Abstract
Lung cancer is among the leading causes of cancer-associated mortality worldwide, with a low 5-year survival rate of 16.1%. However, the underlying molecular mechanisms behind lung cancer tumorigenesis remain largely unknown. Long non-coding RNAs (lncRNAs) have been demonstrated to serve a function in the tumorigenesis of multiple types of cancer. The objective of the present study was to identify the function of a newly identified lncRNA zinc finger E-box-binding homeobox 2 antisense RNA 1 (ZEB2-AS1) in human lung cancer. Results demonstrated that the transcript level of ZEB2-AS1 in human lung cancer was markedly upregulated in vivo and in vitro. The knockdown of ZEB2-AS1 in A549 and NCI-H292 cells, and the overexpression of ZEB2-AS1 in H-125 and H1975 cells, altered colony formation and cell proliferative rate, as examined using colony formation and cell proliferation assays. Western blot analysis revealed that the knockdown of ZEB2-AS1 in A549 and NCI-H292 cells increased the protein levels of B-cell lymphoma 2 (Bcl-2), Bcl-2-associated X protein (Bax), caspase-3 and −9, upregulated the relative activities of caspase-3 and −9, and had no observable effect on caspase-8 activity. Similarly, the overexpression of ZEB2-AS1 in H-125 and H1975 cells resulted in decreased expression of caspase-3, caspase-9, Bcl-2 and Bax. The results identified the effects of lncRNA ZEB2-AS1 on lung cancer progression through promoting its proliferation and inhibiting cell apoptosis, indicating that ZEB2-AS1 may serve as a novel prognostic factor for the diagnosis and treatment of human lung cancer in the clinic.
Keywords: long non-coding RNA, zinc finger E-box-binding homeobox 2 antisense RNA 1, lung cancer, proliferation, apoptosis
Introduction
Lung cancer is one of the leading causes of cancer-associated mortality, with a 5-year-survival rate of 17% worldwide in 2011 (1). Two major types of lung cancer have been identified: Small-cell lung cancer (~15%) and non-small-cell lung cancer (~85%), the latter of which also contains three major histological subtypes: Adenocarcinoma, squamous cell carcinoma and large cell carcinoma (2). Despite the differences between the subtypes of lung cancer, the low survival rate of patients suffering from lung cancer is primarily a result of delayed diagnosis and late detection, resulting in limited treatment options in the late stages of disease (3,4). Therefore, it may be beneficial to identify novel biomarkers to allow for the diagnosis of lung cancer at an early stage.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules that are >200 nucleotides in length, do not possess an open reading frame and do not encode proteins (5). The majority of well-characterized lncRNAs are RNA polymerase II-transcribed, capped and polyadenylated, containing exon-exon splice junctions similar to mRNAs (6,7). It has been demonstrated that lncRNAs serve functions in gene regulation in various biological conditions with distinct underlying molecular mechanisms including the recruitment of transcriptional factors and direct interaction with DNAs or other RNAs (8,9). It has also been reported that lncRNAs have functions in a variety of tumors, including lung cancer. For example, LINC00313 may be used as a diagnostic biomarker of early stage lung adenocarcinoma (10), and AK126698, a newly discovered lncRNA, was demonstrated to confer cisplatin resistance by targeting the Wnt pathway (11). Therefore, the objective of the present study was to identify novel lncRNAs that serve functions in the tumorigenesis of lung cancer.
Zinc finger E-box-binding homeobox 1 (ZEB1) is a transcriptional factor that serves important functions in the process of epithelial-mesenchymal-transition, which is associated with tumorigenesis. It was demonstrated that aberrant expression of ZEB1 was associated with aggressive disease, low differentiation, metastases and poor prognosis in clinical patients with multiple types of cancer (12,13). Zinc finger E-box-binding homeobox 2 antisense RNA 1 (ZEB2-AS1) is a non-coding oncogene identified in human hepatocellular carcinoma (HCC) (14). Li et al (15) demonstrated that the relative transcript level of ZEB2-AS1 was upregulated in HCC in vivo and in vitro and functioned as a prognostic factor for HCC pathogenesis.
The present study aimed to investigate the expression of ZEB2-AS1 in human lung cancer in clinical patients and in cultured lung cancer cells. The detailed function of ZEB2-AS1 in cell proliferation and cell apoptosis was also investigated. The results of the present study indicated that ZEB2-AS1 may function as a prognostic biomarker for lung cancer and may aid the diagnosis and treatment of patients with lung cancer.
Materials and methods
Human tissues
The present study was approved by an Institutional Review Board at the General Department Beijing Chest Hospital, Capital Medical University (Beijing, China). A total of 100 lung cancer tissues and their adjacent non-cancerous tissues were collected from patients (67 males and 33 females; mean age, 62; age range, 48–79) who underwent surgical resection at Department of General Surgery. The tissues were snap-frozen in liquid nitrogen once dissected from the patients and used for subsequent RT-qPCR analysis. The patients selected had not received any chemotherapies or radiotherapies prior to surgical resection. All patients provided informed consent.
Cell culture and transfection
The normal human lung cell line MRC-5 was purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). A total of 5 lung cancer cell lines; H-125, A549, 95D, NCI-H292 and H1975 were obtained from Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (both from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were maintained in an incubator containing 5% CO2 at 37°C. The ZEB2-AS1 expression plasmid was constructed with the pcDNA 3.0 vector (Addgene, Inc., Cambridge, MA, USA). The specific small interfering RNA (siRNA) against ZEB2-AS1 was designed and synthesized by Invitrogen; Thermo Fisher Scientific, Inc., with the sequence, 5′-CAAAGGACACCTTTGGTTACCTGAA-3′. When A549, NCI-H929, H-125 and H1975 cells grew to a confluence of 80%, the transfection was conducted using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Following 6 h of transfection, cell medium was replaced and 48 h later, cells were harvested.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNAs were extracted from fresh frozen samples and cells with TRIzol Reagent (Thermo Fisher Scientific, Inc.) as per the manufacturer's instructions. A total of 1 µg RNA was reverse transcribed into cDNA with First Strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China) with the following protocol: 37°C for 15 min and 85°C for 5 sec. The relative expression of lncRNA ZEB2-AS1 to GAPDH control transcripts was determined using qPCR as per the ABI 7900 Fast Real-Time PCR system (Thermo Fisher Scientific, Inc.). The PCR conditions included: An initial denaturation step of 94°C for 2 min, followed by 30 cycles of 95°C for 30 sec, 59°C for 30 sec, 72°C for 2 min and a final elongation step at 72°C for 10 min. The PCR reaction was normalized to the GAPDH reference gene. The RT-qPCR amplification was performed in triplicate. The relative level of ZEB2-AS1 transcript was determined using the 2−ΔΔCq method (16). The primer sequences were as follows: ZEB2-AS1 forward, 5′-ATGAAGAAGCCGCGAAGTGT-3′ and reverse, 5′-CACACCCTAATACACATGCCCT-3′; GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCCTGTTGCTGTA-3′.
Colony formation assay
A total of A549 (5×105/ml), NCI-H292 (5×105/ml), H-125 and 95D cells (5×105/ml) in 6-well plates were treated with siRNA targeted at ZEB2-AS1 (siZEB2-AS1) or ZEB2-AS1 expressing plasmid and 24 h after treatment, were seeded into 12-well plates (100 cells/well) in triplicate. Following incubation for 10 days at 37°C, the colonies were fixed with pre-iced methanol and stained with crystal violet (1%) at room temperature for 10 min. Colonies were counted using light microscopy (magnification, ×200) and colonies that contained >50 cells were designated as survivors. The following formula was used to calculate the rate of colony formation: Colony formation rate = (number of colonies/number of seeded cells) × 100.
Cell proliferation assay
An MTT cell growth kit (Promega Corporation, Madison, WI, USA) was used to measure cell proliferative abilities according to the manufacturer's protocol. Briefly, A549, NCI-H292, H-125 and H1975 cells were seeded into 96-well plates at an initial concentration of 5×103 cells/well in DMEM supplemented with 10% FBS. Each experimental group of cells was then spread in sextuplicate and the culture medium was replaced every other day. Cell proliferation rate was assessed for 5 consecutive days. At each time-point (1, 2, 3, 4 and 5 days' post-transfection), formazan crystals were dissolved in dimethyl sulfoxide, and the cell proliferation rate was detected using a microplate reader at a wavelength of 490 nm. The absorbance of control cells at day 1 was designated as 1. Other absorbance values were normalized to the control cells at day 1.
Western blot analysis
Cells were seeded into a six-well plate 24 h prior to transfection. A549 and NCI-H292 cells were treated with specific siZEB2-AS1. H-125 and H1975 cells were stimulated with ZEB2-AS1 expression plasmid. Total proteins were collected using NP40 lysis buffer and quantified using a bicinchonic acid assay. An equal quantity of protein from each sample (50 µg/lane) was subjected to 12% SDS-PAGE and electroblotted onto polyvinylidene fluoride membranes. Subsequently, the membrane was blocked with TBS/0.1% Tween-20, supplemented with 5% skimmed milk for 1 h at room temperature and then incubated with primary antibodies at 4°C overnight. Secondary antibodies horseradish peroxidase-conjugated goat anti-mouse IgG (1:5,000; cat. no. ab6717; Abcam, Cambridge, UK) were incubated with the membrane for 1 h at room temperature. Next, proteins were detected using an enhanced chemiluminescence method (EMD Millipore, Billerica, MA, USA). The immunoreactive bands were quantified by the densitometry with ImageJ software (v2.0; National Institutes of Health, Bethesda, MD, USA) when necessary. Primary antibodies against caspase-9 (sc-7885; 1:1,000), caspase-3 (sc271759; 1:1,000), B-cell lymphoma-2 (Bcl-2; sc-578; 1:1,000), GAPDH (sc-47724; 1:1,000) and secondary antibodies (sc-2004; sc-2005; 1:2,000) were all purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Primary antibody against cytoplasmic Bcl-associated X protein (Bax; ab32503; 1:1,000) was purchased from Abcam.
Relative activities of caspases
The activities of caspase-3, −8 and −9 were determined using caspase-3 activity kits, caspase-8 activity kits, caspase-9 activity kits, respectively (Beyotime Institute of Biotechnology, Haimen, China), according to the manufacturers' instructions. Briefly, A549, NCI-H292, H-125 and H1975 cells were transfected with ZEB2-AS1 plasmid or siRNA 48 h prior to the experiment. Subsequently, cell lysates were collected from each group of cells. An equal amount of 10 µl (50 µg) proteins from cell lysates were added into 96-well plates and mixed with an aliquot of 80 µl reaction buffer supplemented with caspase substrate (2 mM). Following a 4-h incubation at 37°C, caspase activities were determined using a microplate reader at an absorbance of 450 nm.
Statistical analysis
All data are presented as the mean ± standard deviation. Each experiment was repeated in triplicate. GraphPad Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) software was used for statistical analysis. Statistical evaluation was performed using Student's t-test or one-way analysis of variance followed by the Student-Newman-Keuls post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of lncRNA ZEB2-AS1 is upregulated in human lung cancer in vivo and in vitro
First, the relative transcript level of lncRNA ZEB2-AS1 in human lung cancer was examined in vivo and in vitro. To this end, a total of 100 patients with lung cancer were included in the present study, and their tumor tissues as well as their adjacent non-cancerous tissues were dissected and collected for the subsequent RT-qPCR analysis. As presented in Fig. 1A, the relative transcript level of ZEB2-AS1 in tumor tissues was significantly increased (3-fold) compared with their adjacent non-cancerous counterparts. A total of 5 lung cancer cell lines and a normal human lung cell line tissue were also assessed by RT-qPCR to detect the expression of ZEB2-AS1. As presented in Fig. 1B, the relative transcript levels of ZEB2-AS1 in lung cancer cells were significantly upregulated, with A549 and NCI-H292 exhibiting the highest ZEB2-AS1 expression (up to 3.4-fold and 3.5-fold, respectively), whereas the expression levels of ZEB2-AS1 in H-125, 95D and H1975 were lower than those in A549 and NCI-H292 cells; however, these levels were significantly increased compared with non-tumor tissue. Thus, A549 and NCI-H292 cells were selected for knockdown assays and H-125 and H1975 cells were selected for overexpression analysis. These results identified that the level of ZEB2-AS1 expression was upregulated in human lung cancer.
Figure 1.
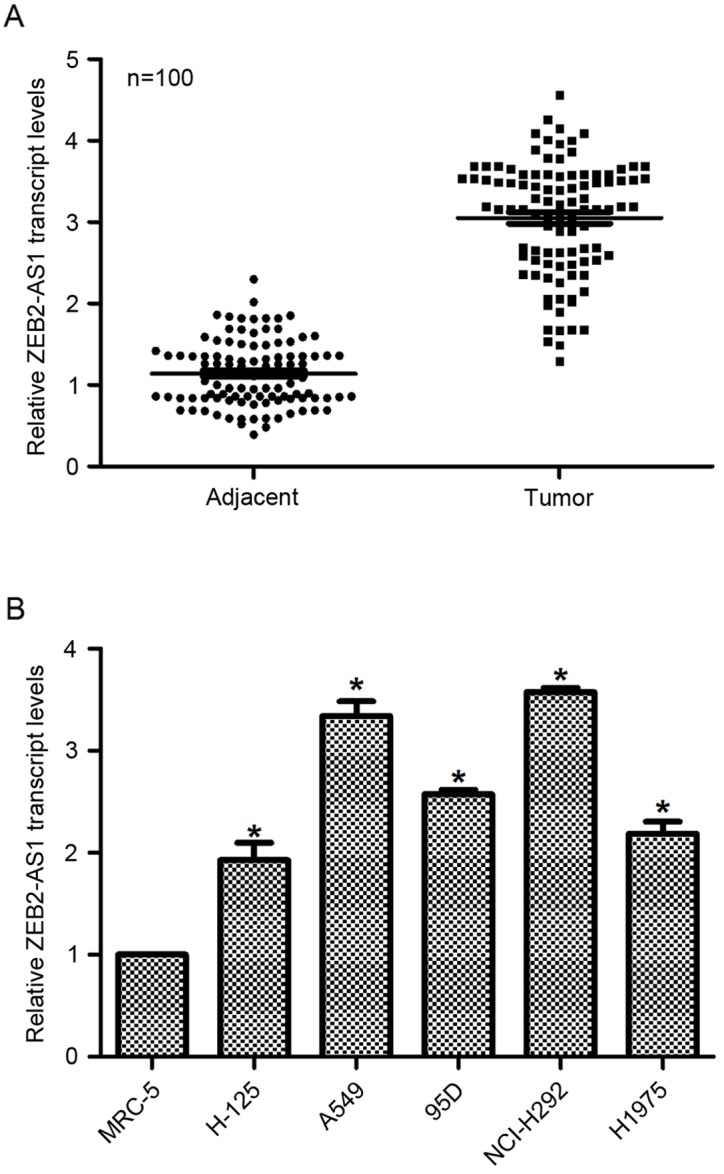
Transcript level of long non-coding RNA ZEB2-AS1 is upregulated in human lung cancer in vivo and in vitro. (A) Tumor tissues as well as their adjacent non-cancerous tissues were collected from 100 patients with lung cancer. Tissues were analyzed using reverse transcription-quantitative polymerase chain reaction to examine the expression of ZEB2-AS1 in vivo. One of the adjacent tissue samples was randomly selected as a control (relative expression, 1). (B) The normal lung MRC-5 and 5 lung cancer H-125, A549, 95D, NCI-H292 and H1975 cell lines were utilized to investigate the expression level of ZEB2-AS1 in vitro. The transcript level of ZEB2-AS1 in MRC-5 cells was set as 1. *P<0.05 vs. MRC-5 cells. ZEB2-AS1, zinc finger E-box-binding homeobox 2 antisense RNA 1.
Expression of lncRNA ZEB2-AS1 is associated with cell proliferation in human lung cancer
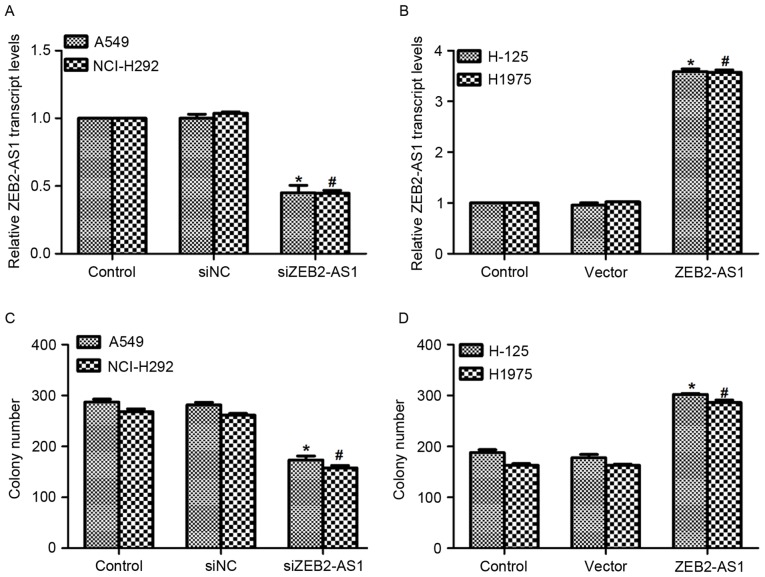
The functions of ZEB2-AS1 in human lung cancer were further evaluated. A specific siRNA against ZEB2-AS1 and an expression plasmid containing ZEB2-AS1 were constructed and transfected into the corresponding cells. As presented in Fig. 2A, when cells were transfected with siZEB2-AS1 for 48 h, the level of ZEB2-AS1 expression was decreased by 55% in A549 cells and 56% in NCI-H292 cells, respectively. Treatment with ZEB2-AS1 expressing plasmid upregulated the transcript level of ZEB2-AS1 3.45-fold inH-125 cells and 3.40-fold in H1975 cells (Fig. 2B). Subsequently, a colony formation assay was performed in all four cell lines. Transfection of A549 cells with siZEB2-AS1 inhibited the colony formation ability of the cells, as evidenced by the significantly decreased colony number compared with controls (Fig. 2C). Approximately 250 colonies were observed in NCI-H292 cells; however, only 152 colonies were counted when siZEB2-AS1 was transfected into NCI-H292 cells (Fig. 2C). Conversely, transfection with the ZEB2-AS1-expressing plasmid promoted colony formation in H-125 and H1975 cells (Fig. 2D). These results demonstrate that overexpression of ZEB2-AS1 in human lung cancer cells increased their colony formation ability.
Figure 2.
Long non-coding RNA ZEB2-AS1 promotes colony formation in human lung cancer. (A) Specific small interfering RNA against ZEB2-AS1 was synthesized and transfected into A549 and NCI-H292 cells, following which the relative transcript level of ZEB2-AS1 was examined. *P<0.05 vs. control in A549 cells; #P<0.05 vs. control in NCI-H292 cells. (B) ZEB2-AS1 expressing plasmid was constructed and transfected into H-125 and H1975 cells. Subsequently, the relative transcript level of ZEB2-AS1 was detected. *P<0.05 vs. control in H-125 cells; #P<0.05, vs. control in H1975 cells. (C) A colony formation assay was performed in A549 and NCI-H292 cells treated with siZEB2-AS1 and the colonies formed were counted under using light microscopy (magnification, ×200). *P<0.05 vs. control in A549 cells; #P<0.05 vs. control in NCI-H292 cells. (D) A colony formation assay was performed in H-125 and H1975 cells transfected with ZEB2-AS1 plasmid and the colonies formed were counted. *P<0.05 vs. control in H-125 cells; #P<0.05 vs. control in H1975 cells. ZEB2-AS1, zinc finger E-box-binding homeobox 2 antisense RNA 1; siNC, negative control small interfering RNA.
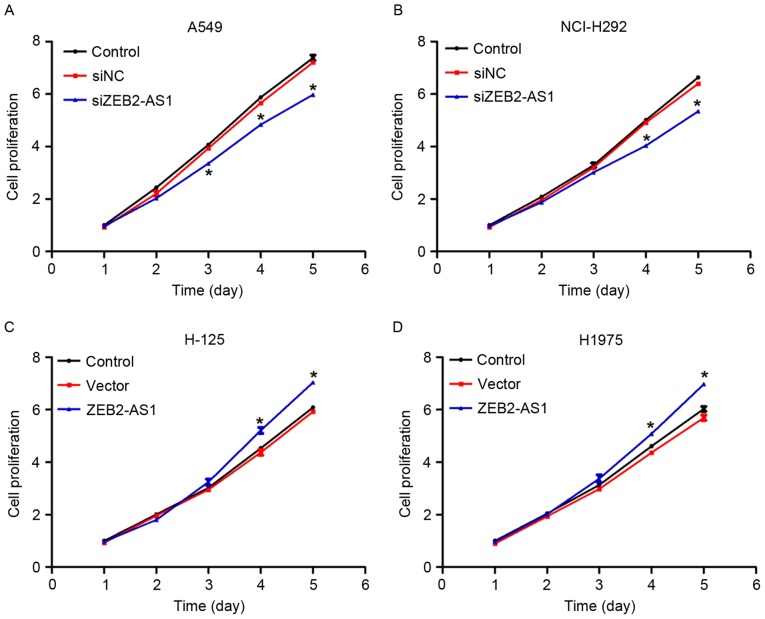
Subsequently, a cell proliferation assay was performed in lung cancer cells. There were no notable differences in the first 2 days between the control groups and the overexpression or siZEB2-AS1-treated group in all four lung cancer cell lines (Fig. 3). However, the cell proliferative rate was decreased by 18, 20 and 22% in siZEB2-AS1-treated A549 cells on days 3, 4 and 5, respectively (Fig. 3A). A similar phenomenon was also identified in NCI-H292 cells transfected with siZEB2-AS1 (Fig. 3B). Similarly, when cells were transfected with ZEB2-AS1-expressing plasmids, cell proliferation was increased by 14 and 10% on the fourth day in H-125 and H1975 cells, respectively, and a further increase was observed on day 5 in the two cell lines (Fig. 3C and D). Together with Fig. 2, these data indicated that ZEB2-AS1 promoted cell proliferation in human lung cancer cell lines in vitro.
Figure 3.
Long non-coding RNA ZEB2-AS1 promotes cell proliferation in human lung cancer cells. (A) A cell proliferation assay was performed in A549 cells treated with siZEB2-AS1 over 5 consecutive days. (B) Cell proliferation assay was performed in NCI-H292 cells treated with siZEB2-AS1 over 5 consecutive days. (C) A cell proliferation assay was performed in H-125 cells transfected with ZEB2-AS1 expressing plasmid over 5 consecutive days. (D) A cell proliferation assay was performed in H1975 cells transfected with ZEB2-AS1 expressing plasmid over 5 consecutive days. *P<0.05 vs. control. ZEB2-AS1, zinc finger E-box-binding homeobox-antisense RNA 1.
Expression of lncRNA ZEB2-AS1 is associated with cell apoptosis in human lung cancer cells
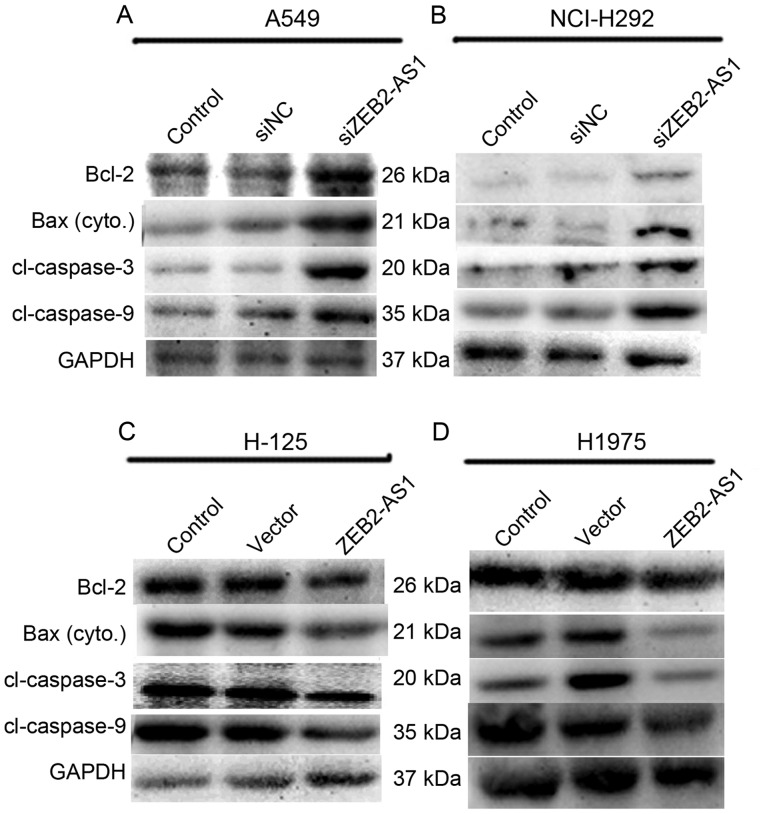
Since ZEB2-AS1 served a significant function in cell proliferation, the effect of ZEB2-AS1 on cell apoptosis was evaluated in the four lung cancer cell lines. Bcl-2, Bax, caspase-3 and −9 may all be considered to be representative of the process of apoptosis, and so were assessed using western blot analysis. As presented in Fig. 4A and B, the expression levels of the four proteins were all markedly increased in A549 and NCI-H292 cells when they were transfected with siZEB2-AS1. When H-125 and H1975 cells were treated with ZEB2-AS1 expressing plasmid, the levels of Bcl-2, Bax, caspase-3 and −9 protein expression were all decreased, whereas that of GAPDH remained unchanged (Fig. 4C and D).
Figure 4.
Long non-coding RNA ZEB2-AS1 promotes cell apoptosis in human lung cancer cells. (A) A549 cells and (B) NCI-H292 cells were transfected with siZEB2-AS1 and total protein was collected for western blot analysis 48 h after treatment. The expression levels of Bcl-2, cytoplasmic Bax, caspase-3 and −9, with molecular weights 26, 21, 20 and 35 kDa, respectively, were investigated. GAPDH was used as an internal control. (C) H-125 and (D) H1975 cells were treated with ZEB2-AS1 plasmid and the expression levels of Bcl-2, Bax, caspase-3 and −9 were detected. ZEB2-AS1, zinc finger E-box-binding homeobox 2 antisense RNA 1; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein.
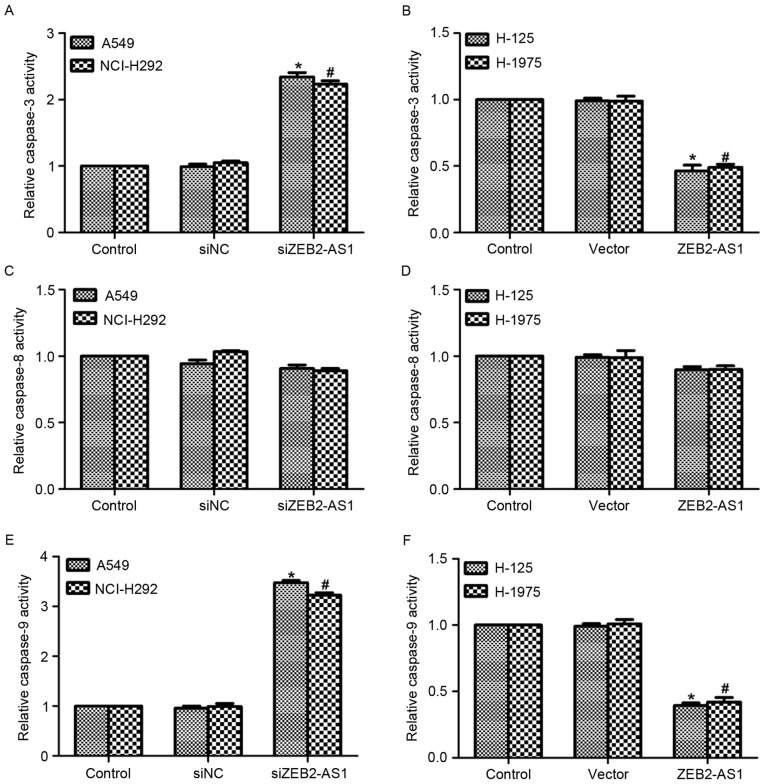
To assess the function of ZEB2-AS1 in lung cancer further, the relative activity of caspase-3, −8 and −9 was detected in the four cell lines. As presented in Fig. 5A, transfection with siZEB2-AS1 in A549 and NCI-H292 cells increased the activity of caspase-3 by 2.2-fold, whereas the relative activity of caspase-3 was decreased by ~50% when cells were treated with ZEB2-AS1 expressing plasmid in the two cell lines (Fig. 5B). On the contrary, the relative activity of caspase-8 remained stable when cells were treated with specific siRNA against ZEB2-AS1 or ZEB2-AS1 expressing plasmid in four lung cancer cell lines (Fig. 5C and D). The relative activity of caspase-9 was also detected in vitro. Similar to the activity of caspase-3, transfection of A549 and NCI-H292 cells with siZEB2-AS1 resulted in increased caspase-9 activity, whereas treatment with ZEB2-AS1 plasmid in H-125 and H1975 cells decreased the activity of caspase-9 (Fig. 5E and F). Taken together, these results indicated that the inhibition of ZEB2-AS1 promoted cell apoptosis in human lung cancer cells.
Figure 5.
Long non-coding RNA ZEB2-AS1 promotes relative activities of caspase-3 and −9 in human lung cancer. (A) The relative activity of caspase-3 was examined in A549 and NCI-H292 cells when the cell lines were treated with siZEB2-AS1 for 48 h. *P<0.05 vs. control in A549 cells; #P<0.05 vs. control in NCI-H292 cells. (B) The relative activity of caspase-3 was examined in H-125 and H1975 cells when the cell lines were transfected with the ZEB2-AS1-overexpression plasmid for 48 h. *P<0.05 vs. control in H-125 cells; #P<0.05 vs. control in H1975 cells. (C) The relative activity of caspase-8 was examined in A549 and NCI-H292 cells following treatment with siZEB2-AS1. (D) The relative activity of caspase-8 was examined in H-125 and H1975 cells when the cell lines were treated with ZEB2-AS1 plasmid. (E) The relative activity of caspase-9 was examined in A549 and NCI-H292 cells following treatment with siZEB2-AS1 for 48 h. *P<0.05 vs. control in A549 cells; #P<0.05 vs. control in NCI-H292 cells. (F) The relative activity of caspase-9 was examined in H-125 and H1975 cells following transfection with ZEB2-AS1-overexpression plasmid for 48 h. *P<0.05 vs. control in H-125 cells; #P<0.05 vs. control in H1975 cells. ZEB2-AS1, zinc finger E-box-binding homeobox 2 antisense RNA 1.
Discussion
Lung cancer is the most common type of cancer and the leading cause of cancer-associated mortality among men and women worldwide in 2005 (17). Although great efforts have been made to improve the diagnosis and treatment of patients with lung cancer, the 5-year survival rate remains low worldwide. Genetic and epigenetic alterations have been widely accepted as the driving factors of cancer (18). Research is presently focusing on the identification of novel biomarkers for the early stages of lung cancer.
ZEB2-AS1 is a recently identified lncRNA that has been demonstrated to perform significant functions in HCC (14,15). It is a non-coding antisense transcript from the promoters of ZEB2, which is reported to function as a transcription factor and is associated with a number of distinct types of cancer (19–21). It was also demonstrated that ZEB2 was overexpressed in human HCC and associated with HCC progression (22). ZEB2-AS1 was also observed to be upregulated in human HCC and served as a prognostic factor (14,15).
In the present study, the overexpression of ZEB2-AS1 in H-125 and H1975 cells promoted their proliferative rate and colony forming ability, consistent with previously published studies (14,15). However, the present study did not evaluate the detailed function of ZEB2-AS1 in cell metastasis, instead investigating the effects of ZEB2-AS1 on cell apoptosis. Li et al (15) identified that knockdown of ZEB2-AS1 in human HCC cells inhibited cell metastasis using Transwell and wound-healing assays, which may represent the next step in research involving lung cancer cell lines. Induction of cell apoptosis in multicellular organisms is one of the most effective ways to eliminate the harmful or unnecessary cells, and its abnormal regulation maybe associated with tumorigenesis (23). Apoptosis is primarily initiated via two pathways: The intrinsic pathway (initiated by stress stimulation), and the extrinsic pathway (initiated by signals from other cells) (24,25). Activation of the two pathways requires activation of initiator caspases (caspase-9 and −8), which then activate effector caspases (caspase-3), following which the cells undergo apoptosis (26,27). There are two hypotheses of the direct initiation of extrinsic pathways in mammals that have been suggested: The TNF-induced model and the Fas-Fas ligand-mediated model (25,28), which are all associated with the activation of caspase-8. The present study demonstrated that the knockdown of ZEB2-AS1 in A549 and NCI-H292 cells promoted cell apoptosis and increased the relative activities of caspase-3 and caspase-9, eliciting no change in caspase-8 activity.
To conclude, the results of the present study demonstrated that the expression of ZEB2-AS1 is upregulated in human lung cancer in vivo and in vitro. Knockdown of ZEB2-AS1 in lung cancer A549 and NCI-H292 cell lines inhibited cell proliferation, whereas overexpression of ZEB2-AS1 in lung cancer H-125 and H1975 cells inhibited cell apoptosis, indicating that ZEB2-AS1 may serve as a prognostic factor for the diagnosis and treatment of patients with lung cancer in the clinic.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Pikor LA, Ramnarine VR, Lam S, Lam WL. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer. 2013;82:179–189. doi: 10.1016/j.lungcan.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 3.Youlden DR, Cramb SM, Baade PD. The international epidemiology of lung cancer: Geographical distribution and secular trends. J Thorac Oncol. 2008;3:819–831. doi: 10.1097/JTO.0b013e31818020eb. [DOI] [PubMed] [Google Scholar]
- 4.Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: Epidemiology, etiology, and prevention. Clin Chest Med. 2011;32:605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 6.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 8.Rinn JL. lncRNAs: Linking RNA to chromatin. Cold Spring Harb Perspect Biol. 2014;6:a018614. doi: 10.1101/cshperspect.a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Qiu M, Xu Y, Mao Q, Wang J, Dong G, Xia W, Yin R, Xu L. Differentially expressed protein-coding genes and long noncoding RNA in early-stage lung cancer. Tumour Biol. 2015;36:9969–9978. doi: 10.1007/s13277-015-3714-6. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham TR, Zhau HE, Odero-Marah VA, Osunkoya AO, Kimbro KS, Tighiouart M, Liu T, Simons JW, O'Regan RM. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008;68:2479–2488. doi: 10.1158/0008-5472.CAN-07-2559. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Tilló E, de Barrios O, Siles L, Amendola PG, Darling DS, Cuatrecasas M, Castells A, Postigo A. ZEB1 promotes invasiveness of colorectal carcinoma cells through the opposing regulation of uPA and PAI-1. Clin Cancer Res. 2013;19:1071–1082. doi: 10.1158/1078-0432.CCR-12-2675. [DOI] [PubMed] [Google Scholar]
- 14.Lan T, Chang L, Wu L, Yuan Y. Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in hepatocellular carcinoma. Mol Med Rep. 2016;14:4606–4612. doi: 10.3892/mmr.2016.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Devesa SS, Bray F, Vizcaino AP, Parkin DM. International lung cancer trends by histologic type: Male: Female differences diminishing and adenocarcinoma rates rising. Int J Cancer. 2005;117:294–299. doi: 10.1002/ijc.21183. [DOI] [PubMed] [Google Scholar]
- 18.Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K, et al. Cancer survival in China, 2003–2005: A population-based study. Int J Cancer. 2015;136:1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 19.Song N, Liu H, Ma X, Zhang S. Placental growth factor promotes ovarian cancer cell invasion via ZEB2. Cell Physiol Biochem. 2016;38:351–358. doi: 10.1159/000438635. [DOI] [PubMed] [Google Scholar]
- 20.Yi X, Shi S, Li X, Zhao L. Expression and clinical significance of ZEB2 and E-cadherin in nasopharyngeal carcinoma. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2015;29:1648–1651. (In Chinese) [PubMed] [Google Scholar]
- 21.Li J, Yuan J, Yuan X, Zhao J, Zhang Z, Weng L, Liu J. MicroRNA-200b inhibits the growth and metastasis of glioma cells via targeting ZEB2. Int J Oncol. 2016;48:541–550. doi: 10.3892/ijo.2015.3267. [DOI] [PubMed] [Google Scholar]
- 22.Pang X, Huang K, Zhang Q, Zhang Y, Niu J. miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor. Oncol Rep. 2015;34:3272–3279. doi: 10.3892/or.2015.4321. [DOI] [PubMed] [Google Scholar]
- 23.Kerr JF, Wyllie AH, Currie AR. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dejean LM, Martinez-Caballero S, Manon S, Kinnally KW. Regulation of the mitochondrial apoptosis-induced channel, MAC, by BCL-2 family proteins. Biochim Biophys Acta. 2006;1762:191–201. doi: 10.1016/j.bbadis.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Wajant H. The Fas signaling pathway: More than a paradigm. Science. 2002;296:1635–1636. doi: 10.1126/science.1071553. [DOI] [PubMed] [Google Scholar]
- 26.Bejarano I, Espino J, Gonzalez-Flores D, Casado JG, Redondo PC, Rosado JA, Barriga C, Pariente JA, Rodríguez AB. Role of calcium signals on hydrogen peroxide-induced apoptosis in human myeloid HL-60 cells. Int J Biomed Sci. 2009;5:246–256. [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy KM, Ranganathan V, Farnsworth ML, Kavallaris M, Lock RB. Bcl-2 inhibits Bax translocation from cytosol to mitochondria during drug-induced apoptosis of human tumor cells. Cell Death Differ. 2000;7:102–111. doi: 10.1038/sj.cdd.4400597. [DOI] [PubMed] [Google Scholar]
- 28.Chen G, Goeddel DV. TNF-R1 signaling: A beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]