Abstract
Zerumbone is a known anti-cancer herbal compound. However, the actual protein target is not fully understood or known. This investigation focus on the association of zerumbone in HCT116 colon cancer cell proliferation and its link with TNF-alpha. The study shows that with the increasing concentration of zerumbone, there was a reduction of HCT116 cells proliferation based on the cell line study and hence higher TNF-alpha inhibition based on the TNF-alpha assay. The study also emphasizes on the computational aspect by investigating the molecular docking analysis of zerumbone against TNF-alpha. The docked complex was further validated using molecular dynamics simulation studies. The docking analysis observed that alpha-beta unsaturated carbonyl scaffold is an important moiety for the anti-cancer activity of zerumbone. Furthermore, the DFT analysis also confirms the reactivity nature of zerumbone based on the frontier molecular orbital analysis.
Introduction
Colorectal or colon cancers occur because of unhealthy lifestyle and old age and in some cases because of genetic factors1,2. It is also the third-leading cause of cancer-related deaths in US3. The risk factors include smoking, obesity, unhealthy diet etc4. The risk factors also include Crohn’s disease and ulcerative colitis which are part of inflammatory bowel disease5.
On the other hand, zerumbone has been tested for various anti-cancer activities including colon cancer. Zerumbone is a cyclic sesquiterpene extracted from the rhizomes of Zingiber zerumbet Smith6. Its rhizomes have been used traditionally for folk medicine and pain relief 7.
Moreover, recent study on zerumbone observed that zerumbone possess unique and potent anticancer activity against colon cancers cells8. It has also shown to inhibit human colonic adeno-carcinoma cell proliferation9. The present investigation deals with the anti-cancer activity of zerumbone against HCT116 cells (colon cancer cell line) and its subsequent role in TNF-alpha inhibition in cancer cell proliferation. The linkage of zerumbone inhibiting TNF-alpha and its role in the proliferation of colon cancer cell has never been reported or study.
The TNF gene, in both the mouse and human, is positioned in the major histocompatibility complex, close to the LT3 gene10,11. Because of this chromosomal arrangement and their sequence homology TNF has also become known as TNF-alpha12. Although the cytotoxic activity of TNF has been well documented, the mechanism of TNF-induced lysis is not well understood13. In fact, the TNF is cytotoxic for many tumoral cell lines, whereas normal cells generally are considered resistant to this action14. In F17 cells, TNF treatment induced a classical form of apoptosis, while TNF induced a necrotic form of cell death in L-M cells13. The TNF-alpha is also a pleiotropic cytokine that activates T-cells via both TNF-Receptor (R-I and R-II) and mediates both apoptotic and survival signals15–17. The TNF-alpha mediates its biological functions predominantly via TNFR-I. Following binding of TNF-alpha to TNFR-I, the TNFR-associated death domain (TRADD) is recruited to TNFR-I forming a platform for downstream signaling18,19. Then, the TNRF-associated factor 2 (TRAF2) and receptor-interacting protein kinase 1 (RIPK1) are recruited to TRADD forming a signaling complex. The TRADD also recruits fas-associated death domain (FADD), which initiates activation of apical caspases resulting in activation of effector caspases, and apoptosis20,21. However, signaling through TNFR1 might also induce apoptosis in a caspase-independent manner22.
The present study involves morphological changes of HCT116 Cells treated with zerumbone in various concentrations and the supernatant was utilize for the inhibition of TNF-alpha using Mouse TNF-alpha ELISA kit. The study was cross examined using molecular docking techniques with zerumbone against TNF-alpha enzyme (PDB ID 5MU8) and the stability of the protein-ligand docked complex confirmed by molecular dynamics simulation. Furthermore, the frontier molecular orbital study was carried out to understand the HOMO and LUMO energies of zerumbone that might contribute in the reactivity of the molecule at the active site of the enzyme.
Materials/Experimental
Cell line study and TNF-alpha assay
Z. zerumbet sample was collected from Moreh, Manipur, India (94.34217° E and 24.35172° N) and given the institutional accession number as IBSD/Z-42-23. Zerumbone was extracted from the oil extract of the fresh rhizomes and identified by FT-IR, Proton and 13 C NMR and mass spectrophotometer.
HCT116 cells were purchased from ATCC (Manassas, VA) and cultured in McCoy’s 5 A medium supplemented with 10% FBS having 100 units/ml penicillin and 100 μg/ml streptomycin solution in a humidified atmosphere of 5% CO2 at 37 °C. Cells were seeded in 24-well plate with a density of 5 × 104 cells/well. To adhere the cells it was incubated for 16 h and treated with cisplatin (25 µg/µL) (Sigma Aldrich, USA) and zerumbone in various concentration (5, 10 and 20 μM) for 24 h. The cells from the media was collected and centrifuged for 15 min at 5000 rpm and the supernatant was utilize for the TNF-alpha assay using Mouse TNF-alpha ELISA kit (Millipore, USA) as per the standard protocol of the manufacturer.
The release of TNF-alpha level in the HCT116 cells was examined for the zerumbone concentrations (5, 10 and 20 μM). Cisplatin, a known anti-cancer drug was used as positive control and the untreated cells were used as the negative control.
The results were expressed as the mean ± SEM for three replicates. The analysis was done in GraphPad Prism 7 (https://www.graphpad.com/). All the graphs and figures were prepared using the mean values in GraphPad Prism 7.
Flexible docking
Human TNF-alpha (PDB ID: 5MU8) was retrieved from Protein Data Bank (http://www.rcsb.org/pdb/explore.do?structureId = 5mu8) and loaded in Molegro Virtual Docker 6.02 (MVD). The binding site was predicted and set at the coordinates (X: −21.51, Y: 18.21, Z: −1.82) (Fig. S1 of supplementary material). The residues surrounding the active site region (Glu 23, Val 150, Phe 144, Leu 26, Tyr 141, Asn 19). On the other hand the 3D conformer of zerumbone and α-humulene was retrieved from PubChem and optimized using Gaussian 09 and loaded in the MVD. MolDock Grid Score was set as the scoring function with a grid resolution of 0.30 Å. Additional evaluation for Internal ES (ElectroStatic) and Internal Hbond (Hydrogen bond) were also set for the simulation. The algorithm was set at a maximum iteration of 1,500 having a maximum evolution size of 50. The docking engine was run for 1000 times for better accuracy and minimum of 1000 poses23. The best pose of zerumbone and α-humulene was considered for the ligand–protein interaction analysis and MD simulation studies.
Binding affinity prediction
The binding affinity of the best docked pose of zerumbone and α-humulene was calculated using the MLR equation implemented in Molegro Data Modeller (Affinity = −19.0155 * C0 + 3.3813 * ′′Cofactor (hbond)′′− 0.594128 * Csp2 − 0.061912 * ′′E-Intra (vdw)′′ + 0.464056 * HBond + 0.483368 * HeavyAtoms + 0.953672 * ′′E-Solvation′′ + 3.0229 * Nplus − 3.9493 * halogen − 1.00763 * N − 3.10696 * OS + 1.61426 * OH).
Molecular dynamics simulation studies
MD simulations were carried out for 5MU8, 5MU8-zerumbone and 5MU8-α-humulene docked complex. MD simulation was carried out using GROMACS version 5.1.2 operated on Ubuntu 16.04. The topology file for PDB ID 5MU8 was processed with OPLS-AA/L force field while 5MU8-zerumbone complex was processed with GROMOS96 43a124. The system was solvated using the equilibrated 3-point solvent model (spc216) and ions were added. The assembled system was relaxed by energy minimization and equilibrated using the NVT and NPT scale for 100 ps. The well-equilibrated was finally run for 30 ns MD production. The generated trajectory file was analyzed for RMSD, RMSF, SASA, Hbond and Radius of gyration.
MM-PBSA calculations
To adhere the molecular docking simulation results and MD simulation trajectory analysis, the binding free-energy (ΔGbind) calculations were carried out for the 30 ns simulated complexes of the 5MU8-Zerumbone complex and 5MU8-α-humulene complex using g_mmpbsa (http://rashmikumari.github.io/g_mmpbsa/). The Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) estimates the free energy interactions as defined by Kollman et al.25. The equation of the ΔGbind is represented by the following equation
| 1 |
The binding free-energy of the 5MU8-Zerumbone and 5MU8-α-humulene complexes were analyzed by taking 100 snapshots at an interval of 100 ps from the last 30 ns MD simulation during its equilibrium phase using g_mmpbsa and the final average binding energy was obtained by executing the MmPbSaStat.py python script.
DFT Calculations
The conformation of the best docking pose of zerumbone and α-humulene was exported and DFT calculations were carried out using Gaussian 0926. Theoretical calculations were carried out at Ground State using DFT/B3LYP/LanL2DLZ. The guess method was set as extended Huckel with mix HOMO and LUMO orbital. Population analysis was performed for Merz-Kollman (ESP) charges for full natural bond order (NBO) analysis. The molecular orbital energies were considered for calculating the band energy gap ΔELUMO-HOMO.
Physicochemical properties and ADME-Toxicity calculation
The Physicochemical property and ADME-Toxicity parameters of zerumbone and α-humulene were computed using SwissADME27 to estimate the pharmacokinetics and their medicinal chemistry property. These parameters were calculated because drug development and discovery process involve assessment of absorption, distribution, metabolism and excretion and their toxic effects27. The newly introduced methods such as BOILED-Egg diagram, iLOGP and Bioavailability Radar map was also calculated for zerumbone and α-humulene. Lastly, LD50 and health effect analysis was carried out using ACD Ilab 2.0 (https://ilab.acdlabs.com/iLab2/) and comparative a chart was plotted to compare the two molecules.
Compliance with Ethical Standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Results and Discussion
The cell line assay of HCT116 observed the death of the cells after 24 hours of treatment with 20–30 μM zerumbone concentration (Fig. 1A,B). The cell lines snap also portrayed that the amount of dead cells in zerumbone (30 μM) treated cells (Fig. 1A(iv)) is more than the cells treated with cisplatin (Fig. 1A(ii)). The study also observed that 20–30 μM of zerumbone was adequate enough to inhibit the TNF-alpha with a higher TNF-alpha release value of 10.61 in case of 20 μM of zerumbone (Table 1). The positive control (cisplatin) has a lower TNF-alpha release value of 6.01 (Table 1) which implies zerumbone as good inhibitor than the known anti-cancer compound. However, zerumbone treated HCT116 in the concentration of 5 μM and 10 μM were unable to kill the cells with more than 35% survival rate in case of 10 μM and 56% survival rate in case of 5 μM zerumbone concentration (Fig. 1B). The TNF-alpha release value was 7.08 and 7.61 respectively for the zerumbone concentration 5 μM and 10 μM (Table 1). In fact, the activity of the zerumbone is because of the alpha-beta unsaturated carbonyl groups which are considered as an active moiety. Additionally, the dose-response curve and bar graph plot from the ELISA assay confirms that 20 μM of zerumbone was good enough to inhibit the TNF-alpha enzyme (Fig. SF2 A and B of supplementary material). Thus zerumbone was found to be active against HCT116 cells leading to apoptosis.
Figure 1.
(A) Morphological changes of HCT116 cells treated with zerumbone (iii, iv, v & vi) and cisplatin (ii) after 24 h hours. Control cells are shown in photos (i) for 24 h. and (B) Graph showing the dose dependent inhibition of cell proliferation. HCT116 cells were treated either with DMSO or with the indicated concentration of zerumbone or cisplatin and the viable cells were measured with the Cell Titer 96 Proliferation Assay (Promega). The data represent the percentage growth compared with DMSO. The data represent the average of 3 independent experiments, with standard deviations.
Table 1.
ELISA assay for TNF-alpha inhibition.
| SN | Compound and Concentration | TNFα inhibition ± SEM |
|---|---|---|
| 1 | Cisplatin (25 µg/µL) | 6.01 ± 0.02 |
| 2 | Zerumbone (2 μM) | 5.80 ± 0.07 |
| 3 | Zerumbone (5 μM) | 7.08 ± 0.30 |
| 4 | Zerumbone (10 μM) | 7.61 ± 0.42 |
| 5 | Zerumbone (15 μM) | 8.42 ± 0.26 |
| 6 | Zerumbone (20 μM) | 10.05 ± 0.15 |
The docking score of the best top five conformational pose of zerumbone is presented in Table 2. Whereas the docking score of the best top five conformational pose α-humulene is presented in Table 3. In both the cases (Tables 2 and 3), the best pose was chosen based on the MolDock score, rerank score, interaction energy and binding affinity. The best pose of zerumbone (Pose No. 545) had an overall score of −138.41 kJ/mol (Table 2). While the best pose of α-humulene had an overall score of −121.78 kJ/mol (Table 3). Thus, indicating that zerumbone is a better inhibitor of TNF-alpha than α-humulene. Interestingly, the structure of zerumbone is 90% similar to α-humulene except that zerumbone has an extra alpha-beta unsaturated carbonyl group which makes zerumbone an active biomolecule. And this may be the reason for zerumbone possessing an overall favourable docking score than α-humulene at the active site of the TNF-alpha enzyme.
Table 2.
Docking score of the top 5 docking poses of zerumbone.
| SN | Docking Pose | MolDock Score | Rerank Score | Interaction Energy | Binding affinity | Overall Score |
|---|---|---|---|---|---|---|
| 1 | Pose No. 545 | −45.46 | −31.12 | −44.01 | −17.82 | −138.41 |
| 2 | Pose No. 968 | −45.76 | −31.13 | −44.30 | −16.66 | −137.85 |
| 3 | Pose No. 481 | −44.63 | −31.44 | −43.18 | −16.66 | −135.91 |
| 4 | Pose No. 97 | −41.70 | −31.69 | −40.26 | −16.66 | −130.31 |
| 5 | Pose No. 676 | −38.61 | −28.58 | −37.15 | −17.82 | −122.16 |
Table 3.
Docking score of the top 5 docking poses of α-humulene.
| SN | Docking Pose | MolDock Score | Rerank Score | Interaction Energy | Binding affinity | Overall Score |
|---|---|---|---|---|---|---|
| 1 | Pose No. 616 | −39.66 | −29.17 | −35.98 | −16.97 | −121.78 |
| 2 | Pose No. 266 | −38.13 | −27.62 | −34.45 | −16.97 | −117.17 |
| 3 | Pose No. 436 | −37.93 | −26.41 | −34.25 | −16.98 | −115.57 |
| 4 | Pose No. 295 | −37.15 | −26.32 | −33.47 | −16.95 | −113.89 |
| 5 | Pose No. 385 | −34.57 | −25.30 | −30.90 | −16.97 | −107.74 |
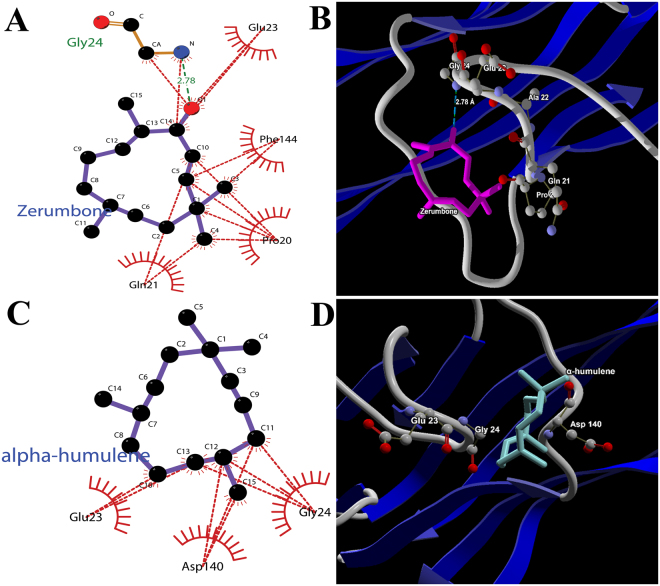
Figures 2A,B depicted the steric and hydrophobic interaction of zerumbone with Pro20, Gln21, Gly23, Gly24 and Phe144 residues of 5MU8. While Fig. 2C,D depicted the interaction of α-humulene with Glu23, Glu24 and Asp140. In case of TNF-alpha-zerumbone docked complex, a hydrogen bond interaction was observed with Gly24 whereas no hydrogen bonding was observed in TNF-alpha- α-humulene docked complex. Thus, suggesting a stronger binding affinity and hence a concrete molecular interaction in case of TNF-alpha-zerumbone docked complex.
Figure 2.
(A) 2D interaction map of zerumbone (B) 3D interaction map of zerumbone (C) 2D interaction map of α-humulene and (D) 3D interaction map of α-humulene at the active site of TNF-alpha enzyme (PDB ID: 5MU8). One hydrogen bond interaction (Gly24) and four hydrophobic interactions (Pro20, Gln21, Glu23 and Phe144) were observed in case of zerumbone with. Whereas only three hydrophobic interactions (Glu23, Glu24 and Asp140) was observed in case of α-humulene. Thus indicating a stronger binding force in zerumbone-TNF-alpha docked complex.
The present report on the association of zerumbone and TNF-alpha using in vitro techniques is the first such report which is supported by computational studies. Moreover, the investigated enzyme is a TNF-α promotion signaling pathway that interferes with the proliferation of colon cancer cells. The strong interaction of zerumbone is because of the alpha-beta unsaturated carbonyl group which is a notable moiety that theater in certain unidentified targets and enzyme28. Similarly, Murakami et al. observed that the absence of alpha-beta unsaturated carbonyl group in compounds such as alpha-humulene, were unable to disrupt various tumour promoter enzymes such as 12-O-Tetradecanoylphorbol-13-acetate (TPA)7. In fact the difference in their structure lies in the carbonyl group present in zerumbone but not in alpha-humulene29. Thus, indicating that the alpha-beta unsaturated carbonyl group is the key component of zerumbone for its biological activity against cancer cell lines and protein targets30. The overall score revealed favourable binding energy and strong bonding which were confirmed from the docking scores with the TNF-α enzyme (Tables 2 and 3).
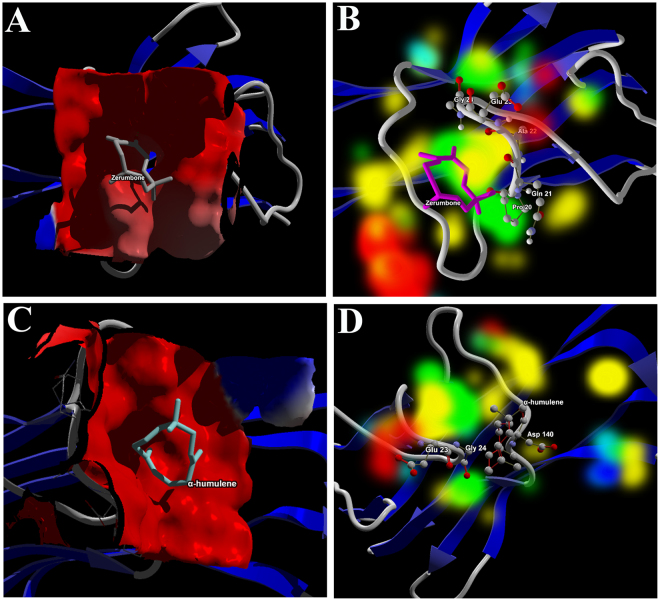
Figure 3 depicts the electrostatic interaction map and energy map of the best docking pose of zerumbone and α-humulene at the binding site of the TNF-alpha (5MU8). It is evident from both the maps that zerumbone possessed fair affinity of energy based interactions compared to α-humulene. As depicted in Fig. 3A,C, zerumbone lies inside the surface of the electrostatic pocket whereas in α-humulene lies above the surface of the electrostatic pocket. Moreover, the 3D hydrophobic interaction map of zerumbone and α-humulene observed that zerumbone lies closer to the hydrophobic pocket compared to α-humulene (Figs SF3 and SF4 of supplementary material)
Figure 3.
Electrostatic interaction map of (A) zerumbone and (C) α-humulene. Energy map of (B) zerumbone and (D) α-humulene at the active site of TNF-alpha enzyme. Figure 3A depicted zerumbone lying inside the surface of the Electrostatic pocket whereas in Fig. 3B, α-humulene lies above the surface of electrostatic pocket.
Additionally, zerumbone is surrounded by a pool of energy based favourable interactions such as hydrogen bond donor/acceptor favourable regions, electrostatic favourable and steric favourable regions (Fig. 3B). However, there are no experimental data available for zerumbone binding to TNF-alpha enzyme based on the available reports. Thus, there is a fair affinity between zerumbone and TNF-alpha.
Figure 4A depicts the RMSD graph processed from the trajectory file of the 30 ns MD production. The 5MU8-zerumbbone complex showed a minor shift at 15 ns, but a stable conformation in the preceding MD production till 30 ns. However, the 5MU8-α-humulene complex observed a major shift and deviations in the first 20 ns MD production as evident from the RMSD graph (Fig. 4A). Hence, the 5MU8-zerumbbone docked complex confirms a stable conformation. On the other hand, Fig. 4B depicts the SASA energy analysis where the graph depicted a uniform hydrophobicity interaction of the protein-ligand complexes during the 30 ns run. However, there were not many variations in the RMSF values and Rg graph between the two zerumbone and α-humulene complexes (Figs SF5 and SF6 of supplementary material). While Figs. 5A, B and C represent the measurement of the hydrogen bond distances of the 5MU8, 5MU8-zerumbone and 5MU8-α-humulene docked complexes. Figure 5B confirms that the docked complex have more hydrogen bond interaction raising up to four numbers thus showing a stable system and a good hydrogen bond interaction.
Figure 4.

Trajectories from the MD Simulation production representing (A) RMSD (root mean squared deviation) and (B) Radius of gyration of the TNF-alpha and TNF-alpha-zerumbone and α-humulene docked complex.
Figure 5.

Hbond interaction number of (A) TNF-aplha (B) TNF-alpha-zerumbone and (C) α-humulene docked complex.
The binding free energy calculation of the protein-ligand interaction from the 100 snapshots extracted every 100 ps from the last 30 ns MD simulation stable trajectory is shown in Table 4. The binding free-energy (ΔGbind) values of the 5MU8-zerumbone complex and 5MU8-α-humulene complex were −52.10 kJ/mol and −40.23 kJ/mol respectively. The ΔGbind values suggest that 5MU8-zerumbone complex have better binding free-energy than the 5MU8-α-humulene complex.
Table 4.
Binding free-energy (ΔGbind) of TNF-alpha-zerumbone and TNF-alpha-α-humulene docked complex calculated using g_mmpbsa.
| Energy parameters (kJ/mol) | Zerumbone (kJ/mol) | α-humulene (kJ/mol) |
|---|---|---|
| van der Waal energy | −66.06 ± 43.07 | −53.12 ± 33.12 |
| Electrostatic energy | −11.80 ± 16.70 | −8.12 ± 11.45 |
| Polar solvation energy | 31.46 ± 34.61 | 24.13 ± 27.32 |
| SASA energy | −5.70 ± 3.80 | −3.12 ± 2.21 |
| Binding energy | −52.10 ± 38.67 | −40.23 ± 34.12 |
In Table 3, the van der Waals energy and electrostatic energy was found higher for the zerumbone complex than α-humulene complex. As it has been seen earlier in docking and interaction analysis study, that zerumbone formed hydrogen bond interaction with Gly24 whereas α-humulene does not form any hydrogen bond interaction contact (Fig. 2C,D). The 5MU8-zerumbone complex proves to be stronger than 5MU8-α-humulene complex. In fact, the hydrogen bond interaction also played an important role in stabilizing the complexes energetically. Overall, all the energy parameters showed significantly favourable in the zerumbone complex thus indicating a favourable binding free-energy than α-humulene complex.
Moreover, the electrostatic energy and SASA energy were also favourable for the binding. These results also suggested that hydrophobic interaction played an important role in protein-ligand binding.
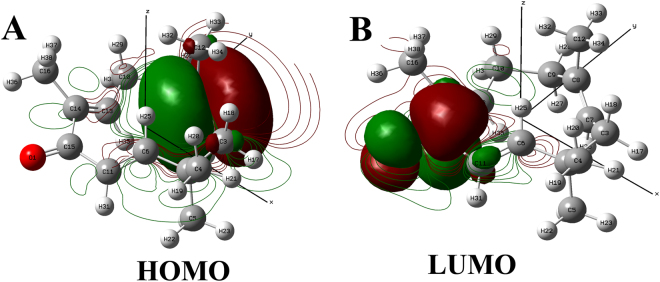
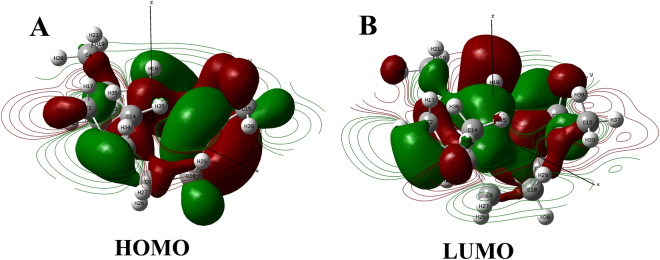
Figures 6 and 7 depict the HOMO and LUMO energies of the best docked pose of zerumbone and α-humulene. While The HOMO and LUMO energies aid in understanding the band gap energy of the docked pose. A low band gap indicates a higher reactivity and thus the compound is believed to be strong enough to bind at the protein’s active site. The band gap energy was (ΔELUMO - HOMO) was −0.12 eV and −0.08 eV for zerumbone and α-humulene which confirms that zerumbone had a lower band gap and will definitely have a strong binding affinity at the active site of TNF-alpha (5MU8). Additionally, the predicted NMR and IR spectra of zerumbone and α-humulene which optimized at DFT/B3LYP/LanL2DLZ level of theory are presented in Figs SF7–SF10 in supplementary material
Figure 6.
Contour map depicting the (A) HOMO and (A) LUMO orbital energies of zerumbone calculated at DFT/B3LYP/ LanL2DLZ level of theory.
Figure 7.
Contour map depicting the (A) HOMO and (A) LUMO orbital energies of α-humulene calculated at DFT/B3LYP/ LanL2DLZ level of theory.
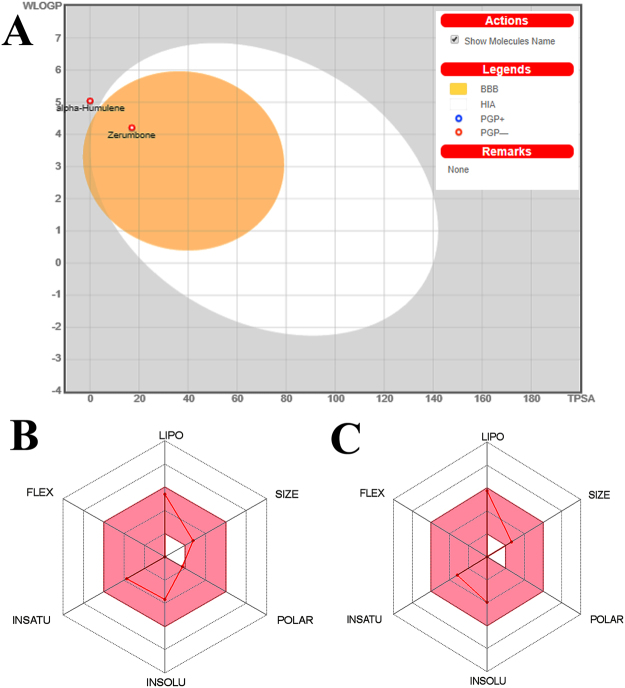
The Boiled egg diagram and bioavailability radar map of zerumbone and α-humulene are presented in Fig. 8. As evident from the Fig. 8 zerumbone has a better oral bioavailability than α-humulene. The physiochemical property analysis also revealed that zerumbone had TPSA of 17.07 Ų compared to 0.00 Ų of α-humulene which indicates zerumbone is a good enzyme inhibitor (Table 5 and Fig. 9A). Table 6 revealed that zerumbone and α-humulene having similar behaviour of ADME parameters with a similar rate of absorption and Blood-Brain Barrier. Figure 9B and Table ST1 of supplementary observed that zerumbone is likely to have some chance of toxic effect on kidney and cardiovascular system. However, Fig. 9C and Table 7 confirms that the LD50 of zerumbone is much higher than α-humulene.
Figure 8.
(A) Boiled egg diagram of zerumbone and α-humulene and Bioavailability Radar map of depicting the LIPO (lipophilicity), SIZE (molecular weight), POLAR (pplarity), INSOLU (insolubility) INSATU (insaturation) and FLEX (rotatable bond flexibility) parameters of (B) zerumbone and (C) α-humulene.
Table 5.
Physiochemical properties of zerumbone and α-humulene.
| Properties | Zerumbone | α-humulene |
|---|---|---|
| 2D Structure |
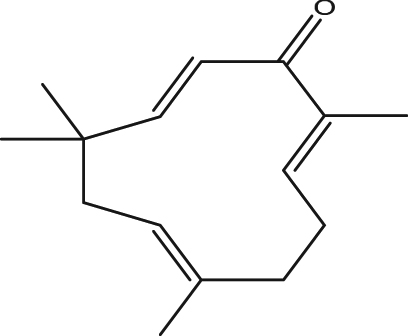
|
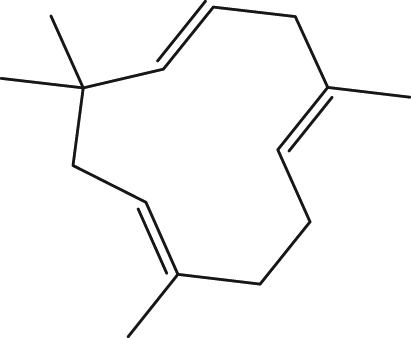
|
| Formula | C15H22O | C15H24 |
| Molecular weight (MW) | 218.33 g/mol | 204.35 g/mol |
| Num. heavy atoms (NHA) | 16 | 15 |
| Num. arom. heavy atoms (NAHA) | 0 | 0 |
| Fraction Csp3 (FCsp3) | 0.53 | 0.60 |
| Num. H-bond acceptors (NH-BA) | 1 | 0 |
| Num. H-bond donors (NH-BD) | 0 | 0 |
| Molar Refractivity (MR) | 70.62 | 70.42 |
| Topological Polar Surface Area (TPSA) | 17.07 Ų | 0.00 Ų |
| XLogP3-AA | 3.9 | 4.5 |
Figure 9.
(A) Physiochemical properties of zerumbone and α-humulene representing the molecular weight (MW), number of heavy atoms (NHA); fraction Csp3 (FCsp3); number of H-bond acceptors (NH-BA); molar refractivity (MR), topological polar surface area (TPSA) and XLogP3-AA. (B) Toxicity analysis of zerumbone and α-humulene on blood, cardiovascular system (CS), gastrointestinal system (GS), kidney, liver and lungs. (C) LD50 analysis of Zerumbone and α-humulene on Mouse/Intraperitoneal (M-IP); Mouse/Oral (M-O); Mouse/Intravenous (M-IV); Mouse/Subcutaneous (M-SC); Rat/Intraperitoneal (R-IP) and Rat/Oral (R-O).
Table 6.
ADME parameters of zerumbone and α-humulene computed using ACD iLab.
| ADME parameters | Zerumbone | α-humulene |
|---|---|---|
| Absorption | 8.43 × 10−4 cm/s | 8.46 × 10−4 cm/s |
| Absorption rate Ka | 0.058 min−1 | 0.058 min−1 |
| Blood-Brain Barrier (BBB) | ||
| LogPS | −1 | −1 |
| LogPB | 0.37 | 0.35 |
| Log(PS*fu, brain) | −2.9 | −3.2 |
| LogBB | 0.37 | 0.35 |
| LogPS | −1 | −1 |
| DBP | 96.92% | 98.59% |
| Vd | 2.23 L/kg | 3.71 L/kg |
| Pgp Substrate | Non-substrate | Non-substrate |
| Pgp Inhibitor | Non-inhibitor | Non-inhibitor |
LogPS: Rate of brain penetration; LogPB: Extent of brain penetration; Log(PS*fu, brain): Brain/plasma equilibration rate; LogBB: Blood-Brain Distribution (rodent); LogPS: BBB Permeability (rat); DBP: Drug binding to plasma proteins (human); Vd: Voume of Distrubtutin (human); Pgp Substrate: P-glycoprotein substrate specificity; Pgp Inhibitor: P-glycoprotein inhibitor specificity.
Table 7.
LD50 analysis of Zerumbone and α-humulene.
| LD50 Species /Administration route | LD50 of Zerumbone (mg/kg) | Reliability | LD50 of α-humulene (mg/kg) | Reliability |
|---|---|---|---|---|
| Mouse/Intraperitoneal | 480 | Moderate(0.66) | 150 | Borderline(0.37) |
| Mouse/Oral | 2700 | Moderate(0.51) | 960 | Borderline(0.38) |
| Mouse/Intravenous | 100 | Moderate(0.51) | 34 | Not Reliable(0.27) |
| Mouse/Subcutaneous | 1400 | Borderline(0.44) | 560 | Moderate(0.55) |
| Rat/Intraperitoneal | 210 | Borderline(0.38) | 110 | Borderline(0.44) |
| Rat/Oral | 3400 | High(0.8) | 1400 | Borderline(0.44) |
Conclusions
The present investigation confirms the inhibition of TNF-alpha by zerumbone and the decrease in HCT116 Cell proliferation in controlling colon cancer. The molecular docking study also confirms the binding mode of zerumbone at the active site of the enzyme. Moreover, the computational investigation such as the binding free-energy analysis on zerumbone and α-humulene point out that the presence of the alpha-beta unsaturated carbonyl group is a major driving force. Because, the compound α-humulene which lacks the alpha-beta unsaturated carbonyl group proved to be a bad inhibitor from the computational investigation as well as from the physiochemical property analysis and ADME-Toxicity analysis. In fact, the strong binding affinity and favourable docking score observed in zerumbone is because of the alpha-beta unsaturated carbonyl group which is also responsible for apoptosis. The 30 ns MD simulation confirmed the conformational stability based on the trajectory analysis and the DFT analysis confirms the reactivity nature of zerumbone based on the frontier molecular orbital analysis. Additionally, the physiochemical property and ADME-Toxicity analysis also confirms that it has good oral bioavailability and higher LD50.
Electronic supplementary material
Acknowledgements
SPS acknowledged the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India for granting the DBT-Post Doctoral Fellowship and CBS thanks Director, IBSD for facilities.
Author Contributions
S.P.S. performed the molecular docking, molecular dynamics simulations and DFT analysis; prepared the figures and tables, wrote the Results and computational investigation sections of the manuscript. K.N., N.I.S. and P.D. prepared the cell line assay and conducted the TNF-alpha assay. C.B.C. T.R.S. and D.S. wrote the Abstract, the Introduction and the Discussion sections. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22362-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bibbins-Domingo, K. et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 315, 2564–75 (2016). [DOI] [PubMed]
- 2.Astin M, Griffin T, Neal RD, Rose P, Hamilton W. The diagnostic value of symptoms for colorectal cancer in primary care: a systematic review. Br. J. Gen. Prac. 2011;61:231–43. doi: 10.3399/bjgp11X572427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics. C.A. Cancer J. Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 4.Adelstein BA, Macaskill P, Chan SF, Katelaris PH, Irwig L. Most bowel cancer symptoms do not indicate colorectal cancer and polyps: a systematic review. BMC Gastroenterol. 2011;11:65. doi: 10.1186/1471-230X-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner H, Stock C, Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ. 2014;348:2467–2467. doi: 10.1136/bmj.g2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitayama T. Attractive reactivity of a natural product, zerumbone. Biosci. Biotechnol. Biochem. 2011;75:199–207. doi: 10.1271/bbb.100532. [DOI] [PubMed] [Google Scholar]
- 7.Murakami AD, et al. Zerumbone, a Southeast Asian ginger sesquiterpene, markedly suppresses free radical generation, proinflammatory protein production, and cancer cell proliferation accompanied by apoptosis: the alpha, beta-unsaturated carbonyl group is a prerequisite. Carcinogenesis. 2002;23:795–802. doi: 10.1093/carcin/23.5.795. [DOI] [PubMed] [Google Scholar]
- 8.Prasannan R, et al. Key cell signaling pathways modulated by zerumbone: role in the prevention and treatment of cancer. Biochem. Pharmacol. 2012;84:1268–1276. doi: 10.1016/j.bcp.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Kim M, et al. Zerumbone, a tropical ginger sesquiterpene, inhibits colon and lung carcinogenesis in mice. Int. J.Cancer. 2009;124:264–271. doi: 10.1002/ijc.23923. [DOI] [PubMed] [Google Scholar]
- 10.Spies T, et al. Genes for the tumor necrosis factors alpha and beta are linked to the human major histocompatibility complex. Proc. Natl. Acad. Sci. USA. 1986;83:8699–8702. doi: 10.1073/pnas.83.22.8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner SM, Mock BA, Hilgers J, Huppi KE, Roeder WD. Mouse lymphotoxin and tumor necrosis factor: structural analysis of the cloned genes, physical linkage, and chromo-soma1 position. J. Immunol. 1987;139:476–483. [PubMed] [Google Scholar]
- 12.Sugarman BJ, et al. Recombinant human tumor necrosis factor-alpha: effects on proliferation of normal and transformed cells in vitro. Science. 1985;22:943–945. doi: 10.1126/science.3933111. [DOI] [PubMed] [Google Scholar]
- 13.Laster SM, Wood JG, Gooding LR. Tumor necrosis factor can induce both apoptic and necrotic forms of cell lysis. J. Immunol. 1988;141:2629–2634. [PubMed] [Google Scholar]
- 14.Robaye B, Mosselmans R, Fiers W, Dumont JE, Galand P. Tumor necrosis factor induces apoptosis (programmed cell death) in normal endothelial cells in vitro. Am. J. Pathol. 1991;138:447–453. [PMC free article] [PubMed] [Google Scholar]
- 15.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109:S81–S96. doi: 10.1016/S0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 17.Li Q, Verma IM. NF-κB regulation in the immune system. Nature Rev. Immunol. 2002;2:725–34. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 18.Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S. Molecular steps of TNF receptor-mediated apoptosis. Curr. Mol. Med. 2001;1:299–306. doi: 10.2174/1566524013363780. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Stark GR. NF-κB-dependent signaling pathways. Exp. Hematol. 2002;30:285–96. doi: 10.1016/S0301-472X(02)00777-4. [DOI] [PubMed] [Google Scholar]
- 21.Vallabhapurapu S, Karin M. Regulation and function of NFkappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, et al. The lysosome-associated apoptosis-inducing protein containing the pleckstrin homology (PH) and FYVE domains (LAPF), representative of a novel family of PH and FYVE domain-containing proteins, induces caspase-independent apoptosis via the lysosomal-mitochondrial pathway. J. Bio. Chem. 2005;280:40985–40995. doi: 10.1074/jbc.M502190200. [DOI] [PubMed] [Google Scholar]
- 23.Gehlhaar, D. K. et al. Proceedings of the Fourth International Conference on Evolutionary Programming, 615-627 (1995).
- 24.Scott WRP, et al. The GROMOS Biomolecular Simulation Package. J. Phys. Chem. A. 1999;103:3596–3607. doi: 10.1021/jp984217f. [DOI] [Google Scholar]
- 25.Kollman PA, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–97. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 26.Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A. 1988;38:3098–3100. doi: 10.1103/PhysRevA.38.3098. [DOI] [PubMed] [Google Scholar]
- 27.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;3(7):42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takada Y, Murakami A, Aggarwal BB. Zerumbone abolishes NF-kappaB and IkappaBalpha kinase activation leading to suppression of antiapoptotic and metastatic gene expression, upregulation of apoptosis, and downregulation of invasion. Oncogene. 2005;24:6957–6969. doi: 10.1038/sj.onc.1208845. [DOI] [PubMed] [Google Scholar]
- 29.Keong YS, et al. Immunomodulatory effects of zerumbone isolated from roots of Zingiber zerumbet. Pak. J. Pharm. Sci. 2010;23:75–82. [PubMed] [Google Scholar]
- 30.Rivas MA, et al. TNF alpha acting on TNFR1 promotes breast cancer growth via p42/P44 MAPK, JNK, Akt and NF-kappa Bdependent pathways. Exp. Cell Res. 2008;314:509–529. doi: 10.1016/j.yexcr.2007.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.