Abstract
The present study assessed the function of epidermal growth factor receptor (EGFR) and its molecular targets in non-small cell lung cancer. The results of the present study demonstrated that EGFR protein and mRNA expression in the normal adjacent tissue specimens was decreased compared with that in the lung cancer tissue samples. Compared with the BEAS-2B normal bronchial epithelial cells, EGFR and phosphorylated (p)-extracellular signal-regulated kinase (ERK) protein expression in the SW-900 and A549 lung cancer cells was increased and microRNA (miR)200a expression in the SW-900 and A549 cells was inhibited compared with the BEAS-2B cells. Downregulating miR200a expression significantly suppressed proliferation and promoted apoptosis and caspase (CASP)3 and CASP9 function in the A549 cells and significantly inhibited EGFR and p-ERK protein expression in the A549 cells, compared with the BEAS-2B cells. The results of the present study indicated that downregulating miR200a significantly suppressed proliferation and promoted apoptosis in A549 cells via the regulation of the EGFR and ERK 1/2 signaling pathways.
Keywords: epidermal growth factor receptor, microRNA 200a, extracellular signal-regulated kinase 1/2, non-small cell lung cancer
Introduction
Lung cancer is a malignant tumor that threatens human health and life, and exhibited the highest global occurrence rate and lethality among all types of cancer in 2012 (1). Its morbidity is increasing in multiple countries (1). At present, the treatment options for patients with lung cancer include surgery, radiotherapy, and chemotherapy. In non-small cell lung cancer (NSCLC), the typical first choice of therapy is operative treatment but, once NSCLC has been definitively diagnosed, surgery is not possible in 75% of patients since lesions have already developed, or the state of their health following surgery would potentially be poorer compared with that prior to surgery (2). Therefore, the patient may opt for chemotherapy. Though chemotherapy may decrease the recurrence rate of NSCLC in patients, only ~30% of patients with NSCLC undergo effective treatment with platinum-based first-line chemotherapy, and chemotherapy resistance is common (3). Furthermore, the 5-year survival rate for patients with NSCLC is only 15% (4). Previous research has indicated that, although novel chemotherapeutics have improved the curative effect in patients with transfer NSCLC, the associated median survival time remains only 8–9 months (5). Therefore, it is crucial to understand the occurrence and development of lung cancer, identify the molecular mechanisms underlying metastasis, and develop effective treatments for patients with malignant lung tumors.
The epidermal growth factor receptor (EGFR) family, also known as the Erb-b2 receptor tyrosine kinases (ERBB), includes four main members: EGFR, ERBB2, ERBB3 and ERBB4. The EGFR family members are allosteric enzymes located at the cell surface (3). The members may be divided into three areas: A ligand-coupling domain that protrudes out of the cytomembrane, an intramembrane tyrosine kinase-activated functional domain and a transmembrane domain. The EGFR family is associated with tumorigenesis and tumor development (6). The coding products of the EGFR family exhibit phosphatase functions, and participate in cytoskeletal protein recombination, thereby facilitating signal transduction, proliferation, apoptosis regulation and malignant transformation in cells (7). Increased EGFR expression has been reported in multiple tumors, including colon, esophageal, breast, lung, ovarian, cervical and pancreatic cancer, and glioma (8). Abnormal EGFR expression is associated with malignant cell proliferation, adhesion, vascularization, metastasis and radiosensitivity (8). EGFR represents one of the most promising therapeutic targets in the study of antineoplastic molecules (9). In previous years, anti-EGFR molecular targeted drugs have received increasing attention (10). A series of antineoplastic drugs targeting EGFR have previously been studied. Cetuximab and gefitinib have undergone clinical trials and acquired improved curative effects (10,11). There are >10 EGFR-targeting inhibitors, and they have acquired antineoplastic curative effects by being used to treat different types of human tumor (2). Therefore, the present study evaluated the function of EGFR and its molecular targets in NSCLC.
Materials and methods
Lung cancer specimens
From July 2015 to January 2016, 12 patients (male, 56.5±6.2 years age) with lung cancer who underwent surgery were enrolled in the present study at the First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China). Normal adjacent tissues specimens were used as controls. The study was approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University and all patients provided written informed consent.
Immunohistochemical analysis
Cancer tissue samples were fixed with 4% paraformaldehyde for 24 h at room temperature, paraffin-embedded, and tumor tissue was cut into 4-µm-thick tissue samples. The sections were deparaffinized, then washed with PBS, and immersed in 3% hydrogen peroxide to block endogenous peroxidase activity at room temperature for 10 min. Sections were incubated with anti-ERGF (cat. no. sc-367974; 1:100; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 1 h at room temperature. Sections were incubated with goat anti-rabbit IgG, horseradish peroxidase-conjugated secondary antibody (cat. no. 7074; 1:500; Cell Signaling Technology, Inc.) for 1 h at room temperature. Tissue samples were observed with a Nikon E200 light microscope (Nikon Corporation, Tokyo, Japan) at magnification, ×20.
Cell culture
The bronchial epithelioid cell line BEAS-2B, and the human lung cancer SW-900 and A549 cell lines were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 50 U/ml penicillin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 50 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA) at 37°C in a humidified atmosphere of 5% CO2.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the lung cancer tissue samples or BEAS-2B, SW-900 and A549 cell lines using TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RT was performed at 94°C for 5 min and 42°C for 30 min with the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). The RT-qPCR exponential phase occurred for 40 cycles to permit quantitative comparison of the complementary DNAs amplified from identical reactions using SYBR Premix Taq (Takara Biotechnology Co., Ltd.). The primer sequences used were as follows: EGFR forward, 5′-TGGAGCTACGGGGTGACCGT-3′ and reverse, 5′-GGTTCAGAGGCTGATTGTGAT-3′; GAPDH forward, 5′-ACCTGACCTGCCGTCTAGAA-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′; microRNA (miR)200a forward, 5′-GCTCACCCTTGCAGGTCTCC-3′ and reverse, 5′-CCCGAAACCCAGCCGCATC-3′; U6 forward, 5′-CGCTTCGGCAGCACATATACTA-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCA-3′. Each cycle was as follows: 94°C for 5 min, followed by 30 cycles of 94°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec. Data analysis was performed using the 2−∆∆Cq method (12). Experiments were repeated in triplicate.
miR transfection
si-miR200a (5′-ACAUCGUUACCAGACAGUGUUA-3′) and miR-negative control (NC; 5′-CAGAUUUUGUGUAGUACAA-3′) were designed and synthesized by Zimmer Company (Shanghai, China). Anti-miR200a (100 ng) and miR-NC (100 ng) were transfected into the A549 cells (1×105) using Lipofectamine 2000® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for 6 h in DMEM, and then the medium was changed to DMEM, according to the manufacturer's protocol.
MTT assay
Following transfection with anti-miR200a and miR-NC for 48 h, A549 cells (1×103) were seeded onto a 96-well plate and cultured in DMEM. Subsequently, 20 µl MTT (5 µg/ml) was added to each well and the cells were cultured for 4 h at 37°C. The medium was then discarded and 150 µl dimethyl sulfoxide was added (Invitrogen; Thermo Fisher Scientific, Inc.). Absorbance was measured using a multi-well spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA) at 490 nm.
Flow cytometry analysis and apoptosis assay
Following transfection with anti-miR200a and miR-NC for 48 h, the A549 cells (1×106) were seeded onto a 6-well plate and cultured in DMEM. An Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection kit (BD Biosciences, San Jose, CA, USA) was used according to the manufacturer's protocol to detect apoptosis. Annexin V-FITC (10 µl) was added to the cells for 15 min. Subsequently, a further 5 µl Annexin V-FITC was added to the cells for 5 min. The apoptosis rate was assessed using a FACSCalibur™ flow cytometer (BD Biosciences) Data was analyzed using FlowJo 7.6.1 (FlowJo LLC, Ashland, OR, USA).
Analysis of caspase (CASP)3/9 activity
Following transfection with anti-miR200a and miR-NC for 48 h, the A549 cells were seeded onto a 6-well plate (1×106) and cultured in DMEM. Ac-DEVD-pNA from a Caspase 3 Activity Assay kit, or Ac-LEHD-pNA from a Caspase 9 Activity Assay kit (both from Beyotime Institute of Biotechnology, Haimen, China) were added to each well and the cells were cultured for 1 h at 37°C. CASP3/9 activity was measured using a multi-well spectrophotometer (BioTek Instruments, Inc.) at 405 nm.
Western blot analysis
Following transfection with anti-miR200a and miR-NC for 48 h, the A549 cells were seeded onto a 6-well plate. Total protein from the A549 cells was subsequently extracted using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with protease inhibitors (Sigma-Aldrich; Merck KGaA). Protein content was quantified using the bicinchoninic acid method (Beyotime Institute of Biotechnology). Protein (50 µg/lane) was fractionated using SDS-PAGE on a 10% gel and subsequently transferred to polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). The PVDF membranes were blocked using 0.5% non-fat milk in TBS containing 0.1% Tween-20 at 37°C for 1 h. The membranes were incubated with anti-ERGF (cat. no. sc-367974; 1:500, Santa Cruz Biotechnology, Inc.), anti-phosphorylated (p)-extracellular signal-regulated kinase (ERK; cat. no. sc-23759-R; 1:500, Santa Cruz Biotechnology, Inc.) and anti-GAPDH (sc-25778, 1:2,000, Santa Cruz Biotechnology, Inc.) antibodies overnight at 4°C. The membrane was washed three times for 10 min each, with TBS containing 0.1% Tween at room temperature and incubated with goat anti-rabbit IgG, horseradish peroxidase-conjugated secondary antibody (cat. no. 7074; 1:5,000, Cell Signaling Technology, Inc.) for 1 h at room temperature. Protein expression was visualized using an enhanced chemiluminescent kit (GE Healthcare, Chicago, IL, USA) and analyzed using Bio-Rad Laboratories Quantity One software 3.0 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Data were presented as the mean ± standard error using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Statistical analysis was performed using the Student's t-test or, when comparing multiple groups, one-way analysis of variance with Bonferroni's multiple comparison test. P<0.05 was considered to indicate a statistically significant difference.
Results
EGFR expression is upregulated in lung cancer tissue
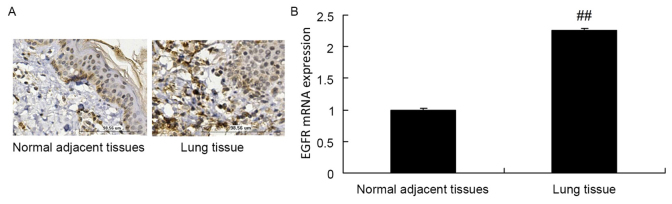
The protein expression of EGFR in normal adjacent tissue specimens was decreased compared with that in lung cancer tissue samples (Fig. 1A). Furthermore, the RT-qPCR results of the present study revealed that the mRNA expression of EGFR in normal adjacent tissue specimens was decreased compared with that in lung cancer tissue samples (Fig. 1B).
Figure 1.
EGFR expression is upregulated in A549 cells. (A) EGFR protein expression in lung cancer and normal adjacent tissue specimens, as assessed using immunohistochemical analysis. Magnification, ×10. (B) EGFR mRNA expression in lung cancer and normal adjacent tissue specimens, as assessed using reverse transcription-quantitative polymerase chain reaction. ##P<0.01 vs. normal adjacent tissue specimens. EGFR, epidermal growth factor receptor.
Expression of EGFR protein in A549 cells
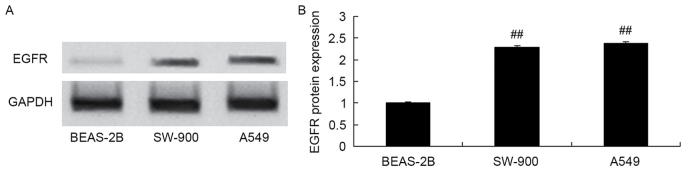
EGFR protein expression was measured in the BEAS-2B, SW-900 and A549 cell lines using western blot analysis. EGFR protein expression in the lung cancer cell lines SW-900 and A549 was increased compared with that in the bronchial epithelioid cell line BEAS-2B (Fig. 2).
Figure 2.
Expression of EGFR protein in A549 cells. EGFR protein expression, as assessed using (A) western blot analysis and (B) statistical analysis of western blotting results. ##P<0.01 vs. the BEAS-2B cell group. EGFR, epidermal growth factor receptor.
Expression of miR200a in A549 cells
RT-qPCR was used to assess miR200a expression in the BEAS-2B, SW-900 and A549 cell lines. The expression of miR200a in the BEAS-2B cells was significantly decreased compared with that in the SW-900 and A549 cells (Fig. 3).
Figure 3.
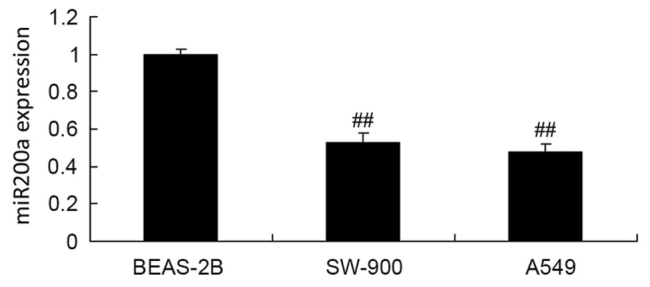
Expression of miR200a in A549 cells. ##P<0.01 vs. the BEAS-2B cell group. miR, microRNA.
Expression of ERK protein in A549 cells
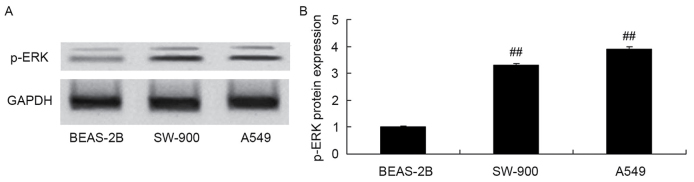
p-ERK protein expression was measured in the BEAS-2B, SW-900 and A549 cells using western blot analysis. Protein expression of p-ERK in SW-900 and A549 cells was significantly increased compared with that in BEAS-2B cells (Fig. 4).
Figure 4.
Expression of ERK protein in A549 cells. p-ERK protein expression, as assessed using (A) western blot analysis and (B) statistical analysis of western blotting results. ##P<0.01 vs. the BEAS-2B cell group. ERK, extracellular signal-regulated kinase; p-, phosphorylated.
Effect of downregulating miR200a expression on A549 cell proliferation
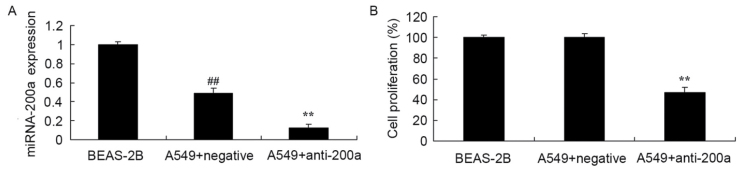
To evaluate the effect of downregulating miR200a expression on A549 cell proliferation, anti-miR200a and miR-NC were transfected into A549 cells. miR200a expression in A549 cells was significantly decreased compared with that in BEAS-2B cells, and miR200a expression in A549 cells transfected with anti-miR200a was significantly suppressed compared with that in A549 cells transfected with miR-NC (Fig. 5A). Proliferation was significantly suppressed in A549 cells in which miR200a was downregulated compared with that in miR-NC-treated A549 cells (Fig. 5B).
Figure 5.
Effect of downregulating miR200a expression on the proliferation of A549 cells. Effect of downregulating miR200a on (A) miR200a expression and (B) the proliferation of A549 cells. ##P<0.01 vs. the BEAS-2B cell group; **P<0.01 vs. the miR-negative control-transfected A549 cells. miR/miRNA, microRNA.
Effect of downregulating miR200a expression on A549 cell apoptosis
Flow cytometry analysis was used to assess the effect of downregulating miR200a on the apoptosis of A549 cells. The apoptosis rate of A549 cells in which miR200a was downregulated was significantly promoted compared with that of miR-NC-treated A549 cells (Fig. 6).
Figure 6.
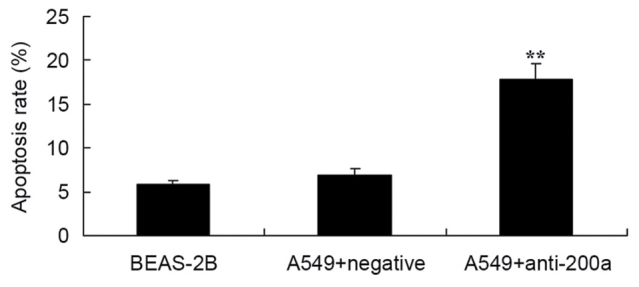
Effect of downregulating miR200a expression on the apoptosis of A549 cells. **P<0.01 vs. the miR-negative control-transfected A549 cells. miR, microRNA.
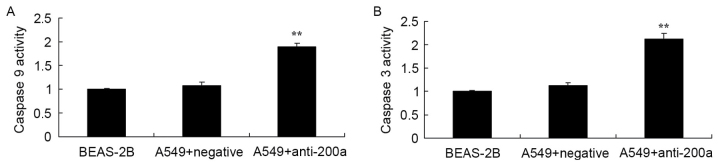
Effect of downregulating miR200a expression on CASP3/9 activity in A549 cells
The present study evaluated the effect of downregulating miR200a expression on CASP3/9 activity in A549 cells. CASP3 and CASP9 activity was significantly increased in A549 cells treated with anti-miR200a compared with that in miR-NC-treated A549 cells (Fig. 7).
Figure 7.
Effect of downregulating miR200a expression on CASP3/9 activity in A549 cells. (A) CASP3 and (B) CASP9 activity in A549 cells. **P<0.01 vs. the miR-negative control-transfected A549 cells. miR, microRNA; CASP, caspase.
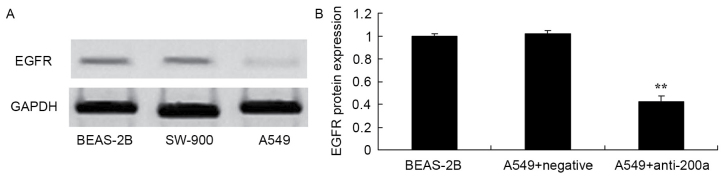
Effect of downregulating miR200a expression on EGFR protein expression in A549 cells
The present study assessed the effect of downregulating miR200a expression on the protein expression of EGFR in A549 cells. EGFR protein expression was significantly inhibited in A549 cells in which miR200a was downregulated compared with that in miR-NC-treated A549 cells (Fig. 8).
Figure 8.
Effect of downregulating miR200a expression on the protein expression of EGFR in A549 cells. EGFR protein expression, as assessed using (A) western blot analysis and (B) statistical analysis. **P<0.01 vs. the miR-negative control-transfected A549 cells. miR, microRNA; EGFR, epidermal growth factor receptor.
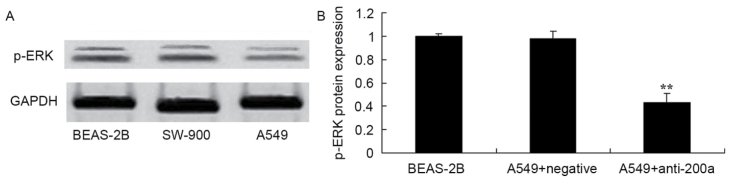
Effect of downregulating miR200a expression on the protein expression of ERK in A549 cells
To assess the effect of downregulating miR200a expression on the protein expression of ERK in A549 cells, p-ERK protein expression in SW-900 and A549 cells was measured using western blot analysis. The protein expression of p-ERK was significantly suppressed in A549 cells in which miR200a was downregulated compared with that in miR-NC-treated A549 cells (Fig. 9).
Figure 9.
Effect of downregulating miR200a expression on the protein expression of ERK in A549 cells. p-ERK protein expression, as assessed using (A) western blot analysis and (B) statistical analysis. **P<0.01 vs. the miR-negative control-transfected A549 cells. miR, microRNA; ERK, extracellular signal-regulated kinase; p-, phosphorylated.
Discussion
Lung cancer was the most common malignant tumor globally in 2012 (13). There are ~1.8 million incident cases (14), and lung cancer-associated mortality occurred in >1.5 million people globally, with the majority of these mortalities being male. NSCLC accounts for >80% of patients with lung cancer. Numerous patients exhibit NSCLC in the middle or advanced stage at the time of diagnosis. By this stage, the 5-year survival rate has decreased (13). Therefore, diagnosing NSCLC early is crucial. At present, imagological examination is a major diagnostic tool in oncology, but increased expense and risks of radiation exposure, among other factors, render it inappropriate for large-scale screening, so it is crucial to be able to accurately screen using inspection, decreased expenses and trauma (15). The importance of miRs has become apparent in oncology (16). Furthermore, as the molecular diagnosis marker of NSCLC, EGFR has received increased attention (15). The present study revealed that mRNA expression of EGFR in normal adjacent tissue specimens was decreased compared with that in lung cancer tissue samples.
miRs are single-stranded, highly conserved, noncoding nucleotide molecules that are 19–25 nt in length. By degrading or inhibiting mRNA targets, miRs participate in multiple important biological processes, including growth, differentiation, proliferation, apoptosis, hormone secretion and neoplasia in animal and plant cells (14). miRs are also associated with sensitivity to tumor drugs. By downregulating miR200C expression, sensitivity to adriamycin is decreased in breast cancer cells (17). In acute promyelocytic leukemia, increased expression of miR125b is involved in the action of current therapeutics (18). In colorectal cancer, upregulating miR224 may increase resistance to methylamine (19). A previous study revealed that NSCLC tissues often possess a deficiency of mixed miR128b (20). miR128B may regulate EGFR gene and protein expression (20). The present study demonstrated that the expression of miR200a in BEAS-2B cells was increased compared with that in SW-900 and A549 cells.
EGFR has attracted attention since it was discovered in 1962 (8). EGFR may combine with ligands to phosphorylate the isogeneous second area of the SRC proto-oncogene protein, induce normal mitosis, and promote homeostasis, cell differentiation and migration; abnormal expression is associated with the proliferation, adhesion, vascularization and metastasis of malignant cells (21). EGFR has gained attention in the study of antineoplastic molecular targeted therapy. The present study demonstrated that downregulating miR200a significantly inhibited EGFR protein expression in A549 cells. Zhen et al (22) reported that overexpressing miR200a significantly downregulated EGFR expression in NSCLC cells.
ERK is a protein kinase that is located in the terminal position in the signaling pathway of mitogen-activated protein kinase (MAPK), and forms p-ERK via phosphorylation (23). p-ERK may influence the transcriptional expression of associated genes, is associated with cell proliferation and serves a crucial function in malignant transformation. Among the multiple components of the MAPK signaling pathway, ERK is crucial (24). ERK helps to regulate cell proliferation and serves a function in multiple physiological and pathological processes, including cell differentiation, period circular regulation and intercellular functional synchronization (24). A previous study revealed that the ERK1/2 signaling pathway is activated in NSCLC; ERK1/2 exhibited increased expression and phosphorylation (25). On entering the nucleus, p-ERK1/2 affects the expression of jun proto-oncogene, nuclear factor κB subunit 1, ELK1, fos proto-oncogene, MYC proto-oncogene, and transcription factors, phosphorylates multiple substrates of nuclear transcription factors, regulates the transcription of associated genes, and serves a key function in malignant transformation (26). The results of the present study suggested that downregulating miR200a significantly suppressed p-ERK protein expression in A549 cells. Liu et al (27) demonstrated that miR200a expression was associated with the neurotrophic receptor tyrosine kinase 2/ERK/protein kinase B signaling pathway in mice exposed to chronic, unpredictable, moderate stress.
To conclude, the present study suggested that downregulating miR200a significantly suppressed proliferation and promoted apoptosis in A549 cells in vitro, partly through the regulation of the EGFR and ERK1/2 signaling pathways, and thereby may facilitate the development of aggressive tumors. The results of the present study suggested that miR200a/EGFR/ERK1/2-based prevention and therapeutics in patients with NSCLC may prove clinically beneficial.
References
- 1.Allendorf DJ, Bordoni RE, Grant SC, Saleh MN, Reddy VB, Jerome ML, Dixon PM, Miley DK, Singh KP, Robert F. Phase I/IIa study of sequential chemotherapy regimen of bendamustine/irinotecan followed by etoposide/carboplatin in untreated patients with extensive disease small cell lung cancer (EDSCLC) Cancer Chemother Pharmacol. 2015;76:949–955. doi: 10.1007/s00280-015-2869-6. [DOI] [PubMed] [Google Scholar]
- 2.Balabko L, Andreev K, Burmann N, Schubert M, Mathews M, Trufa DI, Reppert S, Rau T, Schicht M, Sirbu H, et al. Increased expression of the Th17-IL-6R/pSTAT3/BATF/RorγT-axis in the tumoural region of adenocarcinoma as compared to squamous cell carcinoma of the lung. Sci Rep. 2014;4:7396. doi: 10.1038/srep07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köhler J, Schuler M. LUX-Lung 3: Redundancy, toxicity or a major step forward? Afatinib as front-line therapy for patients with metastatic EGFR-mutated lung cancer. Future Oncol. 2014;10:533–540. doi: 10.2217/fon.14.9. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Zhang B, Li C, Cui C, Yue D, Shi B, Zhang Q, Zhang Z, Zhang X, Wang C. Prognostic value of number of negative lymph node in patients with stage II and IIIa non-small cell lung cancer. Oncotarget. 2017;8:79387–79396. doi: 10.18632/oncotarget.18154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belani CP, Yamamoto N, Bondarenko IM, Poltoratskiy A, Novello S, Tang J, Bycott P, Niethammer AG, Ingrosso A, Kim S, Scagliotti GV. Randomized phase II study of pemetrexed/cisplatin with or without axitinib for non-squamous non-small-cell lung cancer. BMC Cancer. 2014;14:290. doi: 10.1186/1471-2407-14-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen H, Du G, Liu Z, Bao J, Yu Q, Jia C, Liang X, Shan L. Assessment and prognostic analysis of EGFR mutations or/and HER2 overexpression in Uygur's non-small cell lung cancer. Int J Clin Exp Med. 2015;8:22300–22309. [PMC free article] [PubMed] [Google Scholar]
- 7.Li N, Li H, Su F, Li J, Ma X, Gong P. Relationship between epidermal growth factor receptor (EGFR) mutation and serum cyclooxygenase-2 Level, and the synergistic effect of celecoxib and gefitinib on EGFR expression in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8:9010–9020. [PMC free article] [PubMed] [Google Scholar]
- 8.Lococo F, Paci M, Rapicetta C, Rossi T, Sancisi V, Braglia L, Cavuto S, Bisagni A, Bongarzone I, Noonan DM, et al. Preliminary evidence on the diagnostic and molecular role of circulating soluble EGFR in non-small cell lung cancer. Int J Mol Sci. 2015;16:19612–19630. doi: 10.3390/ijms160819612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furcht CM, Muñoz Rojas AR, Nihalani D, Lazzara MJ. Diminished functional role and altered localization of SHP2 in non-small cell lung cancer cells with EGFR-activating mutations. Oncogene. 2013;32:2346–2355. doi: 10.1038/onc.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzger B, Chambeau L, Begon DY, Faber C, Kayser J, Berchem G, Pauly M, Boniver J, Delvenne P, Dicato M, Wenner T. The human epidermal growth factor receptor (EGFR) gene in European patients with advanced colorectal cancer harbors infrequent mutations in its tyrosine kinase domain. BMC Med Genet. 2011;12:144. doi: 10.1186/1471-2350-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Fan J, Li Y, Lin S, Shu P, Ni J, Qin S, Zhang Z. Polymorphisms in epidermal growth factor receptor (EGFR) and AKT1 as possible predictors of clinical outcome in advanced non-small-cell lung cancer patients treated with EGFR tyrosine kinase inhibitors. Tumour Biol. 2016;37:1061–1069. doi: 10.1007/s13277-015-3893-1. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) methods. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Kim JH, Kim HS, Kim BJ. Prognostic value of smoking status in non-small-cell lung cancer patients treated with immune checkpoint inhibitors: A metaanalysis. Oncotarget. 2017;8:93149–93155. doi: 10.18632/oncotarget.18703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimino D, De Pittà C, Orso F, Zampini M, Casara S, Penna E, Quaglino E, Forni M, Damasco C, Pinatel E, et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2013;27:1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- 15.Ardizzoni A, Manegold C, Debruyne C, Gaafar R, Buchholz E, Smit EF, Lianes P, ten Velde G, Bosquee L, Legrand C, et al. European organization for research and treatment of cancer (EORTC) 08957 phase II study of topotecan in combination with cisplatin as second-line treatment of refractory and sensitive small cell lung cancer. Clin Cancer Res. 2003;9:143–150. [PubMed] [Google Scholar]
- 16.Du B, Wang Z, Zhang X, Feng S, Wang G, He J, Zhang B. MicroRNA-545 suppresses cell proliferation by targeting cyclin D1 and CDK4 in lung cancer cells. PLoS One. 2014;9:e88022. doi: 10.1371/journal.pone.0088022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F, Bao G, Kong H, Ge C, Zhang F, et al. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol Cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, He W, Li J, Huang S, Wan X, Luo H, Wu D. MiRNA-125b inhibits proliferation and migration by targeting SphK1 in bladder cancer. Am J Transl Res. 2015;7:2346–2354. [PMC free article] [PubMed] [Google Scholar]
- 19.Liao WT, Li TT, Wang ZG, Wang SY, He MR, Ye YP, Qi L, Cui YM, Wu P, Jiao HL, et al. microRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res. 2013;19:4662–4672. doi: 10.1158/1078-0432.CCR-13-0244. [DOI] [PubMed] [Google Scholar]
- 20.Wang XC, Du LQ, Tian LL, Wu HL, Jiang XY, Zhang H, Li DG, Wang YY, Wu HY, She Y, et al. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer. 2011;72:92–99. doi: 10.1016/j.lungcan.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Lin K, Cheng J, Yang T, Li Y, Zhu B. EGFR-TKI down-regulates PD-L1 in EGFR mutant NSCLC through inhibiting NF-κB. Biochem Biophys Res Commun. 2015;463:95–101. doi: 10.1016/j.bbrc.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 22.Zhen Q, Liu J, Gao L, Liu J, Wang R, Chu W, Zhang Y, Tan G, Zhao X, Lv B. MicroRNA-200a targets EGFR and c-Met to inhibit migration, invasion and gefitinib resistance in non-small cell lung cancer. Cytogenet Genome Res. 2015;146:1–8. doi: 10.1159/000434741. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Pesakhov S, Weng A, Kafka M, Gocek E, Nguyen M, Harrison JS, Danilenko M, Studzinski GP. ERK 5/MAPK pathway has a major role in 1α,25-(OH)2 vitamin D3-induced terminal differentiation of myeloid leukemia cells. J Steroid Biochem Mol Biol. 2014;144:223–227. doi: 10.1016/j.jsbmb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paolo M, Assunta S, Antonio R, Claudia SP, Anna BM, Clorinda S, Francesca C, Fortunato C, Cesare G. Selumetinib in advanced non small cell lung cancer (NSCLC) harbouring KRAS mutation: Endless clinical challenge to KRAS-mutant NSCLC. Rev Recent Clin Trials. 2013;8:93–100. doi: 10.2174/15748871113089990047. [DOI] [PubMed] [Google Scholar]
- 25.Saito N, Mine N, Kufe DW, Von Hoff DD, Kawabe T. CBP501 inhibits EGF-dependent cell migration, invasion and epithelial-to-mesenchymal transition of non-small cell lung cancer cells by blocking KRas to Calmodulin binding. 2017;8:1–74018. doi: 10.18632/oncotarget.18598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wainstein E, Seger R. The dynamic subcellular localization of ERK: Mechanisms of translocation and role in various organelles. Curr Opin Cell Biol. 2016;39:15–20. doi: 10.1016/j.ceb.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Liu BB, Luo L, Liu XL, Geng D, Liu Q, Yi LT. 7-Chlorokynurenic acid (7-CTKA) produces rapid antidepressant-like effects: Through regulating hippocampal microRNA expressions involved in TrkB-ERK/Akt signaling pathways in mice exposed to chronic unpredictable mild stress. Psychopharmacology (Berl) 2015;232:541–550. doi: 10.1007/s00213-014-3690-3. [DOI] [PubMed] [Google Scholar]