Abstract
Fusion of the anaplastic lymphoma receptor tyrosine kinase gene (ALK) with the echinoderm microtubule-associated protein 4 gene (EML4) is the second most common actionable alteration in non-small-cell lung cancer, with a frequency of 5%. Here, we present a case of an EML4-ALK-positive patient with an atypical in-frame insertion from the LTBP1 gene in the canonical junction of variant 1. The patient was a 39-year-old never-smoker female diagnosed with Stage IV lung adenocarcinoma. A core biopsy was negative for EGFR and KRAS mutations but positive for ALK immunohistochemistry and fluorescence in situ hybridization. When submitted to nCounter, the sample showed a 3′/5′ imbalance indicative of an ALK rearrangement, but failed to give a positive signal for any of the variants tested. Finally, a band with a molecular weight higher than expected appeared after reverse transcriptase-polymerase chain reaction analysis. When Sanger sequencing was performed, the band revealed an atypical EML4-ALK fusion gene with an in-frame 129 bp insertion. A 115 bp segment of the insertion corresponded to an intronic region of LTBP1, a gene located in the short arm of chromosome 2, between ALK and EML4. The patient received crizotinib and showed a good therapeutic response that is still ongoing after 12 months. Our result suggests that short in-frame insertions of other genes in the EML4-ALK junction do not affect the sensitivity of the EML4-ALK fusion protein to crizotinib.
Keywords: lung cancer, NSCLC, EML4-ALK, LTBP1, crizotinib, targeted therapy
Introduction
Fusions of the ALK gene with the EML4 gene is the second most common actionable alteration in non-small-cell lung cancer, with a frequency of 5%.1 More than 15 EML4-ALK fusion variants with various breakpoints on the EML4 and ALK genes have been reported. The most frequent is variant 1 (v1, 33%), followed by v3a/3b (29%) and v2 (10%) but other breakpoints and 5′ fusion gene partners different from EML4 have also been described.1–3 Here, we report the first case of a tumor harboring an EML4-ALK fusion with an atypical in-frame insertion from the LTBP1 gene. Together with previous reports, our result suggests that in-frame insertions of other genes in the EML4-ALK junction might be associated with good responses to crizotinib.
Case presentation
A 39-year-old never-smoker female without prior relevant medical history was admitted to the hospital with progressive symptoms of abdominal pain, dyspnea, and bilateral leg edema. A computed tomography scan revealed pericardial effusion; bilateral pleural effusion; a 3 cm mass in the right lung; hilar, mediastinal, and retroperitoneal lymphadenopa-thies; and ascitis (Figure 1). Pericardial and pleural fluids were positive for adenocarcinoma cells, and the patient was diagnosed with lung adenocarcinoma stage IV.
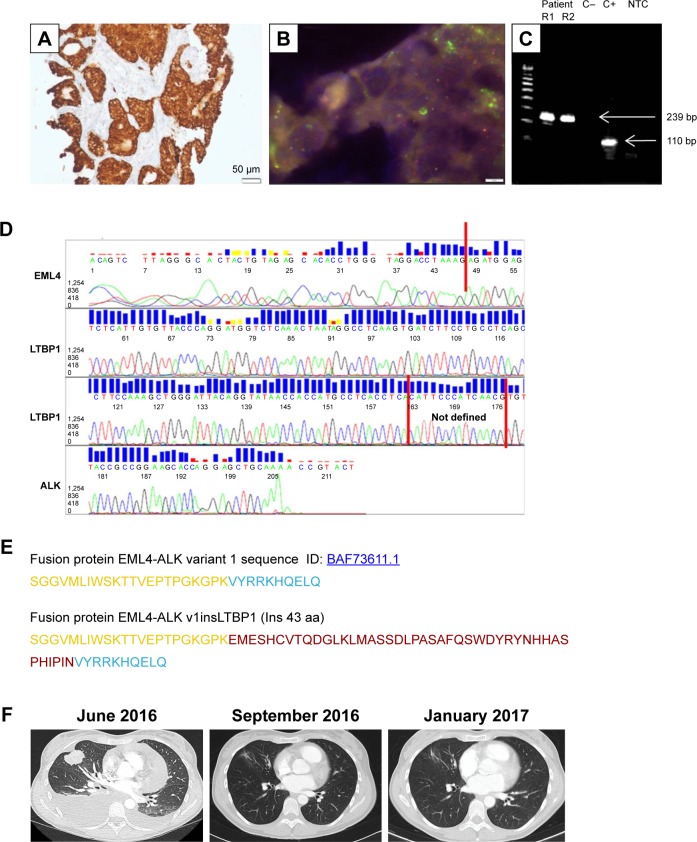
Figure 1.
Molecular testing, characterization, and clinical course of the patient with the atypical fusion variant (v1insLTBP1) with a 129 bp insertion in the EML4-ALK junction.
Notes: (A) Staining with IHC VENTANA clone DF53 (100×), (B) FISH using Vysis LSI ALK dual-color break-apart probe (100×), (C) gel visualization of RT-PCR bands (using primers for v1); R1, replicate 1; R2, replicate 2; C−, negative control; C+, positive control; NTC, non-template control, (D) Sanger sequencing chromatogram, (E) amino acid sequence of the EML4-ALK fusion protein (EML4 in yellow, ALK in blue, new 43 aa in red, (F) thoracic assessment by CT: at diagnosis (June 2016), response to crizotinib after 1 month of treatment (September 2016), and monitoring after 5 months of treatment (January 2017).
Abbreviations: ALK, anaplastic lymphoma receptor tyrosine kinase; CT, computed tomography; EML4, echinoderm microtubule-associated protein 4; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; RT-PCR, reverse transcriptase-polymerase chain reaction.
A core biopsy of the lung mass was performed. Molecular analysis of the EGFR and the KRAS genes revealed absence of mutations. Immunostaining with IHC VENTANA clone DF53 identified overexpression of ALK, and fluorescence in situ hybridization using Vysis LSI ALK dual-color break-apart probe demonstrated an ALK rearrangement. When the sample was analyzed by nCounter, it showed a 3′/5′ imbalance indicative of an ALK rearrangement, but failed to give a positive signal for the EML4-ALK v1, v2, v3, or v5; TFG-ALK_T5:A20; KIF5B-ALK_K17:A20; or KIF5BALK_K24:A20 fusions.4 Finally, a band with a molecular weight higher than expected (239 bp) appeared after reverse transcriptase-polymerase chain reaction analysis (RT-PCR) with primers specific for EML4-ALK v1. No additional bands were apparent. The 239 bp band was submitted to Sanger sequencing revealing an atypical EML4-ALK fusion gene with a 129 bp insertion in the canonical junction of v1. A 115 bp segment of the insertion corresponded to an intronic region of LTBP1, a gene located in the short arm of chromosome 2, between the ALK and EML4 genes. The in silico translation of this new variant, which will be referred to as v1insLTBP1, showed an in-frame insertion of 43 aminoacids (Figure 1).
The patient started crizotinib with good tolerance. The computed tomography scan performed a month later showed a reduction of the primary lesion, disappearance of hilary and reroperitoneal lymphadenopathies, and a reduction of the mediastinal lymph nodes. After 14 months, the patient continues to demonstrate partial response.
Written informed consent has been provided by the patient to have the case details and any accompanying images published.
Discussion
A majority of laboratories determine ALK rearrangements by the two US Food and Drug Administration-approved techniques, fluorescence in situ hybridization and immu-nohistochemistry, and do not test for specific variants due to cost-effectiveness considerations. Consequently, it is difficult to estimate the real frequency of new variants such as the v1insLTBP1, described in this paper. Using an nCounter methodology, we have recently reported that 6/32 (18.8%) ALK rearrangements in a retrospective cohort of positive cases were not EML4-ALK v1, v2, v3, or v5; TFG- ALK_T5:A20 KIF5B-ALK_K17:A20; or KIF5B-ALK_ K24:A20.4 The exact variant of those cases could not be identified either by nCounter or RT-PCR. In addition, as a part of our routine clinical practice, we prospectively test advanced non-small-cell lung cancer patients for ALK translocations by an RT-PCR technique that can identify v1, v2, and v3. We have found 38 positive cases, including the patient with the new variant, suggesting that the frequency of the v1insLTBP1 could be as high as 2.7% (1/38).
The clinical relevance of the different ALK fusion partners and variants is poorly understood, and inconsistent results have been reported. A retrospective study including 55 ALK-positive patients found an association of v1 with a longer progression-free survival (PFS) to crizotinib, while a second study reported a shorter PFS for those carrying v3a/b.3,5 Regarding rare variants, the recently described E6:A18 was intrinsically refractory to crizotinib,2 while a patient with an uncommon 138 bp in-frame insertion from the ATRNL1 gene in v3 derived benefit from this drug.6 The partial response we also observed in the patient with the v1insLTBP1 suggests that in-frame, atypical insertions do not affect the sensitivity of the EML4-ALK fusion protein to crizotinib.
Acknowledgments
The present address for Cristina Teixidó is the Department of Medical Oncology, Hospital Clínic, Barcelona, Spain.
Footnotes
Disclosure
Dr Santiago Viteri reports speaker honoraria from BMS and Roche, advisory board fees from Roche and Boehringer Ingelheim, and meeting inscription/travel expenses fees from Merck Serono. The authors report no other conflicts of interest in this work.
References
- 1.Sasaki T, Rodig SJ, Chirieac LR, Janne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010;46(10):1773–1780. doi: 10.1016/j.ejca.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anai S, Takeshita M, Ando N, et al. A case of lung adenocarcinoma resistant to crizotinib harboring a novel EML4-ALK variant, exon 6 of EML4 fused to exon 18 of ALK. J Thorac Oncol. 2016;11(10):e126–e128. doi: 10.1016/j.jtho.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Woo CG, Seo S, Kim SW, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol. 2017;28(4):791–797. doi: 10.1093/annonc/mdw693. [DOI] [PubMed] [Google Scholar]
- 4.Reguart N, Teixido C, Gimenez-Capitan A, et al. Identification of ALK, ROS1, and RET fusions by a multiplexed mRNA-based assay in formalin-fixed, paraffin-embedded samples from advanced non-small-cell lung cancer patients. Clin Chem. 2017;63(3):751–760. doi: 10.1373/clinchem.2016.265314. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida T, Oya Y, Tanaka K, et al. Differential crizotinib response duration among ALK fusion variants in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3383–3389. doi: 10.1200/JCO.2015.65.8732. [DOI] [PubMed] [Google Scholar]
- 6.Robesova B, Bajerova M, Hausnerova J, Skrickova J, Tomiskova M, Dvorakova D. Identification of atypical ATRNL1 insertion to EML4-ALK fusion gene in NSCLC. Lung Cancer. 2015;87(3):318–320. doi: 10.1016/j.lungcan.2015.01.002. [DOI] [PubMed] [Google Scholar]