Abstract
Renal cell carcinoma (RCC) is insensitive to conventional chemotherapy and radiotherapy, and its prognosis remains poor. Targeted therapy is a novel technique for the treatment of RCC. The present study aimed to determine the precise effects of microRNA-183 (miR-183) on RCC. Reverse transcription-quantitative polymerase chain reaction analysis was conducted in order to analyze the expression level of miR-183 in RCC cell lines. Cell transfection, viability assays and invasion assays were also performed, followed by western blot analysis and a luciferase reporter assay, in order to examine the effects of miR-183. The expression of miR-183 was upregulated in all four RCC cell lines compared with the immortalized normal proximal tubule epithelial HK-2 cell line. The cell viability and invasion assays demonstrated that decreased expression of miR-183 suppressed RCC cell proliferation and invasion in vitro. Furthermore, the present study identified that Dickkopf-related protein-3 (DKK-3) was a direct target of miR-183 in RCC. The results demonstrated that miR-183 functioned as an oncogene by downregulating DKK-3 expression and that it provided a potential diagnostic, prognostic and therapeutic target for RCC.
Keywords: microRNA-183, renal cell carcinoma, Dickkopf-related protein 3, proliferation, metastasis
Introduction
Renal cell carcinoma (RCC) accounts for ~85% of all kidney tumors, and is one of the most common types of cancer of the urinary system (1,2). RCC has several subtypes, including clear cell RCC (ccRCC), papillary RCC, and chromophobe RCC. ccRCC, which is associated with a high risk of metastasis, is the most common subtype of RCC, accounting for 75–80% of all RCC cases (3). RCC is not sensitive to conventional chemotherapy and radiotherapy, and its prognosis remains poor (4–6). Targeted therapy is a novel method for the treatment of RCC (7). However, the detailed molecular mechanism of RCC progression remains poorly understood; therefore, the identification of novel therapeutic targets has an important significance.
MicroRNAs (miRNAs) are a type of endogenous non-coding RNA, which are able to effectively regulate the expression of target genes by binding to the 3′-untranslated region (3′-UTR) or by hybridizing in the coding sequence. They serve important roles in the majority of biological processes, including cell development, proliferation, differentiation and apoptosis (8–10). An increasing volume of evidence has demonstrated that the abnormal expression of miRNAs is a common feature of cancer, and that they serve an important role in the occurrence and development of tumors (11). The miRNA-183 (miR-183) cluster is located on chromosome 7q32, and consists of miR-96, miR-182 and miR-183 (12). Previous studies have revealed that miR-183 may function as an oncogene in the majority of cancer types, including, pancreatic, lung, gastric, breast and colorectal cancer (CRC) (13–17). However, it may also function as a tumor suppressor in certain types of cancer, including cervical cancer (18). Therefore, the role of miR-183 may depend on the type of cancer it is identified in.
The clinical significance of miR-183 has previously been investigated in RCC. For example, Zhang et al (19) identified that the serum expression level of miR-183 was significantly higher in patients with RCC than that in healthy volunteers. Furthermore, its expression level was significantly associated with sensitivity to natural killer cell therapy (19). In order to further investigate the role of miR-183 in the occurrence and development of RCC, TargetScan analysis was conducted to predict the target gene of miR-183, and results demonstrated that Dickkopf-related protein-3 (DKK-3) was a target gene of miR-183. Previously, Ueno et al (20) previously identified that miR-183 was an oncogene targeting DKK-3 in prostate cancer; however, whether there is an association between miR-183 and DKK-3 in RCC remains unknown. The present study focused on the effects of miR-183 on RCC cells in addition to the identification of its direct target gene, DKK-3, in order to illuminate the molecular mechanisms of miR-183 in the initiation and progression of RCC.
Materials and methods
Cell lines and cell culture
The normal human proximal tubule epithelial HK-2 cell line and four human RCC cell lines (ACHN, 786-O, Caki-1 and Caki-2) were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The HK-2 cells were cultured in keratinocyte serum-free medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The four RCC cell lines were cultured in RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). The culture medium was supplemented with 50 µg/ml streptomycin and 50 U/ml penicillin. The cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Cell transfection
The ACHN, 786-O, Caki-1 and Caki-2 cells were seeded onto 24-well plates at a density of 5×104 cells per well and were incubated at 37°C overnight prior to transfection. The miR-183 inhibitor and the negative control (NC) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The primer sequences were as follows: miR-183 inhibitor forward, 5′-AGUGAAUUCUACCAGUGCCAUA-3′ and reverse, 5′-UAUGGCACUGGUAGAAUUCACU-3′; NC forward, 5′-CAGUACUUUUGUGUAGUACAA-3′ and reverse, 5′-UUGUACUACACAAAAGUACUG-3′. Transfection with the miR-183 inhibitor (50 nM) and the respective NC (50 nM) was performed using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Following transfection, the cells were maintained at 37°C for 48 h for additional experiments.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The total RNA, containing miRNA, was extracted from harvested RCC cells using 1 ml TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. RNA was transcribed to cDNA using an EasyScript First-Strand cDNA Synthesis SuperMix kit (TransGen Biotech, Inc., Slough, UK) according to the manufacturer's instructions. qPCR was performed using the SYBR-Green PCR kit (Takara Bio, Inc., Otsu, Japan) on the ABI 7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Reaction conditions included: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec, 57°C for 20 sec, and 72°C for 15 sec. GAPDH or U6 were used as endogenous controls. The relative expression levels were calculated using the 2−ΔΔCq method (21). All RT-qPCR experiments were conducted in triplicate. PCR amplification was performed using the following primers: miR-183 forward, 5′-CGCGGTATGGCACTGGTAGA-3′ and reverse, 5′-AGTGCAGGGTCCGAGGTATTC-3′; DKK-3 forward, 5′-AGGACACGCAGCACAAATTG-3′ and reverse, 5′-CCAGTCTGGTTGTTGGTTATCTT-3′; U6-forward, 5′-AGAGCCTGTGGTGTCCG-3′ and reverse, 5′-CATCTTCAAAGCACTTCCCT-3′; GAPDH-forward, 5′-CATCACCATCTTCCAGGAGCG-3′ and reverse, 5′-TGACCTTGCCCACAGCCTTG-3′.
MTT cell proliferation assay
The MTT assay was conducted to measure the proliferation of RCC cells using MTT kit from Sigma-Aldrich (Merck KGaA, Darmstadt, Germamy), according to the manufacturer's instructions. Following miR-183 inhibitor or mimic transfection, RCC 786-O and ACHN cells were seeded onto 96-well plates (4×103 cells/well) and were incubated at 37°C with 5% CO2. Following incubation for 24, 48 and 72 h post-transfection, cell proliferation was examined. A total of 20 µl MTT solution was added to each well and the cells were subsequently incubated for 4 h prior to measurement. Following removal of the medium, 150 µl dimethyl sulfoxide (DMSO) was added to each well and the plates were agitated at a low speed for 10 min to ensure the purple formazan was fully dissolved. The optical densities of each well were measured at 490 nm using a microplate reader. All experiments were performed in triplicate.
Cell invasion assay
The invasive potential of cancer cells was determined using Transwell chambers and Transwell inserts. In brief, 1×105 RCC 786-O and ACHN cells that had been transfected with a miR-183 inhibitor or mimic were resuspended in 200 µl RPMI-1640 medium without serum and were seeded into the upper chamber. RPMI-1640 medium containing 20% FBS was added to the lower chamber. Following incubation for 48 h at 37°C with 5% CO2, cells that had migrated to the lower surface of filters were fixed with 4% paraformaldehyde at room temperature for 30 min, and then were stained with 0.5% crystal violet at room temperature for 30 min. The number of migrating cells was counted using a confocal microscope in five fields at a magnification of ×200. All experiments were performed in triplicate.
miRNA target prediction and luciferase assay
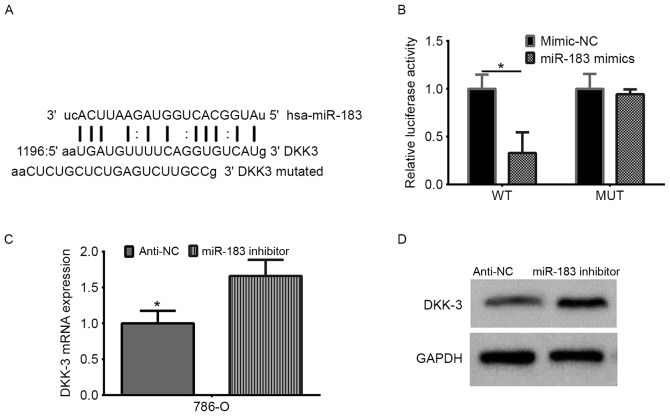
miRNA targets were predicted using the TargetScan algorithms (http://www.targetscan.org), and DKK-3 was predicted to be a target of miR-183. Luciferase reporter plasmids [PmirGLO-DKK-3-3′UTR wild-type (WT) and PmirGLO-DKK-3-3′UTR mutant (MUT)] were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). Transient transfection of miR-183 mimics and/or plasmids were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. 786-O cells were seeded onto 96-well plates (5×103/well) and were co-transfected with 100 ng WT or MUT reporter with 50 nmol/l miR-183 or miR-negative control (NC), according to the manufacturer's instructions. After 48 h, the luciferase activity was measured using the Dual-Luciferase Reporter Assay system (Promega Corporation, Madison, WI, USA). Firefly luciferase activity was normalized to the corresponding Renilla luciferase activity.
Western blot analysis
A total of 48 h after transfection with the miR-183 inhibitor or mimic, the cancer cells were collected and washed three times with phosphate-buffered saline. Following the addition of the cell lysis solution, the cancer cells were centrifuged at 14,000 × g for 15 min at 4°C. Total protein was extracted from RCC cells using a protein extraction kit (Beyotime Institute of Biotechnology, Jiangsu, China), according to the manufacturer's instructions. Protein concentration was quantified using a Bradford protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Protein (50 µg) was separated using 10% SDS-PAGE gels, and then electrophoretically transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Inc.). The membranes were subsequently blocked with 5% skimmed milk for 2 h at room temperature, the primary antibodies were added and incubated overnight at 4°C. The primary antibodies used were as follows: anti-DKK-3 antibody (dilution 1:100; cat. no. 10365-1-AP; Proteintech Group, Inc., Chicago, IL, USA), and anti-GADPH antibody (dilution 1:1,000 dilution; cat. no. ab9484; both from Abcam, Cambridge, UK). Subsequently, a horseradish peroxidase-conjugated secondary antibody (dilution 1:5,000; cat. no. ab6789; Abcam) was added and incubated for 2 h at room temperature. The blots were visualized using an enhanced chemiluminescence kit (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA), and analyzed with Quantity One software version 4.6.2 (Bio-Rad Laboratories, Inc.).
Statistical analysis
All data are presented as mean ± standard deviation. Statistically significant differences between two groups were determined using the two-tailed Student's t-test. One-way analysis of variance (ANOVA) was used for comparisons among multiple groups. The Student-Newman-Keuls test was used as a post hoc test following ANOVA. P<0.05 was considered to indicate a statistically significant difference. All statistical analysis was conducted using the SPSS 18.0 statistical software package (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Expression of miR-183 in RCC cell lines
The expression of miR-183 in RCC cell lines was analyzed using RT-qPCR. In comparison to the immortalized normal proximal tubule epithelial HK-2 cell line, miR-183 expression was significantly upregulated in all four RCC lines (Fig. 1; P<0.05).
Figure 1.
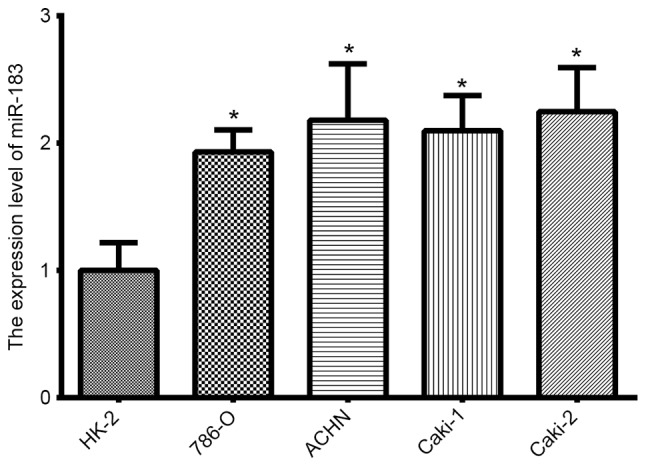
Expression of miR-183 in four renal cancer cell lines (786-O, ACHN, Caki-1 and Caki-2) and the human proximal tubule epithelial HK-2 cell line. Compared with HK-2, miR-183 expression was significantly upregulated in all four renal cancer cell lines (*P<0.05 vs. HK-2, as determined by analysis of variance). miR, microRNA.
Validation of cell transfection efficiency
Silencing experiments were performed in two RCC lines (786-O and ACHN). qPCR was performed to determine the transfection efficiency of the miR-183 inhibitor compared with the negative control. The expression levels of miR-183 were significantly lower in 786-O cells (P<0.05) and ACHN cells (P<0.05) transfected with the miR-183 inhibitor, compared with expression in the respective negative control groups for each of the cell lines (Fig. 2).
Figure 2.
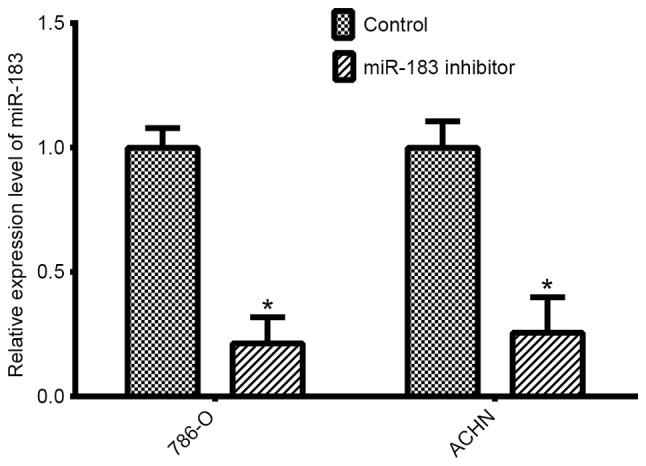
miR-183 was downregulated in 786-O and ACHN cells following transfection with a miR-183 inhibitor (*P<0.05 vs. control, as determined by Student's t-test). miR, microRNA.
Downregulation of miR-183 inhibits 786-O and ACHN cell proliferation
In order to determine whether miR-183 was able to affect RCC cell proliferation, an MTT assay was performed. Following transfection with the miR-183 inhibitor, the proliferation of 786-O cells was decreased by 7.67% (24 h; P<0.05), 10.67% (48 h; P<0.05) and 39.67% (72 h; P<0.05) compared with the negative control group (Fig. 3A). In the ACHN cells, cell proliferation was decreased by 11.33% (24 h; P<0.05), 15.00% (48 h; P<0.05) and 26.00% (72 h; P<0.05) compared with the negative control group (Fig. 3B). These results suggested that downregulation of miR-183 may inhibit RCC cell viability.
Figure 3.
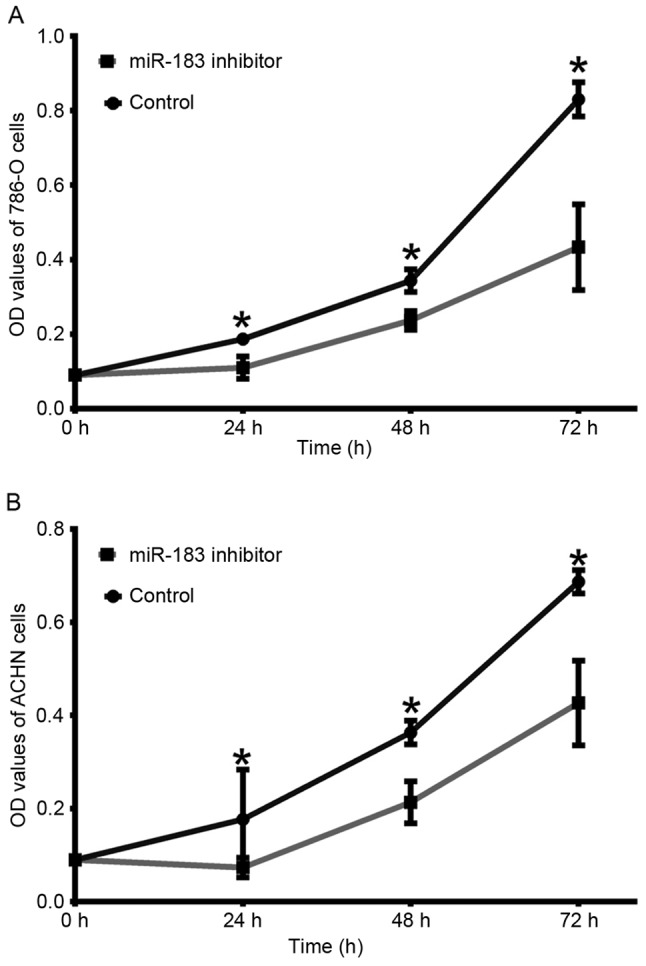
Downregulation of miR-183 inhibits cell proliferation in (A) 786-O and (B) ACHN cell lines (*P<0.05 vs. control at each time-point, as determined by Student's t-test). miR, microRNA; OD, optical density.
Downregulation of miR-183 inhibits the invasion of 786-O and ACHN cells
In order to investigate the role of miR-183 on cell invasion, an invasion assay was performed. As demonstrated in Fig. 4, the number of invasive 786-O and ACHN cells transfected with the miR-183 inhibitor was significantly reduced compared with that in the control groups (both P<0.05). These results indicated that downregulation of miR-183 may decrease the invasion of 786-O and ACHN cells.
Figure 4.
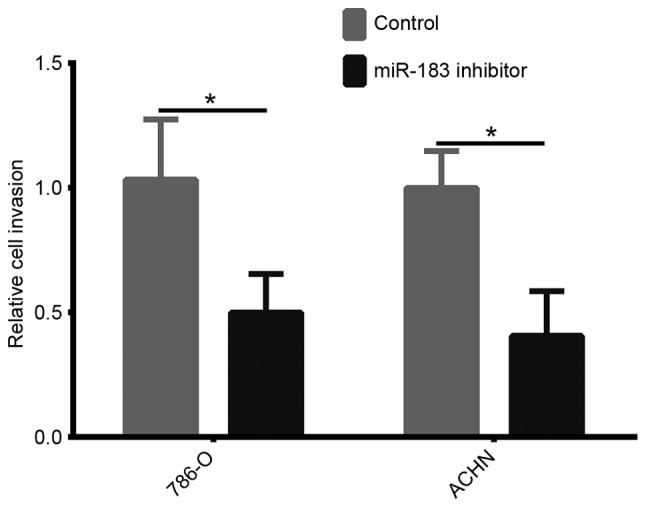
A Transwell assay was performed to determine the invasive capacity of the 786-O and ACHN cells that had been transfected with a miR-183 inhibitor or a negative control (*P<0.05 vs. control, as determined by Student's t-test). miR, microRNA.
DKK-3 is a direct target gene of miR-183
DKK-3 was predicted as a target gene of miR-183 using TargetScan. The 3′-UTR and a mutant 3′-UTR of DKK-3 were used to verify DKK-3 as a target gene of miR-183 (Fig. 5A). The target sequences in the WT DKK-3 3′-UTR and a MUT sequence were cloned into the luciferase reporter. Following co-transfection of the reporters with a miR-183 mimic or mimic-NC into 786-O cells, the luciferase activity was recorded. The data demonstrated that miR-183 induced an evident decrease in the firefly luciferase activity when compared with the mimic-NC group in the WT reporter-transfected cells. However, the same effect was not observed in the MUT group (Fig. 5B; P<0.05). Additionally, the regulatory effect of the miR-183 inhibitor on DKK-3 was investigated by RT-qPCR and western blot analysis in 786-O cells, identifying that the miR-183 inhibitor increased the DKK-3 mRNA and protein levels in 786-O cells (Fig. 5C and D). Taken together, these results indicated that miR-183 directly targets DKK-3 in RCC cells.
Figure 5.
DKK-3 is the direct target of miR-183 in renal cell carcinoma. (A) The miR-183 binding site in the DKK-3 3′-UTR, as predicted by TargetScan. The upper sequence is miR-183, the middle sequence is the target WT, and the bottom sequence is the target-mutated sequence. (B) The firefly luciferase activity in the 786-O cell line was detected following co-transfection of the WT or MUT 3′-UTR (100 ng) with the miR-183 mimic or mimic-NC (50 nmol/l). The firefly luciferase activity of each sample was normalized to Renilla luciferase activity. *P<0.05 vs. mimic NC. (C) Reverse transcription-quantitative polymerase chain reaction analysis of the relative mRNA levels of DKK-3 following miR-183 inhibitor transfection. (*P<0.05 vs. anti-miR-183, as determined by Student's t-test). (D) Western blot analysis of DKK-3 protein expression levels following transfection with a miR-183 inhibitor. DKK-3, Dickkopf-related protein 3; miR, microRNA; WT, wild-type; MUT, mutant; UTR, untranslated region; NC, negative control.
Discussion
As RCC is not sensitive to traditional radiotherapy or chemotherapy, there is a pressing demand to identify more efficient novel treatment strategies, such as targeted therapies. miRNAs are able to regulate gene expression by binding to the 3′-UTR of target genes or by hybridizing in the coding sequence (8). An increasing volume of evidence has demonstrated that miRNAs are abnormally expressed in the majority of cancer types. Furthermore, miRNAs may regulate cancer cell proliferation, invasion and migration, suggesting that they serve an important role in cancer occurrence and progression (9). Therefore, miRNAs have the potential to be used as cancer diagnostic and prognostic biomarkers, as well as target points for cancer therapeutics (8,11).
Previous investigations revealed that miR-183 may function as an oncogene in various types of cancer, including pancreatic, lung, gastric, breast and colorectal cancer (13–17). However, it may also function as a tumor suppressor in certain cancer types, including cervical cancer (18). Therefore, the role of miR-183 may depend on the type of cancer that it is expressed in. A previous study by Huangfu et al (13) identified that miR-183 was able to promote CRC occurrence and progression through regulating the apoptosis and autophagy of CRC cells by targeting the ultraviolet radiation resistance-associated gene. Bi et al (22) also investigated the detailed functional role of miR-183 in CRC and identified that the ATP-binding cassette transporter A1/cholesterol exporter may be one of the downstream targets of miR-183. Zhu et al (15) demonstrated that miR-183 may function as an oncogene in non-small cell lung carcinoma by promoting proliferation, invasion and migration through targeting protein tyrosine phosphatase non-receptor type 4. Additionally, a study undertaken by Miao et al (14) identified that miR-183 may have the potential to promote pancreatic cancer cell proliferation, invasion and metastasis by targeting suppressor of cytokine signaling 6. Li et al (16) demonstrated that miR-183 may have the potential to regulate the proliferation, invasion and apoptosis of gastric cancer cells by regulating programmed cell death 4 (PDCD4), indicating that it may function as an oncogene in gastric cancer. Similarly, Cheng et al (17) identified that miR-183 may function as an oncogene in breast cancer by promoting the proliferation and invasion of breast cancer cells, and that PDCD4 is a target of miR-183.
In contrast to the aforementioned studies, a study undertaken by Fan et al (18) demonstrated that the expression level of miR-183 was significantly lower in cervical tissues than in adjacent normal cervical tissues. Furthermore, they revealed that miR-183 may function as a tumor suppressor by regulating the proliferation, invasion and metastasis of cervical cancer cells, and that matrix metallopeptidase-9 may be one of the downstream targets of miR-183.
The Wnt signaling pathway has been investigated at several molecular levels, as its involvement has been implicated in tumorigenesis and disease progression (23). The DKK family functions as a negative regulator of the Wnt signaling pathway, and therefore serves a tumor-suppressive function (24). Ueno et al have reported that DKK-3 expression was regulated by histone modifications in the DKK-3 promoter region in RCC cells (25). Furthermore, in our previous study, DKK-3 levels were identified to be lower in the tumor tissues compared with the adjacent normal tissues using immunohistochemistry, western blot analysis and qPCR, indicating that DKK-3 may have an associated mechanism in the development of RCC (26). TargetScan identified DKK-3 as a target gene of miR-183, in line with the results of a study undertaken by Ueno et al (20), who had previously identified that miR-183 was an oncogene targeting DKK-3 in prostate cancer (20). However, whether there is an association between miR-183 and DKK-3 in RCC remains to be elucidated.
The clinical significance of miR-183 has previously been investigated in RCC. For example, Zhang et al (19) demonstrated that the serum expression level of miR-183 was significantly higher in patients with RCC than in healthy volunteers. Furthermore, its expression level was significantly associated with sensitivity to natural killer cell therapy (19). The present study expanded the knowledge presently available regarding the expression and function of miR-183 in RCC. Firstly, in comparison to the immortalized normal proximal tubule epithelial HK-2 cell line, miR-183 expression was upregulated in all four RCC cell lines (ACHN, 786-O, Caki-1 and Caki-2). Furthermore, downregulation of miR-183 inhibited RCC cell proliferation and invasion, indicating that miR-183 may function as an oncogene in RCC. Additionally, an important molecular link was identified between miR-183 and DKK-3 in the present study through the prediction of TargetScan that DKK-3 was a direct target gene of miR-183. Furthermore, the results of the luciferase activity assay indicated that miR-183 directly targeted the DKK-3 3′-UTR, as predicted by bioinformatics analysis. Additionally, the regulatory effect of miR-183 on DKK-3 was investigated by RT-qPCR and western blotting in 786-O cells. The data demonstrated that downregulation of miR-183 increased DKK-3 expression at the mRNA and protein levels. These findings revealed that miR-183 may have the potential to regulate DKK-3 expression in vitro, and may have a tumor promoting role in RCC development and progression.
In conclusion, the results of the present study demonstrate that miR-183 functions as an oncogene by downregulating DKK-3 expression, providing a potential diagnostic, prognostic and therapeutic target for RCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the Zhejiang Medical, Health, and Science and Technology Project (grant no. 2016RCA028).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZXL designed the study; ZXL, XG, ZY, and YJJ carried out the experiments and analyzed the data; ZXL, and XG drafted the study. All authors read and approved the final study.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol. 2015;67:519–530. doi: 10.1016/j.eururo.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Patard JJ, Leray E, Rioux-Leclercq N, Cindolo L, Ficarra V, Zisman A, De La Taille A, Tostain J, Artibani W, Abbou CC, et al. Prognostic value of histologic subtypes in renal cell carcinoma: A multicenter experience. J Clin Oncol. 2005;23:2763–2771. doi: 10.1200/JCO.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 4.Lardas M, Stewart F, Scrimgeour D, Hofmann F, Marconi L, Dabestani S, Bex A, Volpe A, Canfield SE, Staehler M, et al. Systematic review of surgical management of nonmetastatic renal cell carcinoma with vena caval thrombus. Eur Urol. 2016;70:265–280. doi: 10.1016/j.eururo.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 5.Ljungberg B, Bensalah K, Canfield S, Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L, Merseburger AS, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67:913–924. doi: 10.1016/j.eururo.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part a: Renal, penile, and testicular tumours. Eur Urol. 2016;70:93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Davis ID, Xie W, Pezaro C, Donskov F, Wells JC, Agarwal N, Srinivas S, Yuasa T, Beuselinck B, Wood LA, et al. Efficacy of second-line targeted therapy for renal cell carcinoma according to change from baseline in international metastatic renal cell carcinoma database consortium prognostic category. Eur Urol. 2017;71:970–978. doi: 10.1016/j.eururo.2016.09.047. [DOI] [PubMed] [Google Scholar]
- 8.Cloney R. Non-coding RNA: Deciphering the rules of microRNA targeting. Nat Rev Genet. 2016;17:718. doi: 10.1038/nrg.2016.148. [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 11.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lumayag S, Haldin CE, Corbett NJ, Wahlin KJ, Cowan C, Turturro S, Larsen PE, Kovacs B, Witmer PD, Valle D, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration; Proc Natl Acad Sci USA; 2013; pp. E507–E516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huangfu L, Liang H, Wang G, Su X, Li L, Du Z, Hu M, Dong Y, Bai X, Liu T, et al. miR-183 regulates autophagy and apoptosis in colorectal cancer through targeting of UVRAG. Oncotarget. 2016;7:4735–4745. doi: 10.18632/oncotarget.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao F, Zhu J, Chen Y, Tang N, Wang X, Li X. MicroRNA-183-5p promotes the proliferation, invasion and metastasis of human pancreatic adenocarcinoma cells. Oncol Lett. 2016;11:134–140. doi: 10.3892/ol.2015.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu C, Deng X, Wu J, Zhang J, Yang H, Fu S, Zhang Y, Han Y, Zou Y, Chen Z, Lin S. MicroRNA-183 promotes migration and invasion of CD133(+)/CD326(+) lung adenocarcinoma initiating cells via PTPN4 inhibition. Tumour Biol. 2016;37:11289–11297. doi: 10.1007/s13277-016-4955-8. [DOI] [PubMed] [Google Scholar]
- 16.Li C, Deng L, Zhi Q, Meng Q, Qian A, Sang H, Li X, Xia J. MicroRNA-183 functions as an oncogene by regulating PDCD4 in gastric cancer. Anticancer Agents Med Chem. 2016;16:447–455. doi: 10.2174/1871520615666150914114237. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Xiang G, Meng Y, Dong R. MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod Biol. 2016;16:225–233. doi: 10.1016/j.repbio.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Fan D, Wang Y, Qi P, Chen Y, Xu P, Yang X, Jin X, Tian X. MicroRNA-183 functions as the tumor suppressor via inhibiting cellular invasion and metastasis by targeting MMP-9 in cervical cancer. Gynecol Oncol. 2016;141:166–174. doi: 10.1016/j.ygyno.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Di W, Dong Y, Lu G, Yu J, Li J, Li P. High serum miR-183 level is associated with poor responsiveness of renal cancer to natural killer cells. Tumour Biol. 2015;36:9245–9249. doi: 10.1007/s13277-015-3604-y. [DOI] [PubMed] [Google Scholar]
- 20.Ueno K, Hirata H, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. microRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer. 2013;108:1659–1667. doi: 10.1038/bjc.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Bi DP, Yin CH, Zhang XY, Yang NN, Xu JY. MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol Rep. 2016;35:2873–2879. doi: 10.3892/or.2016.4631. [DOI] [PubMed] [Google Scholar]
- 23.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 24.Veeck J, Dahl E. Targeting the Wnt pathway in cancer: The emerging role of Dickkopf-3. Biochim Biophys Acta. 2012;1825:18–28. doi: 10.1016/j.bbcan.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Ueno K, Hirata H, Majid S, Chen Y, Zaman MS, Tabatabai ZL, Hinoda Y, Dahiya R. Wnt antagonist DICKKOPF-3 (Dkk-3) induces apoptosis in human renal cell carcinoma. Mol Carcinog. 2011;50:449–457. doi: 10.1002/mc.20729. [DOI] [PubMed] [Google Scholar]
- 26.Guo CC, Zhang XL, Yang B, Geng J, Peng B, Zheng JH. Decreased expression of Dkk1 and Dkk3 in human clear cell renal cell carcinoma. Mol Med Rep. 2014;9:2367–2373. doi: 10.3892/mmr.2014.2077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.