Abstract
Esophageal cancer is one of the most common types of malignant tumors located within the digestive system, with >50% of esophageal cancer cases worldwide occurring in China. Recent studies have demonstrated that long non-coding RNAs (lncRNAs) are frequently dysregulated in cancer; however, few lncRNAs have been characterized in esophageal squamous cell carcinoma (ESCC). In the present study, a novel lncRNA, SET-binding factor 2 (SBF2) antisense RNA1 (SBF2-AS1) was exhibited in ESCC. Expression levels of SBF2-AS1 in ESCC and adjacent non-cancerous tissues were detected using the reverse transcription-quantitative polymerase chain reaction. SBF2-AS1 was knocked down, and proliferation, migration, invasion, apoptosis and the cell cycle were examined in ESCC cells. Results identified that SBF2-AS1 was significantly upregulated in ESCC compared with adjacent non-cancerous tissues (fold increase, 8.82; P=0.023). The SBF2-AS1 expression level was significantly increased in patients who had a smoking (9.927 vs. 4.507; P=0.030) and/or drinking (10.938 vs. 4.232; P=0.032) history. Patients with a large tumor size exhibited increased SBF2-AS1 expression (≥4 vs. <4 cm, 14.898 vs. 5.435; P=0.018). Patients with advanced ESCC exhibited increased upregulation of SBF2-AS1 [tumor-node-metastasis (TNM) I–II vs. TNM III–IV, 1.302 vs. 15.475; P<0.01]. SBF2-AS1 was also silenced using small interfering RNA. Cell proliferative and invasive ability were significantly inhibited (P<0.05) following SBF2-AS1 silencing, the cell cycle was arrested in the G2 phase; however, there was no significant difference in the proportion of apoptotic cells. Gene Set Enrichment Analysis revealed that genes associated with cell cycle biological processes, including the cancer suppressor gene cyclin-dependent kinase 1A (CDKN1A), were significantly associated with SBF2-AS1 in ESCC tissues. Further validation confirmed that CDKN1A expression levels were increased in ECA-109 cells following SBF2-AS1 silencing. The results of the present study demonstrate that SBF2-AS1 is significantly upregulated in ESCC, and that silencing of SBF2-AS1 inhibits the proliferative and invasive ability of ESCC cells. SBF2-AS1 may be a novel biomarker and therefore a potential therapeutic target for ESCC.
Keywords: esophageal squamous cell carcinoma, long non-coding RNA, SET-binding factor 2 antisense RNA1, proliferation
Introduction
Human esophageal cancer (EC) is the third most common digestive system malignancy worldwide, and is associated with a high mortality rate (1). EC is also one of the most common malignant tumors in China, ranking third and fifth in causing male and female malignant tumors, respectively (2). Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EA) are the primary subtypes of EC; however, in China, ESCC accounts for >90% of total cases (3). Despite improved development of multimodal techniques including surgery, chemotherapy and radiotherapy, the survival rate of patients with ESCC remains poor owing to the high incidence of local invasion and distant metastasis (3,4). Therefore, it is imperative to seek novel therapeutic targets and effective biomarkers for ESCC.
Long non-coding RNAs (lncRNA) are non-protein-coding transcripts >200 nucleotides (5,6). On the basis of the proximity to nearby coding genes, lncRNAs are categorized into 5 subclasses: Sense, antisense, intergenic, bidirectional and intronic (7). lncRNAs are involved in every aspect of cellular processes, including cellular development, modulation of apoptosis, metastasis and parental imprinting (8–10). Emerging evidence suggests that the dysregulation of lncRNAs is linked to the development and metastasis in several types of cancer, including EC (11–13). The importance of lncRNAs in ESCC carcinogenesis has been recently evidenced. Hundreds of ESCC-associated lncRNAs were identified, some of which could be used as biomarkers for diagnosing or prognosing ESCC (14–16). ESCC-associated lncRNAs remain to be fully understood but research has progressed in this field: POU class 3 homeobox 3 (POU3F3) promotes the malignant progression of ESCC by binding the enhancer of zeste 2 polycomb repressive complex 2 subunit protein to promote the methylation of POU3F3 (17); colon cancer associated transcript 1 (non-protein coding) could modulate the histone methylation of the promoter of sprouty RTK signaling antagonist 4 by binding polycomb repressive complex 2 (PRC2) and suppressor of variegation 3–9 homolog 1 in the nucleus and promoted the expression of downstream target gene homeobox B13 by adsorbing microRNA-7 in the cytoplasm (18).
In a previous study, the novel lncRNA SET-binding factor 2 (SBF2) antisense RNA1 (SBF2-AS1) was identified and was demonstrated to be upregulated in non-small cell lung cancer (NSCLC), and was also able to promote proliferation of NSCLC cells (19). SBF2-AS1, located at the 11p15.1 locus, is a 2,708-nt antisense RNA to SBF2. A previous study identified that SBF2-AS1 was able to bind with PRC2 and guided PRC2 to the promoter of CDKN1A and therefore decreased CDKN1A expression (19). However, the exact role of SBF2-AS1 in ESCC remains unclear. Therefore, the investigated the expression and biological function(s) of SBF2-AS1 in ESCC in vitro were investigated in the present study.
Materials and methods
Patients and tissue samples
All primary ESCC tissues and paired normal tissues were collected from patients who had undergone surgery at the Department of Thoracic Surgery, Cancer Institute of Jiangsu Province (Nanjing, China) between May 2013 and May 2014. No patients received radiotherapy or chemotherapy prior to surgical resection. Experienced pathologists confirmed all tumors and paired normal tissues, and clinical/pathological characteristics of each patient were collected. Written informed consent was obtained from all patients and the Ethics Committee of Cancer Institute of Jiangsu Province approved the present study.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analyses
Total RNA was extracted from tissues or cultured esophageal cell lines (HesEpiC; ScienCell Research Laboratories, Inc., San Diego, CA, USA), KYSE-410 (Department of Radiology of The First Affiliated Hospital of Nanjing Medical University, Jiangsu, Nanjing, China), ECA-109 (Shanghai Institutes for Biological Science, Shanghai, China) and TE-1 (Shanghai Institutes for Biological Science, Shanghai, China) with TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. The concentration and A260/280 ratio were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.). A 500 ng amount of total RNA was reverse-transcribed in a final volume of 10 µl using random primers under standard conditions using the Prime Script RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol; the reverse transcription reactions were performed at 25°C for 5 min, followed by 42°C for 60 min and 70°C for 5 min. qPCR was performed using the SYBR Green Master Mix (Thermo Fisher Scientific, Inc.) with 0.5 µl cDNA on a Veriti 96 Well Thermal Cycler (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol, to determine relative levels of SBF2-AS1. GAPDH and β-actin were used as internal controls. Reaction mixtures were incubated at 95°C for 10 min, 40 cycles of 95°C for 15 sec, 60°C for 1 min and 72°C for 10 min. The fold changes of individual genes were calculated using the 2−ΔΔCq method (20). The following primer pairs were used for qPCR: SBF2-AS1 forward, 5′-CACGACCCAGAAGGAvGTCTAC-3′, and reverse, 5′-CCCGGTACCTTCCTGTCATA-3′; cyclin-dependent kinase 1A (CDKN1A) forward, 5′-AGACCATGTGGACCTGTCACTG-3′, and reverse, 5′-GTTTGGAGTGGTAGAAATCTGTC-3′; GAPDH forward, 5′-CGCTCTCTGCTCTCCTGTTC-3′, and reverse, 5′-ATCCGTTGACTCCGACCTTCAC-3′; β-actin forward, 5′-CGCTCTCTGCTCCTCCTGTTC-3′, and reverse, 5′-ATCCGTTGACTCCGACCTTCAC-3′.
Nuclear mass separation experiment
The subcellular localization of SBF2-AS1 was detected using the PARIS™ kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Firstly, the cells were washed with ice-cold PBS twice. Subsequently, 1 ml ice-cold cell lysis solution [containing 20 mmol/l HEPES (pH 7.5), 10 mmol/l KCl, 1.5 mmol/l MgCl2, 1 mmol/l EGTA, 1 mmol/l EDTA, 1 mmol/l DTT, 0.1 mmol/l PMSF and 250 mmol/l sucrose] was added to the cells and 3×106 cells were harvested with a cell scraper on ice. The cell lysate was incubated on ice for 5 min. After centrifuging at 500 × g for 5 min at 4°C, the precipitate containing the nuclear RNA was isolated from the supernatant containing the cytoplasmic RNA. Finally, the supernatant was removed. The precipitate was washed with PBS twice and resuspended using Nuclei EZ storage buffer (Thermo Fisher Scientific, Inc.).
Cell culture
KYSE-410 cells were donated by the Department of Radiology of The First Affiliated Hospital of Nanjing Medical University (Jiangsu, Nanjing, China). ECA-109 and TE-1 cells were purchased from Shanghai Institutes for Biological Science (Shanghai, China). Normal human esophageal epithelial cell lines (HEsEpiC) were purchased from ScienCell Research Laboratories, Inc. KYSE-410 cells and HEECs were grown in RPMI-1640 medium (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). ECA-109 and TE-1 cells were grown in Dulbecco's modified Eagle's medium (DMEM; Nanjing KeyGen Biotech Co., Ltd.), supplemented with penicillin (80 U/ml)-streptomycin (0.08 mg/ml) and 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.). All cells were grown at 37°C in a humidified 5% CO2 atmosphere.
siRNA transfection
ESCC cells (70–80% confluence in 6-well plates) were transfected with 100 nM specific small interfering RNA (siRNA) or 100 nM negative control (NC) siRNA using Lipofectamine® RNA interference (RNAi) MAX (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following transfection, cells were cultured for 48 h at 37°C in a humidified 5% CO2 atmosphere. The following siRNA primer sequences were used: SBF2-AS1 sense (204 site), 5′-CAGAAGGAGUCUACUGCUAAG-3′, and antisense, 5′-UAGCAGUAGACUCCUUCUGGG-3′, SBF2-AS1 sense (1021 site), 5′-GCAAGCCUGCAUGGUACAUTT-3′, and antisense, 5′-AUGUACCAUGCAGGCUUGCTT-3′.
Cell proliferation assay
Cell proliferation was assayed using the Cell Counting Kit-8 (CCK-8) assay (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. Transfected cells were plated in 96-well plates (3,000 cells/well). Cell proliferation was measured every 24 h in accordance with the manufacturer's protocol. In brief, 10 µl CCK-8 solution was added to each well and incubated for 2 h at 37°C, and each solution was measured spectrophotometrically at 450 nm as previously described (21).
Clonogenic assay
A total of 100 transfected cells were placed into a fresh 6-well plate and maintained in 2 ml medium containing 10% FBS, replacing the medium every 3 days. After 2 weeks, cells were immobilized with 4% paraformaldehyde at room temperature for 30 min and stained with 0.1% crystal violet at room temperature for 15 min. Visible colonies were counted using light microscopy and a fluorescence microscope (magnification, ×40) (22).
In vitro cell migration and invasion assays
ECA-109 cells transfected with 100 nM si-SBF2-AS1 or si-NC was harvested 24 h after transfection. For migration assays, transfected cells (3×105) were plated in the upper chamber of the Transwell assay inserts (EMD Millipore, Billerica, MA, USA) containing 200 µl serum-free DMEM with a membrane without Matrigel (8-mm pores). Lower chambers were filled with DMEM containing 10% FBS. After 24 h of incubation at 37°C in a 5% CO2 incubator, cells on the filter surface were fixed with 100% methanol at room temperature for 15 min and stained with 0.1% crystal violet at room temperature for 15 min, and images were captured using an Axio Vert.A1 microscope (magnification, ×100; Carl Zeiss AG, Oberkochen, Germany). Cell numbers were calculated using five random fields for each chamber. For invasion assays, transfected cells (5×105) were plated in the upper chamber with a Matrigel-coated membrane (BD Biosciences, Franklin Lakes, NJ, USA) in 500 µl serum-free DMEM with 750 µl 10% FBS in DMEM in the lower chambers. Invasion function was determined using the number of transmigrated cells following 48 h incubation at 37°C in a 5% CO2 incubator.
Flow cytometric analysis
Transfected cells were harvested after 24 h of incubation at 37°C in a 5% CO2 incubator. ECA-109 cells were stained with Annexin V and propidium iodide (PI) using Annexin V-fluorescein isothiocyanate/PI apoptosis detection kits (BD Biosciences) and examined using flow cytometry (FACScan; BD Biosciences). Cells used for cell cycle analysis were stained with propidium oxide using the Cycle Test Plus DNA reagent kit (BD Biosciences) and analyzed using FACScan (BD Biosciences) as described previously (23).
Western blot assay
Proteins were extracted 48 h after transfection. Cells in each well were placed in 100 µl lysis buffer containing 1 mM sodium vanadate, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 2 mM leupeptin, 2 mM aprotinin and 2 mM pepstatin on ice for 30 min. Proteins (60 µl/lane) were resolved using a 10% gel and SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. Membranes were blocked with 5% non-fat dry milk in Tris-buffered saline for 2 h at room temperature. Primary antibodies, including anti-CDKN1A (cat. no. sc-6246; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-cyclin E (cat. no. 20808; 1:1,000; CST Biological Reagents Co., Ltd., Shanghai, China) and anti-β-actin (cat. no. ab8227; 1:1,000; Abcam, Cambridge, UK) were used. Membranes were incubated with primary antibodies at room temperature overnight. Following washing with Tris-buffered saline, the membrane was incubated with goat anti-rabbit immunoglobulin G horseradish peroxidase-conjugated secondary antibody (cat. no. ab6721; 1:10,000; Abcam) for 2 h at room temperature. Finally, blots were visualized using enhanced chemiluminescence detection (Thermo Fisher Scientific, Inc.), as described previously (24).
Bioinformatics analysis
Gene Set Enrichment Analysis (GSEA) was performed using GSEA software (version 2.2.4; Broad Institute, Inc., Massachusetts Institute of Technology, Cambridge, MA, USA), and gene signatures were downloaded from the Molecular Signatures Database (MSigDB, version 4; Broad Institute, Inc.). The MSigDB collects various types of gene sets and included 1,320 canonical pathways derived from the pathway databases including BioCarta, Kyoto Encyclopedia of Genes and Genomes, Pathway Interaction Database and Reactome. The GSE53622 dataset was downloaded from the Gene Expression Omnibus (GEO) and only ESCC tumor samples were used to perform GSEA.
Statistical analysis
Differences between groups were assessed by a paired two-tailed Student's t-test. One-way analysis of variance followed by the Bonferroni post-hoc test or the nonparametric Kruskal-Wallis test was applied to assess the relationship between circPRKCI expression and other characteristics. The strength of the association between continuous variables was tested with the Spearman correlation. All statistical analyses were performed using SPSS 20 software (Abbott Laboratories, Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Association between SBF2-AS1 expression and clinical characteristics
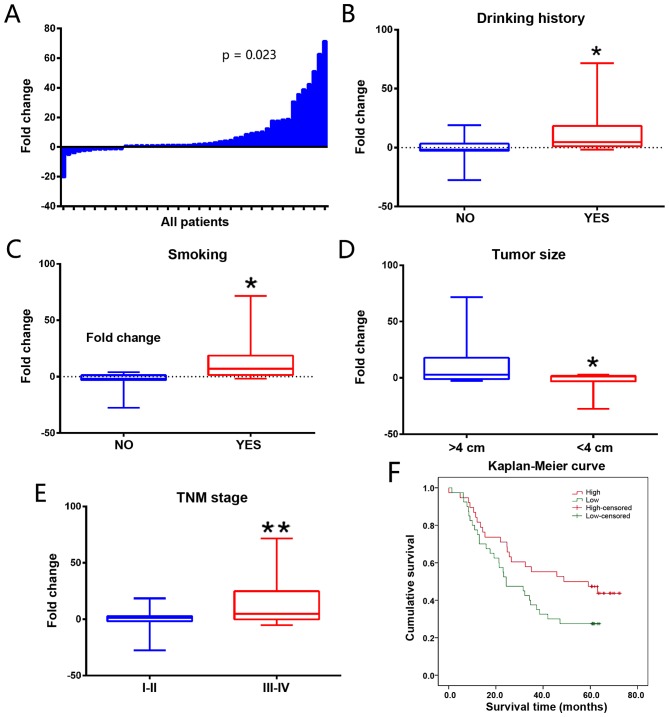
SBF2-AS1 expression levels were detected in 51 paired primary ESCC and adjacent non-cancerous tissues using RT-qPCR. Results demonstrated that SBF2-AS1 was significantly overexpressed in ESCC, with a mean upregulation of 8.82-fold (P=0.023; Fig. 1A). As demonstrated in Table I, drinking history, smoking history, tumor size and TNM stage (P<0.05) were significantly associated with the expression of SBF2-AS1. SBF2-AS1 expression level was also increased in patients with a history of drinking (9.927 vs. 4.507; P=0.030; Fig. 1B) and smoking (10.938 vs. 4.232, P=0.032; Fig. 1C). Patients with a large tumor size had an increased SBF2-AS1 expression (≥4 vs. <4 cm, 14.898 vs. 5.435; P=0.018; Fig. 1D). Patients with ESCC exhibiting a TNM stage of III–IV demonstrated an increased expression level of SBF2-AS1 compared with patients with ESCC exhibiting with a TNM stage of I–II (TNM I–II vs. TNM III–IV, 1.302 vs. 15.475; P<0.01; Fig. 1E). The association between SBF2-AS1 expression level and clinical characteristics is presented in Table I. The association between SBF2-AS1 expression and survival rate of ESCC patients was also evaluated. In an online cohort of 60 ESCC patients, patients with increased SBF2-AS1 expression levels had poorer survival rates compared with those with lower expression levels of SBF2-AS1 [hazard ratio, 1.31; 95% confidence interval (CI), 1.090–1.568; P=0.016; Fig. 1F].
Figure 1.
Analysis of SBF2-AS1 expression in ESCC tissues and associated clinical parameters. (A) SBF2-AS1 was detected in 51 pairs of ESCC tissues using RT-qPCR. Levels of SBF2-AS1 in ESCC tissues were significantly increased compared with those in non-tumor tissues (P=0.023). SBF2-AS1 was upregulated in groups with (B) drinking history (P=0.030), (C) smoking history (P=0.032), (D) tumor size (P=0.018), and (E) TNM stage (P<0.001). (F) Kaplan-Meier estimator curve demonstrates the survival rate of patients with different SBF2-AS1 expression levels. The difference between groups was statistically significant (P=0.016; Mantel-Cox test). *P<0.05, **P<0.01 vs. control groups. Results are presented as the mean ± standard error of the mean. TNM, tumor-node-metastasis; SBF2-AS1, SET-binding factor 2 antisense RNA1; ESCC, esophageal squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Table I.
Association between SBF2-AS1 expression and clinicopathological characteristics.
| Characteristic | Number of patients | Percentage | Fold-change | P-value |
|---|---|---|---|---|
| Age, years | 0.298 | |||
| <60 | 17 | 33.3 | 10.897 | |
| >60 | 34 | 66.7 | 7.78 | |
| Sex | 0.270 | |||
| Male | 35 | 80.3 | 9.861 | |
| Female | 16 | 19.7 | 7.232 | |
| Smoker status | 0.032 | |||
| Yes | 18 | 41.2 | 10.938 | |
| No | 33 | 58.8 | 4.232 | |
| Drinking history | 0.030 | |||
| Yes | 32 | 78.4 | 9.927 | |
| No | 19 | 21.6 | 4.507 | |
| Family history of ESCC | 0.222 | |||
| Yes | 4 | 7.8 | 3.804 | |
| No | 47 | 92.2 | 9.179 | |
| Location of the tumor within the esophagus | 0.102 | |||
| Upper | 9 | 17.6 | 11.612 | |
| Middle | 30 | 58.8 | 9.782 | |
| Lower | 12 | 23.5 | 4.058 | |
| Tumor size, cm | 0.018 | |||
| <4 | 37 | 72.5 | 14.898 | |
| >4 | 14 | 27.5 | 5.435 | |
| Tumor differentiation (according to the differences between tumor cells and normal esophageal epithelial cells) | 0.101 | |||
| Poor (differences, large) | 30 | 58.8 | 13.712 | |
| Moderate (differences, moderate) | 7 | 13.7 | 9.747 | |
| Well (differences, minor) | 14 | 27.5 | 8.163 | |
| Lymph node metastasis | 0.727 | |||
| N0-N1 | 18 | 35.3 | 7.504 | |
| N2-N3 | 33 | 64.7 | 9.422 | |
| TNM stage | <0.010 | |||
| I–II | 22 | 43.1 | 1.208 | |
| III–IV | 29 | 56.9 | 14.485 |
TNM, tumor-node-metastasis; ESCC, esophageal squamous cell carcinoma.
Expression characteristics of SBF2-AS1 in ESCC cell lines
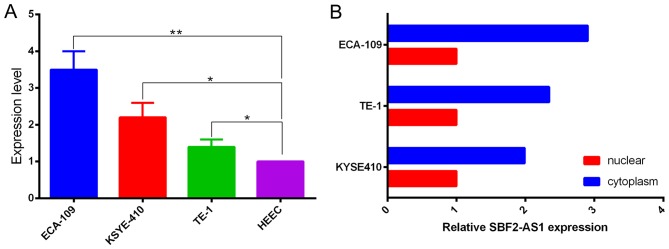
Prior to the functional study of SBF2-AS1 in vitro, expression in ESCC cell lines was measured using RT-qPCR. When normalized to HEEC cell lines, results demonstrated that the expression level of SBF2-AS1 was upregulated in the ESCC cells analyzed (Fig. 2A). The expression level of SBF2-AS1 was highest in ECA-109 cells, therefore ECA-109 was selected as the experimental cell line. Confirmation of the subcellular localization of SBF2-AS1 was achieved by measuring expression levels of SBF2-AS1 in nuclear and cytoplasmic fractions of ECA-109, TE-1 and KYSE410 cell lines. Results demonstrated that SBF2-AS1 was primarily distributed within the cytoplasm (Fig. 2B).
Figure 2.
Expression of SBF2-AS1 in ESCC cell lines demonstrates that SBF2-AS1 promotes proliferation of ESCC cell lines in vitro. (A) Upregulation of SBF2-AS1 in ESCC cells. (B) Localization of SBF2-AS1 in ESCC cells. The majority of SBF2-AS1 was located within the cytoplasm through the nuclear mass separation experiment. *P<0.05, **P<0.01. Results are presented as the mean ± standard error of the mean. SBF2-AS1, SET-binding factor 2 antisense RNA1; ESCC, esophageal squamous cell carcinoma.
SBF2-AS1 promotes the proliferation of ESCC cell lines in vitro
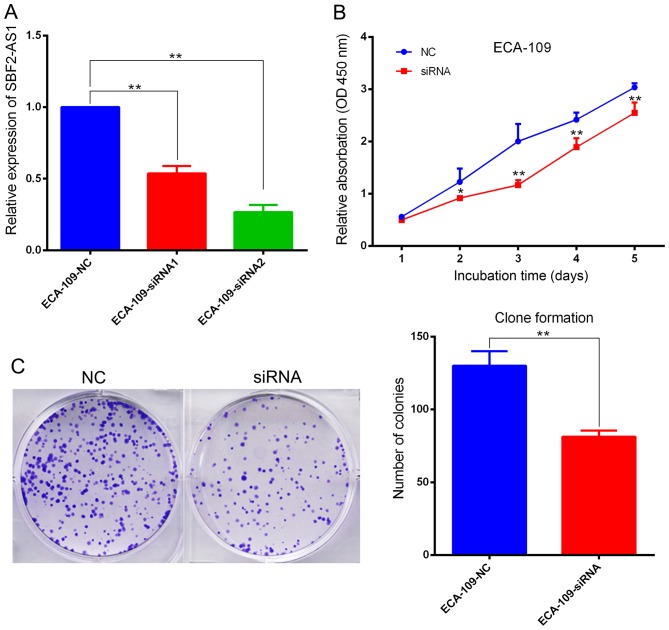
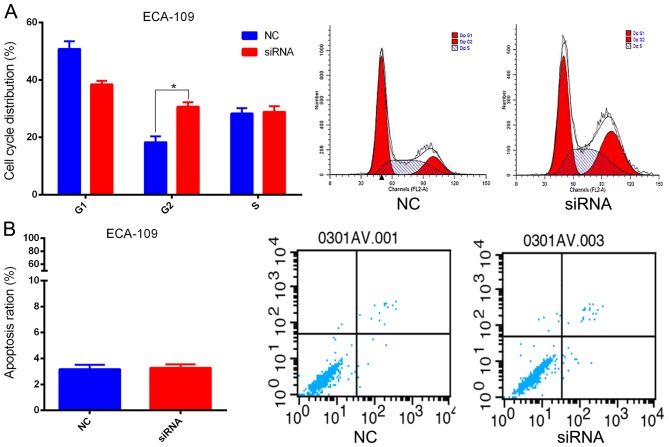
Investigation of the biological function(s) of SBF2-AS1 was achieved by targeting SBF2-AS1 at the 204 (siRNA1) and 1021 (siRNA2) sites using two designer siRNAs as described previously (14). Results indicate that siRNA2 had improved inhibition efficacy, therefore it was used in subsequent cell function experiments (Fig. 3A). ECA-109 cells were transfected with si-SBF2-AS1 or NC siRNA. At 36 h post-treatment, SBF2-AS1 expression was effectively knocked down (Fig. 3A). The CCK-8 assay demonstrated that knockdown of SBF2-AS1 significantly inhibited (P<0.01) cell proliferation in the ECA-109 cell line (Fig. 3B). Furthermore, the colony formation assay revealed that siRNA treatment significantly inhibited (P=0.007) the ability of ESCC cells to form colonies (Fig. 3C). Results generated using flow cytometry demonstrated that cell cycle progression of ECA109 cells was arrested at the G2 phase following treatment with siRNA-2 (Fig. 4A). However, silencing of SBF2-AS1 did not affect the proportion of apoptotic cells (Fig. 4B).
Figure 3.
SBF2-AS1 promotes proliferation of ESCC cell lines in vitro. (A) si-SBF2-AS1 demonstrates increased inhibition efficiency. (B) CCK-8 kit demonstrated that silencing of SBF2-AS1 inhibited cell proliferation of ECA-109 cells. (C) Clone formation complements data generated from the CCK-8 kit. *P<0.05, **P<0.01 vs. NC group. Results are presented as the mean ± standard error of the mean. SBF2-AS1, SET-binding factor 2 antisense RNA1; ESCC, esophageal squamous cell carcinoma; CCK-8, Cell Counting Kit-8; si, small interfering; NC, negative control.
Figure 4.
SBF2-AS1 alters the cell cycle of ESCC cell lines in vitro. (A) ECA-109 cells transfected with si-SBF2-AS1 were blocked in the G2 phase. (B) Flow cytometric analyses indicated that si-SBF2-AS1 did not affect apoptosis. *P<0.05. Results are presented as the mean ± standard error of the mean. si, small interfering; SBF2-AS1, SET-binding factor 2 antisense RNA1; ESCC, esophageal squamous cell carcinoma; NC, negative control.
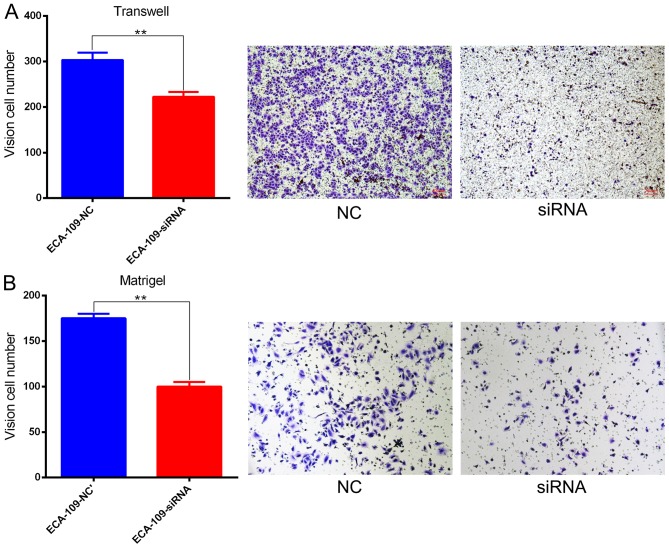
Effect of SBF2-AS1 on ESCC cell migration and invasion
Migration and invasion are important cancer progression events. The potential for SBF2-AS1 to have a direct role in cell invasion within ESCC was investigated using Transwell and Matrigel assays. Results demonstrated that siRNA treatment significantly impaired (P=0.006) migration capacity compared with NC (Fig. 5A). Knockdown of SBF2-AS1 also inhibited the invasive ability of ESCC cells (Fig. 5B).
Figure 5.
SBF2-AS1 promotes invasion and metastasis of ESCC cell lines in vitro. (A) Following silencing of SBF2-AS1, ESCC cells demonstrated a decrease in migratory ability. (B) Knocking down SBF2-AS1 inhibited the invasion of ECA-109 cells. **P<0.01. Results are presented as the mean ± standard error of the mean. SBF2-AS1, SET-binding factor 2 antisense RNA1; ESCC, esophageal squamous cell carcinoma; NC, negative control; siRNA, small interfering RNA.
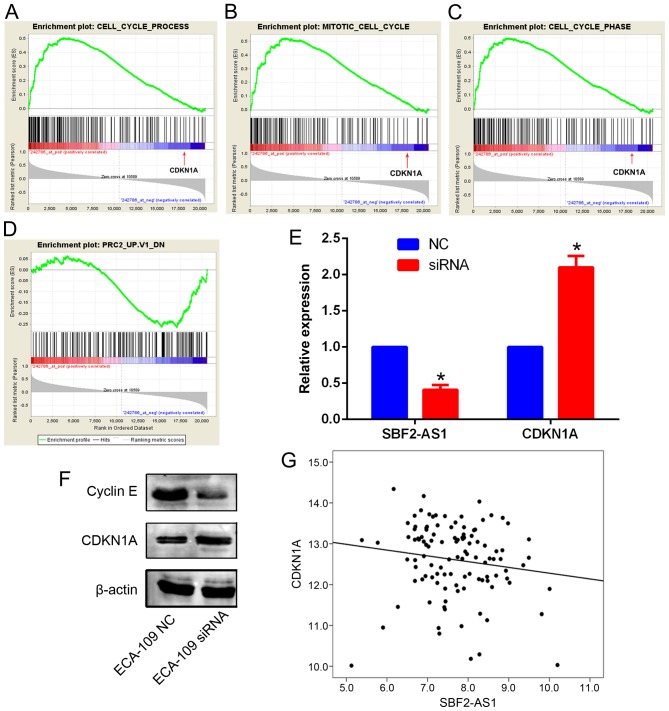
Silencing SBF2-AS1 upregulates CDKN1A expression in ESCC
To explore the potential mechanism of SBF2-AS1 in ESCC GSEA analysis in an ESCC cohort was performed. According to the GSEA results, SBF2-AS1 was significantly associated with genes involved in Gene Ontology items of cell cycle (P=0.032), the mitotic phase of cell cycle (P=0.018) and the cell cycle phase (P=0.027). Notably, CDKN1A was identified in all three gene sets, and was negatively associated with SBF2-AS1 (Fig. 6A-C). In addition, GSEA also revealed that genes that were regulated by PRC2 were significantly associated with SBF2-AS1 (Fig. 6D; P<0.05). Thus, it was hypothesized that SBF2-AS1 may also downregulate CDKN1A expression in ESCC cells. Expression levels of CDKN1A were investigated in ECA-109 cells following the silencing of SBF2-AS1. Results demonstrated that mRNA and protein levels of CDKN1A were significantly increased (P<0.05) in ECA-109 cells (Fig. 6E and F). Western blot analysis further confirmed that cyclin E (CCNE1), a known downstream gene of CDKN1A signaling, was also downregulated following SBF2-AS1 silencing (Fig. 6F). Furthermore, in the third-party ESCC cohort (GSE53622), SBF2-AS1 and CDKN1A were negatively correlated (R=−0.179; P<0.01; Fig. 6G). Therefore, these results indicate that SBF2-AS1 was able to decrease CDKN1A expression in ESCC.
Figure 6.
GSEA results indicate that genes associated with (A) cell cycle process, (B) mitotic cell cycle and (C) cell cycle phase are positively enriched with SBF2-AS1, and among these genes, CDKN1A was significantly negatively associated with SBF2-AS1 (indicated by the red arrow). Genes regulated by PRC2 were negatively associated with SBF2-AS1. (D) Silencing of SBF2-AS1 increased the CDKN1A expression level in ECA-109 cells. (E) Cyclin E expression was decreased following SBF2-AS1 silencing, which is consistent with the (F) upregulation of CDKN1A. (G) SBF2-AS1 was negatively associated with CDKN1A in the GSE53622 dataset (P<0.01 vs. NC groups). *P<0.05. GSEA, Gene Set Enrichment Analysis; SBF2-AS1, SET-binding factor 2 antisense RNA1; CDKN1A, cyclin-dependent kinase 1A; PRC2, Polycomb repressive complex 2; NC, negative control; siRNA, small interfering RNA.
Discussion
Numerous lncRNAs have been identified as critical regulators of gene expression, and serve several key roles in tumorigenesis; therefore, there is a growing interest in the role of lncRNAs in cancer (25). Multiple types of lncRNA have been identified as associated with the development and progression of ESCC, including pist, colon cancer-associated transcript 2, homeobox transcript antisense RNA and metastasis-associated lung adenocarcinoma transcript 1 (26–31). SBF2-AS1 is a novel lncRNA transcribed on chromosome 11p15.1 and, previously, its function and molecular mechanism in NSCLC has been reported (14). However, the function and molecular mechanism of SBF2-AS1 in ESCC remains unclear. In the present study, results demonstrated that the expression level of SBF2-AS1 was associated with smoking and drinking history, tumor size and TNM stage. Previous studies confirmed that smoking and drinking are significantly poor prognostic risk factors of ESCC (32,33). Chang et al (34) revealed that within a Chinese population sex, age, smoking and drinking status are associated with an increased risk of ESCC in 9,805 cases and 10,493 controls. Tai et al (35) reported that women who consumed >158 g alcohol/week had a 20.58-fold increased risk (95% CI, 1.72–245.62; P=0.02) of developing ESCC compared with those who had never consumed alcohol, following adjustment for other covariates. Taghavi et al (36) demonstrated that cigarette smoking was significantly associated with p53 overexpression in ESCC cases (odds ratio, 3.64; 95% CI, 1.32–10.02; P=0.010). Therefore, it is suggested that SBF2-AS1 may be associated with the occurrence and development of EC.
The biological function of SBF2-AS1 in vitro was also analyzed using siRNA-mediated silencing in ECA109 cell lines. Results demonstrated that following the knockdown of SBF2-AS1, ESCC proliferative, migratory and invasive ability was significantly suppressed. Flow cytometric analysis revealed that the cell cycle was arrested in the G2 phase following knockdown of SBF2-AS1; however, the proportion of apoptotic cells was not affected. Evidence indicates that SBF2-AS1 may function as an oncogenic gene in ESCC. In previous studies, it was reported that SBF2-AS2 was able to bind to suppressor of zeste 12 homolog (SUZ12), a core component of PRC2 (19,37–39). The enrichment of SUZ12 subsequently induces the inhibition of trimethylation of histone 3 Lys27 in the promoter region of CDKN1A, and therefore downregulates the expression of CDKN1A (19). Bioinformatical analysis in the present study demonstrated that CDKN1A expression was negatively associated with SBF2-AS1. Furthermore, GSEA revealed that genes that were regulated by PRC2 were also significantly associated with SBF2-AS1. Therefore, the results of the present study confirm that CDKN1A was negatively regulated by SBF2-AS1 at the RNA and protein levels. Nuclear and cytoplasmic fraction data also demonstrated that SBF2-AS1 was distributed in the nucleus and cytoplasm, which indicates that SBF2-AS1 may serve multiple functions at the transcriptional and post-transcriptional levels. According to our previous study (19), SBF2-AS1 may also transcriptionally downregulate the expression of CDKN1A via SUZ12 binding within the nucleus, as confirmed by the present study. CDKN1A, a cyclin-dependent kinase inhibitor, serves an important role in cell cycle arrest, and is therefore regarded as a tumor suppressor gene (40). It is also reported that CCNE1, a known downstream target gene of CDKN1A, was downregulated following SBF2-AS1 silencing; however, the potential post-transcriptional mechanism(s) of SBF2-AS1 require further investigation.
To conclude, the results of the present study demonstrate that SBF2-AS1 is significantly upregulated in ESCC, and silencing SBF2-AS1 inhibits the proliferative and invasive ability of ESCC cells. SBF2-AS1 may be a novel biomarker in the diagnosis of ESCC, and a potential therapeutic target of ESCC; however, further research in this area is required.
Acknowledgements
The present study was funded by the Natural Science Foundation of China (grant nos. 81372321, 81572261, 81472200, 81501977 and 81702377), The Innovation Capability Development Project of Jiangsu Province (grant no. BM2015004) and the Human Resource Summit Grant of Jiangsu Province (grant nos. WSN-023 and WSN-040).
Glossary
Abbreviations
- ESCC
esophageal squamous cell carcinoma
- EA
esophageal adenocarcinoma
- ncRNA
non-coding RNA
- lncRNA
long non-coding RNA
- SBF2-AS1
SET-binding factor 2 antisense RNA 1
- CCK-8
Cell Counting Kit-8
- PRC2
Polycomb repressive complex 2
- CDKN1A
cyclin-dependent kinase inhibitor 1A
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clini. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 4.Kimura H, Konishi K, Arakawa H, Oonishi I, Kaji M, Maeda K, Yabushita K, Tsuji M, Miwa A. Number of lymph node metastases influences survival in patients with thoracic esophageal carcinoma: Therapeutic value of radiation treatment for recurrence. Dis Esophagus. 1999;12:205–208. doi: 10.1046/j.1442-2050.1999.00049.x. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalogue of human long non-coding RNAs: Analysis of their structure, evolution and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amaral PP, Dinger ME, Mattick JS. Non-coding RNAs in homeostasis, disease and stress responses: An evolutionary perspective. Brief Funct Genomics. 2013;12:254–278. doi: 10.1093/bfgp/elt016. [DOI] [PubMed] [Google Scholar]
- 11.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 12.Tong YS, Zhou XL, Wang XW, Wu QQ, Yang TX, Lv J, Yang JS, Zhu B, Cao XF. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med. 2014;12:233. doi: 10.1186/s12967-014-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao Y, Wu W, Shi F, Dalmolin RJ, Yan M, Tian F, Chen X, Chen G, Cao W. Prediction of long noncoding RNA functions with co-expression network in esophageal squamous cell carcinoma. BMC Cancer. 2015;15:168. doi: 10.1186/s12885-015-1179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao W, Wu W, Shi F, Chen X, Wu L, Yang K, Tian F, Zhu M, Chen G, Wang W, et al. Integrated analysis of long noncoding RNA and coding RNA expression in esophageal squamous cell carcinoma. Int J Genomics. 2013;2013:480534. doi: 10.1155/2013/480534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Shi X, Yang W, Lu Z, Wang P, Chen Z, He J. Transcriptome profiling of lncRNA and co-expression networks in esophageal squamous cell carcinoma by RNA sequencing. Tumour Biol. 2016;37:13091–13100. doi: 10.1007/s13277-016-5227-3. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang E, Han L, Yin D, He X, Hong L, Si X, Qiu M, Xu T, De W, Xu L, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:1–3101. doi: 10.1093/nar/gkw1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv J, Qiu M, Xia W, Liu C, Xu Y, Wang J, Leng X, Huang S, Zhu R, Zhao M, et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:75. doi: 10.1186/s13046-016-0352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 21.Qiu M, Xu Y, Wang J, Zhang E, Sun M, Zheng Y, Li M, Xia W, Feng D, Yin R, Xu L. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:e1858. doi: 10.1038/cddis.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Sun Q, Chen C, Yin R, Huang X, Wang X, Shi R, Xu L, Ren B. ZYG11A serves as an oncogene in non-small cell lung cancer and influences CCNE1 expression. Oncotarget. 2016;7:8029–8042. doi: 10.18632/oncotarget.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q, Yin R, Xu L. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol. 2015;36:5523–5528. doi: 10.1007/s13277-015-3220-x. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Zhang Z, Qiu M, Hu J, Fan X, Wang J, Xu L, Yin R. Glypican-5 is a novel metastasis suppressor gene in non-small cell lung cancer. Cancer Lett. 2013;341:265–273. doi: 10.1016/j.canlet.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 26.Wei G, Luo H, Sun Y, Li J, Tian L, Liu W, Liu L, Luo J, He J, Chen R. Transcriptome profiling of esophageal squamous cell carcinoma reveals a long noncoding RNA acting as a tumor suppressor. Oncotarget. 2015;6:17065–17080. doi: 10.18632/oncotarget.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Xu Y, He C, Guo X, Zhang J, He C, Zhang L, Kong M, Chen B, Zhu C. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2015;111:834–839. doi: 10.1002/jso.23888. [DOI] [PubMed] [Google Scholar]
- 28.Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, Zhou GZ, Cao G, Jin L, Xie HW, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 29.Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ, Chen YB, Zhang Y, Jia WH. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675–1682. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Wu Z, Mei Q, Guo M, Fu X, Han W. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saeki H, Ohno S, Araki K, Egashira A, Kawaguchi H, Ikeda Y, Morita M, Kitamura K, Sugimachi K. Alcohol consumption and cigarette smoking in relation to high frequency of p53 protein accumulation in oesophageal squamous cell carcinoma in the Japanese. Br J Cancer. 2000;82:1892–1894. doi: 10.1054/bjoc.1999.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin S, Wang X, Huang C, Liu X, Zhao J, Yu IT, Christiani DC. Consumption of salted meat and its interactions with alcohol drinking and tobacco smoking on esophageal squamous-cell carcinoma. Int J Cancer. 2015;137:582–589. doi: 10.1002/ijc.29406. [DOI] [PubMed] [Google Scholar]
- 34.Chang J, Huang Y, Wei L, Ma B, Miao X, Li Y, Hu Z, Yu D, Jia W, Liu Y, et al. Risk prediction of esophageal squamous-cell carcinoma with common genetic variants and lifestyle factors in Chinese population. Carcinogenesis. 2013;34:1782–1786. doi: 10.1093/carcin/bgt106. [DOI] [PubMed] [Google Scholar]
- 35.Tai SY, Wu IC, Wu DC, Su HJ, Huang JL, Tsai HJ, Lu CY, Lee JM, Wu MT. Cigarette smoking and alcohol drinking and esophageal cancer risk in Taiwanese women. World J Gastroenterol. 2010;16:1518–1521. doi: 10.3748/wjg.v16.i12.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taghavi N, Biramijamal F, Sotoudeh M, Moaven O, Khademi H, Abbaszadegan MR, Malekzadeh R. Association of p53/p21 expression with cigarette smoking and prognosis in esophageal squamous cell carcinoma patients. World J Gastroenterol. 2010;16:4958–4967. doi: 10.3748/wjg.v16.i39.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 39.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long non-coding RNA required for cardiovascular lineage commitment. Cell. 2013;52:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]