Abstract
Several epidemiological studies have reported that polymorphisms in microRNA-196a2 (miR-196a2) were associated with various cancers. However, the results remained unverified and were inconsistent in different cancers. Therefore, we carried out an updated meta-analysis to elaborate the effects of rs11614913 polymorphism on cancer susceptibility. A total of 84 articles with 35,802 cases and 41,541 controls were included to evaluate the association between the miR-196a2 rs11614913 and cancer risk by pooled odds ratios (ORs) and 95% confidence intervals (CIs). The results showed that miR-196a2 rs11614913 polymorphism is associated with cancer susceptibility, especially in lung cancer (homozygote comparison, OR =0.840, 95% CI =0.734–0.961; recessive model, OR =0.858, 95% CI =0.771–0.955), hepatocellular carcinoma (allelic contrast, OR =0.894, 95% CI =0.800–0.998; homozygote comparison, OR =0.900, 95% CI =0.813–0.997; recessive model, OR =0.800, 95% CI =0.678–0.944), and head and neck cancer (allelic contrast, OR =1.076, 95% CI =1.006–1.152; homozygote comparison, OR =1.214, 95% CI =1.043–1.413). In addition, significant association was found among Asian populations (allele model, OR =0.847, 95% CI =0.899–0.997, P=0.038; homozygote model, OR =0.878, 95% CI =0.788–0.977, P=0.017; recessive model, OR =0.895, 95% CI =0.824–0.972, P=0.008) but not in Caucasians. The updated meta-analysis confirmed the previous results that miR-196a2 rs11614913 polymorphism may serve as a risk factor for patients with cancers.
Keywords: miR-196a2, polymorphisms, cancer risk, meta-analysis
Introduction
The rising morbidity and mortality of cancer has drawn extensive attention worldwide, and finding possible risk factors of tumorigenesis has been a priority task for researchers. Recently, an increasing number of studies have focused on associations between miRNA polymorphisms and cancer susceptibility, which indicated that accumulation of genetic variants may be involved in cancer development, including oral cancer,1 lung cancer,2,3 gastric cancer,4 breast cancer,5 glioma,6 non-small cell lung cancer,7 hepatocellular carcinoma,8,9 gallbladder cancer,10 and head and neck cancer (HNC).11 As the molecular mechanism of cancer remains unclear, further exploration of more accurate cancer treatments and prognosis would be of great importance.
MiRNAs are a class of small non-coding RNAs with 18–25 nucleotides in length, which play as oncogenes or anti-oncogenes in the pathogenesis of tumor by targeting multiple genes.12–14 Studies have shown that almost 10%–30% of all human gene expressions have been regulated by mature miRNAs.15 MiRNAs could modulate related genes implicated in cellular processes, including cell differentiation, growth, apoptosis, and immune response.16–18
Hsa-microRNA-196a2 (miR-196a2), initially discovered by Lagos-Quintana et al,19 has been proven to play important roles in various cancers.20,21 Single nucleotide polymorphisms (SNPs) provide new sources of genetic variation, which contribute to potential molecular mechanisms of cancer development.22 SNPs or mutations in miRNA sequence may transform miRNA expression and/or maturation, related to miRNA function by activating the transcription of the primary transcript, pri-miRNA and pre-miRNA processing, and miRNA–mRNA interactions.23 MiR-196a2 rs11614913, as a definitional miRNA polymorphism,24–26 is crucially associated with cancer risk.23,27 It is located in the 3′-untranslated region of the miR-196a2 precursor.28 Hoffman et al5 also showed that miR-196a2 rs11614913 not only influenced the transcription level of mature miR-196a, but also had a biological effect on target gene production. This updated meta-analysis was performed to explore the association between the hsa-miR-196a2 polymorphism and cancer risk and to further estimate the overall cancer risk by pooling all available data.
Materials and methods
Publication search
Two investigators (LYH, HAB) carried out a systematic review on PubMed, Cochrane Library, and Web of Science, by using (“microRNA-196a2” or “miR-196a2”, or “miR-196-a-2” or “miR-196-2” or “miR-196-a” or “rs11614913”), and (“cancer” or “tumor” or “carcinoma” or “neoplasm” or “malignancy”), and (“polymorphism” or “variation” or “susceptibility”) as the search terms in order to identify potentially eligible studies. We based our dates for literature retrieval from January 2008 to September 2017.
Inclusion and exclusion criteria
Relevant studies had to meet the following inclusion criteria: 1) full-text article; 2) evaluation of a link between miRNA polymorphisms and cancer risks; 3) sufficient data for estimating the odds ratio (OR) with 95% CI and a P-value. Studies containing two or more case-control groups were considered as two or more independent studies. Studies that were, 1) review, letters, and comment articles; 2) not for cancer risk; and 3) duplicate samples or publications, were excluded.
Assessment of study quality
The quality of the study was determined by the Newcastle–Ottawa Scale for cohort studies.
Data extraction
Data extraction from the eligible studies were performed independently by two authors (LYH, HAB), based on the inclusion and exclusion criteria. For each publication, the following data were recorded: first author, date of publication, country of origin, ethnicity, type of tumor, source of control groups, total numbers of cases and controls, and genotyping method.
Statistical analysis
The departure of frequencies of miR-196a2 rs11614913 polymorphisms was assessed under the Hardy–Weinberg equilibrium (HWE) for each publication by adopting the goodness-of-fit test (chi-square or Fisher exact test). The association between the miR-196a2 rs11614913 polymorphisms and the risk of cancer was evaluated by calculating pooled OR together with corresponding 95% CI based on the method published by Woolf.29 Also, a P-value<0.05 was considered statistically significant. In addition, we used stratified meta-regression analyses to explore major causes of heterogeneity among the articles. We respectively examined the association between genetic mutants and cancer risk in allelic contrast (T vs C), homozygote comparisons (TT vs CC), heterozygote comparisons (TC vs CC), recessive model (TT vs TC+CC), and dominant model (TT+TC vs CC). Subgroup analyses were performed by ethnicity (Asian and Caucasian), tumor types (if one tumor type contained less than three individual studies, it was combined into “other cancer” subgroups), and source of control (hospital based and population based).
Q tests30 and I2 tests31 were carried out to test the heterogeneity. I2 values describe the percentage of total variation across studies that are due to heterogeneity rather than chance. I2=0% prompts no heterogeneity observed, with 25% identified as low, 50% as moderate, and 75% as high. If I2 was ≥50% or if the P-value of heterogeneity was <0.05, indicating significant heterogeneity among these articles, a random-effect model was used;32 otherwise, a fixed-effect mode was used.33 Sensitivity analyses were conducted to estimate the stability of the meta-analysis result. We adopted Egger’s test to assess potential publication bias by visual inspection of the Funnel plot. A P-value <0.05 was regarded as an indication of potential publication bias.34 All statistical analyses were performed with the Stata software package version 12.0 (Stata Corporation, College Station, TX, USA).
Results
Study identification
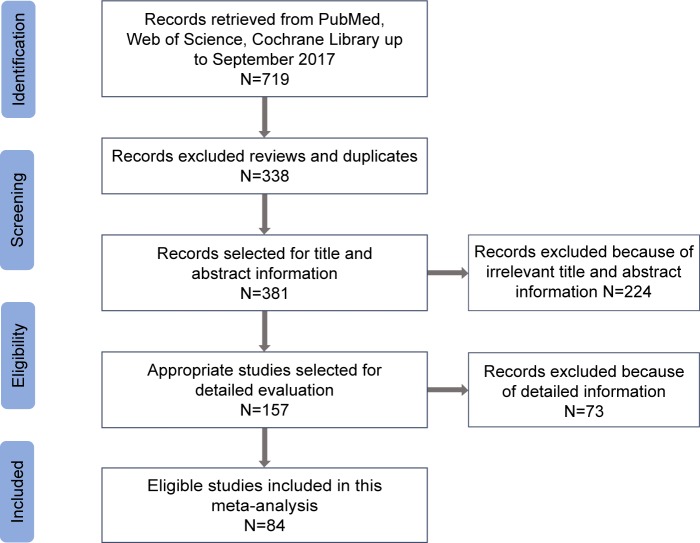
Overall, 84 articles,1–11,26,27,35–100 which were relevant to the search terms, were selected based on the inclusion criteria from PubMed, Cochrane, and Web of Science (Figure 1). These studies with a total of 35,802 cases and 41,541 controls were subjected to further checking. In the present meta-analysis, we excluded 73 articles (36 articles were meta-analysis, 22 articles did not express concern about cancer risk, 11 articles lacked detailed allele frequency data or OR calculation, and four articles were incomplete text). The included study characteristics are provided in Table 1.
Figure 1.
The flow diagram of the included and excluded studies.
Table 1.
Characteristics of studies included in the meta-analysis
| Author | Year | Country | Ethnicity | Cancer type | Genotyping method | Source of control | Case
|
Control
|
HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | CT | CC | TT | CT | CC | ||||||||
| Hu et al7 | 2008 | China | Asian | LC | PCR | PB | 152 | 264 | 140 | 32 | 52 | 23 | 0.827 |
| Hu et al35 | 2009 | China | Asian | BRC | PCR-RFLP | PB | 287 | 483 | 239 | 358 | 517 | 218 | 0.207 |
| Tian et al3 | 2009 | China | Asian | LC | PCR-RFLP | PB | 293 | 512 | 253 | 307 | 519 | 209 | 0.700 |
| Hoffman et al5 | 2009 | USA | Caucasian | BRC | TaqMan | HB | 71 | 229 | 166 | 36 | 209 | 181 | 0.583 |
| Catucci et al36 | 2010 | Italy | Caucasian | BRC | TaqMan | PB | 244 | 842 | 776 | 377 | 1,246 | 1,116 | 0.326 |
| Wang et al38 | 2010 | China | Asian | ESCC | PCR | PB | 48 | 262 | 148 | 111 | 250 | 128 | 0.600 |
| Okubo et al83 | 2010 | Japan | Asian | GC | Gel Pictures | HB | 166 | 281 | 105 | 372 | 592 | 216 | 0.466 |
| Peng et al4 | 2010 | China | Asian | GC | PCR-RFLP | PB | 43 | 94 | 76 | 50 | 107 | 56 | 0.936 |
| Srivastava et al10 | 2010 | India | Asian | GLC | PCR-RFLP | PB | 121 | 97 | 21 | 121 | 94 | 15 | 0.566 |
| Dou et al6 | 2010 | China | Asian | Glioma | PCR-LDR | HB | 189 | 343 | 111 | 208 | 305 | 143 | 0.119 |
| Li et al9 | 2010 | China | Asian | HCC | PCR-RFLP | HB | 82 | 150 | 78 | 78 | 102 | 42 | 0.402 |
| Akkiz et al8 | 2010 | Turkey | Caucasian | HCC | PCR-RFLP | HB | 22 | 86 | 77 | 40 | 87 | 58 | 0.492 |
| Liu et al11 | 2010 | USA | Caucasian | HNC | PCR-RFLP | PB | 194 | 565 | 350 | 202 | 545 | 383 | 0.737 |
| Kim et al110 | 2010 | Korea | Asian | LC | PCR-RFLP | HB | 162 | 305 | 187 | 185 | 300 | 155 | 0.126 |
| Catucci et al36 | 2010 | Germany | Caucasian | BRC | MassARRAY | PB | 216 | 696 | 584 | 157 | 512 | 432 | 0.711 |
| Christensen et al37 | 2010 | USA | Caucasian | HNC | AppliedBiosystems | PB | 0 | 302 | 182 | 0 | 367 | 188 | NA |
| Mittal et al41 | 2011 | India | Asian | BLC | PCR-RFLP | PB | 5 | 131 | 76 | 14 | 127 | 109 | 0.003 |
| Jedlinski et al40 | 2011 | Australia | Caucasian | BRC | PCR | PB | 33 | 86 | 68 | 31 | 82 | 58 | 0.830 |
| Zhan et al42 | 2011 | China | Asian | CRC | PCR-RFLP | HB | 56 | 128 | 68 | 163 | 267 | 113 | 0.849 |
| Zhou et al43 | 2011 | China | Asian | CSCC | PCR-RFLP | PB | 57 | 123 | 46 | 82 | 169 | 58 | 0.077 |
| Vinci et al111 | 2011 | Italy | Caucasian | LC | TaqMan | PB | 12 | 54 | 35 | 10 | 61 | 58 | 0.267 |
| Hong et al2 | 2011 | Korea | Asian | LC | TaqMan | HB | 96 | 224 | 86 | 134 | 198 | 96 | 0.163 |
| George et al39 | 2011 | Italy | Caucasian | PC | PCR-RFLP | PB | 3 | 101 | 55 | 10 | 114 | 106 | 0.002 |
| Linhares et al45 | 2012 | Brazil | Mix | BRC | TaqMan | HB | 117 | 177 | 94 | 96 | 165 | 127 | 0.005 |
| Chen et al44 | 2012 | China | Asian | CRC | PCR-LDR | HB | 35 | 64 | 27 | 107 | 206 | 94 | 0.788 |
| Min et al24 | 2012 | Korea | Asian | CRC | PCR-RFLP | HB | 125 | 201 | 120 | 148 | 254 | 100 | 0.633 |
| Zhu et al47 | 2012 | China | Asian | CRC | TaqMan | HB | 130 | 303 | 140 | 172 | 295 | 121 | 0.790 |
| Hezova et al25 | 2012 | Czech | Caucasian | CRC | TaqMan | HB | 26 | 89 | 82 | 22 | 103 | 87 | 0.291 |
| Zhang et al100 | 2012 | China | Asian | CRC | PCR-RFLP | PB | 172 | 204 | 79 | 185 | 197 | 81 | 0.026 |
| Ahn et al48 | 2013 | Korea | Asian | GC | PCR-RFLP | PB | 119 | 242 | 100 | 128 | 232 | 87 | 0.322 |
| Yoon et al46 | 2012 | Korea | Asian | LC | TaqMan | PB | 99 | 186 | 101 | 24 | 32 | 15 | 0.480 |
| Zhang et al104 | 2012 | China | Asian | BRC | PCR-RFLP | PB | 133 | 93 | 17 | 148 | 89 | 11 | 0.893 |
| Chu et al87 | 2012 | China | Asian | HNC | PCR-RFLP | HB | 136 | 277 | 57 | 132 | 206 | 87 | 0.690 |
| Vinci et al113 | 2013 | Italy | Caucasian | CRC | HRMA | HB | 12 | 86 | 62 | 11 | 84 | 83 | 0.087 |
| Lv et al51 | 2013 | China | Asian | CRC | PCR-RFLP | PB | 114 | 223 | 10 | 91 | 331 | 109 | 0.000 |
| Umar et al112 | 2013 | India | Asian | ESCC | PCR-RFLP | HB | 22 | 121 | 146 | 16 | 122 | 171 | 0.330 |
| Wei et al114 | 2013 | China | Asian | ESCC | SNPscanTM | HB | 106 | 196 | 65 | 113 | 170 | 87 | 0.141 |
| Toraih et al98 | 2016 | Egypt | Caucasian | OSCC | PCR | PB | 32 | 93 | 84 | 10 | 35 | 55 | 0.221 |
| Wang et al53 | 2013 | China | Asian | GC | TaqMan | HB | 226 | 371 | 152 | 232 | 448 | 220 | 0.898 |
| Zhang et al55 | 2013 | China | Asian | HCC | MassARRAY | HB | 294 | 488 | 214 | 328 | 502 | 165 | 0.245 |
| Han et al49 | 2013 | China | Asian | HCC | PCR | PB | 305 | 505 | 207 | 304 | 485 | 220 | 0.310 |
| Tong et al65 | 2013 | China | Asian | ALL | TaqMan | HB | 159 | 308 | 103 | 237 | 307 | 129 | 0.434 |
| Pavlakis et al93 | 2013 | Greece | Caucasian | PCC | PCR-RFLP | HB | 48 | 33 | 12 | 50 | 58 | 14 | 0.647 |
| Pu et al84 | 2014 | China | Asian | GC | PCR-RFLP | HB | 25 | 95 | 39 | 86 | 324 | 101 | 0.000 |
| Bansal et al56 | 2014 | India | Asian | BRC | PCR-RFLP | PB | 12 | 41 | 68 | 21 | 59 | 85 | 0.042 |
| Kupcinskas et al62 | 2014 | Lithuania | Caucasian | CRC | PCR | HB | 27 | 87 | 79 | 54 | 174 | 199 | 0.104 |
| Qu et al64 | 2014 | China | Asian | ESCC | PCR | PB | 48 | 207 | 126 | 82 | 211 | 133 | 0.918 |
| Wang et al66 | 2014 | China | Asian | ESCC | PCR-LDR | PB | 162 | 307 | 128 | 154 | 298 | 145 | 0.970 |
| Dikeakos et al58 | 2014 | Greece | Caucasian | GC | PCR-RFLP | HB | 15 | 46 | 102 | 172 | 229 | 79 | 0.850 |
| Qi et al86 | 2014 | China | Asian | HCC | PCR | HB | 60 | 209 | 45 | 121 | 214 | 71 | 0.156 |
| Chu et al57 | 2014 | China | Asian | HCC | PCR-RFLP | HB | 66 | 81 | 41 | 100 | 167 | 70 | 0.986 |
| Parlayan et al115 | 2014 | Japan | Asian | LC | TaqMan | HB | 38 | 81 | 29 | 146 | 270 | 108 | 0.410 |
| Li et al63 | 2014 | China | Asian | NPC | TaqMan | HB | 322 | 489 | 209 | 270 | 518 | 218 | 0.301 |
| Du et al59,60 | 2014 | China | Asian | RCC | PCR | HB | 121 | 189 | 43 | 109 | 179 | 74 | 0.974 |
| Omrani et al85 | 2014 | Iran | Asian | BRC | PCR-RFLP | PB | 0 | 25 | 78 | 0 | 18 | 218 | NA |
| Kou et al91 | 2014 | China | Asian | HCC | PCR | HB | 37 | 150 | 84 | 103 | 304 | 125 | 0.001 |
| Roy et al94 | 2014 | India | Asian | HNC | AppliedBiosystems | HB | 46 | 187 | 218 | 38 | 168 | 242 | 0.250 |
| Li et al63 | 2014 | China | Asian | HNC | AppliedBiosystems | PB | 322 | 489 | 209 | 270 | 518 | 218 | 0.300 |
| Deng et al67 | 2015 | China | Asian | BLC | PCR-RFLP | PB | 52 | 66 | 41 | 76 | 166 | 56 | 0.040 |
| Qi et al72 | 2015 | China | Asian | BRC | PCR | PB | 168 | 119 | 34 | 185 | 88 | 17 | 0.141 |
| Dikaiakos et al68 | 2015 | Greece | Caucasian | CRC | PCR-RFLP | PB | 69 | 69 | 19 | 117 | 149 | 33 | 0.156 |
| Li et al69 | 2015 | China | Asian | HCC | PCR | HB | 51 | 131 | 84 | 30 | 123 | 113 | 0.689 |
| Li et al69 | 2015 | China | Asian | NHL | PCR-RFLP | PB | 111 | 146 | 61 | 144 | 134 | 42 | 0.225 |
| Nikolic et al71 | 2015 | Serbia | Caucasian | PC | PCR-RFLP | PB | 40 | 161 | 150 | 41 | 147 | 121 | 0.728 |
| He et al90 | 2015 | China | Asian | BRC | MassARRAY | HB | 134 | 223 | 93 | 136 | 233 | 81 | 0.990 |
| Sushma et al97 | 2015 | India | Asian | OSCC | PCR-RFLP | PB | 68 | 10 | 22 | 81 | 15 | 6 | 0.212 |
| Sodhi et al95 | 2015 | India | Asian | LC | PCR-RFLP | PB | 19 | 161 | 70 | 8 | 146 | 101 | 0.000 |
| Jiang et al26 | 2016 | China | Asian | GC | PCR | HB | 300 | 423 | 166 | 290 | 487 | 198 | 0.804 |
| Dai et al74 | 2016 | China | Asian | BRC | MassARRAY | HB | 98 | 265 | 197 | 144 | 284 | 155 | 0.540 |
| Zhao et al82 | 2016 | China | Asian | BRC | TaqMan | PB | 33 | 50 | 31 | 25 | 61 | 28 | 0.449 |
| Song et al79 | 2016 | China | Asian | OC | PCR | PB | 111 | 247 | 121 | 142 | 203 | 86 | 0.385 |
| Shen et al78 | 2016 | China | Asian | ESCC | SNaPshot | PB | 407 | 698 | 295 | 672 | 1,121 | 392 | 0.043 |
| Li et al75 | 2016 | China | Asian | GC | PCR | HB | 75 | 83 | 24 | 92 | 79 | 11 | 0.265 |
| Li et al76 | 2016 | China | Asian | HCC | PCR | HB | 20 | 64 | 25 | 35 | 52 | 18 | 0.861 |
| Xu et al80 | 2016 | China | Asian | HCC | PCR-RFLP | HB | 56 | 128 | 68 | 163 | 267 | 113 | 0.849 |
| Qiu and Liu77 | 2016 | China | Asian | HCC | PCR | PB | 61 | 141 | 68 | 70 | 121 | 46 | 0.626 |
| Jiang et al26 | 2016 | China | Asian | HCC | TaqMan | PB | 159 | 308 | 103 | 237 | 307 | 129 | 0.099 |
| Yin et al81 | 2016 | China | Asian | LC | TaqMan | PB | 149 | 298 | 128 | 178 | 297 | 133 | 0.664 |
| Zhang et al99 | 2016 | China | Asian | HCC | PCR-RFLP | HB | 65 | 85 | 25 | 122 | 138 | 42 | 0.770 |
| Sun et al96 | 2016 | China | Asian | OC | PCR | HB | 39 | 66 | 29 | 77 | 116 | 34 | 0.360 |
| Toraih et al98 | 2016 | Egypt | Caucasian | HCC | PCR | PB | 11 | 31 | 23 | 17 | 53 | 80 | 0.082 |
| Morales et al92 | 2016 | Chile | Mix | BRC | TaqMan | HB | 57 | 191 | 192 | 114 | 351 | 342 | 0.121 |
| Gu and Tu88 | 2016 | China | Asian | GC | PCR | HB | 51 | 96 | 39 | 31 | 98 | 57 | 0.310 |
| Hashemi et al89 | 2016 | Iran | Asian | GC | PCR-RFLP | PB | 17 | 88 | 64 | 12 | 93 | 77 | 0.021 |
Abbreviations: ALL, acute lymphoblastic leukemia; BLC, bladder cancer; BRC, breast cancer; CRC, colorectal cancer; CSCC, cervical cancer; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; GLC, gallbladder cancer; HB, hospital based; HCC, hepatocellular carcinoma; HNC, head and neck cancer; HRMA, high-resolution melting analysis; HWE, Hardy–Weinberg equilibrium of controls; LC, lung cancer; NHL, non-Hodgkin lymphoma; NPC, nasopharyngeal carcinoma; NA, not available; OC, ovarian cancer; OSCC, oral squamous cell carcinomas; PB, population based; PC, prostate cancer; PCC, pancreatic cancer; PCR, polymerase chain reaction; PCR-LDR, polymerase chain reaction-ligation detection reaction; PCR-RFLP, polymerase chain reaction restriction fragment length polymorphism; RCC, renal cell carcinoma.
In total, there were studies on hepatocellular carcinoma (n=14), breast cancer (n=14), colorectal cancer (n=10), gastric cancer (n=10), lung cancer (n=9), esophageal squamous cell carcinoma (ESCC; n=6), HNC (n=5), bladder cancer (n=2), prostate cancer (n=2), oral squamous cell carcinoma (n=2), epithelial ovarian cancer (n=2), renal cell cancer (n=1), glioma (n=1), pancreatic cancer (n=1), cervical cancer (n=1), nasopharyngeal carcinoma (n=1), gallbladder cancer (n=1), acute lymphoblastic leukemia (n=1), and non-Hodgkin lymphoma (n=1). There were 64 studies of Asians and 18 studies of Caucasians.
Among the genotyping methods used in these studies, 57 studies used polymerase chain reaction (including polymerase chain reaction restriction fragment length polymorphism and polymerase chain reaction-ligation detection reaction), 16 studies used Taqman SNP genotyping assay, and others used MassARRAY and DNA sequencing. The controls of 42 studies mainly came from a hospital-based healthy population matched for gender and age, and 42 studies had population-based controls (PB). The distribution of genotypes in the controls of all of the studies was in agreement with HWE (P>0.05).
Quantitative synthesis
In this meta-analysis, we analyzed the hsa-miR-196a2 rs11614913 polymorphism in 84 comparisons with 35,802 cases and 41,541 controls. All the studies were pooled into the meta-analysis, and the results showed that the hsa-miR-196a2 rs11614913 polymorphism was significantly associated with the risk of cancer in the following genetic models: TT vs CC: OR =0.900, 95% CI =0.813–0.987, P=0.043; TT vs TC+CC: OR =0.918, 95% CI =0.851–0.989, P=0.025.
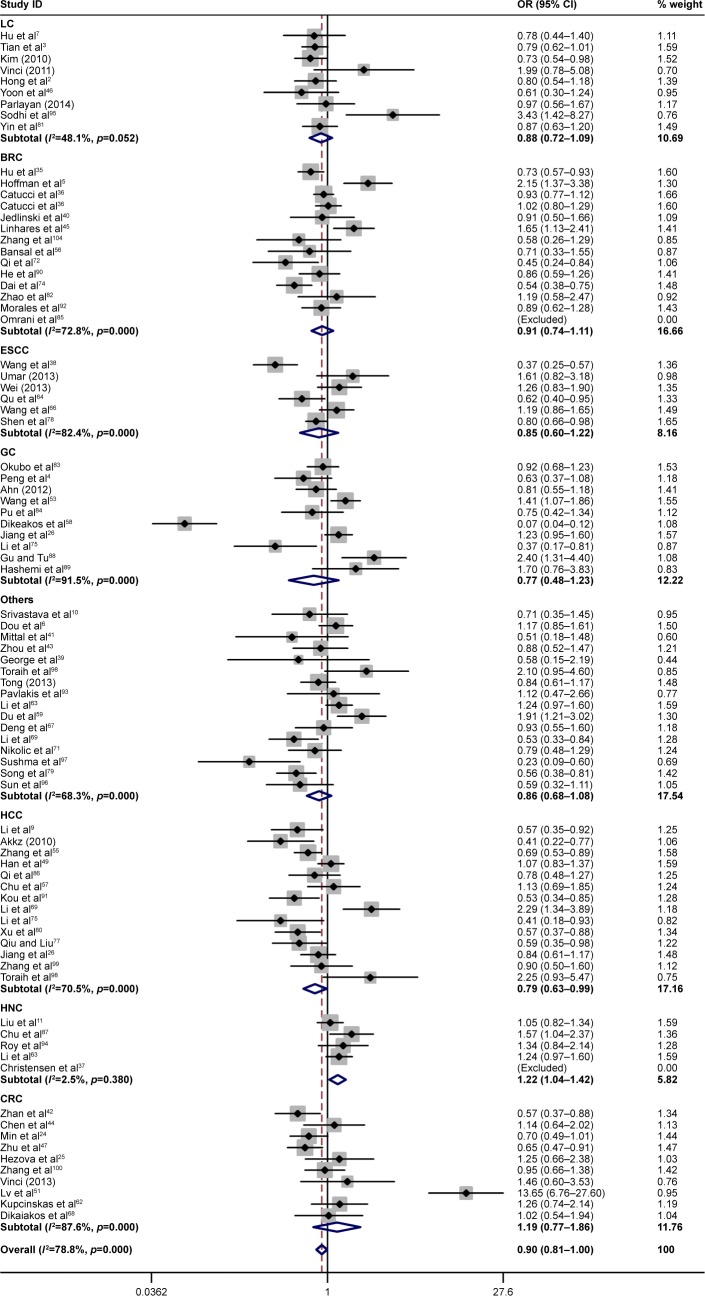
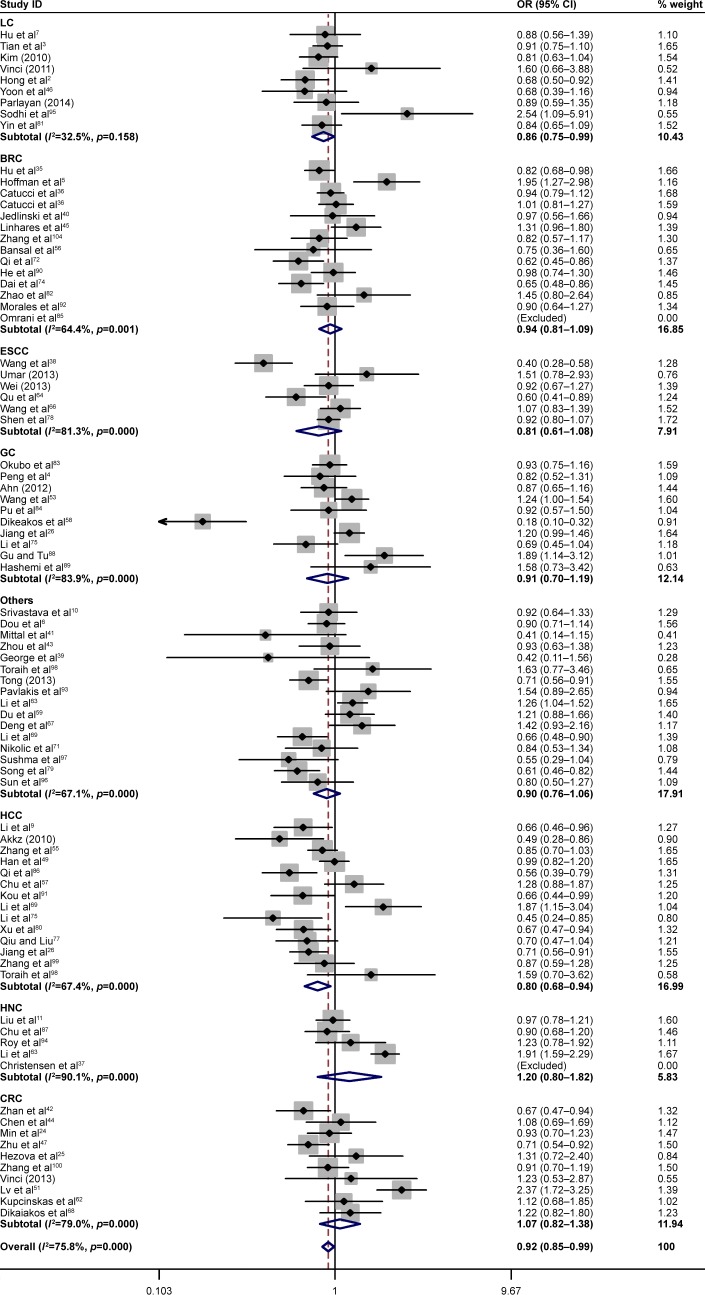
Then, we performed the subgroup analysis of different specific cancer types, genotypes, control sources, and ethnicities (Table 2). In the different cancer types, close association between rs11614913 and cancer risk was found for lung cancer (homozygote comparison, OR =0.840, 95% CI =0.734–0.961, P=0.011; recessive model, OR =0.858, 95% CI =0.771–0.955, P=0.005), hepatocellular carcinoma (allelic contrast, OR =0.894, 95% CI =0.800–0.998, P=0.047; homozygote comparison, OR =0.900, 95% CI =0.813–0.997, P=0.039; recessive model, OR =0.800, 95% CI =0.678–0.944, P=0.008), and HNC (allelic contrast, OR =1.076, 95% CI =1.006–1.152, P=0.033; homozygote comparison, OR =1.214, 95% CI =1.043–1.413, P=0.012; Figures 2 and 3). However, the association between rs11614913 and breast cancer, ESCC, gastric cancer (GC), or colorectal cancer (CRC) is not statistically significant.
Table 2.
Meta-analysis of miR-192a rs11614913 polymorphism with cancer risk
| rs11614913 | na | Case/control | T vs C
|
TT vs CC
|
TC vs CC
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | P–H | I2, % | OR (95% CI) | P-value | P–H | I2, % | OR (95% CI) | P-value | P–H | I2, % | |||
| (A) | ||||||||||||||
| Total | 84 | 35,802/41,541 | 0.958 (0.911–1.008) | 0.096 | 0.000 | 81.30 | 0.900 (0.813–0.987) | 0.043 | 0.000 | 78.80 | 1.005 (0.935–1.079) | 0.902 | 0.000 | 71.60 |
| Genotyping method | ||||||||||||||
| PCR | 57 | 19,301/22,204 | 0.939 (0.871–1.012) | 0.100 | 0.000 | 84.50 | 0.849 (0.732–0.986) | 0.032 | 0.000 | 81.70 | 0.987 (0.883–1.102) | 0.812 | 0.000 | 77.40 |
| Taqman | 16 | 8,565/10,286 | 1.021 (0.940–1.110) | 0.618 | 0.000 | 67.40 | 1.059 (0.894–1.253) | 0.507 | 0.000 | 65.70 | 1.053 (0.977–1.134) | 0.174 | 0.410 | 3.70 |
| Ethnicity | ||||||||||||||
| Asian | 64 | 28,337/31,932 | 0.847 (0.889–0.997) | 0.038 | 0.000 | 77.00 | 0.878 (0.788–0.977) | 0.017 | 0.000 | 76.00 | 1.012 (0.936–1.095) | 0.759 | 0.000 | 66.90 |
| Caucasian | 18 | 7,321/8,414 | 0.997 (0.842–1.181) | 0.971 | 0.000 | 90.30 | 0.974 (0.714–1.329) | 0.870 | 0.000 | 86.10 | 0.963 (0.785–1.180) | 0.714 | 0.000 | 83.90 |
| Cancer type | ||||||||||||||
| BRC | 14 | 7,760/8,811 | 0.972 (0.869–1.088) | 0.626 | 0.000 | 79.70 | 0.972 (0.869–1.088) | 0.341 | 0.000 | 72.80 | 0.979 (0.854–1.121) | 0.754 | 0.001 | 61.50 |
| CRC | 10 | 2,906/4,150 | 1.051 (0.867–1.276) | 0.611 | 0.000 | 86.50 | 1.051 (0.867–1.276) | 0.431 | 0.000 | 87.60 | 1.121 (0.832–1.510) | 0.454 | 0.000 | 81.10 |
| ESCC | 6 | 3,492/4,376 | 0.944 (0.816–1.091) | 0.435 | 0.001 | 76.80 | 0.944 (0.816–1.091) | 0.385 | 0.000 | 82.40 | 1.050 (0.878–1.255) | 0.594 | 0.040 | 57.20 |
| GC | 10 | 3,723/5,256 | 0.857 (0.663–1.109) | 0.241 | 0.000 | 93.80 | 0.857 (0.663–1.109) | 0.276 | 0.000 | 91.50 | 0.778 (0.552–1.098) | 0.153 | 0.000 | 88.70 |
| HCC | 14 | 4,988/5,962 | 0.894 (0.800–0.998) | 0.047 | 0.000 | 72.60 | 0.900 (0.813–0.997) | 0.039 | 0.000 | 70.50 | 0.981 (0.838–1.149) | 0.816 | 0.005 | 56.30 |
| HNC | 5 | 3,534/3,564 | 1.076 (1.006–1.152) | 0.033 | 0.285 | 20.40 | 1.214 (1.043–1.413) | 0.012 | 0.380 | 2.50 | 1.157 (0.922–1.451) | 0.209 | 0.003 | 75.00 |
| LC | 9 | 2,786/3,191 | 0.95 (0.854–1.058) | 0.354 | 0.022 | 55.30 | 0.840 (0.734–0.961) | 0.011 | 0.025 | 48.10 | 0.997 (0.889–1.118) | 0.961 | 0.056 | 47.20 |
| Design | ||||||||||||||
| PB | 42 | 20,691/21,533 | 0.968 (0.907–1.033) | 0.324 | 0.000 | 77.20 | 0.899 (0.777–1.017) | 0.087 | 0.000 | 74.70 | 1.018 (0.928–1.117) | 0.703 | 0.000 | 66.60 |
| HB | 42 | 15,111/20,008 | 0.945 (0.873–1.024) | 0.167 | 0.000 | 84.50 | 0.906 (0.813–0.997) | 0.211 | 0.000 | 81.90 | 0.987 (0.882–1.104) | 0.822 | 0.000 | 75.90 |
| rs11614913 | na | TT vs TC+CC
|
TT+TC vs CC
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | P–H | I2, % | OR (95% CI) | P-value | P–H | I2, % | ||
| (B) | |||||||||
| Total | 84 | 0.918 (0.851–0.989) | 0.025 | 0.000 | 75.80 | 0.974 (0.901–1.052) | 0.498 | 0.000 | 78.40 |
| Genotyping method | |||||||||
| PCR | 57 | 0.880 (0.800–0.9690) | 0.009 | 0.000 | 73.20 | 0.949 (0.842–1.069) | 0.386 | 0.000 | 82.80 |
| Taqman | 16 | 1.000 (0.858–1.166) | 0.996 | 0.000 | 71.90 | 1.063 (0.969–1.165) | 0.195 | 0.095 | 34.10 |
| Ethnicity | |||||||||
| Asian | 64 | 0.895 (0.824–0.972) | 0.008 | 0.000 | 76.50 | 0.972 (0.8396–1.005) | 0.493 | 0.000 | 72.90 |
| Caucasian | 17 | 1.015 (0.820–1.256) | 0.894 | 0.000 | 75.30 | 0.966 (0.766–1.219) | 0.772 | 0.000 | 89.30 |
| Cancer type | |||||||||
| BRC | 14 | 0.943 (0.815–1.091) | 0.429 | 0.001 | 64.40 | 0.967 (0.830–1.126) | 0.663 | 0.000 | 73.30 |
| CRC | 10 | 1.066 (0.823–1.381) | 0.628 | 0.000 | 79.00 | 1.130 (0.826–1.546) | 0.444 | 0.000 | 84.70 |
| ESCC | 6 | 0.813 (0.610–1.085) | 0.160 | 0.000 | 81.30 | 1.000 (0.822–1.216) | 0.997 | 0.008 | 67.80 |
| GC | 10 | 0.910 (0.697–1.189) | 0.489 | 0.000 | 83.90 | 0.763 (0.507–1.148) | 0.194 | 0.000 | 92.90 |
| HCC | 14 | 0.800 (0.678–0.944) | 0.008 | 0.000 | 67.40 | 0.919 (0.776–1.089) | 0.332 | 0.000 | 66.20 |
| HNC | 5 | 1.205 (0.799–1.817) | 0.375 | 0.000 | 90.10 | 1.156 (0.950–1.406) | 0.148 | 0.011 | 69.10 |
| LC | 9 | 0.858 (0.771–0.955) | 0.005 | 0.158 | 32.50 | 0.997 (0.834–1.191) | 0.973 | 0.019 | 56.20 |
| Design | |||||||||
| PB | 42 | 0.924 (0.826–1.034) | 0.170 | 0.000 | 78.10 | 0.988 (0.897–1.087) | 0.800 | 0.000 | 72.40 |
| HB | 42 | 0.912 (0.823–1.010) | 0.078 | 0.000 | 73.90 | 0.955 (0.843–1.081) | 0.465 | 0.000 | 82.70 |
Notes: Random-effects model was used when P-value of Q-test for heterogeneity test (P–H) is <0.05; otherwise, fixed-effect model was used. I2: 0%–25%, no heterogeneity; 25%–50%, modest heterogeneity; ≥50%, high heterogeneity.
Number of studies involved. Bold figures indicate statistically significant (P<0.05).
Abbreviations: BRC, breast cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; HB, hospital based; HCC, hepatocellular carcinoma; HNC, head and neck cancer; LC, lung cancer; OR, odds ratio; PB, population based; PCR, polymerase chain reaction; P–H, P-value of heterogeneity test.
Figure 2.
Forest plots of the association between miR-196a2 rs11614913 polymorphism and cancer risk in different cancer types for homozygote comparison (TT vs CC).
Note: Weights are from random effects analysis.
Abbreviations: BRC, breast cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; HCC, hepatocellular carcinoma; HNC, head and neck cancer; LC, lung cancer; miR-196a2, microRNA-196a2; OR, odds ratio.
Figure 3.
Forest plots of the association between miR-196a2 rs11614913 polymorphism and cancer risk in different cancer types for recessive model (TT vs TC+CC).
Note: Weights are from random effects analysis.
Abbreviations: BRC, breast cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; HCC, hepatocellular carcinoma; HNC, head and neck cancer; LC, lung cancer; miR-196a2, microRNA-196a2; OR, odds ratio.
In ethnic subgroup analysis, a strong association was found between rs11614913 and cancer risk in the allelic contrast (T vs C: OR =0.847, 95% CI =0.899–0.997, P=0.038), the homozygote comparison (TT vs CC: OR =0.878, 95% CI =0.788–0.977, P=0.017), and the recessive model (OR =0.895, 95% CI =0.824–0.972, P=0.008) among Asians, whereas negative results were obtained for Caucasians in all genetic models. Additionally, decreased risk was observed in the polymerase chain reaction (PCR) method for the homozygote comparison (TT vs CC: OR =0.849, 95% CI =0.732–0.986, P=0.032) and the recessive model (TT vs TC+CC: OR =0.880, 95% CI =0.800–0.969, P=0.009), and no significant association of cancer risk was found in Taqman and other methods.
Test of heterogeneity
Among the studies of rs11614913, we found heterogeneity in overall comparisons and subgroup analysis. Moreover, the heterogeneity we evaluated for all genetic models by ethnicity, cancer type, source of controls, as well HWE status was significant. However, we found that heterogeneity could not be explained by the variable ethnicity, cancer type, source of controls, and HWE status (data not shown).
Sensitivity analysis
Sensitivity analysis was conducted to assess the effect by excluding a single study in turn. Sensitivity analysis of the rs11614913 polymorphism in an allelic comparison is presented in Table S1. Overall, we found that no individual study had an influence on the pooled OR. The results demonstrated that the pooled ORs were not materially altered, suggesting the stability of our meta-analysis.
Publication bias
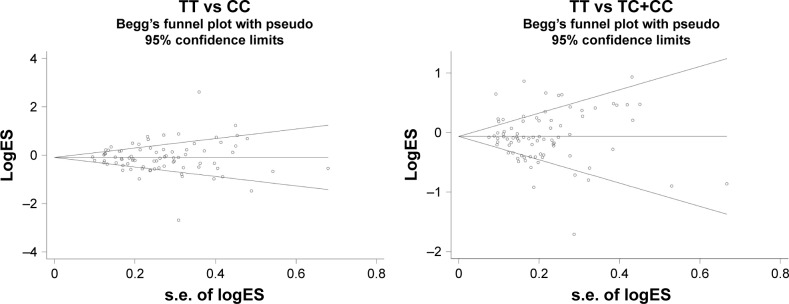
The publication bias of the present meta-analysis was assessed by Begg’s funnel plot and Egger’s test. The funnel plot for the rs11614913 polymorphism in the allelic comparison is presented in Table S2. No evidence of publication bias was noted in Begg’s funnel plot (T vs C [P-value for Begg’s test =0.660], TT vs CC [P-value for Begg’s test =0.971, Figure 4], TC vs CC [P-value for Begg’s test =0.951], TT vs TC+CC [P-value for Begg’s test =0.908, Figure 4], TC+TT vs CC [P-value for Begg’s test =0.592]) and Egger’s test (allele contrast [P=0.923], homozygous model [P=0.822], heterozygous model [P=0.761], recessive model [P=0.899], and dominant model [P=0.401]). The quality of included studies is presented in Table 3.
Figure 4.
Begg’s funnel plot for publication bias of miR-196a2 rs11614913 polymorphism and cancer risk by homozygote comparison and recessive model.
Notes: Each point represents a separate study for the indicated association. LogES represents natural logarithm of OR. Horizontal line means magnitude of the effect. Funnel plot with pseudo 95% confidence limits was used.
Abbreviations: miR-196a2, microRNA-196a2; OR, odds ratio.
Table 3.
Methodological quality of the included studies according to the Newcastle–Ottawa scale
| Author | Adequacy of case definition | Representativeness of the cases | Selection of controls | Definition of controls | Comparability of cases/controls | Ascertainment of exposure | Same method of ascertainment | Non-response rate |
|---|---|---|---|---|---|---|---|---|
| Hu et al7 | * | * | * | * | ** | * | * | NA |
| Hu et al35 | * | * | NA | * | ** | * | * | NA |
| Tian et al3 | * | * | NA | * | * | * | * | NA |
| Hoffman et al5 | * | * | * | * | * | * | * | NA |
| Catucci et al36 | * | * | NA | * | ** | NA | * | NA |
| Wang et al38 | * | * | NA | * | ** | * | * | NA |
| Okubo et al83 | * | * | * | * | ** | * | * | NA |
| Peng et al4 | * | * | NA | * | ** | NA | * | NA |
| Srivastava et al10 | * | * | NA | * | ** | * | * | NA |
| Dou et al6 | * | * | NA | NA | * | NA | * | NA |
| Li et al9 | * | * | * | * | ** | NA | * | NA |
| Akkiz et al8 | * | * | NA | * | ** | NA | * | NA |
| Liu et al11 | * | * | NA | * | * | * | * | NA |
| Kim et al110 | * | * | NA | NA | * | * | * | NA |
| Catucci et al36 | * | * | * | * | ** | * | * | NA |
| Christensen et al37 | * | * | NA | * | ** | * | * | NA |
| Mittal et al41 | * | * | NA | * | ** | * | * | NA |
| Jedlinski et al40 | * | * | * | * | ** | NA | * | NA |
| Zhan et al42 | * | * | NA | * | * | NA | * | NA |
| Zhou et al43 | * | * | NA | * | ** | NA | * | NA |
| Vinci et al111 | * | * | NA | * | ** | * | * | NA |
| Hong et al2 | * | * | NA | * | * | * | * | NA |
| George et al39 | * | * | NA | * | ** | * | * | NA |
| Linhares et al45 | * | * | NA | * | ** | * | * | NA |
| Chen et al44 | * | * | NA | * | ** | NA | * | NA |
| Min et al24 | * | * | NA | * | ** | * | * | NA |
| Zhu et al47 | * | * | NA | * | ** | * | * | NA |
| Hezova et al25 | * | * | NA | * | ** | NA | * | NA |
| Zhang et al100 | * | * | * | * | ** | * | * | NA |
| Ahn et al48 | * | * | NA | * | ** | * | * | NA |
| Yoon et al46 | * | * | NA | * | ** | * | * | NA |
| Zhang et al104 | * | * | * | * | ** | NA | * | NA |
| Chu et al87 | * | * | NA | * | ** | NA | * | NA |
| Vinci et al113 | * | * | * | * | ** | NA | * | NA |
| Lv et al51 | * | * | * | * | ** | NA | * | NA |
| Umar et al112 | * | * | NA | NA | ** | * | * | NA |
| Wei et al114 | * | * | NA | * | ** | * | * | NA |
| Toraih et al98 | * | * | NA | * | ** | * | * | NA |
| Wang et al53 | * | * | NA | * | ** | NA | * | NA |
| Zhang et al55 | * | * | NA | NA | ** | NA | * | NA |
| Han et al49 | * | * | * | * | ** | * | * | NA |
| Tong et al65 | * | * | NA | * | ** | * | * | NA |
| Pavlakis et al93 | * | * | NA | * | ** | * | * | NA |
| Pu et al84 | * | * | * | * | ** | NA | * | NA |
| Bansal et al56 | * | * | NA | * | ** | * | * | NA |
| Kupcinskas et al62 | * | * | * | * | ** | * | * | NA |
| Qu et al64 | * | * | NA | NA | ** | * | * | NA |
| Wang et al66 | * | * | NA | * | ** | * | * | NA |
| Dikeakos et al58 | * | * | NA | * | ** | * | * | NA |
| Qi et al86 | * | * | NA | * | ** | NA | * | NA |
| Chu et al57 | * | * | * | * | * | * | * | NA |
| Parlayan et al115 | * | * | * | * | ** | * | * | NA |
| Li et al63 | * | * | NA | * | ** | * | * | NA |
| Du et al59,60 | * | * | NA | * | * | NA | * | NA |
| Omrani et al85 | * | * | NA | * | ** | * | * | NA |
| Kou et al91 | * | * | * | * | ** | * | * | NA |
| Roy et al94 | * | * | NA | * | ** | * | * | NA |
| Li et al63 | * | * | NA | * | ** | NA | * | NA |
| Deng et al67 | * | * | * | * | ** | NA | * | NA |
| Qi et al72 | * | * | NA | * | ** | NA | * | NA |
| Dikaiakos et al68 | * | * | * | * | * | * | * | NA |
| Li et al69 | * | * | NA | NA | ** | * | * | NA |
| Li et al69 | * | * | NA | NA | ** | * | * | NA |
| Nikolic et al71 | * | * | * | * | ** | * | * | NA |
| He et al90 | * | * | NA | NA | ** | NA | * | NA |
| Sushma et al97 | * | * | NA | * | ** | * | * | NA |
| Sodhi et al95 | * | * | * | * | ** | * | * | NA |
| Jiang et al26 | * | * | NA | * | ** | * | * | NA |
| Dai et al74 | * | * | NA | * | ** | NA | * | NA |
| Zhao et al82 | * | * | NA | * | ** | * | * | NA |
| Song et al79 | * | * | * | * | * | NA | * | NA |
| Shen et al78 | * | * | NA | * | ** | * | * | NA |
| Li et al75 | * | * | NA | * | ** | NA | * | NA |
| Li et al76 | * | * | NA | * | * | * | * | NA |
| Xu et al80 | * | * | NA | NA | * | * | * | NA |
| Qiu and Liu77 | * | * | * | * | * | * | * | NA |
| Jiang et al26 | * | * | * | * | ** | * | * | NA |
| Yin et al81 | * | * | NA | * | * | * | * | NA |
| Zhang et al99 | * | * | * | * | ** | NA | * | NA |
| Sun et al96 | * | * | * | * | * | * | * | NA |
| Toraih et al98 | * | * | NA | * | ** | NA | * | NA |
| Morales et al92 | * | * | NA | * | ** | * | * | NA |
| Gu and Tu88 | * | * | NA | * | * | * | * | NA |
| Hashemi et al89 | * | * | NA | * | ** | * | * | NA |
Notes: This table identified “high”quality choices with a “*”. A study can be awarded a maximum of one “*” for each numbered item within the selection and exposure categories. A maximum of two “**” can be given for comparability.
Abbreviation: NA, not available.
Discussion
MiRNAs are reported as critical posttranscriptional regulators in gene expression and are involved in various diseases. The associations between miR-196a2 rs11614913 polymorphism and susceptibility to different cancers are widely explored. Guo et al101 found that the C allele had the effect of increasing cancer risk in gastric cancer, and Ma et al102 found that TT could decrease the risk of colorectal cancer. Moreover, Wang et al103 and Zhang et al104 showed that the rs11614913 polymorphism has no association with the risk of hepatocellular carcinoma. However, the regulatory effects of miRNA in carcinogenesis remain unclear. Therefore, we performed this updated meta-analysis to explore the molecular mechanisms of the genetic associations between miRNA and SNPs with cancer risk.
MiR-196a2 is composed of two distinct mature miRNAs (miR-196a-3P and miR-196a-5P), which are processed from the same stem loop;105 thus, the potential targets of miR-196a could be influenced by its altered expression patterns. SNPs in miRNAs could potentially affect the processing or target selection of miRNAs,106,107 which is identified as a key factor in oncogenesis, and contributes to regulate the translation or degradation of messenger RNA (mRNA).23 Hoffman et al5 found that the expression of mature miR-196a2 was increased 9.3-fold in cells transfected with pre-miR-196a2-C but upregulated only by 4.4-fold with pre-miR-196a2-T, and that the C allele of rs11614913 increased mature miR-196a2 levels in lung cancer7 and CRC42 tissues. Xu et al108 have shown that miR-196a2 rs11614913 CC is associated with significantly increased expression of mature miR-196a (lower cycle threshold corresponding to a higher expression) in cardiac tissue specimens of congenital heart disease, and the increased miR-196a expression could further decrease mRNA target of HOXB8. These results indicated that the rs11614913 polymorphism may affect the processing of the pre-miRNA to its mature form.
Several meta-analyses have been performed to analyse the SNP of this miRNA that is associated with the cancer risk.104,109 In our present work, we screened out all the studies published to date and included more papers and cancer types than the previously published meta-analyses. For example, Kang et al109 conducted a meta-analysis encompassing the rs11614913 polymorphism in miR-196a2 and cancer risks, which suggested that the rs11614913 polymorphism may contribute to decreased susceptibility to liver cancer (allele model, homozygous model, dominant model, and heterozygous model) and lung cancer (allele model, homozygous model, and recessive model); however, this was not duplicated in our meta-analysis. In this study, we concluded that the rs11614913 polymorphism conferred a decreased susceptibility to lung cancer (homozygote comparison, recessive model) and hepatocellular carcinoma (allelic contrast, homozygote comparison, recessive model) or an increased susceptibility to HNC (allelic contrast, homozygote comparison). Our study had a larger sample size than the previous ones, which might influence the results. In addition, the previous meta-analyses did not evaluate the quality of the included studies.
According to the procedure of seeking for the source of heterogeneity, we performed subgroup studies according to cancer type, ethnicity, and source of control. A strong association was found between rs11614913 and cancer risk in lung cancers, hepatocellular carcinoma, and HNC, but not in breast cancer, gastric cancer, ESCC, or CRC, which was not similar to the findings of previous studies.101–103,109 The present meta-analysis showed that homozygote TT had the effect of decreasing the risk of lung cancer or hepatocellular carcinoma compared with that of CC homozygote or C allele carriers. We conducted another subgroup analysis by population to determine the association between these miRNA polymorphisms and tumorigenesis. The results suggested that individuals with alterative T allele could decrease cancer susceptibility in Asians but not in Caucasians, indicating that the difference of ethnic background and the living environment may also be a risk factor.
To determine the hsa-miR-196a2 rs11614913 polymorphism, PCR, Taqman, and other methods have been adopted. We found that the hsa-miR-196a2 rs11614913 polymorphism significantly decreased cancer risk in homozygous models and the recessive model when using the PCR method, but this result was not shown when selecting Taqman and other methods. Therefore, more effort may be necessary for further progress in SNP analysis. We found sources of heterogeneity in the studies from cancer type and ethnicity suggesting cancer and population playing important roles. When detecting the source of control, we observed significant associations in population-based and hospital-based controls. This may be due to the included studies matching age, gender, and residential area to control selection bias.
Nevertheless, several defects of this meta-analysis should be emphasized. Firstly, although we strictly screened articles and precisely extracted the data, the differences in the selection of subjects could not be eliminated. Secondly, in our meta-analysis, only Asian and Caucasian ethnicities were included, and the impact of the differences in racial descent should not be ignored. Thirdly, potential language bias could not be avoided due to limitation of studies published in English or Chinese. Therefore, it is not possible to avoid potential publication bias in this meta-analysis.
In summary, miR-196a2 rs11614913 polymorphism may contribute to the development of cancer, especially in lung cancer, hepatocellular carcinoma, and HNC. It might be useful as a candidate marker for the diagnosis of these cancers, and could also be a potential protective factor for cancer risks in Asians. Furthermore, more significant studies and investigations with larger populations focusing on cancer types or ethnicities should be performed to confirm the results.
Supplementary materials
Table S1.
Details of the sensitivity analyses of the association between rs11614913 polymorphism and cancer risk homozygous model (TT vs CC) and recessive model (TT vs TC+CC).
| Comparison | Study omitted | Estimate | (95% Conf Interval)
|
|
|---|---|---|---|---|
| Lower CI | Upper CI | |||
| TT vs CC | Hu et al7 | 0.902 | 0.814 | 0.999 |
| Hu et al35 | 0.904 | 0.815 | 1.002 | |
| Tian et al3 | 0.902 | 0.814 | 1.001 | |
| Hoffman et al5 | 0.890 | 0.805 | 0.985 | |
| Catucci et al36 | 0.900 | 0.811 | 1.000 | |
| Wang et al38 | 0.911 | 0.824 | 1.008 | |
| Okubo et al83 | 0.900 | 0.812 | 0.998 | |
| Peng et al4 | 0.904 | 0.816 | 1.002 | |
| Srivastava et al10 | 0.903 | 0.815 | 1.000 | |
| Dou et al6 | 0.897 | 0.809 | 0.994 | |
| Li et al9 | 0.906 | 0.818 | 1.003 | |
| Akkiz et al8 | 0.908 | 0.820 | 1.005 | |
| Liu et al11 | 0.898 | 0.810 | 0.997 | |
| Kim et al101 | 0.904 | 0.815 | 1.002 | |
| Catucci et al36 | 0.899 | 0.810 | 0.997 | |
| Christensen et al37 | 0.900 | 0.813 | 0.997 | |
| Mittal et al41 | 0.904 | 0.816 | 1.001 | |
| Jedlinski et al40 | 0.900 | 0.813 | 0.998 | |
| Zhan et al42 | 0.906 | 0.818 | 1.004 | |
| Zhou et al43 | 0.901 | 0.813 | 0.998 | |
| Vinci et al102 | 0.895 | 0.809 | 0.992 | |
| Hong et al2 | 0.902 | 0.814 | 1.000 | |
| George et al39 | 0.902 | 0.815 | 0.999 | |
| Linhares et al45 | 0.893 | 0.806 | 0.988 | |
| Chen et al44 | 0.898 | 0.811 | 0.995 | |
| Min et al24 | 0.904 | 0.815 | 1.002 | |
| Zhu et al47 | 0.905 | 0.816 | 1.003 | |
| Hezova et al25 | 0.897 | 0.810 | 0.994 | |
| Zhang et al100 | 0.900 | 0.812 | 0.998 | |
| Yoon et al46 | 0.904 | 0.816 | 1.001 | |
| Zhang et al99 | 0.904 | 0.816 | 1.001 | |
| Chu et al87 | 0.894 | 0.807 | 0.990 | |
| Vinci et al105 | 0.897 | 0.810 | 0.994 | |
| Ahn et al103 | 0.902 | 0.814 | 1.000 | |
| Lv et al51 | 0.878 | 0.798 | 0.965 | |
| Umar et al104 | 0.895 | 0.808 | 0.992 | |
| Wei et al106 | 0.896 | 0.809 | 0.993 | |
| Wang et al53 | 0.894 | 0.807 | 0.990 | |
| Zhang et al55 | 0.904 | 0.816 | 1.003 | |
| Han et al49 | 0.898 | 0.810 | 0.996 | |
| Pavlakis et al93 | 0.899 | 0.812 | 0.996 | |
| Tong et al65 | 0.901 | 0.813 | 1.000 | |
| Pu et al84 | 0.902 | 0.814 | 1.000 | |
| Bansal et al56 | 0.902 | 0.815 | 1.000 | |
| Kupcinskas et al62 | 0.897 | 0.809 | 0.994 | |
| Qu et al64 | 0.905 | 0.817 | 1.003 | |
| Wang et al66 | 0.897 | 0.809 | 0.994 | |
| Dikeakos et al58 | 0.925 | 0.843 | 1.015 | |
| Qi et al86 | 0.902 | 0.814 | 1.000 | |
| Chu et al57 | 0.898 | 0.810 | 0.995 | |
| Parlayan et al107 | 0.900 | 0.812 | 0.997 | |
| Li et al63 | 0.896 | 0.808 | 0.993 | |
| Du et al59 | 0.892 | 0.806 | 0.987 | |
| Omrani et al85 | 0.900 | 0.813 | 0.997 | |
| Kou et al91 | 0.907 | 0.819 | 1.004 | |
| Roy et al94 | 0.896 | 0.809 | 0.993 | |
| Li et al63 | 0.896 | 0.808 | 0.993 | |
| Deng et al67 | 0.900 | 0.812 | 0.997 | |
| Qi et al72 | 0.907 | 0.819 | 1.005 | |
| Dikaiakos et al68 | 0.899 | 0.812 | 0.996 | |
| Li et al69 | 0.890 | 0.805 | 0.985 | |
| Li et al69 | 0.907 | 0.819 | 1.004 | |
| Nikolic et al71 | 0.902 | 0.814 | 1.000 | |
| He et al90 | 0.901 | 0.813 | 0.999 | |
| Sushma et al97 | 0.909 | 0.821 | 1.006 | |
| Sodhi et al95 | 0.891 | 0.806 | 0.986 | |
| Jiang et al26 | 0.896 | 0.808 | 0.993 | |
| Toraih et al98 | 0.894 | 0.807 | 0.990 | |
| Dai et al74 | 0.908 | 0.820 | 1.005 | |
| Zhao et al82 | 0.898 | 0.811 | 0.995 | |
| Song et al79 | 0.907 | 0.819 | 1.004 | |
| Shen et al78 | 0.902 | 0.813 | 1.002 | |
| Li et al75 | 0.907 | 0.820 | 1.005 | |
| Li et al76 | 0.906 | 0.819 | 1.004 | |
| Xu et al80 | 0.906 | 0.818 | 1.004 | |
| Qiu et al77 | 0.905 | 0.817 | 1.003 | |
| Jiang et al26 | 0.901 | 0.813 | 1.000 | |
| Yin et al81 | 0.901 | 0.813 | 0.999 | |
| Zhang et al99 | 0.901 | 0.813 | 0.998 | |
| Sun et al96 | 0.904 | 0.817 | 1.002 | |
| Toraih et al98 | 0.894 | 0.808 | 0.990 | |
| Morales et al92 | 0.901 | 0.812 | 0.999 | |
| Gu et al88 | 0.891 | 0.805 | 0.986 | |
| Hashemi et al89 | 0.896 | 0.809 | 0.992 | |
| Combined2–10,25,26,35–107 | 0.900 | 0.813 | 0.997 | |
| TT vs TC+CC | Hu et al7 | 0.918 | 0.851 | 0.991 |
| Hu et al35 | 0.920 | 0.852 | 0.993 | |
| Tian et al3 | 0.918 | 0.850 | 0.991 | |
| Hoffman et al5 | 0.910 | 0.844 | 0.980 | |
| Catucci et al36 | 0.917 | 0.849 | 0.991 | |
| Wang et al38 | 0.928 | 0.862 | 0.999 | |
| Okubo et al83 | 0.917 | 0.850 | 0.991 | |
| Peng et al4 | 0.919 | 0.852 | 0.991 | |
| Srivastava et al10 | 0.918 | 0.850 | 0.990 | |
| Dou et al6 | 0.918 | 0.850 | 0.991 | |
| Li et al9 | 0.922 | 0.854 | 0.994 | |
| Akkiz et al8 | 0.923 | 0.856 | 0.995 | |
| Liu et al11 | 0.917 | 0.849 | 0.990 | |
| Kim et al101 | 0.920 | 0.852 | 0.992 | |
| Catucci et al36 | 0.916 | 0.849 | 0.989 | |
| Christensen et al37 | 0.918 | 0.851 | 0.989 | |
| Mittal et al41 | 0.921 | 0.854 | 0.993 | |
| Jedlinski et al40 | 0.917 | 0.850 | 0.989 | |
| Zhan et al42 | 0.922 | 0.854 | 0.994 | |
| Zhou et al43 | 0.918 | 0.850 | 0.990 | |
| Vinci et al102 | 0.915 | 0.849 | 0.987 | |
| Hong et al2 | 0.922 | 0.854 | 0.994 | |
| George et al39 | 0.920 | 0.853 | 0.992 | |
| Linhares et al45 | 0.913 | 0.847 | 0.985 | |
| Chen et al44 | 0.916 | 0.849 | 0.988 | |
| Min et al24 | 0.918 | 0.850 | 0.990 | |
| Zhu et al47 | 0.921 | 0.854 | 0.994 | |
| Hezova et al25 | 0.915 | 0.848 | 0.987 | |
| Zhang et al100 | 0.918 | 0.850 | 0.991 | |
| Yoon et al46 | 0.920 | 0.853 | 0.993 | |
| Zhang et al99 | 0.919 | 0.852 | 0.992 | |
| Chu et al87 | 0.918 | 0.851 | 0.991 | |
| Vinci et al105 | 0.919 | 0.851 | 0.991 | |
| Ahn et al103 | 0.916 | 0.850 | 0.988 | |
| Lv et al51 | 0.905 | 0.842 | 0.974 | |
| Umar et al104 | 0.914 | 0.848 | 0.986 | |
| Wei et al106 | 0.918 | 0.850 | 0.990 | |
| Wang et al53 | 0.913 | 0.846 | 0.985 | |
| Zhang et al55 | 0.919 | 0.851 | 0.992 | |
| Han et al49 | 0.917 | 0.849 | 0.990 | |
| Pavlakis et al93 | 0.921 | 0.854 | 0.994 | |
| Tong et al65 | 0.913 | 0.847 | 0.985 | |
| Pu et al84 | 0.918 | 0.851 | 0.990 | |
| Bansal et al56 | 0.919 | 0.852 | 0.991 | |
| Kupcinskas et al62 | 0.916 | 0.849 | 0.988 | |
| Qu et al64 | 0.923 | 0.855 | 0.995 | |
| Wang et al66 | 0.916 | 0.848 | 0.988 | |
| Dikeakos et al58 | 0.931 | 0.866 | 1.001 | |
| Qi et al86 | 0.924 | 0.857 | 0.996 | |
| Chu et al57 | 0.914 | 0.847 | 0.986 | |
| Parlayan et al107 | 0.918 | 0.851 | 0.990 | |
| Li et al63 | 0.913 | 0.846 | 0.985 | |
| Du et al59 | 0.914 | 0.847 | 0.986 | |
| Omrani et al85 | 0.918 | 0.851 | 0.989 | |
| Kou et al91 | 0.921 | 0.854 | 0.994 | |
| Roy et al94 | 0.915 | 0.848 | 0.987 | |
| Li et al63 | 0.906 | 0.845 | 0.971 | |
| Deng et al67 | 0.913 | 0.847 | 0.985 | |
| Qi et al72 | 0.923 | 0.856 | 0.995 | |
| Dikaiakos et al68 | 0.914 | 0.848 | 0.987 | |
| Li et al69 | 0.911 | 0.845 | 0.982 | |
| Li et al69 | 0.922 | 0.855 | 0.995 | |
| Nikolic et al71 | 0.919 | 0.852 | 0.991 | |
| He et al90 | 0.917 | 0.850 | 0.990 | |
| Sushma et al97 | 0.921 | 0.855 | 0.994 | |
| Sodhi et al95 | 0.913 | 0.847 | 0.984 | |
| Jiang et al26 | 0.914 | 0.847 | 0.986 | |
| Toraih et al98 | 0.914 | 0.848 | 0.986 | |
| Dai et al74 | 0.922 | 0.855 | 0.995 | |
| Zhao et al82 | 0.914 | 0.848 | 0.986 | |
| Song et al79 | 0.923 | 0.856 | 0.995 | |
| Shen et al78 | 0.918 | 0.849 | 0.992 | |
| Li et al75 | 0.921 | 0.854 | 0.993 | |
| Li et al76 | 0.923 | 0.856 | 0.995 | |
| Xu et al80 | 0.922 | 0.854 | 0.994 | |
| Qiu et al77 | 0.921 | 0.854 | 0.993 | |
| Jiang et al26 | 0.921 | 0.854 | 0.994 | |
| Yin et al81 | 0.919 | 0.851 | 0.992 | |
| Zhang et al99 | 0.918 | 0.851 | 0.991 | |
| Sun et al96 | 0.919 | 0.852 | 0.992 | |
| Toraih et al98 | 0.915 | 0.848 | 0.986 | |
| Morales et al92 | 0.918 | 0.851 | 0.991 | |
| Gu et al88 | 0.911 | 0.845 | 0.982 | |
| Hashemi et al89 | 0.915 | 0.848 | 0.986 | |
| Combined2–10,25,26,35–107 | 0.918 | 0.851 | 0.989 | |
Table S2.
P-values of Begg’s and Egger’s test for the polymorphism rs11614913
| Polymorphism | Comparison | Subgroup | Begg’s test (P>z) |
Egger’s test (P>t) |
|---|---|---|---|---|
| rs11614913 | T vs C | Overall | 0.660 | 0.923 |
| Taqman | 0.368 | 0.723 | ||
| PCR | 0.640 | 0.859 | ||
| Asian | 0.946 | 0.854 | ||
| Caucasian | 0.147 | 0.969 | ||
| HB | 0.509 | 0.386 | ||
| PB | 0.251 | 0.579 | ||
| TT vs CC | Overall | 0.971 | 0.822 | |
| Taqman | 0.719 | 0.606 | ||
| PCR | 0.832 | 0.762 | ||
| Asian | 0.578 | 0.758 | ||
| Caucasian | 0.163 | 0.971 | ||
| HB | 0.721 | 0.489 | ||
| PB | 0.666 | 0.880 | ||
| TC vs CC | Overall | 0.951 | 0.761 | |
| Taqman | 0.418 | 0.289 | ||
| PCR | 0.839 | 0.933 | ||
| Asian | 0.991 | 0.546 | ||
| Caucasian | 0.902 | 0.767 | ||
| HB | 0.721 | 0.601 | ||
| PB | 0.965 | 0.453 | ||
| TT+TC vs CC | Overall | 0.592 | 0.401 | |
| Taqman | 0.418 | 0.613 | ||
| PCR | 0.734 | 0.598 | ||
| Asian | 0.986 | 0.185 | ||
| Caucasian | 0.300 | 0.770 | ||
| HB | 0.737 | 0.543 | ||
| PB | 0.584 | 0.593 | ||
| TT vs TC+CC | Overall | 0.908 | 0.899 | |
| Taqman | 0.719 | 0.440 | ||
| PCR | 0.912 | 0.917 | ||
| Asian | 0.795 | 0.688 | ||
| Caucasian | 0.537 | 0.857 | ||
| HB | 0.673 | 0.503 | ||
| PB | 0.914 | 0.508 |
Abbreviations: HB, hospital based; PB, population based; PCR, polymerase chain reaction.
References
- 1.Liu CJ, Tsai MM, Tu HF, Lui MT, Cheng HW, Lin SC. miR-196a overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Ann Surg Oncol. 2013;20(Suppl 3):S406–S414. doi: 10.1245/s10434-012-2618-6. [DOI] [PubMed] [Google Scholar]
- 2.Hong YS, Kang HJ, Kwak JY, et al. Association between microR-NA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean population. J Prev Med Public Health. 2011;44(3):125–130. doi: 10.3961/jpmph.2011.44.3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiology, Biomarkers & Prev. 2009;18(4):1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 4.Peng S, Kuang Z, Sheng C, Zhang Y, Xu H, Cheng Q. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2010;55(8):2288–2293. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman AE, Zheng T, Yi C, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer research. 2009;69(14):5970–5977. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dou T, Wu Q, Chen X, et al. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol. 2010;136(12):1853–1859. doi: 10.1007/s00432-010-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118(7):2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akkiz H, Bayram S, Bekar A, Akgollu E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepat. 2011;18(7):e399–e407. doi: 10.1111/j.1365-2893.2010.01414.x. [DOI] [PubMed] [Google Scholar]
- 9.Li XD, Li ZG, Song XX, Liu CF. A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. Pathology. 2010;42(7):669–673. doi: 10.3109/00313025.2010.522175. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava K, Srivastava A, Mittal B. Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J Hum Genet. 2010;55(8):495–499. doi: 10.1038/jhg.2010.54. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Li G, Wei S, et al. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer. 2010;116(20):4753–4760. doi: 10.1002/cncr.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42(8):1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC cancer. 2007;7:162. doi: 10.1186/1471-2407-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285(2):116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 21.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 22.Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29(7):1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 23.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nature Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min KT, Kim JW, Jeon YJ, et al. Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog. 2012;51(Suppl 1):E65–E73. doi: 10.1002/mc.21849. [DOI] [PubMed] [Google Scholar]
- 25.Hezova R, Kovarikova A, Bienertova-Vasku J, et al. Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J Gastroenterol. 2012;18(22):2827–2831. doi: 10.3748/wjg.v18.i22.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang J, Jia ZF, Cao DH, Wu YH, Sun ZW, Cao XY. Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Future Oncol. 2016;12(19):2215–2226. doi: 10.2217/fon-2016-0224. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y, Yu CY, Wang JL, Guan J, Chen HY, Fang JY. MicroRNA sequence polymorphisms and the risk of different types of cancer. Sci Rep. 2014;4:3648. doi: 10.1038/srep03648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 29.Woolf B. On estimating the relation between blood group and disease. Ann Hum Genet. 1955;19(4):251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Zhan Y, Chen Z, et al. Directing cellular information flow via CRISPR signal conductors. Nat Methods. 2016;13(11):938–944. doi: 10.1038/nmeth.3994. [DOI] [PubMed] [Google Scholar]
- 31.Vangel MG, Rukhin AL. Maximum likelihood analysis for heteroscedastic one-way random effects ANOVA in interlaboratory studies. Biometrics. 1999;55(1):129–136. doi: 10.1111/j.0006-341x.1999.00129.x. [DOI] [PubMed] [Google Scholar]
- 32.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28(2):105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, Liang J, Wang Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30(1):79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 36.Catucci I, Yang R, Verderio P, et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat. 2010;31(1):E1052–E1057. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- 37.Christensen BC, Avissar-Whiting M, Ouellet LG, et al. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res. 2010;16(14):3713–3720. doi: 10.1158/1078-0432.CCR-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Guo H, Hu H, et al. A functional variation in pre-microRNA-196a is associated with susceptibility of esophageal squamous cell carcinoma risk in Chinese Han. Biomarkers. 2010;15(7):614–618. doi: 10.3109/1354750X.2010.505299. [DOI] [PubMed] [Google Scholar]
- 39.George GP, Gangwar R, Mandal RK, Sankhwar SN, Mittal RD. Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol Biol Rep. 2011;38(3):1609–1615. doi: 10.1007/s11033-010-0270-4. [DOI] [PubMed] [Google Scholar]
- 40.Jedlinski DJ, Gabrovska PN, Weinstein SR, Smith RA, Griffiths LR. Single nucleotide polymorphism in hsa-mir-196a-2 and breast cancer risk: a case control study. Twin Res Hum Genet. 2011;14(5):417–421. doi: 10.1375/twin.14.5.417. [DOI] [PubMed] [Google Scholar]
- 41.Mittal RD, Gangwar R, George GP, Mittal T, Kapoor R. Investigative role of pre-microRNAs in bladder cancer patients: a case-control study in North India. DNA Cell Biol. 2011;30(6):401–406. doi: 10.1089/dna.2010.1159. [DOI] [PubMed] [Google Scholar]
- 42.Zhan JF, Chen LH, Chen ZX, et al. A functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese population. Archiv Med Res. 2011;42(2):144–148. doi: 10.1016/j.arcmed.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Zhou B, Wang K, Wang Y, et al. Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Mol Carcinog. 2011;50(7):499–505. doi: 10.1002/mc.20740. [DOI] [PubMed] [Google Scholar]
- 44.Chen H, Sun LY, Chen LL, Zheng HQ, Zhang QF. A variant in micro RNA-196a2 is not associated with susceptibility to and progression of colorectal cancer in Chinese. Int Med J. 2012;42(6):e115–e119. doi: 10.1111/j.1445-5994.2011.02434.x. [DOI] [PubMed] [Google Scholar]
- 45.Linhares JJ, Azevedo M, Jr, Siufi AA, et al. Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC Med Genet. 2012;13:119. doi: 10.1186/1471-2350-13-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon KA, Yoon H, Park S, et al. The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancer. J Thorac Cardiovasc Surg. 2012;144(4):794–807. doi: 10.1016/j.jtcvs.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 47.Zhu L, Chu H, Gu D, et al. A functional polymorphism in miRNA-196a2 is associated with colorectal cancer risk in a Chinese population. DNA Cell Biol. 2012;31(3):350–354. doi: 10.1089/dna.2011.1348. [DOI] [PubMed] [Google Scholar]
- 48.Ahn DH, Rah H, Choi Y-K, et al. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. 2013;52(S1):39–51. doi: 10.1002/mc.21962. [DOI] [PubMed] [Google Scholar]
- 49.Han Y, Pu R, Han X, et al. Associations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma risk. PLoS One. 2013;8(3):e58564. doi: 10.1371/journal.pone.0058564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong MJ, Choi YY, Jang JA, et al. Association between genetic variants in pre-microRNAs and survival of early-stage NSCLC. J Thorac Oncol. 2013;8(6):703–710. doi: 10.1097/JTO.0b013e318288dc0a. [DOI] [PubMed] [Google Scholar]
- 51.Lv M, Dong W, Li L, et al. Association between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese population. J Cancer Res Clin Oncol. 2013;139(8):1405–1410. doi: 10.1007/s00432-013-1456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song X, Sturgis EM, Liu J, et al. MicroRNA variants increase the risk of HPV-associated squamous cell carcinoma of the oropharynx in never smokers. PLoS One. 2013;8(2):e56622. doi: 10.1371/journal.pone.0056622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Tao G, Wu D, et al. A functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancer. Mol Carcinog. 2013;52(Suppl 1):E87–E95. doi: 10.1002/mc.22017. [DOI] [PubMed] [Google Scholar]
- 54.Yuan Z, Zeng X, Yang D, Wang W, Liu Z. Effects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer risk. PLoS One. 2013;8(4):e61047. doi: 10.1371/journal.pone.0061047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Wang R, Ma Y-Y, et al. Association Between Single Nucleotide Polymorphisms in miRNA196a-2 and miRNA146a and Susceptibility to Hepatocellular Carcinoma in a Chinese Population. Asian Pac J Cancer Prev. 2013;14(11):6427–6431. doi: 10.7314/apjcp.2013.14.11.6427. [DOI] [PubMed] [Google Scholar]
- 56.Bansal C, Sharma KL, Misra S, Srivastava AN, Mittal B, Singh US. Common genetic variants in pre-microRNAs and risk of breast cancer in the North Indian population. Ecancermedicalscience. 2014;8:473. doi: 10.3332/ecancer.2014.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu YH, Hsieh MJ, Chiou HL, et al. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One. 2014;9(2):e89930. doi: 10.1371/journal.pone.0089930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dikeakos P, Theodoropoulos G, Rizos S, Tzanakis N, Zografos G, Gazouli M. Association of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek population. Mol Biol Rep. 2014;41(2):1075–1080. doi: 10.1007/s11033-013-2953-0. [DOI] [PubMed] [Google Scholar]
- 59.Du M, Lu D, Wang Q, et al. Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis. 2014;35(7):1629–1635. doi: 10.1093/carcin/bgu082. [DOI] [PubMed] [Google Scholar]
- 60.Du W, Ma X-L, Zhao C, et al. Associations of Single Nucleotide Polymorphisms in miR-146a, miR-196a, miR-149 and miR-499 with Colorectal Cancer Susceptibility. Asian Pac J Cancer Prev. 2014;15(2):1047–1055. doi: 10.7314/apjcp.2014.15.2.1047. [DOI] [PubMed] [Google Scholar]
- 61.Fan X, Wu Z. Effects of four single nucleotide polymorphisms in microRNA-coding genes on lung cancer risk. Tumour Biol. 2014;35(11):10815–10824. doi: 10.1007/s13277-014-2371-5. [DOI] [PubMed] [Google Scholar]
- 62.Kupcinskas J, Bruzaite I, Juzenas S, et al. Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci Rep. 2014;4:5993. doi: 10.1038/srep05993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P, Yan H, Zhang H, et al. A functional polymorphism in MIR196A2 is associated with risk and progression of nasopharyngeal carcinoma in the Chinese population. Genet Test Mol Biomarkers. Mar. 2014;18(3):149–155. doi: 10.1089/gtmb.2013.0400. [DOI] [PubMed] [Google Scholar]
- 64.Qu Y, Qu H, Luo M, et al. MicroRNAs related polymorphisms and genetic susceptibility to esophageal squamous cell carcinoma. Mol Genet Genom. 2014;289(6):1123–1130. doi: 10.1007/s00438-014-0873-x. [DOI] [PubMed] [Google Scholar]
- 65.Tong N, Xu B, Shi D, et al. Hsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese children. Mutat Res. 2014;759:16–21. doi: 10.1016/j.mrfmmm.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Wang N, Li Y, Zhou RM, et al. Hsa-miR-196a2 functional SNP is associated with the risk of ESCC in individuals under 60 years old. Biomarkers. 2014;19(1):43–48. doi: 10.3109/1354750X.2013.866164. [DOI] [PubMed] [Google Scholar]
- 67.Deng S, Wang W, Li X, Zhang P. Common genetic polymorphisms in pre-microRNAs and risk of bladder cancer. World J Surg Oncol. 2015;13:297. doi: 10.1186/s12957-015-0683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dikaiakos P, Gazouli M, Rizos S, Zografos G, Theodoropoulos GE. Evaluation of genetic variants in miRNAs in patients with colorectal cancer. Cancer Biom. 2015;15(2):157–162. doi: 10.3233/CBM-140449. [DOI] [PubMed] [Google Scholar]
- 69.Li X, Li K, Wu Z. Association of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(8):9560–9566. [PMC free article] [PubMed] [Google Scholar]
- 70.Ni Q, Ji A, Yin J, Wang X, Liu X. Effects of Two Common Polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on Gastric Cancer Susceptibility. Gastroenterol Res Pract. 2015;2015:764163. doi: 10.1155/2015/764163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nikolic Z, Savic Pavicevic D, Vucic N, et al. Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. Exp Mol Pathol. 2015;99(1):145–150. doi: 10.1016/j.yexmp.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 72.Qi P, Wang L, Zhou B, et al. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet Mol Res. 2015;14(2):6289–6296. doi: 10.4238/2015.June.11.2. [DOI] [PubMed] [Google Scholar]
- 73.Wu Y, Hao X, Feng Z, Liu Y. Genetic polymorphisms in miRNAs and susceptibility to colorectal cancer. Cell Biochem Biophysics. 2015;71(1):271–278. doi: 10.1007/s12013-014-0195-y. [DOI] [PubMed] [Google Scholar]
- 74.Dai ZM, Kang HF, Zhang WG, et al. The Associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: A STROBE-Compliant Observational Study. Medicine. 2016;95(7):e2826. doi: 10.1097/MD.0000000000002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J, Cheng G, Wang S. A single-nucleotide polymorphism of miR-196a2T>C rs11614913 Is associated with hepatocellular carcinoma in the Chinese population. Genet Test Mol Biomarkers. 2016;20(4):213–215. doi: 10.1089/gtmb.2015.0271. [DOI] [PubMed] [Google Scholar]
- 76.Li M, Li RJ, Bai H, et al. Association between the pre-miR-196a2 rs11614913 polymorphism and gastric cancer susceptibility in a Chinese population. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15027516. [DOI] [PubMed] [Google Scholar]
- 77.Qiu GP, Liu J. MicroRNA gene polymorphisms in evaluating therapeutic efficacy after transcatheter arterial chemoembolization for primary hepatocellular carcinoma. Genet Test Mol Biomarkers. 2016 doi: 10.1089/gtmb.2016.0073. [DOI] [PubMed] [Google Scholar]
- 78.Shen F, Chen J, Guo S, et al. Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumour Biol. 2016;37(4):4777–4784. doi: 10.1007/s13277-015-4268-3. [DOI] [PubMed] [Google Scholar]
- 79.Song ZS, Wu Y, Zhao HG, et al. Association between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancer. Oncol Lett. 2016;11(1):194–200. doi: 10.3892/ol.2015.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu X, Ling Q, Wang J, et al. Donor miR-196a-2 polymorphism is associated with hepatocellular carcinoma recurrence after liver transplantation in a Han Chinese population. Int J Cancer. 2016;138(3):620–629. doi: 10.1002/ijc.29821. [DOI] [PubMed] [Google Scholar]
- 81.Yin Z, Cui Z, Ren Y, et al. Association between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female population. Lung Cancer. 2016;94:15–21. doi: 10.1016/j.lungcan.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 82.Zhao H, Xu J, Zhao D, et al. Somatic mutation of the SNP rs11614913 and its association with increased MIR 196A2 expression in breast cancer. DNA Cell Biol. 2016;35(2):81–87. doi: 10.1089/dna.2014.2785. [DOI] [PubMed] [Google Scholar]
- 83.Okubo M, Tahara T, Shibata T, et al. Association between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese population. Helicobacter. 2010;15(6):524–531. doi: 10.1111/j.1523-5378.2010.00806.x. [DOI] [PubMed] [Google Scholar]
- 84.Pu JY, Dong W, Zhang L, Liang WB, Yang Y, Lv ML. No association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese population. Iran J Basic Med Sci. 2014;17(2):128–133. [PMC free article] [PubMed] [Google Scholar]
- 85.Omrani M, Hashemi M, Eskandari-Nasab E, et al. hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark Med. 2014;8(2):259–267. doi: 10.2217/bmm.13.118. [DOI] [PubMed] [Google Scholar]
- 86.Qi JH, Wang J, Chen J, et al. High-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patients. BMC Cancer. 2014;14:643. doi: 10.1186/1471-2407-14-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chu YH, Tzeng SL, Lin CW, Chien MH, Chen MK, Yang SF. Impacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancer. PLoS One. 2012;7(6):e39777. doi: 10.1371/journal.pone.0039777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu JY, Tu L. Investigating the role of polymorphisms in miR-146a, -149, and -196a2 in the development of gastric cancer. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15027839. gmr.15027839. [DOI] [PubMed] [Google Scholar]
- 89.Hashemi M, Moradi N, Ziaee SA, et al. Association between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian population. J Adv Res. 2016;7(3):491–498. doi: 10.1016/j.jare.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He B, Pan Y, Xu Y, et al. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol. 2015;36(6):4575–4582. doi: 10.1007/s13277-015-3102-2. [DOI] [PubMed] [Google Scholar]
- 91.Kou JT, Fan H, Han D, et al. Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol Lett. 2014;8(3):1255–1260. doi: 10.3892/ol.2014.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morales S, Gulppi F, Gonzalez-Hormazabal P, et al. Association of single nucleotide polymorphisms in Pre-miR-27a, Pre-miR-196a2, Pre-miR-423, miR-608 and Pre-miR-618 with breast cancer susceptibility in a South American population. BMC Genet. 2016;17(1):109. doi: 10.1186/s12863-016-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pavlakis E, Papaconstantinou I, Gazouli M, et al. MicroRNA gene polymorphisms in pancreatic cancer. Pancreatology. 2013;13(3):273–278. doi: 10.1016/j.pan.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 94.Roy R, De Sarkar N, Ghose S, et al. Genetic variations at microRNA and processing genes and risk of oral cancer. Tumour Bio. 2014;35(4):3409–3414. doi: 10.1007/s13277-013-1450-3. [DOI] [PubMed] [Google Scholar]
- 95.Sodhi KK, Bahl C, Singh N, Behera D, Sharma S. Functional genetic variants in pre-miR-146a and 196a2 genes are associated with risk of lung cancer in North Indians. Future Oncol. 2015;11(15):2159–2173. doi: 10.2217/fon.15.143. [DOI] [PubMed] [Google Scholar]
- 96.Sun XC, Zhang AC, Tong LL, et al. miR-146a and miR-196a2 polymorphisms in ovarian cancer risk. Genet Mol Res. 2016;15(3) doi: 10.4238/gmr.15038468. gmr.15038468. [DOI] [PubMed] [Google Scholar]
- 97.Sushma PS, Jamil K, Kumar PU, Satyanarayana U, Ramakrishna M, Triveni B. Genetic variation in MicroRNAs and risk of oral squamous cell carcinoma in South Indian population. Asian Pac J Cancer Prev. 2015;16(17):7589–7594. doi: 10.7314/apjcp.2015.16.17.7589. [DOI] [PubMed] [Google Scholar]
- 98.Toraih EA, Fawz MS, Elgazzaz MG, Hussein MH, Shehata RH, Daoud HG. Combined genotype analyses of precursor miRNA196a2 and 499a variants with hepatic and renal cancer susceptibility a Preliminary Study. Asian Pac J Cancer Prev. 2016;17(7):3369–3375. [PubMed] [Google Scholar]
- 99.Zhang LH, Hao BB, Zhang CY, Dai XZ, Zhang F. Contributions of polymorphisms in miR146a, miR196a, and miR499 to the development of hepatocellular carcinoma. Genet Mol Res. 2016;15(3) doi: 10.4238/gmr.15038582. [DOI] [PubMed] [Google Scholar]
- 100.Zhang M, Jin M, Yu Y, et al. Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur J Cancer Care (Engl) 2012;21(2):274–280. doi: 10.1111/j.1365-2354.2011.01308.x. [DOI] [PubMed] [Google Scholar]
- 101.Kim MJ, Yoo SS, Choi YY, Park JY. A functional polymorphism in the pre-microRNA-196a2 and the risk of lung cancer in a Korean population. Lung Cancer. 2010;69(1):127–129. doi: 10.1016/j.lungcan.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 102.Vinci S, Gelmini S, Pratesi N, et al. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med. 2011;49(12):2073–2080. doi: 10.1515/CCLM.2011.708. [DOI] [PubMed] [Google Scholar]
- 103.Ahn DH, Rah H, Choi YK, et al. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. 2013;52(Suppl 1):E39–E51. doi: 10.1002/mc.21962. [DOI] [PubMed] [Google Scholar]
- 104.Umar M, Upadhyay R, Prakash G, Kumar S, Ghoshal UC, Mittal B. Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Mol Carcinog. 2013;52(Suppl 1):E10–E18. doi: 10.1002/mc.21931. [DOI] [PubMed] [Google Scholar]
- 105.Vinci S, Gelmini S, Mancini I, et al. Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods. 2013;59(1):138–146. doi: 10.1016/j.ymeth.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 106.Wei J, Zheng L, Liu S, et al. MiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. Human Immunology. 2013;74(9):1199–1205. doi: 10.1016/j.humimm.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 107.Parlayan C, Ikeda S, Sato N, Sawabe M, Muramatsu M, Arai T. Association analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control study. Asian Pacific journal of cancer prevention: APJCP. 2014;15(5):2101–2107. doi: 10.7314/apjcp.2014.15.5.2101. [DOI] [PubMed] [Google Scholar]
- 108.Guo J, Jin M, Zhang M, Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS One. 2012;7(1):e30585. doi: 10.1371/journal.pone.0030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma XP, Zhang T, Peng B, Yu L, Jiang de K. Association between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studies. PLoS One. 2013;8(11):e79584. doi: 10.1371/journal.pone.0079584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang J, Wang Q, Liu H, et al. The association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studies. Mutagenesis. 2012;27(6):779–788. doi: 10.1093/mutage/ges052. [DOI] [PubMed] [Google Scholar]
- 111.Zhang H, Su YL, Yu H, Qian BY. Meta-Analysis of the association between Mir-196a-2 polymorphism and cancer susceptibility. Cancer. 2012;9(1):63–72. doi: 10.3969/j.issn.2095-3941.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011;15(1):14–23. doi: 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature Rev Drug Discov. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu J, Hu Z, Xu Z, et al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum Mut. 2009;30(8):1231–1236. doi: 10.1002/humu.21044. [DOI] [PubMed] [Google Scholar]
- 116.Kang Z, Li Y, He X, et al. Quantitative assessment of the association between miR-196a2 rs11614913 polymorphism and cancer risk: evidence based on 45,816 subjects. Tumour Biol. 2014;35(7):6271–6282. doi: 10.1007/s13277-014-1822-3. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This review was supported by Health Care 3F Project of Shenzhen (Peking University First Hospital-The Second People’s Hospital of Shenzhen, Academician Yinglu Guo’s Team), the Shenzhen Key Medical Discipline Fund, Special Support Funds of Shenzhen for Introduced High-Level Medical Team, Shenzhen Foundation of Science and Technology (JCYJ20150330102720182), and Shenzhen Health and Family Planning Commission Scientific Research Project (201506026, 201601025, and 201606019).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Liu CJ, Tsai MM, Tu HF, Lui MT, Cheng HW, Lin SC. miR-196a overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Ann Surg Oncol. 2013;20(Suppl 3):S406–S414. doi: 10.1245/s10434-012-2618-6. [DOI] [PubMed] [Google Scholar]
- 2.Hong YS, Kang HJ, Kwak JY, et al. Association between microRNA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean population. J Prev Med Public Health. 2011;44(3):125–130. doi: 10.3961/jpmph.2011.44.3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol, Biomarkers & Prev. 2009;18(4):1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 4.Peng S, Kuang Z, Sheng C, Zhang Y, Xu H, Cheng Q. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2010;55(8):2288–2293. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman AE, Zheng T, Yi C, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69(14):5970–5977. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dou T, Wu Q, Chen X, et al. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol. 2010;136(12):1853–1859. doi: 10.1007/s00432-010-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118(7):2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akkiz H, Bayram S, Bekar A, Akgollu E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepat. 2011;18(7):e399–e407. doi: 10.1111/j.1365-2893.2010.01414.x. [DOI] [PubMed] [Google Scholar]
- 9.Li XD, Li ZG, Song XX, Liu CF. A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. Pathology. 2010;42(7):669–673. doi: 10.3109/00313025.2010.522175. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava K, Srivastava A, Mittal B. Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J Hum Genet. 2010;55(8):495–499. doi: 10.1038/jhg.2010.54. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Li G, Wei S, et al. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer. 2010;116(20):4753–4760. doi: 10.1002/cncr.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42(8):1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer. 2007;7:162. doi: 10.1186/1471-2407-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285(2):116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 21.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 22.Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29(7):1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 23.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nature Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min KT, Kim JW, Jeon YJ, et al. Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog. 2012;51(Suppl 1):E65–E73. doi: 10.1002/mc.21849. [DOI] [PubMed] [Google Scholar]
- 25.Hezova R, Kovarikova A, Bienertova-Vasku J, et al. Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J Gastroenterol. 2012;18(22):2827–2831. doi: 10.3748/wjg.v18.i22.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang J, Jia ZF, Cao DH, Wu YH, Sun ZW, Cao XY. Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Future Oncol. 2016;12(19):2215–2226. doi: 10.2217/fon-2016-0224. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y, Yu CY, Wang JL, Guan J, Chen HY, Fang JY. MicroRNA sequence polymorphisms and the risk of different types of cancer. Sci Rep. 2014;4:3648. doi: 10.1038/srep03648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 29.Woolf B. On estimating the relation between blood group and disease. Ann Hum Genet. 1955;19(4):251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Zhan Y, Chen Z, et al. Directing cellular information flow via CRISPR signal conductors. Nat Methods. 2016;13(11):938–944. doi: 10.1038/nmeth.3994. [DOI] [PubMed] [Google Scholar]
- 31.Vangel MG, Rukhin AL. Maximum likelihood analysis for heteroscedastic one-way random effects ANOVA in interlaboratory studies. Biometrics. 1999;55(1):129–136. doi: 10.1111/j.0006-341x.1999.00129.x. [DOI] [PubMed] [Google Scholar]
- 32.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28(2):105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, Liang J, Wang Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30(1):79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 36.Catucci I, Yang R, Verderio P, et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat. 2010;31(1):E1052–E1057. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- 37.Christensen BC, Avissar-Whiting M, Ouellet LG, et al. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res. 2010;16(14):3713–3720. doi: 10.1158/1078-0432.CCR-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Guo H, Hu H, et al. A functional variation in pre-microRNA-196a is associated with susceptibility of esophageal squamous cell carcinoma risk in Chinese Han. Biomarkers. 2010;15(7):614–618. doi: 10.3109/1354750X.2010.505299. [DOI] [PubMed] [Google Scholar]
- 39.George GP, Gangwar R, Mandal RK, Sankhwar SN, Mittal RD. Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol Biol Rep. 2011;38(3):1609–1615. doi: 10.1007/s11033-010-0270-4. [DOI] [PubMed] [Google Scholar]
- 40.Jedlinski DJ, Gabrovska PN, Weinstein SR, Smith RA, Griffiths LR. Single nucleotide polymorphism in hsa-mir-196a-2 and breast cancer risk: a case control study. Twin Res Hum Genet. 2011;14(5):417–421. doi: 10.1375/twin.14.5.417. [DOI] [PubMed] [Google Scholar]
- 41.Mittal RD, Gangwar R, George GP, Mittal T, Kapoor R. Investigative role of pre-microRNAs in bladder cancer patients: a case-control study in North India. DNA Cell Biol. 2011;30(6):401–406. doi: 10.1089/dna.2010.1159. [DOI] [PubMed] [Google Scholar]
- 42.Zhan JF, Chen LH, Chen ZX, et al. A functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese population. Archiv Med Res. 2011;42(2):144–148. doi: 10.1016/j.arcmed.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Zhou B, Wang K, Wang Y, et al. Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Mol Carcinog. 2011;50(7):499–505. doi: 10.1002/mc.20740. [DOI] [PubMed] [Google Scholar]
- 44.Chen H, Sun LY, Chen LL, Zheng HQ, Zhang QF. A variant in microRNA-196a2 is not associated with susceptibility to and progression of colorectal cancer in Chinese. Int Med J. 2012;42(6):e115–e119. doi: 10.1111/j.1445-5994.2011.02434.x. [DOI] [PubMed] [Google Scholar]
- 45.Linhares JJ, Azevedo M, Jr, Siufi AA, et al. Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC Med Genet. 2012;13:119. doi: 10.1186/1471-2350-13-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon KA, Yoon H, Park S, et al. The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancer. J Thorac Cardiovasc Surg. 2012;144(4):794–807. doi: 10.1016/j.jtcvs.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 47.Zhu L, Chu H, Gu D, et al. A functional polymorphism in miRNA-196a2 is associated with colorectal cancer risk in a Chinese population. DNA Cell Biol. 2012;31(3):350–354. doi: 10.1089/dna.2011.1348. [DOI] [PubMed] [Google Scholar]
- 48.Ahn DH, Rah H, Choi Y-K, et al. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. 2013;52(Suppl 1):39–51. doi: 10.1002/mc.21962. [DOI] [PubMed] [Google Scholar]
- 49.Han Y, Pu R, Han X, et al. Associations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma risk. PLoS One. 2013;8(3):e58564. doi: 10.1371/journal.pone.0058564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong MJ, Choi YY, Jang JA, et al. Association between genetic variants in pre-microRNAs and survival of early-stage NSCLC. J Thorac Oncol. 2013;8(6):703–710. doi: 10.1097/JTO.0b013e318288dc0a. [DOI] [PubMed] [Google Scholar]
- 51.Lv M, Dong W, Li L, et al. Association between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese population. J Cancer Res Clin Oncol. 2013;139(8):1405–1410. doi: 10.1007/s00432-013-1456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song X, Sturgis EM, Liu J, et al. MicroRNA variants increase the risk of HPV-associated squamous cell carcinoma of the oropharynx in never smokers. PLoS One. 2013;8(2):e56622. doi: 10.1371/journal.pone.0056622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Tao G, Wu D, et al. A functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancer. Mol Carcinog. 2013;52(Suppl 1):E87–E95. doi: 10.1002/mc.22017. [DOI] [PubMed] [Google Scholar]
- 54.Yuan Z, Zeng X, Yang D, Wang W, Liu Z. Effects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer risk. PLoS One. 2013;8(4):e61047. doi: 10.1371/journal.pone.0061047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Wang R, Ma Y-Y, et al. Association between single nucleotide polymorphisms in miRNA196a-2 and miRNA146a and susceptibility to hepatocellular carcinoma in a Chinese population. Asian Pac J Cancer Prev. 2013;14(11):6427–6431. doi: 10.7314/apjcp.2013.14.11.6427. [DOI] [PubMed] [Google Scholar]
- 56.Bansal C, Sharma KL, Misra S, Srivastava AN, Mittal B, Singh US. Common genetic variants in pre-microRNAs and risk of breast cancer in the North Indian population. Ecancermedicalscience. 2014;8:473. doi: 10.3332/ecancer.2014.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu YH, Hsieh MJ, Chiou HL, et al. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One. 2014;9(2):e89930. doi: 10.1371/journal.pone.0089930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dikeakos P, Theodoropoulos G, Rizos S, Tzanakis N, Zografos G, Gazouli M. Association of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek population. Mol Biol Rep. 2014;41(2):1075–1080. doi: 10.1007/s11033-013-2953-0. [DOI] [PubMed] [Google Scholar]
- 59.Du M, Lu D, Wang Q, et al. Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis. 2014;35(7):1629–1635. doi: 10.1093/carcin/bgu082. [DOI] [PubMed] [Google Scholar]
- 60.Du W, Ma X-L, Zhao C, et al. Associations of single nucleotide polymorphisms in miR-146a, miR-196a, miR-149 and miR-499 with colorectal cancer susceptibility. Asian Pac J Cancer Prev. 2014;15(2):1047–1055. doi: 10.7314/apjcp.2014.15.2.1047. [DOI] [PubMed] [Google Scholar]
- 61.Fan X, Wu Z. Effects of four single nucleotide polymorphisms in microRNA-coding genes on lung cancer risk. Tumour Biol. 2014;35(11):10815–10824. doi: 10.1007/s13277-014-2371-5. [DOI] [PubMed] [Google Scholar]
- 62.Kupcinskas J, Bruzaite I, Juzenas S, et al. Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci Rep. 2014;4:5993. doi: 10.1038/srep05993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P, Yan H, Zhang H, et al. A functional polymorphism in MIR196A2 is associated with risk and progression of nasopharyngeal carcinoma in the Chinese population. Genet Test Mol Biomarkers. 2014;18(3):149–155. doi: 10.1089/gtmb.2013.0400. [DOI] [PubMed] [Google Scholar]
- 64.Qu Y, Qu H, Luo M, et al. MicroRNAs related polymorphisms and genetic susceptibility to esophageal squamous cell carcinoma. Mol Genet Genom. 2014;289(6):1123–1130. doi: 10.1007/s00438-014-0873-x. [DOI] [PubMed] [Google Scholar]
- 65.Tong N, Xu B, Shi D, et al. Hsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese children. Mutat Res. 2014;759:16–21. doi: 10.1016/j.mrfmmm.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Wang N, Li Y, Zhou RM, et al. Hsa-miR-196a2 functional SNP is associated with the risk of ESCC in individuals under 60 years old. Biomarkers. 2014;19(1):43–48. doi: 10.3109/1354750X.2013.866164. [DOI] [PubMed] [Google Scholar]
- 67.Deng S, Wang W, Li X, Zhang P. Common genetic polymorphisms in pre-microRNAs and risk of bladder cancer. World J Surg Oncol. 2015;13:297. doi: 10.1186/s12957-015-0683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dikaiakos P, Gazouli M, Rizos S, Zografos G, Theodoropoulos GE. Evaluation of genetic variants in miRNAs in patients with colorectal cancer. Cancer Biom. 2015;15(2):157–162. doi: 10.3233/CBM-140449. [DOI] [PubMed] [Google Scholar]
- 69.Li X, Li K, Wu Z. Association of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(8):9560–9566. [PMC free article] [PubMed] [Google Scholar]
- 70.Ni Q, Ji A, Yin J, Wang X, Liu X. Effects of two common polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on gastric cancer susceptibility. Gastroenterol Res Pract. 2015;2015:764163. doi: 10.1155/2015/764163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nikolic Z, Savic Pavicevic D, Vucic N, et al. Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. Exp Mol Pathol. 2015;99(1):145–150. doi: 10.1016/j.yexmp.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 72.Qi P, Wang L, Zhou B, et al. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet Mol Res. 2015;14(2):6289–6296. doi: 10.4238/2015.June.11.2. [DOI] [PubMed] [Google Scholar]
- 73.Wu Y, Hao X, Feng Z, Liu Y. Genetic polymorphisms in miRNAs and susceptibility to colorectal cancer. Cell Biochem Biophys. 2015;71(1):271–278. doi: 10.1007/s12013-014-0195-y. [DOI] [PubMed] [Google Scholar]
- 74.Dai ZM, Kang HF, Zhang WG, et al. The Associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: A STROBE-Compliant Observational Study. Medicine. 2016;95(7):e2826. doi: 10.1097/MD.0000000000002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J, Cheng G, Wang S. A single-nucleotide polymorphism of miR-196a2T>C rs11614913 is associated with hepatocellular carcinoma in the Chinese population. Genet Test Mol Biomarkers. 2016;20(4):213–215. doi: 10.1089/gtmb.2015.0271. [DOI] [PubMed] [Google Scholar]
- 76.Li M, Li RJ, Bai H, et al. Association between the pre-miR-196a2 rs11614913 polymorphism and gastric cancer susceptibility in a Chinese population. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15027516. [DOI] [PubMed] [Google Scholar]
- 77.Qiu GP, Liu J. MicroRNA gene polymorphisms in evaluating therapeutic efficacy after transcatheter arterial chemoembolization for primary hepatocellular carcinoma. Genet Test Mol Biomarkers. 2016;20(10):579–586. doi: 10.1089/gtmb.2016.0073. [DOI] [PubMed] [Google Scholar]
- 78.Shen F, Chen J, Guo S, et al. Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumour Biol. 2016;37(4):4777–4784. doi: 10.1007/s13277-015-4268-3. [DOI] [PubMed] [Google Scholar]
- 79.Song ZS, Wu Y, Zhao HG, et al. Association between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancer. Oncol Lett. 2016;11(1):194–200. doi: 10.3892/ol.2015.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu X, Ling Q, Wang J, et al. Donor miR-196a-2 polymorphism is associated with hepatocellular carcinoma recurrence after liver transplantation in a Han Chinese population. Int J Cancer. 2016;138(3):620–629. doi: 10.1002/ijc.29821. [DOI] [PubMed] [Google Scholar]
- 81.Yin Z, Cui Z, Ren Y, et al. Association between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female population. Lung Cancer. 2016;94:15–21. doi: 10.1016/j.lungcan.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 82.Zhao H, Xu J, Zhao D, et al. Somatic mutation of the SNP rs11614913 and its association with increased MIR 196A2 expression in breast cancer. DNA Cell Biol. 2016;35(2):81–87. doi: 10.1089/dna.2014.2785. [DOI] [PubMed] [Google Scholar]
- 83.Okubo M, Tahara T, Shibata T, et al. Association between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese population. Helicobacter. 2010;15(6):524–531. doi: 10.1111/j.1523-5378.2010.00806.x. [DOI] [PubMed] [Google Scholar]
- 84.Pu JY, Dong W, Zhang L, Liang WB, Yang Y, Lv ML. No association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese population. Iran J Basic Med Sci. 2014;17(2):128–133. [PMC free article] [PubMed] [Google Scholar]
- 85.Omrani M, Hashemi M, Eskandari-Nasab E, et al. hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark Med. 2014;8(2):259–267. doi: 10.2217/bmm.13.118. [DOI] [PubMed] [Google Scholar]
- 86.Qi JH, Wang J, Chen J, et al. High-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patients. BMC Cancer. 2014;14:643. doi: 10.1186/1471-2407-14-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chu YH, Tzeng SL, Lin CW, Chien MH, Chen MK, Yang SF. Impacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancer. PLoS One. 2012;7(6):e39777. doi: 10.1371/journal.pone.0039777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu JY, Tu L. Investigating the role of polymorphisms in miR-146a, -149, and -196a2 in the development of gastric cancer. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15027839. gmr.15027839. [DOI] [PubMed] [Google Scholar]
- 89.Hashemi M, Moradi N, Ziaee SA, et al. Association between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian population. J Adv Res. 2016;7(3):491–498. doi: 10.1016/j.jare.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He B, Pan Y, Xu Y, et al. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol. 2015;36(6):4575–4582. doi: 10.1007/s13277-015-3102-2. [DOI] [PubMed] [Google Scholar]
- 91.Kou JT, Fan H, Han D, et al. Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol Lett. 2014;8(3):1255–1260. doi: 10.3892/ol.2014.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morales S, Gulppi F, Gonzalez-Hormazabal P, et al. Association of single nucleotide polymorphisms in Pre-miR-27a, Pre-miR-196a2, Pre-miR-423, miR-608 and Pre-miR-618 with breast cancer susceptibility in a South American population. BMC Genet. 2016;17(1):109. doi: 10.1186/s12863-016-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pavlakis E, Papaconstantinou I, Gazouli M, et al. MicroRNA gene polymorphisms in pancreatic cancer. Pancreatology. 2013;13(3):273–278. doi: 10.1016/j.pan.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 94.Roy R, De Sarkar N, Ghose S, et al. Genetic variations at microRNA and processing genes and risk of oral cancer. Tumour Biol. 2014;35(4):3409–3414. doi: 10.1007/s13277-013-1450-3. [DOI] [PubMed] [Google Scholar]
- 95.Sodhi KK, Bahl C, Singh N, Behera D, Sharma S. Functional genetic variants in pre-miR-146a and 196a2 genes are associated with risk of lung cancer in North Indians. Future Oncol. 2015;11(15):2159–2173. doi: 10.2217/fon.15.143. [DOI] [PubMed] [Google Scholar]
- 96.Sun XC, Zhang AC, Tong LL, et al. miR-146a and miR-196a2 polymorphisms in ovarian cancer risk. Genet Mol Res. 2016;15(3) doi: 10.4238/gmr.15038468. gmr.15038468. [DOI] [PubMed] [Google Scholar]
- 97.Sushma PS, Jamil K, Kumar PU, Satyanarayana U, Ramakrishna M, Triveni B. Genetic variation in MicroRNAs and risk of oral squamous cell carcinoma in South Indian population. Asian Pac J Cancer Prev. 2015;16(17):7589–7594. doi: 10.7314/apjcp.2015.16.17.7589. [DOI] [PubMed] [Google Scholar]
- 98.Toraih EA, Fawz MS, Elgazzaz MG, Hussein MH, Shehata RH, Daoud HG. Combined genotype analyses of precursor miRNA196a2 and 499a variants with hepatic and renal cancer susceptibility a Preliminary Study. Asian Pac J Cancer Prev. 2016;17(7):3369–3375. [PubMed] [Google Scholar]
- 99.Zhang LH, Hao BB, Zhang CY, Dai XZ, Zhang F. Contributions of polymorphisms in miR146a, miR196a, and miR499 to the development of hepatocellular carcinoma. Genet Mol Res. 2016;15(3) doi: 10.4238/gmr.15038582. [DOI] [PubMed] [Google Scholar]
- 100.Zhang M, Jin M, Yu Y, et al. Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur J Cancer Care (Engl) 2012;21(2):274–280. doi: 10.1111/j.1365-2354.2011.01308.x. [DOI] [PubMed] [Google Scholar]
- 101.Guo J, Jin M, Zhang M, Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS One. 2012;7(1):e30585. doi: 10.1371/journal.pone.0030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma XP, Zhang T, Peng B, Yu L, Jiang de K. Association between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studies. PLoS One. 2013;8(11):e79584. doi: 10.1371/journal.pone.0079584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang J, Wang Q, Liu H, et al. The association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studies. Mutagenesis. 2012;27(6):779–788. doi: 10.1093/mutage/ges052. [DOI] [PubMed] [Google Scholar]
- 104.Zhang H, Su YL, Yu H, Qian BY. Meta-analysis of the association between Mir-196a-2 polymorphism and cancer susceptibility. Cancer Biol Med. 2012;9(1):63–72. doi: 10.3969/j.issn.2095-3941.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011;15(1):14–23. doi: 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature Rev Drug Discov. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu J, Hu Z, Xu Z, et al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum Mut. 2009;30(8):1231–1236. doi: 10.1002/humu.21044. [DOI] [PubMed] [Google Scholar]
- 109.Kang Z, Li Y, He X, et al. Quantitative assessment of the association between miR-196a2 rs11614913 polymorphism and cancer risk: evidence based on 45,816 subjects. Tumour Biol. 2014;35(7):6271–6282. doi: 10.1007/s13277-014-1822-3. [DOI] [PubMed] [Google Scholar]
- 110.Kim MJ, Yoo SS, Choi YY, Park JY. A functional polymorphism in the pre-microRNA-196a2 and the risk of lung cancer in a Korean population. Lung cancer. 2010;69(1):127–129. doi: 10.1016/j.lungcan.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 111.Vinci S, Gelmini S, Pratesi N, et al. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clinical chemistry and laboratory medicine. 2011;49(12):2073–2080. doi: 10.1515/CCLM.2011.708. [DOI] [PubMed] [Google Scholar]
- 112.Umar M, Upadhyay R, Prakash G, Kumar S, Ghoshal UC, Mittal B. Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Mol Carcinog. 2013;52(Suppl 1):E10–18. doi: 10.1002/mc.21931. [DOI] [PubMed] [Google Scholar]
- 113.Vinci S, Gelmini S, Mancini I, et al. Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods. 2013;59(1):138–146. doi: 10.1016/j.ymeth.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 114.Wei J, Zheng L, Liu S, et al. MiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. Human immunology. 2013;74(9):1199–1205. doi: 10.1016/j.humimm.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 115.Parlayan C, Ikeda S, Sato N, Sawabe M, Muramatsu M, Arai T. Association analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control study. Asian Pacific journal of cancer prevention: APJCP. 2014;15(5):2101–2107. doi: 10.7314/apjcp.2014.15.5.2101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Details of the sensitivity analyses of the association between rs11614913 polymorphism and cancer risk homozygous model (TT vs CC) and recessive model (TT vs TC+CC).
| Comparison | Study omitted | Estimate | (95% Conf Interval)
|
|
|---|---|---|---|---|
| Lower CI | Upper CI | |||
| TT vs CC | Hu et al7 | 0.902 | 0.814 | 0.999 |
| Hu et al35 | 0.904 | 0.815 | 1.002 | |
| Tian et al3 | 0.902 | 0.814 | 1.001 | |
| Hoffman et al5 | 0.890 | 0.805 | 0.985 | |
| Catucci et al36 | 0.900 | 0.811 | 1.000 | |
| Wang et al38 | 0.911 | 0.824 | 1.008 | |
| Okubo et al83 | 0.900 | 0.812 | 0.998 | |
| Peng et al4 | 0.904 | 0.816 | 1.002 | |
| Srivastava et al10 | 0.903 | 0.815 | 1.000 | |
| Dou et al6 | 0.897 | 0.809 | 0.994 | |
| Li et al9 | 0.906 | 0.818 | 1.003 | |
| Akkiz et al8 | 0.908 | 0.820 | 1.005 | |
| Liu et al11 | 0.898 | 0.810 | 0.997 | |
| Kim et al101 | 0.904 | 0.815 | 1.002 | |
| Catucci et al36 | 0.899 | 0.810 | 0.997 | |
| Christensen et al37 | 0.900 | 0.813 | 0.997 | |
| Mittal et al41 | 0.904 | 0.816 | 1.001 | |
| Jedlinski et al40 | 0.900 | 0.813 | 0.998 | |
| Zhan et al42 | 0.906 | 0.818 | 1.004 | |
| Zhou et al43 | 0.901 | 0.813 | 0.998 | |
| Vinci et al102 | 0.895 | 0.809 | 0.992 | |
| Hong et al2 | 0.902 | 0.814 | 1.000 | |
| George et al39 | 0.902 | 0.815 | 0.999 | |
| Linhares et al45 | 0.893 | 0.806 | 0.988 | |
| Chen et al44 | 0.898 | 0.811 | 0.995 | |
| Min et al24 | 0.904 | 0.815 | 1.002 | |
| Zhu et al47 | 0.905 | 0.816 | 1.003 | |
| Hezova et al25 | 0.897 | 0.810 | 0.994 | |
| Zhang et al100 | 0.900 | 0.812 | 0.998 | |
| Yoon et al46 | 0.904 | 0.816 | 1.001 | |
| Zhang et al99 | 0.904 | 0.816 | 1.001 | |
| Chu et al87 | 0.894 | 0.807 | 0.990 | |
| Vinci et al105 | 0.897 | 0.810 | 0.994 | |
| Ahn et al103 | 0.902 | 0.814 | 1.000 | |
| Lv et al51 | 0.878 | 0.798 | 0.965 | |
| Umar et al104 | 0.895 | 0.808 | 0.992 | |
| Wei et al106 | 0.896 | 0.809 | 0.993 | |
| Wang et al53 | 0.894 | 0.807 | 0.990 | |
| Zhang et al55 | 0.904 | 0.816 | 1.003 | |
| Han et al49 | 0.898 | 0.810 | 0.996 | |
| Pavlakis et al93 | 0.899 | 0.812 | 0.996 | |
| Tong et al65 | 0.901 | 0.813 | 1.000 | |
| Pu et al84 | 0.902 | 0.814 | 1.000 | |
| Bansal et al56 | 0.902 | 0.815 | 1.000 | |
| Kupcinskas et al62 | 0.897 | 0.809 | 0.994 | |
| Qu et al64 | 0.905 | 0.817 | 1.003 | |
| Wang et al66 | 0.897 | 0.809 | 0.994 | |
| Dikeakos et al58 | 0.925 | 0.843 | 1.015 | |
| Qi et al86 | 0.902 | 0.814 | 1.000 | |
| Chu et al57 | 0.898 | 0.810 | 0.995 | |
| Parlayan et al107 | 0.900 | 0.812 | 0.997 | |
| Li et al63 | 0.896 | 0.808 | 0.993 | |
| Du et al59 | 0.892 | 0.806 | 0.987 | |
| Omrani et al85 | 0.900 | 0.813 | 0.997 | |
| Kou et al91 | 0.907 | 0.819 | 1.004 | |
| Roy et al94 | 0.896 | 0.809 | 0.993 | |
| Li et al63 | 0.896 | 0.808 | 0.993 | |
| Deng et al67 | 0.900 | 0.812 | 0.997 | |
| Qi et al72 | 0.907 | 0.819 | 1.005 | |
| Dikaiakos et al68 | 0.899 | 0.812 | 0.996 | |
| Li et al69 | 0.890 | 0.805 | 0.985 | |
| Li et al69 | 0.907 | 0.819 | 1.004 | |
| Nikolic et al71 | 0.902 | 0.814 | 1.000 | |
| He et al90 | 0.901 | 0.813 | 0.999 | |
| Sushma et al97 | 0.909 | 0.821 | 1.006 | |
| Sodhi et al95 | 0.891 | 0.806 | 0.986 | |
| Jiang et al26 | 0.896 | 0.808 | 0.993 | |
| Toraih et al98 | 0.894 | 0.807 | 0.990 | |
| Dai et al74 | 0.908 | 0.820 | 1.005 | |
| Zhao et al82 | 0.898 | 0.811 | 0.995 | |
| Song et al79 | 0.907 | 0.819 | 1.004 | |
| Shen et al78 | 0.902 | 0.813 | 1.002 | |
| Li et al75 | 0.907 | 0.820 | 1.005 | |
| Li et al76 | 0.906 | 0.819 | 1.004 | |
| Xu et al80 | 0.906 | 0.818 | 1.004 | |
| Qiu et al77 | 0.905 | 0.817 | 1.003 | |
| Jiang et al26 | 0.901 | 0.813 | 1.000 | |
| Yin et al81 | 0.901 | 0.813 | 0.999 | |
| Zhang et al99 | 0.901 | 0.813 | 0.998 | |
| Sun et al96 | 0.904 | 0.817 | 1.002 | |
| Toraih et al98 | 0.894 | 0.808 | 0.990 | |
| Morales et al92 | 0.901 | 0.812 | 0.999 | |
| Gu et al88 | 0.891 | 0.805 | 0.986 | |
| Hashemi et al89 | 0.896 | 0.809 | 0.992 | |
| Combined2–10,25,26,35–107 | 0.900 | 0.813 | 0.997 | |
| TT vs TC+CC | Hu et al7 | 0.918 | 0.851 | 0.991 |
| Hu et al35 | 0.920 | 0.852 | 0.993 | |
| Tian et al3 | 0.918 | 0.850 | 0.991 | |
| Hoffman et al5 | 0.910 | 0.844 | 0.980 | |
| Catucci et al36 | 0.917 | 0.849 | 0.991 | |
| Wang et al38 | 0.928 | 0.862 | 0.999 | |
| Okubo et al83 | 0.917 | 0.850 | 0.991 | |
| Peng et al4 | 0.919 | 0.852 | 0.991 | |
| Srivastava et al10 | 0.918 | 0.850 | 0.990 | |
| Dou et al6 | 0.918 | 0.850 | 0.991 | |
| Li et al9 | 0.922 | 0.854 | 0.994 | |
| Akkiz et al8 | 0.923 | 0.856 | 0.995 | |
| Liu et al11 | 0.917 | 0.849 | 0.990 | |
| Kim et al101 | 0.920 | 0.852 | 0.992 | |
| Catucci et al36 | 0.916 | 0.849 | 0.989 | |
| Christensen et al37 | 0.918 | 0.851 | 0.989 | |
| Mittal et al41 | 0.921 | 0.854 | 0.993 | |
| Jedlinski et al40 | 0.917 | 0.850 | 0.989 | |
| Zhan et al42 | 0.922 | 0.854 | 0.994 | |
| Zhou et al43 | 0.918 | 0.850 | 0.990 | |
| Vinci et al102 | 0.915 | 0.849 | 0.987 | |
| Hong et al2 | 0.922 | 0.854 | 0.994 | |
| George et al39 | 0.920 | 0.853 | 0.992 | |
| Linhares et al45 | 0.913 | 0.847 | 0.985 | |
| Chen et al44 | 0.916 | 0.849 | 0.988 | |
| Min et al24 | 0.918 | 0.850 | 0.990 | |
| Zhu et al47 | 0.921 | 0.854 | 0.994 | |
| Hezova et al25 | 0.915 | 0.848 | 0.987 | |
| Zhang et al100 | 0.918 | 0.850 | 0.991 | |
| Yoon et al46 | 0.920 | 0.853 | 0.993 | |
| Zhang et al99 | 0.919 | 0.852 | 0.992 | |
| Chu et al87 | 0.918 | 0.851 | 0.991 | |
| Vinci et al105 | 0.919 | 0.851 | 0.991 | |
| Ahn et al103 | 0.916 | 0.850 | 0.988 | |
| Lv et al51 | 0.905 | 0.842 | 0.974 | |
| Umar et al104 | 0.914 | 0.848 | 0.986 | |
| Wei et al106 | 0.918 | 0.850 | 0.990 | |
| Wang et al53 | 0.913 | 0.846 | 0.985 | |
| Zhang et al55 | 0.919 | 0.851 | 0.992 | |
| Han et al49 | 0.917 | 0.849 | 0.990 | |
| Pavlakis et al93 | 0.921 | 0.854 | 0.994 | |
| Tong et al65 | 0.913 | 0.847 | 0.985 | |
| Pu et al84 | 0.918 | 0.851 | 0.990 | |
| Bansal et al56 | 0.919 | 0.852 | 0.991 | |
| Kupcinskas et al62 | 0.916 | 0.849 | 0.988 | |
| Qu et al64 | 0.923 | 0.855 | 0.995 | |
| Wang et al66 | 0.916 | 0.848 | 0.988 | |
| Dikeakos et al58 | 0.931 | 0.866 | 1.001 | |
| Qi et al86 | 0.924 | 0.857 | 0.996 | |
| Chu et al57 | 0.914 | 0.847 | 0.986 | |
| Parlayan et al107 | 0.918 | 0.851 | 0.990 | |
| Li et al63 | 0.913 | 0.846 | 0.985 | |
| Du et al59 | 0.914 | 0.847 | 0.986 | |
| Omrani et al85 | 0.918 | 0.851 | 0.989 | |
| Kou et al91 | 0.921 | 0.854 | 0.994 | |
| Roy et al94 | 0.915 | 0.848 | 0.987 | |
| Li et al63 | 0.906 | 0.845 | 0.971 | |
| Deng et al67 | 0.913 | 0.847 | 0.985 | |
| Qi et al72 | 0.923 | 0.856 | 0.995 | |
| Dikaiakos et al68 | 0.914 | 0.848 | 0.987 | |
| Li et al69 | 0.911 | 0.845 | 0.982 | |
| Li et al69 | 0.922 | 0.855 | 0.995 | |
| Nikolic et al71 | 0.919 | 0.852 | 0.991 | |
| He et al90 | 0.917 | 0.850 | 0.990 | |
| Sushma et al97 | 0.921 | 0.855 | 0.994 | |
| Sodhi et al95 | 0.913 | 0.847 | 0.984 | |
| Jiang et al26 | 0.914 | 0.847 | 0.986 | |
| Toraih et al98 | 0.914 | 0.848 | 0.986 | |
| Dai et al74 | 0.922 | 0.855 | 0.995 | |
| Zhao et al82 | 0.914 | 0.848 | 0.986 | |
| Song et al79 | 0.923 | 0.856 | 0.995 | |
| Shen et al78 | 0.918 | 0.849 | 0.992 | |
| Li et al75 | 0.921 | 0.854 | 0.993 | |
| Li et al76 | 0.923 | 0.856 | 0.995 | |
| Xu et al80 | 0.922 | 0.854 | 0.994 | |
| Qiu et al77 | 0.921 | 0.854 | 0.993 | |
| Jiang et al26 | 0.921 | 0.854 | 0.994 | |
| Yin et al81 | 0.919 | 0.851 | 0.992 | |
| Zhang et al99 | 0.918 | 0.851 | 0.991 | |
| Sun et al96 | 0.919 | 0.852 | 0.992 | |
| Toraih et al98 | 0.915 | 0.848 | 0.986 | |
| Morales et al92 | 0.918 | 0.851 | 0.991 | |
| Gu et al88 | 0.911 | 0.845 | 0.982 | |
| Hashemi et al89 | 0.915 | 0.848 | 0.986 | |
| Combined2–10,25,26,35–107 | 0.918 | 0.851 | 0.989 | |
Table S2.
P-values of Begg’s and Egger’s test for the polymorphism rs11614913
| Polymorphism | Comparison | Subgroup | Begg’s test (P>z) |
Egger’s test (P>t) |
|---|---|---|---|---|
| rs11614913 | T vs C | Overall | 0.660 | 0.923 |
| Taqman | 0.368 | 0.723 | ||
| PCR | 0.640 | 0.859 | ||
| Asian | 0.946 | 0.854 | ||
| Caucasian | 0.147 | 0.969 | ||
| HB | 0.509 | 0.386 | ||
| PB | 0.251 | 0.579 | ||
| TT vs CC | Overall | 0.971 | 0.822 | |
| Taqman | 0.719 | 0.606 | ||
| PCR | 0.832 | 0.762 | ||
| Asian | 0.578 | 0.758 | ||
| Caucasian | 0.163 | 0.971 | ||
| HB | 0.721 | 0.489 | ||
| PB | 0.666 | 0.880 | ||
| TC vs CC | Overall | 0.951 | 0.761 | |
| Taqman | 0.418 | 0.289 | ||
| PCR | 0.839 | 0.933 | ||
| Asian | 0.991 | 0.546 | ||
| Caucasian | 0.902 | 0.767 | ||
| HB | 0.721 | 0.601 | ||
| PB | 0.965 | 0.453 | ||
| TT+TC vs CC | Overall | 0.592 | 0.401 | |
| Taqman | 0.418 | 0.613 | ||
| PCR | 0.734 | 0.598 | ||
| Asian | 0.986 | 0.185 | ||
| Caucasian | 0.300 | 0.770 | ||
| HB | 0.737 | 0.543 | ||
| PB | 0.584 | 0.593 | ||
| TT vs TC+CC | Overall | 0.908 | 0.899 | |
| Taqman | 0.719 | 0.440 | ||
| PCR | 0.912 | 0.917 | ||
| Asian | 0.795 | 0.688 | ||
| Caucasian | 0.537 | 0.857 | ||
| HB | 0.673 | 0.503 | ||
| PB | 0.914 | 0.508 |
Abbreviations: HB, hospital based; PB, population based; PCR, polymerase chain reaction.