Abstract
Apoptosis is a highly regulated form of programmed cell death, essential to the development and homeostasis of multicellular organisms. Cytochrome c is a central figure in the activation of the apoptotic intrinsic pathway, thereby activating the caspase cascade through its interaction with Apaf-1. Our recent studies have revealed 14-3-3ε (a direct inhibitor of Apaf-1) as a cytosolic cytochrome c target. Here we explore the cytochrome c / 14-3-3ε interaction and show the ability of cytochrome c to block 14-3-3ε-mediated Apaf-1 inhibition, thereby unveiling a novel function for cytochrome c as an indirect activator of caspase-9/3. We have used calorimetry, NMR spectroscopy, site mutagenesis and computational calculations to provide an insight into the structural features of the cytochrome c / 14-3-3ε complex. Overall, these findings suggest an additional cytochrome c-mediated mechanism to modulate apoptosome formation, shedding light onto the rigorous apoptotic regulation network.
Introduction
Apoptosis, a specific form of programmed cell death (PCD), is an essential process for development and homeostasis of multicellular organisms1. There are two main apoptotic pathways in mammalian cells: the extrinsic or death receptor-initiated pathway and the intrinsic or mitochondria-dependent pathway2,3. The last is triggered by signals, including DNA damage, oxidative stress, and growth factor deprivation, which converge on the release of pro-apoptotic proteins from the mitochondria into the cytosol; one such protein is cytochrome c (Cc). Under homeostasis, Cc acts as an electron carrier in the mitochondrial respiratory chain4,5. Upon its release from mitochondria, Cc interacts with WD40 domains of the apoptotic protease activating factor-1 (Apaf-1) in the cytosol, enabling the assembly of the apoptosome6. This protein platform activates caspase-9 and later caspase-3, initiating the caspase cascade, which executes cell death in an orchestrated way7,8.
Recent findings suggest Cc fulfills multiple roles in apoptosis, beyond Apaf-1 activation and apoptosome assembly. Indeed, Cc interacts with various cytosolic and nuclear partners along the onset of PCD9,10. Hence, the full scope of Cc role in apoptosis remains un-elucidated. Recently, our group has reported that Cc inhibits the histone chaperone activity of SET/TAF-Iβ in the nucleus, impairing the formation of core histone-SET/TAF-Iβ complexes under DNA damage11. However, the novel functions of cytosolic Cc stay unveiled, despite a complex network of interactions mediated by Cc during apoptosis has been suggested12.
Therefore, we focused on the interaction between Cc and protein 14-3-3ε, a novel cytosolic Cc target under DNA damage10. This protein belongs to the 14-3-3 family13,14, which comprises seven conserved isoforms (β, γ, ε, η, σ, τ/θ, and ζ), arranged as homo- and heterodimers. Each monomer contains nine α-helices that form a conserved concave groove, used by 14-3-3 proteins to bind their phosphorylated targets15 (Supplementary Figure S1). Furthermore, they are also involved in phosphorylation-independent interactions16–18. 14-3-3 proteins participate in several cell processes related to metabolism, signal transduction, cell cycle control, apoptosis, transcription, and stress responses19–24.
Among 14-3-3ε functions, its ability to inhibit Apaf-1 stands out because it prevents apoptosome assembly and caspase activation25. Such inhibition is enhanced by phosphorylation of Apaf-1 at Ser268 by the p90kDa ribosomal S6 kinase-1 (Rsk-1) when the mitogen-activated protein kinases (MAPK) cascade is active. Hence, the interaction of Cc with 14-3-3ε could modulate such inhibition somehow.
Herein, we show that Cc hinders 14-3-3ε-mediated Apaf-1 inhibition. Indeed, our results indicate a competition between Cc and 14-3-3ε for binding to Apaf-1, which enhances caspase activation. Furthermore, this new regulatory mechanism is modulated by phosphorylation of Apaf-1, which enhances its inhibition by 14-3-3ε. We further show that Cc binds to both the 14-3-3ε concave groove and the convex face, thereby providing a molecular basis for this novel modulation of apoptosome assembly.
Results
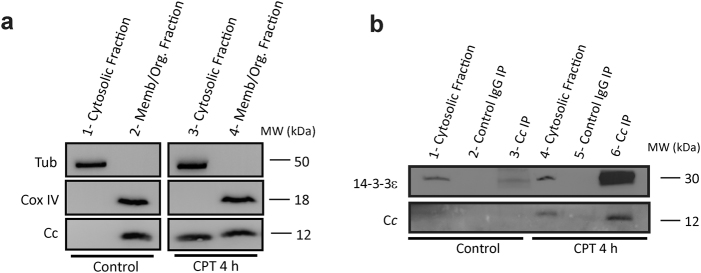
Cc interacts with 14-3-3ε in the cytosol under apoptotic conditions
To elucidate the extra-mitochondrial function of Cc, we explored the interaction of Cc with 14-3-3ε when apoptosis is triggered. To this aim, HeLa cells were treated with the topoisomerase I inhibitor camptothecin (CPT), to induce DNA damage. Then, subcellular fractionation was performed, and Cc was detected in the cytosol (Fig. 1a, lane 3). However, it remained inside the mitochondria under homeostasis (Fig. 1a, lane 2). The in cell Cc / 14-3-3ε interaction was established as immunoprecipitation (IP) of cytosolic proteins associated with Cc yielded intrinsic 14-3-3ε in CPT-treated cells (Fig. 1b, lane 6). To further confirm the IP specificity, untreated and CPT-treated cytosolic lysates were probed with a 14-3-3ε antibody (Fig. 1b, lanes 1 and 4, respectively). Negative controls using IgG (Fig. 1b, lanes 2 and 5) did not display any band. Immunoblotting against the anti-Cc antibody confirmed Cc IP (Fig. 1b, lane 6).
Fig. 1. Cc localization in the cytosol upon cell CPT-treatment.
a Subcellular fractioning showing the Cc location upon cell treatment with 20 μM CPT for 4 h. Purity of fractions was verified by western blot using anti-α-Tubulin and anti-CoxIV antibodies for detecting cytosolic and membrane proteins, respectively. b Cc-IP of cytosolic fractions from non-treated (lane 3) and CPT-treated (lane 6) along with the detection of 14-3-3ε by western blot (upper). Mouse IgG was used as control (lanes 2 and 5). Verification of immunoprecipitation of Cc upon CPT treatment is shown (lower)
Cc blocks 14-3-3ε-mediated caspase inhibition
Following its release into the cytosol, Cc targets Apaf-1 to assemble the apoptosome6. As 14-3-3ε binds Apaf-1 to prevent caspase activation25, we investigated whether Cc modulates Apaf-1 inhibition by 14-3-3ε.
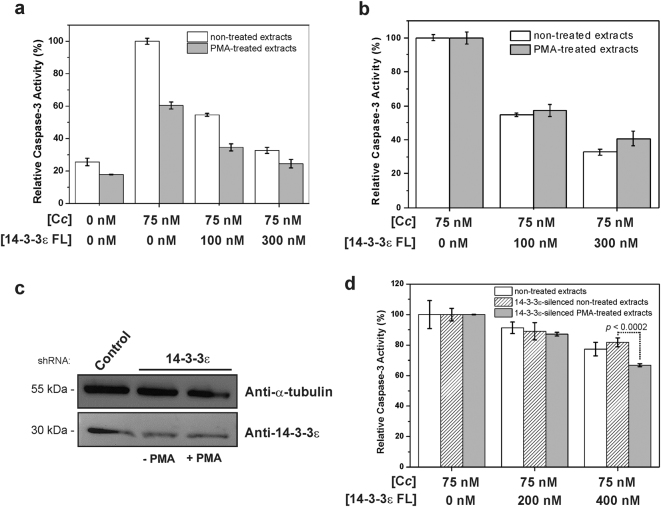
First, we checked the ability of Cc to activate caspase-3 in HEK293T cytosolic cell extracts. Caspase-3 activity was monitored upon Cc addition (Fig. 2a, white columns), resulting in a substantial increase of such activity, as the hemeprotein triggered the apoptosome formation and, subsequently, caspase-9, -3 activation.
Fig. 2. 14-3-3ε FL inhibits caspase-3 activity in HEK293T cytosolic cell extracts.
a Relative caspase-3 activity in non-treated (white columns) and PMA-treated (gray columns) HEK293T cytosolic cell extracts were measured upon addition of recombinant Cc and increasing concentrations of 14-3-3ε FL. b Normalized data shown in a. c HEK293T cells were transfected with empty pSUPER (Control) or with pSUPER-shRNA against 14-3-3ε and immunoblotted with an anti-14-3-3ε antibody to examine the amount of remaining endogenous 14-3-3ε. An immunoblotting with anti-α-tubulin was used as loading control. d Normalized caspase-3 activity in 14-3-3ε-silenced non-treated and PMA-treated cell extracts upon addition of recombinant 14-3-3ε FL. Normalized caspase-3 activity of non-14-3-3ε-silenced, non-treated cell extracts is included as a control
MAPK proteins modulate Apaf-1 activity, leading the activation of Rsk-1, which phosphorylates Apaf-1 at Ser268, favoring the interaction between Apaf-1 and 14-3-3ε, thereby increasing Apaf-1 inhibition25. Therefore, we tested the effect of phorbol 12-myristate 13-acetate (PMA), which triggers the MAPK cascade, on the caspase-3 activity. Apaf-1 phosphorylation under PMA treatment was confirmed by Phos-tagTM SDS PAGE (Supplementary Figure S2). PMA-treated cells extracts showed a lower caspase-3 activity than untreated extracts in the presence of Cc (Fig. 2a, gray columns). This is consistent with Apaf-1 phosphorylation at Ser268, enhancing its inhibition by 14-3-3ε25.
Then, we tested the ability of recombinant 14-3-3ε (herein named as 14-3-3ε full length, 14-3-3ε FL) to inhibit caspase-3 activity in non-treated and PMA-treated cell extracts. Addition of 14-3-3ε FL made extracts to exhibit significantly lower Cc-mediated caspase-3 activity values (Fig. 2a). These were normalized to identify the particular effect of added 14-3-3ε FL but, unexpectedly, the normalization evinced that the inhibition exerted by 14-3-3ε FL was similar in non-treated and PMA-treated extracts (Fig. 2b). One of the plausible explanations for this observation is endogenous 14-3-3ε already abolished the caspase-3 activity by preferentially binding to phosphorylated Apaf-1, thus preventing any additional effect of exogenous 14-3-3ε FL. Additional caspase-3 assays were carried out using extracts from 14-3-3ε knockdown HEK293T cells (Fig. 2c), with the caspase-3 activity in PMA-treated extracts resulting more sensitive than non-treated extracts to the addition of 14-3-3ε FL (Fig. 2d).
In summary, these findings demonstrate how 14-3-3ε FL inhibits caspase activity, and how this inhibitory mechanism can be modulated by Apaf-1 phosphorylation. Nevertheless, the inhibitory effect of 14-3-3ε FL on caspase-3 could be caused by either a direct interaction with Apaf-1 or by hindering the complex between the latter and Cc. Caspase-9 can indeed be phosphorylated by the ERK MAPK pathway, with the resulting modulation of its function and consequently, of caspase-3 activation26.
To discard the direct interaction between Cc and Apaf-1, as well as the ERK-mediated inhibition of caspase-9, we performed caspase-9 activity assays using a recombinant, truncated form of Apaf-1 lacking the Cc-binding WD40 domains (Apaf-1ΔWD40)27. Apaf-1ΔWD40 activates caspase-9 in a constitutive manner in the absence of Cc (Fig. 3). The inability of Cc to alter the Apaf-1ΔWD40-mediated caspase-9 activity was likewise determined (Supplementary Figure S3). Then, the addition of 14-3-3ε FL inhibited caspase-9 activity strongly, due to the binding of Apaf-1ΔWD40 to 14-3-3ε FL (Fig. 3). However, the sequentially titration with Cc restored caspase-9 activity, indicating a competition between Cc and Apaf-1ΔWD40 for the binding site of 14-3-3ε FL, impairing Apaf-1 inhibition. Nuclear magnetic resonance (NMR) experiments corroborated such a competition. Specifically, the ε-methyl group signal of Met80 in reduced Cc substantially broadened upon binding of the hemeprotein to 14-3-3ε (Supplementary Figure S3e). Further addition of Apaf-1ΔWD40 yielded partial recover of the signal intensity, indicating that Apaf-1ΔWD40 competes with Cc for the 14-3-3ε FL binding site. The inability of Apaf-1ΔWD40 to bind Cc was corroborated (Supplementary Figure S3f).
Fig. 3. Cc impairs 14-3-3ε FL-mediated caspase-9 inhibition.
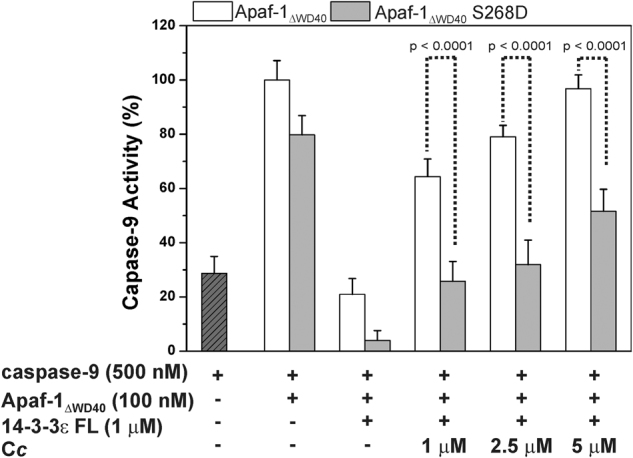
Cc and 14-3-3ε FL were added to caspase-9 and different Apaf-1ΔWD40 mutants
As phosphorylation of Apaf-1 at Ser268 enhances its interaction with 14-3-3ε FL, we tested the ability of Cc to modulate the 14-3-3ε FL / Apaf-1ΔWD40 complex upon phosphorylation. To this end, we designed the recombinant Apaf-1ΔWD40 S268D mutant, which mimics Ser268 phosphorylation. As a control of unspecific effects caused by mutations at such position, the Apaf-1ΔWD40 S268A variant was also tested. Notably, the ability of phosphomimetic Apaf-1ΔWD40 S268D mutant to trigger caspase-9 activity was similar to that exhibited by Apaf-1ΔWD40. Nevertheless, the Apaf-1ΔWD40 S268D-mediated caspase-9 activity was slightly more sensitive to 14-3-3ε (Fig. 3, gray columns). Indeed, the recovery of enzymatic activity by Cc was substantially lower than that measured using Apaf-1ΔWD40 (Fig. 3, gray columns). Such a finding indicates a larger affinity of the phosphorylated Apaf-1 species toward 14-3-3ε that displaces Cc25. On the other hand, the caspase-9 activation in the presence of the S268A mutant behaved in a way similar to that in the presence of Apaf-1ΔWD40 (Supplementary Figure S3b). Hence, the negative charge at position 268 in Apaf-1 is responsible for the increase in affinity of Apaf-1 toward 14-3-3ε FL.
Our results thus support a model in which Cc activates Apaf-1 while blocking the inhibition exerted by 14-3-3ε on the apoptotic factor (Fig. 4).
Fig. 4. Model of Apaf-1 regulation by Cc and 14-3-3ε.

Model of Apaf-1 regulation by Cc and 14-3-3ε. During homeostasis, Apaf-1 is available to interact with 14-3-3ε25. Upon its release into the cytosol, Cc competes with Apaf-1 for the binding to 14-3-3ε, impairing the Apaf-1 / 14-3-3ε complex. Thus, Apaf-1 is available to interact with Cc, thereby activating the caspases cascade6. Upon Apaf-1 phosphorylation, the equilibrium of the reaction is strongly shifted toward the Apaf-1 / 14-3-3ε complex formation. This would hinder the competition performed by Cc, thereby partly impairing the activation of the caspases
Cc recognizes two different binding sites of 14-3-3ε full length
The dimerization state of 14-3-3ε FL was confirmed by analytic ultracentrifugation combined with dynamic light scattering (Supplementary Figure S4a-c). Sedimentation velocity experiments confirmed the dimeric state of 14-3-3ε FL with a molecular mass of 66.4 kDa (Supplementary Figure S4b). Dynamic light scattering measurements corroborated that 14-3-3ε FL behaves as a dimer, with a particle size of 9 ± 2 nm (Supplementary Figure S4c).
Isothermal titration calorimetry (ITC) experiments in Supplementary Figure S5 revealed that Cc binds to 14-3-3ε FL homodimer with a 2:1 stoichiometry and a dissociation constant (KD) of 2.3 μM (Supplementary Table S1). To reveal 14-3-3ε residues implicated in the interaction with Cc, 14-3-3ε FL D21K, K50E, S59E, E92K, D99K, and S187D mutants were designed. Residues were selected according to their role in the interactions of 14-3-3 isoforms with their respective partners28–30 and the impact of each mutation in the electrostatic surface potential of the protein. The dimeric state of these mutants was confirmed by analytic ultracentrifugation and dynamic light scattering (Supplementary Figure S4c and d). According to ITC analysis (Supplementary Table S1 and Figure S5), the six mutants bound to Cc with slightly larger KD values than 14-3-3ε FL WT. Further, the complexes involving 14-3-3ε FL D21K, K50E, S59E, or E92K showed a 1:1 stoichiometry instead of the 2:1 found for the WT species (Supplementary Table S1), suggesting that one of the two Cc-binding sites of 14-3-3ε FL has been compromised in these mutants. Therefore, Asp21, Lys50, Ser59, and Glu92 residues may be essential in Cc recognition.
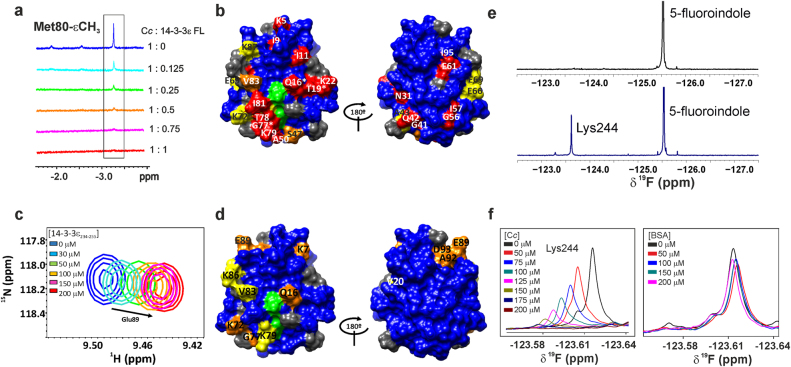
We also performed NMR experiments to further characterize the interaction between reduced Cc and 14-3-3ε FL. We followed Cc signals upon titration of 14-3-3ε FL, which caused an overall broadening of Cc resonances because Cc / 14-3-3ε FL complex formation (Fig. 5a and Supplementary Figure S6a, left). Specifically, the ε-methyl group signal of the Met80 Cc residue was largely broadened at 1:0.5 Cc:14-3-3ε FL ratio (Fig. 5a), in agreement with the stoichiometry inferred from ITC data. Moreover, specific Cc backbone amide groups underwent singularly large changes in their 1H line widths (1H ΔΔν1/2 Binding) upon binding with 14-3-3ε FL (Supplementary Figure S6b). These resonances were expected to be at or in the proximity of the area of Cc involved in the recognition of 14-3-3ε FL. Moreover, several signals displayed substantial chemical-shift perturbations (CSPs) in the presence of 14-3-3ε FL (Supplementary Figure S6a, right). Specifically, Gln16, Thr19, Ser47, Lys72, Gly77 and Val83 signals showed significant CSPs (Δδavg > 0.035 ppm.). Interestingly, the signal corresponding to Val83 (Supplementary Figure S6a, right) seems to be in conformational exchange in the free Cc, fixing its conformation upon the interaction with 14-3-3ε. These effects are consistent with an intermediate exchange rate at the NMR time scale, in agreement with the transient nature of this complex and the KD values in the μM range.
Fig. 5. Binding of Cc to 14-3-3ε as analyzed by NMR.
a 1D 1H NMR spectra of Cc (Fe2+) along the titration with 14-3-3ε FL. The complex formation is monitored by line broadening of the Met80-εCH3 signal, included in the black square. b Mapping of the Cc residues perturbed upon binding to 14-3-3ε FL. The Cc:14-3-3ε ratio is 1:0.75. Residues are colored according to their 1H ΔΔν1/2Binding (Supplementary Figure S6b). Residues with average chemical-shift perturbations (Δδavg) larger than 0.035 p.p.m. are represented in orange. Asterisks stand for residues showing both significant 1H ΔΔν1/2Binding and Δδavg. Residues without substantial 1H ΔΔν1/2Binding and Δδavg are in blue. c Detail of superimposed 2D [1H, 15N] HSQC spectra of 15N-labeled Cc along titration with 14-3-3ε234-255. Signals corresponding to distinct titration steps are colored according to the code in the panel. Arrow indicates direction of CSPs. d CSP map of reduced Cc upon addition of 14-3-3ε234-255. Residues are colored according to Δδavg categories: insignificant Δδavg < 0.020 ppm., blue; small 0.020 ppm. ≤ Δδavg < 0.025 ppm. yellow; and large Δδavg > 0.025 ppm., red. In b, d prolines and non-assigned residues are colored in gray; whereas, the heme group in green. e 1D 19F NMR spectra of free 5-fluorindole (upper) and when it is conjugated with 14-3-3ε234-255 peptide (lower). f 1D 19F NMR spectra of 14-3-3ε234-255 whose Lys244 is covalently bound to 5-fluorindole free and in the titrations with Cc and BSA. 19F CSPs are colored according to the panels
Both broadening and CSPs maps on Cc surface (Fig. 5b) indicate that Cc mainly recognizes 14-3-3ε FL by a surface patch surrounding the heme cleft, as it does in its homeostatic interactions in the electron transport chain31–33.
The C-terminal tails of 14-3-3ε are important in the interaction with Cc
To decipher the role of the unstructured C-terminal tail of 14-3-3ε FL during Cc recognition, the interaction between Cc and the 14-3-3ε core protein, lacking C-terminal tails, was measured by ITC. The recorded thermograms showed very similar thermodynamic parameters for both 14-3-3ε FL and 14-3-3ε core proteins interacting with Cc, although a slight increase in the KD value was observed in the interaction between Cc and the 14-3-3ε core (Supplementary Table S1). This suggests that the C-terminal tails of 14-3-3ε may participate in the interaction with Cc, despite being unessential.
To confirm that the C-terminal tails of 14-3-3ε (named 14-3-3ε234-255) are involved in the interaction with Cc, 2D [1H, 15N] HSQC were recorded along a titration of 14-3-3ε234-255 peptide onto 15N-labeled reduced Cc (Supplementary Figure S6c). Such titration resulted in significant CSPs of the Cc amide signals (Fig. 5c). The resulting interaction surface (Fig. 5d) indicates that 14-3-3ε234-255 binds to the rim of the heme groove of Cc, as with 14-3-3ε FL protein. However, unlike the 14-3-3ε FL interaction, most of the Cc resonances were in fast exchange on the NMR time scale upon binding to the 14-3-3ε234-255 peptide. 1H Δδbind was fitted using the best 1:1 binding model, obtaining a KD of 49 μM (Supplementary Figure S6d). Moreover, ITC experiments in which the 14-3-3ε234-255 peptide was titrated onto Cc yielded a KD value of 16 μM (Supplementary Figure S5 and Table S1). Both NMR and ITC KD values are marginally higher than the value obtained for the interaction between 14-3-3ε FL and Cc. Furthermore, the ITC analysis also confirmed a 1:1 stoichiometry.
Then, to easily determine a residue which could be included in the latter docking calculations, the only one positively charged residue (Lys244) of 14-3-3ε234-255 peptide was tagged with 5-fluorindole molecule through a conjugation reaction. To prevent the reaction of 5-fluorindole with the N-terminal −NH3+ group, the peptide was modified by acetylation. To confirm that the acetylation in the N-terminal residue did not drastically modify the interaction with Cc, ITC experiments were performed with the acetylated 14-3-3ε234-255. Both the non-acetylated and acetylated peptides interacted with Cc in a similar way, despite the KD value showed a slight increase when the peptide was acetylated (Supplementary Table S1).
Proper folding of 14-3-3ε234-255 before and after the conjugation reaction was unambiguously assessed by 1D 1H NMR spectra (Supplementary Figure S6e). Second, 1D 19F NMR spectra of both free 5-fluorindole and fluorinated 14-3-3ε234-255 were registered. The spectrum of fluorinated 14-3-3ε234-255 peptide showed a signal at −125.53 ppm. corresponding to free 5-fluorindole, and another at −123.62 ppm. that was assigned to 5-fluorindole conjugated to the epsilon −NH3+ group of Lys244 (Fig. 5e). The addition of increasing concentration of Cc resulted in significant CSPs of this second signal (Fig. 5f, left panel). Notably, the 19F NMR signal also experienced a significant broadening consistent with the larger correlation time of the peptide bound to Cc.
As a negative control, bovine serum albumin (BSA) protein was titrated into fluorinated 14-3-3ε234-255 peptide, with the negligible resulting 19F CSPs, suggesting that BSA cannot significantly bind to the peptide. (Fig. 5f, right panel). These results agree with Lys244 of 14-3-3ε FL taking part in Cc recognition. Therefore, Lys244 was included as an active residue in further docking calculations (see below).
Structural model of the interaction between Cc and 14-3-3ε full length
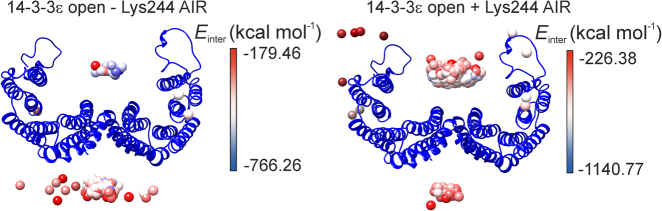
To further understand the molecular features of the interaction between Cc and 14-3-3ε FL, we performed restrain-driven docking computations using HADDOCK software34. For these computations, we used the structural model of 14-3-3ε FL in its open conformation inferred from MD as the input structure (Supplementary Information and Figure S7), to allow the entrance of Cc into the concave groove of the 14-3-3ε dimer. Line widths and CSPs obtained from NMR analysis of the Cc / 14-3-3ε FL complex were used as Cc ambiguous interaction restrains (AIRs). The 14-3-3ε FL residues that resulted in the impairment of one of the Cc-binding sites by ITC were used as 14-3-3ε AIRs. The 500 solutions obtained in the computation showed two main Cc-binding sites on 14-3-3ε FL dimer, in agreement with the stoichiometry inferred from ITC data. These solutions located Cc in both the concave and convex faces of 14-3-3ε; these interacting regions will be referred hereafter to as the concave and convex binding sites, respectively. The convex site resulted more populated, although some of the lowest energy solutions locate within the concave site (Fig. 6, left panel). Statistical classification of solutions yielded three clusters in the convex site of 14-3-3ε and only one in the concave site (Supplementary Figure S8a). Further analyses indicate that electrostatics is the major contributing factor to binding energy in the three clusters (Supplementary Table S2). However, the concave cluster was more energetically favored than the convex clusters, mainly due to a lower electrostatic energy (Eelec). Nevertheless, convex clusters presented lower AIRs violations than the concave cluster.
Fig. 6. HADDOCK molecular docking of Cc and 14-3-3ε FL.
Mass centers of Cc on the structural model of 14-3-3ε FL on its open conformation, including (right) or not (left) Lys244 as an active residue (AIR). Mass centers of Cc are colored as a function of intermolecular energy (Einter) according to the scales represented in their corresponding panels
Furthermore, to study the role of the 14-3-3ε C-terminal tails in the interaction, a docking computation was also performed including Lys244 (located in the C-terminal tail), as an active residue. This computation showed an increase in the number of solutions locating Cc in the concave binding site of 14-3-3ε (Fig. 6, right panel), suggesting that the C-terminal tails contribute to the entrance of Cc into the concave groove of 14-3-3ε. Clustering analysis exhibited three clusters of solutions presenting Cc bound to the concave site of 14-3-3ε, and only one displaying Cc bound to the convex site (Supplementary Figure S8b). Again, electrostatic interactions constituted the major contribution in all clusters, concave site clusters displaying lower energies because of their Eelec term (Supplementary Table S2).
Structural models of Cc / 14-3-3ε FL complex were obtained for computations excluding and including (Supplementary Figure S8c and d, respectively) Lys244 as an active residue of 14-3-3ε. For this, the best two complexes with minimal Einter at each binding site of 14-3-3ε were selected from each computation. Both structural models present a Cc molecule located in the concave groove of 14-3-3ε, contacting at least one of the C-terminal tails of 14-3-3ε dimer. A second Cc molecule is placed at the convex side of 14-3-3ε, specifically at the rim of the dimerization surface thereby binding both 14-3-3 monomers. In addition, the two Cc molecules from both computations present their heme clefts oriented to 14-3-3ε, according to NMR data.
Discussion
Due to the apoptosis significance in multicellular organisms, its regulation relies on a subtle balance between inhibition and activation pathways. In fact, signaling cascades convert continuous stimuli in an all-or-none response to avert physiological fluctuations to trigger a response accidentally35. Feedbacks are needed to lead response ultra-sensitivity: a large stimulus is required to trigger a response, which changes dramatically within a narrow range of stimulus intensity. During homeostasis, apoptotic factors remain inhibited, either blocked by other proteins or sequestered in a particular cell compartment. Along apoptosis, several apoptotic factors are released from the mitochondria to the cytosol. Among these factors is Cc, which promotes the apoptosome assembly through its association with Apaf-1, thereby initiating the caspase cascade6. On the other hand, pro-survival pathways, as the inhibition of Apaf-1 by 14-3-3ε25, reduce the sensitivity to the apoptosis signaling network.
Therefore, interactions between pro-apoptotic and pro-survival proteins, such as that between Cc and 14-3-3ε, are of particular relevance as they provide additional regulation loops. In this study, we have shown that Cc interacts with 14-3-3ε in the cytosol under apoptotic conditions, preventing the binding of 14-3-3ε to a specific site in Apaf-1. Hence, this interaction could be capable of regulating caspase activity. Indeed, Cc enhanced caspase activation in presence of 14-3-3ε inhibitor and a truncated, constitutive form of Apaf-1 unable to bind the hemeprotein. Thus, Cc plays a double role in the activation of caspase cascade directly by interacting with Apaf-1, and indirectly by blocking 14-3-3ε thereby releasing Apaf-1. This double role enables Cc to inhibit pro-survival pathways while apoptotic routes have been triggered (Fig. 4).
Further phenomena modulate the cell fate signaling, including protein phosphorylation events, as those which regulate Cc functions, including its ability to activate caspases36–40. In addition, such a phosphorylation enhances the inhibition of Apaf-1 by 14-3-3ε25, making caspase activation less sensitive to Cc. In fact, activation of MAPK cascade favors the Apaf-1 inhibition by 14-3-3ε. Similarly, the phosphomimetic Apaf-1 mutant is more susceptible to 14-3-3ε-dependent inhibition, which consequently becomes less sensitive to the presence of Cc. Hence, this phosphorylation of Ser268 in Apaf-1 may attenuate the effect of Cc release when caspase activation is undesirable (Fig. 4).
Cc does lack the consensus sequence which 14-3-3ε usually recognizes in its various targets. Our biophysical analysis has revealed that two molecules of Cc can interact with 14-3-3ε at two different sites: the groove of 14-3-3ε where its targets bind, and the convex side of the dimer. Additionally, the data herein suggest that flexible C-terminal 14-3-3ε may recognize Cc without being essential for the binding. These tails are flexible and mostly unstructured, although MD trajectories indicate the presence of residual helical elements. Moreover, NMR data show that Cc recognizes 14-3-3ε by residues surrounding the heme cleft, as reported for other interactions involving the hemeprotein31–33.
In summary, Cc is revealed as a double Apaf-1 activator via two mechanisms: by directly binding to the apoptosis factor and impairing its inhibition by 14-3-3ε. Indeed, such a cooperative behavior of Cc, triggering effector pathways while modulating feedbacks loops make cell response ultrasensitive to its release. The analysis of the various additional interactions of Cc during the onset of programmed cell death recently unveiled may confirm if this conclusion can be extended as a general mechanism by which Cc triggers apoptosis9–12,41.
Materials and methods
Cell cultures
HeLa and Human Embryonic Kidney 293T (HEK293T) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and maintained at 37 °C in a humidified 5% CO2 atmosphere.
For caspase-3 assays, 4.5 × 106 HEK293T cells were cultured in 140 mm Petri dishes in DMEM containing FBS until 70% confluence was reached, and cells were then kept in DMEM without FBS for 4 h. In some experiments (indicated in the main text) cells were treated with 50 ng/mL phorbol-12-myristate-13-acetate (PMA, Sigma-Aldrich) or treated with the same volume of DMSO for 120 min.
shRNA knockdown
In order to knockdown 14-3-3ε expression in HEK293T cells, a construct containing the shRNA sequence (5′-GATCCCCTGTTCAATACTGCTGATTATTCTCCTTCAAGAGAGGAGAATAATCAGCAGTATTGAACATTTTTA-3′) was cloned into a pSUPER vector using BgIII–HindIII restriction sites. An empty pSUPER vector was used as a control. Cells were transfected with the corresponding pSUPER plasmid by calcium phosphate method, as described elsewhere42. Transfected cells were selected by adding puromycin (1.5 μg/mL) to the culture medium. Four days after transfection, cells were collected and cytosolic extracts were obtained.
Subcellular fractionation and extract preparation
For immunoprecipitation assays, 6 × 106 HeLa cells were seeded in 140 mm Petri dishes and treated with an apoptosis inducing agent (20 μM camptothecin, CPT) for 4 h. HeLa cells were fractionated into cytosolic, membrane/organelles and nuclear fractions using a ProteoExtract Subcellular Proteome Extraction Kit (Calbiochem) following the manufacturer’s indications. The purity of subcellular fractions obtained was verified by western blot analysis, using anti-α-tubulin and anti-COX IV for detecting cytosol and mitochondria, respectively.
For caspase-3 activity assays, cytosolic extracts were obtained from HEK293T cells as previously described with briefly modifications43. Briefly, cells were trypsinized and collected by centrifugation (5 min at 1500×g) and washed twice with phosphate buffer saline (PBS) and once with Cellular Extract Buffer (CEB, 20 mM HEPES at pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM Na-EDTA, 1 mM EGTA, 1 mM dithiothreitol [DTT], 100 μM PMSF). Two volumes of CEB were added to cells, and they were incubated for 15 min on ice, allowing the hypotonic swell of cells. The cells were then disrupted by 20–25 strokes with a Douncer homogenizer and lysates were centrifuged at 15,000×g for 15 min at 4 °C to remove organelles.
Immunoprecipitation
Cytosolic fractions from subcellular fractionation of control and CPT-treated HeLa cells were used to immunoprecipitate Cc. Quantity of 300 μg of cytosolic samples were incubated with 50 μL Sepharose 6B (Sigma-Aldrich) for 2 h at 4 °C under agitation as a pre-clearing step to reduce non-specific binding. Samples were centrifuged and 20 μL of rabbit anti-human cytochrome c (Cc) were added to supernatants and incubated overnight at 4 °C under rotation. As a negative control, 300 μg of cytosolic lysates were incubated with 1 μg of anti-mouse IgG under the same conditions. Afterwards, 50 μL Protein A Sepharose (GE Healthcare) was added to each lysate for 4 h at 4 °C under rotation. The protein-Sepharose complexes were washed extensively, collected by centrifugation and boiled in freshly prepared reducing loading buffer. Controls including 30 μg of cytosolic lysates were run concurrently with the immunoprecipitated (IP) samples in the western blot assays.
Apaf-1 immunoprecipitation was performed from total non-treated and PMA-treated lysates. Apaf-1 IP samples were analyzed by western blot using a Phos-tagTM SDS PAGE (Wako) — which reduce the electrophoretic mobility of phosphorylated proteins.
Antibodies
Rabbit anti-human Cc serum was obtained after immunizing male rabbits with full length recombinant Cc suspended in a 0.85% solution of NaCl (20 mg/mL). The suspension was then incorporated into an equal amount of complete (Freund) adjuvant obtained from Difco (BD Biosciences, US). Mouse monoclonal anti-α-Tubulin (catalogue number T8328), as well as secondary horseradish peroxidase (HRP)-conjugated anti-mouse IgG (catalogue number A4416), rabbit polyclonal anti-14-3-3ε (catalogue number SAB4503100), and anti-rabbit IgG (catalogue number A0545) were obtained from Sigma-Aldrich. Rabbit polyclonal antibody to cytochrome c oxidase subunit IV (COX IV) was from Abcam (catalogue number ab16056). Mouse monoclonal anti-Apaf-1 (catalogue number sc-135836) was obtained from Santa Cruz Biotechnology, INC. Mouse monoclonal anti-phosphoserine (Millipore clone A4) was obtained from Merck.
Western blot analysis
For the immunoblot detection of Cc, α-Tubulin, COX IV, and 14-3-3ε in the subcellular fractions or IP samples, protein content was measured using the DCTM protein assay (Bio-Rad Laboratories, US). Proteins were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% gels and then transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore) using a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad). Membranes were blocked in 5% non-fat dry milk in PBS with Tween-20 (TPBS) and immunoblotting was performed with primary antibodies. HRP-conjugated secondary antibodies were used for detection. The immunoreactive bands were detected using Amersham ECL Plus Western Blotting Detection Reagents (GE Healthcare Life Sciences).
Cloning, expression, and purification of recombinant proteins
Cc was expressed in Escherichia coli BL21 (DE3) strain as previously described32. First, 25 mL Luria-Bertani (LB) medium, supplemented with 100 μg/mL ampicillin, were grown overnight at 37 °C under agitation. Volume of 2.5 mL of pre-culture was used to inoculate 2.5 L of LB medium. The culture was incubated at 30 °C under agitation for 24 h. Then, cells were centrifuged at 6000 r.p.m. for 10 min in an Avanti J-25 centrifuge (Beckman Coulter). Following this, cells were re-suspended in 1.5 mM borate buffer (pH 8.5), sonicated for 4 min and, finally, cellular debris was eliminated by centrifugation (20,000 r.p.m. for 30 min). For NMR measurements, 15N-labeled Cc was produced in minimal media with 15NH4Cl as a unique nitrogen source. Cc purification was carried out by ionic chromatography with a carboxy-methylcellulose matrix. Fractions containing Cc were concentrated in an Amicon (3 kDa cutoff) until correct concentration was achieved.
14-3-3ε full length (14-3-3ε FL) was cloned into the pET-28a vector under the T7 promotor using NdeI – XhoI restriction sites and containing a N-terminal hexahistidine tag. For cloning, the following primers were used: 5′-AGCCATATGATGGATGATCGAGAGGAT-3′ and 5′-GTGCTCGAGTCATTCCTGATTTTCGTC-3′. The 14-3-3ε core was cloned with a Stop codon introduced after residue at position 233. The primers used for this mutagenesis were 5′-ACACTATGGACTTCATAGATGCAGGGTGACGGT-3′ and 5′-ACCGTCACCCTGCATCTATGAAGTCCATAGTGT-3′. 14-3-3ε FL D21K, K50E, S59E, E92K, D99K, and S187D mutants were obtained from Mutagenex Inc. (USA). The expression of 14-3-3ε species were performed in the E. coli BL21 (DE3) strain. Cultures were grown in LB medium at 37 °C until an O.D.600 nm of 0.6–0.8 was reached. Protein expression was induced by adding 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) and cultures were grown for 24 h at 30 °C. The cells were collected by centrifugation and re-suspended in 20 mM Tris-HCl (pH 8), 800 mM NaCl, 10 mM imidazole. Cells were then broken by sonication and insoluble debris was removed by centrifugation, as indicated above. 14-3-3ε species were purified by affinity chromatography, using a Ni Sepharose 6 Fast Flow (GE Healthcare) column and an imidazole gradient ranging from 10 mM to 300 mM imidazole. Protein containing fractions were concentrated in an Amicon (3 kDa cutoff) until the desired proper protein concentration was reached.
Proteins were dialyzed against 10 mM sodium phosphate (pH 7.4) buffer for ITC measurements or 5 mM sodium phosphate (pH 6.3) buffer for NMR titrations. For caspase assays, protein was dialyzed against 20 mM HEPES (pH 7.5) buffer.
14-3-3ε C-terminal peptide (14-3-3ε234-255) was purchased from Genosphere Biotechnologies (France).
Apaf-1ΔWD40 S268D and S268A mutants were obtained by mutagenic PCR from pET28a-Apaf-1ΔWD40, using 5′-ACCAGAGACAAGGATGTTACAGATT-3′ and 5′-AATCTGTAACATCCTTGTCTCTGGT-3′ primers for the S268D mutant and 5′-ACCAGAGACAAGGCTGTTACAGATT-3′ and 5′-AATCTGTAACAGCCTTGTCTCTGGT-3′ primers for the S268A mutant. All Apaf-1ΔWD40 species were expressed as previously described with small modifications6. Protein expression was carried out in E. coli BL21 (DE3) at 20 °C after induction with 1 mM IPTG. Cells were broken by freeze/heat cycles and cellular debris was removed by centrifugation. The soluble fraction was purified by affinity chromatography (His·Trap column, GE Healthcare) followed by anionic-exchange (Hi·Trap column, GE Healthcare). Recombinant caspase-9 was expressed in E. coli BL21 (DE3) at 30 °C and purified as indicated above for Apaf-1ΔWD40. Protein quantification was assessed using the Bradford protein assay44.
Caspase-9 assays
Caspase-9 activity was measured as previously described6. Recombinant Apaf-1ΔWD40 species and 14-3-3ε FL were mixed in buffer (40 mM HEPES, pH 7.5, 20 mM KCl, 1 mM DTT) to a final concentration of 100 nM and 1 μM, respectively. Cc was added at different concentrations (ranging from 1 to 5 μM). After incubation for 15 min at room temperature, recombinant caspase-9 was added to a final volume of 100 μL and final concentration of 500 nM. After incubation for another 15 min at 37 °C, the caspase-9 substrate LEHD peptide coupled to 7-amino-4-trifluoromethylcoumarin (Ac-LEHD-AFC, TOCRIS) was added at a final concentration of 50 μM. The increase in fluorescence resulting from Ac-LEHD-AFC cleavage was determined in a Cary Eclipse (Varian) fluorescence spectrophotometer, using an excitation wavelength of 390 nm and an emission wavelength of 510 nm. Each experimental data were the mean ± SD value of at least three independent measurements.
Caspase-3 assays
In vitro caspase-3 assays were performed as formerly described with slight changes45. Recombinant Cc and 14-3-3ε FL were mixed and incubated with 100 μg of cytosolic HEK293T cells extracts for 60 min at 37 °C in 10 mM HEPES (pH 7.5) buffer, supplemented with 25 mM KCl, 200 μM DTT, 200 μM dATP. After incubation, 10 mM HEPES (pH 7.5), 50 mM NaCl, 40 mM β-glycerophosphate, 2 mM MgCl2, 5 mM EGTA, 0.1 mg mL−1 bovine serum albumin, and 0.1% (w/v) 3-[3-cholamidopropyldimethylammonio]-1-propanesulfonate (CHAPS) buffer was added to a final volume of 200 μL, supplemented with 40 μM of acetyl-DEVD-7-amino-4-methylcoumarin (Ac-DEVD-AMD, Enzo Life Sciences), a fluorescent substrate specific to caspase-3/7. The increase in fluorescence resulting from Ac-DEVD-AMC cleavage was determined in a Cary Eclipse (Varian) fluorescence spectrophotometer, using an excitation wavelength of 360 nm and an emission wavelength of 460 nm. To compare the effect of 14-3-3ε in different types of extracts, values for caspase-3 activity before adding recombinant 14-3-3ε FL were normalized. Each experimental data were the mean ± SD value of at least three independent measurements.
ITC measurements
All ITC experiments were performed using an Auto-ITC200 (MicroCal-Malvern Instruments, UK) at 25 °C by titrating 14-3-3ε species and 14-3-3ε peptides with Cc. The reference cell was filled with distilled water. The experiments consisted of 2-μL injections of 300 μM Cc solution in 10 mM sodium phosphate buffer (pH 7.4) into the sample cell, initially containing 20 μM 14-3-3ε solution in the same buffer. All solutions were degassed before titrations were performed. Titrant was injected at appropriate time intervals to ensure the thermal power signal returned to the base line prior to the next injection. To achieve homogeneous mixing in the cell, the stirring speed was maintained constant at 750 r.p.m. The data, specifically the heat per injection normalized per mole of injectant vs. the molar ratio, were analyzed with Origin 7.0 (OriginLab Corp.). Calibration and performance tests of the calorimeter were carried out conducting CaCl2-EDTA titrations with solutions provided by the manufacturer. Control experiments (Cc solution injected into buffer) were performed in order to assess potential unspecific heat effects.
NMR spectroscopy
NMR titration of reduced Cc with 14-3-3ε FL was followed by 1D 1H and 2D [1H, 15N] HSQC spectra on a Bruker Avance 700 MHz at 25 °C. 50 μM of reduced, 15N-labelled Cc in 5 mM sodium phosphate buffer (pH 6.3) was titrated with unlabeled 14-3-3ε in the same buffer. 0.1 M sodium ascorbate and 10% D2O were added in order to ensure the redox state remained constant and to adjust the lock signal, respectively. Water signal was suppressed according to the WATERGATE solvent suppression method46. The interaction of reduced Cc with 14-3-3ε234-255 was performed on a Bruker Avance 600 MHz in the same way as previously described for the titration. Data were processed using TopSpin NMR 2.0 software (Bruker). Line broadening and chemical-shift perturbation analysis were performed using Sparky 3 NMR Assignment Program (T.D. Goddard and D.G. Kneller, University of California – San Francisco, US). CSP titration curves were fitted using the one-site binding model.
To fluorinate the Lys244 side chain of 14-3-3ε234-255, the bioconjugation reaction was performed with 5-fluoroindole compound (Sigma) as previously described47. 500 μM acetylated 14-3-3ε234-255 peptide was mixed with 2 mM formaldehyde solution (37% w/w) and with 6 mM 5-fluoroindole for 24 h at 30 °C. Then, the sample was dialyzed against 10 mM sodium phosphate (pH 6.3) buffer overnight. Proper folding of 14-3-3ε234-255 before and after the reaction was checked by 1D 1H spectra, and the interaction with Cc was followed recording 1D 19F experiments at 25 °C on a Bruker Avance 600 MHz.
Analytical ultracentrifugation
Sedimentation velocity experiments of 14-3-3ε FL WT, D21K, K50E, S59E, E92K, D99K, and S187D were performed at 20 °C in an Optima XL-A Analytical Ultracentrifuge (AU, Beckman Instruments) with an AN50-Ti rotor. These experiments were carried out at 45,000 r.p.m. with 400-μL samples at 37 μM in 5 mM sodium phosphate buffer (pH 6.3) loaded into double sector cells. Radial scans at 280 nm were taken every 10 min and the sedimentation coefficient distribution was calculated by least squares boundary modeling of the sedimentation velocity data using the programme SEDFIT. The experimental coefficients were converted to standard conditions (s20,w).
Molecular dynamic computations
Molecular dynamics (MD) computations were carried out using the AMBER 12 package and the AMBER-2003 force field. For the 14-3-3ε234-255 peptide simulation, an unfolded and outspread conformation was used as initial coordinates. After 100 ns of simulation, the closest structure to the average from the last 40 ns of this computation was attached to the crystallographic structure of 14-3-3ε (PDB: 2BR9)15 at the C-terminal of both monomers. The resulting structure was used as the initial coordinate for the full length protein in the MD computation. With the aim to promote the opening of 14-3-3ε FL and to produce an open conformation, both consensus peptide ligands for 14-3-3 proteins included in the crystallographic structure were removed from the initial coordinates.
All simulations were carried out under periodic boundary conditions using an orthorhombic cell geometry (minimum distance between protein and cell faces was initially set to 10 Å) and PME electrostatics were set with the Ewald summation cutoff at 9 Å. Sodium counter-ions were added to neutralize the charges of the system. The structures were solvated with SPC water molecules. Afterwards, solvent and counter-ions were subjected to 2000 steps of energy minimization followed by 300 ps NPT-MD computations using isotropic molecule position scaling and a pressure relaxation time of 1 ps at 298 K. The density of the system reached a plateau during the first 150 ps. Then, the whole system was energy minimized and submitted to NVT MD computations at 298 K. The SHAKE algorithm48 was used to constrain bonds involving hydrogen atoms. The PTRAJ module of AMBER was used for trajectory analysis.
NMR-driven docking
Restrained docking calculations were performed with the High Ambiguity Driven Docking approach (HADDOCK)49–51, using the 3ZCF.pdb structure of Cc and the resulting open model of 14-3-3ε FL from MD calculations. Dielectric constant was set as distance-dependent. Scaling of ambiguous interaction restraints (AIRs) were fixed to 0.1 for rigid body and to 1.0 for both semi-flexible simulated annealing and final refinement. AIRs for the docking simulation were generated using standard criteria. Random exclusion of AIRs was not employed. Cc residues labeled as active were those showing 1H ΔΔν1/2Binding ≥ 59.2 Hz and a solvent accessibility larger than 50% calculated with NACCESS. These active residues were Gln16, Gln42, Ser47, Ala50, Gly77, Lys79, Ile81, and Val83. Cc residues located at <4 Å from the active ones and showing high-solvent accessible surface (>50%) were labeled as passive residues. 14-3-3ε residues labeled as active were those whose mutation resulted in the impairment of one of the Cc-binding sites: Asp21, Lys50, Ser59, and Glu92. 14-3-3ε residues located at <4 Å from the active ones and showing high-solvent accessible surface (>50%) were labeled as passive residues. For each run, 10,000 rigid-body solutions were generated by energy minimization. The 1000 structures with the lowest AIRs energies were subjected to semi-flexible simulated annealing. The first 500 structures were submitted to a final refinement in explicit water. The 500 best structures were analyzed using standard criteria. AIR violations were calculated using as reference the solution with best HADDOCK score. Structures were represented using UCSF Chimera52.
Dynamic light scattering measurements
Dynamic light scattering experiments were performed to evaluate the dimerization state of 14-3-3ε FL WT and D21K, K50E, S59E, E92K, D99K, and S187D mutants, in 5 mM sodium phosphate at pH 6.3. DLS experiments on 0.1 mg/mL of each 14-3-3ε species were conducted at 25 °C in a Zetasizer Nano ZS (Malvern Instruments).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 Software and experimental data of at least three independent measurements. Statistical significance was estimated using Student's t-test. A p-value of <0.0002 was considered significant.
Electronic supplementary material
Acknowledgements
This work was supported by Spanish Ministry of Economy, Industry and Competitiveness (BFU2015-71017/BMC, SAF2015-64383-P and BFU2013-47064P/BMC MINECO/FEDER, EU), Ramon Areces Foundation (2015-2017), European Social Fund 2007–2013, Andalusian Government (BIO198 and BIO778), Cámara Foundation — University of Seville grant awarded to C.A.E.-R., BIP-cicCartuja and CITIUS platforms.
Authors contributions
C.A.E.-R., A.D.-Q., K.G.-A., I.D.-M., and M.A.D.l.R. designed the research; C.A.E.-R. and A.V.-C. performed and analysed ITC measurements; C.A.E.-R. and M.O. cloned, expressed and purified APAF-1 mutants. C.A.E.-R., K.G.-A., and A.L.-R. carried out cellular experiments. C.A.E.-R., S.G.-C., A.D.-Q., and I.D.-M. performed and analysed NMR data, MD and docking calculations; C.A.E.-R., A.D.-Q., I.D.-M., and M.A.D.l.R. wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Finazzi-Agrò
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41419-018-0408-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Miguel Á. De la Rosa, Phone: +34 954489513, Email: marosa@us.es
Irene Díaz-Moreno, Phone: +34 954489513, Email: idiazmoreno@us.es.
References
- 1.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/S0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 2.Krammer PH. CD95’s deadly mission in the immune system. Nature. 2000;407:789–795. doi: 10.1038/35037728. [DOI] [PubMed] [Google Scholar]
- 3.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, et al. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 5.Díaz-Moreno I, García-Heredia JM, Díaz-Quintana A, De la Rosa MA. Cytochrome c signalosome in mitochondria. Eur. Biophys. J. 2011;40:1301–1315. doi: 10.1007/s00249-011-0774-4. [DOI] [PubMed] [Google Scholar]
- 6.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- 7.Li P, et al. Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/S0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 8.Zou H, Li Y, Liu X, Wang X. An Apaf-1·cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Fábregas J, et al. New Arabidopsis thaliana cytochrome c partners: a look into the elusive role of cytochrome c in programmed cell death in plants. Mol. Cell. Proteom. 2013;12:3666–3676. doi: 10.1074/mcp.M113.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Fábregas J, et al. Structural and functional analysis of novel human cytochrome c targets in apoptosis. Mol. Cell. Proteom. 2014;13:1439–1456. doi: 10.1074/mcp.M113.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Arzola K, et al. Structural basis for inhibition of the histone chaperone activity of SET/TAF-Iβ by cytochrome c. Proc. Natl Acad. Sci. USA. 2015;112:9908–9913. doi: 10.1073/pnas.1508040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Fabregas J, Díaz-Moreno I, González-Arzola K, Díaz-Quintana A, De la Rosa MA. A common signalosome for programmed cell death in humans and plants. Cell Death Dis. 2014;5:e1314. doi: 10.1038/cddis.2014.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore B, Perez V. Physiological and biochemical aspects of nervous integration. In: Carlson F, editor. Specific acidic proteins of the nervous system. Englewood Cliffs, NJ: Prentice-Hall; 1967. pp. 343–359. [Google Scholar]
- 14.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, et al. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl Acad. Sci. USA. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petosa C, et al. 14-3-3ζ binds a phosphorylated raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 1998;273:16305–16310. doi: 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- 17.Masters SC, Pederson KJ, Zhang L, Barbieri JT, Fu H. Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry. 1999;38:5216–5221. doi: 10.1021/bi982492m. [DOI] [PubMed] [Google Scholar]
- 18.Aachmann FL, Fomenko DE, Soragni A, Gladyshev VN, Dikiy A. Solution structure of selenoprotein W and NMR analysis of its interaction with 14-3-3 proteins. J. Biol. Chem. 2007;282:37036–37044. doi: 10.1074/jbc.M705410200. [DOI] [PubMed] [Google Scholar]
- 19.Aitken A, Ellis CA, Harris A, Sellers LA, Toker A. Kinase and neurotransmitters. Nature. 1990;344:594–594. doi: 10.1038/344594a0. [DOI] [PubMed] [Google Scholar]
- 20.Toker A, Ellis CA, Sellers LA, Aitken A. Protein kinase C inhibitor proteins. Eur. J. Biochem. 1990;191:421–429. doi: 10.1111/j.1432-1033.1990.tb19138.x. [DOI] [PubMed] [Google Scholar]
- 21.Aitken A, Howell S, Jones D, Madrazo J, Patel Y. 14-3-3 and are the phosphorylated forms of raf-activating 14-3-3 and in vivo stoichiometric phosphorylation in brain at a Ser-Pro-Glu-Lys motif. J. Biol. Chem. 1996;270:5706–5709. doi: 10.1074/jbc.270.11.5706. [DOI] [PubMed] [Google Scholar]
- 22.Tsuruta F, et al. JNK promotes Bax translocation to mitochondria through phosphorylation of 14‐3‐3 proteins. EMBO J. 2004;23:1889–1899. doi: 10.1038/sj.emboj.7600194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darling DL, Yingling J, Wynshaw‐Boris A. Role of 14–3–3 proteins in eukaryotic signaling and development. Curr. Top Dev. Biol. 2005;68:281–315. doi: 10.1016/S0070-2153(05)68010-6. [DOI] [PubMed] [Google Scholar]
- 24.Paul G, van Heusden H. 14-3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB Life. 2005;57:623–629. doi: 10.1080/15216540500252666. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, et al. Rsk-mediated phosphorylation and 14-3-3ε binding of Apaf-1 suppresses cytochrome c-induced apoptosis. EMBO J. 2012;31:1279–1292. doi: 10.1038/emboj.2011.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allan LY, et al. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat. Cell. Biol. 2003;5:647–654. doi: 10.1038/ncb1005. [DOI] [PubMed] [Google Scholar]
- 27.Palacios-Rodriguez Y, et al. Polypeptide modulators of caspase recruitment domain (CARD)-CARD-mediated protein-protein interactions. J. Biol. Chem. 2011;286:44457–44466. doi: 10.1074/jbc.M111.255364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y. Sphingosine activates protein kinase A type II by a novel cAMP-independent mechanism. J. Biol. Chem. 2005;280:26011–26017. doi: 10.1074/jbc.M409081200. [DOI] [PubMed] [Google Scholar]
- 29.Gu YM, et al. Protein kinase A phosphorylates and regulates dimerization of 14-3-3ζ. FEBS Lett. 2006;580:305–310. doi: 10.1016/j.febslet.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 30.Kim JS, Diebold BA, Babior BM, Knaus UG, Bokoch GM. Regulation of Nox1 activity via protein kinase A-mediated phosphorylation of NoxA1 and 14-3-3 binding. J. Biol. Chem. 2007;282:34787–34800. doi: 10.1074/jbc.M704754200. [DOI] [PubMed] [Google Scholar]
- 31.Moreno-Beltrán B, et al. Cytochrome c1 exhibits two binding sites for cytochrome c in plants. Biochim. Biophys. Acta. 2014;1837:1717–1729. doi: 10.1016/j.bbabio.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 32.Moreno-Beltrán B, et al. Respiratory complexes III and IV can each bind two molecules of cytochrome c at low ionic strength. FEBS Lett. 2015;589:476–483. doi: 10.1016/j.febslet.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Sakamoto K, et al. NMR basis for interprotein electron transfer gating between cytochrome c and cytochrome c oxidase. Proc. Natl Acad. Sci. USA. 2011;108:12271–12276. doi: 10.1073/pnas.1108320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Díaz-Moreno I, Díaz-Quintana A, Ubbink M, De la Rosa MA. Different modes of interaction in cyanobacterial complexes of plastocyanin and cytochrome. F. Biochem. 2005;44:3176–3183. doi: 10.1021/bi047855z. [DOI] [PubMed] [Google Scholar]
- 35.Ferrell JE, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 36.Díaz-Moreno I, García-Heredia JM, Díaz-Quintana A, Teixeira M, De la Rosa MA. Nitration of tyrosines 46 and 48 induces the specific degradation of cytochrome c upon change of the heme iron state to high-spin. Biochim. Biophys. Acta. 2011;1807:1616–1623. doi: 10.1016/j.bbabio.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 37.García-Heredia JM, et al. Specific nitration of tyrosines 46 and 48 makes cytochrome c assemble a non-functional apoptosome. FEBS Lett. 2012;586:154–158. doi: 10.1016/j.febslet.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Guerra-Castellano A, et al. Mimicking tyrosine phosphorylation in human cytochrome c by the evolved tRNA synthetase technique. Chemistry. 2015;21:15004–15012. doi: 10.1002/chem.201502019. [DOI] [PubMed] [Google Scholar]
- 39.Guerra-Castellano A, Díaz-Moreno I, Velázquez-Campoy A, De la Rosa MA, Díaz-Quintana A. Structural and functional characterization of phosphomimetic mutants of cytochrome c at threonine 28 and serine 47. Biochim. Biophys. Acta. 2016;1857:387–395. doi: 10.1016/j.bbabio.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Moreno-Beltrán B, et al. Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48. Proc. Natl Acad. Sci. USA. 2017;114:3041–3050. doi: 10.1073/pnas.1618008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.González-Arzola K, et al. Histone chaperone activity of Arabidopsis thaliana NRP1 is blocked by cytochrome c. Nucleic Acids Res. 2017;45:2150–2165. doi: 10.1093/nar/gkw1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kingston, R. E., Chen, C. A. & Rose, J. K. Calcium phosphate transfection. Curr. Protoc. Mol. Biol.10.1002/0471142727.mb0901s63. (John Wiley & Sons, Inc., 2003). [DOI] [PubMed]
- 43.Pecina P, et al. Phosphomimetic substitution of cytochrome c tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Biochemistry. 2010;49:6705–6714. doi: 10.1021/bi100486s. [DOI] [PubMed] [Google Scholar]
- 44.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 45.Slee EA, et al. Ordering the Cytochrome c–initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9–dependent manner. J. Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 47.Larda ST, Pichugin D, Prosser RS. Site-specific labeling of protein lysine residues and N-terminal amino groups with indoles and indole-derivatives. Bioconjug. Chem. 2015;26:2376–2383. doi: 10.1021/acs.bioconjchem.5b00457. [DOI] [PubMed] [Google Scholar]
- 48.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 49.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: a protein−protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 50.van Dijk ADJ, Boelens R, Bonvin AMJJ. Data-driven docking for the study of biomolecular complexes. FEBS. J. 2005;272:293–312. doi: 10.1111/j.1742-4658.2004.04473.x. [DOI] [PubMed] [Google Scholar]
- 51.de Vries SJ, et al. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.