Abstract
Functional inactivation of human runt-related transcription factor 3 (RUNX3) through mutation or epigenetic silencing has been well-documented in many cancerous entities. In addition to gene mutation and promoter hypermethylation, cytoplasmic mislocalization has emerged as another major manifestation of RUNX3 dysfunction in malignancies including breast, colorectal and gastric cancers. The aim of the present study was to investigate whether patients with non-small cell lung cancer (NSCLC) and different RUNX3 expression patterns would have different overall survival (OS), and the associations between different patterns of clinicopathological parameters and clinical outcome. Expressions of RUNX3 and Ki-67 were immunohistochemically detected in normal lung tissue (n=5) and surgically resected tissues from NSCLC patients (n=188). The optimal cutoff of RUNX3 was determined by X-tile software associated with their survival. Apoptotic index in cancerous tissue was evaluated using the terminal deoxynucleotidyl transferase mediated dUTP-biotin nick end labelling method. The prognostic significance of different expression patterns of RUNX3 was determined by means of Kaplan-Meier survival estimates and log-rank tests. It was revealed that loss of RUNX3 expression in NSCLC was correlated with a low cancerous apoptotic index (P<0.001), shorter OS and worse prognosis (P=0.0142), while no statistical difference of apoptotic index (P=0.73) or survival (P=0.3781) was determined between patient subgroups with different localization of RUNX3 expression, which was quite different from the situation demonstrated in other malignancies. In conclusion, loss of expression rather than cytoplasmic mislocalization of RUNX3 predicted worse outcome in NSCLC, which was quite different from what manifested in other cancer types, and thus, the underlying mechanism may deserve further investigation.
Keywords: runt-related transcription factor 3, cytoplasmic mislocalization, overall survival, prognosis, non-small cell lung cancer
Introduction
Lung cancer is still the leading cause of cancer-related deaths in China and worldwide (1,2). Contributing to approximately 85% of lung cancers and including histological types like adenocarcinoma (ADE), squamous cell carcinoma (SCC), large cell carcinoma and mixed histologies, non-small cell lung cancer (NSCLC) is still one of the major threats to human health. Diagnosis of it often occurs so late that nearly two-thirds have lost their opportunity for radical surgery. Against this relentlessly challenging clinical backdrop, establishment of useful biomarkers to facilitate early diagnosis, therapeutic planning as well as prognostic prediction turns out as great essential.
Human runt-related transcription factor (RUNX) family, encoded by genes RUNX1, RUNX2 and RUNX3, have been proved to play pivotal roles in activating and repressing the transcription of principle regulators of growth, survival and differentiation pathways through binding DNA via partnering with the cofactor, CBFβ/PEBP2β (core-binding factor beta subunit/polyomavirus enhancer-binding protein 2 beta subunit) (3–6). RUNX3, a remarkable biomarker firstly demonstrated in gastric epithelial disorders and cancer, has now emerged to exercise its mainly tumor suppressive activity while partly oncogenic role and interact with other signaling molecules in the context of carcinogenesis in a great variety of cancerous entities (7–11). Besides gastric tumors, RUNX3 had been reported to be epigenetically inactivated in a wide spectrum of malignancies, including bile duct cancer, breast cancer (BC), pancreatic cancer, colorectal cancer (CRC), prostate cancer, lymphoma, and lung cancer as well (12–19). Its protein deficiency, however, usually an aftermath of hemizygous deletion, promoter hypermethylation, histone modification as well as protein mislocalization, often lead to TGF-β1-induced cell growth inhibition and result in reduction of sensitivity to cellular apoptosis, ending up as tumorigenesis (9,20–22).
Several studies (23–25) had demonstrated that nuclear localization of RUNX3 expression was the authentic pattern for RUNX3 protein to exert its function as tumor suppressor gene (TSG), any patterns other than nuclear localization (positive in the nucleus and positive or negative in the cytoplasm) were defined as dysfunctioning patterns and rendered inactivated status of RUNX3, associated with worse outcome and shorter overall survival (OS) in BC, gastric cancer (GC) and CRC. Although importance of gene promoter hypermethylation concerning RUNX3 expression in NSCLC had ever been demonstrated in some studies, seldom report had been focused on clinical significance of patient survival with different expression patterns of RUNX3. We wondered if the situation was ever true like what it was in BC, GC and CRC, that NSCLC patients with nuclear localization of RUNX3 would definitely have better OS than those with non-nuclear localization, and if not, what the possible underlying mechanisms would be.
In the present study, we investigated the expression of RUNX3 and proliferation index Ki-67 immunohistochemically and evaluated the apoptotic index by means of TdT mediated dUTP-biotin nick end labelling (TUNEL) method in 188 NSCLCs and analyzed the patients' OS with different expression patterns of RUNX3 mentioned in other cancer types, and try to interpret its correlations within the clinicopathologic parameters and their clinical significance.
Materials and methods
Patients and tissue samples
Archival formalin-fixed, paraffin-embedded tissue sections from a series of 5 normal lung tissue and 188 patients (128 males and 60 females; age range: 36–78 years old) undergone surgery for NSCLC at the Department of Thoracic Surgery of Fujian Cancer Hospital and Fujian Medical University Cancer Hospital during 2010–2011 were selected. None had received chemo- or radio-therapy prior to tissue collection. The histopathologic features of cancerous specimens and TNM staging was determined according to the 8th AJCC guidelines for NSCLC. Grading of SCC into well-, moderately- and poorly-differentiated types is based on an assessment of the degree of differentiation and cellular atypia via hematoxylin and eosin (H&E) staining. Sheets of cells adopting a pavement-like architecture with prominent intercellular bridges characterize well-differentiated tumors. Keratinization is generally present and may lead to the formation of keratin ‘pearls’. More pronounced cytologic atypia, increased mitotic activity, and frequent areas of necrosis and hemorrhage characterized moderately differentiated tumors. While those with the appearance of anaplastic large cell or small cell as well as even more obviously cytologic atypia and increased mitotic activity is defined as ‘poorly differentiated SCC’. The final follow-up was February 5, 2017 and all patients were available of their survival data. Patients' survival data were censored if they were still alive or dead of disease other than lung cancer at the date of surveillance. The study protocol was approved by the Human Ethics Review Committee of Fujian Cancer Hospital and Fujian Medical University Cancer Hospital, and a signed informed consent was obtained from each patient.
Tissue microarray building
A fresh section was cut from each donor block, stained with H&E, and used as a guide to select the morphologically most-representative regions of the tumor from which to sample the individual core needle biopsies (26,27). A duplicate of 1.0 mm diameter cores were then punched from tumor areas of each donor tissue block and introduced into previously prepared recipient paraffin blocks (12×10 Matrix of 1 mm cores), after having made hosting holes in the blocks with a Tissue Microarrayer (Unitma Co., Ltd., Seoul, Korea). We constructed 4 recipient blocks with a maximum of 10×12 dots. With a microtome, 4 µm sections were cut from the TMA blocks and placed onto 3-aminopropyltriethoxysilane-coated glass slides to generate TMA slides for molecular analyses. Some sections were stained with H&E in a routine manner for histological examination.
Immunohistochemical detection of RUNX3 and Ki-67
Immunohistochemistry was performed with the indirect enzyme-labeled antibody method, as described previously (28,29). For detection of RUNX3, mouse anti-human monoclonal (2B3) RUNX3 antibody (dilution 1:500; Abcam, Cambridge, CA, USA) was used. For detection of Ki-67, rabbit anti-human monoclonal (30–9) anti-Ki-67 antibody purchased from Roche Applied Science (Penzberg, Germany) was used. TMA sections were deparaffinized with toluene and rehydrated in graded alcohols. After autoclaved for 15 min at 120°C in 10 mM citrate buffer (pH 6.0) for antigen retrieval, endogenous peroxidase was inactivated with 0.3% hydrogen peroxide in methanol for 15 min. The sections were then pre-incubated with 500 µg/ml normal goat IgG dissolved in 1% BSA in PBS (pH 7.4) for 1 h, reacted with primary antibodies for 16 h, washed with 0.075% Brij 35 in PBS, and then incubated with HRP-conjugated goat anti-mouse/rabbit (RUNX3/Ki-67) in 1% BSA in PBS for 1 h. After washing with 0.075% Brij 35 in PBS, the sites of HRP were visualized with DAB and H2O2. As a negative control, some sections were reacted with normal mouse IgG instead of the specific antibodies. The stained slides were analyzed under a laser scanning microscope (LSM 5 PASCAL; Carl Zeiss AG, Oberkochen, Germany).
TUNEL staining for apoptotic cells in NSCLCs
To identify nuclei with DNA strand breaks at a cellular level, TUNEL was performed according to the method of Gavrieli et al (30), with a slight modification. Paraffin sections (5 µm) were cut onto silane-coated glass slides, dewaxed with toluene, and rehydrated in an ethanol series. After washing with PBS, the sections were treated with 5 µg/ml of proteinase K in PBS at 37°C for 15 min. The sections were then rinsed once with deionized distilled water and incubated with TdT buffer (25 mM Tris/HCl buffer, pH 6.6, containing 0.2 M potassium cacodylate and 0.25 mg/ml BSA) alone at room temperature for 30 min. After incubation, the slides were reacted with 200 U/ml TdT dissolved in TdT buffer supplemented with 5 µM biotin-16-dUTP, 20 µM dATP, 1.5 mM CoCl2, and 0.1 mM dithiothreitol at 37°C for 1 h. The reaction was terminated by washing with 50 mM Tris/HCl buffer (pH 7.4) for 15 min. Endogenous peroxidase activity was inhibited by immersing the slides in 0.3% H2O2 in methanol at room temperature for 15 min. The signals were detected immunohistochemically with HRP-conjugated goat anti-biotin antibody, as described previously (28,31). For statistical analysis, more than 10,000 cancer cells/patient were counted, and the number of TUNEL-positive cells was expressed per 1,000 of the total cells (mean ± SEM). Data for different groups were compared for statistical difference using Student t-test. P<0.05 was considered to indicate a statistically significant difference.. The IHC and TUNEL scoring was performed by a single pathologist (Y. SHI) following consultation with another pathologist (G. CHEN) and in the absence of information on patient's outcome or pathology.
Statistical analysis
The X-tile software program (v3.6.1; Yale University School of Medicine, New Haven, CT, USA) as described previously (32) was used to determine the best cutoffs of RUNX3 by dichotomizing them into high and low expression subgroups. The SPSS v24.0 statistical software package (SPSS Inc, Chicago, IL, USA) was employed for all analyses. The association between tested marker and different clinicopathologic parameters of the patients, including age, gender, histology, ECOG PS, smoking status, differentiation SCC, lymphatic vessels invasion, nerve invasion, pleural invasion, vascular invasion, T-staging, N-staging, M-staging, TNM-staging, resectibility, depth of invasiveness, postoperative regional relapse, postoperative metastasis, serum CEA level and expression of Ki-67 were evaluated by Pearson's chi-square test, Yates' continuity correction chi-square test, Fisher's exact test or Spearman's rank correlation as appropriate. The Kaplan-Meier method with log-rank test was used to estimate probability of OS. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinicopathological data of patients
As shown in Table I, the diagnosis of 5 normal lung specimen was identically pneumothorax. A total of 3 males and 2 females were enrolled, with an average age of 60.2 years old. The NSCLC patient population included 128 males and 60 females and had a mean age of 58 years old. By histological classification, 75 cases were SCC and 113 were ADE. In the SCC group, the well-, moderately- and poorly differentiated numbers were 4, 57 and 14, respectively. In the ADE group, the predominant growth pattern numbers for lepidic, acinar, papillary, micropapillary and solid were 10, 75, 10, 1 and 17, respectively. As for ECOG performance score the number of <2 and ≥2 in ADE and SCC was 86 and 27, as well as 59 and 16, respectively. In ADE group, 56 were smokers and 57 were nonsmokers, while in SCC group, the number was 60 and 15. Serum CEA level was found abnormal in 55 ADE and 19 SCC patients. Pleural involvement was positive in 92 ADE and 44 SCC patients. Thirteen ADE and 4 SCC patients were positive of vascular invasion. Fifty-four ADE and 31 SCC patients were positive of lymphatic vessel involvement, while others all negative. Nerve invasion was positive in 1 case in ADE patients, while none was found in SCC patients. Number of negative, nuclear, cytoplasmic and whole-cell localization of RUNX3 expression in ADE was 35, 3, 38, 37 and 20, 7, 14, 34 in SCC respectively. Number of nuclear and non-nuclear expression in ADE was 40 and 73 while in SCC was 41 and 34. As for TNM-staging, the number of stage I through IV was 43, 21, 43 and 6 in ADE, and 25, 20, 30 and 0 in SCC. For T-staging, the number of stage 1a, 1b, 1c, 2a, 2b, 3 and 4 was 1, 7, 13, 58, 15, 12 and 7 in ADE, and 1, 6, 5, 24, 8, 14 and 17 in SCC. For N-staging, the number of stage from 0 through 3 was 54, 17, 31 and 11 in ADE, and 43, 15, 15 and 2 in SCC. For M-staging, the number of stage 0, 1a, 1b and 1c was 107, 3, 2 and 1 in ADE, and 75, 0, 0 and 0 in SCC. Twenty-three ADE and 14 SCC patients received postoperative radiotherapy. Fifty-seven ADE and 41 SCC patients received postoperative chemotherapy. Nineteen ADE and 13 SCC patients received postoperative chemoradiation. Postoperative regional relapse was present in 27 ADE and 20 SCC patients, while postoperative distant metastasis was found in 43 ADE and 24 SCC patients. Postoperative follow-up data were available for all patients, and the median follow-up time in ADE and SCC groups were 60.8 and 60.6 months, respectively.
Table I.
Clinicopathological parameters of patients.
| No. of cases (%) | ||
|---|---|---|
| Variables | Adenocarcinoma | SCC |
| Median age (years) | 57.5 | 59.0 |
| Median follow-up (months) | 60.8 | 60.6 |
| Age (years) | ||
| ≤58 | 64 (56.6) | 39 (52.0) |
| >58 | 49 (43.4) | 36 (48.0) |
| Gender | ||
| Male | 63 (55.8) | 65 (86.7) |
| Female | 50 (44.2) | 10 (13.3) |
| Histology | 113 (60.1) | 75 (39.9) |
| ECOG PS | ||
| <2 | 86 (76.1) | 59 (78.7) |
| ≥2 | 27 (23.9) | 16 (21.3) |
| Smoking status | ||
| Yes | 56 (49.6) | 60 (80.0) |
| No | 57 (50.4) | 15 (20.0) |
| Serum CEA (ng/ml) | ||
| <4.7 | 58 (51.3) | 56 (74.7) |
| ≥4.7 | 55 (48.7) | 19 (25.3) |
| Pleural invasion | ||
| Yes | 92 (81.4) | 44 (58.7) |
| No | 21 (18.6) | 31 (41.3) |
| Vascular invasion | ||
| Yes | 13 (11.5) | 4 (5.3) |
| No | 100 (88.5) | 71 (94.7) |
| Lymphatic vessels invasion | ||
| Yes | 54 (47.8) | 31 (41.3) |
| No | 59 (52.2) | 44 (58.7) |
| Nerve invasion | ||
| Yes | 0 (0) | 0 (0) |
| No | 113 (100) | 75 (100) |
| Localization of RUNX3 | ||
| Negative | 35 (31.0) | 20 (26.7) |
| Nucleus | 3 (2.7) | 7 (8.9) |
| Cytoplasm | 38 (33.6) | 14 (18.7) |
| Whole-cell | 37 (32.7) | 34 (45.3) |
| Nuclear RUNX3 expression | ||
| Yes | 40 (35.4) | 41 (54.7) |
| No | 73 (64.6) | 34 (45.3) |
| TNM staging | ||
| I | 43 (38.1) | 25 (33.3) |
| II | 21 (18.6) | 20 (26.7) |
| III | 43 (38.1) | 30 (40.0) |
| IV | 6 (5.2) | 0 (0) |
| T-staging | ||
| 1a | 1 (0.9) | 1 (1.2) |
| 1b | 7 (6.2) | 6 (8.0) |
| 1c | 13 (11.5) | 5 (6.7) |
| 2a | 58 (51.3) | 24 (32.0) |
| 2b | 15 (13.3) | 8 (10.7) |
| 3 | 12 (10.6) | 14 (18.7) |
| 4 | 7 (6.2) | 17 (22.7) |
| N-staging | ||
| 0 | 54 (47.8) | 43 (57.3) |
| 1 | 17 (15.0) | 15 (20.0) |
| 2 | 31 (27.4) | 15 (20.0) |
| 3 | 11 (9.8) | 2 (2.7) |
| M-staging | ||
| 0 | 107 (94.7) | 75 (100) |
| 1a | 3 (2.7) | 0 (0) |
| 1b | 2 (1.8) | 0 (0) |
| 1c | 1 (0.8) | 0 (0) |
| Differentiation (SCC) | ||
| Well | / | 4 (5.3) |
| Moderately | / | 57 (76.0) |
| Poorly | / | 14 (18.7) |
| Predominant growth pattern | ||
| Lepidic | 10 (8.8) | / |
| Acinar | 75 (66.4) | / |
| Papillary | 10 (8.8) | / |
| Micropapillary | 1 (1.0) | / |
| Solid | 17 (15.0) | / |
| PORT | ||
| Yes | 23 (20.4) | 14 (18.7) |
| No | 90 (79.6) | 61 (81.3) |
| Postoperative chemotherapy | ||
| Yes | 57 (50.4) | 41 (54.7) |
| No | 56 (49.6) | 34 (45.3) |
| Postoperative chemoradiation | ||
| Yes | 19 (16.8) | 13 (17.3) |
| No | 94 (83.2) | 62 (82.7) |
| Postoperative regional relapse | ||
| Yes | 27 (23.9) | 20 (26.7) |
| No | 86 (76.1) | 55 (73.3) |
| Postoperative distant metastasis | ||
| Yes | 43 (38.1) | 24 (32.0) |
| No | 70 (61.9) | 51 (68.0) |
ECOG PS, Eastern cooperative oncology group performance score; RUNX3, runt-related transcription factor 3; nuclear expression, including nuclear expression and both nuclear and cytoplasmic expression of RUNX3; non-nuclear expression, including negative and cytoplasmic expression of RUNX3; PORT, postoperative radiotherapy; SCC, squamous cell carcinoma; CEA, carcinoembryonic antigen.
Expression level of RUNX3 in NSCLC tissues and its correlation with clinicopathologic variables and OS
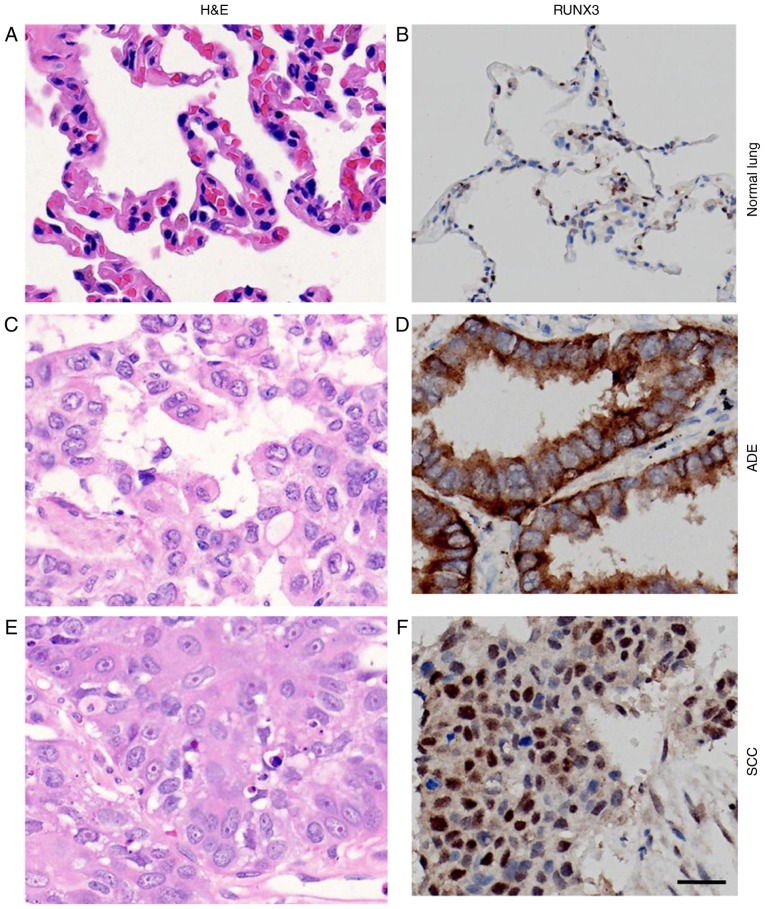
H&E staining of normal lung and NSCLC tissue demonstrated the nomaly of histological status used in our study (Fig. 1A, C, E). RUNX3 was only localized in nuclei of alveolar type II pneumocytes or Clara cells in normal lung tissue (33), while in NSCLCs, however, besides nuclei, it could also be localized in cytoplasm or both (Fig. 1B, D, F). First of all, we divided the cases into subgroups of high and low expression of RUNX3 based on the cutoff point determined by X-tile software, regardless of the localization of its expression and analyzed the correlation between expression levels and clinicopathologic parameters. Later on, we set out to analyze the association between localization of RUNX3 expression and the clinicopathologic parameters, still using the same cutoff to differentiate positive from negative expression. Calculated staining score of immunopositive cells for RUNX3 ranged from 0 to 12 in all tested tissues. According to the X-tile plots (Fig. 2A-C), we categorized the cancerous samples into low (IHC score ≤3) and high (IHC score >3) expression subgroups for RUNX3 based on a cutoff point determined by X-tile software related to survival time and status. As shown in Fig. 2D, the staining score of RUNX3 was significantly higher in normal lung tissue compared to SCC or ADE (12.00 vs. 6.93±0.55 vs. 6.38±0.48, **P<0.01). Among all 188 NSCLC cases, 55 (29%) were negative of RUNX3 expression and the other 133 were positive, either in nucleus, cytoplasm or both. Among the 133 positive individuals, 10 (5%) were localized in nucleus, 52 (28%) in cytoplasm, and 71 (38%) in both nucleus and cytoplasm (Fig. 3A-D, Table I).
Figure 1.
Immunostaining for RUNX3 in normal lung tissue and NSCLCs. (A, C and E) H&E staining and (B, D and F) immunostaining for RUNX3 in (A, B) normal lung tissue, (C, D) ADE tissues and (E, F) SCC tissues (magnification, ×400; scale bar=20 µm). RUNX3, runt-related transcription factor 3; NSCLC, non-small cell lung cancer; H&E, hematoxylin and eosin; ADE, adenocarcinoma; SCC, squamous cell carcinoma.
Figure 2.
(A) X-tile plots of RUNX3 expression for optimal cutoff point (3; P<0.05), which is demarcated by a black circle. (B) The cutoff point was used to separate low RUNX3 expression (blue) from high expression (gray) in the expression frequency histogram of the whole sample set (n=188). (C) Kaplan-Meier curve for evaluating the survival of sample subsets defined by RUNX3 expression <3 (blue line) and >3 (gray line). (D) RUNX3 expression differed in different lung tissues. Data are presented as the mean ± standard deviation. **P<0.01 vs. NOR. RUNX3, runt-related transcription factor 3; ADE, adenocarcinoma; SCC, squamous cell carcinoma; NOR, normal tissues.
Figure 3.
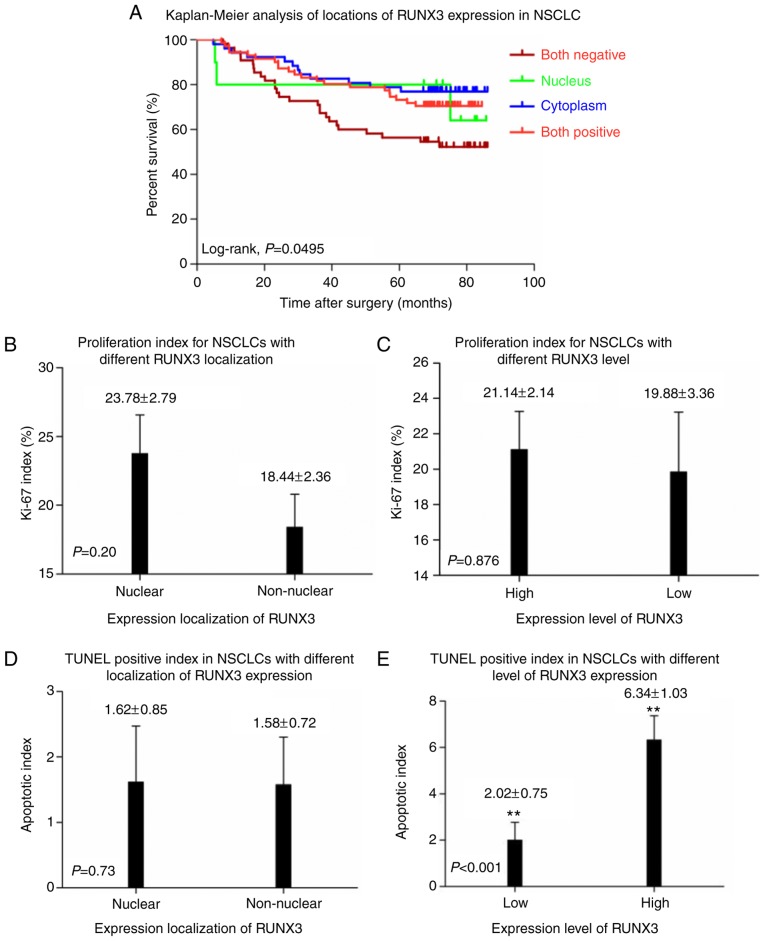
Different localizations of RUNX3 expression; (A) negative, (B) nucleus, (C) cytoplasm, and (D) nucleus and cytoplasm (magnification, ×400; scale bars=20 µm). (E) Kaplan-Meier curves of overall survival in different levels of RUNX3 expression in patients with NSCLC. High expression of RUNX3 (green line) was associated with a better prognostic outcome and longer overall survival while low expression of RUNX3 (red line) was associated with poor prognosis and shorter overall survival (P=0.0142). (F) Kaplan-Meier curves of overall survival in different localization of RUNX3 expression in patients with NSCLC. No statistical significance of overall survival between nuclear expression of RUNX3 (upper, continued line) and non-nuclear expression of RUNX3 (lower, dotted line) was determined (P=0.73). Overall survival, OS; RUNX3, runt-related transcription factor 3; NSCLC, non-small cell lung cancer.
Correlation of RUNX3 expression and clinicopathologic parameters were determined by means of χ2 analysis (Table II), revealing that higher RUNX3 expression was significantly associated with patients with advanced age (P=0.025), lower ECOG PS (P=0.019) as well as absence of postoperative metastasis (P=0.002). In addition, higher RUNX3 expression was observed to have a trend to correlate with absence of LN metastasis (P=0.062) as well as absence of distant metastasis at diagnosis (P=0.083). Eventually, no association had been discovered between expression of RUNX3 and gender (P=0.196), histology (P=0.536), smoking status (P=0.995), cellular differentiation (SCC only, P=1.000), lymphatic vessels invasion (P=0.223), nerve invasion (P=0.492), pleural invasion (P=0.364), vascular invasion (P=0.437), T-staging (P=0.488), mediastinal LN involvement (P=0.464), TNM-staging (P=0.230 or P=0.579), degree of resectibility (P=0.788), depth of invasion (P=0.757), serum CEA level (P=0.160), postoperative regional relapse (P=0.148) or Ki-67 expression (P=0.701). Survival analysis by Kaplan-Meier method with log-rank test indicated that patients with high level of RUNX3 exhibited much better outcome and longer OS than those with low RUNX3 expression (P=0.0142; Fig. 3E), regardless of various localizations of RUNX3 expression.
Table II.
Associations between expression level or localization of RUNX3 and clinicopathological parameters in patients with NSCLC.
| RUNX3b level | RUNX3 localization | |||||||
|---|---|---|---|---|---|---|---|---|
| Parameters | All | H | L | P-value | All | Nuclear | Non-nuclear | P-value |
| Agea (years) | 0.025 | 0.911 | ||||||
| ≤58 | 103 | 63 | 40 | 103 | 44 | 59 | ||
| >58 | 85 | 65 | 20 | 85 | 37 | 48 | ||
| Gender | 0.196 | 0.125 | ||||||
| Female | 60 | 37 | 23 | 60 | 21 | 39 | ||
| Male | 128 | 91 | 37 | 128 | 60 | 68 | ||
| Histology | 0.536 | 0.125 | ||||||
| ADE | 113 | 75 | 38 | 113 | 40 | 73 | ||
| SCC | 75 | 53 | 22 | 75 | 41 | 34 | ||
| ECOG PS | 0.019 | 0.125 | ||||||
| <2 | 145 | 105 | 40 | 145 | 64 | 81 | ||
| ≥2 | 43 | 23 | 20 | 43 | 17 | 26 | ||
| Smoker | 0.995 | 0.360 | ||||||
| Yes | 116 | 79 | 37 | 116 | 53 | 63 | ||
| No | 72 | 49 | 23 | 72 | 28 | 44 | ||
| Differentiation (SCC) | 1.000c | 0.697 | ||||||
| Poorly | 14 | 10 | 4 | 14 | 7 | 7 | ||
| Well+moderately | 61 | 43 | 18 | 61 | 34 | 27 | ||
| Lymphatic vessels invasion | 0.223 | 0.050 | ||||||
| Yes | 85 | 54 | 31 | 85 | 30 | 55 | ||
| No | 103 | 74 | 29 | 103 | 51 | 52 | ||
| Nerve invasion | 0.492c | 0.431c | ||||||
| Yes | 1 | 1 | 0 | 1 | 1 | 0 | ||
| No | 187 | 127 | 60 | 187 | 80 | 107 | ||
| Pleural invasion | 0.364 | 0.236 | ||||||
| Yes | 136 | 90 | 46 | 136 | 55 | 81 | ||
| No | 52 | 38 | 14 | 52 | 26 | 26 | ||
| Vascular invasion | 0.437c | 0.496 | ||||||
| Yes | 17 | 13 | 4 | 17 | 6 | 11 | ||
| No | 171 | 115 | 56 | 171 | 75 | 96 | ||
| T-staging | 0.488 | 0.137 | ||||||
| T1-2 | 138 | 92 | 46 | 138 | 55 | 83 | ||
| T3-4 | 50 | 36 | 14 | 50 | 26 | 24 | ||
| N-staging-1 | 0.062 | 0.034 | ||||||
| N0 | 97 | 72 | 25 | 97 | 49 | 48 | ||
| N1-3 | 91 | 56 | 35 | 91 | 32 | 59 | ||
| N-staging-2 | 0.464 | 0.652 | ||||||
| N0-1 | 129 | 90 | 39 | 129 | 57 | 72 | ||
| N2-3 | 59 | 38 | 21 | 59 | 24 | 35 | ||
| M-staging | 0.083c | 0.701c | ||||||
| M0 | 182 | 126 | 56 | 182 | 79 | 103 | ||
| M1 | 6 | 2 | 4 | 6 | 2 | 4 | ||
| TNM staging-1 | 0.230 | 0.774 | ||||||
| I–II | 109 | 78 | 31 | 109 | 46 | 63 | ||
| III–IV | 79 | 50 | 29 | 79 | 35 | 44 | ||
| TNM staging-2 | 0.579 | 0.602 | ||||||
| I | 68 | 48 | 20 | 68 | 31 | 37 | ||
| II–IV | 120 | 80 | 40 | 120 | 50 | 70 | ||
| Resectibility | 0.788 | 0.868 | ||||||
| R0 | 145 | 98 | 47 | 145 | 62 | 83 | ||
| R1-2 | 43 | 30 | 13 | 43 | 19 | 24 | ||
| Invasiveness | 0.757 | 0.240 | ||||||
| IS/MI | 24 | 17 | 7 | 24 | 13 | 11 | ||
| Invasive | 164 | 111 | 53 | 164 | 68 | 96 | ||
| Serum CEA (µg/ml) | 0.160 | 0.790 | ||||||
| ≤4.7 | 114 | 82 | 32 | 114 | 50 | 64 | ||
| >4.7 | 74 | 46 | 28 | 74 | 31 | 43 | ||
| Postoperative regional relapse | 0.148 | 0.671 | ||||||
| Yes | 47 | 28 | 19 | 47 | 19 | 28 | ||
| No | 141 | 100 | 41 | 141 | 62 | 79 | ||
| Postoperative metastasis | 0.002 | 0.035 | ||||||
| Yes | 67 | 36 | 31 | 67 | 22 | 45 | ||
| No | 121 | 92 | 29 | 121 | 59 | 62 | ||
| Ki-67 | 0.701 | 0.377 | ||||||
| ≤10% | 108 | 73 | 36 | 109 | 44 | 65 | ||
| >10% | 79 | 55 | 24 | 79 | 37 | 42 | ||
NSCLC, non-small cell lung cancer; All, all cases; H, high expression; L, low expression; RUNX3, runt-related transcription factor 3; nuclear expression, including nuclear expression and both nuclear and cytoplasmic expression of RUNX3; non-nuclear expression, including negative and cytoplasmic expression of RUNX3; ADE, adenocarcinoma; SCC, squamous cell carcinoma; ECOG PS, Eastern cooperative oncology group performance score; TNM, tumor-node-metastasis; IS, in situ; MI, minimally invasive; CEA, carcinoembryonic antigen
average value
cutoff point determined by X-tile software
Fisher's exact test (two-sided); χ2 test for all other analyses.
Localization of RUNX3 expression in NSCLC tissues and its correlation with clinicopathologic variables and OS. In accordance with Ito et al (23), after determining the cutoff value of RUNX3 expression, we also categorized all cases into nuclear (positive in the nucleus and positive or negative in the cytoplasm) and non-nuclear groups (including: i) negative in both nucleus and cytoplasm and ii) positive in the cytoplasm while negative in the nucleus) (Fig. 3A-D). As a result, among 188 NSCLC patients, nuclear localization of RUNX3 was observed in 40 cases in ADE histology and 41 in SCC histology, while non-nuclear was 73 in ADE and 34 in SCC (Table I).
As shown in Table II, correlation of localization of RUNX3 expression and clinicopathologic parameters were analyzed and found that non-nuclear localization was significantly associated with ADE histology (P=0.009), lymphatic vessels invasion (P=0.050), lymph node involvement (P=0.034) and postoperative metastasis (P=0.035). No correlation had been determined between localization of RUNX3 expression and age (P=0.911), gender (P=0.125), ECOG PS (P=0.592), smoking status (P=0.360), cellular differentiation (SCC only, P=1.000), nerve invasion (P=0.431), pleural invasion (P=0.236), vascular invasion (P=0.496), T-staging (P=0.137), mediastinal LN involvement (P=0.652), M-staging (P=0.701), TNM-staging (P=0.774 or P=0.602), degree of resectibility (P=0.868), depth of invasion (P=0.240), serum CEA level (P=0.790), postoperative regional relapse (P=0.671) or Ki-67 expression (P=0.377). However, when focusing on the effect that different localization of RUNX3 expression might have on OS via Kaplan-Meier analysis, we found that no statistical significance had been discovered between patient groups of nuclear and non-nuclear localization of RUNX3 expression (P=0.3781; Fig. 3F). It had to be noticed that when categorized into 4 subgroups, i.e., negative, nuclear, cytoplasmic and whole-cell expression of RUNX3, patients with negative expression in both nuclear and cytoplasm showed the worst outcome and shortest OS, while the other 3 subgroups had no difference in OS with one other (Fig. 4A).
Figure 4.
(A) Kaplan-Meier curves of overall survival in different localizations of RUNX3 expression in patients NSCLC: Negative, nucleus, cytoplasm, and nucleus and cytoplasm (P=0.0495). Patients with negative expression exhibited the worst survival, while no statistical significance of overall survival was observed among nuclear, cytoplasmic and whole-cell expression of RUNX3 (P>0.05). In patients with (B) nuclear and non-nuclear expression of RUNX3 or (C) different expression levels of RUNX3, Ki-67 expression demonstrated no statistical difference (P>0.05). (D) In patients with nuclear and non-nuclear expression of RUNX3, TUNEL positive index demonstrated no statistical difference (P=0.73). (E) Apoptotic index in patients with higher expression level of RUNX3 prevailed significantly over those with low expression level of RUNX3 (P<0.001). **P<0.01. Overall survival, OS; NSCLC, non-small cell lung cancer; RUNX3, runt-related transcription factor 3; TUNEL, terminal deoxynucleotidyl transferase mediated dUTP-biotin nick end labelling.
Proliferative and apoptotic evaluation in NSCLC patients with different levels or patterns of RUNX3 expression
As shown in Fig. 4B, C and Table II, IHC score of Ki-67 for nuclear and non-nuclear localization as well as high and low expression of RUNX3 subgroups were 23.78±2.79% vs. 18.44±2.36% and 21.14±2.14% vs. 19.88±3.36%, respectively, and no statistical significance had been determined between them (P=0.377 and 0.701 respectively). No statistical significance had been demonstrated on apoptotic index between patients with nuclear (n=81) and non-nuclear (n=107) localization of RUNX3 expression (1.62±0.85 vs. 1.58±0.72, P=0.73; Fig. 4D). However, in comparison to NSCLC patients with low RUNX3 level (n=129), higher apoptotic index had been observed in patients with high RUNX3 expression (n=59) (6.34±1.03 vs. 2.02±0.75, P<0.001; Fig. 4E).
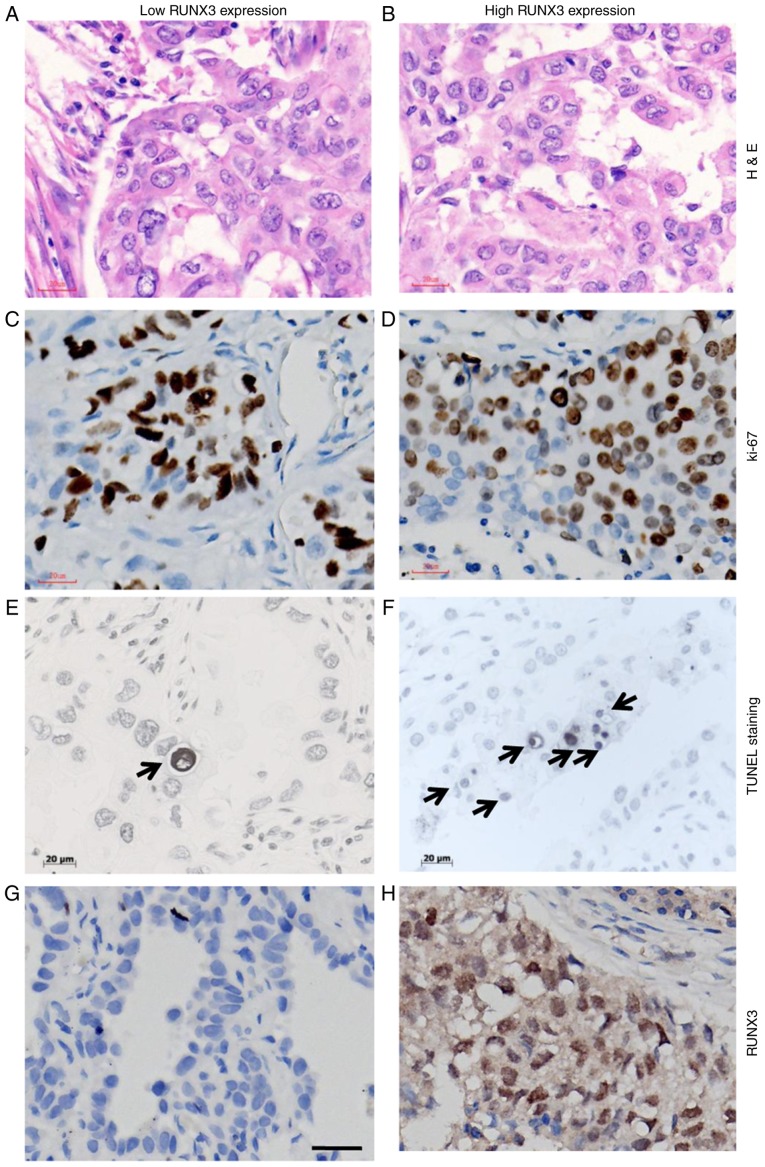
The nomaly of tissue used for IHC staining of Ki-67 and TUNEL evaluation is shown in Fig. 5A and B. Ki-67 staining was illustrated in Fig. 5C and D. As for apoptotic staining by TUNEL method (Fig. 5E and F), cells with the morphological characteristics of apoptosis were identified as chromatin condensation and nuclear fragmentation, which were accompanied by rounding up of the cell, reduction in cellular volume (pyknosis) and retraction of pseudopodes. In addition, formation of crescentic caps of condensed chromatin at the nuclear periphery, and formation of apoptotic bodies could also be observed (Arrows). The immunostaining of RUNX3 with low and high expression, based on which patients were divided into different subgroups is shown in Fig. 5G and H.
Figure 5.
Cellular dynamic parameters between patients with NSCLC and different expression levels of RUNX3. H&E staining for NSCLC tissue with (A) low and (B) high level of RUNX3. Immunostaining for Ki-67 in NSCLC tissue with (C) low and (D) high level of RUNX3. TUNEL staining for apoptotic cells in NSCLC tissue with (E) low and (F) high level of RUNX3, characterized by chromatin condensation and nuclear fragmentation, which were accompanied by cell swelling, reduction in cellular volume (pyknosis) and retraction of pseudopodes. Formation of crescentic caps of condensed chromatin at the nuclear periphery, and formation of apoptotic bodies could also be observed (indicated by arrows). (G) Negative staining for RUNX3; (H) positive staining for RUNX3 in NSCLC. Magnification, ×400; scale bars=20 µm. NSCLC, non-small cell lung cancer; RUNX3, runt-related transcription factor 3; H&E, hematoxylin and eosin; TUNEL, terminal deoxynucleotidyl transferase mediated dUTP-biotin nick end labelling.
Discussion
Loss of expression or protein mislocalization had both been indicated as patterns of dysfunction regarding RUNX3 expression in malignant tumors like GC, CRC and BC (23–25), which was correlated with worse prognosis and shorter OS in these cancer entities. Other studies also demonstrated that methylation-related transcriptional silencing of RUNX3 played pivotal roles in the onset and progression of malignancies like esophageal cancer, hepatocellular carcinoma, pancreatic cancer, prostate cancer, and lung cancer as well (34–36).
Expression RUNX3 is one of the most interesting topics that deserves further study in a variety of cancer types. In order to identify the significance of RUNX3 expression on NSCLC patients, we immunohistochemically detected the expression level as well as expression patterns like protein localization of RUNX3 in 188 patients. In the present study, 4 expression patterns were demonstrated, i.e., negative, nuclear, cytoplasm and whole-cell localization of RUNX3 protein. Obvious difference had been discovered of the OS for these 4 categories of patient population, with the negative subgroup the worst, while other 3 subgroups almost identical in OS (P=0.0495; Fig. 4A).
Since accumulated evidences had revealed that patients with different RUNX3 expression patterns like nuclear and non-nuclear localization in BC, CRC and GC manifested quite different OS, similar analysis was carried out in NSCLC in the present study, demonstrating that non-nuclear localization of RUNX3 (including: i) negative in both nucleus and cytoplasm and ii) positive in the cytoplasm while negative in the nucleus) was significantly associated with ADE histology (P=0.009), lymphatic vessels invasion (P=0.050), lymph node involvement (P=0.034) and postoperative metastasis (P=0.035). However, Kaplan-Meier survival analysis with log-rank test didn't indicate any statistical difference between these two patient cohorts (P=0.3781, Fig. 3F), which was quite different from what Soong et al (25), Kang et al (37) and Ito et al (23) had reported in BC, CRC and GC, that patients with nuclear expression of RUNX3 (positive in the nucleus and positive or negative in the cytoplasm) would definitely have better outcome and longer survival than those with non-nuclear expression.
Since no survival discrepancy had been demonstrated between nuclear and non-nuclear RUNX3 localization in NSCLC, we thus set out to analyze the data by dividing patients into subgroups of high and low RUNX3 expression with the cutoff value determined via X-tile software, irrespective of its expression localization, and evaluated the associations between RUNX3 expression level and clinicopathologic parameters, showing that higher RUNX3 expression was significantly associated with patients with advanced age (P=0.025), lower ECOG PS (P=0.019) and absence of postoperative metastasis (P=0.002). Surprisingly, Kaplan-Meier survival analysis demonstrated that, in comparison to NSCLC patients with lower RUNX3 level, those with higher level of RUNX3 exhibited much better outcome and longer OS (P=0.0142; Figs. 2A-C, 3E), as was quite different from the survival analysis between patient subgroups with different expression localization.
Interestingly, further analyses on the correlations between RUNX3 expression level/pattern and clinicopathologic parameters demonstrating that both expression level and localization of RUNX3 were strongly correlated with postoperative distant metastasis in NSCLC patients, that's, patients with higher level of RUNX3 stood a less chance of metastasis (P=0.002) while those with non-nuclear pattern of RUNX3 expression showed higher probability of distant metastasis after surgery (P=0.035). In addition, in analyzing the correlation between different expression level/pattern of RUNX3 with LN status which was one of the major prognostic factors in NSCLC, we found that non-nuclear RUNX3 expression was obviously correlated with positive LN involvement (N1-3 vs. N0, P=0.034) and marginally correlated with lymphatic vessels invasion (P=0.050), while no statistical relationship had been determined when referred to expression level of RUNX3 (Table II). Further, higher level of RUNX3 was found to be correlated with older patients (P=0.025) and better ECOG PS (P=0.019), while neither of them significantly correlated with expression localization (both P>0.05). Interestingly, non-nuclear RUNX3 expression was majorly found in patients with ADE histology (P=0.009) while expression level of RUNX3 didn't make any difference between ADE and SCC (P=0.536). Our previous study had demonstrated that tissue type, smoking status, ECOG PS, postoperative relapse and postoperative distant metastasis were several prognostic factors correlated with NSCLC patients' OS (28), and expression level of RUNX3 was testified to be closely correlated with the some major prognostic factors of OS like postoperative metastasis, ECOG PS, and etc. Several recent study indicated that loss of RUNX3 expression promoted cell migration (38), cancerous angiogenesis and increased microvessel density (39) in cancers. Taken together, these could partially explain why RUNX3 expression level could be the influencing factor for OS.
As had been well-documented that RUNX3 played a pivotal role in the management of cellular proliferation as well as apoptosis in a variety of cancer cell types, we then set out to evaluate the proliferating as well as apoptotic index in NSCLC patients with different level or localization of RUNX3 expression. No statistical difference in Ki-67 expression had been demonstrated either between patient subgroups of different expression level or between subgroups with different localization of RUNX3 expression (both P>0.05; Figs. 4B and C; 5C and D; Table II). However, as far as the apoptotic index was concerned, compared to those with low level of RUNX3, NSCLC patients with high RUNX3 level exhibited significantly higher apoptotic index (P<0.001; Fig. 4E, 5E and F), while no statistical significance had been determined in relation to the apoptotic index between subgroups of nuclear and non-nuclear localization of RUNX3 expression (P=0.73; Fig. 4D). This could partially explain why NSCLC patients with high RUNX3 would have better outcome than those with low level, for cancer tissue with higher RUNX3 would probably suffer from higher apoptotic index and be more sensitive to therapeutic regimens.
The mechanism of transcription silencing or protein mislocalization of RUNX3 had in part been illustrated as gene mutation, hypermethylation of the promoter or exon of RUNX3 gene as well as alteration of some specific upstream or downstream proteins like src kinase, transforming growth factor beta (TGF-β) and AT motif binding factor 1 (ATBF1) as well. Goh et al (40), demonstrated that overexpression of src kinase resulted in the tyrosine phosphorylation and cytoplasmic localization of RUNX3 in kidney and cervical cancer cell lines, and knockdown of src or inhibition of its kinase activity resulted in re-localization of RUNX3 to the nucleus in these cell lines. Mabuchi and colleagues revealed that just like RUNX3, ATBF1 was also able to shuttle between cytoplasm and nucleus in GC cells and there was a close connection in nuclear localization between them. It's verified that there was a physical association between ATBF1 and RUNX3, both of them could translocate from cytoplasma to nucleus in response to TGF-β signal transduction (41). The statement above could partially illustrate the phenomenon of different localization of RUNX3 in some cancerous types, however, whether the same activity and underlying mechanism would also exist and work in the context of NSCLC or not might still need further verification in the future.
Recently, a great number of studies demonstrated that hypermethylation of RUNX3 promoter was mostly cancer-specific and more frequent in ADE than in SCC histology (13,42) and could be utilized as a molecular diagnostic marker for NSCLC. Fujii et al (43), reported that EZH2 was able to bind directly to the promoter of RUNX3, boosting the methylating level of histone H3 lysine 27 (H3K27), resulting in expression loss of RUNX3 in gastric, breast, prostate, colon and pancreatic cancer cell lines. EZH2 can also direct DNA methylation by recruitment of DNA methyltransferases (DNMTs) to target gene promoters, suggesting that it may be a significant causative factor of aberrent methylation in cancer. Our previous study (28), demonstrated that reduced EZH2 expression in NSCLC was correlated with ADE histology, non-smoking status, low DNA methylation level at CCGG sites and decreased cancerous proliferating activity. Interestingly, further analyses on our previous data indicated that there was obvious difference in expression direction of EZH2 and its catalytic substrate histone H3 lysine 27 among NSCLC patients with different smoking status (data not shown). That is, trend of H3K27me3 and EZH2 expression was in the same direction in the majority of smokers, while in nonsmokers, their expression trend was mostly in the opposite direction, which was partly consistent with Zhang et al's findings (44). Taken together, we hypothesized that the underlying mechanism of different expression level and localization of RUNX3 in NSCLC might probably be connected with the epegenetic markers like H3K27me3 and its methyltransferase EZH2 in part, and further investigation on these markers in different expression patterns of RUNX3 might probably shed some light on the detailed roles that RUNX3 might play in onset as well as progression in patients with NSCLC.
Conclusively, our study indicated loss of expression rather than cytoplasmic mislocalization of RUNX3 predicted worse outcome in NSCLC, and could probably be used as a good biomarker in predicting the prognosis in lung cancer patients.
Acknowledgements
The present study was supported in part by a Grant-in-Aid for Youth Research Project from Health Administration of Fujian Province (grant no. 2013-1-10 to X. Chen), Fujian Provincial Foundation of Natural Science (grant no. 2016J01515 to X. Chen), Miaopu Research Foundation of Fujian Medical University (grant no. 2015MP032 to Y. Deng) and Startup Research Project of Fujian Medical University (grant no. 2016QH040 to Y. Deng).
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF-beta superfamily signaling. Curr Opin Genet Dev. 2003;13:43–47. doi: 10.1016/S0959-437X(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 4.Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int. 2003;27:315–324. doi: 10.1016/S1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- 5.Cameron ER, Blyth K, Hanlon L, Kilbey A, Mackay N, Stewart M, Terry A, Vaillant F, Wotton S, Neil JC. The Runx genes as dominant oncogenes. Blood Cells Mol Dis. 2003;30:194–200. doi: 10.1016/S1079-9796(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 6.Ito Y, Osato M, Ito K. RUNX and cancer. Ann Acad Med Singapore. 2003;32(5 Suppl):S6–S7. [PubMed] [Google Scholar]
- 7.Levanon D, Brenner O, Otto F, Groner Y. Runx3 knockouts and stomach cancer. EMBO Rep. 2003;4:560–564. doi: 10.1038/sj.embor.embor868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levanon D, Glusman G, Bettoun D, Ben-Asher E, Negreanu V, Bernstein Y, Harris-Cerruti C, Brenner O, Eilam R, Lotem J, et al. Phylogenesis and regulated expression of the RUNT domain transcription factors RUNX1 and RUNX3. Blood Cells Mol Dis. 2003;30:161–163. doi: 10.1016/S1079-9796(03)00023-8. [DOI] [PubMed] [Google Scholar]
- 9.Miyazono K, Suzuki H, Imamura T. Regulation of TGF-beta signaling and its roles in progression of tumors. Cancer Sci. 2003;94:230–234. doi: 10.1111/j.1349-7006.2003.tb01425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto F, Stock M, Fliegauf M, Fenaux P, Preudhomme C, Lübbert M. Absence of somatic mutations within the Runt domain of AML2/RUNX3 in acute myeloid leukaemia. Leukemia. 2003;17:1677–1678. doi: 10.1038/sj.leu.2403007. [DOI] [PubMed] [Google Scholar]
- 11.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue Ki, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/S0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 12.Kim R, Trubetskoy A, Suzuki T, Jenkins NA, Copeland NG, Lenz J. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J Virol. 2003;77:2056–2062. doi: 10.1128/JVI.77.3.2056-2062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, Motoyama T. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–592. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anglin I, Passaniti A. Runx protein signaling in human cancers. Cancer Treat Res. 2004;119:189–215. doi: 10.1007/1-4020-7847-1_10. [DOI] [PubMed] [Google Scholar]
- 15.Bae SC, Choi JK. Tumor suppressor activity of RUNX3. Oncogene. 2004;23:4336–4340. doi: 10.1038/sj.onc.1207286. [DOI] [PubMed] [Google Scholar]
- 16.Goel A, Arnold CN, Tassone P, Chang DK, Niedzwiecki D, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR. Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int J Cancer. 2004;112:754–759. doi: 10.1002/ijc.20472. [DOI] [PubMed] [Google Scholar]
- 17.Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202:233–240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 18.Li QL, Kim HR, Kim WJ, Choi JK, Lee YH, Kim HM, Li LS, Kim H, Chang J, Ito Y, et al. Transcriptional silencing of the RUNX3 gene by CpG hypermethylation is associated with lung cancer. Biochem Biophys Res Commun. 2004;314:223–228. doi: 10.1016/j.bbrc.2003.12.079. [DOI] [PubMed] [Google Scholar]
- 19.Wada M, Yazumi S, Takaishi S, Hasegawa K, Sawada M, Tanaka H, Ida H, Sakakura C, Ito K, Ito Y, Chiba T. Frequent loss of RUNX3 gene expression in human bile duct and pancreatic cancer cell lines. Oncogene. 2004;23:2401–2407. doi: 10.1038/sj.onc.1207395. [DOI] [PubMed] [Google Scholar]
- 20.Tamura G. Promoter methylation status of tumor suppressor and tumor-related genes in neoplastic and non-neoplastic gastric epithelia. Histol Histopathol. 2004;19:221–228. doi: 10.14670/HH-19.221. [DOI] [PubMed] [Google Scholar]
- 21.Oshimo Y, Oue N, Mitani Y, Nakayama H, Kitadai Y, Yoshida K, Ito Y, Chayama K, Yasui W. Frequent loss of RUNX3 expression by promoter hypermethylation in gastric carcinoma. Pathobiology. 2004;71:137–143. doi: 10.1159/000076468. [DOI] [PubMed] [Google Scholar]
- 22.Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, Goldenberg D, Negreanu V, Bernstein Y, Levanon D, Jung S, Groner Y. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005;65:7743–7750. doi: 10.1158/0008-5472.CAN-05-0743. [DOI] [PubMed] [Google Scholar]
- 24.Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 2006;66:6512–6520. doi: 10.1158/0008-5472.CAN-06-0369. [DOI] [PubMed] [Google Scholar]
- 25.Soong R, Shah N, Peh BK, Chong PY, Ng SS, Zeps N, Joseph D, Salto-Tellez M, Iacopetta B, Ito Y. The expression of RUNX3 in colorectal cancer is associated with disease stage and patient outcome. Br J Cancer. 2009;100:676–679. doi: 10.1038/sj.bjc.6604899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mello RB, Silva MR, Alves MT, Evison MP, Guimarães MA, Francisco RA, Astolphi RD, Iwamura ES. Tissue microarray analysis applied to bone diagenesis. Sci Rep. 2017;7:39987. doi: 10.1038/srep39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eskaros AR, Egloff SA, Boyd KL, Richardson JE, Hyndman ME, Zijlstra A. Larger core size has superior technical and analytical accuracy in bladder tissue microarray. Lab Invest. 2017;97:335–342. doi: 10.1038/labinvest.2016.151. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Song N, Matsumoto K, Nanashima A, Nagayasu T, Hayashi T, Ying M, Endo D, Wu Z, Koji T. High expression of trimethylated histone H3 at lysine 27 predicts better prognosis in non-small cell lung cancer. Int J Oncol. 2013;43:1467–1480. doi: 10.3892/ijo.2013.2062. [DOI] [PubMed] [Google Scholar]
- 29.Deng Y, Chen X, Ye Y, Shi X, Zhu K, Huang L, Zhang S, Ying M, Lin X. Histological characterisation and prognostic evaluation of 62 gastric neuroendocrine carcinomas. Contemp Oncol (Pozn) 2016;20:311–319. doi: 10.5114/wo.2016.61852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song N, Liu J, An S, Nishino T, Hishikawa Y, Koji T. Immunohistochemical analysis of histone H3 modifications in germ cells during mouse spermatogenesis. Acta Histochem Cytochem. 2011;44:183–190. doi: 10.1267/ahc.11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 33.Araki K, Osaki M, Nagahama Y, Hiramatsu T, Nakamura H, Ohgi S, Ito H. Expression of RUNX3 protein in human lung adenocarcinoma: Implications for tumor progression and prognosis. Cancer Sci. 2005;96:227–231. doi: 10.1111/j.1349-7006.2005.00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C, Li J, Huang T, Duan S, Dai D, Jiang D, Sui X, Li D, Chen Y, Ding F, et al. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget. 2016;7:81255–81267. doi: 10.18632/oncotarget.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whittle MC, Hingorani SR. RUNX3 defines disease behavior in pancreatic ductal adenocarcinoma. Mol Cell Oncol. 2015;3:e1076588. doi: 10.1080/23723556.2015.1076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen F, Liu X, Bai J, Pei D, Zheng J. The emerging role of RUNX3 in cancer metastasis (Review) Oncol Rep. 2016;35:1227–1236. doi: 10.3892/or.2015.4515. [DOI] [PubMed] [Google Scholar]
- 37.Kang KA, Piao MJ, Ryu YS, Maeng YH, Hyun JW. Cytoplasmic localization of RUNX3 via histone deacetylase-mediated SRC expression in oxidative-stressed colon cancer cells. J Cell Physiol. 2017;232:1914–1921. doi: 10.1002/jcp.25746. [DOI] [PubMed] [Google Scholar]
- 38.Chen F, Bai J, Li W, Mei P, Liu H, Li L, Pan Z, Wu Y, Zheng J. RUNX3 suppresses migration, invasion and angiogenesis of human renal cell carcinoma. PLoS One. 2013;8:e56241. doi: 10.1371/journal.pone.0056241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue J, Wu XL, Huang XT, Qu M, Guo F, Sun GY, Zhang PC, Han L, Pan LM. Correlation of RUNX3 expression with microvessel density in colorectal adenocarcinoma tissues and clinical significance. Asian Pac J Trop Med. 2017;10:98–101. doi: 10.1016/j.apjtm.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Goh YM, Cinghu S, Hong ET, Lee YS, Kim JH, Jang JW, Li YH, Chi XZ, Lee KS, Wee H, et al. Src kinase phosphorylates RUNX3 at tyrosine residues and localizes the protein in the cytoplasm. J Biol Chem. 2010;285:10122–10129. doi: 10.1074/jbc.M109.071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mabuchi M, Kataoka H, Miura Y, Kim TS, Kawaguchi M, Ebi M, Tanaka M, Mori Y, Kubota E, Mizushima T, et al. Tumor suppressor, AT motif binding factor 1 (ATBF1), translocates to the nucleus with runt domain transcription factor 3 (RUNX3) in response to TGF-beta signal transduction. Biochem Biophys Res Commun. 2010;398:321–325. doi: 10.1016/j.bbrc.2010.06.090. [DOI] [PubMed] [Google Scholar]
- 42.Lee KS, Lee YS, Lee JM, Ito K, Cinghu S, Kim JH, Jang JW, Li YH, Goh YM, Chi XZ, et al. Runx3 is required for the differentiation of lung epithelial cells and suppression of lung cancer. Oncogene. 2010;29:3349–3361. doi: 10.1038/onc.2010.79. [DOI] [PubMed] [Google Scholar]
- 43.Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283:17324–17332. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Fillmore Brainson C, Koyama S, Redig AJ, Chen T, Li S, Gupta M, Garcia-de-Alba C, Paschini M, Herter-Sprie GS, et al. Lkb1 inactivation drives lung cancer lineage switching governed by Polycomb Repressive Complex 2. Nat Commun. 2017;8:14922. doi: 10.1038/ncomms14922. [DOI] [PMC free article] [PubMed] [Google Scholar]