Abstract
Introduction
Gastric cancer (GC) is a malignant tumor worldwide. As primary pathway for metastasis, the lymphatic system is an important prognostic factor for GC patients. Although the metabolic changes of gastric cancer have been investigated in extensive studies, little effort focused on the metabolic profiling of lymph node metastasis (LNM)-positive or negative GC patients.
Objectives
We performed 1H NMR spectrum of GC tissue samples with and without LNM to identify novel potential metabolic biomarkers in the process of LNM of GC.
Methods
1H NMR-based untargeted metabolomics approach combined with multivariate statistical analyses were used to study the metabolic profiling of tissue samples from LNM-positive GC patients (n = 40), LNM-negative GC patients (n = 40) and normal controls (n = 40).
Results
There was a clear separation between GC patients and normal controls, and 33 differential metabolites were identified in the study. Moreover, GC patients were also well-classified according to LNM-positive or negative. Totally eight distinguishing metabolites were selected in the metabolic profiling of GC patients with LNM-positive or negative, suggesting the metabolic dysfunction in the process of LNM. According to further validation and analysis, especially BCAAs metabolism (leucine, isoleucine, valine), GSH and betaine may be as potential factors of diagnose and prognosis of GC patients with or without LNM.
Conclusion
To our knowledge, this is the first metabolomics study focusing on LNM of GC. The identified distinguishing metabolites showed a promising application on clinical diagnose and therapy prediction, and understanding the mechanism underlying the carcinogenesis, invasion and metastasis of GC.
Electronic supplementary material
The online version of this article (10.1007/s11306-018-1344-x) contains supplementary material, which is available to authorized users.
Keywords: Lymph node metastasis, Gastric cancer, Metabolic profiling, Tissue
Introduction
Gastric cancer (GC) ranks as the fifth most common malignant cancer and is the third mortality of cancer around the world. About 951,600 new GC cases and 723,100 associated deaths were reported worldwide in 2012 (Torre et al. 2015; Song et al. 2017). GC is particularly prevalent in East Asia, especially in China and Japan (Torre et al. 2015; Leung et al. 2008). Hence, the economic burden on GC is greater than other countries (Kanavos 2006). GC is a multistep and multifactorial progression, the 5-year survival rate of early stage of GC (EGC), which just invaded the mucosal or sub-mucosal layer, reaches over 90% after surgery (Jung et al. 2014; Wang et al. 2016). However, EGC is difficult to be found because of its asymptomatic. So many patients are diagnosed at an advanced stage with lymph node metastasis (LNM) or remote metastasis. As primary pathway for metastasis, the lymphatic system is an important prognostic factor for GC patients. LNM is a single indicator in GC patients with Borrmann type I (Chen et al. 2007). The 5-year survival rate of GC patients with LNM drops to less than 30% (Chen et al. 2002). In addition, the prognosis of node-negative GC patients is markedly better than that of LNM-positive ones (Hyung et al. 2002). The recurrence rises to 10.6–14.8% in LNM-positive EGC from 2.7 to 4.4% node-negative EGC, even following curative surgical operation with D1 + b or D2 lymph node resection (Lai et al. 2009; An et al. 2010). Therefore, LNM may be a critical factor for assessment of prognosis and therapy of GC (Deng and Liang 2014). The overall recurrence rate in LNM-positive GC patients is obviously higher than that in LNM-negative patients, and the overall survival of LNM-positive GC patients is significantly shorter than that of LNM-negative patients (Nakamoto et al. 2007; Sarela et al. 2003). Although the incidence and mortality rates of GC have declined in many countries with the economic improvements, the significant challenges of understanding and treating of GC continue to exist, especially in the process of LNM.
Cancer has been shown to be a metabolic disease, metabolism disorder is a significant characteristic (Warburg 1956). Metabolomics, a rapidly expanding field of systems biology, aims to detect, identify and quantify as many low molecular weight metabolites as possible in a cellular or biological system at a given time. Metabolites can reflect the minor alteration at the level of genome, transcriptome and proteome. Therefore, metabolomics is increasingly used for identifying biomarkers for the early diagnosis and understanding the underlying mechanism of various cancers (Wang et al. 2013; Li et al. 2014; Zhang et al. 2016). Nuclear magnetic resonance (NMR) and mass spectrometry (MS) are the most common tools to characterize the metabolites in body fluids, tissues and cells using targeted or untargeted studies (Griffin and Shockcor 2004). Typically, untargeted 1H NMR metabolic profiling, with the advantages of the relative ease of sample preparation and non-destructive analysis has been a popular technique for metabolomics study in various diseases, including arthritis (Weljie et al. 2007), lung cancer (Rocha et al. 2011), melanoma (Wang et al. 2014). Today, most studies focus on identifying biomarkers or exploring the underlying mechanism of GC (Kuligowski et al. 2016; Ramachandran et al. 2016; Gu et al. 2016). However, little attention has been paid to metabolic profiling of LNM, which is particularly important predictor of survival and recurrence in GC (Kunisaki et al. 2009; Lee et al. 2012).
In this study, we demonstrated the metabolic profiling between GC tissues and normal controls, especially the metabolic changes between LNM-positive and LNM-negative GC patients, using an untargeted metabolomics approach based on 1H NMR, coupled with multivariate statistical analyses. We totally identified 33 distinguishing metabolites between GC tissues and normal controls, especially 8 of which performed noticeably well in discriminating LNM-positive from LNM-negative GC patients, including branched-chain amino acids (BCAAs: leucine, isoleucine, valine), glutathione, glycine, betaine, tyrosine and hypoxanthine. To our knowledge, this is the first metabolomics report to identify altered metabolites in LNM-positive GC patients, with the hope of identifying potential biomarkers for early diagnosis, staging and prognostic prediction. We also intended to explore the underlying mechanism of LNM in GC.
Materials and methods
Chemical reagents
Distilled water was prepared by Milli-Q purification system. Deuterium water (D2O, 99.8%) was bought from CIL (Cambridge Isotope Laboratories, USA). High-performance liquid chromatography (HPLC) grade methanol and HPLC-grade chloroform were bought from Merck (Germany). Trimethylsilylpropionic acid-d4 sodium salt (TSP) was bought from Sigma-Aldrich (USA). All other reagents were of analytic grade in our study.
Patients and samples collection
The population recruited in this study totally consisted of 80 patients with primary GC, who underwent surgical resection at the West China Hospital of Sichuan University between 2013 and 2014. The study protocol was approved by the Ethics Committee of the West China Hospital and the written informed consents were obtained before samples collection. Part of the dissected specimens was used for pathological diagnosis, and the other part was immediately frozen in liquid nitrogen and stored at − 80 °C. To ensure the accuracy, the patients, who received any radiation, chemotherapy or had dissected lymph nodes less than 15, were excluded from our study. The pathological diagnosis was confirmed by standard hematoxylin and eosin stain, and each dissected lymph node was stained with H and E. All patients were diagnosed for TNM stages according to the 7th edition of the American Joint Commission on Cancer (AJCC).
Preparation of tissue extraction and 1H NMR spectroscopic analysis
The reagents were ice-cold before use, including distilled water, D2O, methanol and chloroform. To extract the interesting metabolites, the 100–400 mg frozen specimen was minced in liquid nitrogen, weighted and transferred into a 15 mL centrifuge tube. Four milliliter methanol per gram of specimen and 0.85 mL distilled water per gram of specimen were added into the same centrifuge tube and the mixture was vortexed for 1 min. Then 2 mL chloroform per gram of specimen was added and the sample was vortexed again, followed keeping on ice for 30 min. After centrifuging at 1000×g at 4 °C for 30 min, the mixture was separated into three phases, including the bottom lipid phase, the middle protein phase and the top aqueous phase, where the metabolic extraction was contained. Each upper water phase was gently absorbed and transferred into a fresh 1.5 mL centrifuge tube and evaporated under a stream of nitrogen. The dried residue was dissolved in 580 µL of D2O, which contained 30 µM phosphate buffer solution (PBS, pH 7.4), providing the deuterium lock signal for the NMR spectrometer, and 0.5 mM sodium (3-trimethylsilyl)-2,2,3, 3-tetradeuteriopropionate (TSP), providing the chemical shift reference (δ0.0) and internal concentration standard. The suspension was vortexed and centrifuged at 12000×g at 4 °C for 5 min, and about 550 µL solution was loaded into a 5 mm NMR tube for NMR spectroscopy (Beckonert et al. 2007).
1H NMR spectroscopy
The 1H NMR spectra of each samples were recorded on a Bruker Avance II 600 spectrometer (Bruker Biospin, Germany) at 600.13 MHz and 20 °C. One-dimensional NMR spectrum from each sample was acquired using a standard (1D) Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence to observe metabolite signals and 5 s relaxation delay at the water peak position to suppress the residual H2O signal. Sixty four free induction decays (FIDs) were collected into 64 K data points with a spectral width of 12335.5 Hz spectral and an acquisition time of 2.66 s. Therefore, the total pulse recycles delay of 7.66 s.
NMR data processing
The raw NMR data (FIDs) were manually phased adjusted and baseline corrected using MestReNova-6.1.1–6384 software after referencing to TSP chemical shift at 0 ppm. To reduce the complexity of spectral data before statistical analysis, spectral binning was used for producing a data set with manageable proportion. In our study, the spectral regions at 0.5– 9.5 ppm were segmented into 1800 bins with equal width of 0.005 ppm. The spectral regions 3.37–3.34 ppm (methanol signal), 4.94–4.66 ppm (residual water signal) and 7.84–7.62 ppm (chloroform signal) were respectively excluded. In order to get the real metabolites changes in samples, especially for the low concentration metabolites, the integral values of all bins were divided by the value of reference TSP and then normalized to the weight of the measured GC tissue used for metabolites extraction. Moreover, the normalized data was given the same total integration value for each spectra before multivariate data analysis.
Multivariate statistical analysis
The normalized NMR data in the form of excel was imported into statistical software SIMCA-P (Version 11, Umetrics, AB) for multivariate statistical analysis, including principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA), and orthogonal projection to latent structure (OPLS). PCA was firstly performed to see an overview and possible outliers. Then PLS-DA was cross-validated by a permutation analysis (200 times), and the resulting R2 and Q2 values were calculated. OPLS was performed to get well separation between GC cases and controls. The parameters R2 and Q2, indicating the interpretability and predictability of the model respectively, were used to evaluate the quality of the model. The coefficient plot was color-coded with the absolute value of coefficients (r) with Matlab scripts, meaning the red colored metabolites being more significant than the blue colored ones (Feng et al. 2011). The ROC analysis was performed using predicted Y values of PLS-DA or the relative expression of the specific metabolites in SPSS 22.0.
To identify the interesting spectrum peaks, the variable importance in the projection (VIP) > 1 was considered responsible for group discrimination, which was analyzed and taken as a coefficient from OPLS models. Moreover, unpaired Student’s t-test (p < 0.05) to the chemical shifts was also used to evaluate the reliability of each metabolite. The identified metabolites were chosen as distinguishing ones, which both met VIP > 1 and p < 0.05. The corresponding chemical shift and multiplicity of the metabolites were identified according to the previous literatures and the Human Metabolome Database (http://www.hmdb.ca/).
Results
Study population
Based on NMR, a total of 120 tissue specimens were detected in our study, among which, 80 specimens were the matched tumor and control collected from the same patient (n = 40). The clinical characteristics were shown in Table 1. The GC patients were divided into two subgroups, including 40 LNM-negative cases (24 male, 16 female; age range, 39–78 years, mean age, 59 years) and 40 LNM-positive cases (22 male, 18 female; age range, 28–82 years, mean age, 60 years). The remaining ones were normal controls (28 male, 12 female; age range, 28–78 years, mean age, 60 years). The recruited patients were all adenocarcinoma according to histology examination. The TNM stages were determined according to the 7th edition of the American Joint Commission on Cancer (AJCC). The detailed information was presented in Table 1.
Table 1.
Clinical characteristics of gastric cancer patients and normal controls analyzed by 1H-NMR
| LNM-negative GC patients | LNM-positive GC patients | Normal controls | |
|---|---|---|---|
| No. of subjects | 40 | 40 | 40 |
| Age (mean (range)) | 59 (39–78) | 58 (28–82) | 60 (28–78) |
| Gender (male/female) | 24/16 | 22/18 | 28/12 |
| Lauren type | |||
| Intestinal | 18 | 15 | 41 |
| Diffuse | 9 | 10 | |
| NA | 13 | 15 | |
| Histology | Adenocarcinoma (40) | Adenocarcinoma (40) | |
| Pathologic grade | |||
| PD | 21 | 12 | |
| MD | 16 | 26 | |
| NA | 3 | 2 | |
| TNM stage | |||
| I | IA:3; IB:17 | IB:1 | |
| II | IIA:10; IIB:7 | IIA:1; IIB:9 | |
| III | IIIB:3 | IIIA:15; IIIB:9 | |
| IV | IV:5 |
PD poorly differentiated, MD moderately differentiated, NA not applicable
Metabolic profiling of 1H NMR spectrum of GC tissues
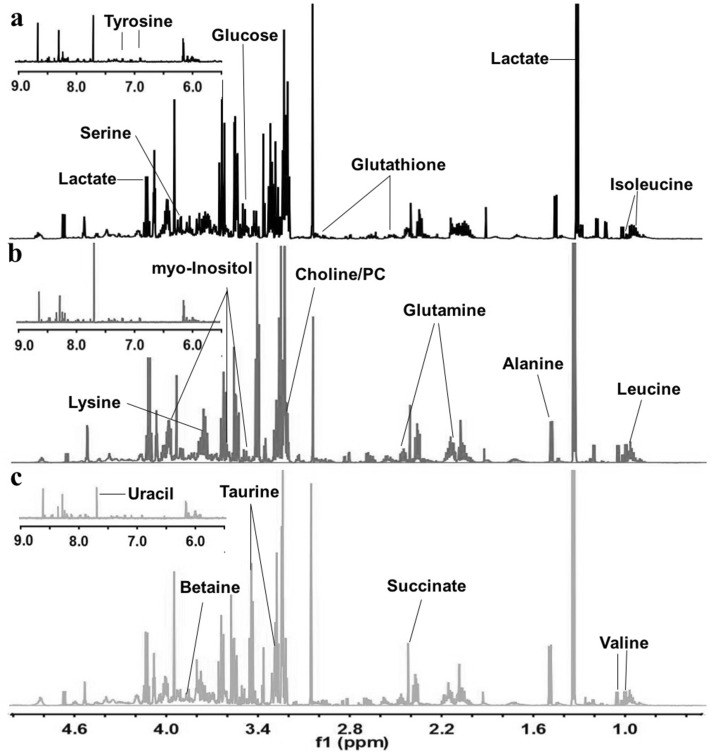
Typical NMR spectra of normal control, LNM-negative GC and LNM-positive GC were depicted in Fig. 1a-c, respectively. Resonance assignments were performed by chemical shift and multiplicity of the metabolites according to the previous literatures and the Human Metabolome Database. An overview of visible differences among the three classes was shown in Fig. 1. Most visible spectra were concentrated on the region between 0.5 and 5 ppm, including BCAA, alanine, lactate, glucose, choline/PC, taurine, betaine, and so on. The peaks from 5.5 to 9 ppm were few, including uracil, fumarate, tyrosine and several unknown signals.
Fig. 1.
Representative tissue 600 MHz 1H NMR spectra. a Normal control specimen, b LNM-negative GC specimen, c LNM-positive GC specimen
Multivariate statistical analysis of LNM-negative GC specimens, LNM-positive GC specimens and normal controls
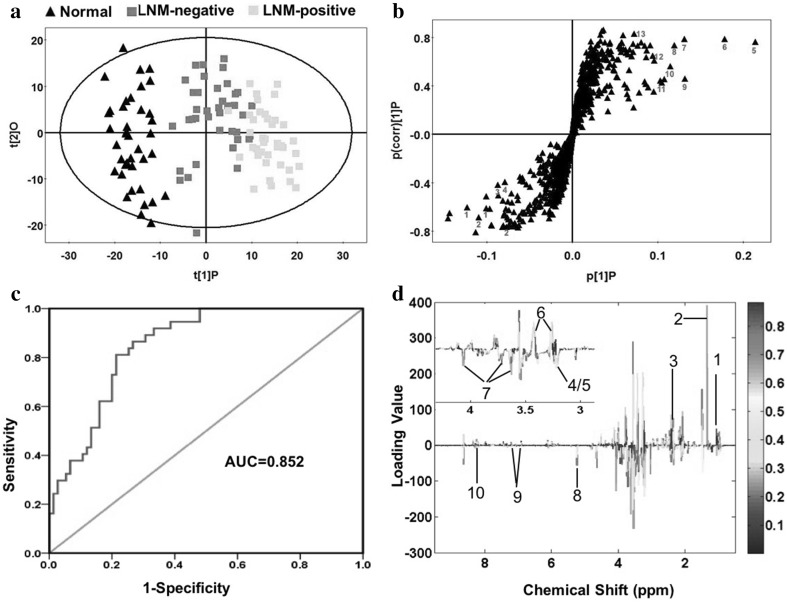
Firstly, PCA was performed using the raw NMR data. The PCA score plot (2PCs, R2X = 0.355, Q2 = 0.146) showed clear classification between GC tissues and normal controls (Fig S1a) without any outlier (Fig S1b). Therefore, all of specimens were kept in the further analysis to obtain maximum information. Then OPLS (R2X = 0.154, R2Y = 0.894, Q2 = 0.823) was performed to evaluate variable importance responsible for separation among LNM-negative GC tissues, LNM-positive GC tissues and normal controls. A well separation between GC and controls was shown in Fig. 2a. In the S-plot (Fig. 2b), the variables far away from the center of the plot were assumed to have a greater contribution to the model separation. The model parameters (R2 = 0.79, Q2 = 0.77) showed good quality of the obtained OPLS model (Fig S1c). A receiver operating characteristic curve (ROC) was carried out using the predicted Y values from OPLS. Area under the curve (AUC) value was 0.852 (Fig. 2c), indicating the OPLS model had good predictive ability. This diagnostic model was just to identify the tissue metabolic biomarkers rather than to replace the established histopathologic diagnostic standard for GC.
Fig. 2.
Metabolic profiling between GC tissues and normal controls. a OPLS scores plot between the GC tissues and normal controls. Black triangles represent normal controls (n = 40); Green boxes represent LNM-negative GC patients (n = 40); Yellow boxes represent LNM-positive GC patients (n = 40). b S-plot of the OPLS model, the variables that lie far away from the center of the plot were assumed to have a greater contribution to the model classification. c ROC analysis was performed using the Y-predicted value determined by the PLS-DA model between the GC tissues and normal controls. d The color map shows the significance of metabolite variations between the two classes. Peaks in the positive direction indicate the increased metabolites in GC tissues in comparison to normal controls. Decreased metabolites in GC tissues are presented as peaks in the negative direction. 1 BCAA, 2 Lactate, 3 Methylamine, 4/5 Choline/PC, 6 Taurine, 7 myo-Inositol, 8 Glucose, 9 Tyrosine, 10 Hypoxanthine
We firstly identified 56 metabolites according corresponding chemical shift and multiplicity. Then the inappropriate ones (VIP < 1, or p > 0.05, or both) were excluded. The remaining 30 and 31 significant class-discriminating metabolites (VIP > 1 and p < 0.05) were respectively chosen for LNM-negative and LNM-positive GC cases, which were summarized in Table 2. Among them, 28 distinguishing metabolites were identified both in the two cases. Color-coded coefficient plots showed the change trend of identified differential metabolites responsible for discriminating GC from controls (Fig. 2d). Higher concentration of BACCs (Isoleucine, Leucine, Valine), Lactate, Methylamine and Taurine in GC specimens compared to the controls were illustrated in the positive quadrant signals. On the other hand, lower concentration of Choline/PC, Taurine, myo-Inositol, Glucose, Tyrosine and Hypoxanthine in GC specimens were represented in the negative quadrant signals.
Table 2.
Differential tissue metabolites among LNM-negative GC patients, LNM-positive GC patients, and normal controls
| Metabolites | Chemical shift (ppm, multiplicity)a | LNM-negative versus normal controls | LNM-positive versus normal controls | ||||
|---|---|---|---|---|---|---|---|
| VIPb | Pc | FCd | VIPb | Pc | FCd | ||
| Isoleucine | 0.945(t) | 2.050 | 0.002 | 1.058 | 1.666 | 0.418 | 1.329 |
| 1.015(d) | 2.318 | 2.97E-12 | 1.710 | 2.539 | 1.92E-20 | 2.144 | |
| Leucine | 0.965(t) | 1.725 | 0.395 | 1.127 | 2.250 | 0.044 | 1.290 |
| Valine | 0.995(d) | 1.814 | 0.327 | 1.052 | 2.238 | 3.06E-04 | 1.203 |
| 1.045(d) | 2.454 | 0.635 | 1.025 | 2.617 | 0.002 | 1.167 | |
| Lactate | 1.33(d) | 2.071 | 4.80E-04 | 1.193 | 2.429 | 2.52E-05 | 1.219 |
| 4.11(q) | 1.675 | 0.005 | 1.140 | 1.953 | 1.99E-04 | 1.165 | |
| Threonine | 1.33(d) | 2.071 | 4.80E-04 | 1.193 | 2.429 | 2.52E-05 | 1.219 |
| 4.24(m) | 2.221 | 3.52E-04 | 1.120 | 1.803 | 9.71E-05 | 1.117 | |
| Alanine | 1.48(d) | 2.563 | 6.27E-06 | 1.297 | 2.521 | 1.23E-07 | 1.423 |
| Citrulline | 1.57(m) | 1.134 | 0.033 | 0.626 | 1.565 | 0.002 | 0.479 |
| VLDL: –CH2–CH2–CH2O | 1.58(br) | 1.681 | 0.006 | 0.596 | 1.786 | 0.005 | 0.617 |
| N-acetyl glycoprotein | 2.05(s) | 1.581 | 0.011 | 1.245 | 1.841 | 0.063 | 1.110 |
| O-acetyl glycoprotein | 2.065(s) | 2.520 | 2.41E-05 | 1.211 | 2.340 | 0.002 | 1.136 |
| Acetic acid | 2.075(s) | 2.409 | 1.23E-23 | 2.012 | 2.441 | 3.21E-26 | 1.993 |
| Glutamine | 2.14(m) | 2.204 | 1.16E-09 | 1.318 | 2.479 | 4.97E-12 | 1.356 |
| 2.455(m) | 2.117 | 0.057 | 1.110 | 2.319 | 0.031 | 1.129 | |
| 3.77(m) | 2.228 | 3.79E-08 | 1.216 | 2.457 | 1.03E-10 | 1.260 | |
| d-ribose | 2.235(s) | 1.168 | 0.004 | 0.673 | 1.364 | 4.33E-04 | 0.611 |
| Acetone | 2.235(s) | 1.168 | 0.004 | 0.673 | 1.364 | 4.33E-04 | 0.611 |
| Lipid, –CH2–C=O | 2.26(br) | 1.736 | 0.001 | 0.814 | 1.674 | 2.33E-04 | 0.809 |
| Pyruvate | 2.375(s) | 2.537 | 1.13E-13 | 1.854 | 2.399 | 6.86E-18 | 2.002 |
| Succinate | 2.405(s) | 2.334 | 0.009 | 1.172 | 2.305 | 0.002 | 1.369 |
| Glutathione | 2.555(m) | 2.592 | 1.76E-12 | 1.537 | 2.464 | 1.31E-12 | 1.618 |
| 2.97(m) | 2.429 | 1.28E-07 | 1.461 | 2.320 | 6.74E-11 | 1.659 | |
| Methylamine | 2.595(s) | 2.329 | 1.58E-12 | 3.554 | 2.315 | 2.51E-15 | 4.335 |
| Choline | 3.2(s) | 1.097 | 0.003 | 0.617 | 0.870 | 0.002 | 0.600 |
| PC (phosphochline) | 3.21(s) | 1.451 | 2.06E-05 | 0.646 | 1.455 | 9.32E-06 | 0.647 |
| Trimethylamine-N-oxide | 3.27(s) | 1.794 | 9.61E-12 | 1.899 | 1.911 | 5.37E-20 | 2.150 |
| Taurine | 3.27(t) | 1.794 | 2.87E-11 | 1.628 | 1.911 | 1.02E-16 | 1.755 |
| 3.425(t) | 1.730 | 2.31E-11 | 2.562 | 1.577 | 4.20E-22 | 2.801 | |
| myo-Inositol | 3.535(dd) | 2.228 | 4.75E-11 | 0.588 | 2.223 | 9.90E-28 | 0.506 |
| 3.63(t) | 1.806 | 7.32E-10 | 0.716 | 1.980 | 7.55E-15 | 0.667 | |
| 4.065(m) | 2.035 | 6.46E-07 | 0.666 | 2.144 | 1.23E-09 | 0.629 | |
| Glucose | 3.535(dd) | 2.228 | 1.83E-07 | 0.588 | 2.223 | 3.91E-10 | 0.506 |
| 5.235(d) | 2.624 | 7.32E-10 | 0.379 | 2.505 | 7.55E-15 | 0.274 | |
| Glycine | 3.565(s) | 2.596 | 8.76E-14 | 2.986 | 2.619 | 1.79E-21 | 3.644 |
| Lysine | 3.77(m) | 2.228 | 1.42E-13 | 1.216 | 2.457 | 9.45E-27 | 1.260 |
| Betaine | 3.89(s) | 2.451 | 3.79E-08 | 0.651 | 2.504 | 1.03E-10 | 0.560 |
| Serine | 3.975(m) | 2.519 | 0.001 | 1.157 | 2.283 | 0.003 | 1.120 |
| Uracil | 7.54(d) | 2.090 | 5.03E-06 | 2.261 | 2.114 | 1.14E-06 | 2.380 |
| 5.8(d) | 2.282 | 2.82E-08 | 4.657 | 2.316 | 3.08E-09 | 5.179 | |
| Fumarate | 6.52(s) | 1.192 | 0.007 | 1.002 | 1.913 | 0.004 | 1.287 |
| Tyrosine | 6.9(d) | 2.607 | 0.001 | 1.324 | 2.716 | 1.21E-06 | 1.492 |
| 7.2(d) | 2.231 | 0.002 | 1.265 | 2.384 | 8.06E-06 | 1.411 | |
| Hypoxanthine | 8.18(s) | 0.539 | 0.746 | 0.964 | 1.040 | 0.001 | 0.632 |
| 8.215(s) | 0.939 | 0.027 | 0.776 | 1.682 | 0.000 | 0.543 | |
aMultiplicity: s singlet, d doublet, t triplet, q quartet, dd doublet of doublets, m multiplet
bVariable importance in the projection was obtained from OPLS model with a threshold of 1.0
cp-value obtained from Student’s t-test
dFold change (FC) was calculated as a binary logarithm of the average mass response (normalized peak area) ratio between LNM-negative versus normal controls or between LNM-positive versus normal controls
Multivariate statistical analysis of LNM-negative GC specimens and LNM-positive GC specimens
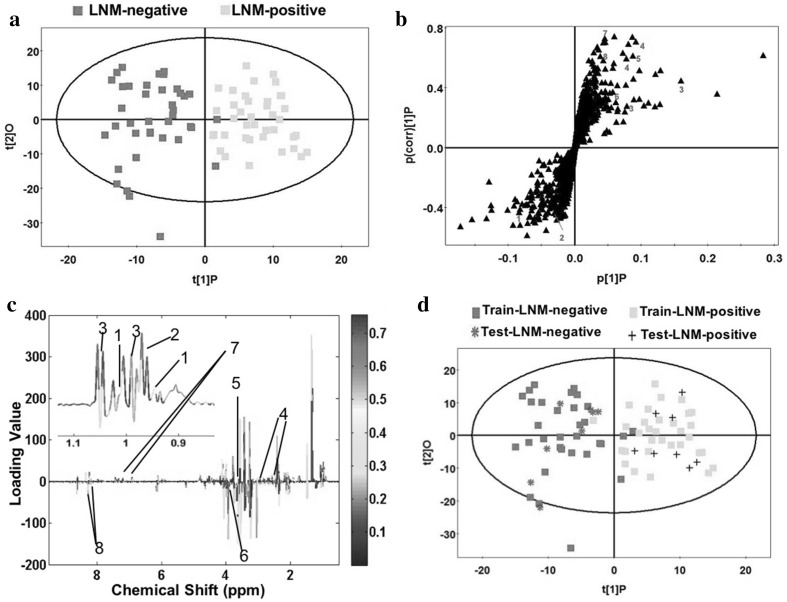
To identify the significantly distinguishing metabolites in discriminating between LNM-negative and LNM-positive GC patients, further multivariate statistical analysis was performed. To our knowledge, this is the first study to characterize the metabolic profile of GC with or without LNM, which was useful in identifying potential biomarkers and understanding the underlying molecular mechanism. A good separation between LNM-negative and LNM-positive GC patients was shown in OPLS score plot (R2X = 0.121, R2Y = 0.75, Q2 = 0.485) in Fig. 3a, in which the two cases were mainly distributed at two sides, with only two specimens misclassified. The corresponding permutation analysis (200 times) was displayed in Fig S1d. Model parameters (R2 = 0.74, Q2 = 0.47) demonstrated the OPLS model was a reliable model with good quality. In the S-plot of the OPLS model (Fig. 3b), the variables that lied far away from the center of the plot were assumed to have a greater contribution to the model classification. To test the predictive ability of established OPLS model in staging GC patients with or without LNM, 80% samples were randomly selected as training set, and the remaining 20% samples were testing set. As a result, the most testing samples were predicted correctly (Fig. 3d), implying LNM was a good indicator for staging GC patients into two subgroups.
Fig. 3.
Discriminating plots of LNM-negative and LNM-positive GC patients. a Scores plot of OPLS model. Green boxes represent LNM-negative GC patients (n = 40); Yellow boxes represent LNM-positive GC patients (n = 40). b S-plot of the OPLS model, the variables that lie far away from the center of the plot were assumed to have a greater contribution to the model classification. c The color map shows the significance of metabolite variations between the two classes. Peaks in the positive direction indicate the increased metabolites in LNM-positive GC patients in comparison to LNM-negative GC patients. Decreased metabolites in LNM-positive GC patients are presented as peaks in the negative direction. 1 Isoleucine, 2 Leucine, 3 Valine, 4 Glutathione, 5 Glycine, 6 Betaine, 7 Tyrosine, 8 Hypoxanthine. d Scores plot of OPLS prediction model. Eighty percentage of samples were applied to construct the model, and then used it to predict the remaining 20% of samples
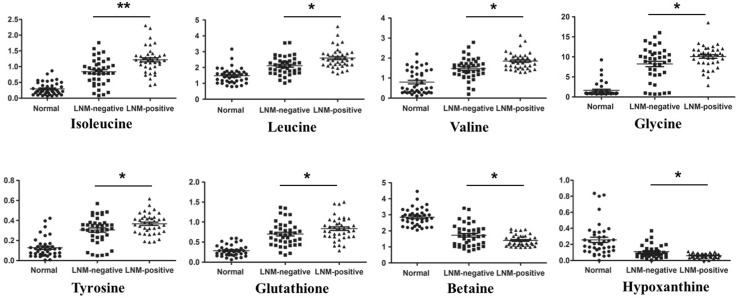
To further identified the potential biomarkers in the process of LNM in GC, 8 tissue metabolites were selected with VIP > 1 and p < 0.05, which were listed in Table 3. Compared with that of LNM-negative GC patients, BCAAs, Glutathione, Glycine and Tyrosine were increased in LNM-positive GC patients, on the other hand, Betaine and Hypoxanthine were significantly decreased. In order to clearly show these differences, scatter plots were conducted for illustrating the relative concentration (Fig. 4). The identified differential metabolites between LNM-negative and LNM-positive were also displayed in color-coded coefficient plots (Fig. 3c). Higher concentration of differential metabolites in LNM-positive GC patients compared to the LNM-negative ones were located in the positive quadrant, otherwise, lower concentrations of metabolites in LNM-positive GC patients were shown in negative quadrant.
Table 3.
Differential metabolites between LNM-negative GC patients and LNM-positive GC patients
| Metabolites | Chemical shift (ppm, multiplicity)a | LNM-positive versus LNM-negative | ||
|---|---|---|---|---|
| VIPb | Pc | FCd | ||
| Isoleucine | 0.945(t) | 1.059 | 0.023 | 1.257 |
| 1.015(d) | 3.302 | 1.73E-04 | 1.254 | |
| Leucine | 0.965(t) | 2.846 | 0.010 | 1.145 |
| Valine | 0.995(d) | 2.544 | 0.010 | 1.143 |
| 1.045(d) | 3.086 | 0.016 | 1.138 | |
| Glutathione | 2.555(m) | 1.727 | 0.322 | 1.052 |
| 2.97(m) | 1.524 | 0.046 | 1.135 | |
| Glycine | 3.565(s) | 1.622 | 0.004 | 1.220 |
| Betaine | 3.89(s) | 2.081 | 0.025 | 0.860 |
| Tyrosine | 6.9(d) | 3.275 | 0.045 | 1.126 |
| 7.2(d) | 2.023 | 0.062 | 1.115 | |
| Hypoxanthine | 8.18(s) | 1.082 | 0.002 | 0.656 |
| 8.215(s) | 1.895 | 0.005 | 0.700 | |
aMultiplicity: s singlet, d doublet, t triplet, m multiplet
bVariable importance in the projection was obtained from OPLS model with a threshold of 1.0
cp-value obtained from Student’s t-test
dFold change (FC) was calculated as a binary logarithm of the average mass response (normalized peak area) ratio between LNM-positive versus LNM-negative
Fig. 4.
Scatter plots illustrating discrimination among normal controls, LNM-negative and LNM-positive GC patients. The Y axis represents relative abundance of NMR signals (normalized to the total peaks). *p < 0.05; **p < 0.01 from LNM-positive GC versus LNM-negative GC
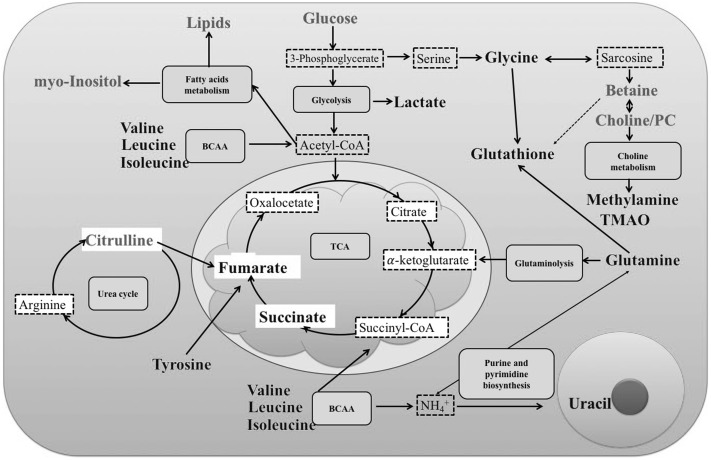
We totally assigned 33 differential metabolites between GC patients and controls in Table 2. After further analyze, eight metabolites were chosen as potential biomarkers for use in discriminating LNM-positive GC patients from LNM-negative ones. To better understand the relationship of the identified metabolites, the related metabolic pathways were conducted in Fig. 5. The altered metabolic pathways included glycolysis, fatty acids metabolism, choline metabolism, glutaminolysis, purine and pyrimidine biosynthesis, urea cycle and TCA. Furthermore, the metabolic pathways including the selected 8 metabolites should be played more attention, such as BCAAs metabolism, Choline metabolism, which may play more important role in the process of LNM in GC.
Fig. 5.
Disturbed metabolic pathways of the relevant metabolites between GC patients and normal controls. Green: lower concentration in GC patients than in normal controls. Red: higher concentration in GC patients than in normal controls
Discussion
Lymph node metastasis was an important prognostic factor for GC patients, especially ones with EGC. The patients with LNM-positive had a poorer overall survival compared with those with LNM-negative (Cao et al. 2011; Cheong et al. 2006). In the present study, we analyzed the metabolic profiling of human GC tissue specimens, and identified 33 distinguishing metabolites between GC patients and controls, 28 of which were reported in our previous work (Wang et al. 2016), the other five differently metabolites (Threonine, Alanine, Pyruvate, Taurine and Betaine) were new identified in our study. This might be duo to the difference in the source and quantity of the used specimens. Furthermore, to confirm the potential biomarkers in the process of LNM, further metabolic profiling of LNM-positive or negative patients was performed, 8 differential metabolites were selected with VIP > 1 and p < 0.05. To our knowledge, the study is the first to display the differences of LNM-positive or negative GC patients based on untargeted metabolomics. The identified metabolites may be as the potential factors of diagnose and prognosis of LNM-positive or negative GC patients. Moreover, they also were valuable in understanding the molecular mechanism in the process of LNM.
There were a growing evidences implicating the dysregulation of BCAA metabolism in many disorders like metabolic syndrome, hepatic disease and cancer (Newgard et al. 2009; Kawaguchi et al. 2011; Tonjes et al. 2013). As essential amino acids, BCAAs accounted for over 20–40% of total dietary protein (Brosnan and Brosnan 2006; Harper et al. 1984), which were used to make protein and as nitrogen donors for nonessential amino acid and nucleotide synthesis in proliferating cells, especially in cancer cells. BCAAs were largely transferred into cancer tissues from the blood to supply amino groups (Baracos and Mackenzie 2006), resulting in higher levels of BCAAs in cancer contrast to normal controls. In our previous work (Wang et al. 2016), the levels of BCAAs were also higher in GC tissue specimens. In the present study, BCAAs were not only increased in GC tissues compared with normal controls (Table 2), but also up-regulated in LNM-positive GC patients compared with LNM-negative ones (Table 3, VIP > 1 and p < 0.05). To identify the diagnostic potential of BCAAs between LNM-positive and negative GC patients, ROC curve was performed (Fig S2). AUC was used to assess the potential diagnostic value: AUC (Isoleucine) = 0.745 (95% CI 0.636–0.854), AUC (Leucine) = 0.725 (95% CI 0.614–0.835), AUC (Valine) = 0.741 (95% CI 0.632–0.850), AUC (Combination) = 0.769 (95% CI 0.666–0.873). These results indicated BCAAs may be as predictor in the process of LNM in GC.
BCAA metabolism played a central role in diverse physiological and pathological process, especially in cancer development (Tonjes et al. 2013; Mayers et al. 2014, 2016). The initial step of BCAA catabolism was transferring amino groups to α-ketoglutarate by branched-chain aminotransferase (cytosolic BCAT1 or mitochondrial BCAT2), generating glutamate and the respective α-ketoacids (α-ketoisocaproic acid (KIC), α-keto-β-methylvaleric acid (KMV) and α-ketoisovaleric acid (KIV)) (Fig S3a). Through further enzymatic reactions, α-ketoacids were converted to acetyl-CoA and succinyl-CoA, which entered into TCA cycle and were oxidized to supply macromolecule precursors and energy. As an important enzyme, BCAT1 was up-regulated in gliomas, breast cancer and myeloid leukaemia (Tonjes et al. 2013; Zhang and Han 2017; Hattori et al. 2017). Knockdown of BCAT1 repressed the cell proliferation and invasiveness in vitro and inhibited the tumor growth in the xenograft model. In our results, BCAAs were increased in LNM-positive GC patients. Moreover, BCAT1 was also up-regulated in LNM-positive GC tissue specimens compared with controls (Fig S3b). LNM-positive GC patients may need more BCAAs from blood to tissue. The elevated expression of BCAT1 played an important role in initiating the catabolism of BCAAs to serve as macromolecule precursors and energy source. Based on previous literatures and our results, BCAAs may be as a useful diagnostic biomarker of GC patients and distinguishing LNM-positive GC patients from LNM-negative ones. Furthermore, BCAT1 may be as attractive targets for treating LNM-positive GC patients. Certainly, further studies must be performed to investigate the function and mechanism of BCAAs and BCAT1 underlying in GC occurrence and progression.
Cancer cells reprogram metabolism to fulfill their rapid proliferation (Moncada et al. 2012), especially glucose and glutamine supply energy, carbon and nitrogen source for macromolecule synthesis to support cell growth through glycolysis and glutaminolysis (Warburg 1956). In this study, the glucose levels significantly decreased and lactate, as the end product of glycolysis, apparently increased in the LNM-negative and LNM-positive GC specimens compared with controls, which was not surprised because of the well-known Warburg effect (Vander Heiden et al. 2009). While, the Warburg effect was not significantly disturbed between LNM-positive GC patients and LNM-negative ones. Therefore, glycolysis may play a central role in GC tumorigenesis, not in the process of LNM. The similar increased glycolysis was also found in our previous work. Beyond the elevated glycolysis, cancer cells depended more on glutamine (Pan et al. 2015), which can be converted into TCA cycle to generate ATP and intermediates for macromolecule synthesis such as proteins, lipids and nucleotides (DeBerardinis and Cheng 2010). Glutamine was up-regulated in GC tissue specimens compared with controls in our study, which showed GC cells needed more glutamine to support their rapid growth as alternative energy, carbon and nitrogen sources. Therefore, targeting glutamine metabolism for cancer treatment had received increasing attention (Cervantes-Madrid et al. 2017; Dequanter et al. 2017). Although the levels of glutamine didn’t change significantly between LNM-positive GC patients and LNM-negative ones, glutathione (GSH), as a product of glutamine and glutamate metabolism, was increased in LNM-positives ones, which indicated antioxidant capacity was obviously enhanced in LNM-positives GC cells compared with LNM-negative ones. Recently, oxidative stress abnormality has been well acknowledged as a hallmark of cancer. The level of reactive oxygen (ROS) was commonly increased in cancer cells, and some types of cancer cells also demonstrated enhanced sensitivity to ROS. GSH is a key antioxidant, the level of which was elevated in lung, head and neck, ovarian and breast cancer (Gamcsik et al. 2012). The increased level of GSH widely enhanced the antioxidant capacity and resistance to oxidative stress. Therefore, the LNM-positive GC cells with higher level of GSH showed elevated antioxidant capacity and more easily survived from oxidative stress, they were more malignant and metastatic ability. A number of studies have reported GSH would be a reliable marker in solid tumors and hematological cancers (Gamcsik et al. 2012; Kearns et al. 2001). Base on our results and these previous reports, we thought GSH might play an important role in the process of lymph node metastasis of GC.
In our study, choline and betaine were both reduced in GC tissues compared with controls. Especially the level of betaine, as an oxidative metabolite of choline (Ji and Kaplowitz 2003), was lower in LNM-positive GC patients in contrast with LNM-negative ones and controls. Choline and betaine, as major methyl donors in one-carbon metabolism (Zeisel and Blusztajn 1994), may be involved in carcinogenesis, such as colorectal, breast, lung and liver cancer (Lu et al. 2015; Zhang et al. 2013; Ying et al. 2013; Zhou et al. 2017). A recent meta-analysis also demonstrated that the food intake with choline plus betaine (100 mg/day) could lower the cancer incidence by 11% (Sun et al. 2016). Some studies showed one-carbon metabolism was involved in the epigenetic modulation of gene expression and biosynthesis of nucleotides, which may be the cause of being implicated in carcinogenesis (Kim 1999; Pandey et al. 2014). In addition, betaine was involved in the methionine cycle, supplying homocysteine through the transmethylation pathway, which was further used for synthesis of glutathione via transsulfuration (Craig 2004). As mentioned above, GSH could enhance the antioxidant capacity and protect cells from ROS. With the lower level in LNM-positive patients, betaine may be a potential diagnostic biomarker distinguishing LNM-positive GC patients from LNM-negative ones. Our findings together with the previous studies (Sun et al. 2016; Du et al. 2016) proved that betaine might play a critical role in the process of LNM of GC. Therefore, dietary choline and betaine intake with a fixed dose may alleviate GC. While, the actual effect needs to be further investigated.
In addition, we also recognized some limitations in our study. Firstly, our study was performed in a relatively small size in each group. The indication of the robustness of OPLS models was relatively small: R2X = 0.121, the maximum fold change is of about 35% (Table 3). Therefore, the interesting potential biomarkers from our study should be verified in the other larger patient cohorts. Secondly, the 1H NMR metabolomics platform used in our study was less sensitive than mass spectrometry and only covered a limited number of metabolite targets. which may lost some information about the process of LNM of GC. In addition, two signals of some metabolites were statistically not relevant, such as isoleucine and Hypoxanthine (Table 2), glutathione (Table 3), which should be verified in better metabolomics platform. Lastly, our understanding of these distinguishing metabolites remained at rudimentary levels. More future studies should be focused on elucidating the mechanisms underlying the process of LNM of GC before the clinical application of the candidate metabolites.
Conclusion
In summary, utilizing untargeted 1H NMR spectroscopy combined with multivariate statistical analysis, we globally analyzed the tissue metabolic profiling of GC patients compared with normal controls, especially between LNM-positive and negative GC patients. Totally 33 distinguishing metabolites were identified between GC patients and controls. More importantly, a panel of 8 differential metabolites was selected according to the LNM of GC. According to further validation and analysis, especially BCAAs metabolism, GSH and betaine might be as potential factors of diagnose and prognosis of LNM-positive or negative GC patients. In our knowledge, this is the first metabolomics study focusing on LNM of GC. On the basis of this research, we believed that the identified distinguishing metabolites might show a promising application on monitoring the carcinogenesis, invasion and metastasis of GC. Certainly, future more functional studies and a larger scale of tissue specimen analysis were needed to demonstrate the potential clinical application and the underlying mechanism of GC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank West China Hospital of Sichuan University for samples collection. We acknowledge all physicians and nurses who participated in the recruiting participants. We thank Analytical and Testing Center of Sichuan University for metabolomics measurements. We thank Hongmin Yuan and Ming Fan for their help in revising the manuscript. Finally, we are grateful to all participants involved in this study.
Author contributions
HLZ and HJW designed the work, provided the financial support, analyzed the 1H NMR data and drafted the manuscript. LZC, WL, ZFW, YY and XL collected the tissues samples and performed the experiments. All of authors read and approved the final manuscript.
Funding
This work was supported by the Project of the National Natural Science Foundation of China (No. 81502096), the Project of the Natural Science Foundation of Henan province (162300410037), the Key Scientific Research Project Plan of Colleges and Universities in Henan province (16A310024) and Basic Research Foundation of Henan University (yqpy20170042)
Compliance with ethical standards
Conflict of interest
The all authors declare that they have no conflicts of interests.
Ethical approval
This study was approved by the ethical committee of West China Hospital of Sichuan University.
Informed consent
Informed written consent was obtained from each participant before specimen collection.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s11306-018-1344-x) contains supplementary material, which is available to authorized users.
References
- An JY, Heo GU, Cheong JH, Hyung WJ, Choi SH, Noh SH. Assessment of open versus laparoscopy-assisted gastrectomy in lymph node-positive early gastric cancer: A retrospective cohort analysis. Journal of Surgical Oncology. 2010;102(1):77–81. doi: 10.1002/jso.21554. [DOI] [PubMed] [Google Scholar]
- Baracos VE, Mackenzie ML. Investigations of branched-chain amino acids and their metabolites in animal models of cancer. Journal of Nutrition. 2006;136(1 Suppl):237s242 s. doi: 10.1093/jn/136.1.237S. [DOI] [PubMed] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature Protocols. 2007;2(11):2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- Brosnan JT, Brosnan ME. Branched-chain amino acids: Enzyme and substrate regulation. Journal of Nutrition. 2006;136(1 Suppl):207s211s. doi: 10.1093/jn/136.1.207S. [DOI] [PubMed] [Google Scholar]
- Cao L, Hu X, Zhang Y, Huang G. Adverse prognosis of clustered-cell versus single-cell micrometastases in pN0 early gastric cancer. Journal of Surgical Oncology. 2011;103(1):53–56. doi: 10.1002/jso.21755. [DOI] [PubMed] [Google Scholar]
- Cervantes-Madrid D, Dominguez-Gomez G, Gonzalez-Fierro A, Perez-Cardenas E, Taja-Chayeb L, Trejo-Becerril C, et al. Feasibility and antitumor efficacy in vivo, of simultaneously targeting glycolysis, glutaminolysis and fatty acid synthesis using lonidamine, 6-diazo-5-oxo-L-norleucine and orlistat in colon cancer. Oncology Letters. 2017;13(3):1905–1910. doi: 10.3892/ol.2017.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Wu CW, Lo SS, Hsieh MC, Lui WY, Shen KH. Peritoneal carcinomatosis and lymph node metastasis are prognostic indicators in patients with Borrmann type IV gastric carcinoma. Hepato-Gastroenterology. 2002;49(45):874–877. [PubMed] [Google Scholar]
- Chen JH, Wu CW, Lo SS, Li AF, Hsieh MC, Shen KH, et al. Lymph node metastasis as a single predictor in patients with Borrmann type I gastric cancer. Hepato-Gastroenterology. 2007;54(75):981–984. [PubMed] [Google Scholar]
- Cheong JH, Hyung WJ, Shen JG, Song C, Kim J, Choi SH, et al. The N ratio predicts recurrence and poor prognosis in patients with node-positive early gastric cancer. Annals of Surgical Oncology. 2006;13(3):377–385. doi: 10.1245/ASO.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Craig SA. Betaine in human nutrition. American Journal of Clinical Nutrition. 2004;80(3):539–549. doi: 10.1093/ajcn/80.3.539. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29(3):313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JY, Liang H. Clinical significance of lymph node metastasis in gastric cancer. World Journal of Gastroenterology. 2014;20(14):3967–3975. doi: 10.3748/wjg.v20.i14.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequanter D, Dok R, Nuyts S. Basal oxidative stress ratio of head and neck squamous cell carcinomas correlates with nodal metastatic spread in patients under therapy. Onco Targets and Therapy. 2017;10:259–263. doi: 10.2147/OTT.S118980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du YF, Luo WP, Lin FY, Lian ZQ, Mo XF, Yan B, et al. Dietary choline and betaine intake, choline-metabolising genetic polymorphisms and breast cancer risk: A case-control study in China. British Journal of Nutrition. 2016;116(6):961–968. doi: 10.1017/S0007114516002956. [DOI] [PubMed] [Google Scholar]
- Feng J, Liu H, Bhakoo KK, Lu L, Chen Z. A metabonomic analysis of organ specific response to USPIO administration. Biomaterials. 2011;32(27):6558–6569. doi: 10.1016/j.biomaterials.2011.05.035. [DOI] [PubMed] [Google Scholar]
- Gamcsik MP, Kasibhatla MS, Teeter SD, Colvin OM. Glutathione levels in human tumors. Biomarkers. 2012;17(8):671–691. doi: 10.3109/1354750X.2012.715672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nature Reviews Cancer. 2004;4(7):551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- Gu J, Hu X, Shao W, Ji T, Yang W, Zhuo H, et al. Metabolomic analysis reveals altered metabolic pathways in a rat model of gastric carcinogenesis. Oncotarget. 2016;7(37):60053–60073. doi: 10.18632/oncotarget.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annual Review of Nutrition. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature. 2017;545(7655):500–504. doi: 10.1038/nature22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyung WJ, Lee JH, Choi SH, Min JS, Noh SH. Prognostic impact of lymphatic and/or blood vessel invasion in patients with node-negative advanced gastric cancer. Annals of Surgical Oncology. 2002;9(6):562–567. doi: 10.1007/BF02573892. [DOI] [PubMed] [Google Scholar]
- Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124(5):1488–1499. doi: 10.1016/S0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- Jung J, Jung Y, Bang EJ, Cho SI, Jang YJ, Kwak JM, et al. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR-based metabolomic profiling. Annals of Surgical Oncology. 2014;21(Suppl 4):S736742. doi: 10.1245/s10434-014-3886-0. [DOI] [PubMed] [Google Scholar]
- Kanavos P. The rising burden of cancer in the developing world. Annals of Oncology. 2006;17(Suppl 8):viii15viii23. doi: 10.1093/annonc/mdl983. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Izumi N, Charlton MR, Sata M. Branched-chain amino acids as pharmacological nutrients in chronic liver disease. Hepatology. 2011;54(3):1063–1070. doi: 10.1002/hep.24412. [DOI] [PubMed] [Google Scholar]
- Kearns PR, Pieters R, Rottier MM, Pearson AD, Hall AG. Raised blast glutathione levels are associated with an increased risk of relapse in childhood acute lymphocytic leukemia. Blood. 2001;97(2):393–398. doi: 10.1182/blood.V97.2.393. [DOI] [PubMed] [Google Scholar]
- Kim YI. Folate and carcinogenesis: Evidence, mechanisms, and implications. Journal of Nutritional Biochemistry. 1999;10(2):66–88. doi: 10.1016/S0955-2863(98)00074-6. [DOI] [PubMed] [Google Scholar]
- Kuligowski J, Sanjuan-Herraez D, Vazquez-Sanchez MA, Brunet-Vega A, Pericay C, Ramirez-Lazaro MJ, et al. Metabolomic analysis of gastric cancer progression within the correa’s cascade using ultraperformance liquid chromatography-mass spectrometry. Journal of Proteome Research. 2016;15(8):2729–2738. doi: 10.1021/acs.jproteome.6b00281. [DOI] [PubMed] [Google Scholar]
- Kunisaki C, Takahashi M, Nagahori Y, Fukushima T, Makino H, Takagawa R, et al. Risk factors for lymph node metastasis in histologically poorly differentiated type early gastric cancer. Endoscopy. 2009;41(6):498–503. doi: 10.1055/s-0029-1214758. [DOI] [PubMed] [Google Scholar]
- Lai JF, Kim S, Kim K, Li C, Oh SJ, Hyung WJ, et al. Prediction of recurrence of early gastric cancer after curative resection. Annals of Surgical Oncology. 2009;16(7):1896–1902. doi: 10.1245/s10434-009-0473-x. [DOI] [PubMed] [Google Scholar]
- Lee JH, Choi MG, Min BH, Noh JH, Sohn TS, Bae JM, et al. Predictive factors for lymph node metastasis in patients with poorly differentiated early gastric cancer. British Journal of Surgery. 2012;99(12):1688–1692. doi: 10.1002/bjs.8934. [DOI] [PubMed] [Google Scholar]
- Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, et al. Screening for gastric cancer in Asia: Current evidence and practice. Lancet Oncology. 2008;9(3):279–287. doi: 10.1016/S1470-2045(08)70072-X. [DOI] [PubMed] [Google Scholar]
- Li Y, Song X, Zhao X, Zou L, Xu G. Serum metabolic profiling study of lung cancer using ultra high performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Journal of Chromatography B. 2014;966:147–153. doi: 10.1016/j.jchromb.2014.04.047. [DOI] [PubMed] [Google Scholar]
- Lu MS, Fang YJ, Pan ZZ, Zhong X, Zheng MC, Chen YM, et al. Choline and betaine intake and colorectal cancer risk in Chinese population: A case-control study. PLoS One. 2015;10(3):e0118661. doi: 10.1371/journal.pone.0118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353(6304):1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nature Medicine. 2014;20(10):1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada S, Higgs EA, Colombo SL. Fulfilling the metabolic requirements for cell proliferation. Biochemical Journal. 2012;446(1):1–7. doi: 10.1042/BJ20120427. [DOI] [PubMed] [Google Scholar]
- Nakamoto J, Torisu R, Aoki R, Kimura Y, Yasuda M, Shiota K, et al. Clinicopathological evaluation of biological behavior of submucosal invasive gastric carcinomas: Relationship among lymph node metastasis, mucin phenotype and proliferative activity. Journal of Medical Investigation. 2007;54(1–2):99–108. doi: 10.2152/jmi.54.99. [DOI] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Gao L, Wu G, Shen G, Xie S, Wen H, et al. Elevated expression of glutaminase confers glucose utilization via glutaminolysis in prostate cancer. Biochemical and Biophysical Research Communication. 2015;456(1):452–458. doi: 10.1016/j.bbrc.2014.11.105. [DOI] [PubMed] [Google Scholar]
- Pandey S, Garg P, Lee S, Choung HW, Choung YH, Choung PH, et al. Nucleotide biosynthesis arrest by silencing SHMT1 function via vitamin B6-coupled vector and effects on tumor growth inhibition. Biomaterials. 2014;35(34):9332–9342. doi: 10.1016/j.biomaterials.2014.07.045. [DOI] [PubMed] [Google Scholar]
- Ramachandran GK, Yong WP, Yeow CH. Identification of gastric cancer biomarkers using 1H nuclear magnetic resonance spectrometry. PLoS ONE. 2016;11(9):e0162222. doi: 10.1371/journal.pone.0162222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha CM, Carrola J, Barros AS, Gil AM, Goodfellow BJ, Carreira IM, et al. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of blood plasma. Journal of Proteome Research. 2011;10(9):4314–4324. doi: 10.1021/pr200550p. [DOI] [PubMed] [Google Scholar]
- Sarela AI, Turnbull AD, Coit DG, Klimstra D, Brennan MF, Karpeh MS. Accurate lymph node staging is of greater prognostic importance than subclassification of the T2 category for gastric adenocarcinoma. Annals of Surgical Oncology. 2003;10(7):783–791. doi: 10.1245/ASO.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Song Y, Wang J, Cheng Z, Gao P, Sun J, Chen X, et al. Quantitative global proteome and lysine succinylome analyses provide insights into metabolic regulation and lymph node metastasis in gastric cancer. Scientific Reports. 2017;7:42053. doi: 10.1038/srep42053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Li X, Ren A, Du M, Du H, Shu Y, et al. Choline and betaine consumption lowers cancer risk: A meta-analysis of epidemiologic studies. Scientific Reports. 2016;6:35547. doi: 10.1038/srep35547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nature Medicine. 2013;19(7):901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Zhang H, Deng P, Chen J, Zhou B, et al. 1H NMR-based metabolic profiling of human rectal cancer tissue. Molecular Cancer. 2013;12(1):121. doi: 10.1186/1476-4598-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang H, Deng P, Liu C, Li D, Jie H, et al. Tissue metabolic profiling of human gastric cancer assessed by 1H NMR. BMC Cancer. 2016;16:371. doi: 10.1186/s12885-016-2356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu M, Feng J, Liu M, Hu JZ. 1H NMR Metabolomics study of metastatic melanoma in C57BL/6J mouse spleen. Metabolomics. 2014;10(6):1129–1144. doi: 10.1007/s11306-014-0652-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Weljie AM, Dowlatabadi R, Miller BJ, Vogel HJ, Jirik FR. An inflammatory arthritis-associated metabolite biomarker pattern revealed by 1H NMR spectroscopy. Journal of Proteome Research. 2007;6(9):3456–3464. doi: 10.1021/pr070123j. [DOI] [PubMed] [Google Scholar]
- Ying J, Rahbar MH, Hallman DM, Hernandez LM, Spitz MR, Forman MR, et al. Associations between dietary intake of choline and betaine and lung cancer risk. PLoS ONE. 2013;8(2):e54561. doi: 10.1371/journal.pone.0054561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel SH, Blusztajn JK. Choline and human nutrition. Annual Review of Nutrition. 1994;14:269–296. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
- Zhang CX, Pan MX, Li B, Wang L, Mo XF, Chen YM, et al. Choline and betaine intake is inversely associated with breast cancer risk: A two-stage case-control study in China. Cancer Science. 2013;104(2):250–258. doi: 10.1111/cas.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qiao L, Li X, Wan Y, Yang L, Wang H. Tissue metabolic profiling of lymph node metastasis of colorectal cancer assessed by 1H NMR. Oncology Reports. 2016;36(6):3436–3448. doi: 10.3892/or.2016.5175. [DOI] [PubMed] [Google Scholar]
- Zhang L, Han J. Branched-chain amino acid transaminase 1 (BCAT1) promotes the growth of breast cancer cells through improving mTOR-mediated mitochondrial biogenesis and function. Biochemical and Biophysical Research Communication. 2017;486(2):224–231. doi: 10.1016/j.bbrc.2017.02.101. [DOI] [PubMed] [Google Scholar]
- Zhou RF, Chen XL, Zhou ZG, Zhang YJ, Lan QY, Liao GC, et al. Higher dietary intakes of choline and betaine are associated with a lower risk of primary liver cancer: A case-control study. Scientific Reports. 2017;7(1):679. doi: 10.1038/s41598-017-00773-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.