Abstract
This scientific commentary refers to ‘A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy’, by Simons et al. (doi:10.1093/brain/awx314).
This scientific commentary refers to ‘A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy’, by Simons et al. (doi:10.1093/brain/awx314).
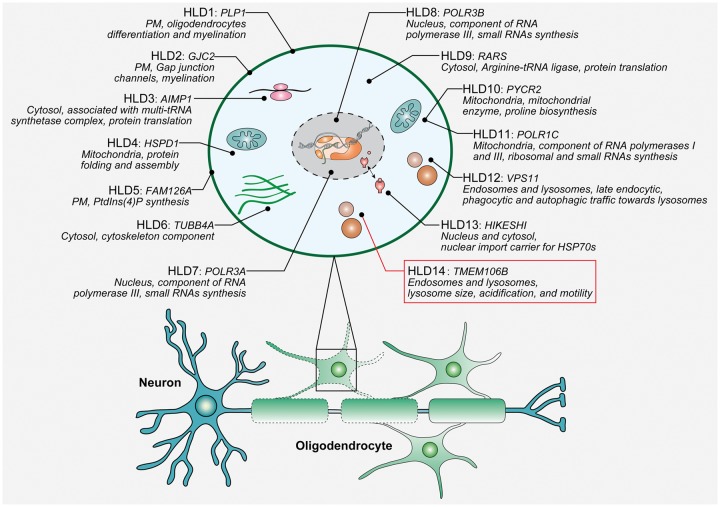
Leukodystrophies are a group of rare genetic disorders that affect the CNS by disrupting the growth or maintenance of the myelin sheath that insulates nerve cells. A classification system based on the pathological mechanisms responsible for the white matter pathology was recently proposed, reserving the term hypomyelinating leukodystrophies (HLDs) for those diseases with a primary or predominant involvement of oligodendrocytes and/or myelin and a permanent deficiency in the formation or deposition of myelin (in contrast to demyelinating leukodystrophies in which the integrity of myelin is disrupted after its formation) (van der Knaap and Bugiani, 2017). HLDs are genetically and clinically diverse; however, most patients present in the neonatal or infantile period with a combination of hypotonia and nystagmus and a range of possible additional symptoms including developmental delay, ataxia, spasticity intellectual disability. Pelizaeus-Merzbacher disease (PMD) is the archetypical HLD caused by mutations in the gene encoding proteolipid protein 1 (PLP1), a primary constituent of myelin. However, more than a dozen additional HLD genes have been identified with a wide range of functions involved in different cellular processes: from RNA and protein synthesis to endolysosomal trafficking (Fig. 1) (Baskin et al., 2016; Charzewska et al., 2016; Edvardson et al., 2016). In this issue of Brain, Simons and co-workers reveal an intriguing connection between TMEM106B, a relatively unknown transmembrane protein localized to the lysosomal membrane, and HLD through the identification of a recurrent de novo TMEM106B mutation in four families (Simons et al., 2017).
Figure 1.
Overview of known disease genes for classical hypomyelinating leukodystrophies (HLDs). For each HLD-associated protein, the primary subcellular localization is reported as well as its primary known function(s) (Baskin et al., 2016; Charzewska et al., 2016; Edvardson et al., 2016). Nomenclature and numbering of HLD1 through HLD13 is in accordance with the OMIM database (https://www.omim.org/), while the newly identified HLD gene, TMEM106B, was temporarily assigned the acronym HLD14. PM = plasma membrane.
Two unrelated patients recruited and studied on different continents by independent research groups formed the basis for the discovery. Researchers from the Care4Rare Canada Consortium and the Amsterdam Database of Unclassified Leukoencephalopathies in The Netherlands each diagnosed a patient with PMD-like disease based on the presence of nystagmus and hypotonia shortly after birth, delayed motor development, and prominent hypomyelination on brain MRI; however, genetic testing excluded mutations in PLP1. As a result of the childhood onset of disease and absence of family history, trio exome sequencing was performed in both families and, remarkably, the same de novo mutation c.754G>A (p.Asp252Asn) in TMEM106B was identified in both patients. Through effective use of the GeneMatcher website, which enables connections between researchers dealing with ‘unsolved exomes’, the researchers noted the strong overlap in clinical presentation and identical gene mutation, suggesting a potential causal role for this mutation in their patients. The study of exome data from 10 additional trios from The Netherlands and one unrelated patient from Canada, identified another two unrelated patients carrying the same c.754G>A mutation. Each of the four unrelated patients had the classical clinical presentation of hypomyelination with early-onset nystagmus, hypotonia and delayed motor development with variable degrees of intellectual disability and epilepsy. In one family the mutation was found to be transmitted from the father, who is a mosaic for the p.Asp252Asn mutation, to the affected child. The father expresses approximately 25% mutant TMEM106B, according to quantification of expression in leucocytes. While this presumably led to nystagmus and developmental delay in infancy, the currently 65-year-old male has normal cognition and no obvious neurological abnormalities.
TMEM106B was first reported in 2010 as a genetic risk factor for frontotemporal lobar degeneration with pathologically confirmed TDP-43 pathology (FTLD-TDP), a neurodegenerative disease characterized by the preferential atrophy of the frontal and temporal lobes (Nicholson and Rademakers, 2016). Subsequent studies provided strong support for TMEM106B as a disease modifier, especially in patients with FTLD-TDP with loss-of-function mutations in progranulin (GRN), a neurotrophic growth factor that is processed into possibly functionally active granulin peptides within lysosomes. While the basis for the risk/protective effect of TMEM106B is still being studied, available data suggest that an increase in TMEM106B levels is cytotoxic and is associated with increases in lysosomal size and reduced lysosomal acidification, leading to the disruption of endolysosomal- and autophagic-lysosomal degradation (Nicholson and Rademakers, 2016). Lowering TMEM106B levels has therefore been suggested as a potential therapeutic avenue in patients with GRN mutations, and it was recently reported that Tmem106b deletion can normalize lysosomal protein levels in Grn−/− mice and rescue FTLD-related behavioural abnormalities and retinal degeneration in this model (Klein et al., 2017). However, TMEM106B knockdown may not be completely without consequences. Studies in neuronal cultures suggested mild effects on lysosomal trafficking, and the activity of several lysosomal enzymes was reduced in Tmem106b−/− mice, arguing for a tight regulation of TMEM106B in vivo (Nicholson and Rademakers, 2016; Klein et al., 2017). In line with these observations, relatively mild effects on the expression levels of TMEM106B were observed in individuals carrying the TMEM106B risk (increased expression) or TMEM106B protective (decreased expression) alleles (Nicholson and Rademakers, 2016). Intriguingly, the same TMEM106B variant(s) implicated in FTLD-TDP were recently identified in an unbiased screen for genetic modifiers of healthy brain ageing, with increased inflammation, neuronal loss, and cognitive deficits in brain specimens of TMEM106B risk allele carriers (Rhinn and Abeliovich, 2017). This study suggested an inappropriate polarization of microglia and other innate immune myeloid cells toward a pro-inflammatory state in TMEM106B risk allele carriers, yet the authors did not rule out a function for TMEM106B in neurons.
The current study by Simons and colleagues in this issue of Brain (Simons et al., 2017) is the first to link TMEM106B to oligodendrocytes and myelination, unveiling an unexplored area of research into TMEM106B function and disease mechanisms. Unfortunately, the effect of the specific p.Asp252Asn mutation on TMEM106B expression and/or function was not studied in vitro or in patient material, and discussion of the potential disease mechanism is consequently speculative at this time. The close vicinity of the p.Asp252Asn mutation to one of the sites requiring complex glycosylation for proper TMEM106B transport, sorting and probably function (Nicholson and Rademakers, 2016), combined with the fact that all patients carried the exact same de novo mutation supports the hypothesis of a loss-of-function disease mechanism, although a gain of toxic function associated with the specific mutation cannot yet be excluded. Since all patients were heterozygous for the mutation, a dominant-negative disease mechanism may in fact be at play. The majority of HLD genes are transmitted as autosomal recessive disorders or are x-linked (PLP1) with the notable exception of TUBB4A, in which the heterozygous p.Asp249Asn mutation was shown to cause HLD with atrophy of the basal ganglia and cerebellum. In the latter case, a dominant-negative effect of the mutation presumably led to the loss or inefficient dimerization of microtubules (Simons et al., 2013; Charzewska et al., 2016).
Given that PLP1 is one of the main structural components of the myelin sheath, and that PLP1 mutations are known to cause PMD with overlapping disease phenotypes to those described in association with the new TMEM106B mutation, it is tempting to speculate that mutant TMEM106B could potentially interfere with the highly regulated endocytosis and/or exocytosis of PLP1, thereby affecting its spatial and temporal expression (Saher and Stumpf, 2015). PLP1 is synthesized in oligodendrocytes in the rough endoplasmic reticulum (ER) and then transported to the Golgi and plasma membrane in lipid raft-like membrane domains, where it is integrated into the developing myelin sheet. In the absence of neuronal signals, PLP1 is internalized and stored in late endosomes and lysosomes from where it can be rapidly recruited to the sites of membrane growth when needed (Feldmann et al., 2011). In PMD, point mutations in PLP1 interfere with its trafficking, resulting in accumulation of PLP1 within the ER/Golgi. By contrast, overexpression of PLP1 due to duplications leads to excessive
Glossary
Hypomyelinating leukodystrophies (HLD): Genetically determined white matter diseases caused by a primary deficit in myelin deposition. Multiple HLD genes have been identified (Fig. 1).
TMEM106B: Type I transmembrane protein mainly localized to late endosomes and lysosomes. Common variants at the TMEM106B locus have been implicated in frontotemporal dementia with TDP-43 pathology.
PLP1 accumulation in the late endosomes and lysosomes, illustrating that multiple trafficking deficits and sites of PLP1 accumulation can be toxic (Saher and Stumpf, 2015; van der Knaap and Bugiani, 2017).
Regardless of the specific mechanism associated with the recurrent TMEM106B mutation, the addition of TMEM106B to the list of known HLD genes reinforces the connection between lysosomes and myelination. Currently available data further suggest that TMEM106B levels are tightly regulated and that either too much or too little TMEM106B may have devastating consequences. Future mechanistic studies of this newly discovered p.Asp252Asn mutation will undoubtedly provide much-needed insights into the normal function of TMEM106B within lysosomes. This would appear to be the critical next step towards the development of therapies or disease-modifying treatments not only for HLDs but also for patients with FTLD-TDP with and without GRN mutations.
Funding
X.Z. is supported by a postdoctoral fellowship from The Bluefield Project to Cure Frontotemporal Dementia. R.R. is funded by NIH grants R35 NS097261, UG3/UH3 NS0103870 P50 AG016574 and P01 NS084974 and the Consortium for Frontotemporal Dementia (CFR).
References
- Baskin JM, Wu XD, Christiano R, Oh MS, Schauder CM, Gazzerro E, et al. The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat Cell Biol 2016; 18: 132–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charzewska A, Wierzba J, Izycka-Swieszewska E, Bekiesinska-Figatowska M, Jurek M, Gintowt A, et al. Hypomyelinating leukodystrophies—a molecular insight into the white matter pathology. Clin Genet 2016; 90: 293–304 [DOI] [PubMed] [Google Scholar]
- Edvardson S, Kose S, Jalas C, Fattal-Valevski A, Watanabe A, Ogawa Y, et al. Leukoencephalopathy and early death associated with an Ashkenazi-Jewish founder mutation in the Hikeshi gene. J Med Genet 2016; 53: 132–7 [DOI] [PubMed] [Google Scholar]
- Feldmann A, Amphornrat J, Schonherr M, Winterstein C, Mobius W, Ruhwedel T, et al. Transport of the major myelin proteolipid protein is directed by VAMP3 and VAMP7. J Neurosci 2011; 31: 5659–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ZA, Takahashi H, Ma M, Stagi M, Zhou M, Lam TT, et al. Loss of TMEM106B Ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 2017; 95: 281–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson AM, Rademakers R. What we know about TMEM106B in neurodegeneration. Acta Neuropathol 2016; 132: 639–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn H, Abeliovich A. Differential aging analysis in human cerebral cortex identifies variants in TMEM106B and GRN that regulate aging phenotypes. Cell Syst 2017; 4: 404–15 [DOI] [PubMed] [Google Scholar]
- Saher G, Stumpf SK. Cholesterol in myelin biogenesis and hypomyelinating disorders. Biochim Biophys Acta 2015; 1851: 1083–94 [DOI] [PubMed] [Google Scholar]
- Simons C, Dyment D, Bent SJ, Crawford J, D'Hooghe M, Kohlschütter A, et al. A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy. Brain2017; 140: 3105–111. doi: 10.1093/brain/awx314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C, Wolf NI, McNeil N, Caldovic L, Devaney JM, Takanohashi A, et al. A de novo mutation in the beta-tubulin gene TUBB4A results in the leukoencephalopathy hypomyelination with atrophy of the basal ganglia and cerebellum. Am J Hum Genet 2013; 92: 767–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap MS, Bugiani M. Leukodystrophies: a proposed classification system based on pathological changes and pathogenetic mechanisms. Acta Neuropathol 2017; 134: 351–82 [DOI] [PMC free article] [PubMed] [Google Scholar]