Abstract
Peanut is an important oilseed and food legume cultivated as a rain-fed crop in semi-arid tropics. Drought and high salinity are the major abiotic stresses limiting the peanut productivity in this region. Development of drought and salt tolerant peanut varieties with improved yield potential using biotechnological approach is highly desirable to improve the peanut productivity in marginal geographies. As abiotic stress tolerance and yield represent complex traits, engineering of regulatory genes to produce abiotic stress-resilient transgenic crops appears to be a viable approach. In the present study, we developed transgenic peanut plants expressing an Arabidopsis homeodomain-leucine zipper transcription factor (AtHDG11) under stress inducible rd29A promoter. A stress-inducible expression of AtHDG11 in three independent homozygous transgenic peanut lines resulted in improved drought and salt tolerance through up-regulation of known stress responsive genes (LEA, HSP70, Cu/Zn SOD, APX, P5CS, NCED1, RRS5, ERF1, NAC4, MIPS, Aquaporin, TIP, ELIP) in the stress gene network, antioxidative enzymes, free proline along with improved water use efficiency traits such as longer root system, reduced stomatal density, higher chlorophyll content, increased specific leaf area, improved photosynthetic rates, and increased intrinsic instantaneous WUE. Transgenic peanut plants displayed high yield compared to non-transgenic plants under both drought and salt stress conditions. Holistically, our study demonstrates the potentiality of stress-induced expression of AtHDG11 to improve the drought, salt tolerance in peanut.
Keywords: peanut, AtHDG11 (Arabidopsis Homeodomain globarous11), drought stress, high-salinity stress, water use efficiency, yield potential
Introduction
Peanut or groundnut (Arachis hypogaea L.) is globally important legume crop belonging to the Fabaceae family. Grown for its oil content, feed for both human, and livestock, it forms a major revenue source for marginal farmers as well as for the commercial producers. About 60% of the world peanut production comes from the semi-arid tropics (SAT) such as Africa, Asia, North, and South America, where extremes of temperature, drought, and soil salinity are predominant (Mace et al., 2006; Cuc et al., 2008). Such unfavorable environmental conditions severely affect the plant growth and productivity. Drought and salt stress are the serious constraints affecting both productivity and quality of peanut in SAT regions (Wright and Nageswara Rao, 1994; Reddy and Anbumozhi, 2003; Krishna et al., 2015). It is estimated that drought stress alone can cause an annual loss of 6 million tons worth about 520 million USD (Sharma and Lavanya, 2002; ICRISAT, 2006; Bhatnagar-Mathur et al., 2007). Soil salinity in the SAT regions reduces the plants ability to uptake minerals that inhibits growth and development of peanut, which is known as “saline induced water deficit effect” or “osmotic effect” (Singh and Abrol, 1985; Nautiyal et al., 1989; Singh et al., 1989; Janila et al., 1999).
Development of peanut genotypes with improved drought resistance and WUE (water use efficiency) is highly desirable compared with those varieties developed only with drought tolerance, as always there would be a yield advantage in earlier case (Sharma and Lavanya, 2002). WUE and yield are inter-related complex traits influenced by the interaction of a plentitude of genes related to physiological drought-resistance and yield-associated traits (Araus et al., 2008). As a result, simultaneous selection of several traits for WUE, drought resistance, yield along with elimination of undesirable genomic regions that were co-transferred during breeding programs is difficult task and requires tremendous effort and time (Richards, 1996; Yeo, 1998)., Further, transferring desired traits from wild to cultivated Arachis species through classical and molecular breeding has been limited owing to ploidy nature and cross-pollination incompatibility barriers (Janila et al., 2013). Marker assisted selection of useful alleles/QTLs could enhance selection power regardless of breeding constraints.
Genetic engineering or transgenic technology is considered as a versatile and alternative technology to overcome the limitations associated with classical and modern breeding technologies. Significant progress has been achieved in unraveling regulatory mechanisms underlying the drought stress responses and a large number of drought tolerant genes have been identified in a model plant Arabidopsis as well as in wide range of plant species (Hauser et al., 2011). Using the identified stress-responsive genes with putative regulatory roles (transcription factors) from the model plants to develop stress resilient crops has proven to be an efficient strategy compared to traditional breeding (Molinar, 2012; Basu et al., 2016). The effective use of transcription factors (TFs) for the development of stress-resilient peanut crop have already been witnessed in previous studies such as heterologous overexpression of individual TFs AtDREB1A (Bhatnagar-Mathur et al., 2007; Sarkar et al., 2016), MuNAC4 (Pandurangaiah et al., 2014), and simultaneous expression of AtDREB2A, AtHB7, and AtABF3 (Pruthvi et al., 2014); Alfin1, PgHSF4, and PDH45 (Ramu et al., 2016). Though drought and salt tolerance mechanisms share common genes, few genes were found specifically involve in salt tolerance. The potential source of salt tolerance genes are haplophytes and their heterologous expression pattern with possible mechanism was thoroughly discussed (Mishra and Tanna, 2017). The constitutive expression of WRKY3 transcription factor has drastically improved salt tolerance in tomato along with moderate tolerance to drought (Hichri et al., 2017). Apart from transcription factor several other genes like NHX, HKT, and P5CS were proved efficient for salt tolerance in different genetic backgrounds (Mian et al., 2011; Zhang et al., 2015). To date only a small number of genes directly improving plant WUE and the associated drought tolerance were identified such as ABA-responsive barley gene HVA1 (Sivamani et al., 2000), a putative transcription factor ASR1 (Jeanneau et al., 2002), a gene associated with stomatal movement MRP5-1 (Ghasem et al., 2009), a gene encoding an NADP-malic enzyme (Laporte et al., 2002), an AP2/ERF-like transcription factor gene HRD (Karaba et al., 2007), and a DREB1A TF gene (Bhatnagar-Mathur et al., 2007). Stress inducible expression system was found better when compared to strong constitutive in improving abiotic stress tolerance through genetic engineering approach (Ben-Saad et al., 2015; Tiwari et al., 2016).
In a recent study Yu et al. (2008) identified and ascertained that overexpression of Arabidopsis HDG11 gene, a HD-START family transcription factor, confers drought resistance in Arabidopsis and tobacco by exhibiting drought tolerant traits such as increased root growth, reduced stomatal density. Further over expression of AtHDG11 under a constitutive promoter has shown to confer salt, drought stress tolerance, and improved yield in tall fescue, sweet potato, rice, cotton, poplar, wheat, and Chinese Kale (Cao et al., 2009; Ruan et al., 2012; Yu et al., 2013, 2015; Li et al., 2016; Zhu et al., 2016). The HDG11 transcription factor is a developmental regulator gene that has insignificant role in stress response (Yu et al., 2008). Constitutive overexpression of HDG11 in transgenic plants accumulates high amount of HDG11 protein in the nucleus and regulates the expression of several other stress induced genes which are having HD binding sites in their promoters (Yu et al., 2008). However, no experimental data is available on the performance of AtHDG11 under abiotic stress inducible promoter.
In the present study, we describe the successful transformation and heterologous overexpression of AtHDG11 gene under the control of a stress inducible Atrd29A promoter in a popular peanut variety JL-24 and discuss the performance under drought and salt stress conditions.
Materials and methods
Plant material and cloning of AtHDG11 gene
The seeds of Arabidopsis thalina ecotype columbia SALK T-DNA insertion mutant line (SALK_003819) were procured from Arabidopsis Biological Resource Centre, Japan (Yu et al., 2008). The seeds were surface sterilized with 50% liquid bleach containing of 0.05% Tween 20 and sown in a mixture of vermiculite: perlite: peat moss (2:2:1). Pots were kept in the dark for 3 days, at 4°C for stratification and subsequently transferred to normal growth conditions. Thereafter, plants were grown at 22°C under long-day conditions (16-h-light/8-h-dark). Leaves were collected from 30 days old plant and total RNA was isolated using “Nucleospin RNAII kit” (Macherey Nagel). cDNA was prepared using oligo-dT primer, following the procedure described by “Revert Aid premium first strand cDNA synthesis kit” (Life Technologies). For amplification of HDG11 gene, 1 μL of the reverse transcription reaction was used with Platinum Pfx polymerase (Invitrogen) according to the manufacturer's instructions. The 2.1 kb cDNA fragment of AtHDG11 (At1g73360) was amplified from cDNA using gene-specific primers. The amplified cDNA fragment was column purified (Qiagen) and cloned into pTZ57R/T (MBIFermentas) vector following manufacturer's protocol.
Construction of plant transformation vector with AtHDG11
The 2.1 kb of AtHDG11 coding region was released from pTZ57R/T vector using XhoI and NotI restriction enzymes (NEB) and ligated into pGreen0029-binary vector between stress inducible promoter Atrd29A-and-polyA terminator. The resulting recombinant vector Atrd29::AtHDG11:PolyA was mobilized along with the helper vector pSoup into EHA105 strain of Agrobacterium tumefaciens by electroporation (Eppendorf) method. Recombinant colony of A. tumefaciens strain carrying the construct was selected using colony PCR method. Selected colony was cultured in YEP broth (50 μg/mL) and rifampicin (10 μg/mL) and allowed to grow at 28°C on a shaker with 180 rpm. When the OD (A600) reached 0.4–0.5, the bacterial culture was removed and centrifuged at 5000 rpm for 10 min at 4°C, the pellet was dissolved in 50 mL of half strength MS media with 100 μM of acetosyringone and the suspension was used for co-cultivation. PGreen0029-binary vector without any transgene served as an empty vector control.
Plant transformation
Peanut (Arachis hypogaea L. cv. JL24) seeds were obtained from Regional Agriculture Research Station, Kadiri, Andhra Pradesh, India. Mature seeds were surface sterilized with tween-20 for 5–8 min then with 0.1% (w/v) aqueous mercuric chloride (HgCl2) for 7 min followed by 5–6 rinses with sterile double distilled water. Blot dried seeds were inoculated on half strength MS media for obtaining aseptic seedlings. Cotyledonary node (CN) explants were prepared from 6 days old aseptic seedlings as explained by Beena et al. (2005). The CN explants were independently immersed in the bacterial suspensions carrying the construct and empty vector for 5 min and blot dried by placing between the sterile filter paper to remove excess of bacterial suspension and explants were transferred to the regeneration media. Regeneration protocol was followed as discussed by Anuradha et al. (2006).
Analysis of transformants
Kanamycin analysis
All the T1 and T2 generation seeds were collected from transgenic plants, surface sterilized with 1% (w/v) bavistin and allowed to grow on half strength MS media consisting of 200 mg/L kanamycin, along with the non-transgenic seeds for 10–15 days at 28°C. Healthy plants with green leaves and having good root and shoot ratio were selected as positives.
Molecular analysis of putative transgenic plants for stable transgene integration and segregation
Genomic DNA was isolated from T1, T2, and T3 putative transgenic and non-transgenic plants using CTAB method (Doyle and Doyle, 1987). PCR analysis was carried out with different combinations of gene, promoter, and nptII specific primers for detection of cloned components. PCR amplification was carried out in a 25 μL reactions containing 100 ng of genomic DNA as a template. The PCR was initiated by a hot start at 94°C for 5 min, 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The amplicons were resolved on 0.8% agarose gel electrophoresis.
Southern analysis was carried out using 20 μg of purified genomic DNA from selected T2 PCR confirmed transgenics along with non-transgenic plants. DNA was digested with XhoI enzyme, electrophoresed, and blotted onto a positively charged nylon membrane (Hybond-N+). Radiolabeled PCR product of AtHDG11 gene was used as a probe. Hybridization and washing were carried out at 58°C as described by Sambrook et al. (1989).
Screening for drought and salt stress tolerance by outdoor lysimetric dry-down experiment
The abiotic stress response in selected T3 transgenic plants along with NT plants were carried out under field conditions using the lysimetric dry-down conditions as described by Jagana et al. (2012). Selected peanut transgenic lines were grown in the PVC lysimeters (120 × 20 cm) consisting of 50 kg soil. Plants were watered regularly for 1 month to gain full field capacity. Drought stress was imposed in 1 month old plants by withholding the water and salt stress was imposed by adding 250 mM salt (NaCl) solution to the cylinders. Drought and salt stress were continued for 30 days. Field capacity of drought stressed plants was regularly monitored by weighing the plants using an electrical balance connected with a pulley. Leaf samples were collected with 10 days interval (10, 20 and 30 days) for drought and 5 days interval (5, 10, and 15 days) for salt stress; after stress imposition. Thirty days after imposition of drought stress, NT, and transgenic plants were rehydrated by adding the water to their field capacity and allowed for rehydration. Rehydration capacities of non-transgenic and transgenic plants were measured based on their recovery from drought stress and samples were collected after 5 days of recovery.
Quantitative real time PCR (qRT-PCR)
The total RNA for qRT-PCR analysis was isolated using Nucleospin RNAII kit (Macherey Nagel) from leaf tissues of transgenic (T3 generation) and non-transgenic (NT) plants. After DNAse I (Fermentas) treatment, RNA was used for cDNA synthesis using RevertAid™ premium First strand cDNA synthesis Kit (Fermentas). Real-time PCR was carried out using FastStart universal SYBR green master mix (Roche Applied Science) as instructed in the manual with AtHDG11 gene specific primers F-5′-ATC CAA CAA CAC ACG CTC AA-3′ and R-5′-AGA AGG ATC TTC ACC GCT CA−3′. ADH3 (EG529529) was used as an internal control with F-5′-GCTTCAAGAGCAGGTCACAAGT-3′ and R-5′-GAGACATCCTCCTTCGTGCATA-3′ as primers. Amplicons were analyzed on Roche Light Cycler 480 II Real-Time PCR System according to the manufacturer's instructions. The average values of three biological samples were used to calculate the relative fold expression. The relative fold in expression levels were calculated using qbase+Software (Hellemans et al., 2007) by normalizing with Actin reference gene (Condori et al., 2011).
Known stress responsive genes (Supplementary Table 1) expression was measured in homozygous peanut transgenic lines under drought and salt stress conditions using qRT-PCR. RNA samples were isolated from the NT and transgenic lines on 10th day after drought stress imposition and day 5th after salt stress and their respective control well-watered plants grown in the lysimetric experiment. Total RNA isolation, cDNA synthesis and qRT-PCR were performed as mentioned in the previous section.
Chlorophyll estimation
Chlorophyll content of non-transgenic type and transgenic plants grown in the lysimetric experiment under well-water, drought, and salt stress conditions was measured using SPAD chlorophyll meter (SPAD-502, Konica Minolta, Japan) according to Rao et al. (2001) and Qin et al. (2013). SPAD chlorophyll meter readings () were taken from the fully expanded third leaf of non-transgenic and transgenic lines at a 10 days interval (10, 20, and 30 days) for drought and 5 days interval (5, 10, and 15 days) for salt stress.
Specific leaf area (SLA)
Specific Leaf Area of NT and transgenic plants grown in the lysimetric experiment under well-water, drought, and salt stress conditions was measured using a leaf area meter (LI-3000C LI-COR Biosciences) according to Sebahattin and Necdet (2007). SLA readings were taken from the fully expanded third leaf of NT and transgenic lines at 10 days interval (10, 20, and 30) for drought and 5 days interval (5, 10, and 15) for salt stress. To determine the leaf dry weight, leaf samples were oven dried at 80°C for 48 h. SLA was calculated using the following formula.
Relative water content (RWC)
Relative Water Content in the leaves of NT and transgenic plants was measured according to the (Gonzales and Gonzales-Vilar, 2001). Four leaflets from the top leaves of the main stem were used for the measurement of RWC. Initially, fresh weights (FW) of the harvested leaves were recorded within 15 min, then leaf samples were floated on 20 mL distilled water for 5 h to attain full turgidity and turgid weight (TW) was recorded. Then leaflets were oven dried at 80°C for 48 h and dry weight (DW) of the leaflets were recorded. The leaf relative water content was calculated using the following formula.
Determination of stomatal size and density
Stomata size [Stomatal Length (STL LEN) & Stomatal width (STL WID)] and Stomatal density (STL DEN) of transgenic and NT plants were determined from the fully expanded third leaf of NT and transgenic lines after 10 days of drought and 5 days after salt stress using leaf surface imprint method (Yu et al., 2008). The leaf imprints were collected and placed on glass slide and observed under Olympus microscope (BX51, Olympus). Stomatal density and size were determined using OLYMPUS Image Analysis software.
Determination of photosynthetic rate, transpiration rate, and water use efficiency
Gas exchange parameters such as photosynthetic rate (P) (μmolm−2 s−1) and Transpiration rate (T) (mmol−2 s−1) were measured in NT and transgenic plants using a portable photosynthetic system Li-6400 (LI-COR, USA) according to (Babitha et al., 2015). Three measurements were taken from the fully expanded third leaf of NT and transgenic lines at 10 days interval (10, 20, and 30) for drought and 5 days interval (5, 10, and 15) for salt stress in the morning (9–11 a.m.). Water Use Efficiency (WUE) was derived from P/T ratio.
Measuring root parameters
After the stress period, plants were harvested carefully without disturbing the root system from PVC lysimeter tubes. Shoot and root lengths were measured in centimeters (cm) using standard scale. Fresh weights of root and shoots, number of lateral roots and, nodules were also recorded. Further, shoots and roots were oven dried at 60°C for 48 h and weights were recorded for transgenic and NT plants.
Yield: Numbers of pods and seeds per plant were counted after harvesting the plants. Dry weight was taken after complete drying of the pods in an oven at 60°C and expressed in terms of grams per plant.
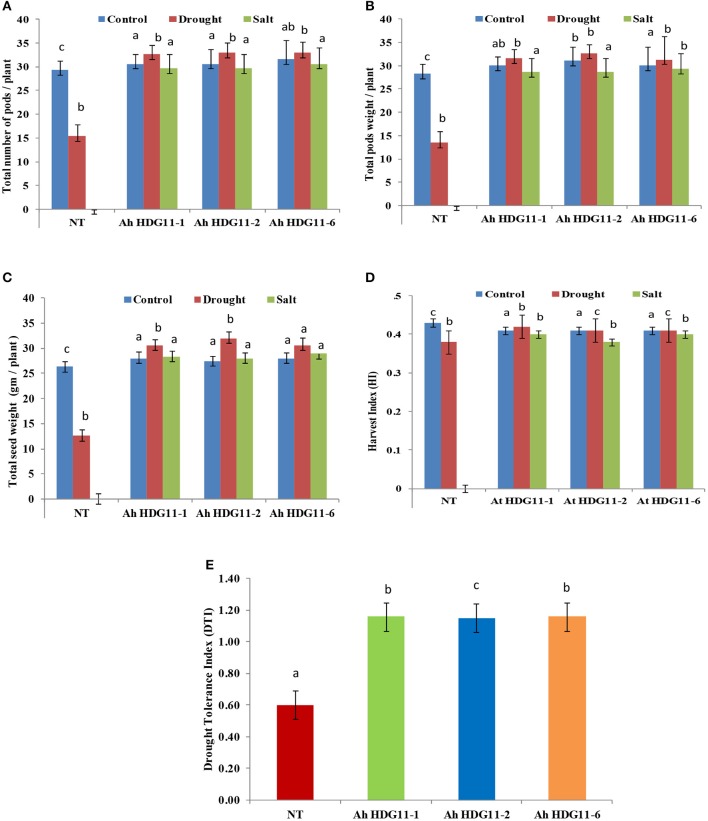
Drought Tolerance Index: Drought tolerance index (DTI) of NT and transgenic plants grown in the lysimetric experiment under well-water and drought stress conditions was measured based on the pod yield according to Nautiyal et al. (2002), using following formula.
Harvest index (HI): Harvest index was calculated using the following formula as described by Painawadee et al. (2009).
Measurement of free proline and antioxidative enzymes
Estimation of proline in the leaves of NT and transgenic plants grown in the lysimetric experiment under well-water, drought, and salt stress conditions was measured according to Bates et al. (1973). Antioxidative enzymes such as superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX) were quantified in the NT and transgenic plants grown in the lysimetric experiment under well-water, drought, and salt stress conditions according the method described by Elavarthi and Martin (2010).
Lipid peroxidation (MDA) and electrolyte leakage
Leaf samples from the stressed and control samples from transgenic and NT plants were used for the measurement of malondialdehyde (MDA) concentration (a product of lipid peroxidation) as described by Quan et al. (2004). Leaf discs were punched from the stress treated and control samples and membrane electrolyte leakage was assayed as described by Shou et al. (2004).
Statistical analysis
Chi square (X2), probability (P) values, correlation analysis and ANOVA (Duncan method) were done by using Software IBM SPSS Statistics v. 2.0. Graph data represents mean± S.E (n = 3). Cluster analyses were performed and heat maps were generated to group the physiological and biochemical traits based on the degree of correlation with R Statistical Environment (R Core Team, 2012).
Results
Developing transgenic construct
Full length cDNA clone of AtHDG11 was successfully isolated from A. thalina mutant line SALK_003819 and cloned initially into pTZ57R/T vector using XhoI and NotI restriction enzymes and later into plant transformation vector pGreen0029 in between At rd29A promoter and polyA terminator. Conformation of the individual clones in the construct was done through PCR analysis, restriction digestion, and sequencing of the HDG11 gene.
Development of transgenic peanut plants
Five hundred cotyledonary node (CNN) explants in five batches were co-cultivated with Agrobacterium strain (EHA105) harboring AtHDG11 (Supplementary Figure 1A) by exposing the proximal cut ends of explants in the bacterial suspension. Following infiltration, the explants were transferred to shoot induction media (SIM) (Supplementary Figure 1B) supplemented with kanamycin (200 mg/L) monosulphate and cefotaxime (10 μg/mL) as selection agent. The well-differentiated 312 shoot-lets (62.4%) were transferred to shoot elongation medium (SEM) comprising kanamycin (200 mg/L) and cefotaxime (10 μg/mL). Shoot-lets were further sub-cultured on SEM with 2 week growth interval to ensure further selection of transgenics. Well-developed 262 shoot-lets (52.4%) were transferred to root induction medium containing NAA (0.8 mg/L) without antibiotic. Finally 125 plants were recovered (25%) with fully developed root system that were placed in sterile vermiculite containing pots. After hardening for 2 weeks, 62 plant-lets (12.4%), were transferred to earthen-pots containing sterile soil and sand mixture (3:1) and grown under greenhouse conditions. The survived 14 putative transgenic plants (2.8%) were subjected for functional validation using various biochemical and physiological parameters.
Molecular characterization and transgene inheritance analysis
Integration and segregation analysis of transgene in T1& T2 generation was carried out using PCR and southern blot analysis. PCR analysis in 14 (T0) putative transgenics using different combinations of primers has revealed, eight plants were positive for all components of the transgenic construct (Supplementary Figure 2A and Figure 2B). Eight (T0) putative transgenic peanut plants were advanced to T1 through selfing and the progeny populations (T2) were analyzed for transgene integration and stability through PCR and transgene inheritance analysis was carried out in eight populations to know the zygosity nature of transgenic lines. Southern blot analysis carried out in seven independent T2 transgenic lines and integration of single copy of transgene has been confirmed in three transgenic lines (Supplementary Figure 2D lane: 1–3) and multiple copies of integration in three lines (Supplementary Figure 2D lane: 5–7), while no transgene integration was found in one line (Supplementary Figure 2D lane: 4). The single copy integrated T2 individual transgenic lines were advanced to T3 generation by selfing. The χ2 analysis of transgene inheritance was presented in Table 1.
Table 1.
Segregation analysis of AtHDG11 gene in self progenies (T1) derived from AtHDG11 transgenic plants.
| T1-Generation | Total number of seeds | Kan & PCR +Ve | Kan & PCR –Ve | Ratio | Test Ratio | X2 | P |
|---|---|---|---|---|---|---|---|
| Plant-1 | 11 | 8 | 3 | 2.6 | 3:1 | 0.030 | 0.862 |
| Plant-2 | 12 | 9 | 3 | 3 | 3:1 | 0.00 | 1 |
| Plant-3 | No seeds | – | – | − | 0 | 0 | 0 |
| Plant-4 | 11 | 8 | 3 | 2.6 | 3:1 | 0.030 | 0.862 |
| Plant-5 | 10 | 7 | 3 | 2.3 | 3:1 | 0.133 | 0.715 |
| Plant-6 | 6 | 4 | 2 | 2 | 3:1 | 0.22 | 0.637 |
| Plant-7 | No seeds | – | – | − | 0 | 0 | 0 |
| Plant-8 | 7 | 5 | 2 | 2.5 | 3:1 | 0.048 | 0.827 |
Transgenic peanut plants over expressing HDG11 showed enhanced abiotic stress tolerance
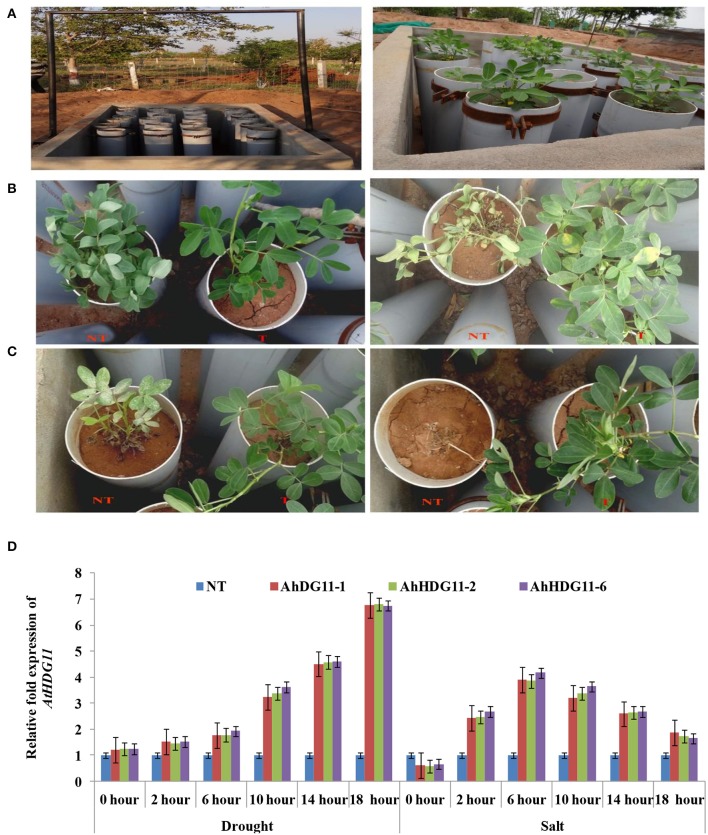
Progeny plants derived from single copy integrated transgenics were designated as Ah-HDG11-1, Ah-HDG11-2, and Ah-HDG11-6. These transgenics were characterized for their abiotic stress tolerance in an outdoor lysemetric experiment using various physiological and biochemical parameters (Figure 1A). No significant phenotypic variation was observed between transgenic and non-transgenic (NT) plants under well-watered conditions (data not shown). However, upon imposition of drought stress NT plants exhibited visual wilting symptoms on day 10th, while on the other hand transgenic lines, displayed trivial visual wilting symptoms only on day 30th of post drought stress imposition (Figure 1B). To evaluate the post stress recovery capacity, transgenic lines, and non-transgenic plants were re-watered to their saturated filed capacity and monitored the recovery. All the three independent transgenic lines recovered with a survival rate of 97.8% from drought stress, whereas the NT plants showed a survival rate of only 5%.
Figure 1.
Screening of non-transgenic and transgenic plants for drought and salt tolerance using lysimetric experiment. (A) Lysimetric experiment setup. Representative pictures of stress-induced phenotype of non-transgenic (NT) and transgenic (T) lines under (B) drought stress,15 days of stress imposition (left panel) and 30 days of stress imposition (right panel) (C) salt stress, 7 days of stress imposition (left panel) and 15 days of stress imposition (right panel). (D) qRT-PCR analysis of the T3 generation peanut transgenic lines for the AtHDG11 transcript at various time points under induced drought and salt lysimetric experiments to check the stress-inducible expression of transgene AtHDG11 under rd29A promoter.
To study the role of AtHDG11 in regulating the salinity response, the transgenic lines and NT plants were exposed to high salinity stress conditions. On exposure to 250 mM NaCl, NT plants showed visual symptoms of chlorosis, severe growth retardation on 5th day after stress imposition, and eventually died after 15 days (Figure 1C). In contrast, transgenic lines did not exhibit salt-induced stress symptoms even after 15 days of salt stress imposition. Transgenic plants recorded increase in fresh and dry weights of shoot and root compared to NT plants, both under drought and salt stress conditions (Figures 2A–C, 4B,C). These results indicate that AtHDG11 promotes growth and tolerance under drought and salt stress conditions in transgenic peanut plants.
Figure 2.
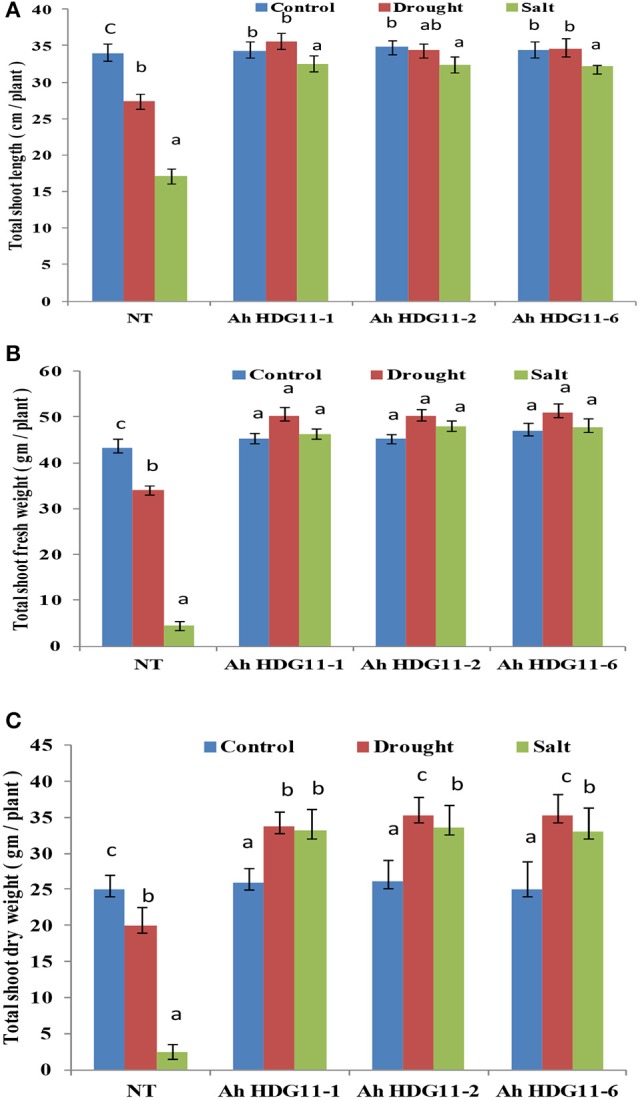
Post-harvest shoot growth and biomass analysis of three transgenic lines AhHDG11-1, AhHDG11-2, AhHDG11-6 along with the control untransformed parent JL-24 (NT) under control (well-watered), drought and salt stress from the confined outdoor lysimetric experiment. (A) Shoot length (cm/plant). (B) Shoot fresh weights (gm/plant). (C) Shoot dry weights (gm/plant). Alphabets in lowercase indicates the statistical significance in differences between WT and transgenics at p < 0.05.
Drought and salt stress-inducible expression of transgene AtHDG11
To validate the stress inducible nature of rd29a promoter, a qRT-PCR experiment was carried out with the samples recovered from lysimetric dry-down stress experiments at different time intervals. As shown in the Figure 1D, a gradual increase in the expression of At HDG11 transcript was observed in the transgenic lines under drought and salt stress, while no transcripts were detected in the NT. The data clearly indicates the tight regulation of transgene expression during the drought and salt stress conditions.
Transgenic plants displayed improved yield under drought and salt stress conditions
Yield in terms of pod number, pod, and seed weight were measured in non-transgenic (NT) as well as in transgenic lines of Ah-HDG11-1, Ah-HDG11-2, and Ah-HDG11-6 under well-watered (WW), drought, and salt-stress conditions (Figures 3A–D). Pod yield has drastically reduced (15 pods/plant) in NT plants compared to transgenics (30–32/plant) under drought and high salinity stress (Figure 3A). Considerable increase in the weight of the seed in transgenic lines has been noticed. No significant variation in yield and weight of the seeds was observed among the transgenic lines.
Figure 3.
Yield analysis of three transgenic lines AhHDG11-1, AhHDG11-2, AhHDG11-6 along with the control untransformed parent JL-24 (NT) under control (well-watered), drought, and salt stress from the confined outdoor lysimetric experiment. (A) Total number of matured pods/plant. (B) Pod weight (gm/plant). (C) Seed weight (gm/plant). (D) Harvest index. (E) Drought Tolerance Index (DTI). Alphabets in lowercase indicates the statistical significance in differences between WT and transgenics at p < 0.05.
Transgenic plants exhibited improved water-use efficiency (WUE) traits
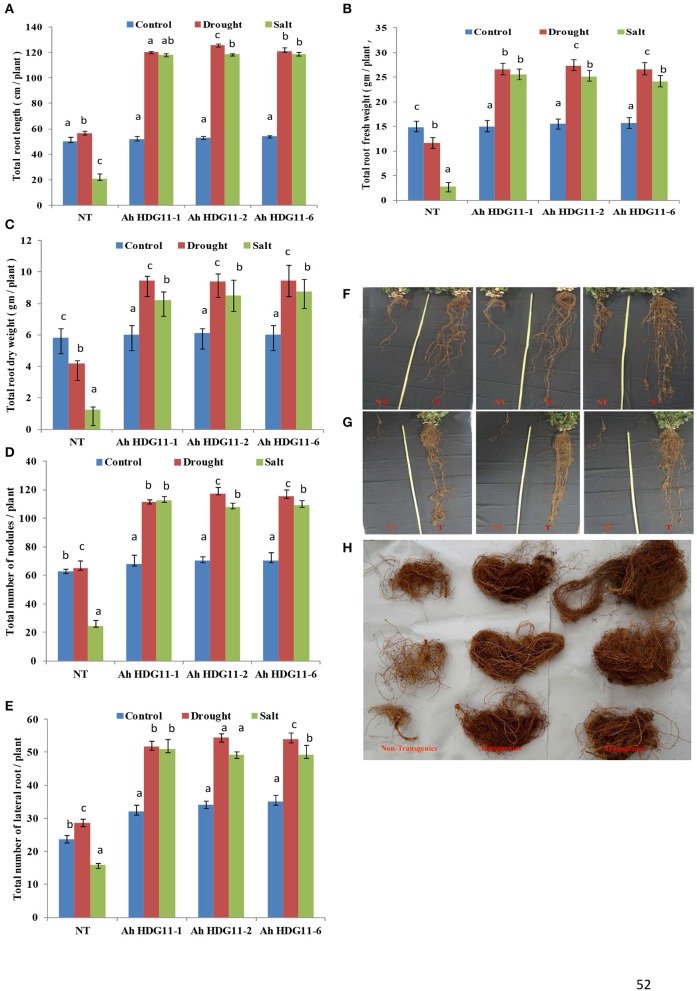
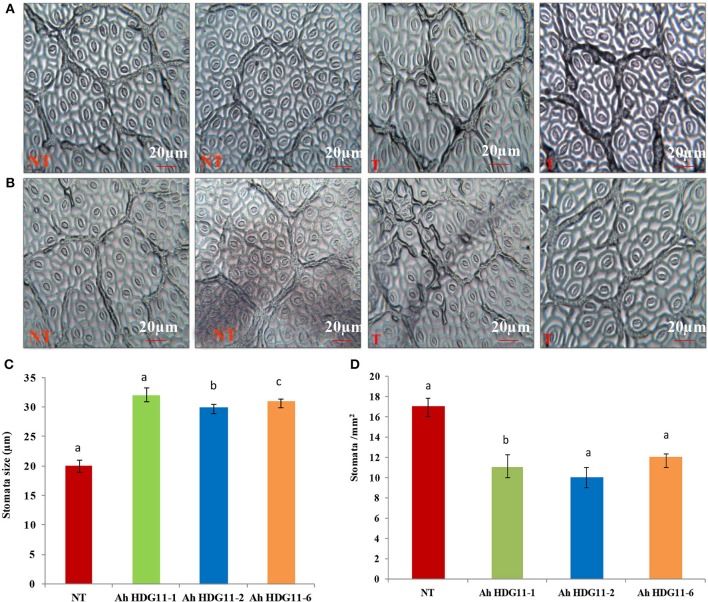
Transgenic peanut lines over expressing AtHDG11 developed deeper root system with increased number of lateral roots and root nodules (Figures 4A,D,E) both under drought and salt-stress conditions. As shown in the (Figures 4F,G) transgenic lines showed larger root systems (50–75%), root biomass (60–70%) (Figure 4H), and root nodules (55–65%) compared to NT plants. Stoma plays a key role in keeping the oxidative stress at pace in plants by regulating the transpiration. In order to understand the role of stomata in enhanced WUE of transgenic plants, we analyzed the stomatal density in the transgenic and NT plants both under drought and salt stress conditions. The adaxial stomatal density and size of the 3rd fully expanded leaf were determined by leaf surface imprint method. The average stomatal densities of the selected transgenic lines were decreased by 50–60% compared with that of the NT plants (Figures 5A,B,D). However, the stomatal size (length and width) was increased by 20% in the transgenic plants compared to NT (Figure 5C).
Figure 4.
Post-harvest root morphology, growth, and biomass analysis of three transgenic lines AhHDG11-1, AhHDG11-2 and AhHDG11-6 along with the control untransformed parent JL-24 (NT) under control (well-watered), drought and salt stress from the confined outdoor lysimetric experiment. (A) Root length (cm/plant). (B) Root fresh weight (gm/plant). (C) Root dry weight (gm/plant). (D) Root nodules. (E) Lateral roots. (F) Phenotype of roots under drought stress. (G) Phenotype of roots under salt stress. (H) Root mass of Non-transgenic (NT) and transgenic plants (T). Alphabets in lowercase indicates the statistical significance in differences between WT and transgenics at p < 0.05.
Figure 5.
Stomatal analysis of three transgenic lines AhHDG11-1, AhHDG11-2, AhHDG11-6 along with the control untransformed parent JL-24 (NT) under control (well-watered), drought, and salt stress from the confined outdoor lysimetric experiment. Adaxial epidermal imprint image of (A) Drought stressed plants. (B) Salt stressed plants. (C) Stomatal width (μm). (D) Stomatal density (Stomata/mm2).
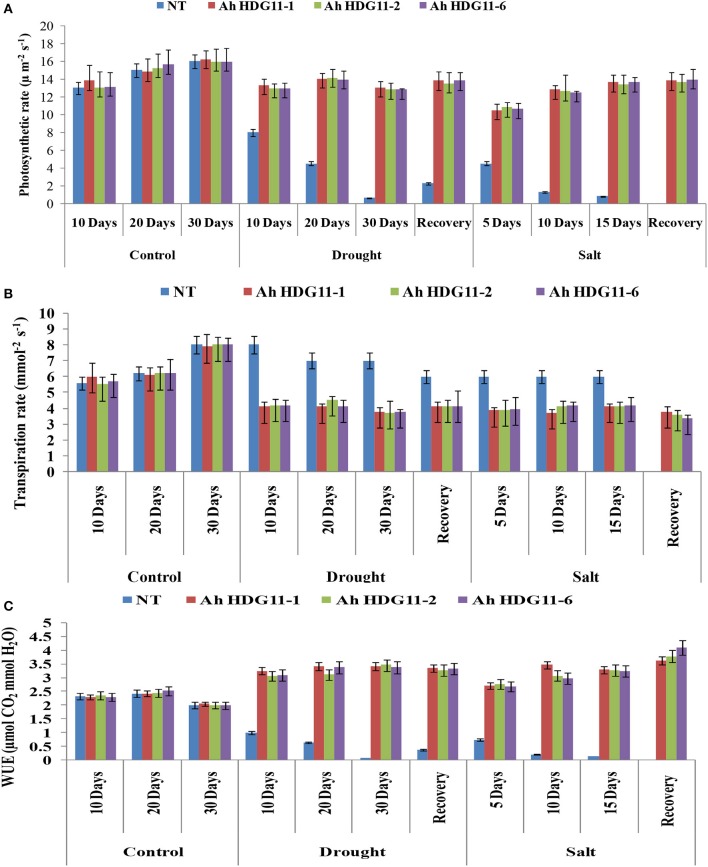
In order to know whether the reduced stomatal density affected the photosynthesis or not, the photosynthesis and transpiration rates of NT and transgenic peanut lines were measured with a 10 days interval (10, 20, and 30th days) after drought stress imposition, 5, 10, and 15th day of salt stress stressed plants and 5 days after recovery from the stress. Under well-water conditions both transgenic and non-transgenic lines recorded similar photosynthetic rates, however, under drought and salt stress conditions photosynthetic rates were drastically reduced in non-transgenic plants. Interestingly, though the number of stomata decreased in transgenic lines, they recorded 30–50% higher photosynthetic rates compared to non-transgenic lines and maintained higher photosynthetic rates even under severe drought and salt stress conditions (Figure 6A). In contrast, transgenic lines under drought and salt stress conditions recorded lower transpiration rates compared to non-transgenic plants (Figure 6B). As a result transgenic plants recorded higher WUE rates under both drought and salt stress conditions (Figure 6C).
Figure 6.
Gas exchange and Water Use Efficiency Analysis of transgenic lines AhHDG11-1, AhHDG11-2, AhHDG11-6 along with the Non-transgenic (NT) plants. (A) Photosynthesis rate (B) Transpiration rate (C) Water Use Efficiency.
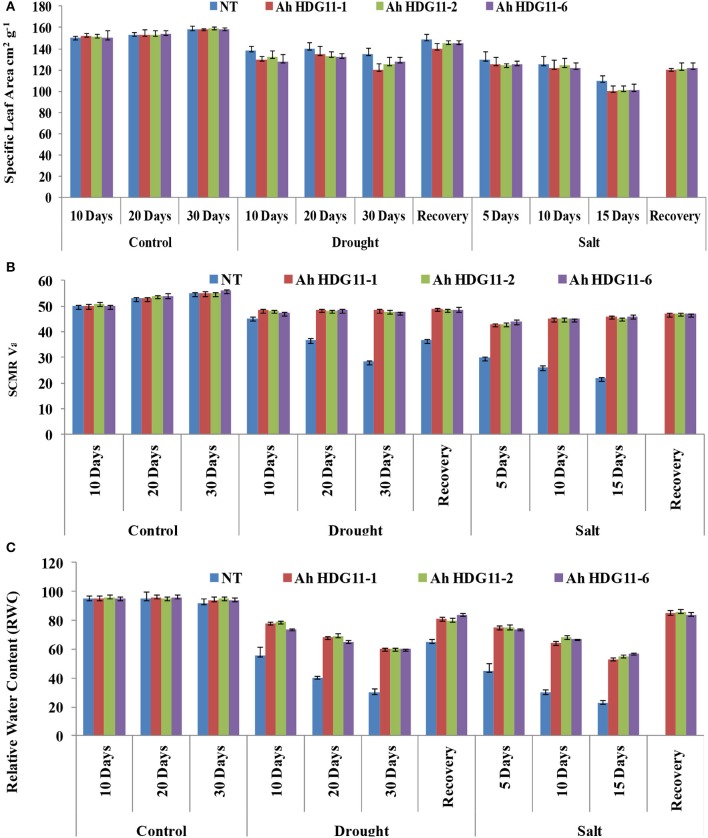
Traits such as SLA and SCMR were used as indirect surrogates for measuring WUE. The SLA and SCMR readings were measured from drought and salt stress imposed plants for every 5 days interval and 5 days after recovery from the stress. Transgenic lines of Ah-HDG11-1, Ah-HDG11-2, and Ah-HDG11-6 recorded higher SCMR values compared to NT plants during drought and salt stress (Figure 7B). SCMR values increased with duration of both drought and salt stress and highest SCMR values were recorded in transgenic lines of Ah-HDG11-1. Relatively, high SCMR values were observed in drought stress recovered transgenic lines of Ah-HDG11-1, Ah-HDG11-2, and Ah-HDG11-6 compared to NT plants under WW conditions. Low SLA values were recorded in transgenic lines of Ah-HDG11-1, Ah-HDG11-2, and Ah-HDG11-6 (Figure 7A) under drought and salt stress conditions compared to their control plants and NT plants. SLA was decreased with increased duration of stress.
Figure 7.
Physiological analysis of transgenic lines AhHDG11-1, AhHDG11-2, AhHDG11-6 along with the Non-transgenic (NT) plants. (A) Specific Leaf Area (SLA) (B) Chlorophyll content (SCMR) (C) Relative Water Content (RWC).
To investigate the current water content in the transgenic and non-transgenic plants relative to their maximal water content they can hold at full turgidity under drought and salt stress conditions, relative water content (RWC) was measured. A sharp decline in the RWC was noticed in both non-transgenic as well as in transgenic plants with increase in the severity of drought and salt stress. However, transgenic lines Ah-AtHDG11-1, Ah-AtHDG11-2, and Ah-AtHDG11-6 maintained higher RWC (78% on day 10, 68%on day 20, and 60% on day 30th) of drought stress in their leaves (Figure 7C). Whereas, non-transgenic plant leaves had RWC of (55% on day 10, 40% on day 20, and 30% on day 30th) of drought stress. Up on release of drought stress, fast recovery of RWC observed in transgenic plants (85% RWC) compared to non-transgenic plants (65% RWC) (Figure 7C). Similar pattern was also observed in the salt-stress imposed transgenic and non-transgenic plants (Figure 7C).
The correlation between the traits contributing to WUE in drought and salt stress are listed in Table 2. Root traits (RL, RFW, and RDW), chlorophyll content (in relative SPAD units), RWC, TE, HI, DTI, STL LEN, and STL WID under drought and salt stress, were positively correlated with each other. Apparently, SLA under drought stress negatively correlated with root traits (RL−0.61; RFW−0.63; RDW−0.61), chlorophyll content (−0.64), RWC(−0.61), TE (−0.62), HI (−0.56), DTI (−0.62), STL LEN (−0.69), and STL WID (−0.69), while SLA in drought stress positively correlated with stomatal density (STL DEN 0.64), and negatively correlated with STL DEN (−0.75) in salt stress conditions. Stomatal density in drought and salt stresses negatively correlated with all WUE traits, except with SLA in drought stress (0.64) (Table 2).
Table 2.
Correlation coefficient (r) between various morpho-physiological traits contributing to WUE in peanut AtHDG11 transgenic lines under drought and salt stress.
| RL (D) | RL (S) | RFW (D) | RFW (S) | RDW (D) | RDW (S) | SPAD(D) | SPAD(S) | RWC(D) | RWC (S) | SLA(D) | SLA(S) | TE (D) | TE (S) | HI (D) | HI (S) | DTI (D) | DTI (S) | STL DEN (D) | STL DEN (S) | STL LEN (D) | STL LEN (S) | STL WID (D) | STL WID (S) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RL (D) | 1 | ||||||||||||||||||||||||
| RL (S) | 0.99** | 1 | |||||||||||||||||||||||
| RFW (D) | 0.99** | 0.99** | 1 | ||||||||||||||||||||||
| RFW (S) | 0.99** | 0.99** | 0.99** | 1 | |||||||||||||||||||||
| RDW (D) | 0.99** | 0.99** | 0.99** | 0.99** | 1 | ||||||||||||||||||||
| RDW (S) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 1 | |||||||||||||||||||
| SPAD(D) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 1 | ||||||||||||||||||
| SPAD(S) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 1 | |||||||||||||||||
| RWC(D) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 1 | ||||||||||||||||
| RWC (S) | 0.98* | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 1 | |||||||||||||||
| SLA(D) | −0.61 | −0.61 | −0.63 | −0.63 | −0.61 | −0.57 | −0.64 | −0.62 | −0.61 | −0.53 | 1 | ||||||||||||||
| SLA(S) | 0.78 | 0.78 | 0.75 | 0.76 | 0.78 | 0.8 | 0.75 | 0.76 | 0.77 | 0.82 | 0 | 1 | |||||||||||||
| TE (D) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.98* | −0.62 | 0.76 | 1 | ||||||||||||
| TE (S) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.98* | 0.99** | 0.99** | 0.99** | 0.97* | −0.68 | 0.72 | 0.99** | 1 | |||||||||||
| HI (D) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | −0.56 | 0.82 | 0.99** | 0.98* | 1 | ||||||||||
| HI (S) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 1** | 0.99** | −0.61 | 0.77 | 0.99** | 0.99** | 0.99** | 1 | |||||||||
| DTI (D) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | −0.62 | 0.77 | 0.99** | 0.99** | 0.99** | 0.99** | 1 | ||||||||
| DTI (S) | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | 0.99** | −0.62 | 0.77 | 0.99** | 0.99** | 0.99** | 0.99** | 1** | 1 | |||||||
| STL DEN (D) | −0.98* | −0.97* | −0.96* | −0.96* | −0.96* | −0.96* | −0.96* | −0.95* | −0.96* | −0.94 | 0.64 | −0.75 | −0.98* | −0.97* | −0.97* | −0.96* | −0.96* | −0.96* | 1 | ||||||
| STL DEN (S) | −0.98* | −0.97* | −0.96* | −0.96* | −0.96* | −0.96* | −0.96* | −0.95* | −0.96* | −0.94 | 0.64 | −0.75 | −0.98* | −0.97* | −0.97* | −0.96* | −0.96* | −0.96* | 1** | 1 | |||||
| STL LEN (D) | 0.98* | 0.99** | 0.99** | 0.99** | 0.99** | 0.98* | 0.99** | 0.99** | 0.99** | 0.98* | −0.69 | 0.71 | 0.99** | 0.99** | 0.98* | 0.99** | 0.99** | 0.99** | −0.96* | −0.96* | 1 | ||||
| STL LEN (S) | 0.98* | 0.99** | 0.99** | 0.99** | 0.99** | 0.98* | 0.99** | 0.99** | 0.99** | 0.98 | −0.69 | 0.71 | 0.99** | 0.99** | 0.98* | 0.99** | 0.99** | 0.99** | −0.96* | −0.96* | 1** | 1 | |||
| STL WID (D) | 0.97* | 0.98* | 0.99** | 0.99** | 0.98* | 0.98* | 0.99** | 0.99** | 0.98* | 0.97* | −0.69 | 0.68 | 0.98* | 0.98* | 0.97* | 0.98* | 0.99** | 0.99** | −0.93 | −0.93 | 0.99** | 0.99** | 1 | ||
| STL WID (S) | 0.97* | 0.98* | 0.99** | 0.99** | 0.98* | 0.98* | 0.99** | 0.99** | 0.98* | 0.97* | −0.69 | 0.68 | 0.98* | 0.98* | 0.97* | 0.98* | 0.99** | 0.99** | −0.93 | −0.93 | 0.99** | 0.99** | 0.99** | 1** | 1 |
Correlation is significant at the 0.05 level.
Correlation is significant at the 0.01 level.
Transgenic plants exhibited improved oxidative stress tolerance and high proline accumulation
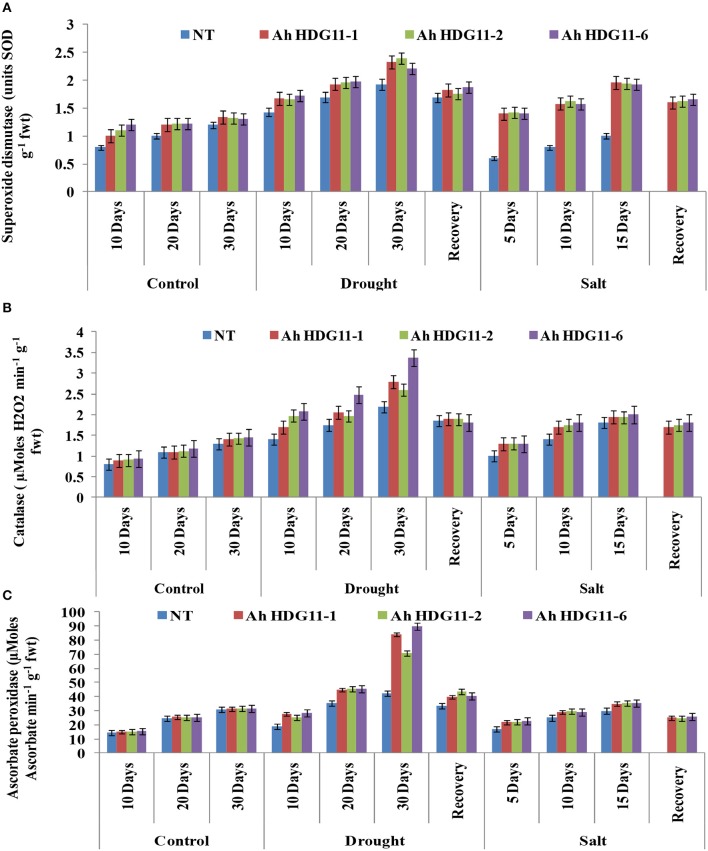
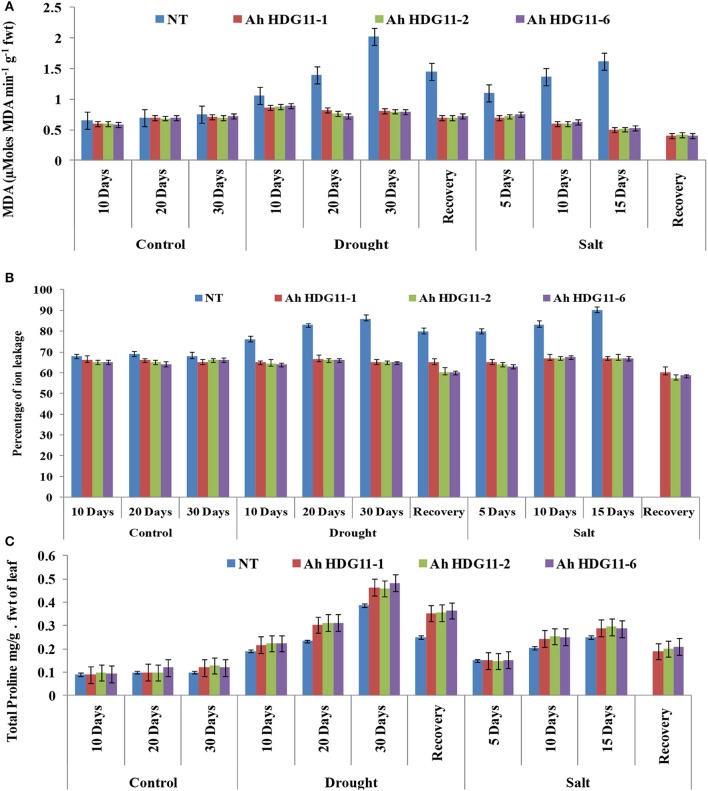
Drought and salt stress damage cells and changes in lipid composition and lipid peroxidation. Malondialdehyde (MDA), a naturally occurring product of lipid peroxidation and it is the important indicator for the oxidative damage of tissues. Therefore, to measure the drought and salt stress tolerance of transgenic plants we measured MDA content of transgenic and non-transgenic plants as an indicator of lipid peroxidation. Drought and salt stressed non-transgenic plants recorded greater MDA contents than transgenic plants. MDA contents increased with the duration of both the stresses (Figure 9A).
Electrolyte leakage indicates the cell membrane stability under abiotic stress conditions. A significance difference in electrolyte leakage was observed across the transgenic lines and non-transgenic (NT) plants under stress conditions. Electrolyte leakage was high (90%) in the NT plants and it is directly proportional to the severity of drought stress, while in the transgenic lines it was slightly less (60–70%) (Figure 9B). When compared to drought, salt stress caused more damage to the membrane resulting in considerable ion leakage in NT plants. Further, the electrolyte leakage increased in NT plants, with increase in severity of the salt stress. Whereas, the transgenic lines of Ah-HDG11-1, Ah-HDG11-2, and Ah-HDG11-6 maintained high membrane stability both under drought and salt stress.
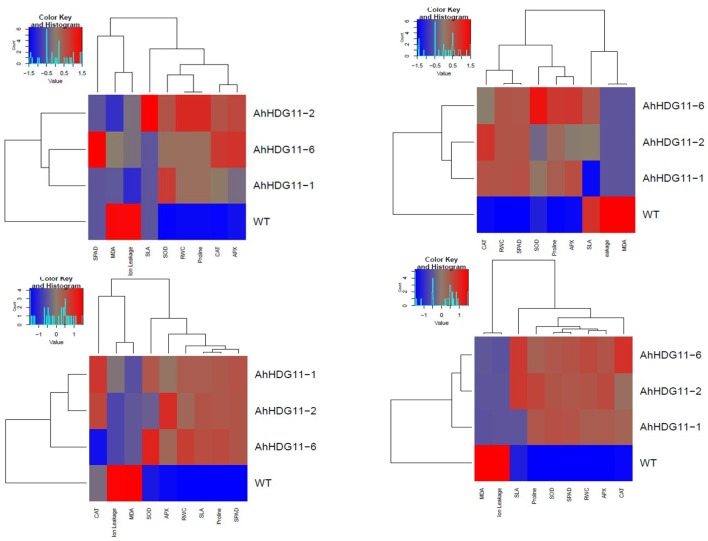
Further, to gain insights into the oxidative stress tolerance mechanism of transgenic plants, reactive oxygen species (ROS) scavenging activities were measured in both transgenic plants and the non-transgenic controls. Antioxidative enzymes SOD, CAT, and APX are the major enzymes responsible for scavenging ROS in plants under stress. A sharp increase in SOD, APX, and CAT activity were detected in transgenic lines of Ah-AtHDG11-1, Ah-AtHDG11-2, and Ah-AtHDG11-6 upon drought and salt stress and enzyme activities were found to be increased with the increase in severity of the stress (Figure 8). The heat map hierarchical clustering algorithm (Figure 11) clearly suggests that SOD, APX, and CAT enzyme activities under drought and salt stress condition were positively correlated with each other and negatively correlated with MDA and electrolyte leakage. Antioxidative enzyme activities have strong positive correlation with RL, chlorophyll content, and pod yield, RWC, TE, HI, DTI, STL LEN, and STL WID. However, SLA, STL DEN were negatively correlated with SOD, APX, and CAT enzyme activities under drought and salt stress conditions.
Figure 8.
Physiological and biochemical analysis of transgenic lines AhHDG11-1, AhHDG11-2, AhHDG11-6 along with the Non-transgenic (NT) plants. (A) Superoxide Dismutase (SOD) (B) Catalase (CAT) (C) Ascorbate Peroxidase (APX).
Figure 11.
Overall assessments of physiological and biochemical responses in non-transgenic and transgenic lines during well-water (A), drought stress (B), re-watering (C), and salt stress (D). The relative values of physiological, biochemical parameters for the heat-map analysis. Scale: for the drought and salt tolerance capabilities, from brightest blue equal lowest to brightest red equal highest capabilities; for physiological and biochemical parameters, from brightest blue equals most decreased to brightest red equals most increased.
A common phenomenon of abiotic stress response is accumulation of osmolyte proline. During drought and salt stress conditions, a three-fold increase in the proline was observed in the transgenic lines of Ah-HDG11-1, Ah-HDG11-2, and Ah-HDG11-6 compared to NT (Figure 9C).
Figure 9.
Biochemical analysis of transgenic lines AhHDG11-1, AhHDG11-2, AhHDG11-6 along with the Non-transgenic (NT) plants. (A) Malondialdehyde (MDA) (B) Percentage of ions leakage (C) Proline (Pro).
Elevated levels of known stress responsive genes observed in the transgenic plants
To figure out the mechanisms of drought and salt tolerance in transgenic plants, expression levels of known stress responsive genes in the stress gene network which codes for functional proteins ELIP (Early light induced proteins), HSP 70 (heat shock protein 70), Cu/Zn SOD (superoxide dismutase), APX (ascorbate peroxidase), P5CS (Δ1-pyrroline-5-carboxylate synthetase), LEA (Late embryogenic abundant protein), TIP (Tonoplast intrinsic protein), Drought protein, MIPS (Major Intrinsic Proteins), AhAq1 (Aquaporin), AhNCED1 (9-cis-epoxycarotenoid dioxygenase1) and few of the stress responsive regulatory genes such as AhERF1(ethylene-responsive element binding factor), AhNAC4 (NAM, ATAF, and CUC) family proteins 4), and AhRRS5 (nucleotide binding site leucine rich repeat 5) were monitored using qRT-PCR. These stress responsive genes were selected based on their expression in earlier studies in HDG11 transgenic plants i.e., Cu/Zn SOD, APX, P5CS, and RRS5 whose expression levels were up-regulated in the transgenic plants (Yu et al., 2008). As revealed by the qRT-PCR data Figure 10, expression levels of known stress responsive genes were elevated in the transgenic and non-transgenic plants under drought and salt stress. However, the level of induction was more in the transgenic plants and expression levels of these stress responsive genes were correlated with the expression of HDG11 transgenic in the transgenic plants. The obtained data clearly indicates all of these known stress responsive genes probably directly or indirectly regulated by the AtHDG11 protein.
Figure 10.
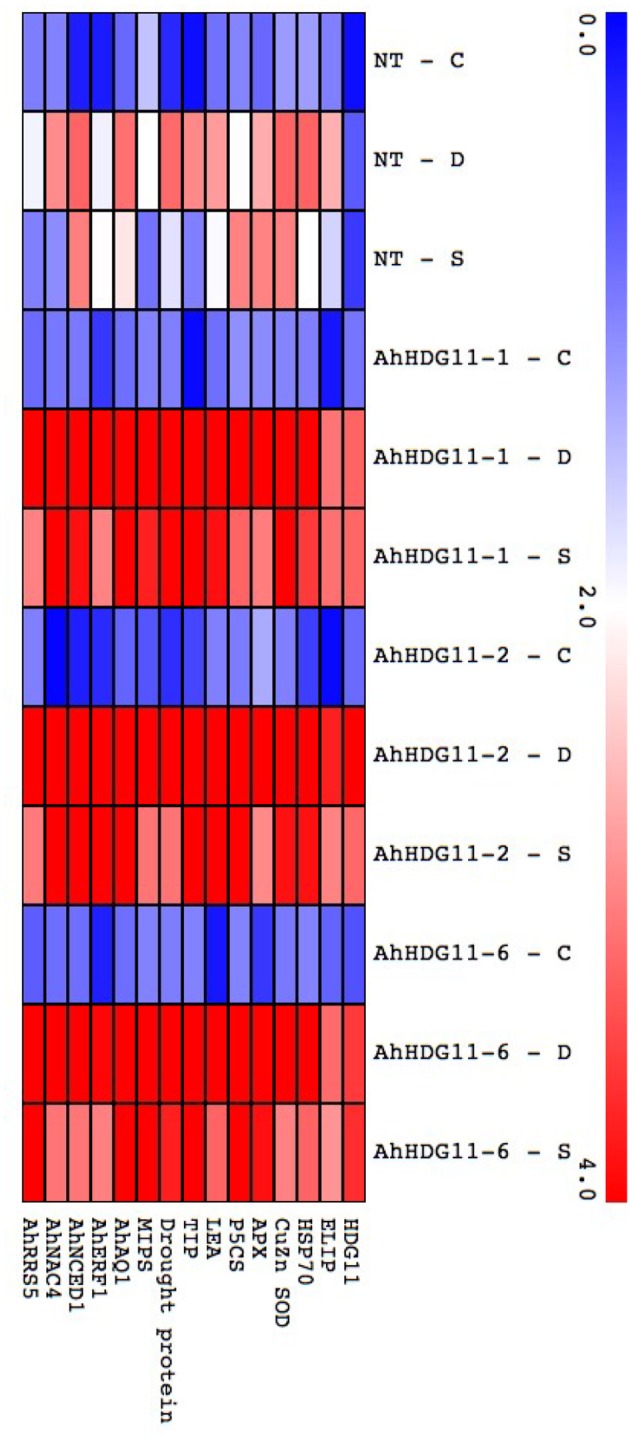
Gene expression analysis of known stress responsive genes in non-transgenic (NT) and Transgenic lines. qRT-PCR analysis of T3 generation peanut transgenic lines for the expression of known stress responsive genes under induced drought and salt lysimetric experiments. Heat map showing the expression of known stress responsive genes of stress gene network in non-transgenic (NT) and transgenic plants under well-water, drought and salt stress conditions. The log2 fold change scale is indicated in the heat map.
Discussion
Soil moisture stress at various growth stages of peanut often limit the quality of production in SAT regions. A number of efforts were made to improve the traits such as cell turgor, positive carbon balance, cellular level tolerance and plant type for imparting stress tolerance and crop improvement in peanut under stress conditions (Rao et al., 2012). Lack of genetic and genomic resources has made molecular and traditional approaches faint in getting improved peanut varieties. One of the promising alternative ways is genetic manipulation. Since, these traits were controlled by several genes, transcription factors could be ideal candidates for genetic manipulation, toward crop improvement.
The present research describes stress inducible overexpression of AtHDG11 in transgenic peanut plants that imparts drought and salt stress tolerance along with improved productivity. In earlier studies, AtHDG11 transgene was expressed under constitutive promoter “CaMV35S”, hence, regardless of environmental cues plants produced extensive root systems and reduced stomatal density (Ruan et al., 2012; Yu et al., 2013, 2015; Li et al., 2016). Under favorable conditions when all the requirements are sufficiently supplied to the plant, larger root system is not required, and this plant type might pose metabolic load on yield potential (Blum, 2005). Therefore, in the present study AtHDG11 gene expression was tightly regulated under a stress inducible promoter rd29A. The qRT-PCR data (Figure 1D) indicates that transgene expression was triggered by rd29A promoter under both drought and salt stress conditions.
A strong and efficient expression system is more advantageous for efficient gene functioning of desired traits in transgenic crops. Most of the studies that use constitutive promoter like CaMV35S cause adverse physiological effects and undesirable phenotypic characters in transgenic crops. Constitutive expression of transgene under unwanted conditions may also cause metabolic stress in transgenic plants. In recent past, several studies focused on stress inducible promoter of rd29a, rab16a, Os ABA2, OS NCED3, BADH etc. for achieving maximum stress tolerance in different transgenic crops (Shinozaki and Shinozaki, 1993; Zhang et al., 2008; Rai et al., 2009; Sun et al., 2010). Recently a maize ZmGAPP promoter (D8) was reported as suitable for alien gene expression under salt tolerance (Hou et al., 2016). Yi et al. (2010), reported, an abiotic stress inducible promoter Wsi18 that functions in grain development of rice. In the present study transgenic peanut lines displayed several beneficial physio-morphological characters such as delayed wilting symptoms, better shoot and root ratio, improved biomass under drought and salt stress conditions. The resulted phenotypic data clearly indicates that stress regulated expression of AtHDG11 is sufficient enough to obtain tolerant phenotype only under stress conditions, but not under ideal growth conditions. Analog phenotypes were observed in peanut transgenic carrying transgens under rd29A regulation (Kasuga et al., 2004; Bhatnagar-Mathur et al., 2007; Vadez et al., 2007; Devi et al., 2011; Jagana et al., 2012; Sarkar et al., 2014; Ramu et al., 2016).
Root architecture is an important agronomic trait that plays a major role in drought tolerance mechanism (Smith and De Smet, 2012) of crop plants. The greater water uptake capability of a plant depends on the greater development of lateral roots and deeper penetration ability of tap roots (De dorlodot et al., 2007; Comas et al., 2013). In this present study, transgenic peanut lines carrying HDG11 exhibited extensive root system (50–60% more) with increased root nodules and biomass than NT. It has been established that HDG11 directly binds to HD binding sites in the promoters of JA biosynthetic genes i.e., AOS, AOC3, OPR3, and OPCL1 and increases the production of JA content in the root. The elevated levels of JA could further induce a cascade of auxin signaling genes thatpromote the lateral root formation besides up-regulating cell-wall-loosening proteins, that induce the primary root elongation (Xu et al., 2014; Cai et al., 2015). Deep and profuse root systems not only enhances drought tolerance, but also boosts the nutrient uptake and exclude the excess salt from the root cells through membrane transporters (Vadez et al., 2007). Thus, HDG11 transgenic plants not only displayed drought tolerance but also showed salt tolerance.
Leaf water potential is a primary physiological index of plant water status and so considered the trait as an indicator to know the efficacy of the transgene. Higher water potential in the transgenic plants even under severe stress conditions could have possible by the extraction of water from the deeper layers of the soil due to profuse root growth and reduced transpiration associated with fewer stomata (Figure 6B). The elevated expression levels of three of the important aquaporin genes TIP (Regon et al., 2014), MIPS (Forrest and Bhave, 2007), and AhAq1 (Pan et al., 2009) in the transgenic plants in the present study could be the reason for the maintenance of high RWC in the leaves of transgenic plants. Plant aquaporins play an important role in maintaining the plant water relations (Maurel et al., 1993, 1997; Tyerman et al., 2002).
Stomata play a key role in gas exchange and water uptake with a strong impact on traits associated with transpiration, carbon assimilation, and WUE. The average stomatal density in HDG11 transgenic peanut plants was reduced by 30% while stomata size increased by 20% compared with that of the NT plants under drought and salt stress conditions. The reduced stomatal density apparently contributed to the reduction in water loss that is evidenced by decreased transpiration rates in transgenic plants (Figure 6B). However, the reduced stomatal density does not affect the photosynthetic rates of transgenic plants (Figure 6A). The WUE which is a ratio of the rate of carbon assimilation to the rate of transpiration drastically increased in the transgenic plants (Figure 6C). Higher intrinsic instantaneous WUE data in the present study could be due to the low stomatal conductance and higher carboxilation capacity of the transgenic plants under drought and salt stress conditions (Gilbert et al., 2011).
Soil Plant Analytical Development (SPAD) chlorophyll meter readings (SCMR) and Specific leaf area (SLA) are easily measureable non-destructive surrogate traits of TE, used for selection of drought tolerant traits. Transgenic peanut lines recorded high SCMR and slightly reduced SLA under drought and salt stress conditions (Figures 7A,B). SCMR is an indication of the light-transmittance characteristics of the leaf which is dependent on the leaf chlorophyll content and density (Richardson et al., 2002; Arunyanark et al., 2008). Leaf chlorophyll content per unit area positively correlates with the photosynthetic activity of the leaf. Peanut transgenic lines with high SCMR under drought and salt stress could maintain a higher rate of photosynthesis per unit leaf area compared to non-transgenic plants. SLA indicates thickness of the leaves, and low SLA of transgenic lines in the present study specifies thicker leaves and more chlorophyll content per unit area. A similar phenotype of thick leaves with tightly packed mesophyll cell and higher photosynthesis rate under drought stress has been reported for the Arabidopsis ERECTA gene, a putative leucine-rich-repeat receptor protein kinase gene (Masle et al., 2005). In the present study the expression of a nucleotide binding site leucine rich repeat 5 (AhRRS5) which is a homolog of ERECTA showed enhanced expression in the transgenic plants under drought and salt stress condition (Figure 10). As reported earlier, Arabidopsis ERECTA gene was trans activated by HDG11 protein (Yu et al., 2008). Thus, in this study, the increase in the leaf thickness in the transgenics might be associated with the up regulation of ERECTA like genes. The increase in shoot biomass and reduction in SLA supports the possibility for greater assimilation in the transgenic lines under drought and salt stress. Previous studies on peanut suggest that SLA and SCMR correlates with WUE. As reported earlier in peanut (Rao et al., 2001; Sheshshayee et al., 2006; Songsri et al., 2009) a strong positive correlation between WUE and SCMR, and a negative correlation between SLA and SCMR have been observed in the present study. Therefore, high SCMR and low SLA of HDG11 transgenic lines specify the greater WUE of transgenic peanut lines compared to non-transgenic plants.
One of the well-studied compatible solute is the amino acid proline, which is a multi-functional molecule that plays a major role in plant abiotic stress defense. It is actively involved in osmoregulation, scavenging of free radicals, and as a molecular chaperone for stabilizing protein structure, thus protects plant cells from the damaging effects of various environmental stresses (Parvaiz and Satyawati, 2008; Verbruggen and Hermans, 2008; Szabados and Savouré, 2010; Koyro et al., 2012; De Carvalho et al., 2013). A high level of proline content (30–40%) and elevated levels of P5CS gene expression was recorded in peanut transgenic lines compared to NT both under drought and salt stresses (Figures 9C, 10). Transgenic peanut plants not only accumulated high solutes, but also exhibited higher membrane integrity under severe stress conditions. Transgenic peanut lines overexpressing HDG11 showed significantly reduced electrolyte leakage and MDA levels (Figure 9A), thus display greater membrane stability (Figure 9B) compared to NT under drought and salt stress conditions. In totality, the obtained data clearly indicates that membrane is better protected from stress-induced ROS toxicity. One of the most important defense mechanisms against abiotic stress is the antioxidant system, which detoxifies ROS molecules such as hydrogen peroxide, superoxide, and singlet oxygen and keeps an adequate cellular redox balance (Noctor and Foyer, 1998; Asada, 1999). Supporting this, higher levels of antioxidative enzymes such as SOD, APX, and CAT were recorded in the HDG11-overexpressing peanutplants (Figure 8). Further, the antioxidative enzyme activities were in consonance with APX and Cu/Zn SOD gene expression levels in the transgenic plants (Figure 10), therefore protecting the plants from oxidative damage by enhancing ROS scavenging capability. The results obtained in the present investigation were in agreement with the altered antioxidative system in the transgenic tobacco (Yu et al., 2008), rice (Yu et al., 2013), tall fescue (Cao et al., 2009), sweet potato (Ruan et al., 2012), cotton and poplar (Yu et al., 2015), and wheat (Li et al., 2016).
In the present study we observed that shoot and root lengths, biomass were significantly (30–40%) higher in transgenic lines compared to non-transgenic plants (Figures 4A–C,F, 2A–C). Transgenic peanut lines also displayed greater pod number, pod, and seed weight, HI compared to NT under drought and salt stress conditions (Figures 3A–D). Present data of peanut transgenic are in consistent with the data presented in a previous study of transgenic rice over expressing HDG11 that resulted in greater yield under drought stress conditions. A well-equipped WUE traits and fine-tuned defense mechanisms might be contributing to the enhanced biomass and yield in the transgenic peanut plants under drought and salt conditions (Asif et al., 2011; Qin et al., 2011; Yu et al., 2013; Bhauso et al., 2014; Manjulatha et al., 2014; Pruthvi et al., 2014).
Present study has successfully demonstrated the stable integration and stress inducible overexpression of AtHDG11 in peanut with improved drought and salt tolerance. Transgenic plants displayed impressed biochemical and physiological traits like (1) Efficient water extraction from deeper layers of soil demonstrated by enhanced root lengths and later roots; (2) Minimized water loss through the stomata evidenced by reduced stomatal density; (3) Enhanced WUE traits, elevated photosynthetic rates to minimize the yield loss under stress conditions; (4) Cellular stress tolerance with increased accumulation proline, LEA, ELIPs, and efficient ROS detoxification mechanism.
Author contributions
JB, VP, SK, and KG: Performed the experiments; TC, CA, SP, and CP: Analyzed the data; CP: Conceived and designed the experiments; JB, TC, and CP: Wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the financial support (No.SR/SO/PS-62/08 dated 24/9/2009) from Department of Science and Technology (DST), New Delhi, India. The authors thankful to Prof. Arjula R. Reddy, Emeritus Professor, Department of Plant Sciences, University of Hyderabad, Hyderabad for his inputs in the form of critical comments for the improvement of manuscript. The authors thankful to Dr. R. V. Jayanth Kasyap, Asst. Professor in English, Yogi Vemana University, Kadapa, India for verifying the linguistic correctness.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2018.00034/full#supplementary-material
References
- Anuradha T. S., Jami S. K., Datla R. S., Kirti P. B. (2006). Genetic transformation of peanut (Arachis hypogaea L.) using cotyledonary node as explant and a promoter less gusnptII fusion gene based vector. J. Biosci. 31, 235–246. 10.1007/BF02703916 [DOI] [PubMed] [Google Scholar]
- Araus J. L., Slafer G. A., Royo C., Serret M. D. (2008). Breeding for yield potential and stress adaptation in cereals. Crit. Rev. Plant Sci. 27, 377–412. 10.1080/07352680802467736 [DOI] [Google Scholar]
- Arunyanark A., Jogloy S., Akkasaeng C., Vorasoot N., Kesmala T., Rao R. C. N., et al. (2008). Chlorophyll stability is an indicator of drought tolerance in peanut. J. Agron. Crop Sci. 194, 113–125. 10.1111/j.1439-037X.2008.00299.x [DOI] [Google Scholar]
- Asada K. (1999). The water-water cycle in chloroplasts: scavenging of active oxygen and dissipation of excess photons. Annu. Rev. plant Physiol. plant Mol. Biol. 50, 601–639. 10.1146/annurev.arplant.50.1.601 [DOI] [PubMed] [Google Scholar]
- Asif M. A., Zafar Y., Iqbal J., Iqbal M. M., Rashid U., Ali G. M., et al. (2011). Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea L.) improves salt and drought tolerance. Mol. Biotechnol. 49, 250–256. 10.1007/s12033-011-9399-1 [DOI] [PubMed] [Google Scholar]
- Babitha K. C., Vemanna R. S., Nataraja K. N., Udayakumar M. (2015). Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS ONE 10:e0137098. 10.1371/journal.pone.0137098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Ramegowda V., Kumar A., Pereira A. (2016). Plant adaptation to drought stress. F1000 Res. 5, 1–10. 10.12688/f1000research.7678.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates L. S., Walden R. P., Teare I. D. (1973). Rapid determination of free proline for water stress studies. Plant Soil 39, 205–207. 10.1007/BF00018060 [DOI] [Google Scholar]
- Beena M. R., Jami S. K., Srinivasan T., Swathi T. A., Padmaja G., Kirti P. B. (2005). An efficient direct shoot regeneration from cotyledonary node explants of peanut (Arachis hypogaea L. Cv. JL-24). Indian J. Plant 10, 205–210. Available online at: http://inorderbox.com/siteimages/publication_file/4/ijpp-10-3-001.pdf [Google Scholar]
- Ben-Saad R., Meynard D., Ben-Romdhane W., Mieulet D., Verdeil J. L., Al-Doss A., et al. (2015). The promoter of the AlSAP gene from the halophyte grass Aeluropus littoralis directs a stress-inducible expression pattern in transgenic rice plants. Plant Cell Rep. 34, 1791–1806. 10.1007/s00299-015-1825-6 [DOI] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P., Devi M. J., Reddy D. S., Lavanya M., Vadez V., Serraj R. (2007). Stress-inducible expression of AtDREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 26, 2071–2082. 10.1007/s00299-007-0406-8 [DOI] [PubMed] [Google Scholar]
- Bhauso T. D., Radhakrishnan T., Kumar A., Mishra G. P., Dobaria J. R., Rajam M. V. (2014). Over-expression of bacterial mtlD gene confers enhanced tolerance to salt-stress and water-deficit stress in transgenic peanut (Arachis hypogaea) through accumulation of mannitol. Aust. J. Plant Physiol. 8, 413–421. 10.1155/2014/125967 [DOI] [Google Scholar]
- Blum A. (2005). Drought resistance, water-use efficiency, and yield potential-are they compatible, dissonant, or mutually exclusive? Aus. J. Agric. Res. 56, 1159–1168. 10.1071/AR05069 [DOI] [Google Scholar]
- Cai X. T., Xu P., Wang Y., Xiang C. B. (2015). Activated expression of AtEDT1/HDG11 promotes lateral root formation in Arabidopsis mutant edt1 by upregulating jasmonate biosynthesis. J. Integr. Plant Biol. 57, 1017–1030. 10.1111/jipb.12347 [DOI] [PubMed] [Google Scholar]
- Cao Y. J., Wei Q., Liao Y., Ling H., Li S. X., Xiang C. B., et al. (2009). Ectopic over expression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Rep. 28, 579–588. 10.1007/s00299-008-0659-x [DOI] [PubMed] [Google Scholar]
- Comas L., Becker S., Cruz V. M. V., Byrne P. F., Dierig D. A. (2013). Root traits contributing to plant productivity under drought. Front. Plant Sci. 4:442. 10.3389/fpls.2013.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condori J., Nopo-Olazabal C., Medrano G., Medina-Bolivar F. (2011). Selection of reference genes for qPCR in hairy root cultures of peanut. BMC Res. Notes 4:392. 10.1186/1756-0500-4-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuc L. M., Mace E. S., Crouch J. H., Quang V. D., Long T. D., Varshney R. K. (2008). Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea). BMC Plant Biol. 8:55. 10.1186/1471-2229-8-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho K., de Campos M. K. F., Domingues D. S., Pereira L. F. P., Vieira L. G. E. (2013). The accumulation of endogenous proline induces changes in gene expression of several antioxidant enzymes in leaves of transgenic Swingle citrumelo. Mol. Biol. Rep. 40, 3269–3279. 10.1007/s11033-012-2402-5 [DOI] [PubMed] [Google Scholar]
- De dorlodot S., Forster B., Pages L., Price A., Tuberosa R., Draye X. (2007). Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12, 474–481. 10.1016/j.tplants.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Devi M. J., Bhatnagar-Mathur P., Sharma K. K., Serraj R., Anwar S. Y., Vadez V. (2011). Relationships between transpiration efficiency and its surrogate traits in the rd29A:DREB1A transgenic lines of groundnut. J. Agron. Crop Sci. 197, 272–283. 10.1111/j.1439-037X.2011.00464.x [DOI] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Elavarthi S., Martin B. (2010). Spectrophotometric assays for antioxidant enzymes in plants, in Plant Stress Tolerance: Methods and Protocols, ed Sunkar R. (New Yor, NY: Springer; ), 273–280. [DOI] [PubMed] [Google Scholar]
- Forrest K. L., Bhave M. (2007). Major intrinsic proteins (MIPs) in plants: a complex gene family with major impacts on plant phenotype. Funct. Integr. Genomics 7, 263–289. 10.1007/s10142-007-0049-4 [DOI] [PubMed] [Google Scholar]
- Ghasem H. S., Matthew R., John B., Boyer J. (2009). Conceptual framework for drought phenotyping during molecular breeding. Trends Plant Sci. 9, 488–496. 10.1016/j.tplants.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Gilbert M. E., Zwieniecki M. A., Holbrook N. M. (2011). Independent variation in photosynthetic capacity and stomatal conductance leads to differences in intrinsic water use efficiency in 11 soybean genotypes before and during mild drought. J. Exp. Bot. 62, 2875–2887. 10.1093/jxb/erq461 [DOI] [PubMed] [Google Scholar]
- Gonzales L., Gonzales-Vilar M. (2001). Determination of relative water content, in Handbook of Plant Ecophysiology Techniques, ed Reigosa M. J. (Dordrecht: Kluwer Academic Publishers; ), 207–212. [Google Scholar]
- Hauser F., Waadt R., Schroeder J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. 10.1016/j.cub.2011.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri I., Muhovski Y., ŽiŽková E., Dobrev P. I., Gharbi E., Franco-Zorrilla J. M., et al. (2017). The Solanum lycopersicum WRKY3 transcription factor SlWRKY3 is involved in salt stress tolerance in tomato. Front. Plant Sci. 8:1343. 10.3389/fpls.2017.01343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Jiang P., Qi S., Zhang K., He Q., Xu C., et al. (2016). Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize type-II H+-pyrophosphatase gene by deletion analysis in transgenic tobacco plants. PLoS ONE 11:e0154041. 10.1371/journal.pone.0154041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICRISAT (2006). Groundnut. Available online at: http://www.icrisat.org/what-we-do/crops/GroundNut/GroundNut.htm
- Jagana S. R., Vadez V., Bhatnagar-Mathur P., Narasu M. L., Sharma K. K. (2012). Better root:shoot ratio conferred enhanced harvest index in transgenic groundnut over expressing the rd29A:DREB1A gene under intermittent drought stress in an outdoor lysimetric dry-down trial. J. SAT Agric. Res. 10, 1–7. Available online at: http://ejournal.icrisat.org/Volume10/Groundnut/KKS.pdf [Google Scholar]
- Janila P., Nigam S. N., Pandey M. K., Nagesh P., Varshney R. K. (2013). Groundnut improvement: use of genetic and genomic tools. Front. Plant Sci. 4:23. 10.3389/fpls.2013.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janila P., Rao T. N., Kumar A. A. (1999). Germination and early seedling growth of groundnut (Arachis hypogaea L.) varieties under salt stress. Ann. Agril. Res. 20, 180–182. [Google Scholar]
- Jeanneau M., Gerentes D., Foueillassar X., Zivy M., Vidal J., Toppan P. (2002). Improvement of drought tolerance in maize: towards the functional validation of the Zm-Asr1 gene and increase of water use efficiency by over-expressing C4-PEPC. Biochimie 84, 1127–1135. 10.1016/S0300-9084(02)00024-X [DOI] [PubMed] [Google Scholar]
- Karaba A., Dixit S., Greco R., Aharoni A., Trijatmiko K. R., Marsch-Martinez N., et al. (2007). Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc. Natl. Acad. Sci. U.S.A. 104, 15270–15275. 10.1073/pnas.0707294104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Miura S., Shinozaki K., Yamaguchi-Shinozaki K. A. (2004). Combination of the Arabidopsis DREB1A gene and stress-Inducible rd29A promoter improved drought and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. 10.1093/pcp/pch037 [DOI] [PubMed] [Google Scholar]
- Koyro H. W., Tabassum H., Bernhard H., Ajmal Khana M. (2012). Photosynthetic and growth responses of a perennial halophytic grass Panicum turgidum to increasing NaCl concentrations. Environ. Exp. Bot. 91, 22–29. 10.1016/j.envexpbot.2013.02.007 [DOI] [Google Scholar]
- Krishna G., Singh B. K., Kim E. K., Morya V. K., Ramteke P. R. (2015). Progress in genetic engineering of peanut (Arachis hypogaea L.). - a review. Plant Biotechnol. J. 13, 147–162. 10.1111/pbi.12339 [DOI] [PubMed] [Google Scholar]
- Laporte M. M., Shen B., Tarczynski M. C. (2002). Engineering for drought avoidance: expression of maize NADP-malic enzyme in tobacco results in altered stomatal function. J. Exp. Bot. 53, 699–705. 10.1093/jexbot/53.369.699 [DOI] [PubMed] [Google Scholar]
- Li L., Zheng M., Deng G., Liang J., Zhang H., Pan Z., et al. (2016). Overexpression of AtHDG11 enhanced drought tolerance in wheat (Triticum aestivum L.). Mol. Breed. 36:23 10.1007/s11032-016-0447-1 [DOI] [Google Scholar]
- Mace E. S., Phong D. T., Upadhaya H. D., Chandra S., Crouch J. H. (2006). SSR analysis of cultivated groundnut (Arachis hypogaea L.) germplasm resistant to rust and late leaf spot diseases. Euphytica 152, 317–30. 10.1007/s10681-006-9218-0 [DOI] [Google Scholar]
- Manjulatha M., Sreevathsa R., Sudhakar C., Prasad T. G., Tuteja N., Udayakumar M. (2014). Overexpression of a pea DNA helicase (PDH45) in peanut (Arachis hypogaea L.) confers improvement of cellular level tolerance and productivity under drought stress. Mol. Biotechnol. 56, 111–125. 10.1007/s12033-013-9687-z [DOI] [PubMed] [Google Scholar]
- Masle J., Gilmore S. R., Farquhar G. D. (2005). The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436, 866–870. 10.1038/nature03835 [DOI] [PubMed] [Google Scholar]
- Maurel C., Tacnet F., Güclü J., Guern J., Ripoche P. (1997). Purified vesicles of tobacco cell vacuolar and plasma membranes exhibit dramatically different water permeability and water channel activity. Proc. Natl. Acad. Sci. U.S.A. 94, 7103–7108. 10.1073/pnas.94.13.7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C., Kado R., Guern J., Chrispeels M. (1993). The vacuolar membrane protein gamma-TIP creates water specific channels in Xenopus oocytes. EMBO J. 12, 2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian A., Oomen R. J. F. J., Isayenkov S., Sentenac H., Maathuis F. J. M., Véry A.-A. (2011). Over-expression of an Na+ and K+−permeable HKT transporter in barley improves salt tolerance. Plant J. 68, 468–479. 10.1111/j.1365-313X.2011.04701.x [DOI] [PubMed] [Google Scholar]
- Mishra A., Tanna B. (2017). Halophytes: potential resources for salt stress tolerance genes and promoters. Front. Plant Sci. 8:829. 10.3389/fpls.2017.00829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinar R. (2012). Traditional Plant Breeding vs. Genetic Engineering-a Primer. Available online at: http://www.westernfarmpress.com/management/traditional-plant-breeding-vs-genetic-engineering-primer
- Nautiyal P. C., Nageswara Rao R. C., Joshi Y. C. (2002). Moisture-deficit-induced changes in leaf water content, leaf carbon exchange rate and biomass production in groundnut cultivars differing in specific leaf area. Field Crops Res. 74, 67–79. 10.1016/S0378-4290(01)00199-X [DOI] [Google Scholar]
- Nautiyal P. C., Ravindra V., Joshi Y. C. (1989). Germination and early seedling growth of some groundnut cultivars under salt stress. Indian J. Plant Physiol. 32, 251–253. [Google Scholar]
- Noctor G., Foyer C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. 10.1146/annurev.arplant.49.1.249 [DOI] [PubMed] [Google Scholar]
- Painawadee M. S., Jogloy T., Kesmala C., Akkasaeng Patanothai A. (2009). Identification of traits related to drought resistance in peanut (Arachis hypogaea L.). Asian J. Plant Sci. 8, 120–128. 10.3923/ajps.2009.120.128 [DOI] [Google Scholar]
- Pan L., Yang Q., Jiang Y., Yu S. (2009). Identification of AhAQ1 encoding a putative aquaporin whose expression is regulated by salt stress in peanut (Arachis hypogaea L.). Mol. Plant Breed. 7, 867–872. 10.5376/lgg.2010.01.0005 [DOI] [Google Scholar]
- Pandurangaiah M., Rao G. L., Sudhakarbabu O., Nareshkumar A., Kiranmai K., Lokesh U., et al. (2014). Overexpression of horsegram (Macrotyloma uniflorum Lam.Verdc.) NAC transcriptional factor (MuNAC4) in groundnut confers enhanced drought tolerance. Mol. Biotechnol. 56, 758–769. 10.1007/s12033-014-9754-0 [DOI] [PubMed] [Google Scholar]
- Parvaiz A., Satyawati S. (2008). Salt stress and phyto-biochemical responses of plants – a review. Soil Environ. 54, 89–99. 10.17221/2774-PSE [DOI] [Google Scholar]
- Pruthvi V., Narasimhan R., Nataraja K. N. (2014). Simultaneous expression of abiotic stress responsive transcription factors, AtDREB2A, AtHB7 and AtABF3 improves salinity and drought tolerance in peanut (Arachis hypogaea L.). PLoS ONE 9:e111152. 10.1371/journal.pone.0111152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Gu Q., Kuppu S., Sun L., Zhu X., Mishra N., et al. (2013). Expression of the Arabidopsis vacuolar H+ -pyrophosphatase gene AVP1 in peanut to improve drought and salt tolerance. Plant Biotechnol. Rep. 7, 345–355. 10.1007/s11816-012-0269-5 [DOI] [PubMed] [Google Scholar]
- Qin H., Gu Q., Zhang J., Sun L., Kuppu S., Zhang Y., et al. (2011). Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol. 52, 1904–1914. 10.1093/pcp/pcr125 [DOI] [PubMed] [Google Scholar]
- Quan R., Shang M., Zhang H., Zhao Y., Zhang J. (2004). Engineering of enhanced glycine betaine synthesis improves drought tolerance in maize. Plant Biotechnol. J. 2, 477–486. 10.1111/j.1467-7652.2004.00093.x [DOI] [PubMed] [Google Scholar]
- R Core Team (2012). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: Available online at: http://www.R-project.org/ [Google Scholar]
- Rai M., He C., Wu R. (2009). Comparative functional analysis of three abiotic stress-inducible promoters in transgenic rice. Transgenic Res. 18, 787–799. 10.1007/s11248-009-9263-2 [DOI] [PubMed] [Google Scholar]
- Ramu V. S., Swetha T. N., Sheela S. H., Babitha C. K., Rohini S., Reddy M. K., et al. (2016). Simultaneous expression of regulatory genes associated with specific drought-adaptive traits improves drought adaptation in peanut. Plant Biotechnol. J. 14, 1008–1020. 10.1111/pbi.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. C. N., Sheshshayee M. S., Karaba N. N., Sreevathsa R., Rama N., Kumaraswamy S., et al. (2012). Groundnut genetic approaches to enhance adaptation of groundnut (Arachis hypogaea, L.) to drought, in Improving Crop Productivity in Sustainable Agriculture (Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; ), 303–359. [Google Scholar]
- Rao R. C. N., Talwar H. S., Wright G. C. (2001). Rapid assessment of specific leaf area and leaf N in peanut (Arachis hypogaea L.) using chlorophyll meter. J. Agron. Crop Sci. 189, 175–182. [Google Scholar]
- Reddy T. Y., Anbumozhi V. R. V. (2003). Physiological responses of groundnut (Arachis hypogea L.) to drought stress and its amelioration. A critical review. Plant Growth Regul. 41, 75–88. 10.1023/A:1027353430164 [DOI] [Google Scholar]
- Regon P., Panda P., Kshetrimayum E., Panda S. K. (2014). Genome-wide comparative analysis of tonoplast intrinsic protein (TIP) genes in plants. Funct. Integr. Genomics 14:617. 10.1007/s10142-014-0389-9 [DOI] [PubMed] [Google Scholar]
- Richards R. A. (1996). Defining selection criteria to improve yield under drought. Plant Growth Regul. 20, 157–166. 10.1007/BF00024012 [DOI] [Google Scholar]
- Richardson A. D., Duigan S. P., Berlyn G. P. (2002). An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol. 153, 185–194. 10.1046/j.0028-646X.2001.00289.x [DOI] [Google Scholar]
- Ruan L., Chen L. J., Chen Y. H., He J. L., Zhang W., Gao Z. L., et al. (2012). Expression of Arabidopsis HOMEODOMAIN GLABROUS 11 enhances tolerance to drought stress in transgenic sweet potato plants. J. Plant Biol. 55, 151–158. 10.1007/s12374-011-9198-z [DOI] [Google Scholar]
- Sambrook J., Fritschi E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sarkar T., Thankappan R., Kumar A., Mishra G. P., Dobaria J. R. (2016). Stress inducible expression of AtDREB1A transcription factor in transgenic peanut (Arachis hypogaea L.) conferred tolerance to soil-moisture deficit stress. Front. Plant Sci. 7:935. 10.3389/fpls.2016.00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T., Thankappan R., Kumar A., Mishra G. P., Dobaria J. R. (2014). Heterologous expression of the AtDREB1A gene in transgenic peanut-conferred tolerance to drought and salinity stresses. PLoS ONE 9:e110507. 10.1371/journal.pone.0110507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebahattin A., Necdet C. (2007). Effects of temperature and light intensity on growth of fodder beet (Beta vulgaris L. var. crassa mansf.). Bangladesh J. Bot. 36, 1–12. 10.3329/bjb.v36i1.1542 [DOI] [Google Scholar]
- Sharma K. K., Lavanya M. (2002). Recent developments in transgenics for abiotic stress in legumes of the semi-arid tropics. JIRCAS Working Rep. 23, 61–73. Available online at: http://oar.icrisat.org/id/eprint/3611 [Google Scholar]
- Sheshshayee M. S., Bindumadhava H., Rachaputi N. R., Prasad T. G., Udayakumar M., Wright G. C., et al. (2006). Leaf chlorophyll concentration relates to transpiration efficiency in peanut. Ann. Appl. Biol. 148, 7–15. 10.1111/j.1744-7348.2005.00033.x [DOI] [Google Scholar]
- Shinozaki Y., Shinozaki K. (1993). Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 236, 331–340. [DOI] [PubMed] [Google Scholar]
- Shou H., Bordallo P., Fan J. B., Yeakley J. M., Bibikova M., Sheen J., et al. (2004). Expression of an active tobacco mitogen-activated protein kinase enhances freezing tolerance in transgenic maize. Proc. Natl. Acad. Sci. U.S.A. 101, 3298–3303. 10.1073/pnas.0308095100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B. G., Rao P. V., Hiremath S. M. (1989). Effect of salinity on germination and seedling growth in three varieties of groundnut (Arachis hypogaea L.). J. Res. APAU 17, 432–433. [Google Scholar]
- Singh S. B., Abrol I. P. (1985). Effect of soil sodicity on growth, yield and chemical composition of groundnut. Plant Soil 84, 123–127. 10.1007/BF02197873 [DOI] [Google Scholar]
- Sivamani E., Bahieldin A., Wraith J. M., Al-Niemi T., Dyer W. E., Ho T. H. D. (2000). Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci. 155, 1–9. 10.1016/S0168-9452(99)00247-2 [DOI] [PubMed] [Google Scholar]
- Smith S., De Smet I. (2012). Root system architecture: insights from Arabidopsis and cereal crops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1441–1452. 10.1098/rstb.2011.0234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songsri P., Jogloy S., Holbrook C. C., Kesmala T., Vorasoot N., Akkasaeng C., et al. (2009). Association of root, specific leaf area and SPAD chlorophyll meter reading to water use efficiency of peanut under different available soil water. Agric. Water Manage. 96, 790–798. 10.1016/j.agwat.2008.10.009 [DOI] [Google Scholar]
- Sun Q. H., Gao F., Zhao L., Li K. P., Zhang J. R. (2010). Identification of a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila. BMC Plant Biol. 10:90. 10.1186/1471-2229-10-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L., Savouré A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. 10.1016/j.tplants.2009.11.009 [DOI] [PubMed] [Google Scholar]
- Tiwari V., Patel M. K., Chaturvedi A. K., Mishra A., Jha B. (2016). Functional characterization of the tau class Glutathione-S-Transferases gene (SbGSTU) promoter of Salicornia brachiata under salinity and osmotic Stress. PLoS ONE 11:e0148494. 10.1371/journal.pone.0148494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman S., Niemietz C., Bramley H. (2002). Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant Cell Environ. 25, 173–194. 10.1046/j.0016-8025.2001.00791.x [DOI] [PubMed] [Google Scholar]
- Vadez V., Krishnamurthy L., Kashiwagi J. W., Kholova J., Devi J. M., Sharma K. K. (2007). Exploiting the functionality of root systems for dry, saline, and nutrient deficient environments in a changing climate. J. Semi Arid Trop. Agric. Res. 4, 1–61. Available online at: http://oar.icrisat.org/id/eprint/2510 [Google Scholar]
- Verbruggen N., Hermans C. (2008). Proline accumulation in plants: a review. Amino Acids 35, 753–759. 10.1007/s00726-008-0061-6 [DOI] [PubMed] [Google Scholar]
- Wright G. C., Nageswara Rao R. C. (1994). Groundnut water relations, in The Groundnut Crop. A Scientific Basis for Improvement, ed Smartt J. (London: Chapman and Hall; ), 281–325. [Google Scholar]
- Xu P., Cai X. T., Wang Y., Xing L., Chen Q., Xiang C. B. (2014). HDG11 up regulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J. Exp. Bot. 65, 4285–4295. 10.1093/jxb/eru202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo A. (1998). Molecular biology of salt tolerance in the context of whole-plant physiology. J. Exp. Bot. 49, 913–929. 10.1093/jxb/49.323.915 [DOI] [Google Scholar]
- Yi N., Kim Y. S., Jeong M. H., Oh S. J., Jeong J. S., Park S. H., et al. (2010). Functional analysis of six drought-inducible promoters in transgenic rice plants throughout all stages of plant growth. Planta 232, 743–754. 10.1007/s00425-010-1212-z [DOI] [PubMed] [Google Scholar]
- Yu L. H., Chen X., Wang Z., Wang S. M., Wang Q. S., Zhu S. G., et al. (2013). Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 162, 1378–1391. 10.1104/pp.113.217596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. H., Wu S. J., Peng Y. S., Liu R. N., Chen X., Zhao P., et al. (2015). Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14, 72–84. 10.1111/pbi.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Chen X., Hong Y. Y., Wang Y., Xu P., Ke S. D., Xiang C. B., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151. 10.1105/tpc.108.058263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. M., Zhang H. M., Liu Z. H., Li H. C., Cuo X. L., Li G. L. (2015). The wheat NHX antiporter gene TaNHX2 confers salt tolerance in transgenicalfalfa by increasing the retention capacity of intracellular potassium. Plant Mol. Biol. 87, 317–327. 10.1007/s11103-014-0278-6 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yin H., Li D., Zhu W., Li Q. (2008). Functional analysis of BADH gene promoter from Suaeda liaotungensis K. Plant Cell Rep. 27, 585–592. 10.1007/s00299-007-0459-8 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Sun B., Xu X., Chen H., Zou L., Chen G., et al. (2016). Overexpression of AtEDT1/HDG11 in chinese kale (Brassica Oleracea Var. Alboglabra) enhances drought and osmotic stress tolerance. Front. Plant Sci. 7:1285. 10.3389/fpls.2016.01285 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.