Abstract
To investigate the effect of microRNA (miR)-155 on colon cancer chemoresistance to cisplatine and its mechanism. Reverse transcription quantitative polymerase chain reaction was used to measure the levels of miR-155 and forkhead box O3 (FOXO3) in colon cancer specimens and cell lines. Overexpression of miR-155 and miR-155 inhibitor were transfected into colon cancer cell lines to investigate its role of chemoresistance to cisplatin in colon cancer. MTS assays were used to analyse cell viability in vitro. In vivo tumor formation assays were performed in C57BL/6 wild type and miR-155 knockout mice (miR-155-/-). A luciferase reporter assay was used to measure the translation of FOXO3. Additionally, the expression of FOXO3 was detected by western blot analysis. It was identified that miR-155 was markedly upregulated in colon cancer tissue and cell lines. Overexpression of miR-155 enhanced colon cancer cell chemoresistance to cisplatin in vitro and tumorigenesis in vivo. In addition, overexpression of miR-155 was associated with decreased levels of FOXO3, primarily through inhibiting the expression of FOXO3 to increase colon cancer resistanec to cisplatin. The present study demonstrated that miR-155 increased colon cancer drug resistance and decreased FOXO3 expression in vivo and in vitro. This may provide a novel method for the treatment of drug-resistant colon cancer.
Keywords: colon cancer, microRNA-155, forkhead box O3, chemoresistance, cisplatin
Introduction
Colorectal cancer (CRC) was the third leading cause of cancer-associated mortality worldwide in 2015, despite the advancements in the diagnosis and treatment (1). A total of ~50% of patients with colon cancer will develop liver metastasis and the 5-year survival rate for metastatic colon cancer is only 10–15% in the United States of America (2,3). Metastasis is a major cause of mortality in patients with colon cancer and is considered incurable due to a lack of effective therapy (4). To identify the novel melecules serving key roles in colon cancer, a number of studies focused on determining the microRNA (miRNA/miR) associated with this type of cancer (5–7).
Forkhead box O3 (FOXO3) is one of the most comprehensively characterized members of the FOXO family of transcription factors. It is a tumor suppressor, and is a potent transcriptional activator which triggers the expression of a program of genes involved in cell cycle arrest, DNA repair, hypoxia and apoptosis (8,9). It has been identified that the FOXO3 transcription factor serves as a target of anti-tumor drugs in different types of cancer, including breast cancer, chronic myeloid leukemia and colon cancer (10,11). It also affects the sensitivity of colon cancer cells to cisplatin (11). FOXO3 activity may be regulated by post-translational modifications, including phosphorylation (12), but the molecular mechanisms regulating the expression of the FOXO3 expression remain unclear.
miRs are naturally-occurring endogenous single-stranded RNA usually measuring 18–24 nucleotides in length (13). A total of >700 miRs have been identified in the human genome. A function of microRNA is the downregulation of target proteins expression by mRNA cleavage and decay or translational repression (14). miRs have been implicated in the regulation of gene expression essential for organ development, cellular differentiation, homeostasis and those involved in tumor occurance (15–17). miRNA expression is often dysregulated in cancer tissues and they may function either as tumor suppressors or oncogenes (oncomiRs) (18). miR-155 has been demonstrated to be an oncogenic miRNA in certain tumors, including liposarcoma and leukaemia (19–21). miR-155 transgenic mice have also been demonstrated to develop acute lymphocytic lymphoma or leukaemia (22). miR-155 is upregulated in gastric cancer and serves as a prognosis biomarker in these patients (23). Increased miR-155 expression is involved in poor prognosis in pancreatic and lung cancers (24,25), while in breast cancer miR-155 contributes to the progression of invasion (26,27). There are numerous target genes of miR-155, including inositol polyphosphate-5-phosphatase D and CCAAT/enhancer binding protein β, suppressor of cytokine signaling 1 and FOXO3 (28). However, the function of miR-155 in colon cancer is not understood, although it has been identified that miR-155 is upregulated in colon cancer (29,30). It remains unknown if miR-155 targets FOXO3 to effect colon cancer cell viability.
In the present study, clinical tissues from patients with colon cancer were collected and miR-155 and FOXO3 expression levels were detected. It was identified that the expression levels of miR-155 and FOXO3 were increased and decreased, respectively, in colon cancer tissues and human cell lines. miR-155 was transfected into colon carcinoma HT29 and SW620 cell lines, and it was determined that FOXO3 expression was decreased at the protein level in a dose-dependent manner, suggesting that they are negatively associated. The sensitivity of colon carcinoma cells to chemotherapy drugs cisplatin and paclitaxel was detected, and it was demonstrated that miR-155 increased chemoresistance. The present study suggests a novel pathway and target for colon cancer therapy.
Materials and methods
Cell lines and reagents
Human colon carcinoma HT29 and SW620 cell lines were purchased from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) and were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Zhejiang Tianhang Biotechnology Co. Ltd, China), 100 units/ml penicillin and 100 g/ml streptomycin at 37°C in a humidified chamber with 5% CO2. Samples from patients with colon cancer and para-carcinoma tissues were collected by radical colon resection from the People's Hospital of Rizhao City (Rizhao, China). The present study was approved by the Ethical Committee of the People's Hospital of Rizhao (Rizhao, China), all patients were informed and written informed consent was obtained. Cisplatin was purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). miR-155-5p mimic (C-3006 47-05-0005) and scrambled control oligonucleotide (CN-001000-01-05; sequence, 5′-CCCUAUCACGAUUAGCAUUAAUU-3′) were purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). The CellTiter 96® Aqueous Non-Radioactive Cell Proliferation Assay kit was purchased from Promega Corporation (Madison, WI, USA). All the PCR primers were synthetized by Invitrogen (Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated using TRIzol® (Life Technologies; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For mRNA detection, cDNA for PCR was obtained by RT-PCR following the manufacturer's protocol of a Toyobo SYBR-Green RT-PCR Master Mix kit (Toyobo Life Science, Osaka, Japan). cDNA was subjected to qPCR using the SYBR-Green PCR reagents kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) on a ABI StepOnePlus (ABI) instrument (Thermo Fisher Scientific, Inc.). For miR-155 detection, polyAs were added into RNA using Escherichia coli polyA polymerase (Fermentas; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA). A total of 2 µg tailed total RNA was reverse transcribed with miR reverse transcriptase primer (1 µM; Shanghai Genepharma Co., Ltd., Shanghai, China). qPCR was performed with miR-155 forward (F) and reverse (R) primers, and the thermocycler conditions were as follows: 95°C for 10 min; then 41 cycles at 95°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec. All primers used for qPCR analysis were synthesized by Invitrogen (Thermo Fisher Scientific, Inc.), as follows: FoxO3 F, AGTGGATGGTGCGCTGTGT; FoxO3 R, CTGTGCAGGGACAGGTTGT (31); GAPDH F, TGTGTCCGTCGTGGATCTGA; GAPDH R, TTGCTGTTGAAGTCGCAGGAG; miR reverse transcriptase primer, GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTTTTTTTTN; miR-155 F, TTAATGCTAATCGTGATAGGGGT; miR-155 R, GCTGTCAACGATACGCTACGTAACG (20); U6 F, CTCGCTTCGGCAGCACA; and U6 R, AACGCTTCACGAATTTGCGT. The relative amount of target mRNA was determined using the comparative threshold (Cq) method by normalizing target mRNA Cq values to those of GAPDH or U6 (32).
Plasmid construction and stable/transient transfection of miR-155
To construct the human FOXO3 recombined plasmid, the FOXO3 gene (NCBI Reference Sequence: NM_001455.3) was cloned into pCMV-tag2a vector (Agilent Technologies; Thermo Fisher Scientific, Inc.). A human genomic fragment of 65 bp containing the miR-155 precursor DNA sequence (NCBI Reference Sequence: NR_030784.1) was cloned into the pcDNA3.1(−)-myc-his vector (Invitrogen; Thermo Fisher Scientific, Inc.). The recombinant plasmid was pcDNA3.1-miR-155. The primers for construction were as follows; underlined nucleotides represent BamH I and HindIII sites: miR-155 P1, GATCCCTGTTAATGCTAATCGTGATAGGGGTTTTTGCCTCCAACTGACTCCTACATATTAGCATTAACAGA; miR-155 P2, AGCTTCTGTTAATGCTAATATGTAGGAGTCAGTTGGAGGCAAAAACCCCTATCACGATTAGCATTAACAGG; FOXO3-P1, ATTAGGATCCATGGCAGAGGCACCGGCTTC; and FOXO3-P2, GCAAAAGCTTTCCTGGCACCCAGCTCTGAG. To generate a cell line stably expressing miR-155, HT29 and SW620 cells were transfected with 200 ng pcDNA3.1-miR-155 using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Following 48 h transfection, following 800 mg/ml G418 selection, the single clone that over-expressed miR-155 was identified. For miR155 transient transfection, miR-155 mimics (Invitrogen; Thermo Fisher Scientific, Inc.) were used to transfect the 2 cell lines.
Luciferase assays
The online database search TargetScanHuman 6.2 (http://www.targetscan.org/), microRNA (http://www.microrna.org) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php) were used to predict a potential target of miR-155 (Date of access, January 2016). The wild type (WT) or mutant 3′-UTR of FOXO3 containing the putative miR-155 binding sites was synthesized and inserted into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation) to generate the recombinant constructs, pmirGLO-FOXO3-3′UTR-WT and pmirGLO-FOXO3-3′UTR-M, respectively. miR-155 mimics or miR-inhibitor using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and incubated for 48 h. The primers for 3′UTR WT and mutant were as follow; FOXO3-3′UTR-WT P1, AAACTCTTTGCATAAAAAGCATTAGGCATAT; FOXO3-3′UTR-WT P2, CTAGATATGCCTAATGCTTTTTATGCAAAGAGTTT; FOXO3-3′UTR-M P1, AAACTCTTTGCATAAAAACGATAAGGCATAT; and FOXO3-3′UTR-M P2, CTAGATATGCCTUAUGGTTTTTATGCAAAGAGTTT. Underlined nucleotides represent PmeI and XbalI sites, respectively. Italicized nucleotides represent the miR-155 target sequence. Bold italicized nucleotides represent the sites of mutation. Cells were harvested 48 h following transfection for the measurement of luciferase activity with a Dual-Luciferase Reporter Assay system (Promega Corporation) according to manufacturer's protocol. Renilla luciferase activity was applied as the method of normalization for firefly luciferase activity.
Cell proliferation assays
The effect of miR-155-5p on cisplatin chemotherapy sensitivity of colon cancer cell lines was detected using Cell Titer 96® Aqueous Non-Radioactive Cell Proliferation Assay kit (Promega Corporation) according to manufacturer's protocol. Cells grown in regular media were plated on 96-well plates (5,000 cells/well) were treated with cisplatin (0, 10, 50, 100 and 200 µM) at 37°C in a 5% CO2 humidified atmosphere for 48 h. Subsequently, 20 µl combined MTS/phenazine methosulfate solution was pipetted into each well and incubated for 4 h at 37°C. A total of 25 µl 10% SDS (Boster Biological Thecnology, Pleasanton, CA, USA) was added to each well to stop the reaction. Absorbance was detected at 490 nm using a 96-well plate reader.
Western blot analysis
Tissues from the CT-26 tumors (2×106 cells/mouse; purchased from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences, Shanghai, China) were collected and lysed cells by lysis buffer (Tris 20 mM, NaCl 150 mM, 1% Triton X-100 and 1% cocktail protease inhibitors). All of the protein extraction process was carried out according to the product specifiation. The BCA protein assay kit (Beyotime Institute of Biotechnology, Haimen, China) was applied to detected the protein concentration. Then total protein (30 µg) were separated by electrophoresis on a 12% SDS-PAGE (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and transferred onto nitrocellulose membranes (Merck KGaA). The membranes were blocked in 5 % non-fat milk solution for 2–4 h at 4°C, and then washed twice with TBST solution (0.1% Tween-20). Membranes were incubated overnight at 4°C with the following antibodies. FOXO3 polyclonal antibodies (1:1,000; Abcam, Cambridge, UK; cat. no. ab23683) or β-actin (1:3,000, cat. no. BM0626, Specificity: monoclonal mouse; Boster Biological Technology, USA) monocolonal antibodies were used as primary antibodies, followed by goat anti-rabbit (cat. no. A21020; dilution: 1:1,000; AmyJet Scientific, Wuhan, China) or goat anti-mouse secondary antibodies (cat. no. A21010; dilution: 1:1,000) conjugated with horseradish peroxidase (ProteinTech Groups, Inc., Chicago, IL, USA) incubated for 2 h at 4°C. The protein analysis was performed using Millipore ECL Western Blotting Substrate on a UVP ChemiDoc-It imaging system (Chemidoc-IT 510; Visionworks LS, UVP, LLC, USA).
Caspase 3 activity detection
Colon cancer cells treated with or without cisplatin were were seeded at a density of 2×105 cells/well in a 6-well plate with 2 ml culture medium. Following 24 h incubation, cells were treated with 200 µM cisplatin to induce apoptosis. Casapse-3 activity was measured using a Caspase 3 Fluorometric activity assay kit (cat no. C1115; Beyotime Institute of Biotechnology) following the manufacturer's protocol.
Flow cytometry
Colon cancer cells treated with or without cisplatin were harvested by 500 µl 0.25% trypsin and then centrifugation 5 min at room temperature (241.5 × g). A total of 500 µl binding buffer (including precooled 70% ethanol and 0.5 mmol/l EDTA) was added to each tube and incubated overnight at 4°C, then intracellularly stained with phycoerythrin-anti human Ki67 mAb (cat no. 350503; dilution, 1:20; BioLegend, Inc., San Diego, CA, USA) in staining buffer (PBS containing 0.5% BSA) for 30 min at room temperature in the dark. Samples were washed twice with PBS, and analyzed by flow cytometry using a BD C6 auri flow cytometry and Expo 32-ADC v. 1.2B software (both BD Biosciences, Franklin Lakes, NJ, USA).
Animal model
Pathogen-free 10 weeks old male WT C57BL/6 mice (mean weight:24.26±3.04 g, n=7) and pathogen-free 10-week-old male miR-155 knockout mice (miR-155-/-) (mean weight, 24.75±2.93 g; n=6) were obtained from Jackson Laboratory (Ben Harbour, ME, USA). The animals were housed under standard conditions (18–29°C, 0.03%) in a 12 h light/dark cycle with access to water and food (ad libitum). The mouse colon cancer CT26 cell line (2×106 cells per mouse) was inoculated into the backs of the mice by hypodermic inoculation (matrix glue was administered to the mice in order to prevent the spread of the tumor, and multiple tumors were not observed). A total of 2 days following tumor cell inoculation, mice were treated with cisplatin (300 µM) every 2 days for 10 days by intraperitoneal injection. The tumor sizes were measured every 3 days using calipers from 1st day of drug treatment to the 30th day, and tumor volume was calculated as: Tumor volume=[length (a) × width (b)2)/2 (33). The data are presented as the mean volume ± standard deviation (SD). The Ethical Committee of the People's Hospital of Rizhao approved all animal experimental procedures.
Statistical analysis
SPSS software (version 16; SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. All data were calculated as the mean ± standard deviation. A one-way analysis of variance and Student Newman-Keuls post-hoc test were used to analyze the results between treated and control groups, and an unpaired two-tailed Student's t-test was used to compare two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-155 is negatively associated with FOXO3 in colon cancer tissues
To investigate miR-155 expression in colon cancer and para-carcinoma tissues, samples (colon cancer, n=25; para-carcinoma, n=20) from Rizhao People's Hospital were collected. Total RNA of tissues was extracted, and qPCR was performed. The results indicated that miR-155 expression was increased in colon cancer tissues compared with para-carcinoma tissues, while FOXO3 expression was decreased in colon cancer tissues compared with para-carcinoma tissues (Fig. 1A and B). The plasmid pcDNA3.1-miR-155 was transfected into colon cancer HT29 and SW620 cell lines, and the mRNA expression of FOXO3 was also detected. FOXO3 mRNA levels decreased following miR-155 enhancement compared with the control group (Fig. 1C-E).
Figure 1.
miR-155 and FOXO3 expression in colon cancer and para-carcinoma tissue. (A) Expression of miR-155 in colon cancer and para-carcinoma tissue. (B) Expression of FOXO3 in colon cancer and para-carcinoma tissue. (C) Expression of miR-155 in HT-29 cells following transfection. (D) Expression of miR-155 in SW620 cells following transfection. (E) Expression of FOXO3 in HT-29 and SW620 cells following pcDNA3.1-miR-155 transfection. miR, microRNA; FOXO3, forkhead box O3.
FOXO3 is a direct target of miR-155 in colon cancer cells
Based on an online database search for targets of miRNAs that were identified in previous studies (34,35), it was suggested that FOXO3 is a potential target of miR-155. Then, whether miR-155 mediated FOXO3 was determined. pmirGLO-UTR WT or M plasmids were constructed and transfected into HT-29 cells. It was demonstrated that reduced firefly luciferase expression indicated the binding of miR-155 to the cloned miRNA target sequence, and the mutant UTR group was not altered (Fig. 2A). This suggested that miR-155 may bind to position 1497–1503 of FOXO3 3′UTR and FOXO3 be directly targeted by miR-155 in human colon cancer cells (Fig. 2B).
Figure 2.
Validation of FOXO3 as a direct target of miR-155 in HT-29 cells. (A) Results of dual-luciferase assay in HT-29 cells transfected with pmirGLO-FOXO3-3′ UTR-WT or pmirGLO-FOXO3-3′UTR M reporter together with miR-155 mimics or miR control. (B) Predicted miR155-binding sites in the 3′UTR of FOXO3. Data were expressed in mean ± standard devation. miR, microRNA; FOXO3, forkhead box O3; 3′UTR, 3′untranslated region; WT, wild type; M, mutated.
miR-155 decreases chemosensitivity to cisplatin in colon cancer cells
Different concentrations of cisplatin (0, 10, 50, 100 and 200 µM) were used to treat HT29 and SW620 cells for 48 h. The viability of cells was detected using a Non-Radioactive Cell Proliferation Assay kit. HT29 cells demonstrated increased sensitivity compared with HT29-miR-155 cells, which stabley expressed high levels of miR-155 (Fig. 3A). The same result was exhibited in SW620 cells (Fig. 3B). Antigen Ki-67 is a nuclear protein that is associated with cellular proliferation and is a cellular marker for proliferation (36). Ki67 was detected in HT29, SW620, HT29-miR-155 and SW620-miR-155 cells by flow cytometry. The results indicated that ki67 expression was increased in miR-155 high-expressed cells compared control cells (Fig. 3C, D and E). These results suggests that the miR-155-mediated decrease of chemosensitivity to cisplatin in colon cancer cells may occur through targeting FOXO3.
Figure 3.
Effect of miR-155 on colon cancer cell sensitivity to the chemotherapy drug cisplatin. (A) miR-155 increased the viability of HT-29 cells following cisplatin treatment. (B) miR-155 increased the viability of SW620 cells following cisplatin treatment. (C) Ki67 was detected in HT-29 and HT-29-miR-155 cells treated with cisplatin by flow cytometry. (D) Ki67 was detected in SW620- and SW620-miR-155 cells treated with cisplatin by flow cytometry. (E) Ki67 percentages in colon cancer cells. miR, microRNA; PE, phycoerythrin; Iso, control group.
miR-155 blocks the caspase 3 activity induced by cisplatin
Cisplatin may induce apoptosis in HT-29 and SW620 cells (37–39). miR-155 mimics were transfected into these two cell lines to obtain HT-29-miR-155 and SW620-miR-155 cells. Colon cancer cells were treated with cisplatin for 48 h and harvested for caspase 3 activity detection. High miR-155 expression decreased the caspase 3 activity induced by cisplatin in HT-29 and SW620 cells (Fig. 4A and B).
Figure 4.
Caspase 3 activity detection in colon cancer cells. (A) Caspase 3 activity in HT-29 and HT-29-miR-155 cells treated with cisplatin. (B) Caspase 3 activity in SW620 and SW620-miR-155 cells treated with cisplatin. (C) Caspase 3 activity in HT-29 and HT-29-miR-155 cells treated with cisplatin and FOXO3. (D) Caspase 3 activity in SW620 and SW620-miR-155 treated with cisplatin and FOXO3. NC, negative control; miR, microRNA; FOXO3, forkhead box O3.
FOXO transcription factors have been indicated to regulate apoptosis and cell cycle-associated genes (40–42). When FOXO3 and miR-155 were co-transfected into colon cancer cells, it was demonstrated that cisplatin efficiently induced caspase 3 activity (Fig. 4C and D). These data suggested that miR-155 decreased colon cancer sensitivity to cisplatin by targeting FOXO3.
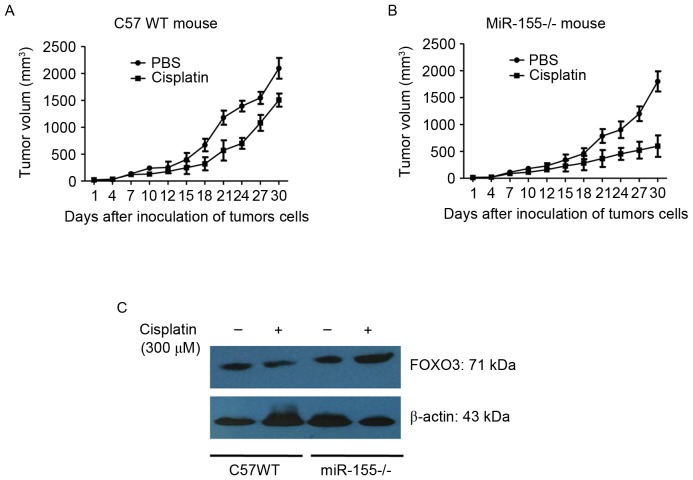
miR-155 promotes colon cancer growth in mice
C57BL/6 (WT) and mir-155 knockout mice (miR-155-/-) were divided into 4 groups at random. CT26 cells were inoculated into the backs of the mice. The tumor-bearing mice were treated with 300 µM cisplatin for 10 days. Tumor length (a) and width (b) were measured using vernier calipers. Tumor volume was calculated using the formula: V (mm3)=(a × b2)/2.
The results showed that: the tumor weight was (0.7184±0.1547) g and (0.6728±0.1415) g in the control group of C57 WT mouse and miR-155-/- mouse, respectively, and the tumor weight was (0.5649±0.1252) g and (0.4937±0.1096) g in the cisplatin group of C57 WT mouse and miR-155-/- mouse, respectively. It was observed that tumor growth was significantly reduced in the miR-155-/- mice compared with the WT mice (Fig. 5A and B). It was also identified that FOXO3 expression in tumor tissues of miR-155-/- was increased compared with that of the WT mice (Fig. 5C). The data additionally suggested that miR-155 enhanced tumor growth and chemoresistance by targeting the FOXO3 gene.
Figure 5.
Tumor volume in WT and miR-155-/- mice with or without cisplatin treatment. (A) Volume of CT26 in WT mice treated with PBS or cisplatin. (B) Volume of CT26 in miR-155-/- mice treated with PBS or cisplatin. (C) FOXO3 expression in tumor tissues in WT and miR-155-/- mice. miR, microRNA; WT, wild type; FOXO3, forkhead box O3.
Discussion
Although the mechanism by miR-155 regulation is not well understood, it has been suggested that the oncogenic microRNA miR-155 is enhanced in numerous types of cancer (43–46). However, miR-155 expression in colon cancer has not been identified. In the present study, to the best of our knowledge, the effect of miR-155 targeting FOXO3 gene in colon cancer cells was demonstrated for the first time. Tissues from patients with colon cancer were collected for miR-155 detection. The results indicated that miR-155 was increased in the colon cancer tissues, but not in para-carcinoma tissues. This result was in accordance with similar previous studies that have detected miR-155 expression in colon cancer (29,30).
As a prominent oncomiR, miR-155 may target the mismatch repair genes MutL Homolog 1, MutS Homolog (MSH) 2 and MSH6 and contribute to the microsatellite instability (MSI) phenotype in colon cancer (47). It downregulates B-cell lymphoma (Bcl)-6 and causes V-Myc avian myelocytomatosis viral oncogene homolog and Cyclin D1 upregulation, which promotes cell proliferation (48). In renal cancer cells, miR-155 may function as an oncogene by targeting BTB domain and CNC homolog (49). In the present study, miR-155 targets were predicted using TargetScanHuman 6.2, and it was identified that FOXO3 3′UTR was able to bind with miR-155. This prediction was associated with the results revealed by Zhang et al (50). In the present study, FOXO3 expression in colon cancer was negatively associated with miR-155 expression. To additionally demonstrate the interaction between FOXO3 and miR-155, a recombinant plasmid pmirGLO-FOXO3-3′UTR-WT expressing the FOXO3 3′UTR wild type and a pmirGLO-FOXO3-3′UTR-M plasmid with the FOXO3 3′UTR with a small number of mutant bases, were constructed. Then, miR-155 mimics and inhibitors were co-transfected into HT-29 cells and luciferase reporter assays demonstrated that miR-155 directly bound to the FOXO3 3′UTR.
The functions of miR-155 in various types of cancer are different: It has been suggested that miR-155 is associated with the development of liver, leukemia, breast, lung and stomach tumors (51–53). Conversely, miR-155 may serve to prevent cancer in transgenic mice by promoting proper immune function. To clarify the function of miR-155 in colon cancer, the effect of miR-155 on chemoresistance was evaluated in the present study. Cisplatin was used to treat HT-29, SW620, HT-29-miR-155 and SW620-miR-155 cells. miR-155 increased the resistance of colon cancer cells to cisplatin. This result is consistent with results from Yu et al (54). However, the novel observation of the present study was that in colon cancer cells, miR-155 increased chemoresistance by targeting FOXO3. In the miR-155-/- mice, CT26 cell growth was inhibited, and these cells demonstrated an increased sensitivity to cisplatin compared with the WT mice.
To conclude, the present study identified that miR-155 may promote colon cancer growth and increase colon cancer cells chemoresistance to cisplatin by directly targeting FOXO3. This suggests a novel pathway for the treatment and cure of colon cancer in the future.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, et al. Modulation of mismatch repair and genomic stability by miR-155; Proc Natl Acad Sci USA; 2010; pp. 6982–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geng L, Chaudhuri A, Talmon G, Wisecarver JL, Are C., Brattain M, Wang J. MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene. 2014;33:5332–5340. doi: 10.1038/onc.2013.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eker B, Ozaslan E, Karaca H, Berk V, Bozkurt O, Inanc M, Duran AO, Ozkan M. Factors affecting prognosis in metastatic colorectal cancer patients. Asian Pac J Cancer Prev. 2015;16:3015–3021. doi: 10.7314/APJCP.2015.16.7.3015. [DOI] [PubMed] [Google Scholar]
- 5.Ozawa T, Kandimalla R, Gao F, Nozawa H, Hata K, Nagata H, Okada S, Izumi D, Baba H, Fleshman J, et al. A microrna signature associated with metastasis of T1 colorectal tumors to lymph nodes. Gastroenterology. 2017 Nov 30; doi: 10.1053/j.gastro.2017.11.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bu P, Wang L, Chen KY, Srinivasan T, Murthy PK, Tung KL, Varanko AK, Chen HJ, Ai Y, King S, et al. A miR-34a-numb feedforward loop triggered by inflammation regulates asymmetric stem cell division in intestine and colon cancer. Cell Stem Cell. 2016;18:189–202. doi: 10.1016/j.stem.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654–665. doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Cai M, Gong Z, Zhang B, Li Y, Guan L, Hou X, Li Q, Liu G, Xue Z, et al. Geminin facilitates FoxO3 deacetylation to promote breast cancer cell metastasis. J Clin Invest. 2017;127:2159–2175. doi: 10.1172/JCI90077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan L, Zhang L, Gong Z, Hou X, Xu Y, Feng X, Wang H, You H. FoxO3 inactivation promotes human cholangiocarcinoma tumorigenesis and chemoresistance through Keap1-Nrf2 signaling. Hepatology. 2016;63:1914–1927. doi: 10.1002/hep.28496. [DOI] [PubMed] [Google Scholar]
- 10.Essafi A, Fernández de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, Medema RH, Lam EW. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–2329. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- 11.Fernández de Mattos S, Villalonga P, Clardy J, Lam EW. FOXO 3a mediates the cytotoxic effects of cisplatin in colon cancer cells. Mol Cancer Ther. 2008;7:3237–3246. doi: 10.1158/1535-7163.MCT-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai KL, Sun YJ, Huang CY, Yang JY, Hung MC, Hsiao CD. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 2007;35:6984–6994. doi: 10.1093/nar/gkm703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond SM. RNAi, microRNAs, and human disease. Cancer Chemother Pharmacol. 2006;58(Suppl 1):S63–S68. doi: 10.1007/s00280-006-0318-2. [DOI] [PubMed] [Google Scholar]
- 14.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 15.Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705–713. doi: 10.1016/j.biopha.2016.09.099. [DOI] [PubMed] [Google Scholar]
- 16.Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: Pathways and clinical implications. Dis Model Mech. 2017;10:197–214. doi: 10.1242/dmm.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagaraju GP, Madanraj AS, Aliya S, Rajitha B, Alese OB, Kariali E, Alam A, El-Rayes BF. MicroRNAs as biomarkers and prospective therapeutic targets in colon and pancreatic cancers. Tumour Biol. 2016;37:97–104. doi: 10.1007/s13277-015-4346-6. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Zhang P, Bill K, Liu J, Young E, Peng T, Bolshakov S, Hoffman A, Song Y, Demicco EG, Terrada DL, et al. miR-155 is a liposarcoma oncogene that targets casein kinase-1α and enhancees β-catenin signaling. Cancer Res. 2012;72:1751–1762. doi: 10.1158/0008-5472.CAN-11-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson ED, Wasylyk C, Ye T, Jung AC, Wasylyk B. The oncogenic MicroRNA Hsa-miR-155-5p targets the transcription factor ELK3 and links it to the hypoxia response. PLoS One. 2014;9:e113050. doi: 10.1371/journal.pone.0113050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice; Proc Natl Acad Sci USA; 2006; pp. 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Tang Y, Wu JH, Liu FH. Role of microRNAs in diagnosis and treatment of the pathogenesis of gastric cancer. Int J Clin Exp Med. 2014;7:5947–5957. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Saito Y, Suzuki H, Matsuura M, Sato A, Kasai Y, Yamada K, Saito H, Hibi T. MicroRNAs in hepatobiliary and pancreatic cancers. Front Genet. 2011;2:66. doi: 10.3389/fgene.2011.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 26.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neilsen PM, Noll JE, Mattiske S, Bracken CP, Gregory PA, Schulz RB, Lim SP, Kumar R, Suetani RJ, Goodall GJ, Callen DF. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene. 2013;32:2992–3000. doi: 10.1038/onc.2012.305. [DOI] [PubMed] [Google Scholar]
- 28.Willimott S, Wagner SD. miR-125b and miR-155 contribute to BCL2 repression and proliferation in response to CD40 ligand (CD154) in human leukemic B-cells. J Biol Chem. 2012;287:2608–2617. doi: 10.1074/jbc.M111.285718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Zhang P, Li Y, Liu G, Zhou B, Zhan L, Zhou Z, Sun X. The quantitative analysis by stem-loop real-time PCR revealed the microRNA-34a, microRNA-155 and microRNA-200c overexpression in human colorectal cancer. Med Oncol. 2012;29:3113–3118. doi: 10.1007/s12032-012-0241-9. [DOI] [PubMed] [Google Scholar]
- 30.Kara M, Yumrutas O, Ozcan O, Celik OI, Bozgeyik E, Bozgeyik I, Tasdemir S. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567:81–86. doi: 10.1016/j.gene.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 31.Renault VM, Thekkat PU, Hoang KL, White JL, Brady CA, Kenzelmann Broz D, Venturelli OS, Johnson TM, Oskoui PR, Xuan Z, et al. The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor. Oncogene. 2011;30:3207–3221. doi: 10.1038/onc.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Naito S, von Eschenbach AC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46:4109–4115. [PubMed] [Google Scholar]
- 34.Min M, Peng L, Yang Y, Guo M, Wang W, Sun G. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a. Inflamm Bowel Dis. 2014;20:652–659. doi: 10.1097/MIB.0000000000000009. [DOI] [PubMed] [Google Scholar]
- 35.Ling N, Gu J, Lei Z, Li M, Zhao J, Zhang HT, Li X. microRNA-155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncol Rep. 2013;30:2111–2118. doi: 10.3892/or.2013.2685. [DOI] [PubMed] [Google Scholar]
- 36.Scholzen T, Gerdes J. The Ki-67 protein: From the known and the unknown. J Cell Physiol. 2010;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 37.Hu XJ, Xie MY, Kluxen FM, Diel P. Genistein modulates the anti-tumor activity of cisplatin in MCF-7 breast and HT-29 colon cancer cells. Arch Toxicol. 2014;88:625–635. doi: 10.1007/s00204-013-1184-4. [DOI] [PubMed] [Google Scholar]
- 38.Serova M, Calvo F, Lokiec F, Koeppel F, Poindessous V, Larsen AK, Laar ES, Waters SJ, Cvitkovic E, Raymond E. Characterizations of irofulven cytotoxicity in combination with cisplatin and oxaliplatin in human colon, breast, and ovarian cancer cells. Cancer Chemother Pharmacol. 2006;57:491–499. doi: 10.1007/s00280-005-0063-y. [DOI] [PubMed] [Google Scholar]
- 39.Lacour S, Micheau O, Hammann A, Drouineaud V, Tschopp J, Solary E, Dimanche-Boitrel MT. Chemotherapy enhances TNF-related apoptosis-inducing ligand DISC assembly in HT29 human colon cancer cells. Oncogene. 2003;22:1807–1816. doi: 10.1038/sj.onc.1206127. [DOI] [PubMed] [Google Scholar]
- 40.Burgering BM. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–2262. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 41.Arden KC. FoxO: Linking new signaling pathways. Mol Cell. 2004;14:416–418. doi: 10.1016/S1097-2765(04)00213-8. [DOI] [PubMed] [Google Scholar]
- 42.Chen Q, Ganapathy S, Singh KP, Shankar S, Srivastava RK. Resveratrol induces growth arrest and apoptosis through activation of FOXO transcription factors in prostate cancer cells. PLoS One. 2010;5:e15288. doi: 10.1371/journal.pone.0015288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lympho cytic leukemias; Proc Natl Acad Sci USA; 2004; pp. 11755–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tili E, Croce C, Michaille J. miR-155: On the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 45.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 46.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets; Proc Natl Acad Sci USA; 2006; pp. 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, et al. Modulation of mismatch repair and genomic stability by miR-155; Proc Natl Acad Sci USA; 2010; pp. 6982–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandhu SK, Volinia S, Costinean S, Galasso M, Neinast R, Santhanam R, Parthun MR, Perrotti D, Marcucci G, Garzon R, Croce CM. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcrip tional activity of B-cell lymphoma 6 (BCL6) in the Emu-miR-155 transgenic mouse model; Proc Nat Acad Sci USA; 2012; pp. 20047–20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S, Chen T, Zhong Z, Wang Y, Li Y, Zhao X. microRNA-155 silencing inhibits proliferation and migration and induces apoptosis by upregulating BACH1 in renal cancer cells. Mol Med Rep. 2012;5:949–954. doi: 10.3892/mmr.2012.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang P, Bill K, Liu J, Young E, Peng T, Bolshakov S, Hoffman A, Song Y, Demicco EG, Terrada DL, et al. miR-155 is a liposarcoma oncogene that targets casein kinase-1α and enhances b-catenin signaling. Cancer Res. 2012;72:1751–1762. doi: 10.1158/0008-5472.CAN-11-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lv L, An X, Li H, Ma L. Effect of miR-155 knockdown on the reversal of doxorubicin resistance in human lung cancer A549/dox cells. Oncol Lett. 2016;11:1161–1166. doi: 10.3892/ol.2015.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: An ancient regulator of the immune system. Immunol Rev. 2013;253:146–157. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 53.Higgs G, Slack F. The multiple roles of microRNA-155 in onco genesis. J Clin Bioinforma. 2013;3:17. doi: 10.1186/2043-9113-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu DD, Lv MM, Chen WX, Zhong SL, Zhang XH, Chen L, Ma TF, Tang JH, Zhao JH. Role of miR-155 in drug resistance of breast cancer. Tumour Biol. 2015;36:1395–1401. doi: 10.1007/s13277-015-3263-z. [DOI] [PubMed] [Google Scholar]