Abstract
Ductal carcinoma in situ (DCIS) is defined as a proliferation of neoplastic cells within the duct of the mammary gland that have not invaded into the surrounding stroma. DCIS is considered a precursor to invasive ductal carcinoma (IDC); however, approximately half of DCIS may progress to IDC, if left untreated. Current research has shown that the genomic and transcriptomic changes are present in DCIS before the emergence of invasive disease, indicating that the malignant nature of the DCIS is defined before invasion. However, important questions remain surrounding the specific changes and processes required for malignant progression and identification of prognostic indicators of aggressiveness. miRNAs are small regulatory RNAs that can modulate gene expression by complementary binding to target mRNAs and inducing translational repression and/or mRNA degradation. In the past decade, research has shown that miRNA expression is dysregulated in IDC and that these changes are already present at the DCIS stage. Therefore, changes in miRNA expression may provide the necessary information to identify a clinical indicator of the aggressiveness of DCIS. Herein, we review the miRNA signatures identified in DCIS, describe how these signatures may be used to predict the aggressiveness of DCIS, and discuss future perspectives for DCIS biomarker discovery.
Breast cancer is the most diagnosed nonskin cancer and the second leading cause of cancer death among women in the United States. Most breast cancers are invasive, meaning they have invaded through the walls of the ducts or lobules of the mammary gland, and are referred to as either invasive ductal carcinoma (IDC) or invasive lobular carcinoma, respectively. It was estimated that there were approximately 231,000 new cases of invasive breast cancer (IBC) and 60,000 additional cases of the precursor lesion carcinoma in situ in 2015.1 Carcinoma in situ is an abnormal proliferation of cells that appear morphologically malignant within the lumen of the mammary duct [ductal carcinoma in situ (DCIS)] or lobules [lobular carcinoma in situ (LCIS)] but have not invaded into the surrounding tissues. DCIS is the most common type of in situ breast cancer and currently accounts for 83% of all in situ diagnoses, whereas LCIS is rarer and accounts for approximately only 13%. Rates of DCIS diagnosis have dramatically increased during the past three decades, because of increased rates of screening by mammography.1 A woman diagnosed with DCIS is 8 to 10 times more likely to be subsequently diagnosed with IBC within her lifetime.2 Although diagnosis of DCIS indicates an increased risk,3 DCIS is widely considered a nonobligate precursor to IDC and approximately half of DCISs will not progress, even if left untreated.4 Currently, there is no way to identify the DCIS lesions that will progress to IDC from those that will not, which often leads to aggressive and potentially unnecessary interventions, including surgery, radiation, and/or hormonal therapy. In recent years, it has been established that the molecular changes present in breast cancer tissues at the preinvasive stage5, 6 are virtually indistinguishable from the molecular changes present in IDC. In fact, most cellular and molecular changes occur in the transition from normal epithelium to DCIS.7, 8 It is unclear which specific cellular and molecular changes are required for progression of DCIS to IDC, and a prognostic indicator of progression has yet to be identified.
miRNA Profiling in Breast Cancer
miRNAs are small (17 to 22 nucleotides) noncoding RNAs that post-transcriptionally regulate gene expression through the RNA interference pathway. The biogenesis of most miRNAs involves transcription by RNA polymerase II into primary miRNAs9, 10 and subsequent processing in the nucleus into shorter hairpin structures, called pre-miRNAs by the enzyme Drosha. The pre-miRNAs are then transported out of the nucleus and into the cytoplasm, where they are further processed into mature miRNAs by the enzyme Dicer.10 Mature miRNAs are loaded into a ribonucleoprotein complex, called the RNA-induced silencing complex. In the canonical site-targeting process, the RNA-induced silencing complex guides the mature miRNA to a target mRNA, where sequence-specific binding to the 3′-untranslated region occurs. The binding results in either translational inhibition or mRNA degradation.11 The most recent update of miRBase (http://www.miRBase.org, last accessed September 27, 2017) has cataloged 1881 human miRNA genes that produce 2588 mature miRNAs.12 A single miRNA may regulate thousands of mRNAs; likewise, a single mRNA may be targeted by hundreds of miRNAs, establishing miRNAs as the largest class of gene regulators.13 Through this mechanism, miRNAs have become important factors involved in the regulation of most cellular and developmental processes.14 Likewise, because of their broad regulatory activity, miRNAs are involved in many pathologic processes, including cancer development and progression.15 In the context of cancer, miRNAs may function as either tumor suppressors or oncogenes and assist in the promotion or suppression of cancer growth and progression. Aberrant miRNA expression has been described across many cancer types, including breast cancer, with global down-regulation of miRNA expression seen as a common trend.16, 17
The first indication that miRNAs may be involved in cancer promotion was the discovery that miR-15 and miR-16 are deleted in most (68%) patients with B-cell chronic lymphocytic leukemia18 and that miRNA genes are commonly found at fragile sites in the genome, which are frequently lost, mutated, amplified, or rearranged.19 Since these initial studies, a plethora of research using loss-of-function and gain-of-function approaches in human cancer cells, mouse xenografts, transgenic mouse models, and knockout mouse models has demonstrated that miRNAs play key roles in all of the common hallmarks of cancer, including initiation, progression, and metastasis.20
The first study to demonstrate the power of miRNA expression analysis in cancer tissues showed that miRNA profiling of just 217 miRNAs across >300 cancer specimens may distinguish tumor tissues from normal tissues and classify tumors by common origin and unknown origin; it also showed that miRNAs classified tumors better than expression profiling of >16,000 mRNAs.16 The first study to examine miRNA expression changes in IBC measured 245 miRNAs by microarray analysis in 76 breast tumor tissues and pooled normal tissues. This study showed that miRNA expression is deregulated in IBC, with miR-10b, miR-125b, miR-145 being the most significantly down-regulated and miR-21 and miR-155 being the most significantly up-regulated. This miRNA profile can distinguish IBC tissues from normal tissues and correlated with pathologic features, such as estrogen and progesterone receptor expression, tumor stage, vascular invasion, or proliferation index. Furthermore, a distinct miRNA signature was identified in luminal IBC, with up-regulation of miR-191 and miR-26 and down-regulation of miR-206.6 The first integrative molecular analysis by Blenkiron et al21 examined miRNA expression, mRNA expression, and genomic changes in breast cancer in 93 primary tumors, 21 cell lines, and 5 normal samples. Using a bead-based flow-cytometric miRNA profiling method, this study demonstrated that miRNAs may classify breast tumors by their molecular subtypes, such as luminal A, luminal B, basal-like, human epidermal growth factor receptor 2+, and normal like, and were associated with certain clinicopathological features. Specifically, a set of nine miRNAs (miR-100, miR-99a, miR-130a, miR-126, miR-136, miR-146b, miR-15b, miR-107, and miR-103) can distinguish luminal A from luminal B tumors. In addition, the changes in miRNA expression were somewhat explained by correlative changes in DNA copy number gains or losses.21 These studies have established that miRNA expression is altered in IBC tissue compared with normal tissues. Numerous studies have followed that provide further evidence to refine the concept that miRNA signatures have great potential to serve as biomarkers of breast cancer, for detecting the tumor at an early stage, further classifying breast cancer subtypes, and monitoring therapeutic responses.22
miRNA Profiling in DCIS Tissues
The first study to look specifically at miRNA expression changes in breast epithelial cells profiled miRNAs in normal tissues, IBC tumor tissues, and cell lines, and confirmed their results by fluorescent in situ hybridization on formalin-fixed, paraffin-embedded normal and matching tumor tissue specimens. The expression of miR-145 and miR-205 was confined to the myoepithelial cell layer in normal tissues and lost in the matching tumor specimens, whereas expression of let-7a, miR-21, miR-141, and miR-214 was confined to the luminal epithelial cell layer. This study also identified early aberrant miR-145 expression in atypical ductal hyperplasia (ADH), an intermediate stage of preinvasive breast cancer, and DCIS lesions.23 The first study to specifically examine the molecular changes in breast epithelial cells from preinvasive DCIS tissues used an integrative approach to profile both the miRNA expression by quantitative RT-PCR and mRNA expression by microarray. This was performed in epithelial cells microdissected from normal tissues obtained from reduction mammoplasty (n = 9) procedures from women with no history of cancer and paired samples of histologically normal (n = 8) and DCIS (n = 8). Thirty-five miRNAs were differentially expressed among the three groups, with 29 miRNAs differentially expressed, 15 miRNAs overexpressed, and 14 miRNAs underexpressed, between DCIS and histologically normal samples (Table 1). Of these miRNAs, 18 of 29 (62%) were previously implicated in IBC, and concordant with the earlier reports,6 this showed that many miRNA expression changes are present in DCIS before the development of IBC. Interestingly, 11 miRNAs were differentially expressed between the reduction mammoplasty and histologically normal samples, indicating that miRNA expression changes are present in normal-appearing tissues before the emergence of neoplasia. Examination of the mRNA expression profiles from these same tissue samples showed that 420 mRNAs were differentially expressed among the histologically normal and DCIS samples. The miRNA and mRNA expression profiles were integrated with miRNA target prediction, with 113 miRNA/mRNA functional pairs identified with high confidence. Several target pairs were validated in breast cancer cell lines in vitro, and inhibition of miR-182 (highly expressed in DCIS) increased expression of chromobox 7 and E-cadherin (positively regulated by chromobox 7), which is generally lost during breast cancer progression.5
Table 1.
Summary of miRNA Expression Profiling Studies in Preinvasive Breast Cancer
| Sample type | Analysis method | Major findings | miRNA expression∗ |
Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| Normal-ADH | Normal-DCIS | DCIS-IBC | |||||||
| Microdissected epithelial cells: RM (n = 9), paired normal (n = 8) and DCIS (n = 8) | RT-qPCR, TaqMan low-density array | 99 miRNAs were differentially expressed between DCIS and normal mammary epithelial cells | ↑miR-7 ↑miR-18a ↑miR-21 ↑miR-93 ↑miR-99b ↑mir-181b ↑miR-182 ↑miR-183 ↑miR-191 ↑miR-193b ↑miR-200b/c ↑miR-324-5p ↑miR-365 ↑miR-425-5p ↑miR-449b |
↓let-7c ↓miR-10b ↓miR-99a ↓mir-125b ↓miR-127 ↓miR-130a ↓miR-145 ↓miR-195 ↓miR-204 ↓miR-376a ↓miR-382 ↓miR-410 ↓miR-511 |
Hannafon et al5 | ||||
| Bulk extracted tissues: normal (n = 11), DCIS (n = 17), IBC (n = 151) | Solexa (Cambridge, UK) next-generation sequencing (in house) | Normal breast tissues may be distinguished from DCIS and IBC by increased miR-21 and decreased miR-98 and let-7 levels; most changes in miRNA expression already present in DCIS | ↑miR-21 ↑miR-142-3p ↑miR-142-5p |
↓miR-22 ↓miR-98 cluster (miR-125a, miR-99a, let-7a) ↓miR-451 ↓miR-144 ↓miR-143* ↓miR-320 ↓miR-378 ↓miR-497 |
↑miR-142-3p | ↓miR-125a ↓miR-451 ↓miR-144 ↓miR-145 ↓miR-143 ↓miR-378 |
Farazi et al24 | ||
| Subset of samples from Farazi et al, 201124: normal (n = 6), DCIS (n = 8), IBC (n = 80) | Reanalysis of Solexa next-generation sequencing-used only the most reliable results | 66 miRNAs were deregulated in the normal-DCIS† transition, whereas only nine were different between the DCIS-IBC transition; most changes in miRNA expression already present in DCIS | ↑miR-21 ↑miR-200c ↑miR-16 ↑miR-142-5p/3p ↑miR-374a ↑miR-26b ↑miR-29b ↑miR-183 ↑miR-96 ↑miR-106b ↑miR-182 ↑miR-361-5p |
↓miR-320 ↓miR-378 ↓miR-127-3p ↓let-7b, c, d ↓miR-376a/c ↓miR-193a/b ↓miR-99a ↓miR-22 ↓miR-423-5p ↓miR-145 ↓miR-125b ↓miR-497 |
↑let-7d ↑miR-181a ↑miR-210 ↑miR-221 |
↓miR-10b ↓miR-126 ↓miR-143 ↓miR-218 ↓miR-335-5p |
Volinia et al25 | ||
| Laser capture–microdissected FFPE tissues: normal (n = 8), ADH (n = 4), DCIS (n = 6), IBC (n = 7) | miRNA microarray | ↑miR-21 ↑miR-200b ↑miR-15b ↑miR-183 ↑miR-30d |
↓miR-1275 ↓miR-638 ↓miR-572 ↓miR-671-5p |
↑miR-556-3p | ↓miR-557 ↓miR-1207-5p ↓miR-874 |
Chen et al26 | |||
| Bulk-extracted fresh-frozen tissues: normal (n = 186), DCIS (n = 18), IBC (n = 1338) | miRNA microarray | 70 miRNAs were deregulated from normal-to-DCIS tissues; no miRNAs were deregulated in DCIS-to-IBC transition; breast cancer subtype–specific miRNA expression patterns were observed | ↑miR-21-5p ↑miR-96-5p ↑miR-106b-5p ↑miR-142-5p/3p ↑miR-155-5p ↑miR-183-5p ↑miR-200b/c-3p ↑miR-342-3p ↑miR-425-5p ↑miR-429-3p |
↓let-7b/c-5p ↓miR-22-3p ↓miR-99a-5p ↓miR-100-5p ↓miR-125b-5p ↓miR-140-3p ↓miR-143-5p ↓miR-145-5p/3p ↓miR-193a-5p ↓miR-193b-3p ↓miR-378a-3p ↓miR-497-5p ↓miR-652-3p |
↑miR-106b-5p ↑miR-142 ↑miR-342-3p ↑miR-425-5p |
↓let-7c-5p ↓miR-125b-5p ↓miR-140-3p ↓miR-145-5p/3p ↓miR-193a-5p ↓miR-378a-3p |
Haakensen et al27 | ||
↑, Increased; ↓, decreased; ADH, atypical ductal hyperplasia; DCIS, ductal carcinoma in situ; FFPE, formalin-fixed, paraffin-embedded; IBC, invasive breast cancer; RM, reduction mammoplasty; RT-qPCR, quantitative RT-PCR.
Fold change ≥1.5.
Top 12 differentially expressed miRNAs are listed.
Next-generation deep sequencing was used to develop a more comprehensive miRNA expression profile of normal (n = 11), DCIS (n = 17), and IBC (n = 151) specimens.24 Overall, normal breast tissues may be distinguished from DCIS and IBC tissues by increased miR-21 expression and decreased levels of multiple miRNAs, including miR-98 and let-7, with most changes in miRNA expression already apparent in the DCIS samples. In addition, miR-423 and miR-375 levels were higher in patients who developed metastases, and triple-negative breast cancers may be distinguished from other tumor subtypes by increased expression of the miR-17 to miR-92 cluster. A reanalysis of this next-generation sequencing data by excluding those with low-complexity runs and including only a subset of samples with the most reliable sequencing results, from normal (n = 6), DCIS (n = 8), and IBC (n = 80) samples,25 showed that 66 miRNAs were deregulated in the normal-to-DCIS transition (Table 126, 27), whereas only 9 were changed on DCIS-to-IBC transition. This, again, demonstrated that most changes in miRNA expression were already present in the DCIS samples. The study also examined breast cancer subtype–specific miRNA expression changes and found that certain miRNAs exhibited subtype-specific expression patterns, including increased expression of miR-190b in estrogen receptor–positive/human epidermal growth factor receptor 2− disease and decreased expression of miR-96 and miR-148a in estrogen receptor–positive/human epidermal growth factor receptor 2+ disease. The most reliable subtype-specific changes occurred in triple-negative tumors, with increased expression of the miR-17 to miR-92 cluster, miR-15/16, miR-128, and miR-200c and decreased expression of miR-143/145 and miR-199b. The association of clinical parameters and miRNA expression was also assessed, including time to metastasis, which was associated with miR-127-3p, miR-210, miR-185, miR-143*, and let-7b, whereas overall survival was associated with miR-210, miR-221, and miR-652. In addition, expression of miR-210, miR-21, miR-106b*, miR-197, and let-7i correlated with both parameters.25
Proliferative breast lesions, such as ADH, represent an intermediate stage of preinvasive ductal breast cancer that possesses many of the same cytologic and architectural features of DCIS, but may or may not progress to DCIS and IBC. Similar to DCIS, diagnosis of ADH increases the relative risk of developing breast cancer; however, only approximately 15% of patients with ADH will progress to invasive disease.28, 29 To determine how early changes in miRNA expression occur in proliferative breast disease, Chen et al26 analyzed archived formalin-fixed, paraffin-embedded specimens by laser-capture microdissection of normal (n = 8), ADH (n = 4), DCIS (n = 6), and IDC (n = 7) samples. Expression of miR-21, miR-200b/c, miR-141, and miR-183 was consistently up-regulated, whereas miR-557 expression was consistently down-regulated, in ADH, DCIS, and IDC, compared with normal tissues. The authors noted that the most significant changes in miRNA expression occurred during the transition of normal to ADH, suggesting that miRNA dysregulation is an early event that precedes development of DCIS in breast tumorigenesis.26
One of the largest and most recent studies to investigate early miRNA dysregulation in preinvasive breast cancer used miRNA microarray data from two large data sets, the Molecular Taxonomy of Breast Cancer International Consortium30 and the Akershus University Hospital (AHUS),31 consisting of 1542 breast tissue samples in total: normal (n = 186), DCIS (n = 18), and IBC (n = 1338).27 Cross validation was conducted using a previous data set from Volinia et al,25 as discussed earlier in this review. In this study, 70 miRNAs were differentially expressed between normal and DCIS in both data sets, and 27 of 70 were expressed in the same direction as the data set of Volinia et al25 and were proposed as a DCIS signature27 (Table 1). Of these 27 miRNAs, 14 were also differentially expressed in benign tumors in the same direction, including miR-21, let-7, and the miR-200 family. No miRNAs were consistently identified as differentially expressed in the DCIS relative to the invasive samples, demonstrating again that miRNA dysregulation occurs at an early stage of breast cancer development. To investigate the subtype-specific signatures of miRNAs differentially expressed from DCIS to each subtype of IBC, the samples were stratified on the basis of their molecular signatures. Among the different invasive subtypes, expression of seven miRNAs was consistently down-regulated, including let-7c-5p, miR-125b-5p, miR-140-3p, miR-145-3p, miR-145-5p, miR-193a-5p, and miR-378a-3p, consistent with a tumor-suppressive function. Expression of four miRNAs was consistently up-regulated, including miR-106b-5p, miR-142, miR-342-3p, and miR-425-5p. These candidate miRNAs are potentially responsible for the transformation from DCIS to the various invasive subtypes and provide a set of potentially clinically relevant biomarkers indicative of invasive transition. However, researchers should practice caution in their interpretation of these results because of the small DCIS sample size, thus reducing the statistical power, and the complex heterogeneity of breast cancer. A discrepancy was noted in this study with respect to miR-210; although Volinia et al25 found that expression of miR-210, a marker of poor prognosis, was down-regulated in the normal-to-DCIS transition, then up-regulated in the invasive transition, this study found that expression of miR-210 was up-regulated in DCIS compared with normal samples and not significantly altered in any invasive subtype. These findings bring into question the prognostic utility of miR-210 in breast cancer.
miRNA Profiles of DCIS in Liquid Biopsy Specimens
Although identifying the dysregulated miRNAs involved in the development and progression of DCIS is crucial to our understanding of the process of breast tumorigenesis, these miRNAs may also serve as prognostic biomarkers of the disease. Because of their small size and stability, many studies have reported that circulating miRNAs are detectable and elevated in the serum or plasma of patients with various cancers, including lung cancer,32 ovarian cancer,33 prostate cancer, colorectal cancer,34 and breast cancer.35, 36 Ng et al37 identified a set of circulating miRNA biomarkers and validated them across a case-control cohort of breast cancer, controls, and other types of cancers. The biomarkers were then blindly validated across an additional independent data set of breast cancer cases and controls. miRNA profiling from tumor and corresponding plasma samples from breast cancer patients and matched healthy controls found that the level of eight miRNAs was up-regulated and one was down-regulated across the breast tumor tissues and in the circulation (Table 1).37 The combined expression of miR-145 and miR-451 performed as the best biomarkers for discriminating breast cancer from normal controls, and any other types of cancer, with an 88% positive predictive value. Moreover, the combined miR-145 and miR-451 expression from only the DCIS samples of the breast cancer cohort had good diagnostic power, with a positive predictive value of 96%. These results are the first to evaluate circulating miRNA levels in patients with DCIS and suggest that the combined expression of miR-145 and miR-451 may be useful for the diagnosis of early-stage breast cancer. Although these results remain to be validated across a larger cohort of DCIS patients, there are several advantages to miRNA profiling for biomarker development. For example, screening much fewer miRNAs, rather than many circulating mRNAs, may be more efficient at distinguishing normal from cancer cases. Furthermore, unlike circulating mRNAs, miRNAs, primarily because of their small size and their association with extracellular vesicles or proteins, are more stable and remain intact in the circulation. However, when examining circulating miRNA expression, identifying those from the diseased tissue of origin remains a conceptual challenge, suggesting that alternative fluid samples, such as tissue-specific biofluids, may be superior sources for biomarker examination. The mammary gland is unique in that it can be directly sampled through the collection of nipple aspirate or ductal lavage fluids. Although slightly more invasive than a simple blood draw, the collection of these biofluids is less invasive compared with tissue biopsy specimens. Although their utility for cytologic diagnostic evaluation is low,38 these fluids remain a rich resource for detecting and evaluating potential molecular biomarkers to identify cancerous breast lesions at the earliest stage possible.39 A recent study evaluated the miRNA profiles in ductal lavage fluids from patients with DCIS alone or IDC with concurrent DCIS compared with the fluid from the unaffected breast in 22 patients with unilateral breast tumors.40 Seventeen miRNAs were differentially expressed in the ductal fluids from the invasive breast compared with the uninvolved breast. Among subjects with both DCIS and IDC, 14 miRNAs were differentially expressed, suggesting that the miRNAs released from cancer cells at an earlier stage of carcinogenesis may differ from those released at a later stage. However, a larger sample set is required to further evaluate this hypothesis. This study demonstrates the feasibility of examining miRNAs in breast fluid samples and their potential for biomarker identification, an approach that is particularly amenable to evaluation of subjects with DCIS.
Summary and Future Perspectives
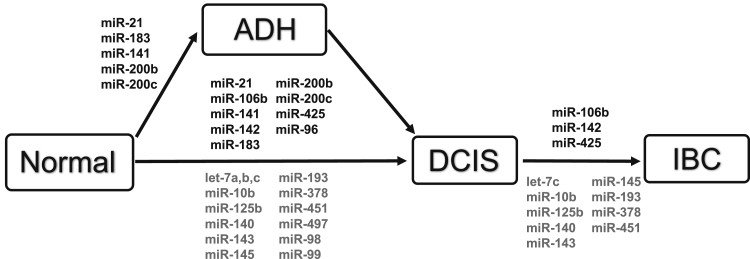
Most studies published thus far, and summarized in this review (Figure 1), are in agreement that miRNA expression changes occur early in breast cancer development. However, in most of these studies, DCIS samples are overall underrepresented, thus lending to a low statistical power. Furthermore, only a single study was identified that examined miRNA expression in ADH lesions compared with DCIS. However, it was observed that many of the miRNA expression changes present in DCIS were also already present in the histologically normal epithelial cells from the affected breast of patients with DCIS, when compared with the epithelial cells of healthy normal control samples.5 This observation suggests that miRNA expression changes may occur before the emergence of hyperplastic cells.
Figure 1.
Summary of reported miRNA expression changes present in the transition of normal breast epithelium to invasive breast cancer. miRNAs that were reported in two or more independent studies or present in more than one transition are presented. Increased miRNA expression (black text above the horizontal arrows) and decreased miRNA expression (gray text below the horizontal arrows) are shown. ADH, atypical ductal hyperplasia; DCIS, ductal carcinoma in situ; IBC, invasive breast cancer.
Commonly, few changes in miRNA expression are found in the transition of DCIS to invasive disease, which is perhaps surprising, considering the importance of this step in the progression of benign disease to an invasive cancer. However, it has been suggested that the invasive potential of DCIS may not be dependent on the intrinsic changes occurring within the luminal epithelial cells, but rather the invasive potential of DCIS may be heavily influenced by extrinsic factors found in the tumor stromal microenvironment. These extrinsic factors include the extracellular matrix, the myoepithelial cell layer, immune cells, and cancer-associated fibroblasts, which may all work together to compromise the natural barriers and allow the invasion of these neoplastic cells.41 The role of miRNAs in affecting these extrinsic events is not well understood. Recent work has demonstrated, however, that the interaction of tumor-associated myoepithelial and DCIS cells can promote the invasive progression of DCIS cells by stimulating transforming growth factor-β signaling, which, in turn, activates oncogenic miR-10b-5p and results in the down-regulation of RB1-inducible coiled-coil protein 1 (RB1CC1), an miR-10b-5p tumor suppressor target gene.42 However, further investigations are required to determine the underlying role of miRNAs in the interaction of DCIS cells with myoepithelial cells and their microenvironment, which may promote DCIS progression.
Recent developments in cancer biology have demonstrated that nanometer-sized (50 to 100 nm) secretory vesicles, called exosomes, may be important players in breast cancer progression. Exosomes can mediate cell-to-cell communication in the tumor microenvironment by encapsulating biologically active molecules, such as lipids, proteins, and nucleic acids (including miRNAs), and transferring these molecules to surrounding cells and into the circulation.43 Breast cancer cells, in a hypoxic environment, have been shown to release more exosomes than normal epithelial cells,44 which suggests their potential as a driving force for breast cancer progression. In addition, exosomes can transfer oncogenic factors that modify the tumor microenvironment and, therefore, may significantly contribute to cancer progression and invasion.45, 46, 47, 48 However, the factors present in exosomes from DCIS cells and how they may interact with the stromal microenvironment and promote progression to IBC have not been characterized.
We, and others, have recently shown that certain miRNAs are highly enriched and exclusively present in exosomes derived from breast cancer cells compared with normal epithelial cell–derived exosomes.49, 50 These miRNA species are also highly enriched in plasma exosomes from patient-derived xenograft mouse models and breast cancer patients.50 This work indicates that changes in the miRNA contents of exosomes occur during malignant transformation; however, whether these changes occur at an early stage has not been examined. Furthermore, whether these changes can be used as markers of DCIS progression remains to be determined.
Conclusions
In conclusion, our understanding of the biology involved in determining the invasive potential of DCIS is lacking, as is a robust marker able to predict the emergence of invasive disease. This gap in knowledge is primarily because of the continued focus on the invasive and metastatic stages of breast cancer progression and a lack of effort in understanding the underlying biology of the preinvasive component of breast cancer. Furthermore, current research has continually demonstrated that the molecular changes, including miRNAs, in DCIS cells are already present before invasion, indicating that the malignant nature of the DCIS is predefined. Therefore, a renewed focus on the role of the extrinsic microenvironment and secreted factors will greatly improve our understanding of DCIS progression and our ability to predict the aggressiveness of DCIS to invasive disease.
Acknowledgments
B.N.H. conceived of, wrote, and revised the manuscript; W.-Q.D. wrote and revised the manuscript; both authors read and approved the final manuscript.
Footnotes
Supported by the National Institute of General Medical Sciences grant U54GM104938 (W.-Q.D.), the Presbyterian Health Foundation (W.-Q.D.), and the Oklahoma Center for Advancement of Science and Technology grant HR17-052 (B.N.H.).
Disclosures: None declared.
Contributor Information
Bethany N. Hannafon, Email: bethany-hannafon@ouhsc.edu.
Wei-Qun Ding, Email: weiqun-ding@ouhsc.edu.
References
- 1.American Cancer Society . American Cancer Society; Atlanta: 2015. Breast Cancer Facts & Figures 2015-2016. [Google Scholar]
- 2.Lopez-Garcia M.A., Geyer F.C., Lacroix-Triki M., Marchio C., Reis-Filho J.S. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 2010;57:171–192. doi: 10.1111/j.1365-2559.2010.03568.x. [DOI] [PubMed] [Google Scholar]
- 3.Collins L.C., Tamimi R.M., Baer H.J., Connolly J.L., Colditz G.A., Schnitt S.J. Outcome of patients with ductal carcinoma in situ untreated after diagnostic biopsy: results from the Nurses' Health Study. Cancer. 2005;103:1778–1784. doi: 10.1002/cncr.20979. [DOI] [PubMed] [Google Scholar]
- 4.Allegra C.J., Aberle D.R., Ganschow P., Hahn S.M., Lee C.N., Millon-Underwood S., Pike M.C., Reed S.D., Saftlas A.F., Scarvalone S.A., Schwartz A.M., Slomski C., Yothers G., Zon R. NIH state-of-the-science conference statement: diagnosis and management of ductal carcinoma in situ (DCIS) NIH Consens State Sci Statements. 2009;26:1–27. [PubMed] [Google Scholar]
- 5.Hannafon B.N., Sebastiani P., de las Morenas A., Lu J., Rosenberg C.L. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 2011;13:R24. doi: 10.1186/bcr2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iorio M.V., Ferracin M., Liu C.G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., Menard S., Palazzo J.P., Rosenberg A., Musiani P., Volinia S., Nenci I., Calin G.A., Querzoli P., Negrini M., Croce C.M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 7.Ma X.J., Salunga R., Tuggle J.T., Gaudet J., Enright E., McQuary P., Payette T., Pistone M., Stecker K., Zhang B.M., Zhou Y.X., Varnholt H., Smith B., Gadd M., Chatfield E., Kessler J., Baer T.M., Erlander M.G., Sgroi D.C. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter D., Lahti-Domenici J., Keshaviah A., Bae Y.K., Argani P., Marks J., Richardson A., Cooper A., Strausberg R., Riggins G.J., Schnitt S., Gabrielson E., Gelman R., Polyak K. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- 9.Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 13.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 14.Taft R.J., Pang K.C., Mercer T.R., Dinger M., Mattick J.S. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 15.Melo S.A., Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Downing J.R., Jacks T., Horvitz H.R., Golub T.R. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Guo Y., Chen Z., Zhang L., Zhou F., Shi S., Feng X., Li B., Meng X., Ma X., Luo M., Shao K., Li N., Qiu B., Mitchelson K., Cheng J., He J. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 18.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C.M. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C.M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics: a comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blenkiron C., Goldstein L.D., Thorne N.P., Spiteri I., Chin S.F., Dunning M.J., Barbosa-Morais N.L., Teschendorff A.E., Green A.R., Ellis I.O., Tavare S., Caldas C., Miska E.A. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amorim M., Salta S., Henrique R., Jeronimo C. Decoding the usefulness of non-coding RNAs as breast cancer markers. J Transl Med. 2016;14:265. doi: 10.1186/s12967-016-1025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sempere L.F., Christensen M., Silahtaroglu A., Bak M., Heath C.V., Schwartz G., Wells W., Kauppinen S., Cole C.N. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 24.Farazi T.A., Horlings H.M., Ten Hoeve J.J., Mihailovic A., Halfwerk H., Morozov P., Brown M., Hafner M., Reyal F., van Kouwenhove M., Kreike B., Sie D., Hovestadt V., Wessels L.F., van de Vijver M.J., Tuschl T. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volinia S., Galasso M., Sana M.E., Wise T.F., Palatini J., Huebner K., Croce C.M. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci U S A. 2012;109:3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L., Li Y., Fu Y., Peng J., Mo M.H., Stamatakos M., Teal C.B., Brem R.F., Stojadinovic A., Grinkemeyer M., McCaffrey T.A., Man Y.G., Fu S.W. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PLoS One. 2013;8:e54213. doi: 10.1371/journal.pone.0054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haakensen V.D., Nygaard V., Greger L., Aure M.R., Fromm B., Bukholm I.R., Luders T., Chin S.F., Git A., Caldas C., Kristensen V.N., Brazma A., Borresen-Dale A.L., Hovig E., Helland A. Subtype-specific micro-RNA expression signatures in breast cancer progression. Int J Cancer. 2016;139:1117–1128. doi: 10.1002/ijc.30142. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann L.C., Sellers T.A., Frost M.H., Lingle W.L., Degnim A.C., Ghosh K., Vierkant R.A., Maloney S.D., Pankratz V.S., Hillman D.W., Suman V.J., Johnson J., Blake C., Tlsty T., Vachon C.M., Melton L.J., 3rd, Visscher D.W. Benign breast disease and the risk of breast cancer. N Engl J Med. 2005;353:229–237. doi: 10.1056/NEJMoa044383. [DOI] [PubMed] [Google Scholar]
- 29.Degnim A.C., Visscher D.W., Berman H.K., Frost M.H., Sellers T.A., Vierkant R.A., Maloney S.D., Pankratz V.S., de Groen P.C., Lingle W.L., Ghosh K., Penheiter L., Tlsty T., Melton L.J., 3rd, Reynolds C.A., Hartmann L.C. Stratification of breast cancer risk in women with atypia: a Mayo cohort study. J Clin Oncol. 2007;25:2671–2677. doi: 10.1200/JCO.2006.09.0217. [DOI] [PubMed] [Google Scholar]
- 30.Dvinge H., Git A., Graf S., Salmon-Divon M., Curtis C., Sottoriva A., Zhao Y., Hirst M., Armisen J., Miska E.A., Chin S.F., Provenzano E., Turashvili G., Green A., Ellis I., Aparicio S., Caldas C. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 31.Tahiri A., Leivonen S.K., Luders T., Steinfeld I., Ragle Aure M., Geisler J., Makela R., Nord S., Riis M.L., Yakhini Z., Kleivi Sahlberg K., Borresen-Dale A.L., Perala M., Bukholm I.R., Kristensen V.N. Deregulation of cancer-related miRNAs is a common event in both benign and malignant human breast tumors. Carcinogenesis. 2014;35:76–85. doi: 10.1093/carcin/bgt333. [DOI] [PubMed] [Google Scholar]
- 32.Lin P.Y., Yang P.C. Circulating miRNA signature for early diagnosis of lung cancer. EMBO Mol Med. 2011;3:436–437. doi: 10.1002/emmm.201100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resnick K.E., Alder H., Hagan J.P., Richardson D.L., Croce C.M., Cohn D.E. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 34.Ng E.K., Chong W.W., Jin H., Lam E.K., Shin V.Y., Yu J., Poon T.C., Ng S.S., Sung J.J. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 35.Heneghan H.M., Miller N., Lowery A.J., Sweeney K.J., Newell J., Kerin M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 36.Zhao H., Shen J., Medico L., Wang D., Ambrosone C.B., Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng E.K., Li R., Shin V.Y., Jin H.C., Leung C.P., Ma E.S., Pang R., Chua D., Chu K.M., Law W.L., Law S.Y., Poon R.T., Kwong A. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan S.A., Wolfman J.A., Segal L., Benjamin S., Nayar R., Wiley E.L., Bryk M., Morrow M. Ductal lavage findings in women with mammographic microcalcifications undergoing biopsy. Ann Surg Oncol. 2005;12:689–696. doi: 10.1245/ASO.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Zhao J., Yu X., Lange J., Kuerer H., Krishnamurthy S., Schilling E., Khan S.A., Sukumar S., Chan D.W. Identification of biomarkers for breast cancer in nipple aspiration and ductal lavage fluid. Clin Cancer Res. 2005;11:8312–8320. doi: 10.1158/1078-0432.CCR-05-1538. [DOI] [PubMed] [Google Scholar]
- 40.Do Canto L.M., Marian C., Willey S., Sidawy M., Da Cunha P.A., Rone J.D., Li X., Gusev Y., Haddad B.R. MicroRNA analysis of breast ductal fluid in breast cancer patients. Int J Oncol. 2016;48:2071–2078. doi: 10.3892/ijo.2016.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeong J., Thike A.A., Tan P.H., Iqbal J. Identifying progression predictors of breast ductal carcinoma in situ. J Clin Pathol. 2017;70:102–108. doi: 10.1136/jclinpath-2016-204154. [DOI] [PubMed] [Google Scholar]
- 42.Lo P.K., Zhang Y., Yao Y., Wolfson B., Yu J., Han S.Y., Duru N., Zhou Q. Tumor-associated myoepithelial cells promote the invasive progression of ductal carcinoma in situ through activation of TGFbeta signaling. J Biol Chem. 2017;292:11466–11484. doi: 10.1074/jbc.M117.775080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannafon B.N., Ding W.Q. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King H.W., Michael M.Z., Gleadle J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maji S., Chaudhary P., Akopova I., Nguyen P.M., Hare R.J., Gryczynski I., Vishwanatha J.K. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol Cancer Res. 2017;15:93–105. doi: 10.1158/1541-7786.MCR-16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao D., Barry S., Kmetz D., Egger M., Pan J., Rai S.N., Qu J., McMasters K.M., Hao H. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016;376:318–327. doi: 10.1016/j.canlet.2016.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L., Zhang S., Yao J., Lowery F.J., Zhang Q., Huang W.C., Li P., Li M., Wang X., Zhang C., Wang H., Ellis K., Cheerathodi M., McCarty J.H., Palmieri D., Saunus J., Lakhani S., Huang S., Sahin A.A., Aldape K.D., Steeg P.S., Yu D. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez M., Silva J., Herrera A., Herrera M., Pena C., Martin P., Gil-Calderon B., Larriba M.J., Coronado M.J., Soldevilla B., Turrion V.S., Provencio M., Sanchez A., Bonilla F., Garcia-Barberan V. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget. 2015;6:40575–40587. doi: 10.18632/oncotarget.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pigati L., Yaddanapudi S.C., Iyengar R., Kim D.J., Hearn S.A., Danforth D., Hastings M.L., Duelli D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hannafon B.N., Trigoso Y.D., Calloway C.L., Zhao Y.D., Lum D.H., Welm A.L., Zhao Z.J., Blick K.E., Dooley W.C., Ding W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]