Abstract
Background
Many neuronal and glial diseases have been associated with changes in metabolism. Therefore, metabolic reprogramming has become an important area of research to better understand disease at the cellular level, as well as to identify targets for treatment. Model systems are ideal for interrogating metabolic questions in a tissue dependent context. However, while new tools have been developed to study metabolism in cultured cells there has been less progress towards studies in vivo and ex vivo.
New Method
We have developed a method using newly designed tissue restraints to adapt the Agilent XFe96 metabolic analyzer for whole brain analysis. These restraints create a chamber for Drosophila brains and other small model system tissues to reside undisrupted, while still remaining in the zone for measurements by sensor probes.
Results
This method generates reproducible oxygen consumption and extracellular acidification rate data for Drosophila larval and adult brains. Single brains are effectively treated with inhibitors and expected metabolic readings are observed. Measuring metabolic changes, such as glycolytic rate, in transgenic larval brains demonstrates the potential for studying how genotype affects metabolism.
Comparison with Existing Methods and Conclusions
Current methodology either utilizes whole animal chambers to measure respiration, not allowing for targeted tissue analysis, or uses technically challenging MRI technology for in vivo analysis that is not suitable for smaller model systems. This new method allows for novel metabolic investigation of intact brains and other tissues ex vivo in a quick, and simplistic way with the potential for large-scale studies.
Keywords: Drosophila melanogaster, metabolism, oxygen consumption, extracellular acidification, metabolic reprogramming, XFe96 analyzer, ex vivo
1. Introduction
An increasing number of neurological diseases and disorders with observed changes in metabolism have been identified in the last ten years. Mutations in metabolic enzymes were identified in Glioblastoma (GB) in 2008 (Parsons, Jones, Zhang, & Lin, 2008; Yan et al., 2009), and since more effective treatments have been developed by targeting metabolic vulnerabilities (Cairns, Harris, & Mak, 2011; Hanahan & Weinberg, 2011; Ward & Thompson, 2012; Wolf et al., 2011; Zhao, Butler, & Tan, 2013). Metabolic dysregulation in neurodegenerative disorders, such as Alzheimer’s disease, Huntington’s disease, and Parkinson’s disease have also been identified (Atamna & Frey, 2007; Cai et al., 2012). Mitochondrial dysfunction and altered metabolism have been associated with mental health disorders, including major depressive disorder (MDD) (Brody et al., 1999), and schizophrenia (Prabakaran et al. 2004). This is not a new medical phenomenon, but instead reflects the progress made in the field of metabolism, including new tools for studying metabolic change.
Investigating metabolism in tissues following disease or genetic alteration has become a promising direction for new, targeted treatment options (Vander Heiden, 2011). Many new tools and instruments have emerged for studying metabolism in cells, but there has been less advancement in methods developed to analyze metabolic changes at the whole organ/tissue level. We are interested in the metabolic changes that occur in the brain, both glia and neurons, following genetic alterations. Using a small model system, such as Drosophila, we can investigate brains of transgenic animals expressing disease genes, specifically in glial cells and/or neurons. It is important to isolate the brain for this type of analysis since transgenes will only be expressed in this tissue, and slight changes in metabolism will likely not be detectable when measuring the whole animal. While methods exist to study metabolic reprogramming in cell culture, this technology does not enable the whole organ metabolism to be captured; missing interactions between different cell types that may contribute to the overall energy utilization of the tissue.
Current methodology either utilizes whole animal chambers to measure respiration, not allowing for targeted tissue analysis (Frappell, Blevin, & Baudinette, 1989; Lighton, 2000; Withers, 2001), or uses technically challenging MRI technology for in vivo analysis that is not suitable for smaller model systems (Liu et al., 2011). A user-friendly technology was developed in the early 2000’s by Seahorse Biosciences that non-invasively measures metabolic activity in cell culture (Ferrick, Neilson, & Beeson, 2008). The XF Extracellular Flux Analyzer measures oxygen consumption and extracellular acidification simultaneously and is capable of delivering four separate injections of drugs and/or inhibitors to challenge cells metabolic response. These two parameters report on mitochondrial respiration and glycolytic respiration, respectively. Drug treatments provide further insight into a cells energy utilization, including fatty acid oxidation, nutrient preference, and the ability to shift metabolic program. This metabolic analyzer has greatly impacted the field of metabolism allowing for fast, reproducible results studying primary and established cell lines, including cancer, cardiac, and neuronal cell lines (Hardie et al., 2017; Kwang Kim et al., 2015; Xu et al., 2017). This instrument, however, cannot measure whole tissues (other than spheroids) in the 96-well format- a format that is crucial for the accurate measurement of small tissues. The XFe24 platform has larger wells that measure an increased volume of media above the cell sample. This does not allow for the sensitivity required to detect small metabolic changes in single brain samples. A requirement of this technology is a sample’s ability to adhere to the bottom of the well during analysis. While non-adherent cells are easily plated using poly-lysine or other coating agents, the geometry of the brain and other whole tissues have difficulty adhering. This challenge has made this technology inaccessible for studies on whole brains in Drosophila. The ability to measure whole brains ex vivo, while still not an in vivo analysis, would provide close insight into how the organ is functioning at the metabolic level. Studies have shown that dissected Drosophila brains can be kept alive for hours in the proper media and under certain conditions (Williamson & Hiesinger, 2010). This suggests that short metabolic assays will yield biologically relevant data from live tissue.
In this study, we have designed and produced micro-tissue restraints (Tipping & Waters, 2017) that, when placed in the well of an XFe96 cell plate, hold the tissue in place for proper measurement. We have used this new technology to develop a method for measuring the energy utilization of whole Drosophila larvae and adult brains. We are able to chemically challenge these brains using the drug delivery ports of the XFe96 cartridge and observe changes in the metabolism of these organs. We also interrogated larval brains expressing genetic mutations that resulted in significantly different metabolic effects from wild type larval brains.
This new methodology can be used more broadly, extending to other insects and model systems, such as C. elegans (Fig. 5B). Small model systems have used the XFe24 platform for metabolic analysis (Gibert, McGee, & Ward, 2013), but utilizing this new method will decrease the sample size required for assays and increase the sensitivity. This method can be applied beyond the brain to investigate other tissues in small model systems (Fig. 5A).
Fig. 5.
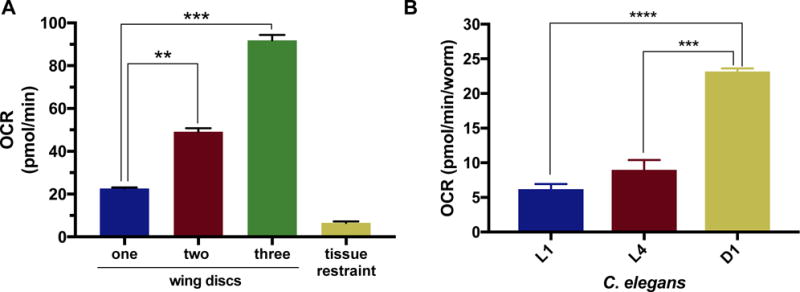
Utilizing the method beyond Drosophila brains.
A) Basal OCR levels of Oregon-R larval imaginal wing discs at the 6th cycle. One (blue), two (red) or three (green) wing discs were measured per well, (p-values: 0.001, 0.0003), n=4. B) Basal OCR levels of C. elegans at the 6th cycle measurement using the XFe96. L1, L4 and D1 worms were analyzed and reported in pmol/min/worm, (p-values: 1.01 × 106 and, 4.75 × 105, respectively), L1:n= 14, L4:n= 13, D1:n=21. A and B were analyzed by the XFe96 metabolic analyzer using the same procedure established for Drosophila brains, with slight modifications reported in methods.
2. Materials and Methods
2.1 Drosophila. melanogaster stocks
The following D. melanogaster stocks were obtained from the Bloomington Drosophila Stock Center (Indiana University): w[1118]; P{w[+mC]=UAS-Idh.R195H.FLAG}3 [RRID:BDSC_56203], IMP-L3 (LDH) RNAi transgenic fly: y[1]v[1];P{TRiP.HMS00039}attP2 [RRID:BDSC_33640], w[1118]; P{w[+m*]=GAL4}repo/TM3, Sb[1] [RRID:BDSC_7415], Oregon-R-C [RRID:BDSC_5], Canton-S [RRID:BDSC_64349]. All stocks were reared at 60% humidity and 25°C in a cornmeal, molasses and yeast based food. Late third instar wandering larva were used for larval brain studies. Two to five day old adult male and female flies were used for adult brain studies.
2.2 C. elegans cultivation
N2 Bristol (strain VC2010) was used as wild-type and grown on NGM agar plates (For 1 liter in water: 3g NaCl, 17g agar, 2.5g Peptone, and 1ml 1M CaCl2, 1ml 5mg/ml cholesterol in ethanol, 1 ml 1M MgSO4, and 25ml 1M KPO4 added after autoclaving) seeded with E. coli (strain OP50).
2.3 C. elegans synchronization for metabolic analysis
Gravid wild-type worms (egg bound) adults were treated with hypochlorite solution (for 5ml in water: 0.5ml 5N NaOH, 1ml household bleach containing 5% sodium hypochlorite) for no longer than 3 minutes and washed 3 times with M9 media (for 1 liter in water: 3g Kh2PO4, 6g NaHPO4, 5g NaCl, 1ml 1M MgSO4, filter sterilized) to collect embryos. Embryos were placed in M9 media at 20˚C overnight with agitation and allowed to hatch as starved L1s. L1 worms were then counted and plated on seeded NGM agar plates (as above) and grown to L4 stage. L4 worms were visually inspected and picked from the plate into M9 and washed 3 times in M9 to remove E. coli carried over from the plate. These L4 worms were then counted and added to the cell plates for analysis. Basal OCR rates were measured in M9 for 10 cycles according to the protocol described in Koopman et al., with each cycle consisting of two-minutes mixing, 30-seconds waiting, and two-minutes measuring (Koopman et al., 2016). After analysis, worms per well were counted again. For each well OCR was divided by number of worms to generate a pmol/min/worm oxygen consumption rate.
2.4 Tissue restraint design
Tissue restraints were manufactured by the Instrument Design and Fabrication Core Facility at Arizona State University. The mesh screen is nylon with pores of 0.0039 inches in diameter, and the plastic ring is made from the acetal homopolymer, delrin. The plastic ring has an outer diameter of 0.146 inches, and inner diameter of 0.1335 inches. The thickness of the ring is 0.0125 inches, and the height is 0.016 inches. Further details for manufacturing these tissue restraints will be provided upon request. Provisional patent no. 62/491,431.
2.5 Dissecting larval and adult Drosophila brains, and larval wing imaginal discs
Larval brains
Late third-instar larvae were selected from the vial wall and washed with 1xPBS (phosphate buffered solution). Larvae were then grasped at their mid-section and by their mouth hooks with forceps, and gently pulled apart. This causes the mouth hooks to detach from the brain and imaginal discs. Imaginal discs and other tissue are removed from the brain to isolate the brain stem and lobes. The brains were then transferred to the cell plate using forceps, and immersed in XFe96 assay media.
Adult brains
Adult flies were anesthetized on a CO2 pad briefly. Flies were sexed and selected for brain dissection. Using forceps, flies were positioned in a spot plate containing 1x PBS on their back and grasped under the first set of legs. Another set of forceps was used to grasp the proboscis. The head was removed by pulling up on the proboscis. Once the head was removed microscope was refocused and head was kept in focal plane using forceps. Holding the proboscis with one set of forceps, another set of forceps was used to grasp and remove the compound eye without disrupting the brain tissue beneath. This was repeated for the second compound eye. Remaining tissue, including the proboscis, was removed from the brain. The brains were then transferred to the cell plate using forceps, and immersed in XFe96 assay media.
Wing imaginal discs
Late third-instar larvae were dissected as described for larval brains. Wing discs were then pulled away from the cuticle from non-head body segment, and additional tissue removed. The discs were then transferred to the cell plate using forceps, and immersed in XFe96 assay media.
2.6 Protocol for analyzing whole brains/tissues with Agilent Seahorse XFe96 Analyzer
The Agilent Seahorse XFe96 metabolic analyzer was placed in an incubator set to 12°C, and analyzer was set to 25°C with the heat on (running Wave software version 2.4). An Agilent Seahorse XFe96 cartridge (Agilent, 102416-100) was hydrated with 200 μl of calibrant solution (Agilent, 100840-000) overnight at 25°C. The next day, brains and tissues were dissected in phosphate buffered solution (PBS) and added to an Agilent 96-well cell plate (Agilent, 102416-100) containing 50 μl of Agilent Seahorse assay media with supplements required for specific assay (methods 2.7, 2.8). Tissue was sunk to the bottom of the well, centered in the middle between the three raised spheres. Forceps were used to lower the tissue restraint such that the plastic ring is facing towards the bottom of the well and the nylon screen is facing the top of the well. A probe was used to gently push the edge of the tissue restraint down toward the bottom of the well until the restraint did not move or float in the well. 130 μl of assay media was added to each well; resulting in a total of 180 μl final in each well. Cell plate was placed on the tray of the XFe96 analyzer. The instrument was used as is for typical cell assays with all cycle procedures consisting of one-minute mixing, zero-minutes waiting, and three-minutes measuring.
2.7 Basal OCR and ECAR measurements using the XFe96
Basal levels of oxygen consumption (OCR) and extracellular acidification (ECAR) were measured for a minimum of six cycles. Tissue restraints were measured without tissue as a control. Agilent Seahorse XF assay medium (Agilent, 102365-100) supplemented with 10mM glucose, and 10mM sodium pyruvate was used for all basal measurement assays. Standard error of the mean (SEM) was used in analyzing metabolic measurement levels. Statistical significance was determined using the Holm-Sidak method with alpha=0.05.
2.8 Treatment assays using the XFe96
Oligomycin treatment
Analysis of mitochondrial respiration was conducted in assay medium described in 2.7. Basal brain OCR and ECAR was measured for 12 cycles prior to oligomycin injections. 20 μl of 100 μM oligomycin was added to injection port A, resulting in a final concentration of 10 μM of oligomycin per well. Standard error of the mean (SEM) was used in analyzing metabolic measurement levels. Statistical significance was determined using the Holm-Sidak method with alpha=0.05.
Glycostress Assay
Analysis of a tissue’s capacity to perform glycolysis when mitochondrial respiration is blocked was conducted in base medium (Agilent, 103334-100 base medium) supplemented with 1 mM glutamine. 20 μl of 50 mM glucose was added to port A and injected at the 6th cycle, resulting in a final concentration of 5mM glucose. 22 μl of 100 μM oligomycin was added to port B and injected at the 11th cycle, resulting in a final concentration of 10 μM oligomycin. 25 μl of 1 M 2- deoxyglutarate (2-DG) was added to port C and injected at the 23rd cycle, resulting in 100mM 2-DG. Standard error of the mean (SEM) was used in analyzing metabolic measurement levels. Statistical significance was determined using the Holm-Sidak method with alpha=0.05.
Glycolytic Rate Assay
Analysis of a tissue’s glycolytic rate with mitochondrial-produced acidification subtracted was conducted using base medium without phenol red (Agilent, 103335-100), supplemented with 5mM Hepes (Agilent, 103337-100), 2 mM glutamine, 10mM glucose and 1mM sodium pyruvate. 20 μl of 50 μM rotenone and antimycin-A was added to port A and injected at the 7th cycle, resulting in a final concentration of 5 μM rotenone and antimycin-A. 22 μl of 1M 2- deoxyglutarate (2-DG) was added to port B and injected at the 12th cycle, resulting in a final concentration of 100mM 2-DG. The software package included with this kit analyzes the oxygen consumption and extracellular acidification rates, while factoring in the buffer capacity of the media. It also calculates the acidification caused by the mitochondria and substracts this from the data. This method produces the proton efflux rate (PER). Standard error of the mean (SEM) was used in analyzing metabolic measurement levels. Statistical significance was determined using the Holm-Sidak method with alpha=0.05.
2.9 Normalization of metabolic measurements by protein concentration
Metabolic levels were normalized by protein concentration when indicated. Protein normalization was conducted using the Pierce 660nm protein assay reagent (Thermo Fisher Scientific, 22660). Each sample was homogenized in default lysis buffer (50 mM Tris (pH 7.5), 125 mM NaCl, 5% glycerol, 0.2% IGEPAL, 1.5 mM MgCl2, 1 mM DTT, 25 mM NaF, 1 mM Na3VO4, 1 mM EDTA and 2 × Complete protease inhibitor (Roche, Indianapolis, IN)) on ice, incubated on ice for 15 minutes, centrifuged at full speed for 15 minutes at 4°C. Supernatant was collected and measured as indicated in the Pierce 660nm protein assay reagent manual. Samples were assayed using a Biotek Cytation 3 plate reader. Brain OCR values were divided by μg amount of protein measuring to determine the normalized pmol/min/μg rate of oxygen consumption. Standard error of the mean (SEM) was used in analyzing metabolic measurement levels. Statistical significance was determined using the Holm-Sidak method with alpha=0.05.
2.10 Normalization of metabolic measurements by volume
Metabolic levels were normalized by volume when indicated. Media was removed from well and brains were washed with PBS before fixing at room temperature for 20 minutes in 4% formaldehyde. Brains were washed three times in PBS with 0.1% Tween (PBST) for 5 minutes, permeabilized for 20 minutes in .3% Triton in PBS, and washed once with PBS for 5 minutes. Brains were incubated with Alex Fluor™ 647 Phalloidin (1:500) (Thermo Fisher Scientific, A22287) in equal parts PBST and Western Blocking Reagent (Millipore Sigma, 11921673001) overnight at 4°C. Brains were washed five times in PBST for 5 minutes at room temperature, and one final wash with PBS for 1 minute. Brains were mounted in prolong gold with DAPI, and coverslip was added over a bridge formed with two broken coverslips to avoid crushing the brain. Z-stack images of 2 μm slices were taken with a Zeiss LSM 700 confocal microscope, and processed using Fiji to obtain volume in cubic microns. Standard error of the mean (SEM) was used in analyzing metabolic measurement levels. Statistical significance was determined using the Holm-Sidak method with alpha=0.05.
2.11 Whole animal respirometry
We measured whole animal metabolic rates using indirect calorimetry. Our system was configured for stop flow respirometry to measure the decreases in oxygen concentration and increases in carbon dioxide concentration within a sealed chamber containing either individual ants or groups of flies (n=3). Animals (adult flies or larva) were cold anesthetized, placed in the chamber, equilibrated with room temperature, and a baseline flow of ultra zero air from a compressed gas cylinder (30 mL min−1; 20.95% O2, N2 balance) was used to purge the chamber of room air for three minutes. Chambers were sealed for 15 minutes so that changes in gas concentration would accumulate. Air flow rate was regulated with an Omega MFC, gas stream switched between baseline and chamber with a Sable Systems multiplexer, oxygen concentration measured with a Sable Systems FoxBox, carbon dioxide concentration with a LiCor 7000 and data collected using a Sable Systems UI-2 analog to digital converter with Expedata software sampling at 1 Hz. The analysis of respirometry data files included automatic drift correction and integration of O2 depletion and CO2 accumulation peaks (Lighton, 2008). The wet mass of insects following respirometry was measured to the nearest 0.01 mg (Mettler Toledo XS22SDU Analytical Balance). Standard error of the mean (SEM) was used in analyzing metabolic measurement levels. Statistical significance was determined using the Holm-Sidak method with alpha=0.05.
3. Results and Discussion
3.1 Adapting the XFe96 metabolic analyzer for measurement of whole organs and tissues
The Agilent Seahorse XFe96 Analyzer (Fig. 1A) was designed for the measurement of cells, and, with the use of spheroid plates, organoids and other three dimensional cultures. Essential to the method of the analyzer is a sample’s ability to stay at the bottom the cell plate’s well (Fig. 1A), centered between three spheres (Fig. 1A) in which the sensor probe (Fig. 1B) rests upon. To measure whole brains from model systems, such as Drosophila melanogaster, a device is needed to restrain the tissue at the bottom of the well. Movement of the tissue when the probe moves up and down to mix and measure results in inaccurate, and imprecise results. After several prototypes and modifications, we have designed a micro-tissue restraint that fits exactly in the 96-well plate, holds the brain in place without disrupting the tissue, and is nearly inert (Fig. 1C). This tissue restraint is composed of nylon mesh adhered to an acetal homopolymer thermoplastic ring. This polymer is used in tissue-engineering bioreactors and surgical implants, chosen for its machining characteristics, strength, and thermal stability that renders it innocuous (Penick, Solchaga, Berilla, & Welter, 2005). The nylon mesh faces the sensor probes (Fig. 1Ci) with the plastic ring facing towards the sample (Fig. 1Cii). The plastic ring sits on the three spheres on the bottom of the plate (Fig. 1A) creating a small chamber where the brain can freely move while staying within the zone of measurement (Fig. 1Ciii).
Fig. 1.
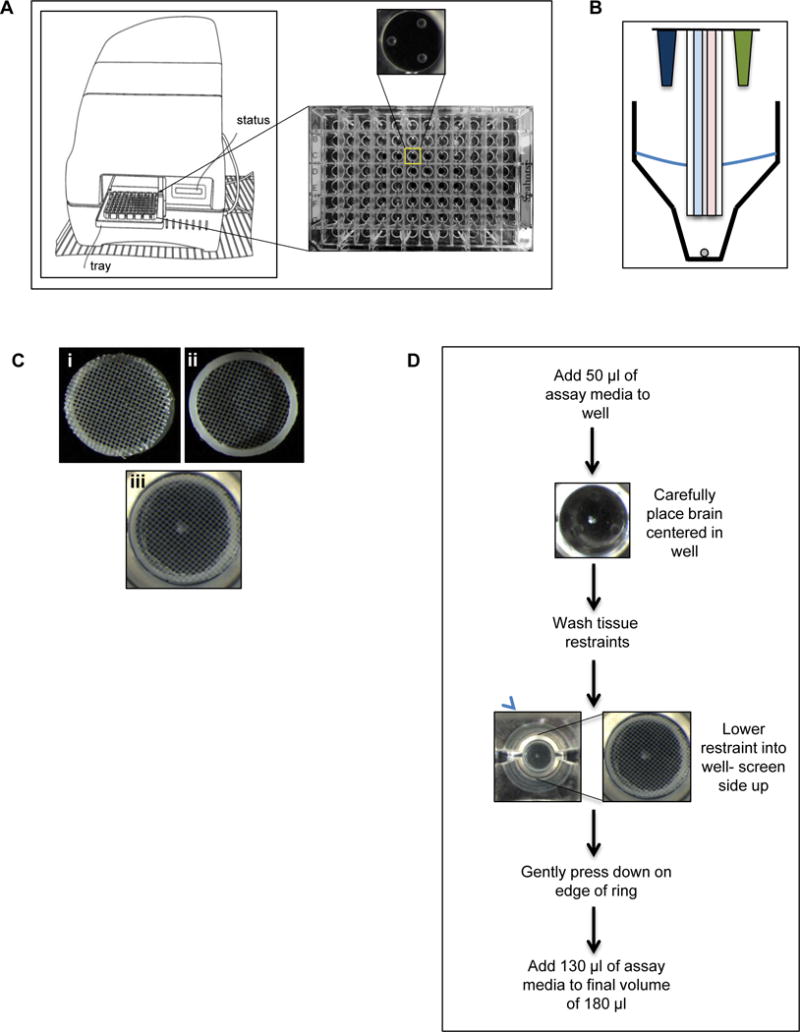
Design and implementation of a tissue restraint for analyzing metabolism using the XFe96 metabolic analyzer.
A) Cartoon of the XFe96 metabolic analyzer and cell plate. Zoomed in view of the bottom of a single well depicting the three raised spheres that the sensor probes rest on. B) Cartoon of the XFe96 probes (blue: oxygen consumption detection, pink: extracellular acidification detection) with injection ports (dark blue and green), and tissue being measured (grey circle). C) Images of tissue restraint, i) nylon screen facing up (correct orientation), ii) bottom view of restraint with plastic ring facing up, and iii) tissue restraint in well of XFe96 cell plate in correct orientation holding down a Drosophila larval brain. D) Protocol for analyzing Drosophila brain with the XFe96 metabolic analyzer using the tissue restraint.
The protocol for using the tissue restraint to measure metabolism of brains in the XFe96 Analyzer (Fig. 1D) involves adding a small volume of assay media to ease the sinking and positioning of the tissue, followed by lowering the restraint into the well. This must be done carefully to avoid disrupting the position of the brain, and to ensure the nylon mesh is facing upward and the plastic ring touches the bottom of the well. Once in place the restraint is anchored in the well by gently pushing the edges with a probe to immobilize the restraint. The volume of assay media is then increased to prepare for use in the analyzer.
3.2 Stable and reproducible metabolic measurements of Drosophila larval brains
To validate our method utilizing tissue restraints with the XFe96 instrument to analyze whole brains, we measured the oxygen consumption rate (OCR) in pico-moles per minute (pmol/min) of Drosophila third instar larval brains over a 40-minute period. Each well contained one brain from either a male or female larva for two wild type, commonly used lab strains, Canton-S (red) and Oregon-R (blue) (Fig. 2A). Each data point is the result of a one-minute mix and three-minute measure cycle. The OCR readings consistently stabilized by the 6th cycle measurement in all samples; brains maintained 6th cycle levels past two hours of analysis (data not shown). Canton-S and Oregon-R larval brain oxygen consumption rates were not significantly different, as expected (Fig 2A. and B.). The tissue restraint was analyzed in the absence of tissue and consumed a negligible amount of oxygen (yellow) (Fig. 2A and B). Extracellular acidification rate (ECAR) in milli-pH per minute (mpH/min) was measured simultaneously, and also stabilized at the 6th cycle measurement, was not significantly different between wild type strains, and measured low levels for wells containing only the tissue restraint (Fig. 2C). All brains analyzed were dissected from late third instar wandering larvae, however slight size differences are possible. Although the measurements of these brains were very similar, as noted by the small standard error of the mean (SEM), we assessed the variability of the brains based on size by protein normalization after XFe96 analysis. Protein normalization resulted in a standard deviation of 3.07 pmol/min/μg between five individual brain OCR measurements (Fig. 2D). Measurements from these same brains, un-normalized, resulted in standard deviation of 26.36 pmol/min. Normalized and un-normalized, the standard deviation is 20% of the average OCR reading. This suggests that variability in the readings is due to slight individual metabolic differences, and is not a result of brain size. These findings demonstrate that our novel tissue restraint allows for an adapted method utilizing the XFe96 to precisely measure metabolism in whole larval brains at the basal level. The OCR and ECAR readings are robust, suggesting that differences in metabolism due to genotype or drug treatment will be detectable using this method. Both observed OCR and ECAR levels demonstrate that tissue restraints do not significantly contribute to brain metabolism measurements, and that individual sample differences are slight enough that normalization may not be required depending on assay design.
Fig. 2.
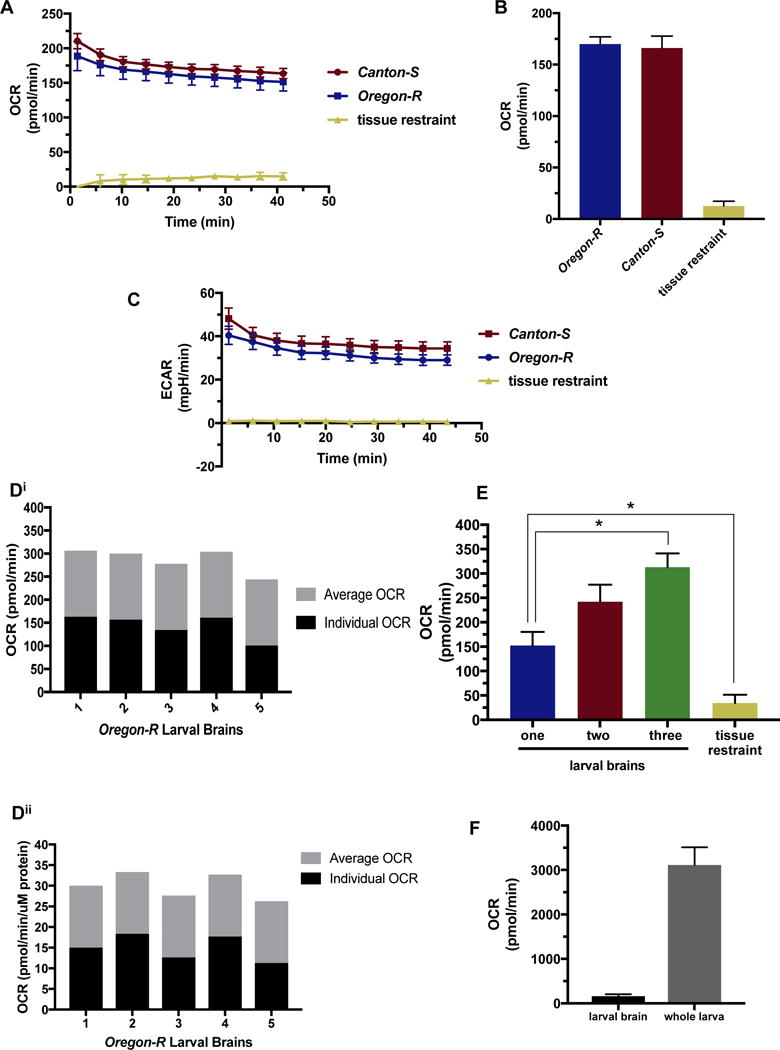
Metabolic analysis of Drosophila larval brains.
A, B, C) Oregon-R (blue) and Canton-S (red) wild-type Drosophila strain larval brains were analyzed using the XFe96 metabolic analyzer. Tissue restraints were measured as background control (yellow) A) Single larval brain basal level oxygen consumption (OCR) was measured in pico-moles per minute (pmol/min) for 10-cycles using a one-minute mix and three-minute measure procedure for each cycle, n= 14 for brains, n=6 for restraint control. B) Average single larval brain OCR measurements for the 6th cycle. This is the point at which the OCR stabilizes for our specimen, n= 14 for brains, n=6 for restraint control. C) Single larval brain basal level extracellular acidification (ECAR) was measured in milli-pH per minute (mpH/min) for 10-cycles using a one-minute mix and three-minute measure procedure for each cycle, n= 14 for brains, n=6 for restraint control. Di) Single Oregon- R larval brain OCR not normalized. Black bars represents individual OCR, and gray bars indicated average of all brains depicted in figure. Dii) Oregon- R larval brains from Di normalized to μg protein. Black bars represents individual OCR, and gray bars indicated average of all brains depicted in figure. E) Incremental differences in basal OCR (pmol/min) analyzing one (blue), two (red), and three (green) Oregon-R larval brains per well at the sixth cycle reading, n= 14 for brain wells, n=6 for restraint control (yellow). p-values of 0.015 and 0.022, respectively. F) Single Oregon-R larval brain OCR measuring with XFe96 compared to respiration of whole Oregon-R larva measured using stop flow respirometry, n= 14 larval brain, n= 9 whole larva.
As the assay media is a DMEM based recipe, we were concerned that measurements from Drosophila tissue may report readings that were not biologically significant due to the differences between hemolymph and assay media composition. To assess this we analyzed the oxygen consumption rates of Oregon-R larval brains using Schneider media instead of XFe96 assay media. Schneider media recipes were originally developed to closely mimic the composition of hemolymph (Schneider, 1964). OCR readings were not significantly different between brains measured in Schneider media and brains measured in XFe96 media assay, although Schneider media contains half the concentration of sugar (supplemental Fig. 1A). When sugar was added to the Schneider media to the same concentration as the assay media this difference was decreased (supplemental Fig. 1A). This suggests that using XFe96 assay media will provide biologically relevant results. Analysis of ECAR measurements was not possible since this media contains sodium bicarbonate and fluctuations in acidification are thus undetectable. Schneider media is composed of trehalose, the main energy supplying metabolite in Drosophila (Friedman, 1978; Lee & Park, 2004; Pasco & Léopold, 2012). Since we do not observe significant differences in oxygen consumption rates in media using trehalose versus glucose, we hypothesize that in this ex vivo setting this particular carbohydrate substitution does not alter respiration rates. We therefore conclude that our ex vivo conditions model in vivo biological interactions as closely as possible. However, one must still use this method and analyze the data carefully with thoughtful consideration to the fact that the brain is no longer communicating with tissues outside the central nervous system.
3.3 Single larval brains provide more accurate readings than multiple brains using this method
To further confirm the validity of these readings, we analyzed basal OCR levels of wells containing one-, two-, or three-third instar larval brains. We hypothesized that there would be an incremental increase in OCR as more brains were assayed per well. While we do observe incremental increases between one- and two-brain wells, we also observe significantly lower OCR readings than we would have expected from wells with three brains (p=0.008) (Fig. 2E). Numerous biological replicates of this experiment suggest two possibilities for these results. First, the slight variability between individuals may be magnified when combining levels from three metabolically unique individuals, and second, the difficulty in positioning three brains in the center of the well where measurements are taken. Visualization of these wells after assay completion often showed the three-brain wells with lowest readings containing one brain that was no longer fully in the area of measurement. While this result demonstrates that there are increases in OCR levels as more tissue is added, it also reveals that this method is best adapted for single brain samples, where brain positioning in the area of measurement is easiest and most reproducible.
3.4 Drosophila larval brain OCR levels scale to whole larva levels
It has been reported using magnetic resonance imaging (MRI) that the human brain accounts for ~20% of total body oxygen consumption (Jain, Langham, & Wehrli, 2010). While this has not been studied in Drosophila, we were interested to compare larval brain OCR levels with whole larva respiration levels. Our results suggest a decreased energy utilization, with larval brains consuming ~5% of the total larval consumption (Fig. 2F). This is significantly different from the reported scaling in humans and may reflect a biologically relevant difference.
3.5 Drosophila larval brain metabolism is altered by drug treatment
The power of using the XFe96 platform with whole brains is not only to assess basal metabolism, but also to investigate how drug treatments alter metabolism. We injected Oregon-R single larval brains with 10 μM oligomycin (blue) after the 12th cycle measurement (Fig. 3A). Oligomycin is an inhibitor of mitochondrial ATP synthase and thus significantly decreases oxygen consumption (Lardy, Johnson, & McMurray, 1958). We initially questioned whether the larval blood brain barrier would allow for drug uptake. However, OCR results from oligomycin optimization experiments containing step-wise dilutions of drug revealed concentration dependent biological responses (Supplmental Fig 2A.). The lowest oligomycin concentration tested (1 μM) resulted in no change in OCR, and the highest concentration (20 μM) caused rapid decline in OCR to levels indicative of loss of mitochondrial respiration (Supplmental Fig 2A.). While we cannot precisely report the amount of drug that passes the blood brain barrier, we can confidently state that the drug is having a dose-dependent affect on the tissue.
Fig. 3.
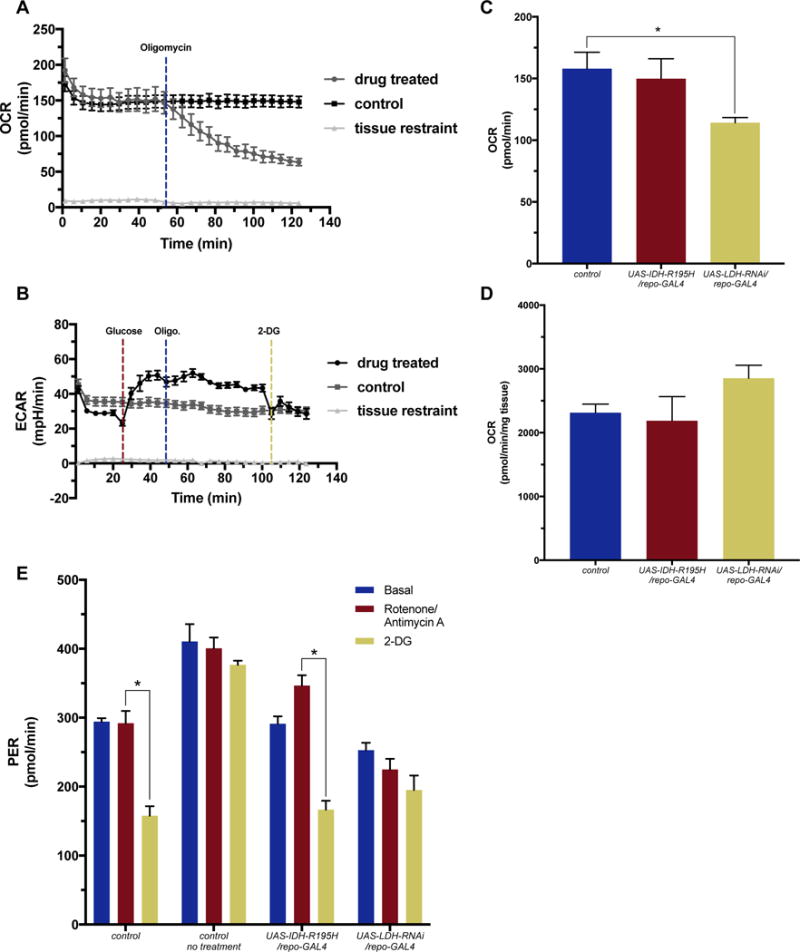
Measured changes in metabolism following drug treatment in single Drosophila larval brain.
A) OCR levels of Oregon-R Drosophila larval brains injected with 10 μM oligomycin (blue) after the 12th cycle results in a decrease in oxygen consumption, n=6. B) ECAR levels from glycolytic stress test performed on Oregon-R larval brains. 5 mM glucose (red) was injected at the 6th cycle, 10 μM oligomycin (blue) was injected at the 11th cycle, and 100 mM 2- deoxyglutarate (2-DG) (yellow) was injected at the 23rd cycle, n=11. Control larval brain wells and tissue restraint wells were injected with equal volumes of assay media, n=16. C) Basal OCR levels repo-GAL4 control driver line (blue), UAS-IDH-R195H/repo-GAL4 (red) and UAS-LDH-RNAi/repo-GAL4 (yellow) larval brains, n=6. Basal OCR levels are significantly lower in LDH knock-down brains compared to control brains (p-value=0.033). OCR values have been normalized using protein concentration. D) OCR levels in pico-moles per minute per milligram of tissue (pmol/min/mg tissue) in whole larva using using stop flow respirometry was measured in larva described in C, n=9 for repo-GAL4 control, UAS-IDH-R195H/repo-GAL4; n=6 for UAS-LDH-RNAi/repo-GAL4. E) Proton efflux rate (PER) measured in pmol/min for larval brains from larva described in C. Basal levels (blue) were measured for six cycles following 5 μM rotenone and antimycin-A (red) injected at the 7th cycle, and 100 mM 2- deoxyglutarate (2-DG) (yellow) injected at the 12th cycle. Data depicted on graph were from the 6th, 11th and 16th cycle, n=4. repo-GAL4 control driver line brains were treated (control) for comparison, and also untreated (control no treatment) for base line readings, n=4. Treatment of repo-GAL4 control driver line, and UAS-IDH-R195H/repo-GAL4 larval brains with 2-DG resulted in a significant decrease in PER levels, p-values= 0.011 and 0.011, respectively).
Our results demonstrate the decrease in OCR over 60-minutes of readings compared to our control Oregon-R larval brains that were injected with assay media. OCR levels do not drop to zero due to non-mitochondrial oxygen consumption. While OCR levels are expected to decrease when treated with oligomycin, ECAR levels should increase if cells are able to shift to a glycolytic metabolism in an attempt to generate energy. This shift is commonly seen in cancerous cells in the presence of oxygen and is known as the Warburg effect (Warburg, 1956; Ward & Thompson, 2012). To investigate the Drosophila larval brain’s ability to shift metabolic program we conducted a glycolytic stress test, where we incubate the brains in glucose-free media for five cycles (20 minutes), followed by injection of 5 mM glucose (red) (Fig. 3B). ECAR levels increase due to this bolus of glucose (Fig. 3B). At the 11th cycle 10 μM oligomycin (blue) is added in an attempt to further increase ECAR levels. We do not observe an additional ECAR level increase when treated with oligomycin (Fig. 3B). Increased concentrations of oligomycin damage the tissue and result in decreased levels (Supplemental Fig. 2A). A final injection of 2-deoxyglutarate (2-DG) is added at the 23rd cycle measurement. 2-DG is a glucose analog that bind to hexokinase and effectively halts glycolysis (Wick, Drury, Nakada, & Wolfe, 1957). We observe decreased ECAR levels that return to basal after four cycles (Fig. 3B). We are confident that 2-DG is able to pass through the blood brain barrier, as we have previously observed uptake of this drug by fluorescent microscopy when incubated with a fluorescently labeled 2-DG. These results suggest that wild type Drosophila larval brains do not have the capacity to shift to glycolysis. In fact, overtime treatment with oligomycin causes the ECAR levels to drop slightly as some cells are most likely dying (Fig. 3B). However, testing glycolytic stress in brains from altered genetic backgrounds may be used to investigate genes associated with metabolism reprogramming.
3.6 Investigating varying responses to metabolic challenges in brains harboring genetic alterations
Analyzing brains from wild type larvae can provide insight into general brain metabolism. More interesting, is challenging transgenic larvae to observe varying responses based on their capacity for metabolic reprogramming. To observe this, we generated larvae that expressed a transgenic construct under the control of the UAS-GAL4 binary system (Brand & Perrimon, 1993). The upstream activation sequence (UAS) will only be activated upon GAL4 protein binding, allowing for controlled expression of a transgene in a specific tissue. We used the repo-GAL4 driver line, which expresses the GAL4 protein in cells under the control of the reversed polarity promoter. This promoter is only expressed in glial cells, and thus would allow targeted expression of our transgene in the brain (Halter et al., 1995). After protein normalization, repo-GAL4 control larval brains (blue) and UAS-IDH-R195H/repo-GAL4 (red), an over-expression construct of a missense mutation in isocitrate dehydrogenase (IDH) implicated in human gliomas (Parsons et al., 2008), showed similar basal OCR levels at the 6th cycle measurement (Fig. 3C). However, knockdown of lactate dehydrogenase (LDH in humans, IMP-L3 in Drosophila) (UAS-LDH-RNAi/repo-GAL4), the enzyme that converts pyruvate to lactate in Drosophila (Abu-Shumays & Fristrom, 1997), resulted in significantly decreased OCR levels (Fig. 3C). This result could only be observed when measuring the brain, and not when measuring whole larva oxygen consumption (Fig. 3D). Whole larva OCR is not significantly different for either transgenic animal, although knockdown of LDH is slightly higher. This demonstrates the importance of measuring a specific tissue ex vivo. Model systems are commonly manipulated to genetically alter the tissue or cells of choice to allow for targeted studies. If one would like to analyze metabolism, whole animal methods do not possess the sensitivity that testing just the cells affected affords.
We further investigated these transgenic larval brains by challenging them with drug treatments aimed to assess their glycolytic rate. This assay subtracts the extracellular acidification from mitochondrial respiration by treatment with rotenone (Turrens & Boveris, 1980) and antimycin-A (Potter & Reif, 1952), inhibitors of the electron transport chain, and also factors in the buffering capacity of the assay media. The algorithm in the glycolytic rate software package utilizes the OCR rates to account for and subtract acidification resulting from the TCA cycle. Thus the proton efflux rates (PER) reported reflect acidification of the media caused by glycolysis and not the TCA cycle. Basal PER levels were calculated using media injected repo-GAL4 brains (control no treatment) as a control. repo-GAL4 brains were also treated (control) for comparison with UAS-IDH-R195H/repo-GAL4, and UAS-LDH-RNAi/repo-GAL4 treated brains. PER levels after treatment with 5 μM rotenone and antimycin-A (red) directly correlates to lactic acid production and increased glycolysis. Control and LDH knockdown brains were unable to shift to glycolysis after inhibition of mitochondrial respiration, however IDH point mutants had a slight increase in PER level, though not significant (Fig. 3E). Treatment with 2-DG (yellow) should abolish glycolysis and affect the PER levels of those cells using glycolytic respiration. Both repo-GAL4 (control) and IDH point mutant brains are significantly affected by 2-DG treatment, while LDH knockdown brains are not significantly affected (Fig. 3E). Since LDH is required to generate lactate, the metabolite being measured in this assay, the results from UAS-LDH-RNAi/repo-GAL4 brains reflect the cells inability to produce lactate and display a similar trend to control no treatment brains. Control treated brains are similar to Oregon-R glycolytic stress results that reveal no increase in glycolysis. However, this assay allows us to identify that these cells are already utilizing glycolytic respiration to some extent. UAS-IDH-R195H/repo-GAL4 brains were able to enhance their dependence on glycolysis slightly, and also demonstrated their normal utilization for glycolytic respiration, similar to repo-GAL4 controls. Together this data suggests that this method will allow for the interrogation of genes that play a role in metabolic reprogramming or the basic study of how genotype affects metabolism.
The results of an LDH knock-down in glial cells was expected to inhibit production of lactate even when mitochondrial respiration was challenged. However, the ability to detect lactate in the media as an acidifying factor was unknown. While lactate production can be measured in Drosophila cultured cells, and increases are detected upon mitochondrial challenge or gene mutation (Freije, Mandal, & Banerjee, 2012), it is less clear whether lactate produced by glial cells would enter the extracellular environment and be detected in the media. As reported in Volkenhoff et al., glial cells are able to produce lactate, but this lactate is then utilized by neurons (Volkenhoff et al., 2015). From this analysis we were able to detect decreases in PER reflecting decreased lactate levels and media acidification in wild type brains, but not LDH deficient brains. This data would suggest that some lactate does enter the media and we can observe changes in these levels due to genetic alterations. However, with the knowledge that neurons are actively utilizing this lactate, PER data needs to be analyzed carefully.
When compared to LDH, the role of the IDH-R195H mutation in glial cell metabolism is more uncertain. Since we did not detect significant changes in PER data between controls and IDH mutant brains, we performed the same experiment at 29°C. This increase in temperature enhances GAL4 expression and thus increases IDH mutant expression in larval glial cells. Parental flies were crossed at 29°C, and larvae were raised at this temperature until reaching third instar larval stages. The XFe96 was also increased to 29°C during the assay. The results still did not show a significant difference between IDH mutant brains and control brains in either basal OCR (Supplemental Fig. 3Ai) or when challenged and measured for PER (Supplemental Fig. 3Aii). There are reports suggesting that expression of this missense protein product cause a shift to glutaminolysis (Ohka et al., 2014). This would not be reflected in this type of experimentation, as acidification rates would not increase –as we have reported here. This method could be used to determine substrate utilization in mutants that have become dependent on alternative metabolic pathways, opening new avenues of exploration.
3.7 Stable and Reproducible metabolic measurements of adult Drosophila brains
We conducted similar basal measurement studies with adult brains to verify that this method will work for both larvae and adults; allowing for developmental studies and greater flexibility in analyzing brain metabolism. Basal OCR levels for young adult Oregon-R brains were slightly increased compared to larval brains (Fig. 4A and C), but also stabilized by the 6th cycle measurement (Fig. 4A). ECAR levels were slightly lower than larval brains (Fig. 4B), suggesting the metabolic program of developing larval brains may be more glycolytic than adult brains. This would support data that shows larvae rely on glycolysis to increase body mass quickly during development (Tennessen, Baker, Lam, Evans, & Thummel, 2011). As observed with larval brains, normalization of OCR levels in adult brains using volume (Fig. 4Ci and Cii) and protein concentration (Supplemental Fig. 4Ai and Aii) methods result measurements with no significant difference in the percent standard deviation between normalized and un-normalized brains. We further investigated whether the sex of the adult fly altered the OCR levels observed at the 6th cycle measurement. There was no significant difference between female and male similarly aged young adult brains (Fig. 4D).
Fig. 4.
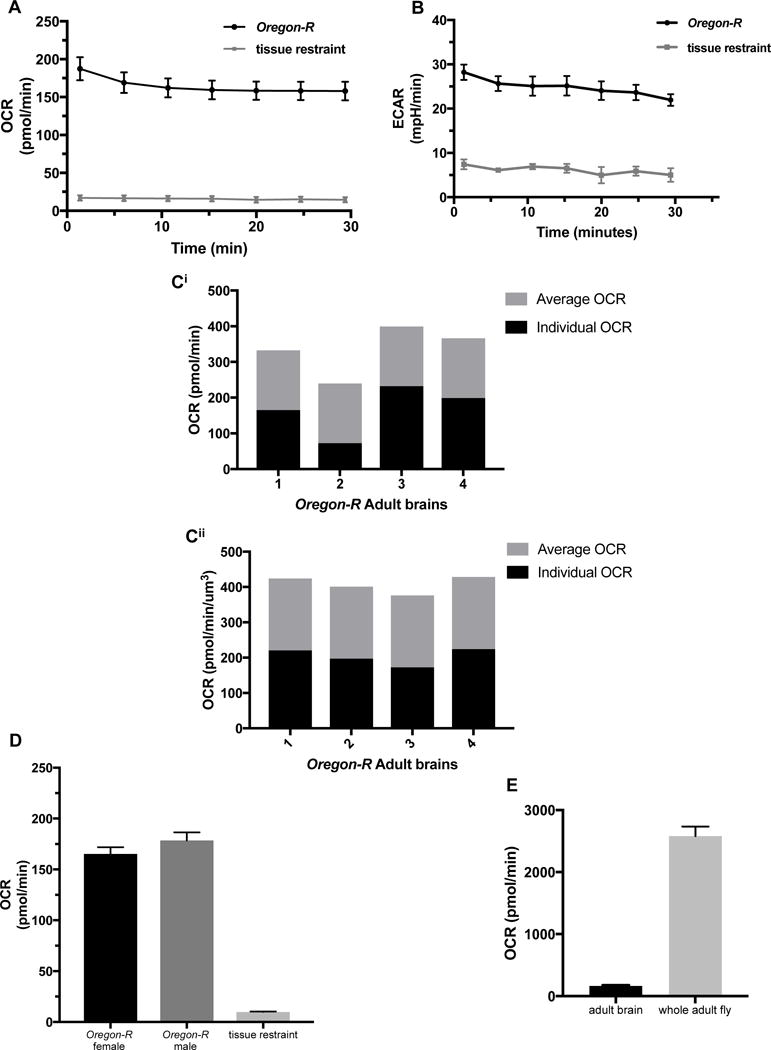
Metabolic analysis of single Drosophila adult brains.
A) Basal OCR levels of Oregon-R adult brains for 7 cycles using one-minute mix and three-minute measure procedure, n=8. B) Basal ECAR levels of Oregon-R adult brains for 7 cycles using one-minute mix and three-minute measure procedure, n=7. Ci) OCR levels of single Oregon-R adult brains. Black bars represents individual OCR, and gray bars indicated average of all brains depicted in figure. Cii) Oregon-R adult brains from Ci normalized using volume; reported in pmol/min/μm3. Black bars represents individual OCR, and gray bars indicated average of all brains depicted in figure. D) Basal OCR levels of Oregon-R male and female adult brains measured at the 6th cycle, n=5. E) Basal OCR levels of Oregon-R adult brains at the 6th cycle using the XFe96 metabolic analyzer (n=5) compared to respiration of the whole Oregon-R adult fly measured using stop flow respirometry (n=9).
3.8 Drosophila adult brain OCR levels scale to whole animal levels similar to larva
As we tested larvae, we wanted to similarly investigate how OCR levels in the adult brain compared to the whole fly. While larval brains consumed ~5% of the overall oxygen, adult brains consume ~6%. This result is not significantly different, and strengthens the validity of a potential difference in energy utilization scaling in human and insects.
3.9 Broader impact of tissue restraint utilization with the XFe96 analyzer
We have reported data that validates this novel adaptation of the XFe96 analyzer for use with Drosophila brains. However, this method can also be applied to other tissues and model systems. To demonstrate its versatility we have analyzed Drosophila imaginal wing discs, which are commonly investigated in developmental studies, and in clonal gene analysis. Basal OCR levels from one, two or three of these small tissues were measured, and the 6th cycle measurement is reported in figure 5A. Expected incremental increases between one- and two-disc groups were observed, however, significantly higher OCR levels were obtained for three-disc groups than expected –a phenomenon also observed with brains. These tissues are much smaller and can easily fit in the measurement area of the well, unlike the brains. However, we hypothesize that since the two-wing disc wells are using paired samples from the same larva their values will be very close to one another and report an incremental increase, while three-wing disc wells combine discs from more than one larva and may have greater variation. This variation could explain values that are significantly different from expected rates.
Beyond Drosophila other model organisms can use this analytical platform. Previous studies have used the XFe96 to measure metabolism of C. elegans (Koopman et al., 2016). We measured larval stage 1 (L1), larval stage 4 (L4), and young adult day 1 (D1) worms using our micro-tissue restraints to demonstrate the versatility of this method, and to further validate our approach (Fig. 5B). Worms were washed in M9 media and counted in 2μl of M9 media. Volume was corrected for loading of 3-10 worms in each well of the XFe96 cell plate in M9 media. Basal OCR levels were measured over 10 cycles using the protocol described in Koopman et al. and methods. After the assay was completed OCR levels were normalized per worm. L1 (blue) and L4 (red) did not have significantly different OCR levels (6.20 and 8.99 pmol/min/worm, respectively), but D1 worms (yellow) OCR level was significantly higher (23.18 pmol/min/worm). These findings agree with the Koopman et al. study, although our OCR levels were slightly higher than reported. We suspect that temperature stabilization resulted in this difference. Since the reported protocol was published, a new version of the XFe96 WAVE software was released. This software allows for better stabilization of lower temperatures. By keeping the instrument in a cooler environment we are able to set the heated temperature to 20°C throughout the analysis. Use of the micro-tissue restraints also allows for quicker load to assay working time, minimizing the need for gravity-based tissue sinking methods.
4. Conclusions
In conclusion, we have validated a method for analyzing the metabolism of whole brains and other small organs ex vivo, enabling measurements of cells working together in a tissue dependent context. This method relies on the use of tissue restraints specifically designed to create a chamber for the brain to move freely, yet stay within the zone of measurement, when using the XFe96 metabolic analyzer. The procedure is fast, simplistic and allows for the measurement of multiple samples at a time. We report precise, and reproducible data with larval and adult Drosophila brains, Drosophila wing imaginal discs, and C. elegans. This method could easily be adapted to other small model systems and have a much broader impact. The ability to challenge these tissues with drug treatments to investigate metabolic sensitivities and dependencies will be useful in our continued effort to better understand disease through model systems, to interrogate gene functions through transgenic animals, to identify metabolic targets for therapeutic treatment, and to study the role of metabolism in behavior and social interaction.
Supplementary Material
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018637) were used in this study. Research reported in this publication was supported by the Institutional Development Award (IDeA) Network for Biomedical Research Excellence from the National Institute of General Medical Sciences of the National Institute of Health [grant number P20GM103430], and by the Rhode Island Foundation.
Footnotes
Disclosures
These results have not been previously published in any journal. M. Tipping and J.S. Waters hold provisional patent (no. 62/491,431) for micro-tissue restrains used in this reported method.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- Abu-Shumays RL, Fristrom JW. IMP-L3, a 20-hydroxyecdysone-responsive gene encodes Drosophila lactate dehydrogenase: structural characterization and developmental studies. Developmental Genetics. 1997;20(1):11–22. doi: 10.1002/(SICI)1520-6408(1997)20:1<11::AID-DVG2>3.0.CO;2-C. http://doi.org/10.1002/(SICI)1520-6408(1997)20:1<11::AID-DVG2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Atamna H, Frey WH. Mechanisms of mitochondrial dysfunction and energy deficiency in Alzheimer’s disease. Mitochondrion. 2007 doi: 10.1016/j.mito.2007.06.001. http://doi.org/10.1016/j.mito.2007.06.001. [DOI] [PubMed]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England) 1993;118(2):401–15. doi: 10.1242/dev.118.2.401. http://doi.org/10.1101/lm.1331809. [DOI] [PubMed] [Google Scholar]
- Brody AL, Saxena S, Silverman DH, Alborzian S, Fairbanks LA, Phelps ME, Baxter LR., Jr Brain metabolic changes in major depressive disorder from pre- to post-treatment with paroxetine. Psychiatry Res. 1999;91(3):127–139. doi: 10.1016/s0925-4927(99)00034-7. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10641577. [DOI] [PubMed] [Google Scholar]
- Cai H, Cong W, Ji S, Rothman S, Maudsley S, Martin B. Metabolic dysfunction in Alzheimer’s disease and related neurodegenerative disorders. Current Alzheimer Research. 2012;9(1):5–17. doi: 10.2174/156720512799015064. http://doi.org/10.2174/156720512799015064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature Publishing Group. 2011;11(2):85–95. doi: 10.1038/nrc2981. http://doi.org/10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discovery Today. 2008 doi: 10.1016/j.drudis.2007.12.008. http://doi.org/10.1016/j.drudis.2007.12.008. [DOI] [PubMed]
- Frappell PB, Blevin HA, Baudinette RV. Understanding respirometry chambers: What goes in must come out. Journal of Theoretical Biology. 1989;138(4):479–494. doi: 10.1016/s0022-5193(89)80046-3. http://doi.org/10.1016/S0022-5193(89)80046-3. [DOI] [PubMed] [Google Scholar]
- Freije WA, Mandal S, Banerjee U. Expression Profiling of Attenuated Mitochondrial Function Identifies Retrograde Signals in Drosophila. G3: Genes|Genomes|Genetics. 2012;2(8):843–851. doi: 10.1534/g3.112.002584. http://doi.org/10.1534/g3.112.002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S. Trehalose regulation, one aspect of metabolic homeostasis. Annual Review of Entomology. 1978;23(3):389–407. http://doi.org/10.1146/annurev.en.23.010178.002133. [Google Scholar]
- Gibert Y, McGee SL, Ward AC. Metabolic profile analysis of zebrafish embryos. Journal of Visualized Experiments: JoVE. 2013;(71):e4300. doi: 10.3791/4300. http://doi.org/10.3791/4300. [DOI] [PMC free article] [PubMed]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development (Cambridge, England) 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Ha1. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Vol. 144, Cell. 2011. p. 646–74. llmarks of cancer: The next generation. Cell. 2011 doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hardie RA, Van Dam E, Cowley M, Han TL, Balaban S, Pajic M, Saunders DN. Mitochondrial mutations and metabolic adaptation in pancreatic cancer. Cancer & Metabolism. 2017:1–15. doi: 10.1186/s40170-017-0164-1. http://doi.org/10.1186/s40170-017-0164-1. [DOI] [PMC free article] [PubMed]
- Jain V, Langham MC, Wehrli FW. MRI estimation of global brain oxygen consumption rate. Journal of Cerebral Blood Flow & Metabolism. 2010;30(9):1598–1607. doi: 10.1038/jcbfm.2010.49. http://doi.org/10.1038/jcbfm.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman M, Michels H, Dancy BM, Kamble R, Mouchiroud L, Auwerx J, Houtkooper RH. A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nature Protocols. 2016;11(10):1798–1816. doi: 10.1038/nprot.2016.106. http://doi.org/10.1038/nprot.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwang Kim K, Abelman S, Yano N, Ribeiro JR, Singh RK, Tipping M, Moore RG. Tetrathiomolybdate inhibits mitochondrial complex IV and mediates degradation of hypoxia-inducible factor-1α in cancer cells. Scientific Reports. 2015 Aug;5:14296. doi: 10.1038/srep14296. http://doi.org/10.1038/srep14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardy HA, Johnson D, McMurray WC. Antibiotics as tools for metabolic studies. I. A survey of toxic antibiotics in respiratory, phosphorylative and glycolytic systems. Archives of Biochemistry and Biophysics. 1958;78(2):587–597. doi: 10.1016/0003-9861(58)90383-7. http://doi.org/10.1016/0003-9861(58)90383-7. [DOI] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167(1):311–323. doi: 10.1534/genetics.167.1.311. http://doi.org/10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighton JRB. Measuring Metabolic Rates. Spatial Epidemiology: Methods and Applications. 2000 Sep;:87–103. http://doi.org/10.1093/ISBN.
- Lighton JRB. Measuring Metabolic Rate:s A Manual for Scientists. Oxford: Oxford University Press; 2008. [Google Scholar]
- Liu S, Shah SJ, Wilmes LJ, Feiner J, Kodibagkar VD, Wendland MF, Rollins MD. Quantitative tissue oxygen measurement in multiple organs using 19F MRI in a rat model. Magnetic Resonance in Medicine. 2011;66(6):1722–1730. doi: 10.1002/mrm.22968. http://doi.org/10.1002/mrm.22968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohka F, Ito M, Ranjit M, Senga T, Motomura A, Motomura K, Natsume A. Quantitative metabolome analysis profiles activation of glutaminolysis in glioma with IDH1 mutation. Tumor Biology. 2014;35(6):5911–5920. doi: 10.1007/s13277-014-1784-5. http://doi.org/10.1007/s13277-014-1784-5. [DOI] [PubMed] [Google Scholar]
- Parsons D, Jones S, Zhang X, Lin J. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;1807(2008):1807–1812. doi: 10.1126/science.1164382. http://doi.org/10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasco MY, Léopold P. High sugar-induced insulin resistance in Drosophila relies on the Lipocalin Neural Lazarillo. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0036583. http://doi.org/10.1371/journal.pone.0036583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penick KJ, Solchaga LA, Berilla JA, Welter JF. Performance of polyoxymethylene plastic (POM) as a component of a tissue engineering bioreactor. Journal of Biomedical Materials Research - Part A. 2005;75(1):168–174. doi: 10.1002/jbm.a.30351. http://doi.org/10.1002/jbm.a.30351. [DOI] [PubMed] [Google Scholar]
- Potter VR, Reif AE. Inhibition of an electron transport component by antimycin A. The Journal of Biological Chemistry. 1952;194(1):287–297. [PubMed] [Google Scholar]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JTJ, Griffin JL, Bahn S. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Molecular Psychiatry. 2004;9(7):684–697. doi: 10.1038/sj.mp.4001511. 643. http://doi.org/10.1038/sj.mp.4001532. [DOI] [PubMed] [Google Scholar]
- Schneider I. Differentiation of larval drosophila eye???antennal Discs in Vitro. Journal of Experimental Zoology. 1964;156(1):91–103. doi: 10.1002/jez.1401560107. http://doi.org/10.1002/jez.1401560107. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metabolism. 2011;13(2):139–148. doi: 10.1016/j.cmet.2011.01.005. http://doi.org/10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipping M, Waters J. Method and device for restraining contents of a cell plate. USA: 2017. [Google Scholar]
- Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. (Drug Discovery).Nature Reviews. 2011;10(9):671–684. doi: 10.1038/nrd3504. http://doi.org/10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- Volkenhoff A, Weiler A, Letzel M, Stehling M, Klämbt C, Schirmeier S. Glial glycolysis is essential for neuronal survival in drosophila. Cell Metabolism. 2015;22(3):437–447. doi: 10.1016/j.cmet.2015.07.006. http://doi.org/10.1016/j.cmet.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the Origin of Cancer Cells. Science. 1956;123(3191):309–14. doi: 10.1126/science.123.3191.309. http://doi.org/10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. http://doi.org/10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WICK AN, DRURY DR, NAKADA HI, WOLFE JB. Localization of the primary metabolic block produced by 2-deoxyglucose. The Journal of Biological Chemistry. 1957;224(2):963–969. http://doi.org/10.1126/science.1321497. [PubMed] [Google Scholar]
- Williamson WR, Hiesinger PR. Preparation of developing and adult Drosophila brains and retinae for live imaging. Journal of Visualized Experiments: JoVE. 2010;(37):1–5. doi: 10.3791/1936. http://doi.org/10.3791/1936. [DOI] [PMC free article] [PubMed]
- Withers PC. Design, calibration and calculation for flow-through respirometry systems. Australian Journal of Zoology. 2001 http://doi.org/10.1071/ZO00057.
- Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, Guha A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. The Journal of Experimental Medicine. 2011;208(2):313–26. doi: 10.1084/jem.20101470. http://doi.org/10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhong Fang S, Li Xin X, Jia Xin L, Min W, Xiao Xuan W, Fang Y. Glutamate Impairs Mitochondria Aerobic Respiration Capacity and Enhances Glycolysis in Cultured Rat Astrocytes *. Biomed Environ Sci. 2017;30(1):44–51. doi: 10.3967/bes2017.005. http://doi.org/10.3967/bes2017.005. [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Bigner DD. IDH1 and IDH 2 Mutations in Gliomas. New England Journal of Medicine. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. http://doi.org/10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death & Disease. 2013;4(3):e532. doi: 10.1038/cddis.2013.60. http://doi.org/10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


