Abstract
Non-small cell lung cancer (NSCLC) accounts for ~80% of all types of lung cancer, which has the highest morbidity and mortality of all types of cancer worldwide. It is important to identify novel biomarkers and the molecular mechanism of NSCLC to improve current treatments of NSCLC. The present study aimed to investigate the effect of miR-148b expression on the proliferation, epithelial-mesenchymal transition (EMT) and radiosensitivity of NSCLC cells. It was demonstrated that miR-148b expression was significantly decreased in NSCLC tissues and cell lines. A549 cells were then transfected with a miR-148b mimic and a miR-148b inhibitor. Transfection with the miR-148b mimic decreased proliferation whereas transfection with the miR-148b inhibitor increased the proliferation of A549 cells. Additionally, the miR-148b mimic increased E-cadherin expression and decreased N-cadherin and vimentin expression. By contrast, transfection with the miR-148b inhibitor decreased E-cadherin expression and increased N-cadherin and vimentin expression. Irradiation-induced cell death was significantly promoted by the miR-148b mimic but inhibited by the miR-148b inhibitor. The miR-148b mimic significantly decreased the expression of Rho-associated protein kinase 1 (ROCK1) and it was demonstrated that overexpression of ROCK1 significantly inhibited the effects of miR-148b on cell proliferation, the EMT and irradiation-induced cell death. Therefore, the current study revealed that miR-148b inhibited NSCLC cell proliferation and the EMT, and increased the radiosensitivity of NSCLC cells by inhibiting ROCK1 expression. Therefore, miR-148b/ROCK1 signaling may be a novel therapeutic target to inhibit the growth of NSCLC cells and enhance the effects of radiotherapy to treat patients with NSCLC.
Keywords: microRNA-148b, Rho-associated protein kinase 1, non-small cell lung carcinoma, cell proliferation, epithelial-mesenchymal transition
Introduction
Lung cancer is one of the most common causes of cancer-associated mortality worldwide (1). Non-small cell lung cancer (NSCLC) accounts for ~80% of all lung cancer cases (2). In 2014, there were 160,000 mortalities resulting from lung cancer, accounting for 20% of all cancer-associated mortalities in the United States (3). Out of all lung cancer-associated mortalities, ~80% occurred in patients with NSCLC (3–5) and the main types of NSCLC include adenocarcinoma and squamous cell carcinoma (6). Recently, the therapeutic strategies available to treat NSCLC have advanced greatly; however, patients with advanced NSCLC have a poor prognosis and the 5-year survival rate of patients with NSCLC is still only <5% (7,8). By contrast, the 5-year survival rate for early-stage NSCLC following curative resection is 30–60% (9). Therefore, it is important to identify novel biomarkers and determine the molecular mechanisms of NSCLC to improve the early diagnosis and treatment of patients with NSCLC, thus improving their prognosis (10).
microRNAs (miRNAs or miRs) are a class of small, regulatory, non-coding RNAs that are 20–24 nucleotides long. They are involved in various biological events, including cell growth, differentiation, apoptosis and migration, as well as pathological processes, including the development of tumors, and metabolic and neurodegenerative illnesses (11–14). It is estimated that one-third of all mammalian genes are directly or indirectly regulated by miRNAs. These directly bind to the 3′-untranslated region of target mRNA, causing mRNA destabilization and degradation, and consequently altering the expression of target proteins (14,15). miRNA deregulation serves an important role in the pathogenesis of different tumors (16,17). Furthermore, it has been hypothesized that miRNA levels change prior to the phenotypic changes that occur during cancer progression (18). The detection and quantification of miRNAs is easily performed using standard diagnostic biological material, including formalin-fixed paraffin-embedded samples, blood, serum and sputum (19).
It is important to identify novel miRNAs and determine their roles in tumor development to elucidate the mechanism of cancer progression and aid in the development of novel methods to diagnose and treat cancer (20). It has been demonstrated that a variety of miRNAs are deregulated during NSCLC progression (21,22). It has been demonstrated that miR-148b levels are reduced in plasma samples taken from patients with NSCLC (23). Furthermore, it has been proposed that miR-148b may be a novel biomarker in NSCLC (9,24). However, the molecular mechanism underlying the role of miR-148b in NSCLC remains unclear.
The present study aimed to investigate the effect of miR-148b on cell proliferation, the epithelial-mesenchymal transition (EMT) and radiosensitivity in NSCLC cells. It was revealed that miR-148b expression was reduced in NSCLC tissues and cell lines, inhibited NSCLC cell proliferation and the EMT and increased radiosensitivity in NSCLC cells by regulating the expression Rho-associated protein kinase 1 (ROCK1).
Materials and methods
Clinical specimens
A total of 16 cases (mean age, 53; 9 males and 7 females) of NSCLC were retrieved from the Departments of Oncology 2 Division and Respiratory Medicine, Foshan Nanhai District People's Hospital (Foshan, China) between January and March 2016. Patients did not receive any chemo- or radiotherapy prior to surgery to resect clinical specimens. Tumor tissues and adjacent non-tumor tissues were resected from patients. The present study was approved by the Ethical Review Committee of People's Hospital of Nanhai District Guangdong Province (Foshan, China) and complied with the Declaration of Helsinki. Informed consent was obtained from all patients.
Cell culture
The human bronchial cell line (HBE1) and NSCLC cell lines, including H1299, H1650, H460 and A549, were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100 U/ml penicillin in an incubator with 5% CO2 at 37°C. The culture medium was replenished every 2–3 days and the cells were passaged at 1:6 every 4 days following trypsinization with 0.05% trypsin-EDTA. Cells were cultured with plasmids for 3 days.
Cell transfection
The miR-148b mimic (5′-UACUAGACAUCGCAUACACUA-3′; 5′-GCAUAUACUAUGUCAUGACUU-3′), NC-mimic (5′-UUCUCCGAACGUGUCACGUTT-3′; 5′-ACGUGACACGUUCGGAGAATT-3′); miR-148b inhibitor (5′-ACAAAGUUCUGUGAUGCACUGA-3′) and anti-NC (5′-CAGUACUUUUGUGUAGUACAA-3′) were synthesized commercially using pCMV-miR by Guangzhou RiboBio Co., Ltd., Guangzhou, China). The ROCK1 sequence was cloned into the pCMV vector. The expression vector pCMV (Invitrogen; Thermo Fisher Scientific, Inc.) was used to construct the ROCK1 expression vector. The genomic sequence of ROCK1 (NC_000018.10) was cloned from 293 cell cDNA and the PCR products were digested by EcoRI and BamHI. PCR amplication was performed using a High Yield PCR EcoDry™ Premix (Takara Biotechnology Co., Ltd., Dalian, China). Thermocycling conditions were as follows: Initial denaturation at 95°C for 10 min followed by 40 cycles at 95°C for 1 min, annealing at 53°C for 1 min, extension at 72°C for 1 min and final extension at 72°C for 5 min. The primer sequences for ROCK1 were as follows: Forward 5′-TGGATCCATGATGGCTCTGGGCGCAGCGGGAG-3′ and reverse, 5′-CGAATTCTTAGTGTCTCTGACAAGTGTGAAGCCTAGAAG-3′. The amplified product was then subcloned into the pCMV vector. A549 cells were transfected with plasmids. Transient transfection of 100 nM miR-148b mimic, 100 nM NC-mimic, 100 nM miR-148b inhibitor, 100 nM anti-NC, and 100 nM pCMV-ROCK1 was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer's protocols. A total of 6 h following transfection, the cell growth medium was removed and cells were incubated in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 5% FBS for another 24–72 h. A total of 48 h following transfection, RT-qPCR was performed to measure the level of miR-148b, 24–72 h following transfection, cell proliferation was determined and 72 h following transfection, the expression of EMT markers, apoptosis and radiosensitivity were evaluated.
Cell proliferation
Cell proliferation was determined using the Cell Counting Kit-8 assay kit (Beyotime Institute of Biotechnology, Haimen, China) following the manufacturer's protocols. A total of 4×104 cells were seeded in the plates and transfected with miR-148b mimic, NC-mimic, miR-148b inhibitor, anti-NC, with or without pCMV-ROCK1 for 24–72 h. Absorbance at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells of all transfection groups using the PARIS™ kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA templates were synthesized by MultiScribe Reverse Transcriptase (42°C for 15 min, 75°C for 3 min; Applied Biosystems; Thermo Fisher Scientific, Inc.) and qPCR was conducted using the Maxima SYBR Green/ROX qPCR Master Mix Assays (Fermentas; Thermo Fisher Scientific, Inc.) in an Applied Biosystems 7500 detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was performed as follows: Initial denaturation at 95°C for 10 min followed by 40 cycles at 95°C for 1 min, annealing at 53°C for 1 min, extension at 72°C for 1 min and final extension at 72°C for 5 min. U6 and β-actin were used as loading controls. Relative expression levels were normalized to the expression of β-actin mRNA using the 2−ΔΔCq method (25). Primer sequences used in the current study were as follows: E-cadherin, forward, 5′-CTGCTGCAGGTCTCCTCTTG-3′ and reverse, 5′-TGTCGACCGGTGCAATCTTC-3′; Vimentin, forward, 5′-AAGGCGAGGAGAGCAGGATT-3′ and reverse 5′-GGTCATCGTGATGCTGAGAAG-3′; N-cadherin, forward, 5′-ACAGTGGCCACCTACAAAGG-3′ and reverse, 5′-TGATCCCTCAGGAACTGTCC-3′; ROCK1, forward, 5′-ATGAGTTTATTCCTACACTCTACCACTTTC-3′ and reverse, 5′-TAACATGGCATCTTCGACACTCTAG-3′; β-actin, forward, 5′-CCTGGGCATGGAGTCCTGTG-3′ and reverse, 5′-TCTTCATTGTGCTGGGTGCC-3′; miR-148b, forward, 5′-TCAGTGCATCACAGAACTTTGTAA-3′ and reverse, 5′-GCTGTCAACGATACGCTACGT-3′; and U6, forward, 5′-CGCTTCGGCAGCACATATAC-3′ and reverse, 5′-TTCACGAATTTGCGTGTCAT-3′. Individual experiments were performed in triplicate and results were presented as a proportion of the control.
Western blot analysis
Cells were lysed using radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific, Inc.) supplemented with protease inhibitor cocktails (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Following protein extraction, protein concentration was determined using a bicinchoninic acid assay (Thermo Fisher Scientific, Inc.). Total protein samples (2 µg/lane) were separated by 10% SDS-PAGE and transferred onto PVDF membranes (EMD Millipore, Billerica, MA, USA). Membranes were blocked with 8% skimmed milk in Tris-buffered saline Tween-20 (TBST) buffer at 37°C for 1 h and subsequently incubated overnight at 4°C with primary antibodies (β-actin; cat no. 4970, 1:1,000; ROCK1, cat no. 4035, 1:1,000; both Cell Signaling Technology, Inc. Danvers, MA, USA). Following four washes (10 min/wash) in TBST, membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (1:1,000; cat no. 31460; Thermo Fisher Scientific, Inc.) at 37°C for 30 min. Bands were visualized with an enhanced chemiluminescence kit (cat no. 32106; Thermo Fisher Scientific, Inc.) and images were captured using ChemiDoc™ (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and analysed using the Quantity One 4.6 (Bio-Rad Laboratories, Inc.).
Radiosensitivity
Cells were exposed to 0, 2, 4, 6, 8 and 10 Gy irradiation using the Primus K linear accelerator (Siemens AG, Munich, Germany) and a clonogenic assay was then conducted. Following 12 days incubation, the colonies formed were fixed with 100% methanol at room temperature for 5 min and stained with 1% crystal violet at room temperature for 2 h. Colonies of >50 cells were scored as survivors. The cells were then harvested and assessed using a multifunctional microplate reader at 546 nm. The clonogenic fractions of irradiated cells were normalized to the plating efficiencies of the non-irradiated controls.
Apoptosis
Following treatment, cells were fixed with 10% paraformaldehyde for 10 min at room temperature and apoptosis was measured using a terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assay kit (Roche Diagnostics, Basel, Switzerland) following the manufacturer's protocol. Detection and analysis of apoptosis were performed using a FACScalibur Flow Cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Antifade Mounting Medium (Beyotime Institute of Biotechnology) was used and six random fields of view was observed. Triplicate individual experiments were performed and results were provided as a proportion of the control.
Statistical analysis
All data are presented as the mean ± standard error of the mean and data analysis was performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). The significance among ≥2 groups was analyzed using one-way analysis of variance followed by a Tukey test. Differences between two groups were analyzed using Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-148b levels are decreased in NSCLC tissues and cell lines
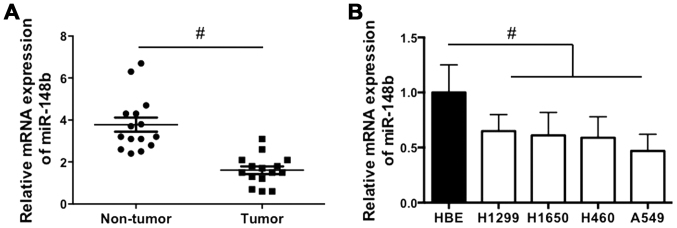
The expression of miR-148b in NSCLC tissues and cell lines was examined. It was demonstrated that miR-148b expression was significantly decreased in NSCLC tissues compared with corresponding adjacent normal lung tissues (P<0.05; Fig. 1A). In addition, miR-148b expression was significantly decreased in all NSCLC cell lines compared with the HBE cell line (all P<0.05; Fig. 1B). The results indicate that miR-148b expression is increased in NSCLC tissue.
Figure 1.
Relative miR-148b expression in NSCLC tissues and cell lines. (A) The expression of miR-148b in NSCLC tumor tissues and adjacent normal lung tissues. (B) The expression of miR-148b in the normal HBE and NSCLC cell lines (H460, H1299, H1650 and A549). #P<0.05. HBE, human bronchial epithelial; NSCLC, non-small cell lung cancer; miR-148b, microRNA-148b.
miR-148b inhibits the proliferation of A549 cells
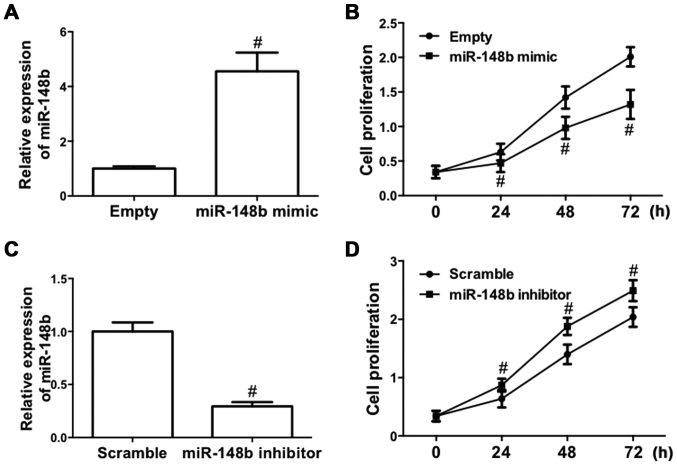
To determine the role of miR-148b in NSCLC, the effect of deregulated expression of miR-148b on cell proliferation in A549 cells was evaluated, as A549 cells are one of the most common cell models used for the study of tumor biology in NSCLC (26). A549 cells were transfected with a miR-148b mimic or inhibitor. The results demonstrated that transfection of the miR-148b mimic significantly increased miR-148b expression (P<0.05; Fig. 2A). Furthermore, following transfection with miR-148b mimic, the proliferation of A549 cells was significantly inhibited (P<0.05; Fig. 2B). By contrast, transfection of miR-148b inhibitor significantly decreased miR-148b expression (P<0.05; Fig. 2C) and significantly increased the proliferation of A549 cells (P<0.05; Fig. 2D). These results demonstrate that miR-148b increases the proliferation of NSCLC cells.
Figure 2.
Effect of miR-148b on the proliferation of A549 cells. (A and B) A549 cells were transfected with miR-148b mimic. (A) Relative expression of miR-148b and (B) proliferation of A549 cells detected by the CCK-8 assay. (C and D) A549 cells were transfected with miR-148b inhibitor. (C) Relative expression of miR-148b and (D) proliferation of A549 cells detected by the CCK-8 assay. #P<0.05 vs. control. CCK-8, Cell Counting Kit-8; miR-148b, microRNA 148b.
miR-148b inhibits the EMT in A549 cells
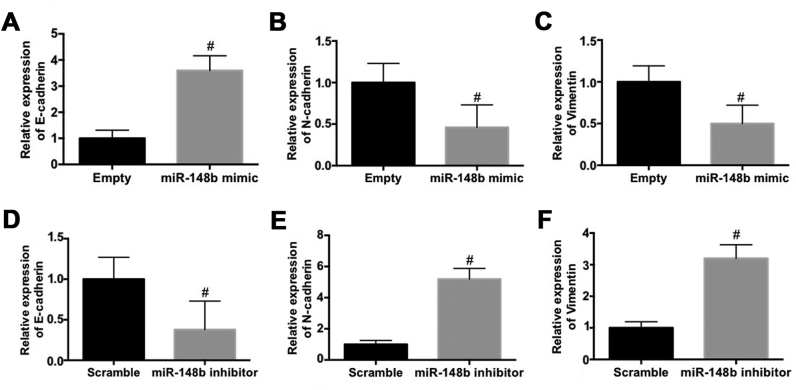
The effect of miR-148b on the expression EMT biomarkers in A549 cells was evaluated. The mRNA expression of E-cadherin was significantly increased (P<0.05; Fig. 3A) whereas the mRNA expression of N-cadherin (Fig. 3B) and vimentin (Fig. 3C) were significantly decreased (P<0.05) following transfection with miR-148b mimic. By contrast, transfection with the miR-148b inhibitor significantly decreased the mRNA expression of E-cadherin (P<0.05; Fig. 3D) and significantly increased the mRNA expression of N-cadherin (P<0.05; Fig. 3E) and vimentin (P<0.05; Fig. 3F). These results demonstrate that miR-148b inhibits the EMT and the invasiveness of A549 cells.
Figure 3.
Effect of miR-148b on epithelial-mesenchymal transition markers in A549 cells. (A-C) A549 cells were transfected with the miR-148b mimic. The relative mRNA expression of (A) E-cadherin, (B) N-cadherin and (C) vimentin was determined. (D-F) A549 cells were transfected with the miR-148b inhibitor. Relative mRNA expression of (D) E-cadherin, (E) N-cadherin and (F) vimentin was identified. #P<0.05 vs. control. miR-148b, microRNA 148b.
miR-148b enhances radiosensitivity and promotes apoptosis in A549 cells
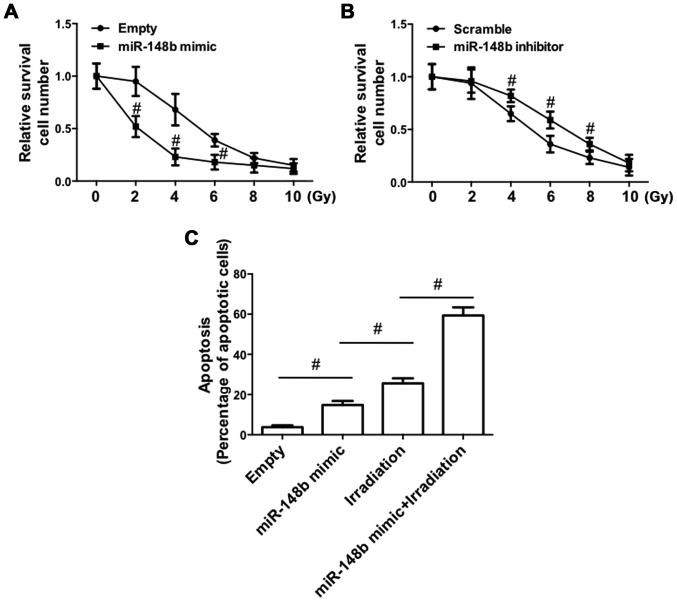
The effect of the deregulated expression of miR-148b on irradiation-induced cell death was determined. Transfection with the miR-148b mimic significantly promoted irradiation-induced cell death (P<0.05; Fig. 4A). By contrast, irradiation-induced cell death was significantly inhibited following transfection with the miR-148b inhibitor (P<0.05; Fig. 4B). Furthermore, irradiation of A549 cells transfected with the miR-148b mimic significantly increased the percentage of apoptotic cells, compared with negative controls (Fig. 4C). The miR-148b mimic promoted irradiation-induced apoptosis, indicating that miR-148b expression enhances radiosensitivity.
Figure 4.
Effect of miR-148b expression on radiosensitivity in A549 cells. A549 cells were transfected with miR-148b mimic or miR-148b inhibitor and then subjected to 0, 2, 4, 6, 8 or 10 Gy of 60 Co-γ ionizing radiation. (A and B) A total of 1×103 cells/well were seeded in 6-well plates and cultured for 10–14 weeks, and cells were stained with crystal violet (0.5 in 20% ethanol). Subsequently, cells were harvested and absorbance was determined using a multifunctional microplate reader at 546 nm. Relative cell survival number was then determined. (C) Cellular apoptosis was determined by terminal deoxynucleotidyl transferase dUTP nick-end labeling staining and analyzed by flow cytometry. #P<0.05 vs. control. miR-148b, microRNA 148-b.
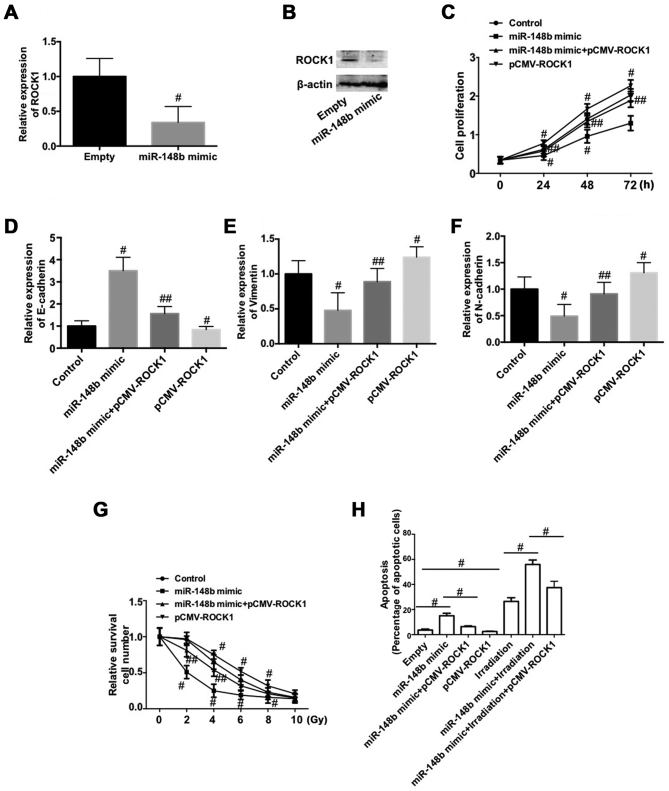
Downregulation of ROCK1 is involved in the miR-148b-induced inhibition of cell proliferation and EMT, and the increase in radiosensitivity in A549 cells
To determine the mechanism by which miR-148b decreases the proliferation of NSCLC cells, inhibits the EMT and increases radiosensitivity, the expression of ROCK1 was determined following transfection with miR-148b mimic. The expression of ROCK1 mRNA and protein was significantly decreased following transfection with miR-148b mimic (P<0.05; Fig. 5A and B). To test whether the downregulation of ROCK1 is involved in the negative role of miR-148b in NSCLC, A549 cells were co-transfected with miR-148b and pCMV-ROCK1. The results demonstrate that overexpression of ROCK1 significantly reversed the miR-148b-induced decrease in cell proliferation (P<0.05; Fig. 5C). Furthermore, overexpression of ROCK1 per se significantly reduced cell proliferation compared with the control (P<0.05; Fig. 5C). The miR-148b-induced increase in E-cadherin expression (Fig. 5D) and decreases in vimentin (Fig. 5E), and N-cadherin (Fig. 5F) expression were significantly inhibited following overexpression of ROCK1 (all P<0.05). Additionally, overexpression of ROCK1 significantly decreased the expression of E-cadherin and increased the expression of vimentin and N-cadherin, compared with the control (P<0.05). Overexpression of ROCK1 inhibited the irradiation-induced decrease in A549 cell survival (P<0.05) and significantly reversed the miR-148b-induced decrease in cell survival following irradiation (P<0.05; Fig. 5G). ROCK1 overexpression significantly reversed the increase in apoptosis that occurred following transfection with miR-148b mimic (P<0.05; Fig. 5H) and significantly reversed the miR-148b-induced increase in apoptosis that occurred following irradiation (P<0.05; Fig. 5H). These data demonstrate that the downregulation of ROCK1 is involved in the miR-148b-induced inhibition of cell proliferation and the EMT, and increases the radiosensitivity of A549 cells.
Figure 5.
Role of ROCK1 in the miR-148b on the proliferation, epithelial-mesenchymal transition and radiosensitivity of A549 cells. (A and B) A549 cells were transfected with miR-148b mimic. Relative (A) mRNA and (B) protein expression of ROCK1. (C-H) A549 cells were co-transfected with miR-148b mimic and plasmid-expressing ROCK1. (C) Cell proliferation detected by Cell Counting Kit-8 assay. The relative mRNA expression of (D) E-cadherin, (E) vimentin and (F) N-cadherin were measured. (G and H) Cells were treated with 0, 2, 4, 6, 8 or 10 Gy 60 Co-γ ionizing radiation. (G) Radiosensitivity was evaluated by crystal violet staining and flow cytometry. #P<0.05 vs. control; ##P<0.05 vs. miR-148b mimic. (H) Cellular apoptosis was determined by terminal deoxynucleotidyl transferase dUTP nick-end labeling staining and analyzed by flow cytometry. ROCK1, Rho-associated protein kinase 1; miR-148b, microRNA-148b.
Discussion
The present study investigated the role of miR-148b in the proliferation of NSCLC cells, the EMT and radiosensitivity in order to elucidate its potential molecular mechanisms of action. It has been previously reported that miR-148b serves a role in regulating several types of tumors (27–29). Furthermore, it has been demonstrated that miR-148b expression is reduced in the plasma samples of patients with NSCLC (23). In addition, miR-148b is a potential prognostic biomarker and predictor of the response to radiotherapy in patients with NSCLC (9,24,29). However, the mechanism by which miR-148b regulates the proliferation and radiosensitivity of NSCLC cells remains unclear.
The present study demonstrated that miR-148b expression is reduced in NSCLC tumor tissue and cell lines. Furthermore, the overexpression of miR-148b decreased the proliferation of A549 cells, whereas downregulation of miR-148b increased cell proliferation, confirming that miR-148b decreases the proliferation of NSCLC cells. Subsequently, the effect of miR-148b expression on the EMT in NSCLC cells was determined. The EMT is a process that is critical for inducing the metastatic progression of cancer by regulating the migration and invasion of cells (30,31). During the EMT, epithelial cells lose their characteristics and switch to a mesenchymal phenotype, consequently exhibiting increased migratory and invasive capabilities and promoting tumor progression (32,33). The molecular mechanism of the EMT involves the alteration of various crucial regulators, including the downregulation of E-cadherin and upregulation of the mesenchymal markers N-cadherin and vimentin (34). The present study demonstrated that miR-148b overexpression inhibited the EMT, as indicated by the upregulation of E-cadherin, and downregulation of N-cadherin and vimentin expression that occurred. Furthermore, downregulation of miR-148b expression decreased E-cadherin expression but increased N-cadherin and vimentin expression. The results indicate that miR-148b inhibits the EMT in NSCLC cells.
The development of radioresistance in NSCLC cells limits the effectiveness of radiotherapy to treat patients with NSCLC (35). Therefore, the present study examined the effect of miR-148b expression on irradiation-induced cell death. It was confirmed that miR-148b overexpression increased the sensitivity of NSCLC cells to irradiation. Furthermore, miR-148b overexpression significantly increased irradiation-induced apoptosis. Overall, the results indicated that miR-148b induces radiosensitivity in NSCLC cells.
The present study aimed to identify the protein that mediated the effect of miR-148b on NSCLC cells. The results demonstrated that miR-148b overexpression markedly decreased the expression of ROCK1. Furthermore, overexpression of ROCK1 significantly inhibited the effects of miR-148b on A549 cell proliferation, EMT and irradiation-induced cell apoptosis. ROCK1 is a Rho-GTPase effector that regulates key aspects of the actin cytoskeleton and is essential for cell cycle progression, senescence and tumorigenesis (36). ROCK1 is required for NSCLC anchorage-independent growth and invasion (37). A previous study has revealed that miR-148a suppresses the EMT by targeting ROCK1 in NSCLC cells (38). Additionally, it has been demonstrated that miR-148b targets ROCK1 expression in breast cancer and hepatocellular carcinoma cells (39). Based on these results and the results of the present study, it was indicated that the downregulation of ROCK1 mediates the inhibitory effect of miR-148b on NSCLC cells. Therefore, ROCK1 may be a target of miR-148b in different types of cancer.
In conclusion, the results of the present study determined that miR-148b inhibits the proliferation of NSCLC cells and the EMT, and increases radiosensitivity by inhibiting ROCK1. Furthermore, miR-148b/ROCK1 signaling was identified as a novel therapeutic target for the inhibition of NSCLC and the enhancement of radiotherapy to treat patients with NSCLC.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Murray N. Reality check for pemetrexed and maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2014;32:482–483. doi: 10.1200/JCO.2013.53.3448. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippma SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claassens L, van Meerbeeck J, Coens C, Quinten C, Ghislain I, Sloan EK, Wang XS, Velikova G, Bottomley A. Health-related quality of life in non-small-cell lung cancer: An update of a systematic review on methodologic issues in randomized controlled trials. J Clin Oncol. 2011;29:2104–2120. doi: 10.1200/JCO.2010.32.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 6.Shtivelman E, Hensing T, Simon GR, Dennis PA, Otterson GA, Bueno R, Salgia R. Molecular pathways and therapeutic targets in lung cancer. Oncotarget. 2014;5:1392–1433. doi: 10.18632/oncotarget.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scagliotti GV, Novello S. Adjuvant therapy in completely resected non-small-cell lung cancer. Curr Oncol Rep. 2003;5:318–325. doi: 10.1007/s11912-003-0074-y. [DOI] [PubMed] [Google Scholar]
- 8.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Chen YY, Li SQ, Huang C, Qin YZ. Expression of miR-148/152 family as potential biomarkers in non-small-cell lung cancer. Med Sci Monit. 2015;21:1155–1161. doi: 10.12659/MSM.892940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Socinski MA. Seeking new options for the treatment of small-cell lung cancer. Lung cance. 2005;50(Suppl 1):S25–S26. doi: 10.1016/S0169-5002(05)81558-2. [DOI] [PubMed] [Google Scholar]
- 11.Ranganathan K, Sivasankar V. MicroRNAs-Biology and clinical applications. J Oral Maxillofac Pathol. 2014;18:229–234. doi: 10.4103/0973-029X.140762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li MH, Fu SB, Xiao HS. Genome-wide analysis of microRNA and mRNA expression signatures in cancer. Acta Pharmacol Sin. 2015;36:1200–1211. doi: 10.1038/aps.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutnam ZJ, Yang BB. The involvement of microRNAs in malignant transformation. Histol Histopathol. 2012;27:1263–1270. doi: 10.14670/HH-27.1263. [DOI] [PubMed] [Google Scholar]
- 14.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300:10–19. doi: 10.1016/j.canlet.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol. 2015;21:8527–8540. doi: 10.3748/wjg.v21.i28.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skrzypski M, Dziadziuszko R, Jassem J. MicroRNA in lung cancer diagnostics and treatment. Mutat Res. 2011;717:25–31. doi: 10.1016/j.mrfmmm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, Wang K, Liu L, Chen Z, Meng Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7:334–345. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Chen Z. MicroRNA-19a functions as an oncogenic microRNA in non-small cell lung cancer by targeting the suppressor of cytokine signaling 1 and mediating STAT3 activation. Int J Mol Med. 2015;35:839–846. doi: 10.3892/ijmm.2015.2071. [DOI] [PubMed] [Google Scholar]
- 22.Sun W, Ma Y, Chen P, Wang D. MicroRNA-10a silencing reverses cisplatin resistance in the A549/cisplatin human lung cancer cell line via the transforming growth factor-beta/Smad2/STAT3/STAT5 pathway. Mol Med Rep. 2015;11:3854–3859. doi: 10.3892/mmr.2015.3181. [DOI] [PubMed] [Google Scholar]
- 23.Huang MX. Down-expression of circulating micro ribonucleic acid (miRNA)-148/152 family in plasma samples of non-small cell lung cancer patients. J Cancer Res Ther. 2016;12:671–675. doi: 10.4103/0973-1482.150420. [DOI] [PubMed] [Google Scholar]
- 24.Yang JS, Li BJ, Lu HW, Chen Y, Lu C, Zhu RX, Liu SH, Yi QT, Li J, Song CH. Serum miR-152, miR-148a, miR-148b and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36:3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Xie WY, Zhou XD, Yang J, Chen LX, Ran DH. Inhibition of autophagy enhances heat-induced apoptosis in human non-small cell lung cancer cells through ER stress pathways. Arch Biochem Biophys. 2016;607:55–66. doi: 10.1016/j.abb.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 27.Orso F, Quirico L, Virga F, Penna E, Dettori D, Cimino D, Coppo R, Grassi E, Elia AR, Brusa D, et al. miR-214 and miR-148b Targeting Inhibits dissemination of melanoma and breast cancer. Cancer Res. 2016;76:5151–5162. doi: 10.1158/0008-5472.CAN-15-1322. [DOI] [PubMed] [Google Scholar]
- 28.Song Y, Sun J, Xu Y, Liu J, Gao P, Chen X, Zhao J, Wang Z. Microarray analysis of long non-coding RNAs related to microRNA-148b in gastric cancer. Neoplasma. 2017;64:199–208. doi: 10.4149/neo_2017_205. [DOI] [PubMed] [Google Scholar]
- 29.Wang R, Ye F, Zhen Q, Song T, Tan G, Chu W, Zhang Y, Lv B, Zhao X, Liu J. MicroRNA-148b is a potential prognostic biomarker and predictor of response to radiotherapy in non-small-cell lung cancer. J Physiol Biochem. 2016;72:337–343. doi: 10.1007/s13105-016-0485-5. [DOI] [PubMed] [Google Scholar]
- 30.Kharbanda A, Rajabi H, Jin C, Alam M, Wong KK, Kufe D. MUC1-C confers EMT and KRAS independence in mutant KRAS lung cancer cells. Oncotarget. 2014;5:8893–8905. doi: 10.18632/oncotarget.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 2011;71:245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karamitopoulou E, Zlobec I, Panayiotides I, Patsouris ES, Peros G, Rallis G, Lapas C, Karakitsos P, Terracciano LM, Lugli A. Systematic analysis of proteins from different signaling pathways in the tumor center and the invasive front of colorectal cancer. Human Pathol. 2011;42:1888–1896. doi: 10.1016/j.humpath.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 34.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 35.You S, Li R, Park D, Xie M, Sica GL, Cao Y, Xiao ZQ, Deng X. Disruption of STAT3 by niclosamide reverses radioresistance of human lung cancer. Mol Cancer Ther. 2014;13:606–616. doi: 10.1158/1535-7163.MCT-13-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumper S, Mardakheh FK, McCarthy A, Yeo M, Stamp GW, Paul A, Worboys J, Sadok A, Jørgensen C, Guichard S, Marshall CJ. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. ELife. 2016;5:e12994. doi: 10.7554/eLife.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vigil D, Kim TY, Plachco A, Garton AJ, Castaldo L, Pachter JA, Dong H, Chen X, Tokar B, Campbell SL, Der CJ. ROCK1 and ROCK2 are required for non-small cell lung cancer anchorage-independent growth and invasion. Cancer Res. 2012;72:5338–5347. doi: 10.1158/0008-5472.CAN-11-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Song Y, Wang Y, Luo J, Yu W. MicroRNA-148a suppresses epithelial-to-mesenchymal transition by targeting ROCK1 in non-small cell lung cancer cells. Mol Cell Biochem. 2013;380:277–282. doi: 10.1007/s11010-013-1682-y. [DOI] [PubMed] [Google Scholar]
- 39.Cimino D, De Pittà C, Orso F, Zampini M, Casara S, Penna E, Quaglino E, Forni M, Damasco C, Pinatel E, et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS and CSF1. FASEB J. 2013;27:1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]