Abstract
Golgi Protein 73 (GP73) is a newly identified diagnostic and prognostic marker for liver cancer. GP73 is highly expressed in liver cancer tissues, however, the mechanism of its overexpression in tumors remains unknown. In the present study, the effect of hepatitis B virus (HBV) on GP73 expression was investigated in HepG2 cells, which are negative for HBV, and in HepG2.2.12 cells, which are integrated with HBV, using reverse transcription-quantitative polymerase chain reaction and western blot analysis. In addition, the cells were transfected with plasmid constructs overexpressing hepatitis B virus protein X (HBx), hypoxia-inducible factor (HIF)-1α, or HIF-2α in order to examine their roles in GP73 expression. The results demonstrated that HBV upregulated the expression of GP73 and HIF-2α in liver cancer cells. HIF-2α induced the expression of GP73 in HepG2 cells and was positively correlated with GP73 expression in liver cancer tissues. By contrast, HBx and HIF-1α did not induce GP73 expression in liver cancer cells. In summary, HBV may upregulate the expression of GP73 by activating the HIF-2α signaling pathway. The present results may illuminate the mechanism by which GP73 is overexpressed in liver cancer tissues.
Keywords: hepatitis B virus, Golgi protein 73, hepatitis B virus protein X, hypoxia-inducible factor-1α, hypoxia-inducible factor-2α, hepatocellular carcinoma
Introduction
In recent years, the rapid development of genetic technologies, proteomics, and tumor immunology has identified several new biomarkers for liver cancer (1). Golgi protein 73 (GP73) is one of the most promising serum markers for liver cancer diagnosis. GP73, also named Golgi phosphoprotein 2 (GOLPH2) and Golgi membrane protein 1 (GOLM1), is termed GP73 due to its molecular mass of 73 kDa in SDS-PAGE (2,3). It is a transmembrane protein in the Golgi apparatus and highly expressed in liver cancer tissues and patient serum. It has been predicted that GP73 may become a marker for early diagnosis of liver cancer with sensitivity superior to α-fetoprotein (AFP) (4,5).
Currently, the mechanism underlying high GP73 expression in liver cancer tissues remains unclear. Kladney et al (6) demonstrated that GP73 protein expression in liver cancer cells was significantly elevated during hepatitis B virus (HBV) replication, indicating that HBV replication may activate GP73 protein expression. Currently, the mechanism of HBV-induced GP73 protein overexpression is not clear. Because hepatitis B virus protein X (HBx), hypoxia-inducible factor (HIF)-1α and HIF-2α are important factors promoting liver cancer occurrence and development, it was postulated that upregulation of GP73 expression may be related to these three factors. Therefore, the present study investigated the role of HBx, HIF-1α and HIF-2α in inducing GP73 expression in liver cancer tissues.
Materials and methods
Clinicopathological patient data
All human studies were approved by the Human Ethics Committee of the Center Hospital of Huanggang (Huanggang, China). All hepatocellular carcinoma (HCC) patients included in the present study tested negative for hepatitis C virus and human immunodeficiency virus. All tissue samples were collected from patients prior to medical treatment. HBV-positive HCC tissues and their paired peritumoral tissues (n=52), and HBV-negative HCC tissues (n=10) were sectioned for immunohistochemical analysis. Tissues were collected from the Center Hospital of Huanggang (Huanggang, China) between June 2008 and June 2011. Peritumoral tissues were obtained at least 2 cm away from the primary tumor site. Clinical data and tumor characteristics are presented in Table I.
Table I.
Clinicopathological features of 52 patients with hepatocellular carcinoma.
| Variable | Patients |
|---|---|
| Mean age (range) | 42 (25–67) |
| Gender | |
| Male | 45 (86.5%) |
| Female | 7 (13.5%) |
| Cirrhosis | |
| Presence | 31 (59.6%) |
| Absence | 21 (40.4%) |
| Tumor size | |
| <5 cm | 36 (69.2%) |
| ≥5 cm | 16 (30.8%) |
| Vascular invasion | |
| Presence | 6 (11.5%) |
| Absence | 46 (88.5%) |
| Tumor number | |
| Single | 44 (84.6%) |
| Multiple | 8 (15.4%) |
| Tumor differentiation | |
| Well | 11 (21.2%) |
| Moderate | 28 (53.8%) |
| Poor | 13 (25.0%) |
Reagents and cell lines
Mouse anti-human GP73 monoclonal antibodies (sc-365817), mouse anti-human HIF-1α monoclonal antibodies (cat. no. sc-53546), mouse anti-human HIF-2α monoclonal antibodies (cat. no. s-13596), and mouse monoclonal antibodies raised against baculovirus-expressed recombinant HepBx (cat. no. sc-57760) were purchased from Santa Cruz Biotechnology, Inc., (Dallas, TX, USA). The human hepatoblastoma cell line HepG2 was purchased from American Type Culture Collection (Manassas, VA, USA). HepG2.2.15 cell line was kindly provided by Professor Yinping Lu (Department of Infectious Disease, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology). Cells were grown in 6-well plates with Dulbecco's modified Eagle's medium (DMEM; GE Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum (FBS), 2 mmol/l L-glutamine (all GE Healthcare Life Sciences) at 37°C in 5% CO2. HIF-1α-overexpressing plasmid, HIF-2α-overexpressing plasmid, and the corresponding control plasmid (pcDNA3.1), as well as HBx-overexpression plasmid and its control plasmid (pEGFP-N1) were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). HIF-2α small interfering (si)RNA (cat. no. sc-35316) and negative control siRNA (cat. no. sc-37007) were purchased from Santa Cruz Biotechnology, Inc.
HepG2 cells at 60–70% confluence were transfected with 2.0 µg plasmids or 100 pmol siRNA using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. Transfected cells were incubated at 37°C for 6 h, and were then cultured for a further 16 h with fresh DMEM medium containing 10% FBS (all GE Healthcare Life Sciences).
Immunohistochemistry
Immunohistochemical analysis was performed as previously described (7,8). The levels of GP73 protein were scored according to the number of cells exhibiting cytoplasmic staining using the classification system published by Sai et al (8). Cytoplasmic staining in <25% of tumor cells was considered low expression, while staining in >25% of tumor cells was considered high expression (8). The levels of HIF-2α protein were scored according to the number of cells exhibiting cytoplasmic and nuclear staining using the classification system published by Yang et al (7). Nuclear or cytoplasmic staining in <50% of tumor cells was considered low expression, while staining in >50% of tumor cells was considered high expression (7). Liver fibrosis was scored on a 0–4 scale according to the METAVIR scoring system (9).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting
The methods of Yang et al (10) were followed for these experiments. TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to isolate total RNA from 150–200 mg of 52 freshly frozen HCC tumors and their adjacent liver tissues. RNA extraction, cDNA synthesis, qPCR reactions, and western blotting were performed as previously reported (10). The primer sequences used in the current study are listed in Table II.
Table II.
Polymerase chain reaction primers and conditions.
| Gene | Primer sequence (5′-3′) | Temperature (°C) | Product size (bp) |
|---|---|---|---|
| GP73 | GTGGCCTGCATCATCGTCTT | 60.7 | 167 |
| CTGCTTCTCCAGCTCTCCCT | |||
| HIF-1α | CATCTCCATCTCCTACCCACA | 58.3 | 105 |
| CTTTTCCTGCTCTGTTTGGTG | |||
| HIF-2α | TCATGCGACTGGCAATCAGC | 61.3 | 141 |
| GTCACCACGGCAATGAAACC | |||
| HBx | CGTCCTTTGTCTACGTCCCG | 59.4 | 408 |
| AAGTTGCATGGTGCTGGTGA | |||
| β-actin | AGTTGCGTTACACCCTTTCTTGAC | 63.9 | 171 |
| GCTCGCTCCAACCGACTGC |
GP73, Golgi protein 73; HIF, hypoxia inducible factor; HBx, hepatitis B virus protein X.
Promoter analysis
The TRANSFAC software (http://www.gene-regulation.com/pub/programs.html#match) was used to identify potential HIF-2α binding sites in the promoter region of GP73 by following the software's instructions, and as previously reported (10).
Statistical analysis
Paired t-test statistical analyses were performed using SPSS software (version 20.0; IBM Corp., Armonk, NY, USA). Results are expressed as mean ± standard deviation of three independent experiments. Linear associations were evaluated using Spearman's correlation coefficients. P<0.05 was considered to indicate a statistically significant difference.
Results
GP73 is overexpressed in HCC tissues
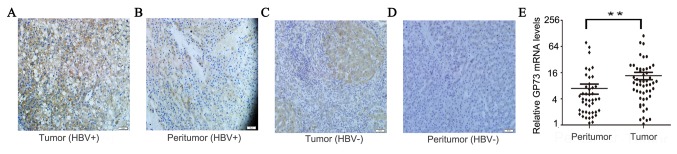
GP73 was overexpressed in HBV-positive HCC tissues, as evidenced by high expression of GP73 in 73.1% (38/52) of tumor tissues but only 36.5% (19/52) of peritumoral tissues (Fig. 1A and B). In HBV-negative HCC, GP73 was overexpressed in 40.0% (4/10) of tumor tissues, which was lower compared with HBV-positive HCC tissues (P=0.04; Fig. 1C and D). In peritumoral tissues, the GP73 level in patients of F2-4 groups which with significant fibrosis was significantly higher (54.5%; 12/22) compared with patients of F0-1 groups with no/minimal fibrosis (23.3%; 7/30), which was consistent with results from previously published studies (data not shown) (11). The positive GP73 staining was located in the cytoplasm (Fig. 1). In addition, the mRNA expression levels of GP73 were also demonstrated to be increased in tumor tissues compared with peritumoral normal tissues (Fig. 1E). In summary, these results demonstrated that both mRNA and protein expression levels of GP73 were increased in HBV-positive HCC tissues.
Figure 1.
Expression of GP73 in HCC tissues. Representative images of GP73 staining in (A) HBV-positive HCC tissues and (B) their matched peritumoral tissues. (C) HBV-negative HCC tissues and (D) their matched peritumoral tissues. (E) GP73 mRNA levels in HBV-positive HCC tissues and their matched peritumoral tissues. **P<0.01. GP73, Golgi protein 73; HCC, hepatocellular carcinoma; HBV, hepatitis B virus.
HBV enhances the expression of GP73
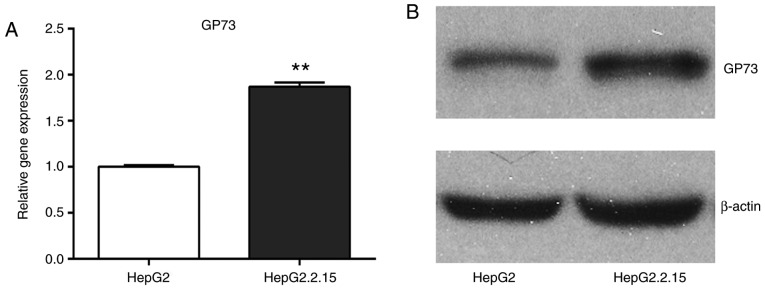
To investigate the mechanisms that lead to the high expression of GP73 in HCC tissues, first the effect of HBV on GP73 expression was examined because HBV is a common cause of liver cancer. The mRNA and protein expression levels of GP73 were compared between the HepG2 cells, which are negative for HBV, and the HepG2.2.15 cells, which were stably transfected with a complete HBV genome. The results demonstrated that GP73 mRNA and protein levels were significantly higher in the HepG2.2.15 cells compared with the HepG2 cells (Fig. 2). Therefore, it was hypothesized that HBV may be a positive regulator of GP73.
Figure 2.
HBV enhances the expression of GP73. (A) Reverse transcription-quantitative polymerase chain reaction analysis of GP73 mRNA expression levels in HepG2 cells and HepG2.2.15 cells. The GP73 expression levels were normalized to β-actin and expressed relative to the HepG2 cells. Results are expressed as the mean ± standard deviation of three independent experiments. **P<0.01. (B) Western blot analysis of GP73 protein expression in HepG2 cells and HepG2.2.15 cells. HBV, hepatitis B virus; GP73, Golgi protein 73.
HIF-2α is involved in HBV-induced upregulation of GP73
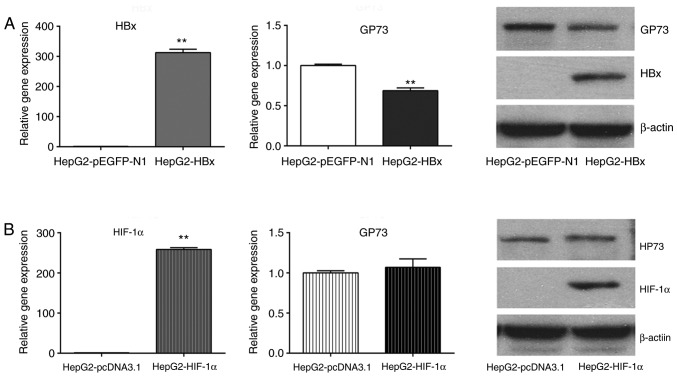
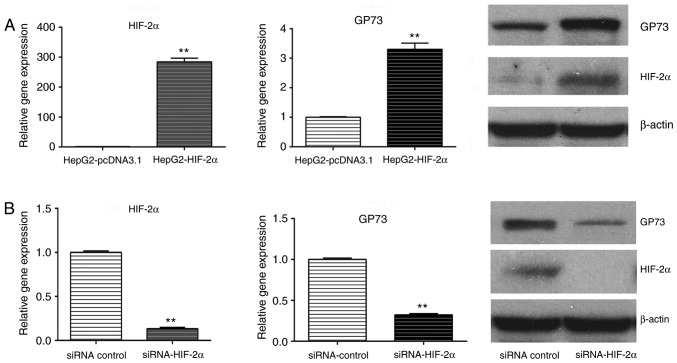
To further investigate the mechanism of HBV-mediated upregulation of GP73, the effect of HBx, HIF-1α and HIF-2α was examined on GP73 expression. The results demonstrated that transfection of HepG2 cells with a HBx-overexpressing plasmid reduced the mRNA and protein levels of GP73 (Fig. 3A), while transfection with a HIF-1α-overexpressing plasmid did not alter GP73 mRNA and protein levels (Fig. 3B). By contrast, transfection with the HIF-2α-overexpressing plasmid increased the levels of GP73 mRNA and protein (Fig. 4A). Similarly, GP73 mRNA and protein expression was downregulated following HIF-2α siRNA silencing (Fig. 4B). Using the TRANSFAC software, two potential HIF-2α binding sites were identified in the promoter region of GP73 (Fig. 5). HIF-2α may enhance the expression of GP73 through binding with the hypoxia response elements (CGTG) in its promoter region. Our previous research demonstrated that HBV could activate HIF-2α signaling (12). The present results indicate that HIF-2α may be involved in HBV-induced upregulation of GP73.
Figure 3.
Effect of HBx and HIF-1α on GP73 expression. (A) HepG2 cells were transfected with either empty vector control (pEGFP-N1) or a HBx-overexpressing plasmid. HBx overexpression was confirmed by RT-qPCR, and GP73 levels were examined by RT-qPCR and western blotting. (B) HepG2 cells were transfected with either empty vector control (pcDNA3.1) or a HIF-1α-overexpressing plasmid. HIF-1α overexpression was confirmed by RT-qPCR, and GP73 levels were examined by RT-qPCR and western blotting. β-actin was used as internal control. Results are expressed as the mean ± standard deviation of three independent experiments. **P<0.01. HBx, hepatitis B virus protein X; HIF, hypoxia-inducible factor; GP73, Golgi protein 73; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Figure 4.
Effect of HIF-2α on GP73 expression. (A) HepG2 cells were transfected with either empty vector control (pcDNA3.1) or a HIF-2α-overexpressing plasmid. HIF-2α overexpression was confirmed by RT-qPCR, and GP73 levels were examined by RT-qPCR and western blotting. (B) HepG2 cells were transfected with either a negative control or HIF-2α-targeting siRNA, and HIF-2α silencing was confirmed by RT-qPCR. GP73 levels were examined by RT-qPCR and western blotting. β-actin was used as internal control. Results are expressed as the mean ± standard deviation of three independent experiments. **P<0.01. HIF, hypoxia-inducible factor; GP73, Golgi protein 73; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA.
Figure 5.

Nucleotide sequence of the promoter region of the human GP73 gene (accession no. NM177937.2). The numbers indicate the base location relative to the start site of the first exon that is labelled with an up-arrow. Letters in grey boxes represent putative binding sites for HIF-2α and hypoxia response elements (CGTG) that were identified by TRANSFAC analysis. GP73, Golgi protein 73.
HIF-2α and GP73 expression are positively correlated in HCC tissues
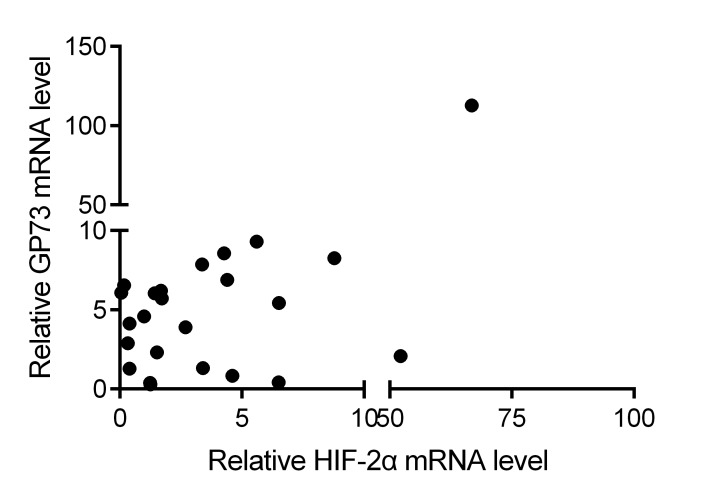
Further, the correlation between the expression levels of HIF-2α and GP73 was determined in HCC tissues. The results demonstrated that HIF-2α mRNA expression was positively correlated with GP73 mRNA expression in HCC tissues (r=0.427; P<0.001 by Spearman's correlation; Fig. 6). These data further indicate that HIF-2α may be a factor that contributes to the upregulation of GP73 in liver cancer.
Figure 6.
Overlay scatters indicate the correlation between GP73 mRNA levels and HIF-2α mRNA levels analyzed by real-time PCR.
Discussion
In 2000, Kladney et al (13) discovered the Golgi apparatus protein GP73 while studying the pathogenesis of human giant-cell hepatitis. Subsequent studies demonstrated that GP73 expression is closely related to liver diseases. The majority of liver cells in normal tissues do not express GP73, and only a small number of liver cells express GP73 at low levels. However, GP73 expression is significantly increased in hepatitis and hepatocirrhosis tissues, with the highest expression in liver cancer tissues (13). In the present study, it was demonstrated that the expression of GP73 in liver cancer tissues was significantly higher compared with adjacent normal tissues, which is consistent with previous reports.
GP73 is an integral membrane protein in the cis Golgi capsule. Under pathogenic states, GP73 can be released from the cis Golgi capsule and localize to the cytoplasm and cell surface (2,6). The 55th amino acid in GP73 can be enzymatically digested by proprotein convertases, including furin protease, transported through endosomes, and released into the extracellular space and the blood stream as sGP73 (2,6). It has been discovered that sGP73 is highly expressed in the serum from patients with specific types of tumors, including liver cancer, cholangiocarcinoma, and lung adenocarcinoma. Thus, sGP73 can be regarded as a serum marker for early liver cancer diagnosis (2,6).
Currently, a definitive mechanism for the high expression of GP73 in hepatitis, hepatocirrhosis, and liver cancer remains unclear. The ex vivo experiments have confirmed the upregulation of GP73 following HBV infection in liver cancer cells (6). Therefore, it was hypothesized that HBV infection, the main inducer of hepatitis, hepatocirrhosis and liver cancer, may have a role in inducing GP73 expression.
HBx is a major HBV coding protein and serves a significant role in HBV-induced hepatocarcinogenesis and increased serum AFP level in liver cancer patients (14–18). However, in the present study, founder results demonstrated that overepxression of HBx decreased rather than increased GP73 expression in liver cancer cells. The GP73 mRNA and protein levels in the HBx-transfected HepG2 cells were 69±2 and 51±5% of the levels in the control group, respectively, indicating that HBV-mediated GP73 upregulation did not occur through the HBx pathway.
Next, the potential roles of HIF-1α and HIF-2α in inducing GP73 expression were explored. HIF-1α and HIF-2α are important transcription factors in HCC (19–22). Under normoxic conditions, the HIF-α subunits are hydroxylated in key proline residues by the von Hippel-Lindau (VHL) protein complex, followed by proteasome degradation. Under hypoxic conditions, the low level of oxygen inhibits the activity of hydroxylase, leading to the stabilization of HIF-α subunits (19–22). Accumulated HIF-α subunits translocate to nuclei and dimerize with HIF-1β to form a functional transcription factor capable of DNA binding at the hypoxia response elements (HREs) and the transcriptional activation of target genes that are involved in cell survival, tumor angiogenesis, metastasis, and resistance to radiation and chemotherapy (19–22). In addition to the hypoxic microenvironment, the stability of HIF-α proteins is modulated by HBV, as our previous study demonstrated that HBV induced HIF-2α expression by its encoded protein HBx though binding to pVHL and activating the nuclear factor (NF)-κB signaling pathway (12). Although HIF-1α and HIF-2α have similar structure and common HREs, their target genes are different (23–25). HIF-1 preferentially induces genes that encode glycolytic enzymes, such as phosphofructokinase and lactate dehydrogenase A. By contrast, HIF-2 induces genes that are involved in invasion, including the matrix metalloproteinase (MMP) 2 and 13, and the stem cell factor OCT-3/4 (23–25). In the present study, it was demonstrated that HIF-2α, but not HIF-1α, induced the expression of GP73 in liver cancer cells. GP73 expression was upregulated following HIF-2α overexpression and was downregulated following HIF-2α silencing. In addition, two potential HIF-2α binding sites were identified in the promoter region of GP73 by bioinformatics analysis. In human HCC tissues, the expression of HIF-2α was positively correlated with GP73 expression. These results indicated that HIF-2α can activate GP3 expression in liver cancer cells.
The present study identified that HBV induced GP73 expression in liver cancer cells through HIF-2α signaling activation. HIF-2α may upregulate GP73 expression in liver cancer cells by directly binding to and activating its promoter. These results provided a possible mechanism explaining GP73 upregulation in liver cancers. More detailed investigations on the molecular mechanisms of GP73 expression in the future will contribute to understanding its functional implication in diseases and evaluating its role as a novel biomarker for liver cancer.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81402041) and the 2016–2017 Special Fund for the Medical Colleges of Health and Family Planning of Hubei Province (grant no. WJ2016-YZ-10).
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tunissiolli NM, Castanhole-Nunes MMU, Biselli-Chicote PM, Pavarino EC, da Silva RF, da Silva RC, Goloni-Bertollo EM. Hepatocellular carcinoma: A comprehensive review of biomarkers, clinical aspects, and therapy. Asian Pac J Cancer Prev. 2017;18:863–872. doi: 10.22034/APJCP.2017.18.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao G, Dong F, Xu X, Hu A, Hu Y. Diagnostic value of serum Golgi protein 73 for HBV-related primary hepatic carcinoma. Int J Clin Exp Pathol. 2015;8:11379–11385. [PMC free article] [PubMed] [Google Scholar]
- 3.Ismail MM, Morsi HK, Abdulateef NA, Noaman MK, Abou El-Ella GA. Evaluation of prothrombin induced by vitamin K absence, macrophage migration inhibitory factor and Golgi protein-73 versus alpha fetoprotein for hepatocellular carcinoma diagnosis and surveillance. Scand J Clin Lab Invest. 2017;77:175–183. doi: 10.1080/00365513.2017.1286684. [DOI] [PubMed] [Google Scholar]
- 4.Dai M, Chen X, Liu X, Peng Z, Meng J, Dai S. Diagnostic value of the combination of Golgi protein 73 and alpha-fetoprotein in hepatocellular carcinoma: A meta-analysis. PLoS One. 2015;10:e0140067. doi: 10.1371/journal.pone.0140067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waidely E, Al-Yuobi AR, Bashammakh AS, El-Shahawi MS, Leblanc RM. Serum protein biomarkers relevant to hepatocellular carcinoma and their detection. Analyst. 2016;141:36–44. doi: 10.1039/C5AN01884F. [DOI] [PubMed] [Google Scholar]
- 6.Kladney RD, Cui X, Bulla GA, Brunt EM, Fimmel CJ. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology. 2002;35:1431–1440. doi: 10.1053/jhep.2002.32525. [DOI] [PubMed] [Google Scholar]
- 7.Yang SL, Liu LP, Niu L, Sun YF, Yang XR, Fan J, Ren JW, Chen GG, Lai PB. Downregulation and pro-apoptotic effect of hypoxia-inducible factor 2 alpha in hepatocellular carcinoma. Oncotarget. 2016;7:34571–34581. doi: 10.18632/oncotarget.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sai W, Wang L, Zheng W, Yang J, Pan L, Cai Y, Qiu L, Zhang H, Wu W, Yao D. Abnormal expression of Golgi protein 73 in clinical values and their role in HBV-related hepatocellular carcinoma diagnosis and prognosis. Hepat Mon. 2015;15:e32918. doi: 10.5812/hepatmon.32918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR cooperative study group. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 10.Yang SL, Ren QG, Zhang T, Pan X, Wen L, Hu JL, Yu C, He QJ. Hepatitis B virus X protein and hypoxiainducible factor-1α stimulate Notch gene expression in liver cancer cells. Oncol Rep. 2017;37:348–356. doi: 10.3892/or.2016.5211. [DOI] [PubMed] [Google Scholar]
- 11.Wei H, Li B, Zhang R, Hao X, Huang Y, Qiao Y, Hou J, Li X, Li X. Serum GP73,a marker for evaluating progression in patients with chronic HBV infections. PLoS One. 2013;8:e53862. doi: 10.1371/journal.pone.0053862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu JL, Liu LP, Yang SL, Fang X, Wen L, Ren QG, Yu C. Hepatitis B virus induces hypoxia-inducible factor-2α expression through hepatitis B virus X protein. Oncol Rep. 2016;35:1443–1448. doi: 10.3892/or.2015.4480. [DOI] [PubMed] [Google Scholar]
- 13.Kladney RD, Bulla GA, Guo L, Mason AL, Tollefson AE, Simon DJ, Koutoubi Z, Fimmel CJ. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249:53–65. doi: 10.1016/S0378-1119(00)00136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong F, You H, Tang R, Zheng K. The regulation of proteins associated with the cytoskeleton by hepatitis B virus X protein during hepatocarcinogenesis. Oncol Lett. 2017;13:2514–2520. doi: 10.3892/ol.2017.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou Z, Xu X, Fu X, Tao S, Zhou J, Liu S, Tan D. HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am J Cancer Res. 2017;7:845–856. [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Dong Z, Yang P, Wang X, Jin G, Yu H, Chen L, Li L, Tang L, Bai S, et al. Hepatitis B virus X protein stimulates high mobility group box 1 secretion and enhances hepatocellular carcinoma metastasis. Cancer Lett. 2017;394:22–32. doi: 10.1016/j.canlet.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Zhu M, Li W, Lu Y, Dong X, Lin B, Chen Y, Zhang X, Guo J, Li M. HBx drives alpha fetoprotein expression to promote initiation of liver cancer stem cells through activating PI3K/AKT signal pathway. Int J Cancer. 2017;140:1346–1355. doi: 10.1002/ijc.30553. [DOI] [PubMed] [Google Scholar]
- 18.Zhu M, Lu Y, Li W, Guo J, Dong X, Lin B, Chen Y, Xie X, Li M. Hepatitis B virus X protein driven alpha fetoprotein expression to promote malignant behaviors of normal liver cells and hepatoma cells. J Cancer. 2016;7:935–946. doi: 10.7150/jca.13628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geis T, Döring C, Popp R, Grossmann N, Fleming I, Hansmann ML, Dehne N, Brüne B. HIF-2alpha-dependent PAI-1 induction contributes to angiogenesis in hepatocellular carcinoma. Exp Cell Res. 2015;331:46–57. doi: 10.1016/j.yexcr.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Guo XF, Wang AY, Liu J. HIFs-MiR-33a-Twsit1 axis can regulate invasiveness of hepatocellular cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:3011–3016. [PubMed] [Google Scholar]
- 21.Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478:1067–1073. doi: 10.1016/j.bbrc.2016.08.065. [DOI] [PubMed] [Google Scholar]
- 22.Lee JH, Hur W, Hong SW, Kim JH, Kim SM, Lee EB, Yoon SK. ELK3 promotes the migration and invasion of liver cancer stem cells by targeting HIF-1α. Oncol Rep. 2017;37:813–822. doi: 10.3892/or.2016.5293. [DOI] [PubMed] [Google Scholar]
- 23.Yu T, Tang B, Sun X. Development of inhibitors targeting hypoxia-inducible factor 1 and 2 for cancer therapy. Yonsei Med J. 2017;58:489–496. doi: 10.3349/ymj.2017.58.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin D, Wu J. Hypoxia inducible factor in hepatocellular carcinoma: A therapeutic target. World J Gastroenterol. 2015;21:12171–12178. doi: 10.3748/wjg.v21.i42.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]