Version Changes
Revised. Amendments from Version 1
Raw images of all western blots were added in a PDF format as well as TIFFs, to enable all readers to view the raw data files. The data itself is unchanged.
Abstract
Malate dehydrogenases (Mdhs) reversibly convert malate to oxaloacetate and serve as important enzymes in several metabolic pathways. In the yeast Saccharomyces cerevisiae there are three Mdh isozymes, localized to different compartments in the cell. In order to identify specifically the Mdh2 isozyme, GenScript USA produced three different antibodies that we further tested by western blot. All three antibodies recognized the S. cerevisiae Mdh2 with different background and specificity properties. One of the antibodies had a relatively low background and high specificity and thus can be used for specific identification of Mdh2 in various experimental settings.
Keywords: Malate dehydrogenase, Saccharomyces cerevisiae, antibody, western blot
Introduction
Malate dehydrogenases (Mdhs) catalyse the interconversion of malate and oxaloacetate using NAD + or NADH 1. In the yeast Saccharomyces cerevisiae there are three known isozymes: Mdh1 is located in mitochondria, Mdh2 is mostly cytosolic, and Mdh3 is localized to peroxisomes. The residues of the three isozymes are between 43–50% identical 2. It is thus very important for purposes of isolation and identification to utilize specific antibodies that will recognize only one specific isozyme. Using three Mdh2 peptides, which were specifically designed to unique regions in the Mdh2 protein, GenScript USA produced three different antibodies that should have high specificity for S. cerevisiae Mdh2 relative to Mdh1 and Mdh3. We then tested all three antibodies by western blotting and found one with specific binding. Due to its specificity, this antibody has the potential to also work in other experimental assays such as immunoprecipitation, immunohistochemistry and ELISA (Enzyme-Linked Immunosorbent Assay).
Methods
Antibody details
Three antibodies for S. cerevisae Mdh2 were custom produced for us by GenScript USA : 1. Purified antibody, Anti-peptide #1, item number: U1684BK300_2, LOT number: A416120074, catalogue number: SC1195. 2. Purified antibody, Anti-peptide #2, item number: U1684BK300_5, LOT number: A416120094, catalogue number: SC1195. 3. Purified antibody, Anti-peptide #3, item number: U1684BK300_8, LOT number: A416120072, catalogue number: SC1195.
All three antibodies were raised in New Zealand Rabbit and are polyclonal. The immunogen for all three antibodies is conjugated KLH Peptide. To find specific Mdh2 peptides, the three S. cerevisiae malate dehydrogenases (Mdh1, Mdh2 and Mdh3) were aligned and analysed by GenScript, using their Antigen Design Tool. From this analysis, three peptides (corresponding to the three antibodies) were selected: #1: CHPQSRNSGIERRIM; #2: CINIESGLTPRVNSM; #3: MPHSVTPSIEQDSLC. The cysteines in the N’ terminus (peptides number 1 and 2) or C’ terminus (peptide number 3) were added for KLH conjugation.
Yeast strains and strain construction
Yeast strains are all based on the BY4741 laboratory strain 3. Manipulations were performed using a standard PEG/LiAC transformation protocol 4. The GFP-Mdh1, GFP-Mdh2, GFP-Mdh3 and OE-mCherry-Mdh3 strains were picked from the SWAT N’ GFP or N’ mCherry yeast libraries that were recently prepared in our lab 5. All Strains with a fluorescent tag, including the strains that were picked from the SWAT libraries were verified using PCR (one primer from within the endogenous Open Reading Frame (ORF) and one from within the tag sequence) as well as by fluorescent microscopy. Primers for creating strains with deletions or C’ tagging of genes (Δ mdh1, Δ mdh2, Δ mdh3 and Mdh2-mCherry) were designed using the Primers-4-Yeast web tool 6. All deletions were verified using primers from within the endogenous ORF. All primers are summarized in Table 1.
Table 1. A list of primers used in this study.
| Gene | Primer Name | Sequence | Description |
|---|---|---|---|
| MDH1 | MDH1 KO pFA6 F | AAAAAAAACAAAAGGAAAAGGAAGGATACCATATACAATGCGGATCCCCGGGTTAATTAA | Primer for KO of gene using pFA6 plasmids. |
| MDH1 | MDH1 KO pFA6 R | TTCCCTATTTTTCACTCTATTTCTGATCTTGAACAATCTAGAATTCGAGCTCGTTTAAAC | Primer for KO of gene using pFA6 plasmids. |
| MDH1 | MDH1 N' tag CHK R | TGGAACGGTAGAATTGACTG | Primer for checking N' tagging of the gene. |
| MDH1 | MDH1 WT CHK F | TCCAACCCAGTCAATTCTAC | Primer for testing the presence of the gene. |
| MDH1 | MDH1 WT CHK R | GTTAGCGAATTTAGCACCAG | Primer for testing the presence of the gene. |
| MDH2 | MDH2 KO pFA6 F | CAAAAGTTCAATACAATATCATAAAAGTTATAGTAACATGCGGATCCCCGGGTTAATTAA | Primer for KO of gene using pFA6 plasmids. |
| MDH2 | MDH2 KO pFA6 R | CAATTTGCTGCATTCTTATGCTTCGGTCCGATGCTCATTAGAATTCGAGCTCGTTTAAAC | Primer for KO of gene using pFA6 plasmids. |
| MDH2 | MDH2 N' tag CHK R | TGGAAATGACAAGAACGAAG | Primer for checking N' tagging of the gene. |
| MDH2 | MDH2 WT CHK F | AACTGTTTGCATAACGCTTC | Primer for testing the presence of the gene. |
| MDH2 | MDH2 WT CHK R | CATGGAGTTAACACGAGGAG | Primer for testing the presence of the gene. |
| MDH2 | MDH2 C' tag CHK F | ATTCCGTTGTTTTCACAGTC | Primer for checking C' tagging of the gene. |
| MDH2 | MDH2 C' tag pFA6 F | TAAGGGCTTGGAATTCGTTGCATCGAGATCTGCATCATCTCGGATCCCCGGGTTAATTAA | Primer for C' tagging of gene using pFA6 plasmids. |
| MDH2 | MDH2 C' tag pFA6 R | CAATTTGCTGCATTCTTATGCTTCGGTCCGATGCTCATTAGAATTCGAGCTCGTTTAAAC | Primer for C' tagging of gene using pFA6 plasmids. |
| MDH3 | MDH3 KO pFA6 F | TGCAAAAGAAAATAAAAAGAGACAAACAATCATAAACATGCGGATCCCCGGGTTAATTAA | Primer for KO of gene using pFA6 plasmids. |
| MDH3 | MDH3 KO pFA6 R | AGTATAGAGTTAAGAAAAATATAAAAATTGAAGTAGCTCAGAATTCGAGCTCGTTTAAAC | Primer for KO of gene using pFA6 plasmids. |
| MDH3 | MDH3 N' tag CHK R | TTCTTCAAAGTTTCCACAGC | Primer for checking N' tagging of the gene. |
| MDH3 | MDH3 WT CHK F | ATTCAGGGGAAACCATTATC | Primer for testing the presence of the gene. |
| MDH3 | MDH3 WT CHK R | TCGATGGATACTACGCTACC | Primer for testing the presence of the gene. |
| MDH1/2/3 | N 'tag CHK F | CGACAGAGAATTCATCGATG | Primer for checking N' tagging. Using pym plasmid |
| MDH1/2/3 | F2-Rev-Com | TTAATTAACCCGGGGATCCG | Reverse complementary to the F1/F2 primer of the
Longtine's pFA6a plasmids. |
Western blot analysis
Yeast proteins were extracted by the NaOH protocol as previously described 7 and resolved on polyacrylamide gels with the following modifications:
-
1.
Liquid cultures were grown over night at 30°C in glucose containing media, and then were diluted and incubated for an additional 4–6 hours to get to mid-log phase. Cells that were grown in a different carbon source than glucose were grown in synthetic media with 0.1% glucose (6.7 g/l yeast nitrogen base and 0.1% glucose) for the overnight incubation, then diluted in synthetic media containing galactose (6.7 g/l yeast nitrogen base and 2% galactose) or oleic acid (6.7 g/l yeast nitrogen base, 0.2% oleic acid and 0.1% Tween-80) and incubated for an additional 4–6 hours. Cells that were grown in glucose were grown in yeast extract peptone dextrose (YPD) rich medium (1% Yeast Extract, 2% peptone, 2% glucose) for the whole process.
-
2.
The extractions were incubated for 10 minutes at 70°C and 15 µl supernatant was loaded per lane of the gel.
The gels were transferred to nitrocellulose membrane blots, blocked for 1 hour in SEA block diluted in TBS-T (1:5) at room temperature (RT), and probed with primary rabbit/mouse antibodies against Mdh2, Histone H3, Actin and mCherry ( Table 2) for 1 hour at RT. Final concentrations of the primary antibodies were: 1 µg/ml for anti-Mdh2 peptides #1 and #3, 0.5 µg/ml for anti-Mdh2 peptide #2, anti-mCherry and anti-Actin and 0.2 µg/ml for anti-Histone H3. The membranes were washed 3 times without incubation in TBS-T, followed by 3x3 minute washes in TBS-T. The membranes were then probed with a secondary goat anti-rabbit/mouse antibody conjugated to IRDye800 or to IRDye680 ( Table 2) for 30 minutes at RT and washed as before. Final concentrations of the secondary antibodies were 0.1 µg/ml for IRDye 800CW Goat anti-Rabbit IgG and IRDye 680RD Goat anti-mouse IgG, and 0.2 µg/ml for Goat anti-mouse HRP conjugated. Membranes were scanned for infrared signal using the Odyssey Imaging System (Li-Cor). For detecting the anti-Actin primary antibody ( Table 2), a goat anti-mouse HRP conjugated secondary antibody ( Table 2) was used. The membrane was then washed with ECL reagents and scanned using imageQuant LAS 4000 system (GE Healthcare). Images were transferred to ImageJ 1.51s for slight adjustments of contrast and brightness. For further information about the western blot reagents see Table 3.
Table 2. Primary and secondary antibodies.
| Antibody | Manufacturer | Catalogue
Number |
RRID |
|---|---|---|---|
| Rabbit anti-Mdh2 peptide #1 (Ab1) | GenScript | SC1195 | |
| Rabbit anti-Mdh2 peptide #2 (Ab2) | GenScript | SC1195 | |
| Rabbit anti-Mdh2 peptide #3 (Ab3) | GenScript | SC1195 | |
| Mouse anti-mCherry | Abcam | ab125096 | AB_11133266 |
| Rabbit anti-Histone H3 | Abcam | ab1791 | AB_302613 |
| Mouse anti-Actin | Abcam | ab8224 | AB_449644 |
| IRDye 800CW Goat anti-Rabbit IgG | LI-COR, Inc | 926-32211 | AB_621843 |
| IRDye 680RD Goat anti-mouse IgG | LI-COR, Inc | 926-68070 | AB_10956588 |
| Goat anti-mouse HRP conjugated | Abcam | ab6789 | AB_955439 |
Table 3. Details of reagents for Western Blot.
| Process | Reagent | Manufacturer | Catalogue Number | Concentration |
|---|---|---|---|---|
| Protein extraction | Sodium hydroxide (NaOH) | Merck
Chemicals |
1064981000 | 0.2 M |
| 5 X SDS sample buffer | β-mercapto-ethanol | Sigma | 02390-25ML | 5% |
| Bromophenol Blue | Amresco | 0449-50G | 0.5% | |
| Tris pH 6.8 | MP Biomedicals | 819620 | 0.312 M | |
| Glycerol | J.T.Baker | 2136-01 | 25% | |
| SDS | Sigma | L4509-500G | 10% | |
| Mdh2 Antibodies Diluent | BSA (in wash buffer) | MP Biomedicals | 160069 | 1% |
| Other antibodies Diluent | Skim milk powder (in wash buffer) | Tnuva-423 | 48703 | 5% |
| 10 X TBS-T Wash buffer | Tris 1 M pH 7.9 | MP Biomedicals | 819620 | 10% |
| Tween 20 | Sigma | P1379-500ML | 0.5% | |
| 0.5 M EDTA pH 8.0 | J.T.BAKER | 8993-01 | 0.5% | |
| NaCl | J.T.BAKER | 0277 | 1.5 M | |
| Blocking | SEA block (in wash buffer) | Thermo scientific | 37527 | 20% |
| Chemiluminescence | Amersham ECL western blot
reagents |
GE Healthcare | RPN2106 | Proprietary |
Controls
Several controls were used in this study. Loading controls for the total amount of protein were performed using anti-Histone H3 or anti-Actin antibodies. A Δ mdh2 strain, alongside Δ mdh1 and Δ mdh3 strains, were used to verify the specificity of the Mdh2 antibodies. An anti-mCherry antibody was used as a control for the bands detected in Mdh2-mCherry or Tef2-mCherry-Mdh3 (over expression) compared to the bands detected by the anti-Mdh2 antibodies. An anti-GFP antibody was used as a control for the bands detected in GFP-Mdh1, GFP-Mdh2 and GFP-Mdh3 with anti Mdh2 antibody peptide #3.
Results
The three GenScript antibodies detect Mdh2 from cells grown on oleic acid
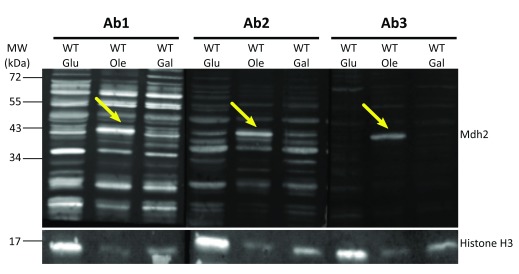
For first examination of the three antibodies, we grew control cells in three different carbon sources: glucose, galactose and oleic acid. Since transcription of S. cerevisiae Mdh2 is repressed by glucose 8, we hypothesized that the antibodies will not detect Mdh2 in extracts of cells grown in glucose, but will detect it in the galactose and oleic acid samples. We then performed protein extraction using the NaOH approach, and examined the three GenScript anti-Mdh2 antibodies.
Indeed, all three antibodies could not detect a specific band at the correct size of Mdh2 (41 KDa) in protein extractions from cells grown in glucose ( Figure 1, Glu). Although the general protein levels of the control strain grown in oleic acid were much lower than from the levels of controls grown in glucose, a prominent band at the correct size for Mdh2 was detected by all three antibodies. In galactose there was no prominent band at the correct size, although this could be due to the general low levels of protein in this condition, as can be seen by the Histone H3 loading control. Endogenous Mdh2 was also recognized in protein extractions from cells grown on other carbon sources such as EtOH (not shown). This suggests that the upregulation of Mdh2 in cells grown in oleic acid is due to the removal of the glucose repression and not because of an oleic acid-specific up-regulation.
Figure 1. Three new antibodies recognize Mdh2 in cells grown in oleic acid.
Western blot analysis was performed on protein extracts of control cells (BY4741) grown in glucose (Glu), oleic acid (Ole) or galactose (Gal) using antibodies 1–3 (Ab1, Ab2, Ab3). Native Mdh2 (41 KDa) is detected by all three anti-Mdh2 antibodies in protein extractions from cells grown in oleic acid as indicated by yellow arrows. Histone H3 was used as a loading control. In total we detected seven times a band at the size of Mdh2 when cells were grown on Oleate: One time using antibody1, three times using antibody 2 and three times using antibody 3.
Antibody 3 has high specificity for Mdh2
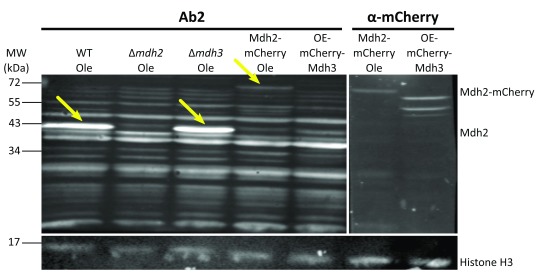
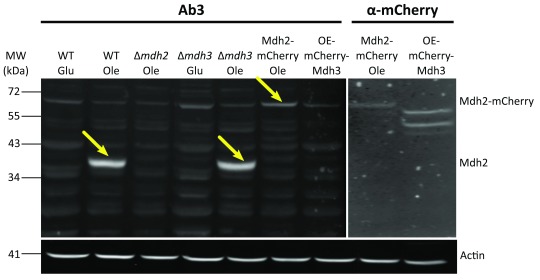
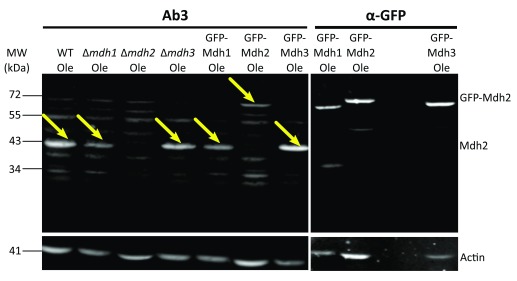
After the first comparison, we focused on antibodies 2 and 3 as they displayed the lowest background signal. We verified the affinity and specificity of the antibodies using several strains, in addition to the control strain: Δ mdh1, Δ mdh2, Δ mdh3, as well as strains that had a mCherry tag fused to either Mdh2 or Mdh3 or GFP tag fused to Mdh1, Mdh2 or Mdh3. Both antibody 2 and antibody 3 detected endogenous Mdh2 in wild type cells and in Δ mdh3 cells grown in oleic acid ( Figure 2 and Figure 3), but were specific to Mdh2 as they did not cross react in the Δ mdh2 strain. Another way to demonstrate specificity is to tag Mdh2 with mCherry, thus shifting only this specific isoform in size. Indeed, under these conditions both antibodies 2 and 3 detected a higher protein form only (Mdh2 tagged with mCherry = ∼70 kDa), though with lower affinity. Reciprocally, neither antibody detected over expressed mCherry-Mdh3 (expressed under a Tef2 promotor), although it was highly expressed as could be verified by an anti-mCherry antibody. In addition, antibody 3 detected endogenous Mdh2 in Δ mdh1 cells grown in oleic acid, as well as a bigger band size only in GFP-Mdh2 tagged cells (Mdh2 tagged with GFP = ∼68 kDa) ( Figure 4). Although both antibodies fit the requirement of specific identification of the yeast Mdh2, we have decided to use antibody 3, as it has a better specificity as seen from the lower background signal ( Figure 3).
Figure 2. Antibody 2 displays isoform specificity.
Western blot analysis was performed on protein extractions from different strains grown on oleic acid (Ole) or in glucose (only for OE-mCherry-Mdh3) using antibody 2 and anti-mCherry antibody. Antibody 2 recognizes the native Mdh2 in control cells and in Δ mdh3 cells grown in oleic acid, but does not recognize it in Δ mdh2 strain (second lane). The antibody also recognizes Mdh2 tagged with mCherry ( ∼70 kDa), but not the over-expressed and mCherry tagged Mdh3. The last one can be recognized in the control with the anti-mCherry. Histone H3 was used as a loading control. Yellow arrows indicate bands corresponding to Mdh2. (OE = over expression). We saw that antibody 2 specifically recognized Mdh2 and not Mdh3 in three experiments.
Figure 3. Antibody 3 has the best specificity to Mdh2.
Western blot analysis was done on protein extractions from different strains grown on oleic acid (Ole) or glucose (Glu) (OE-mCherry-Mdh3 was grown in glucose) using antibody 3 and anti-mCherry. Similarly to antibody 2, antibody 3 does not recognize Mdh2 in Δ mdh2 strain or in cells grown on glucose. This antibody has a good specificity to Mdh2, as seen by the low background signal. Actin was used as a loading control. Yellow arrows indicate bands corresponding to Mdh2. (OE = over expression). We saw that antibody 3 specifically recognizes Mdh2 in three experiments.
Figure 4. Antibody 3 recognizes Mdh2 in Δ mdh1 and Δ mdh3 cells extractions.
Western blot analysis was done on protein extractions from different strains grown on oleic acid (Ole) using antibody 3 and an anti-GFP antibody. As shown in Figure 3, this antibody does not recognize Mdh2 in Δ mdh2 strain or in cells grown on glucose but does recognize Mdh2 in Δ mdh3 cells grown on oleic acid. Here we show that antibody 3 also recognizes Mdh2 in Δ mdh1 cells grown on oleic acid. Actin was used as a loading control. Yellow arrows indicate bands corresponding to Mdh2. We saw that antibody 3 specifically recognizes Mdh2 and not Mdh3 in two experiments and that antibody 3 does not recognize Mdh1 in one experiment.
Copyright: © 2018 Gabay-Maskit S et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Conclusion
Three antibodies against endogenous S. cerevisiae Mdh2 were prepared by GenScript, using specific peptides of Mdh2 as the immunogens. The antibodies were checked by western blot and were shown to have the ability to detect endogenous Mdh2, when cells are grown under conditions in which Mdh2 is expressed. Although two of the three antibodies demonstrated the specificity qualities required from such an antibody, antibody 3 had the best specificity qualities, and the lowest background signal. We thus recommend antibody 3 as the best option to detect endogenous S. cerevisiae Mdh2. This antibody can be used in western blot, as shown in this manuscript, and should be tested for other uses (e.g. ELISA, FACS, IP).
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2018 Gabay-Maskit S et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
Dataset 1: Raw images of all western blots included in figures presented. 10.5256/f1000research.13396.d212121 9
Acknowledgements
We would like to thank Pierce Feng from GenScript for help in antigen design and for rapid turnover time from order to delivery.
Funding Statement
This work was supported by a European Union ERC CoG #646604 and an SFB 1190 from the DFG. MS is an incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 2 approved]
References
- 1. McAlister-Henn L, Steffan JS, Minard KI, et al. : Expression and function of a mislocalized form of peroxisomal malate dehydrogenase (MDH3) in yeast. J Biol Chem. 1995;270(36):21220–21225. 10.1074/jbc.270.36.21220 [DOI] [PubMed] [Google Scholar]
- 2. Small WC, McAlister-Henn L: Metabolic effects of altering redundant targeting signals for yeast mitochondrial malate dehydrogenase. Arch Biochem Biophys. 1997;344(1):53–60. 10.1006/abbi.1997.0179 [DOI] [PubMed] [Google Scholar]
- 3. Brachmann CB, Davies A, Cost GJ, et al. : Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. [DOI] [PubMed] [Google Scholar]
- 4. Gietz RD, Woods RA: Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- 5. Yofe I, Weill U, Meurer M, et al. : One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods. 2016;13(4):371–378. 10.1038/nmeth.3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yofe I, Schuldiner M: Primers-4-Yeast: a comprehensive web tool for planning primers for Saccharomyces cerevisiae. Yeast. 2014;31(2):77–80. 10.1002/yea.2998 [DOI] [PubMed] [Google Scholar]
- 7. Kushnirov VV: Rapid and reliable protein extraction from yeast. Yeast. 2000;16(9):857–860. [DOI] [PubMed] [Google Scholar]
- 8. Minard KI, McAlister-Henn L: Glucose-induced degradation of the MDH2 isozyme of malate dehydrogenase in yeast. J Biol Chem. 1992;267(24):17458–17464. [PubMed] [Google Scholar]
- 9. Gabay-Maskit S, Schuldiner M, Zalckvar E: Dataset 1 in: Validation of a yeast malate dehydrogenase 2 (Mdh2) antibody tested for use in western blots. F1000Research. 2018. 10.5256/f1000research.13396.d212121 [DOI] [PMC free article] [PubMed] [Google Scholar]