B. However, as we explain above, the rate of release of eIF2-GTP from eIF2B appears to be too slow to be physiologically relevant. Also, the available data in the literature,34,35 as well as the results of Jennings and co-authors,29 indicate that the eIF2B·TC complex is relatively stable and eIF2B needs to be displaced from the TC.
Thus, there is now genetic and biochemical evidence in support of (i) the transfer of eIF2-GDP from eIF5 directly to eIF2B, (ii) the role of eIF2B in both nucleotide exchange and Met-tRNAi binding, with the TC (and not eIF2-GTP) being the product of eIF2B catalysis, and (iii) the transfer of the TC from eIF2B to eIF5. Therefore, eIF2 appears to be channeled in a full cycle from the ribosome to eIF2B, and back to the ribosome. The thermodynamic analysis presented below fully supports this conclusion.
Thermodynamic Description of the eIF2B Catalytic Cycle
We sought to combine the recent structural insights into the mechanism of eIF2B action and regulation described above with binding and rate constants derived from decades of research to build a complete thermodynamic description of the entire process and correlate it to respective structures (Figure 3). All experimental, calculated, and estimated KDs used to generate Figure 3 are listed in Table 1. The complete thermodynamic description of the eIF2B-catalyzed recycling of eIF2-GDP to the TC was built as follows.
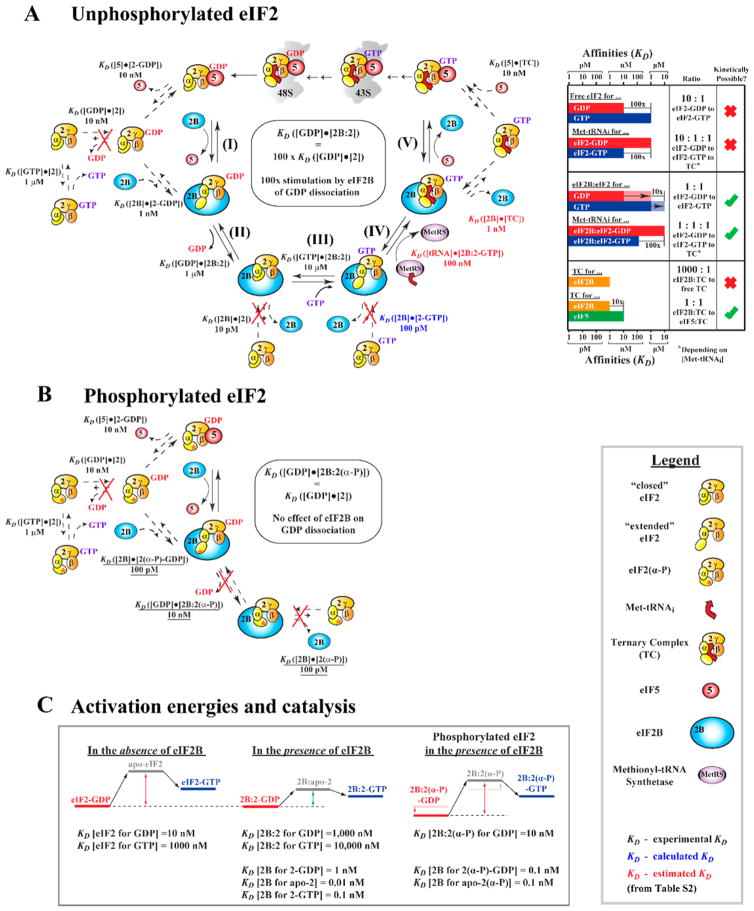
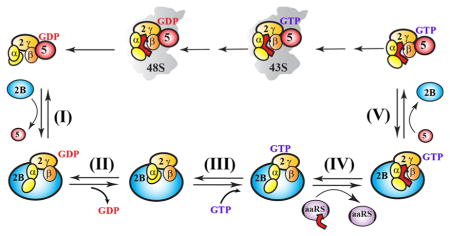
Figure 3.
Thermodynamic description of the eIF2B catalytic cycle. (A) Thermodynamic description of the pathway of eIF2B-catalyzed recycling of eIF2-GDP to the TC and ribosome binding. Solid arrows indicate progression along the path, with steps labeled as follows: (I) displacement of eIF5 from eIF2-GDP by eIF2B, (II) eIF2B-promoted dissociation of GDP from eIF2, (III) binding of GTP to eIF2B-bound apo-eIF2, (IV) binding of Met-tRNAi to form the TC on eIF2B, and (V) displacement of eIF2B from the TC by eIF5. Dashed arrows represent steps off the main pathway. Red crosses mark steps that can be safely ignored because of either off rates being too slow on the time scale of translation or the equilibrium being shifted too far away from the product. Dissociation constants (KDs) are provided for each step and represent either experimentally determined values reported in the literature (black) or calculated (blue) or estimated (red) values based on thermodynamic coupling considerations (see Table 1). The ratios of KDs and resulting equilibria for the relevant steps are shown in the table on the right. (B) Thermodynamic description of how phosphorylation blocks catalysis by inhibiting eIF2B-mediated nucleotide exchange. The color and overall scheme are identical to those presented in panel A. KDs affected by phosphorylation are underlined. Because the affinity of eIF2B for apo-eIF2(α-P) (reaction intermediate) is the same as that for eIF2(α-P)-GDP (substrate), the enzymatic driving force is removed and eIF2B has no effect on GDP dissociation. (C) Activation energies for nucleotide exchange on eIF2 when free, in the presence of eIF2B, and when phosphorylated.
Table 1.
Reference KDs of Interactions Relevant to the eIF2 Catalytic Cycle
| binding partner A | binding partner B | KD (nM) | source | KD for Figure 3 (nM) | ref |
|---|---|---|---|---|---|
| EIF2 | GTP | 1700 | experimental | 1000 | 24 |
| 1600 | experimental | 20 | |||
| GDP | 20 | experimental | 10 | 24 | |
| 5 | experimental | 20 | |||
| EIF2B | apo-eIF2 | 0.05 | experimental | 0.01 | 25 |
| apo-eIF2(α-P) | 0.5 | experimental | 0.1 | 25 | |
| eIF2-GDP | 10 | theoreticala | 1 | 20 | |
| 40 (KM) | experimental | 27 | |||
| 2 | calculatedb | 26 | |||
| 0.2 | experimental | 25 | |||
| eIF2(α-P)-GDP | 0.3 | experimental | 0.1 | 27 | |
| 0.3 | experimental | 25 | |||
| eIF2-GTP | 0.1 | calculatedc | 0.1 | ||
| TC | 1 | estimatedd | 1 | ||
| eIF2B·eIF2 | GDP | 2000 | theoreticala | 1000 | 20 |
| GTP | 20000 | theoreticale | 10000 | 20 | |
| eIF5 | eIF2-GDP | 20 | experimental | 10 | 51 |
| TC | 20 | experimental | 10 | 51 | |
| apo-eIF2 | 20 | calculatedf | N/A | ||
| eIF2·Met-tRNAi | GTP | 200 | experimental | 100 | 24 |
| Met-tRNAi | eIF2-GTP | 10 | experimental | 10 | 24 |
| eIF2B·eIF2-GTP | 100 | estimatedd | 100 | ||
| eIF5·apo-eIF2 | GDP | 10 | calculatedf | 10 | 19, 21 |
| eIF5·apo-eIF2·Met-tRNAi | GTP | 100 | calculatedg | 100 |
Value not derived from experimental data but manually selected and used to fit data.
Approximately 1/10 of that of eIF5 for eIF2-GDP, based on the competition assay presented in ref 26.
See relationship R2.
See Estimation of KD([tRNA]·[2B:2-GTP]) and KD([2B]·[TC]).
The KD of eIF2B·eIF2 for GTP was found by ref 20 to be 10 times that of eIF2B·eIF2 for GDP, which they estimated to be 2000 nM (see footnote a).
See relationship R5.
See relationship R6.
Known KDs that were consistent among different reports were rounded to multiples of ten and used to populate the pathway (Figure 3), while ensuring that after rounding up, they are internally consistent with thermodynamic coupling.28 As described above, thermodynamically coupled interactions exert reciprocal effects on each other, which stems from the Conservation of Energy Principle. For instance, if eIF2B lowers the affinity of eIF2 for GDP ~100-fold, GDP must in turn lower the affinity of eIF2 for eIF2B ~100-fold.
In cases in which reported KDs varied by more than 2–3-fold between different sources, the KD value was selected on the basis of buffer conditions in the reports (e.g., physiological Mg2+ concentration, salt, and temperature were preferred) and thermodynamic coupling with other KDs.
In cases in which no KDs have been reported, thermodynamic coupling was used to calculate the KD from known KDs, or at least to set upper and lower limits for the KD.
Because the relevant rates in translation are on the order of seconds2,3,6,52 and each eIF2 molecule is recycled every few seconds,53 steps whose rates were on the order of once per minute or slower were considered not physiologically relevant, unless special circumstances make them relevant.
The following relationships among KDs (R1–R6) were used:
-
(R1)Binding of eIF2B to eIF2 weakens GDP binding 100-fold, and vice versa.20
-
(R2)Binding of eIF2B to eIF2 weakens GTP binding 10-fold, and vice versa.20
From this relation, we can calculate that KD([2B]·[2-GTP]) is 10 times greater than KD([2B]·[2]), or 100 pM, if KD([2B]·[2]) is 10 pM.
-
(R3)The affinity of Met-tRNAi for eIF2B·eIF2-GTP and the affinity of eIF2B for TC are not known. However, binding of Met-tRNAi to eIF2-GTP and binding of eIF2B to eIF2-GTP are thermodynamically coupled, allowing the following determination to be made:Rearranging this equation yields
Using 10 nM for KD([tRNA]·[2-GTP]) and 100 pM for KD([2B]·[2-GTP]), we can calculate that their ratio is 100:1. From the equation given above, the ratio of KD([tRNA]·[2B:2-GTP]) to KD([2B]·[TC]) is also 100:1. Therefore, we are able to determine that Met-tRNAi binds to eIF2B·eIF2-GTP roughly 100-fold weaker than eIF2B binds to the TC.
-
(R4)Because the affinity of eIF2B for eIF2(α-P) is the same as the affinity of eIF2B for eIF2(α-P)-GDP25using thermodynamic coupling we can obtain the following relation
-
(R5)No effect21 or a two-fold effect19 of eIF5 on dissociation of GDP from eIF2 was observed. Therefore, GDP’s affinity for eIF5·eIF2 should be similar to its affinity for eIF2 (KD ~ 10 nM). Then, on the basis of thermodynamic coupling, eIF5’s affinity for apo-eIF2 is similar to its affinity for eIF2-GDP.
-
(R6)
Because the affinities of eIF5 for apo-eIF2, eIF2-GDP, and TC are all similar (KD ~ 10 nM), it is unlikely that binding of eIF5 to eIF2 is strongly dependent on which nucleotide is bound to eIF2. In this case, we can surmise that eIF5’s affinity for eIF2-GTP is likely similar to its affinity for apo-eIF2, eIF2-GDP, and TC (KD ~ 10 nM). Therefore, by thermodynamic coupling, we can expect the affinity of GTP for eIF5·apo-eIF2·Met-tRNAi to be similar to the affinity of GTP for apo-eIF2·Met-tRNAi (KD ~ 100 nM).
As a note, with regard to representations in Figure 3B, the KDs for binding of a nucleotide to eIF2(α-P) are the same as those for binding to eIF2. A difference is seen only when eIF2 is bound to eIF2B. Therefore, in Figure 3B, we use the same names for them as in Figure 3A, to emphasize their equivalence.
Estimation of KD([tRNA]·[2B:2-GTP]) and KD([2B]·[TC])
Given the affinities of Met-tRNAi for eIF2-GTP (KD ~ 10 nM), of eIF2B for eIF2-GTP (KD ~ 100 pM), and of eIF5 for TC (KD ~ 10 nM), and also relationship R3, we can estimate a likely bound on the affinity of Met-tRNAi for eIF2B·eIF2-GTP and a corresponding bound on the affinity of eIF2B for the TC. We estimate the KD of Met-tRNAi for eIF2B·eIF2-GTP to be ~100 nM and certainly between 10 nM and 1 μM. Correspondingly, the KD of eIF2B for the TC should be ~1 nM and certainly between 100 pM and 10 nM. The rationale is as follows.
If Met-tRNAi’s affinity were the same for eIF2B·eIF2-GTP and eIF2-GTP or higher for eIF2B·eIF2-GTP than for eIF2-GTP, then by relationship R3 this would mean that eIF2B’s affinity for the TC would be the same as or higher than its affinity for eIF2-GTP alone, which is already very high (KD = 100 pM). This would mean that the affinity of eIF2B for TC would be at least 100-fold higher than the affinity of eIF5 for the TC (i.e., KD ≲ 100 pM vs ~10 nM). Such a disparity would make it hard for eIF5 to displace eIF2B from the TC, a necessary next step in catalysis. Spontaneous dissociation of the TC from eIF2B would also be too slow, as is the case for dissociation of eIF2-GTP from eIF2B. Thus, we can conclude that Met-tRNAi’s affinity must be lower for eIF2B·eIF2-GTP than for eIF2-GTP. This conclusion is fully supported by published reports that the rate of dissociation of TC from eIF2B is faster than that of eIF2-GTP34 and that the affinity of Met-tRNAi for eIF2-GTP is lower in the presence of eIF2B29. While the authors of the latter report calculated KDs for binding of Met-tRNAi to eIF2-GTP in the presence of eIF2B, those are only apparent KDs, because the eIF2B concentrations used were not always saturating.29
If Met-tRNAi’s affinity were much more than 10-fold lower for eIF2B·eIF2-GTP than for eIF2-GTP, that would make the KD much greater than 100 nM. As the cellular concentration of Met-tRNAi is ~300 nM,54 this would shift the equilibrium away from the formation of the product, TC. Thus, we can conclude that Met-tRNAi’s affinity is unlikely to be much more than 10-fold lower for eIF2B·eIF2-GTP than for eIF2-GTP. Then, again by relationship R3, this would mean that the KD of eIF2B for the TC would be unlikely to be much more than 1 nM.
A KD of Met-tRNAi for eIF2B·eIF2-GTP of ~100 nM is consistent with the report that half-maximal stimulation of nucleotide exchange was achieved with 60 nM Met-tRNAi.34 It has also been shown that the rates of dissociation of eIF2-GDP and the TC from eIF2B are similar,34 which is consistent with the KD of eIF2B for the TC being similar to that of eIF2B for eIF2-GDP (~1 nM). Finally, the 10-fold lower affinity of the TC for eIF5 (10 nM) compared to that for eIF2B (1 nM) is balanced by the fact that the cellular concentration of eIF5 is roughly ten-fold higher than that of eIF2B, leading to an equilibrium optimally primed for regulation.
The thermodynamic scheme shown in Figure 3 has some obvious important implications.
(1) eIF2 is indeed channeled from the translation initiation complex (IC) to eIF5, to eIF2B, to eIF5 again, and back to the IC. Because the KDs of eIF5 and eIF2B for eIF2, eIF2-GDP, eIF2-GTP, and the TC are all ≤20 nM, there cannot be a meaningful amount of free eIF2 in the cell unless there is more eIF2 than eIF5 and eIF2B combined. Because the eIF2 and eIF5 concentrations are comparable and that of eIF2B varies between 1/10 and 1/2 of that of eIF2 (see, e.g., the PaxDb protein abundance database49), the excess of eIF2 seems unlikely in most cell types. Furthermore, off rates of eIF2 from eIF5 and eIF2B are known to be on the order of ≤1 min−1, or the complexes have been observed using analytical centrifugation, size exclusion chromatography, or pull-down experiments and are therefore stable for at least minutes (see, e.g., 20, 24, 25, 34, and 35). Because the process of eIF2 recycling occurs on the time scale of seconds,53 the spontaneous dissociation of these complexes is too slow to be physiologically relevant.
Of the slow off rates presented above, two are on the catalytic pathway and thus present a challenge for eIF2B-catalyzed nucleotide exchange: that for dissociation of eIF5 from eIF2-GDP (step I in Figure 3A) and that for dissociation of eIF2B from TC (step V in Figure 3A). As described above, the issue of the tight binding of eIF5 to eIF2-GDP has been identified previously, with eIF5 acting as a GDI, and eIF2B acting as a GDF, displacing eIF5 from eIF2-GDP by forming an unstable transient eIF5·eIF2-GDP·eIF2B complex leading to a faster eIF5 dissociation rate.19,26,46
The stability of the eIF2B·TC complex35 also indicates that the rate of displacement of eIF2B from the TC by eIF5 (Figure 3A, step V) must be faster than the spontaneous rate of release of the TC from eIF2B. Therefore, there needs to be a “GEF displacing factor” (“GEF-DF”), which we hypothesize is most likely eIF5 and/or the MFC (see Channeling of eIF2). The relative instability of the eIF5·eIF2-GDP·eIF2B complex also offers a mechanism for acceleration of displacement of eIF2B from the TC by eIF5. On the basis of thermodynamic coupling (see above), simultaneous binding of eIF5 and eIF2B to eIF2 equally destabilizes the eIF2B–eIF2 and eIF5–eIF2 interactions. While eIF5 may have a lower or comparable affinity for the TC compared to that of eIF2B, the cellular concentration of eIF5 is higher than that of eIF2B (see, e.g., the PaxDb protein abundance database49). In competing with eIF2B for TC, eIF5 could bind to eIF2β-NTT in eIF2B·TC complexes when eIF2β-NTT is transiently released from eIF2Bε-CTD. It is also possible that eIF5 and eIF2B can bind to different portions of eIF2β-NTT simultaneously, destabilizing each other’s binding. The DWEAR motif in eIF519 could also play a role. Therefore, the concept of GDI, GDF, and “GEF-DF” functions of eIF5 and eIF2B should be viewed in a broader sense. eIF2B and eIF5 displace each other as eIF2-GDP is channeled from eIF5 to eIF2B. eIF2-GDP is recycled by eIF2B to the TC, which is then channeled back to eIF5, the MFC, and the ribosome for a new round of translation initiation.
If there are cases in which the eIF2 concentration in a cell exceeds the sum of the eIF5 and eIF2B concentrations, one might expect a weakening in the competition between eIF2B and eIF5 for eIF2-GDP, and thus in channeling at that step. While excess eIF2 would make it easier for eIF2B to bind free eIF2-GDP, without competing with eIF5, it could be problematic at the end of the cycle: little free eIF5 could make it difficult for eIF5 to compete with eIF2B for the TC. It has been reported that in mammals, MFCs containing eIF2-GDPNP have affinity for Met-tRNAi that is 2-fold greater than that of free eIF2-GDPNP, whereas the affinity of MFCs containing eIF2-GDP for Met-tRNAi is much lower than that of free eIF2-GDP.21 Therefore, the MFC discriminates against formation of aberrant complexes containing eIF2-GDP and Met-tRNAi. While the MFC could be formed independent of the presence or absence of GDP, GDPNP, or Met-tRNAi,21 the MFC and the 43S PIC, or at least the 43S PIC, must also have a higher affinity for the TC than for eIF2-GDP (or there would be no driving force for the formation of the active TC-containing complexes over inactive ones containing eIF2-GDP and no Met-tRNAi). Therefore, the MFC, or the 43S PIC, should still be able to pick a TC directly from eIF2B, even in excess free eIF2-GDP.
(2) eIF2B-bound eIF2 exists in an ~1:1 ratio between eIF2-GDP and eIF2-GTP complexes. eIF2B destabilizes binding of GDP to eIF2 ~100-fold compared to free eIF2: from 10 nM to ~1 μM (the basis for its catalytic effect on nucleotide exchange). It also destabilizes binding of GTP to eIF2 approximately ten-fold from 1 to 10 μM. Therefore, as previously reported,20 eIF2B-bound eIF2 equilibrates to an ~1:1 ratio between eIF2-GDP and eIF2-GTP complexes, because the affinity of eIF2B·eIF2 for GDP is approximately ten-fold higher and the concentration of GDP approximately ten-fold lower (steps II and III in Figure 3A). Given the increased rate of dissociation of GDP from eIF2B·eIF2, equilibration should be fast enough to be physiologically relevant, and because these KDs are lower than the cellular concentrations of GDP and GTP, there should be little eIF2B·apo-eIF2. In contrast, in the absence of eIF2B, the equilibrium is 10:1 in favor of eIF2-GDP over eIF2-GTP, and even that ratio cannot be reached because of the slow dissociation of GDP from eIF2, compared to the time scale of translation.
(3) The eIF2B complexes with eIF2-GDP, eIF2-GTP, and TC could be at an ~1:1:1 equilibrium. Given that the KD of Met-tRNAi for eIF2B·eIF2 is ~100 nM (Figure 3A and Table 1) and the concentration of Met-tRNAi is ~300 nM,54 comparable to that of eIF2B, the equilibrium between eIF2B·eIF2-GTP and eIF2B·TC should not be shifted much toward eIF2B·TC. If the eIF2B complexes with eIF2-GDP, eIF2-GTP, and TC are near a 1:1:1 equilibrium, the important practical implication is that the equilibrium will be dependent on, and potentially regulated by, the GTP:GDP ratio in the cell, as well as by the concentration of charged Met-tRNAi.
The eIF2B-catalyzed regeneration of TC and its delivery to the 43S IC are thus dependent on the concentration of the eIF2B·TC complex and the rate of transfer of the TC from eIF2B to eIF5, which are determined by the GTP:GDP ratio in the cell, the Met-tRNAi concentration, the relative affinities of eIF2B for eIF2-GDP, eIF2-GTP, and the TC, and the relative affinities of eIF5 and eIF2B for eIF2-GDP and the TC. These parameters are in turn dependent on, and can be regulated by, phosphorylation of eIF2, eIF2B, and eIF5 by a number of kinases, the energy state of the cell, and direct binding of a ligand to eIF2B.
The thermodynamic model shown in Figure 3 also illustrates the limitations of the commonly used in vitro eIF2B activity assay using exchange of eIF2-bound labeled GDP (typically [3H]GDP or BODIPY-GDP) with unlabeled GDP, as we had suggested earlier.3 The assay measures the slower of two rates: the off rate of eIF2-GDP from eIF2B (rate of substrate release) or the off rate of GDP from eIF2B·eIF2 (the first step in the actual catalysis). Under normal conditions, the release of eIF2-GDP from eIF2B is slower and is thus effectively the rate being measured (Figure 3A). If eIF2B fails to stimulate the release of GDP from eIF2, as is the case with eIF2(α-P)-GDP, then GDP dissociation is actually being measured and the results of the assay are informative. Thus, this assay measuring the rate of exchange of labeled and unlabeled GDP is able to reproduce the inhibition of eIF2B activity by eIF2α phosphorylation, because formation of the stable eIF2B·eIF2(α-P)-GDP complex is the end point of the inhibition and the slow off rate of eIF2(α-P)-GDP from eIF2B is what leads to competitive inhibition of eIF2B (Figure 3B). This is the likely reason (along with its relative simplicity) why the assay has been used almost exclusively to measure eIF2B activity for the past two decades. However, it fails to reproduce the in vivo effects of a vast number of CACH/VWM mutations in eIF2B, phosphorylation of eIF2Bε by some kinases, or the requirement for the eIF2Bβ and eIF2Bγ subunits for eIF2B activity in vivo (reviewed in ref 3). The [3H]GDP/GDP exchange assay is necessarily insensitive to changes in the GDI activity of eIF5 or the GDF activity of eIF2B (Figure 3A, step I), as pointed out recently,26 or to changes in the relative affinities of eIF5 and eIF2B for the TC (Figure 3A, step V), i.e., the “GEF-DF” role of eIF5. Therefore, in cases in which the GDP dissociation rate does not become limiting, reliable information about eIF2B activity can be obtained only using in vivo phenotypes or using a complete in vitro reconstituted system for recycling of eIF2-GDP to the TC that includes not only eIF2B and Met-tRNAi but also eIF5, as well as physiological concentrations of GTP and GDP.
Even when used to measure inhibition by eIF2(α-P)-GDP, the in vitro exchange assay would not quantitatively reflect the level of inhibition observed in vivo when a fraction of eIF2 is phosphorylated, unless the ratio of eIF2B to eIF2(α-P)-GDP in the in vitro assay is the same as that in vivo (which is not easy to achieve, if at all possible). For instance, if 10% of eIF2 in the cell is phosphorylated and the eIF2B:eIF2 ratio is 1:5, then approximately half of eIF2B will be bound to eIF2(α-P)-GDP and inhibited. However, mixing in vitro eIF2B with eIF2 that is 10% phosphorylated in a 1:10 ratio would lead to nearly all of eIF2B being bound to eIF2(α-P)-GDP and inhibited.
It is interesting that the [3H]GDP/GDP exchange assay measures a significant GDP dissociation rate with free eIF2Bε,14 and even eIF2Bε-CTD (the catalytic domain).40 Therefore, eIF2Bε alone stimulates the rate of dissociation of GDP from eIF2. If the rate of dissociation of eIF2-GTP from eIF2Bε is fast enough, then eIF2Bε could act as a “simple” GEF, releasing eIF2-GTP as the product of catalysis, and one would be able to use GTP instead of GDP to observe efficient GDP exchange in vitro (not efficient for WT eIF2B, which releases eIF2-GTP at a rate that is ~10-fold slower than that of eIF2-GDP). Why then is eIF2Bε not sufficient to perform the eIF2B function in vivo? The explanation is that the entire system has evolved for channeling. eIF2Bε is unable to efficiently compete with eIF5 for binding to eIF2-GDP.26 Furthermore, it appears that Met-tRNAi is channeled from the methionyl-tRNA synthetase to eIF2B (reviewed in ref 3), and eIF2Bδ may promote binding of Met-tRNAi to eIF2 because its essential function in S. cerevisiae can be overcome by tRNAi overexpression.16
In summary, we provide the first complete thermodynamic description of the process of eIF2 recycling. The evidence leads to the conclusion that eIF2 is channeled from the ribosome (as an eIF5·eIF2-GDP complex) to eIF2B, and then back to eIF5 and the ribosome (as the TC). The system appears to have evolved to be sensitive to, and regulated by, multiple factors, including post-translational modifications of eIF2, eIF2B, and eIF5, as well as directly by the energy balance in the cell. We offer a solid thermodynamic framework for analysis of the vast amount of available data. New experiments for obtaining a complete set of all relevant binding constants along the eIF2 recycling pathway will be required to test the predictions stemming from our analysis and to fill the remaining gaps in our knowledge of the mechanisms of eIF2B catalysis and regulation.
Acknowledgments
Funding
This work was supported by National Institutes of Health Grant GM095720 to A.M.
The authors thank Alan Hinnebusch and Boriana Marintcheva for helpful discussions.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 4.Dever TE, Dar AC, Sicheri F. The eIF2α kinases. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Plainview, NY: 2007. pp. 319–344. [Google Scholar]
- 5.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 6.Hinnebusch AG, Dever TE, Asano K. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Plainview, NY: 2007. pp. 225–268. [Google Scholar]
- 7.Ron D, Harding HP. eIF2α phosphorylation in cellular stress responses and disease. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Plainview, NY: 2007. pp. 345–368. [Google Scholar]
- 8.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 9.Bogorad AM, Xia B, Sandor DG, Mamonov AB, Cafarella TR, Jehle S, Vajda S, Kozakov D, Marintchev A. Insights into the architecture of the eIF2Balpha/beta/delta regulatory subcomplex. Biochemistry. 2014;53:3432–3445. doi: 10.1021/bi500346u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordiyenko Y, Schmidt C, Jennings MD, Matak-Vinkovic D, Pavitt GD, Robinson CV. eIF2B is a decameric guanine nucleotide exchange factor with a gamma2epsilon2 tetrameric core. Nat Commun. 2014;5:3902. doi: 10.1038/ncomms4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashiwagi K, Takahashi M, Nishimoto M, Hiyama TB, Higo T, Umehara T, Sakamoto K, Ito T, Yokoyama S. Crystal structure of eukaryotic translation initiation factor 2B. Nature. 2016;531:122–125. doi: 10.1038/nature16991. [DOI] [PubMed] [Google Scholar]
- 12.Kuhle B, Eulig NK, Ficner R. Architecture of the eIF2B regulatory subcomplex and its implications for the regulation of guanine nucleotide exchange on eIF2. Nucleic Acids Res. 2015;43:9994–10014. doi: 10.1093/nar/gkv930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wortham NC, Martinez M, Gordiyenko Y, Robinson CV, Proud CG. Analysis of the subunit organization of the eIF2B complex reveals new insights into its structure and regulation. FASEB J. 2014;28:2225–2237. doi: 10.1096/fj.13-243329. [DOI] [PubMed] [Google Scholar]
- 14.Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson FL, Nika J, Rippel S, Hannig EM. Minimum requirements for the function of eukaryotic translation initiation factor 2. Genetics. 2001;158:123–132. doi: 10.1093/genetics/158.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dev K, Qiu H, Dong J, Zhang F, Barthlme D, Hinnebusch AG. The beta/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol Cell Biol. 2010;30:5218–5233. doi: 10.1128/MCB.00265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavitt GD, Yang W, Hinnebusch AG. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol Cell Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erickson FL, Hannig EM. Ligand interactions with eukaryotic translation initiation factor 2: role of the gamma-subunit. EMBO J. 1996;15:6311–6320. [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings MD, Pavitt GD. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature. 2010;465:378–381. doi: 10.1038/nature09003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panniers R, Rowlands AG, Henshaw EC. The effect of Mg2+ and guanine nucleotide exchange factor on the binding of guanine nucleotides to eukaryotic initiation factor 2. J Biol Chem. 1988;263:5519–5525. [PubMed] [Google Scholar]
- 21.Sokabe M, Fraser CS, Hershey JW. The human translation initiation multi-factor complex promotes methionyl-tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res. 2012;40:905–913. doi: 10.1093/nar/gkr772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 23.Sprang SR, Coleman DE. Invasion of the nucleotide snatchers: structural insights into the mechanism of G protein GEFs. Cell. 1998;95:155–158. doi: 10.1016/s0092-8674(00)81746-8. [DOI] [PubMed] [Google Scholar]
- 24.Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol. 2004;335:923–936. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 25.Goss DJ, Parkhurst LJ, Mehta HB, Woodley CL, Wahba AJ. Studies on the role of eukaryotic nucleotide exchange factor in polypeptide chain initiation. J Biol Chem. 1984;259:7374–7377. [PubMed] [Google Scholar]
- 26.Jennings MD, Zhou Y, Mohammad-Qureshi SS, Bennett D, Pavitt GD. eIF2B promotes eIF5 dissociation from eIF2*GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev. 2013;27:2696–2707. doi: 10.1101/gad.231514.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowlands AG, Panniers R, Henshaw EC. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem. 1988;263:5526–5533. [PubMed] [Google Scholar]
- 28.Fersht A. Structure and Mechanism in Protein Science. W. H. Freeman and Co; 1999. [Google Scholar]
- 29.Jennings MD, Kershaw CJ, Adomavicius T, Pavitt GD. Fail-safe control of translation initiation by dissociation of eIF2alpha phosphorylated ternary complexes. eLife. 2017;6:e24542. doi: 10.7554/eLife.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gromadski KB, Wieden HJ, Rodnina MV. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry. 2002;41:162–169. doi: 10.1021/bi015712w. [DOI] [PubMed] [Google Scholar]
- 31.Crechet JB, Parmeggiani A. Characterization of the elongation factors from calf brain. 2 Functional properties of EF-1 alpha, the action of physiological ligands and kirromycin. Eur J Biochem. 1986;161:647–653. doi: 10.1111/j.1432-1033.1986.tb10489.x. [DOI] [PubMed] [Google Scholar]
- 32.Crechet JB, Parmeggiani A. Characterization of the elongation factors from calf brain. 3 Properties of the GTPase activity of EF-1 alpha and mode of action of kirromycin. Eur J Biochem. 1986;161:655–660. doi: 10.1111/j.1432-1033.1986.tb10490.x. [DOI] [PubMed] [Google Scholar]
- 33.Janssen GM, Möller W. Elongation factor 1 beta gamma from Artemia. Purification and properties of its subunits. Eur J Biochem. 1988;171:119–129. doi: 10.1111/j.1432-1033.1988.tb13766.x. [DOI] [PubMed] [Google Scholar]
- 34.Gross M, Rubino MS, Hessefort SM. The conversion of eIF-2. GDP to eIF-2.GTP by eIF-2B requires Met-tRNA(fMet) Biochem Biophys Res Commun. 1991;181:1500–1507. doi: 10.1016/0006-291x(91)92109-w. [DOI] [PubMed] [Google Scholar]
- 35.Salimans M, Goumans H, Amesz H, Benne R, Voorma HO. Regulation of protein synthesis in eukaryotes. Mode of action of eRF, an eIF-2-recycling factor from rabbit reticulocytes involved in GDP/GTP exchange. Eur J Biochem. 1984;145:91–98. doi: 10.1111/j.1432-1033.1984.tb08526.x. [DOI] [PubMed] [Google Scholar]
- 36.Dholakia JN, Wahba AJ. Mechanism of the nucleotide exchange reaction in eukaryotic polypeptide chain initiation. Characterization of the guanine nucleotide exchange factor as a GTP-binding protein. J Biol Chem. 1989;264:546–550. [PubMed] [Google Scholar]
- 37.Siekierka J, Mauser L, Ochoa S. Mechanism of polypeptide chain initiation in eukaryotes and its control by phosphorylation of the alpha subunit of initiation factor 2. Proc Natl Acad Sci U S A. 1982;79:2537–2540. doi: 10.1073/pnas.79.8.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams DD, Price NT, Loughlin AJ, Proud CG. Characterization of the mammalian initiation factor eIF2B complex as a GDP dissociation stimulator protein. J Biol Chem. 2001;276:24697–24703. doi: 10.1074/jbc.M011788200. [DOI] [PubMed] [Google Scholar]
- 39.Nika J, Yang W, Pavitt GD, Hinnebusch AG, Hannig EM. Purification and kinetic analysis of eIF2B from Saccharomyces cerevisiae. J Biol Chem. 2000;275:26011–26017. doi: 10.1074/jbc.M003718200. [DOI] [PubMed] [Google Scholar]
- 40.Gomez E, Mohammad SS, Pavitt GD. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 2002;21:5292–5301. doi: 10.1093/emboj/cdf515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dholakia JN, Mueser TC, Woodley CL, Parkhurst LJ, Wahba AJ. The association of NADPH with the guanine nucleotide exchange factor from rabbit reticulocytes: a role of pyridine dinucleotides in eukaryotic polypeptide chain initiation. Proc Natl Acad Sci U S A. 1986;83:6746–6750. doi: 10.1073/pnas.83.18.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimball SR, Jefferson LS. Allosteric regulation of eukaryotic initiation factor eIF-2B by adenine nucleotides. Biochem Biophys Res Commun. 1995;212:1074–1081. doi: 10.1006/bbrc.1995.2079. [DOI] [PubMed] [Google Scholar]
- 43.Webb BL, Proud CG. Eukaryotic initiation factor 2B (eIF2B) Int J Biochem Cell Biol. 1997;29:1127–1131. doi: 10.1016/s1357-2725(97)00039-3. [DOI] [PubMed] [Google Scholar]
- 44.Kimball SR, Fabian JR, Pavitt GD, Hinnebusch AG, Jefferson LS. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2alpha. Role of the alpha- and delta-subunits of eiF2b. J Biol Chem. 1998;273:12841–12845. doi: 10.1074/jbc.273.21.12841. [DOI] [PubMed] [Google Scholar]
- 45.Bogorad AM, Lin KY, Marintchev A. Novel mechanisms of eIF2B action and regulation by eIF2α phosphorylation. Nucleic Acids Res. 2017;45:11962–11979. doi: 10.1093/nar/gkx845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, Asano K. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 2006;25:4537–4546. doi: 10.1038/sj.emboj.7601339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang M, Weiss M, Simonovic M, Haertinger G, Schrimpf SP, Hengartner MO, von Mering C. PaxDb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics. 2012;11:492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panniers R, Henshaw EC. A GDP/GTP exchange factor essential for eukaryotic initiation factor 2 cycling in Ehrlich ascites tumor cells and its regulation by eukaryotic initiation factor 2 phosphorylation. J Biol Chem. 1983;258:7928–7934. [PubMed] [Google Scholar]
- 54.Smith DW. Reticulocyte transfer RNA and hemoglobin synthesis. Science. 1975;190:529–535. doi: 10.1126/science.1103288. [DOI] [PubMed] [Google Scholar]